Abstract
Objective: Angiogenesis plays a vital role in tumor growth and metastasis. Here, we aimed to find novel efficient antiangiogenic molecules targeting vascular endothelial growth factor A (VEGFA ) at the transcriptional level to treat triple-negative breast cancer (TNBC).
Methods: We used a cell-based seryl tRNA synthetase (SerRS) promoter-driven dual-luciferase reporter system to screen an in-house library of 384 naturally occurring small molecules and their derivatives to find candidate molecules that could upregulate the expression of SerRS, a potent transcriptional repressor of VEGFA. The levels of SerRS and VEGFA were examined by quantitative RT-PCR (qRT-PCR), western blotting, and/or ELISAs in TNBC cells after candidate molecule administration. Zebrafish, the Matrigel plug angiogenesis assay in mice, the TNBC allograft, and xenograft mouse models were used to evaluate the in vivo anti-angiogenic and anti-cancer activities. Furthermore, the potential direct targets of the candidates were identified by proteomics and biochemical studies.
Results: We found the most active compound was 3-(4-methoxyphenyl) quinolin-4(1H)-one (MEQ), an isoflavone derivative. In TNBC cells, MEQ treatment resulted in increased SerRS mRNA (P < 0.001) and protein levels and downregulated VEGFA production. Both the vascular development of zebrafish and Matrigel plug angiogenesis in mice were inhibited by MEQ. MEQ also suppressed the angiogenesis in TNBC allografts and xenografts in mice, resulting in inhibited tumor growth and prolonged overall survival (P < 0.05). Finally, we found that MEQ regulated SerRS transcription by interacting with MTA2 (Metastasis Associated 1 Family Member 2).
Conclusions: Our findings suggested that the MTA2/SerRS/VEGFA axis is a drug-treatable anti-angiogenic target, and MEQ is a promising anti-tumor molecule that merits further investigation for clinical applications.
Keywords: Isoflavone, MTA2, SerRS, tumor angiogenesis, VEGFA
Introduction
It is estimated that over 2 million women in the world are diagnosed with breast cancer every year1. Among these, at least 15% are classified as triple-negative1. Triple-negative breast cancer (TNBC) is characterized by younger age occurrence, higher rates of relapse, greater metastatic potential, and shorter overall survival compared with the hormone receptor-positive/human epidermal growth factor 2 receptor (HER2) negative (70% of patients) and HER2 positive (15%–20% of patients) phenotypes2,3. Based on the molecular pathogenesis of these two cancer subtypes, multiple effective targeted therapies have been developed, such as endocrine agents for estrogen receptor (ER)-positive or progesterone receptor (PR)-positive breast cancers, anti-HER2 antibodies (such as trastuzumab and pertuzumab), and small-molecule tyrosine kinase inhibitors (such as lapatinib and neratinib) for HER2-positive types4. However, TNBC lacks the expression of both the ER and PR and does not overexpress HER25. And the specific molecular pathophysiology of TNBC remains poorly understood. Therefore, there is currently no effective, specifically targeted therapy available for TNBC4.
Targeting angiogenesis is a valuable strategy to inhibit solid tumor development and progression because solid tumors (like TNBC) cannot grow beyond a few millimeters in size or spread without a blood supply6. Tumor cells can induce the blood supply by secreting angiogenic factors, like VEGFA, to stimulate angiogenesis7. Bevacizumab, an anti-angiogenic inhibitor targeting VEGFA, is approved to treat several tumor types, including TNBC8. However, due to the relative poor tissue penetration of bevacizumab for its large size, the addition of bevacizumab to traditional chemotherapy drugs did not result in a statistically significant increase in the numeric rate of pathologic complete response (pCR)9,10. Also, the remaining and still active VEGFA after bevacizumab treatment may still be enough to induce angiogenesis. Several adverse effects of bevacizumab, such as headache, hypertension, proteinuria, taste alteration, and rectal hemorrhage have been reported10,11. Thus, novel small molecule drugs with good cell and tissue permeability, if able to effectively reduce the VEGFA level in tumor tissue and having few side effects, could be promising therapeutic agents for TNBC.
We previously reported that seryl tRNA synthetase (SerRS), which transfers L-serine to its specific tRNA for protein biosynthesis in the cytoplasm, was also a potent transcriptional repressor of VEGFA12. Furthermore, SerRS counteracts the effects of c-Myc and NFκB12,13 in modulating VEGFA expression. So, increasing SerRS expression could be a promising strategy to inhibit abnormal VEGFA expression and tumor angiogenesis. Although gene therapy is a method to directly introduce more SerRS protein in cells, the technology is still undeveloped and has potential risks for clinical applications14,15. Identifying small molecular drugs targeting SerRS expression may therefore be a more promising anti-angiogenesis strategy for the treatment of TNBC.
Natural small molecules isolated from traditional Chinese herbs are presently considered as optimal cancer drug candidates for their structural diversity, good cell permeability, low toxicity, and nonimmunogenic profiles16. Because of their evolutionary advantage, for high throughput screening, a library of natural small molecules is a better resource to discover molecules with fewer side effects than a synthetic compound library17,18. Derivatives based on natural small molecules also can possess improved pharmaceutical characteristics and minimal side effects. Based on these considerations, we established an in-house library consisting of 384 natural small molecules and their derivatives, which was screened with a TNBC cell-based and SerRS promoter-driven luciferase reporter system. We identified MEQ as a novel anti-angiogenesis agent showing robust activity to inhibit VEGFA and TNBC progression by targeting SerRS expression in both in vitro and in vivo model systems.
Materials and methods
Cell culture
MDA-MB-231 and 4T1 cells were purchased from the American Type Tissue Collection (Manassas, VA, USA). Both MDA-MB-231 cells and 4T1 cells were cultured in Dulbecco’s Modified Eagle’s Medium (Biological Industries, Cromwell, CT, USA) supplemented with 10% fetal bovine serum (Biological Industries) under 5% CO2 at 37 °C.
Chemicals
Our in-house small molecule library (384 naturally occurring small molecules or their derivatives) was established by the State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University, China. All compounds were dissolved in dimethyl sulfoxide (DMSO) and stored at −20 °C at a concentration of 20 mM.
Dual-luciferase vector construction and the dual-luciferase reporter assay
The gene sequence of the SerRS promoter was amplified by the polymerase chain reaction (PCR) from the HEK 293 cell line using a forward primer: 5′-TCAGGTACC AGTAGAGACGGGATTTTGTCATG-3′ and reverse primer: 5′-TCACTCGAGCAAATCCAGATCCAGCACCATCTTC-3′ with Kpn I and Xho I restriction enzyme sites. The fragment was then subcloned into the pGL4.11 [luc2P] vector. A stably-transfected MDA-MB-231 cell line containing a SerRS promoter-driven firefly luciferase [pGL4.11 (luc2P)] and Renilla (pRL-SV40) was constructed for the purpose of screening for molecules that activated the promoter of SerRS. Stably-transfected MDA-MB-231 cells were seeded into 24-well plates, incubated for 24 h, exposed to experimental compounds or DMSO as a control for 48 h, and the luciferase activities measured (Promega, Madison, WI, USA).
Drug toxicity assay
BALB/c mice (6–8 weeks of age) were conditioned for 1 week in our pathogen-free animal facility. BALB/c mice (6–8 weeks of age, n = 3) were treated with 100 mg/kg MEQ or DMSO (intraperitoneal injection, twice a day) and their weights measured for 4 weeks (once every 4 days) before sacrifice. Blood samples were taken by eyeball extirpation, and the hearts, livers, spleens, lungs, kidneys, and brains were harvested after sacrifice. Blood tests were performed with a hematology analyzer (Celltac E; Nihon Kohden, Tokyo, Japan).
Immunoblotting
Cell or tissue lysates were prepared with lysis buffer [200 mM Tris-Cl (pH 7.5), 1.5 M NaCl, 20 mM EDTA, 10% Triton X-100, 10 mM Na3VO4·12 H2O, and 1% protease inhibitor cocktail (Sigma-Aldrich St. Louis, MO, USA)] on ice. Protein concentrations of samples were determined using a Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA). Primary antibodies were as follows: anti-SerRS (1:1,000; made in the laboratory), anti-VEGFA (1:1,000; Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-β-actin (1:5,000; Santa Cruz Biotechnology), and anti-MTA2 (1:1,000, Santa Cruz Biotechnology). All antibodies recognized both human and mouse proteins. After SDS-PAGE and western blotting, the bands were detected by chemiluminescence using an ECL kit (Thermo Fisher Scientific).
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from the indicated cells, zebrafish tissues, or mouse tumors using TRIzol reagent (Yeasen Biotechnology, Shanghai, China). One μg of total RNA from each sample was reverse transcribed into cDNA using M-MLV Reverse Transcriptase (Promega). All real-time qPCRs were performed using a LightCycler 96 (Roche, Basel, Switzerland) Real-Time PCR system with Hieff qPCR SYBR Green Master Mix (Yeasen Biotech). The following primers were used for the qPCR reactions: 5′-AAGAAAGCAGCAGCAAGAGACG-3′ and 5′-CATGCGAGGAGACAGGAACATC-3′ for human SARS1; 5′-GAGGGCAGAATCATCACGAAG-3′ and 5′-TGTGCTGT AGGAAGCTCATCTCTC-3′ for human VEGFA; 5′-CGTCA CCAACTGGGACGA-3′ and 5′-ATGGGGGAGGGCATACC-3′ for human ACTB; 5′-TGCGACCACTCGTGTCATCT-3′ and 5′-GAGACTGTGGAGGGCGTGTC-3′ for zebrafish sars1; 5′-GGCTCTCCTCCATCTGTCTGC-3′ and 5′-CAGTGGTT TTCTTTCTTTGCTTTG-3′ for zebrafish vegfa; 5′-TCACCACC ACAGCCGAAAGAG-3′ and 5′-GTCAGCAATGCCAGGGTA CAT-3′ for zebrafish actb1; 5′-ACCTGGTGGTGATGGTAG ATGG-3′ and 5′-AAGGGGGTGTAGATTGGAGTGT-3′ for mouse Sars; 5′-GTCCGATTGAGACCCTGGTG-3′ and 5′-TTG ACCCTTTCCCTTTCCTCG-3′ for mouse Vegfa; and 5′-GGCT GTATTCCCCTCCATCG-3′ and 5′-GCACAGGGTGCTCCT CAG-3′ for mouse Actb. The PCR reaction program was the following: 95 °C for 10 min, followed by 45 cycles of 95 °C for 20 s, and 60 °C for 1 min, ending with 95 °C for 2 min. All experiments were carried out in triplicate. The results are expressed as 2-ΔΔCT values and the SEM, and were in triplicate. The mRNA levels of SerRS and VEGFA were normalized to that of β-actin. Student’s t-test was used to analyze the difference between two groups.
Enzyme-linked immunosorbent assay (ELISA)
Supernatants from MDA-MB-231 cells incubated with MEQ or DMSO for 48 h were collected. For the detection and quantification of secreted VEGFA, an ELISA was used according to the manufacturer’s instructions with signal detected at 450 nm (DLDEVELOP, Wuxi, China). Total protein was measured for normalization using a Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific/Pierce).
Zebrafish angiogenesis model
Tg (Fli1a: EGFP) transgenic zebrafish were raised in a constant temperature (28.5 °C) room with a 14:10 h day/night cycle. Male and female zebrafish were randomly selected for copulation with a ratio of 3:2 and separated for 12 h before mating. Dead or unfertilized embryos were removed and those in good condition were kept in E3 embryo medium (5 mmol/L NaCl, 0.17 mmol/L KCl, 0.33 mmol/L CaCl2, and 0.33 mmol/L MgSO4, pH 7.2) at 28.5 °C. E3 embryo medium was changed to E3 plus 0.003% 1-phenyl-2-thiourea at 12 h post-fertilization. At the same time, the embryos were transferred to 24-well plates with 10 embryos per well and incubated with 10 μM MEQ or 0.1% DMSO. After 72 h of treatment, images of larval intersegmental vessels were captured using a DP72 digital camera mounted on an SZX16 dissecting microscope (Olympus, Tokyo, Japan).
Matrigel plug assay
Matrigel (BD Biosciences, San Jose, CA, USA) was extracted from Engelbreth-Holm-Swarm mouse sarcomas. It consisted of extracellular matrix components and growth factors. Matrigel was stored at −20 °C and thawed at 4 °C before use. A total of 210 μL of a mixture of Matrigel (Matrigel:DMEM = 1:2), 1 × 106 4T1 cells, and 0.2 μL MEQ (10 mM) or 0.2 μL DMSO were injected subcutaneously into BALB/c mice. After 7 days, the Matrigel plug was removed and new blood vessel formation in the Matrigel plug was analyzed by immunofluorescence for CD31 and immunohistochemical (IHC) staining for SerRS and VEGFA.
Animal studies
All mice were evaluated and noted to be healthy and free of disease before tumor implantation. Female BALB/c mice (6 weeks) and BALB/c NOD-SCID mice (6 weeks) were from SPF Biotechnology (Beijing, China). All animal experiments were approved by the Animal Care and Use Committee at Nankai University (Tianjin, China). After 1 week acclimation in the pathogen-free animal facility, 1 × 105 4T1 cells and 1 × 105 MDA-MB-231 cells were injected into the #2 mammary fat pads of BALB/c mice and NOD-SCID mice, respectively. Mice were randomly divided into three groups (BALB/c mice) or two groups (NOD-SCID mice), and 1 week after the tumor cells injection, the control group received DMSO (the same volume as MEQ), while the other groups received MEQ at either 100 mg/kg or 200 mg/kg by intraperitoneal injection every other day. The volume (V) of each tumor was measured every 4 days and calculated using the formula: V = length × width2/2. Tumors were excised for analysis at the end of the expriments.
Immunofluorescence (IF), hematoxylin and eosin (H&E), and IHC staining
Paraffin sections of Matrigel plugs were stained with a primary antibody against CD31 (1:50; Abcam, Cambridge, UK), goat-anti-rabbit Alexa 594 secondary antibody (ZSGB-Biotech, Beijing, China), and 4′,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich) for nuclear staining and the IF was assessed. Sections were photographed using an FV-1000 laser scanning confocal biological microscope (Olympus). H&E staining of paraffin-embedded tumor sections from both mouse TNBC models and organs from the drug toxicity test mouse model was conducted for overall morphological observations. Tumors were also analyzed using SerRS (1:500; antiserum made in the laboratory), VEGFA (1:50; Santa Cruz Biotechnology), CD31 (1:50; Abcam), Ki67 (1:200; Abcam), and cleaved caspase-3 (CC3, 1:200; Cell Signaling Technology, Danvers, MA, USA) primary antibodies for immunohistochemical analyses. The DAPI was used for labeling the nucleus. Numbers of immune positive cells were quantified using ImageJ software, version 1.50 (National Institutes of Health, Bethesda, MD, USA). All images used the same exposure conditions.
The shRNAs
DNAs expressing a short-hairpin RNA (shRNA) designed against the human SerRS (5′-AAAAGGCATAGGGACCCATC ATTGATTGGATCCAATCAATGATGGGTCCCTATG CC-3′) and human MTA2 (5′-AAAAGCAGATCGACCAGTT TCTTGTTTGGATCCAAACAAGAAACTGGTCGAT CTGC-3′) genes were inserted into the pLV-H1-ef1α-puro plasmid.
Streptavidin-biotin affinity pull-down assay
MDA-MB-231 cells were harvested, and lysates prepared and incubated with free biotin (MedChemExpress, Monmouth Junction, NJ, USA) or MEQ-biotin (10 μM) with rotation for 5 h at 4 °C. Recombinant streptavidin agarose beads were then added and incubated with rotation at 4 °C overnight. The beads were washed three times and bead-bound proteins separated by SDS-PAGE. Protein bands were visualized by silver staining. Bands between 70 kDa and 120 kDa were cut out for mass spectrometric analyses. MTA2 was detected using western blotting of the same streptavidin agarose bead-binding proteins.
Statistical analysis
Data are presented as the mean ± SEM. Statistical analysis was performed using an unpaired Student’s t-test for comparisons of two groups. Analysis of variance (ANOVA) was used for multiple comparisons. P < 0.05, P < 0.01, or P < 0.001 were considered statistically significant and indicated by *, **, or ***, respectively. All statistics were performed using Prism 5 software (GraphPad, San Diego, CA, USA).
Results
Screening the natural small molecule library for compounds that activated the promoter of SerRS
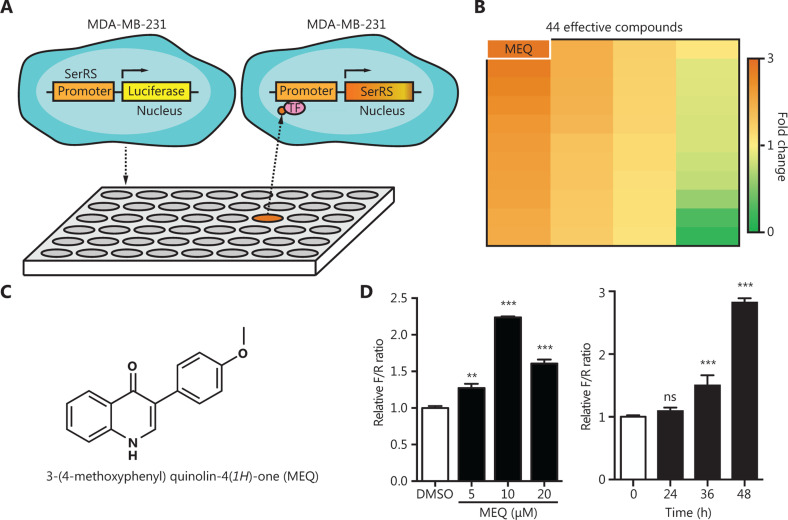
High-throughput screening has been developed as an efficient method for drug discovery. We used the TNBC cell line, MDA-MB-231, which was stably-transfected with a SerRS promoter-driven firefly luciferase gene and the Renilla luciferase gene, to screen a small molecule library containing 384 naturally occurring small molecules and their derivatives from our in-house drug library, each at a concentration of 10 μM (Figure 1A). At 48 h post-incubation, we used the firefly/Renilla ratio to determine the effect of these molecules on the SerRS promoter. Compared with the control group treated with DMSO, 34 compounds showed activation of the SerRS promoter, while 10 compounds showed the opposite activity (Figure 1B). Among the 34 small molecules that activated the SerRS promoter, MEQ, an isoflavone derivative synthesized by the method reported by Santosh Rajput19, exhibited the strongest activity (Figure 1B and 1C). MEQ activated the SerRS promoter at less than 10 μM without cellular toxicity and reached maximum activation at 48 h in MDA-MB-231 cells (P < 0.001) (Figure 1D).
Figure 1.
High throughput screening for compounds that act on the SerRS promoter. (A) Scheme of the dual luciferase reporter-based high throughput screening. (B) Heat map of the luciferase/Renilla ratios for 44 compounds responsive to the SerRS promoter. (C) The molecular structure of MEQ. (D) Relative firefly/Renilla (F/R) ratios for MEQ at indicated dosages and time points. Data are plotted as means ± SEM (n = 3; ns, not significant; **P < 0.01; ***P < 0.001 using Student’s t-test.
MEQ inhibited VEGFA expression by upregulating SerRS transcription in MDA-MB-231 cells
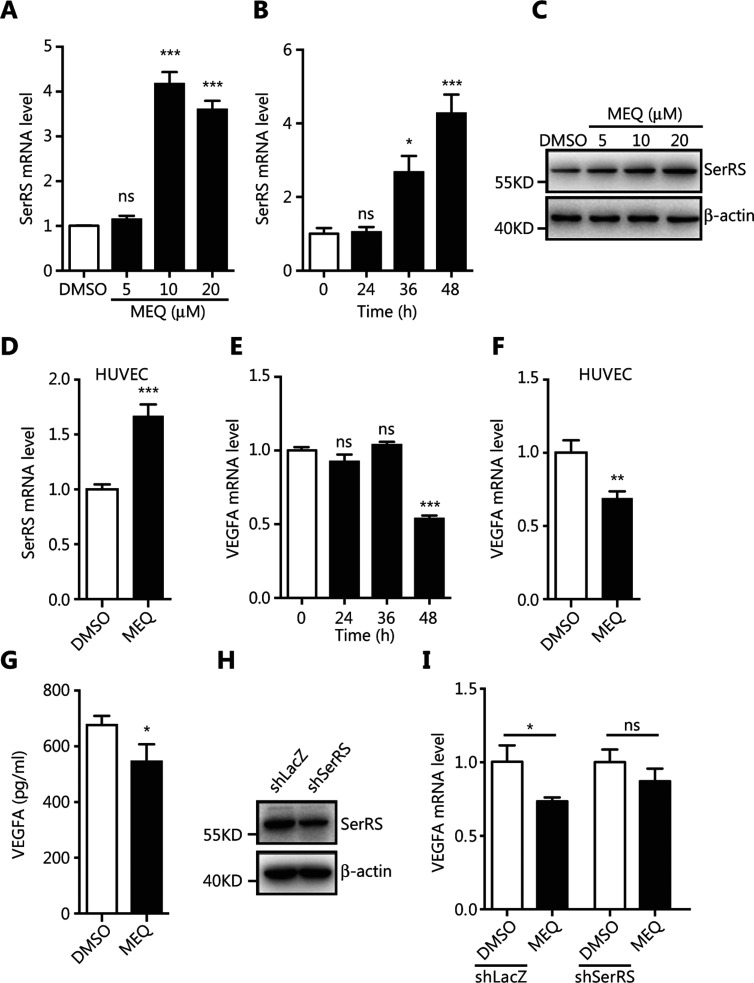
To verify that MEQ upregulated the expression of endogenous SerRS, we treated MDA-MB-231 cells with MEQ at different dosages and for different time periods before SerRS expression was examined by qRT-PCR and western blotting. Consistent with the results from the reporter system, MEQ upregulated the expression of SerRS at 10 μM within 48 h (P < 0.001) (Figure 2A–2C). In addition, MEQ also enhanced SerRS expression in human umbilical vein endothelial cells (HUVEC) (P < 0.001) (Figure 2D). Given that SerRS is a potent transcriptional repressor of VEGFA12,13, we further examined the mRNA and protein levels of VEGFA in MEQ-treated MDA-MB-231 and HUVEC cells. Figure 2E shows that 10 M of MEQ reduced VEGFA mRNA by 50% within 48 h in MDA-MB-231 cells (P < 0.001), similar to the results in HUVEC cells (P < 0.01) (Figure 2F). Consistently, MEQ treatment also reduced secreted VEGFA (P < 0.05) (Figure 2G).
Figure 2.
MEQ inhibits VEGFA expression by upregulating SerRS in cultured cells. (A, B) The qRT-PCR shows SerRS expression in MDA-MB-231 cells treated with MEQ at indicated dosages or with dimethyl sulfoxide (DMSO) as a vehicle control for 48 h (A) or treated with 10 μM MEQ for the indicated time periods (B). (C) Western blotting to show SerRS protein levels in MDA-MB-231 cells treated with the indicated dosages of MEQ or DMSO for 48 h. (D) The qRT-PCR shows SerRS expression in human umbilical vein endothelial cells (HUVECs) cells treated with 10 μM of MEQ for 48 h. (E, F) The qRT-PCR shows vascular endothelial growth factor A (VEGFA) expression in MDA-MB-231 cells treated with MEQ for the indicated time periods (E) and in HUVEC cells treated with 10 μM of MEQ for 48 h (F). (G) ELISA analyses of secreted VEGFA from MDA-MB-231 cells treated with 10 μM MEQ for 48 h. (H) Western blotting shows the knockdown efficiency of shRNA against SerRS (shSerRS) with a shRNA against lacZ gene (shLacZ) as a negative control. (I) The qRT-PCR shows VEGFA expression in shSerRS or shLacZ transfected MDA-MB-231 cells treated with 10 μM MEQ for 48 h. All bar graphs are plotted as the mean ± SEM (n = 3; ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001 using Student’s t-test.
To further investigate if the MEQ-induced decrease in VEGFA expression was through its regulation of SerRS, we knocked down SerRS in MDA-MB-231 cells by shRNA (Figure 2H). After the knockdown, MEQ could no longer inhibit VEGFA expression in SerRS-silenced cells (Figure 2I), confirming that MEQ suppressed VEGFA expression via its regulation of SerRS.
MEQ inhibited angiogenesis in vivo
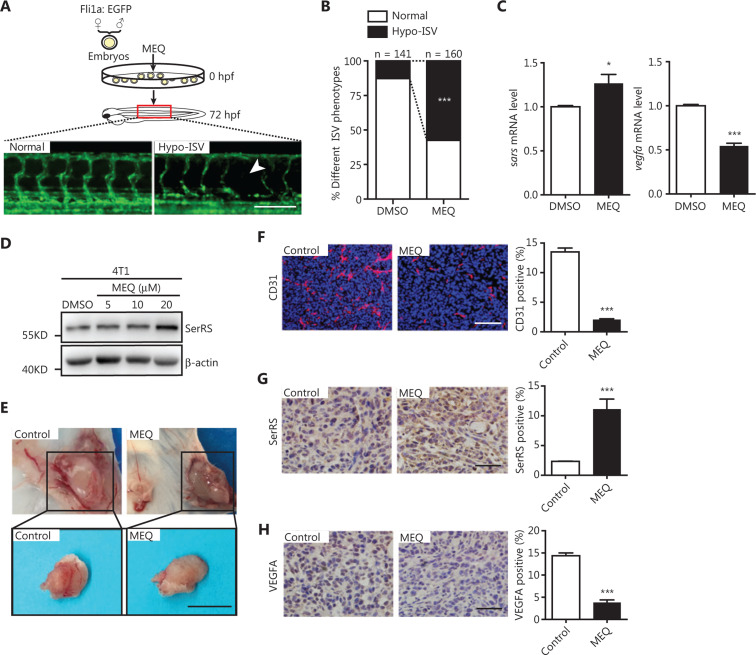
To investigate the effect of MEQ on angiogenesis in vivo, we first utilized the transgenic zebrafish Tg [Fli1a: EGFP] with EGFP specifically expressed in vascular endothelial cells20. We determined that the safe dosage of MEQ that zebrafish could tolerate was below 2.5 μM (Supplementary Figure S1). As expected, when treated with 2.5 μM MEQ, more zebrafish showed hypo-intersegmental vessel (Hypo-ISV) branches than those treated with DMSO (P < 0.001) (Figure 3A and 3B). We observed increased SerRS expression (P < 0.05) and decreased Vegfa expression (P < 0.001) in MEQ-treated zebrafish (Figure 3C), suggesting that MEQ induced SerRS expression to negatively regulate vascular development in fish.
Figure 3.
MEQ inhibits angiogenesis in vivo. (A, B) The embryos of transgenic zebrafish (Tg [Fli1a: EGFP]) at 1-2 cell stage were injected with MEQ and vascular development was monitored at 72 h post-fertilization (A) and the percentage of fish with deficiency in intersegmental vessels (Hypo-ISV) were analyzed (B). ***P < 0.001 using the χ2-test. (C) The qRT-PCR results show the expression of Sars (i.e., fish SerRS) and Vegfa in the zebrafish treated with MEQ or dimethyl sulfoxide (DMSO). Data are plotted as the mean ± SEM (n = 3; *P < 0.05; ***P < 0.001 using Student’s t-test). (D) Western blotting shows SerRS protein levels induced by MEQ in 4T1 cells. (E) Representative images of Matrigel plugs formed by 4T1 cells treated with MEQ or DMSO control. Scale bar represents 1 cm. (F–H) Immunofluorescence of CD31(F), immunochemical staining of SerRS (G) and VEGFA (H) in the Matrigel plugs and the quantification of positively-stained cells are plotted as the mean ± SEM (n = 5; *P < 0.05; ***P < 0.001 using Student’s t-test. Scale bars represent 100 μm.
To further investigate if MEQ functioned as a tumor angiogenesis inhibitor in mammals, we next performed a Matrigel plug angiogenesis assay in mice21. The 4T1 cells, mouse breast cancer cells responsive to MEQ (Figure 3D), were mixed with 10 μM of MEQ and embedded in Matrigel before being injected subcutaneously into Balb/c mice. At 1 week post-inoculation, the Matrigel plugs containing tumor cells were dissected for angiogenesis examination (Figure 3E). We observed greatly inhibited blood vessel formation in the MEQ-treated group compared to the control group (P < 0.001) (Figure 3E and 3F). IHC staining showed that MEQ significantly increased SerRS expression and reduced VEGFA expression in the cells of the Matrigel plugs (P < 0.001) (Figure 3G and 3H). Taken together, these results strongly suggested that MEQ robustly suppressed angiogenesis in vivo by enhancing SerRS expression.
MEQ suppressed the progression of breast cancer in a mouse allograft tumor model
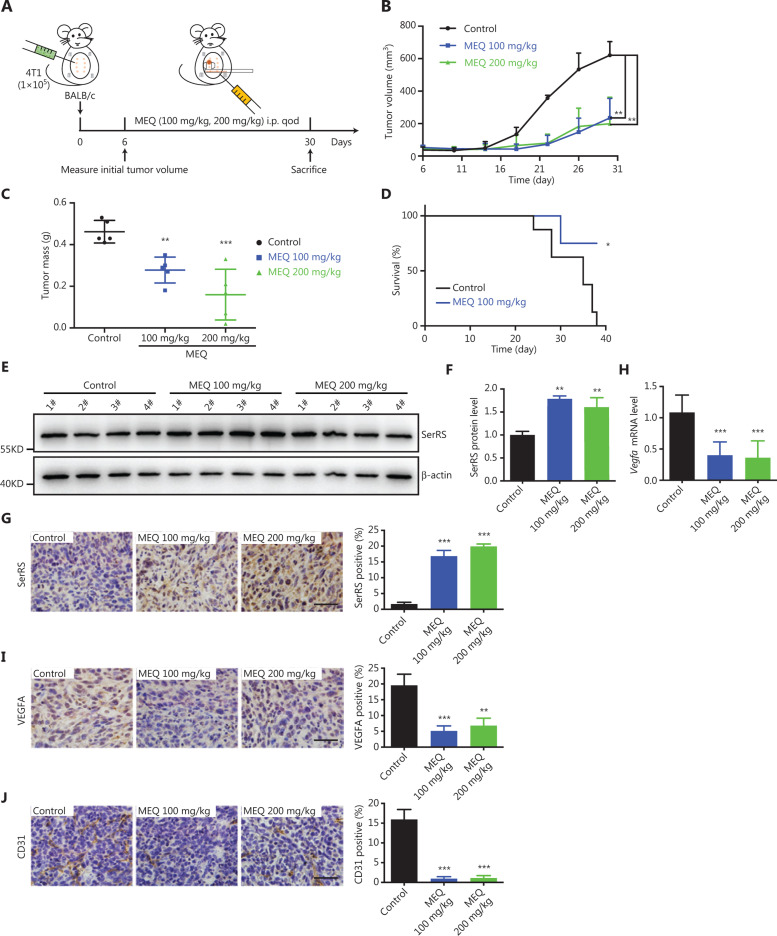
To further evaluate the anti-tumor effects of MEQ in vivo, we used a breast cancer allograft mouse model. The 4T1 highly malignant murine breast cancer cells were subcutaneously inoculated into female Balb/c mice. Six days after inoculation, the mice were treated with MEQ (100 mg/kg and 200 mg/kg) or DMSO every 2 days (Figure 4A). We observed greatly inhibited tumor growth in MEQ-treated mice (Figure 4B and 4C) and prolonged survival of tumor-bearing mice upon MEQ treatment (P < 0.05) (Figure 4D). MEQ-enhanced SerRS expression was also observed in tumor tissues by western blotting and IHC staining (Figure 4E–4G). VEGFA expression was also reduced by MEQ in tumor tissues (Figure 4H and 4I). As a result, dramatically inhibited tumor angiogenesis shown by IHC staining for CD31 was also observed in MEQ-treated mice (P < 0.001) (Figure 4J). Accordingly, poor tumor vascularization inhibited cell proliferation (Supplementary Figure 2A) and triggered more cell apoptosis (Supplementary Figure 2B).
Figure 4.
MEQ suppressed the progression of breast cancer allografts in mice. (A) Scheme of mouse experiments based on the breast cancer allograft model. qod: quaque omni die. (B) Tumor growth curve. (n = 5, **P < 0.01 using the Student’s t-test.). (C) Tumor weights at 30 days post-inoculation. (n = 5, **P < 0.01; ***P < 0.001 using Student’s t-test). (D) Kaplan-Meier survival curves of tumor-bearing Balb/c mice treated with MEQ (100 mg/kg) or dimethyl sulfoxide control (n = 8, *P < 0.05 using the log-rank test). (E, F) Western blotting to show SerRS protein levels in tumor tissues (E) and the densitometry quantification. (F) Data are plotted as the mean ± SEM (n = 4; **P < 0.01; ***P < 0.001 using Student’s t-test). (G) Immunohistochemical staining of SerRS in tumor tissue and its quantification (n = 5; ***P < 0.001 using Student’s t-test). (H) The qRT-PCR shows VEGFA mRNA levels of tumor tissues. (I, J) Immunohistochemical staining of VEGFA (I) and CD31 (J) in tumor tissues and quantification of positive cell numbers plotted as the mean ± SEM (n = 5; **P < 0.01; ***P < 0.001 using Student’s t-test). All scale bars represent 100 μm.
To exclude the possibility that MEQ-inhibited tumor progression was due to its high toxicity in mice, we measured the weight change of BALB/c mice during MEQ treatment and found no statistical difference when compared with the control groups (Supplementary Figure S3A). We also performed hemogram tests for mice treated with MEQ (Supplementary Table S1), and found that every index was within the normal range and comparable to the control group. Furthermore, we examined H&E-stained sections of major organs including the heart, liver, spleen, lung, kidney, and brain, and found no pathological damage to the organs of MEQ-treated mice (Supplementary Figure S3B). Together, these results suggested a low toxicity of MEQ. Furthermore, its inhibitory role on tumor progression was due to regulation of the SerRS-VEGFA pathway.
MEQ inhibited the growth of human TNBC xenografts in mice
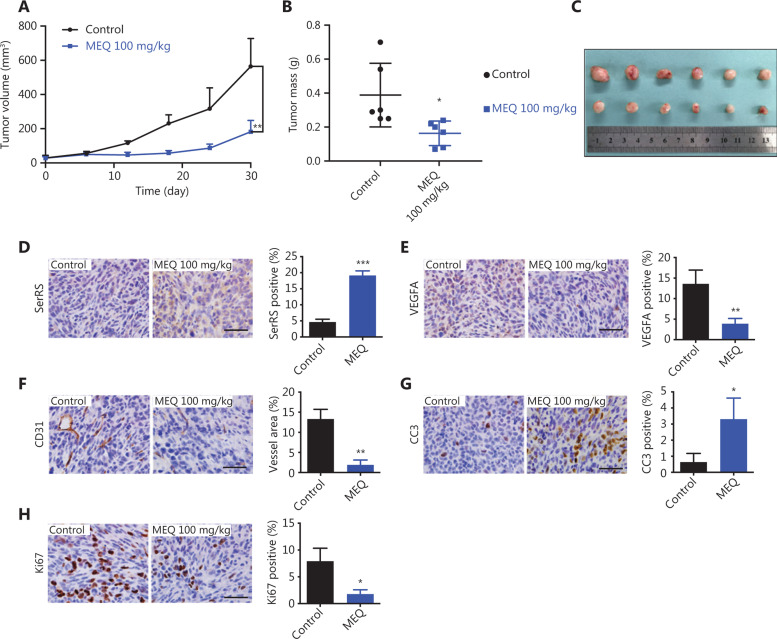
The MDA-MB-231 cell line-derived xenograft (CDX) mouse model can represent TNBC characteristics and is frequently used in Phase II clinical trials to test the efficacy of anti-cancer drugs22,23. Thus, we further examined the effect of MEQ on human TNBC xenografts in immunodeficient NOD-SCID mice. Consistently, we observed that MEQ strongly suppressed the growth of MDA-MB-231 xenografts (Figure 5A–5C). In addition, MEQ treatment also resulted in upregulated SerRS (P < 0.001) and downregulated VEGFA (P < 0.01) in TNBC xenografts (Figure 5D and 5E). Accordingly, CD31 staining showed dramatically inhibited angiogenesis in MEQ-treated xenografts (P < 0.01) (Figure 5F), leading to increased tumor cell apoptosis and inhibited cell proliferation as measured by IHC staining of CC3 (P < 0.05) and Ki-67 (P < 0.05), respectively (Figure 5G and 5H).
Figure 5.
MEQ inhibits the growth of human triple-negative breast cancer (TNBC) xenografts in mice. (A) The growth curves of MDA-MB-231 xenografts in mice treated with MEQ or the dimethyl sulfoxide control. Data are plotted as the mean ± SD. (B, C) At 30 days post-inoculation, the xenografts were dissected and weighted, n = 5. D-H Immunohistochemical staining for SerRS (D), VEGFA (E), CD31 (F), CC3 (G), and Ki67 (H) in the xenografts; n = 5. All scale bars represent 100 μm and all bar graphs are plotted as the mean ± SEM (ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001 using Student’s t-test).
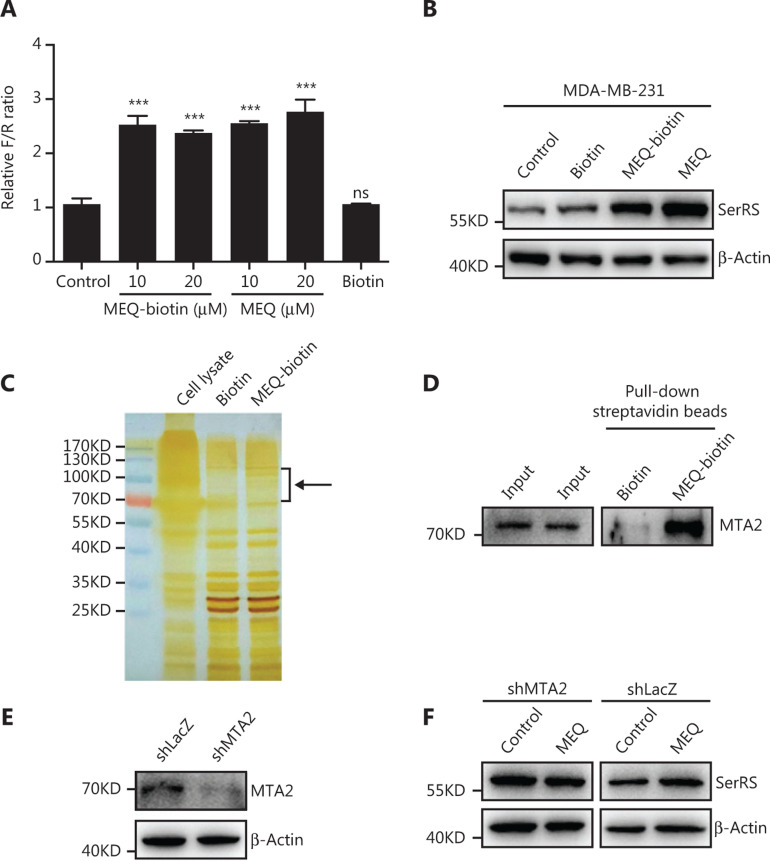
MEQ activated SerRS expression by targeting MTA2
To further investigate the direct target of MEQ, through which MEQ enhances SerRS transcription, we utilized a streptavidin-biotin affinity pull-down assay. First, we linked biotin to the nitrogen of MEQ (Supplementary Figures S4–S6), which did not affect its function in activating SerRS expression (Figure 6A and 6B). Next, we incubated the biotin-labelled MEQ (MEQ-biotin) with MDA-MB-231 cell lysates and affinity purified the complex using streptavidin-conjugated beads. Following SDS-PAGE and silver staining, we observed a few specific protein bands between 70 kDa and 120 kDa in MEQ-biotin purified elutes compared with that in biotin purified elutes (Figure 6C). Mass spectrometric analysis of these specific bands suggested that MTA2, a cofactor in the NuRD transcriptional regulatory complex, was a candidate target of MEQ. We further confirmed the interaction between MTA2 and MEQ by western blot analysis on biotin-MEQ pull-down samples (Figure 6D). To further investigate the involvement of MTA2 in MEQ-activated SerRS expression, we knocked down MTA2 by shRNA (Figure 6E). When MTA2 was silenced, MEQ could no longer upregulate SerRS expression in MDA-MB-231 cells (Figure 6F), strongly suggesting that MTA2 was a key mediator of MEQ, which induced SerRS expression and subsequently inhibited tumor angiogenesis.
Figure 6.
MEQ targets MTA2 to regulate SerRS expression. (A) Dual-luciferase reporter assay for biotin-conjugated MEQ (MEQ-biotin). Values are the mean ± SEM. ns, not significant, ***P < 0.001 using analysis of variance. (B) Western blots show that MEQ-biotin is as active as MEQ in regulating SerRS expression. (C) MEQ-interacting proteins were pulled down from the MDA-MB-231 cell lysate with streptavidin beads and analyzed by SDS-PAGE and silver staining. The black arrow indicates protein bands that may specifically interact with MEQ. (D) Western blotting to confirm that MTA2 was co-purified with MEQ-biotin. (E) Western blotting to show the knockdown efficiency of shRNA against MTA2 (shMTA2) in MDA-MB-231 cells. (F) Western blotting to show the protein levels of MEQ required to induce SerRS expression in MTA2-silenced (shMTA2) or control (shLacZ) MDA-MB-231 cells.
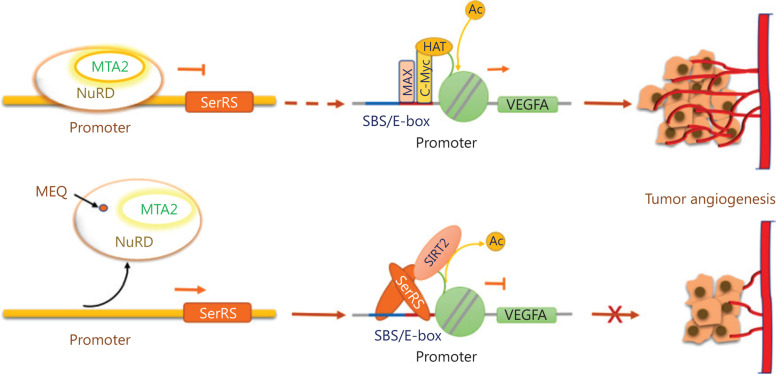
Taken together, these results suggested that MEQ interacted with MTA2, which is a nonenzymatic subunit in the nucleosome remodeling and histone deacetylase (NuRD) complex, to upregulate SerRS expression, and inhibit the expression of VEGFA and TNBC angiogenesis (Figure 7).
Figure 7.
Mechanistic model to show how MEQ inhibits triple-negative breast cancer (TNBC) angiogenesis by regulating the MTA2/SerRS/VEGFA axis. The MTA2-containing NuRD deacetylase complex binds to the SerRS promoter to suppress its transcription, which allows more SerRS competitor, i.e., c-Myc proteins, to bind to the VEGFA promoter and activate VEGFA expression and tumor angiogenesis. MEQ interacts with MTA2 and may cause the release of the NuRD complex from the SerRS promoter, resulting in elevated expression of SerRS, which competes off the c-Myc and binds to the VEGFA promoter to suppress its transcription and inhibit tumor angiogenesis.
Discussion
Angiogenesis plays an important role in the growth and progression of TNBC. Several groups have reported that VEGFA is expressed at significantly higher levels in TNBC patient tumors compared to those of non-TNBC patients24–26. Thus, drugs targeting the VEGFA pathway could be an effective treatment for TNBC. However, VEGFA-targeted drugs, such as the bevacizumab neutralizing antibody, have only shown a slight benefit for TNBC patients at the cost of adverse side effects10,11. Other small inhibitors targeting receptor tyrosine kinase (RTK) have relatively low general response rates in conjunction with particular toxicity profiles27. Recent reports showed that drugs targeting VEGFA itself or its receptors have always resulted in drug resistance, primarily due to the activation of alternative proangiogenic mechanisms in the tumors28–30. Thus, targeting a transcriptional factor that regulates multiple factors and signaling pathways important for angiogenesis could be a better solution. We found that nuclear-localized SerRS was a robust transcriptional repressor of VEGFA, through direct competition with c-Myc and NFκB for binding to the VEGFA promoter12,13. In addition, our previous studies showed that SerRS triggered the senescence of cervical tumor cells in mice by shortening telomeres31. Because MEQ treatment can increase the expression of SerRS in MDA-MB-231 cells, this compound could be a potential and multifunctional drug against TNBC and potentially against other cancers.
MEQ is an isoflavone derivative that can act as a phytoestrogen in mammals32. There is controversy about whether isoflavones are risk factors or effective treatments for breast cancer33–39. Most studies have shown that the carcinogenic character of isoflavone is due to its binding to estrogen receptors (ER)40,41. Because TNBC is ER negative, it might be one of the reasons why MEQ is effective in our TNBC models. Consistent with the anti-angiogenesis and anti-tumor functions of MEQ to TNBC, other researchers have reported that isoflavones have anti-proliferative properties in TNBC, both in vitro and in vivo40,42–44. Specifically, isoflavones can decrease microvascular density, reduce circulating levels of VEGF, and increase endostatin levels in rats with mammary tumors45. Thus, our finding that MEQ inhibited the expression of VEGFA in TNBC through the MTA2/SerRS/VEGFA pathway revealed the underlying mechanism for isoflavones and their derivatives, allowing the optimization of this natural molecule for cancer treatment.
Isoflavones, like formononetin, has increased benefits with fewer side effects when used as a combination drug with chemotherapy or as a postoperative adjuvant therapy46,47. We also found that MEQ displayed few adverse effects in mice, suggesting that MEQ may have clinical applications in combination with other chemotherapeutic drugs in TNBC treatment. Besides the anti-angiogenesis function of MEQ, we also found that MEQ inhibited TNBC proliferation and promoted apoptosis in vivo, similar to other isoflavones and their derivatives40,42–44.
The NuRD complex has been found to interact with different factors to promote cancer development48–50. The potential target of MEQ, MTA2, as one nonenzymatic subunit in the NuRD complex, confers functional specificity for the NuRD complex by associating with different transcription factors or targeting distinct gene loci50. Moreover, MTA2 is found to be overexpressed in human breast cancer51, and the VEGF signaling pathway was able to be modulated by targeting MTA2 expression in non small cell lung cancer cells52. Thus, characterizing drugs that target NuRD subunits for cancer therapy should be more specific and effective than those targeting pan-epigenetic modification enzymes. Our study is the first to show the connection between MTA2 and SerRS in modulating VEGFA expression in MDA-MB-231 cells. Our finding that MTA2 could be targeted by MEQ also provides a way to manipulate MTA2-mediated gene expression, which could be developed for cancer treatment. Further studies are needed to clarify the exact mechanism of MEQ in regulating MTA2 function.
In conclusion, our study identified an anti-angiogenesis compound, MEQ, which inhibited the progression of TNBC by targeting the MTA2/SerRS/VEGFA pathway.
Supporting Information
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (Grant Nos. 81772974, 81972882, and 81874297), the Bilateral Inter-Governmental S&T Cooperation Project from Ministry of Science and Technology of China (Grant No. 2018YFE0114300), and the 1.3.5 Project for Disciplines of Excellence, West China Hospital, Sichuan University. We thank Dr. Scott McKercher (The Scripps Research Institute, La Jolla, CA, USA) and Dr. Phillip Bryant (Children’s Hospital of Philadelphia, PA, USA), and Dr. Cameron R. McKay (Nankai University School of Medicine, Tianjin, China) for revising the manuscript.
Conflict of interest statement
No potential conflicts of interest are disclosed.
References
- 1.Warburg O. On the origin of cancer cells. Science. 1956;123:309–14. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 2.DeSantis CE, Ma J, Goding Sauer A, Newman LA, Jemal A. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J Clin. 2017;67:439–48. doi: 10.3322/caac.21412. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 4.Waks AG, Winer EP. Breast cancer treatment: a review. J Am Med Assoc. 2019;321:288–300. doi: 10.1001/jama.2018.19323. [DOI] [PubMed] [Google Scholar]
- 5.Ismail-Khan R, Bui MM. A review of triple-negative breast cancer. Cancer Control. 2010;17:173–6. doi: 10.1177/107327481001700305. [DOI] [PubMed] [Google Scholar]
- 6.Hillen F, Griffioen AW. Tumour vascularization: sprouting angiogenesis and beyond. Cancer Metastasis Rev. 2007;26:489–502. doi: 10.1007/s10555-007-9094-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lacal PM, Graziani G. Therapeutic implication of vascular endothelial growth factor receptor-1 (VEGFR-1) targeting in cancer cells and tumor microenvironment by competitive and non-competitive inhibitors. Pharmacol Res. 2018;136:97–107. doi: 10.1016/j.phrs.2018.08.023. [DOI] [PubMed] [Google Scholar]
- 8.Smith IE, Pierga J-Y, Biganzoli L, Cortés-Funes H, Thomssen C, Pivot X, et al. First-line bevacizumab plus taxane-based chemotherapy for locally recurrent or metastatic breast cancer: safety and efficacy in an open-label study in 2251 patients. Ann Oncol. 2010;22:595–602. doi: 10.1093/annonc/mdq430. [DOI] [PubMed] [Google Scholar]
- 9.Samant RS, Shevde LA. Recent advances in anti-angiogenic therapy of cancer. Oncotarget. 2011;2:122–34. doi: 10.18632/oncotarget.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sikov WM, Berry DA, Perou CM, Singh B, Cirrincione CT, Tolaney SM, et al. Impact of the addition of carboplatin and/or bevacizumab to neoadjuvant once-per-week paclitaxel followed by dose-dense doxorubicin and cyclophosphamide on pathologic complete response rates in stage ii to iii triple-negative breast cancer: CALGB 40603 (alliance) J Clin Oncol. 2015;33:13. doi: 10.1200/JCO.2014.57.0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.von Minckwitz G, Schneeweiss A, Loibl S, Salat C, Denkert C, Rezai M, et al. Neoadjuvant carboplatin in patients with triple-negative and HER2-positive early breast cancer (GeparSixto; GBG 66): a randomised phase 2 trial. Lancet Oncol. 2014;15:747–56. doi: 10.1016/S1470-2045(14)70160-3. [DOI] [PubMed] [Google Scholar]
- 12.Shi Y, Xu X, Zhang Q, Fu G, Mo Z, Wang GS, et al. tRNA synthetase counteracts c-MYC to develop functional vasculature. eLife. 2014;3:e02349. doi: 10.7554/eLife.02349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu C-Y, Wang P-C, Tsai H-J. Competitive binding between Seryl-tRNA synthetase/YY1 complex and NFKB1 at the distal segment results in differential regulation of human VEGFA promoter activity during angiogenesis. Nucleic Acids Res. 2016;45:2423–37. doi: 10.1093/nar/gkw1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaufmann KB, Buning H, Galy A, Schambach A, Grez M. Gene therapy on the move. EMBO Mol Med. 2013;5:1642–61. doi: 10.1002/emmm.201202287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baruteau J, Waddington SN, Alexander IE, Gissen P. Gene therapy for monogenic liver diseases: clinical successes, current challenges and future prospects. J Inherit Metab Dis. 2017;40:497–517. doi: 10.1007/s10545-017-0053-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santos R, Ursu O, Gaulton A, Bento AP, Donadi RS, Bologa CG, et al. A comprehensive map of molecular drug targets. Nat Rev Drug Discov. 2017;16:19. doi: 10.1038/nrd.2016.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan E, Tan M, Xin J, Sudarsanam S, Johnson DE. Interactions between traditional Chinese medicines and Western therapeutics. Curr Opin Drug Discov Devel. 2010;13:50–65. [PubMed] [Google Scholar]
- 18.Maplestone RA, Stone MJ, Williams DH. The evolutionary role of secondary metabolites – a review. Gene. 1992;115:151–7. doi: 10.1016/0378-1119(92)90553-2. [DOI] [PubMed] [Google Scholar]
- 19.Rajput S, Gardner CR, Failes TW, Arndt GM, Black DS, Kumar N. Synthesis and anticancer evaluation of 3-substituted quinolin-4-ones and 2,3-dihydroquinolin-4-ones. Bioorg Med Chem. 2014;22:105–15. doi: 10.1016/j.bmc.2013.11.047. [DOI] [PubMed] [Google Scholar]
- 20.Tobia C, Gariano G, Guerra J, Presta M. Zebrafish embryo intersegmental vessels: a tool for investigating sprouting angiogenesis. Methods Mol Biol. 2015;1214:173–84. doi: 10.1007/978-1-4939-1462-3_10. [DOI] [PubMed] [Google Scholar]
- 21.Akhtar N, Dickerson EB, Auerbach R. The sponge/Matrigel angiogenesis assay. Angiogenesis. 2002;5:75–80. doi: 10.1023/a:1021507031486. [DOI] [PubMed] [Google Scholar]
- 22.Voskoglou-Nomikos T, Pater JL, Seymour L. Clinical predictive value of the in vitro cell line, human xenograft, and mouse allograft preclinical cancer models. Clin Cancer Res. 2003;9:4227–39. [PubMed] [Google Scholar]
- 23.Lallo A, Schenk MW, Frese KK, Blackhall F, Dive C. Circulating tumor cells and CDX models as a tool for preclinical drug development. Transl Lung Cancer Res. 2017;6:397–408. doi: 10.21037/tlcr.2017.08.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bahhnassy A, Mohanad M, Shaarawy S, Ismail MF, El-Bastawisy A, Ashmawy AM, et al. Transforming growth factor-beta, insulin-like growth factor I/insulin-like growth factor I receptor and vascular endothelial growth factor-A: prognostic and predictive markers in triple-negative and non-triple-negative breast cancer. Mol Med Rep. 2015;12:851–64. doi: 10.3892/mmr.2015.3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bahnassy A, Mohanad M, Ismail MF, Shaarawy S, El-Bastawisy A, Zekri AR. Molecular biomarkers for prediction of response to treatment and survival in triple negative breast cancer patients from Egypt. Exp Mol Pathol. 2015;99:303–11. doi: 10.1016/j.yexmp.2015.07.014. [DOI] [PubMed] [Google Scholar]
- 26.Bender RJ, Mac Gabhann F. Expression of VEGF and semaphorin genes define subgroups of triple negative breast cancer. PLoS One. 2013;8:e61788. doi: 10.1371/journal.pone.0061788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qin S, Li A, Yi M, Yu S, Zhang M, Wu K. Recent advances on anti-angiogenesis receptor tyrosine kinase inhibitors in cancer therapy. J Hematol Oncol. 2019;12:27. doi: 10.1186/s13045-019-0718-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kopetz S, Hoff PM, Morris JS, Wolff RA, Eng C, Glover KY, et al. Phase II trial of infusional fluorouracil, irinotecan, and bevacizumab for metastatic colorectal cancer: efficacy and circulating angiogenic biomarkers associated with therapeutic resistance. J Clin Oncol. 2010;28:453–9. doi: 10.1200/JCO.2009.24.8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lieu CH, Tran H, Jiang ZQ, Mao M, Overman MJ, Lin E, et al. The association of alternate VEGF ligands with resistance to anti-VEGF therapy in metastatic colorectal cancer. PLoS One. 2013;8:e77117. doi: 10.1371/journal.pone.0077117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vasudev NS, Reynolds AR. Anti-angiogenic therapy for cancer: current progress, unresolved questions and future directions. Angiogenesis. 2014;17:471–94. doi: 10.1007/s10456-014-9420-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, Li X, Cao M, Jiang Y, Yan J, Liu Z, et al. Seryl TRNA synthetase cooperates with POT1 to regulate telomere length and cellular senescence. Signal Transduct Target Ther. 2019;4:1–11. doi: 10.1038/s41392-019-0078-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lecomte S, Demay F, Ferriere F, Pakdel F. Phytochemicals targeting estrogen receptors: beneficial rather than adverse effects? Int J Mol Sci. 2017;18:1381. doi: 10.3390/ijms18071381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ong SKL, Shanmugam MK, Fan L, Fraser SE, Arfuso F, Ahn KS, et al. Focus on formononetin: anticancer potential and molecular targets. Cancers (Basel). 2019;11:611. doi: 10.3390/cancers11050611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jordan VC. Avoiding the bad and enhancing the good of soy supplements in breast cancer. J Natl Cancer Inst. 2014;106:dju233. doi: 10.1093/jnci/dju233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y, Meeran SM, Patel SN, Chen H, Hardy TM, Tollefsbol TO. Epigenetic reactivation of estrogen receptor-alpha (ERalpha) by genistein enhances hormonal therapy sensitivity in eralpha-negative breast cancer. Mol Cancer. 2013;12:9. doi: 10.1186/1476-4598-12-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bondesson M, Gustafsson JA. Does consuming isoflavones reduce or increase breast cancer risk? Genome Med. 2010;2:90. doi: 10.1186/gm211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koch L. Nutrition: high isoflavone intake delays puberty onset and may reduce breast cancer risk in girls. Nat Rev Endocrinol. 2010;6:595. doi: 10.1038/nrendo.2010.157. [DOI] [PubMed] [Google Scholar]
- 38.Kang X, Zhang Q, Wang S, Huang X, Jin S. Effect of soy isoflavones on breast cancer recurrence and death for patients receiving adjuvant endocrine therapy. Can Med Assoc J. 2010;182:1857–62. doi: 10.1503/cmaj.091298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shu XO, Zheng Y, Cai H, Gu K, Chen Z, Zheng W, et al. Soy food intake and breast cancer survival. J Am Med Assoc. 2009;302:2437–43. doi: 10.1001/jama.2009.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taylor CK, Levy RM, Elliott JC, Burnett BP. The effect of genistein aglycone on cancer and cancer risk: a review of in vitro, preclinical, and clinical studies. Nutr Rev. 2009;67:398–415. doi: 10.1111/j.1753-4887.2009.00213.x. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y, Chen WF, Lai WP, Wong MS. Soy isoflavones and their bone protective effects. Inflammopharmacology. 2008;16:213–5. doi: 10.1007/s10787-008-8018-7. [DOI] [PubMed] [Google Scholar]
- 42.Chen J, Zeng J, Xin M, Huang W, Chen X. Formononetin induces cell cycle arrest of human breast cancer cells via IGF1/PI3K/AKT pathways in vitro and in vivo. Horm Metab Res. 2011;43:681–6. doi: 10.1055/s-0031-1286306. [DOI] [PubMed] [Google Scholar]
- 43.Chen J, Sun L. Formononetin-induced apoptosis by activation of RAS/P38 mitogen-activated protein kinase in estrogen receptor-positive human breast cancer cells. Horm Metab Res. 2012;44:943–8. doi: 10.1055/s-0032-1321818. [DOI] [PubMed] [Google Scholar]
- 44.Chen J, Zhao X, Ye Y, Wang Y, Tian J. Estrogen receptor beta-mediated proliferative inhibition and apoptosis in human breast cancer by calycosin and formononetin. Cell Physiol Biochem. 2013;32:1790–7. doi: 10.1159/000356612. [DOI] [PubMed] [Google Scholar]
- 45.Kang X, Jin S, Zhang Q. Antitumor and antiangiogenic activity of soy phytoestrogen on 7,12-dimethylbenz[alpha]anthracene-induced mammary tumors following ovariectomy in Sprague-Dawley rats. J Food Sci. 2009;74:H237–42. doi: 10.1111/j.1750-3841.2009.01278.x. [DOI] [PubMed] [Google Scholar]
- 46.Park S, Bazer FW, Lim W, Song G. The O-methylated isoflavone, formononetin, inhibits human ovarian cancer cell proliferation by sub G0/G1 cell phase arrest through PI3K/AKT and ERK1/2 inactivation. J Cell Biochem. 2018;119:7377–87. doi: 10.1002/jcb.27041. [DOI] [PubMed] [Google Scholar]
- 47.Zhang X, Ni Q, Wang Y, Fan H, Li Y. Synergistic anticancer effects of formononetin and temozolomide on glioma C6 cells. Biol Pharm Bull. 2018;41:1194–202. doi: 10.1248/bpb.b18-00002. [DOI] [PubMed] [Google Scholar]
- 48.Sen N, Gui B, Kumar R. Physiological functions of MTA family of proteins. Cancer Metastasis Rev. 2014;33:869–77. doi: 10.1007/s10555-014-9514-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumar R. Functions and clinical relevance of MTA proteins in human cancer. Preface. Cancer Metastasis Rev. 2014;33:835. doi: 10.1007/s10555-014-9509-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lai AY, Wade PA. Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat Rev Cancer. 2011;11:588–96. doi: 10.1038/nrc3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Covington KR, Fuqua SA. Role of MTA2 in human cancer. Cancer Metastasis Rev. 2014;33:921–8. doi: 10.1007/s10555-014-9518-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang B, Tao F, Zhang H. Metastasis-associated protein 2 promotes the metastasis of non-small cell lung carcinoma by regulating the ERK/AKT and VEGF signaling pathways. Mol Med Rep. 2018;17:4899–908. doi: 10.3892/mmr.2018.8535. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.