Abstract
The conserved N-glycan on Asn297 of immunoglobulin G (IgG) has significant impacts on antibody effector functions, and is a frequent target for antibody engineering. Chemoenzymatic synthesis has emerged as a strategy for producing antibodies with homogenous glycosylation and improved effector functions. Central to this strategy is the use of enzymes with activity on the Asn297 glycan. EndoS and EndoS2, produced by Streptococcus pyogenes, are endoglycosidases with remarkable specificity for Asn297 glycosylation, making them ideal tools for chemoenzymatic synthesis. Although both enzymes are specific for IgG, EndoS2 recognizes a wider range of glycans than EndoS. Recent progress has been made in understanding the structural basis for their activities on antibodies. In this review, we examine the molecular mechanism of glycosidic bond cleavage by these enzymes and how specific point mutations convert them into glycosynthases. We also discuss the structural basis for differences in the glycan repertoire that IgG-active endoglycosidases recognize, which focuses on the structure of the loops within the glycoside hydrolase (GH) domain. Finally, we discuss the important contributions of carbohydrate binding modules (CBMs) to endoglycosidase activity, and how CBMs work in concert with GH domains to produce optimal activity on IgG.
Keywords: endoglycosidase, glycosynthase, IgG, structure, antibody, EndoS
Introduction
Glycosylation is a common post-translational modification of proteins in many biological systems and refers to the addition of glycans to the side chains of proteins, creating glycoproteins. These modifications can impact proteins substantially, influencing their folding patterns, stability, serum half-life and function (Helenius and Aebi 2001, Petrescu et al. 2006; Marth and Grewal, 2008; Hart and Copeland 2010). They are particularly important in the immune system, contributing to lymphocyte development, cell adhesion, immune signaling, and host-pathogen interactions (Varki 1993; Haltiwanger and Lowe 2004; Nimmerjahn and Ravetch 2008; Jefferis 2009; Alhorn et al. 2010; Dalziel et al. 2014; Macauley et al. 2014). Perhaps no carbohydrate is better studied than the conserved N-linked glycan attached to the Asn297 residue of the fragment crystallizable (Fc) region of immunoglobulin G (IgG) antibodies (Figure 1a). The composition of this complex biantennary glycan has important effects on the structure and dynamics of the antibody Fc, which in turn critically modulate its effector functions (Jennewein and Alter 2017). For instance, removal of the entire Asn297-linked glycan results in almost complete loss of Fc γ receptor (FcγR) binding and complement fixation (Lu et al. 2015; Subedi Ganesh and Barb 2015). However, the absence of the core fucose moiety alone increases binding to the activating receptor FcγRIIIa ~ 100-fold, producing markedly increased antibody-dependent cellular cytotoxicity (Shields et al. 2002; Ferrara et al. 2011). The degree of galactosylation and sialylation can also modulate FcγR binding to a lesser extent (Nimmerjahn et al. 2007; Thomann et al. 2016). Additionally, α(2,6)-sialylation may have anti-inflammatory properties through interactions with DC-SIGN (Anthony et al. 2008a, 2008b), although other groups have reported conflicting findings, suggesting other cell surface lectins may be involved (Yu et al. 2013; Campbell et al. 2014; Sharma et al. 2014).
Fig. 1.
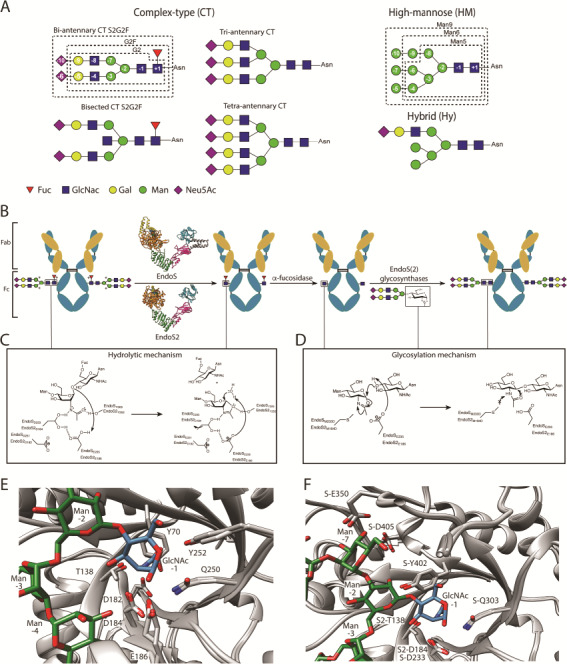
(a) The different types of N-glycosylation. (b) Schematic representation of the chemoenzymatic synthesis of homogenous IgGs using EndoS(2) WT, EndoS(2) glycosynthase mutants and glycan oxazoline derivatives. Asterisks (*) denote carbohydrates that can be variable. Representation of structural details of EndoS and EndoS2 are displayed in Figure 2. (c) Catalytic mechanism proposed for hydrolysis by EndoS(2). (d) Glycosylation mechanism of EndoS(2) mutants. (e) Structural superposition of the catalytic sites of EndoS (4NUY) and EndoS2 (6MDV), demonstrating conservation of catalytic residues, using EndoS2 numbering. (f) Residue mutations in the binding site of EndoS(2) to obtain efficient glycosynthases are labeled: EndoSD233M, EndoSD233Q/Q303L, EndoSD233Q/E350Q, EndoSD233Q/Y402F/D405Q, EndoS2D184M and EndoS2T138Q.
IgG antibodies are an important and rapidly growing part of the clinical arsenal, and now glyco-engineered antibodies are making it to market. Since 2013, the United States Food and Drug Administration has approved three afucosylated antibodies, predicted to be more potent than their fucosylated counterparts through enhanced FcγR binding. Most glyco-engineered antibodies are produced using fucosylation inhibitors or cell lines genetically altered to decrease fucosylation (Pereira et al. 2018). While this strategy has been successful, it still results in variable and heterogeneous glycosylation that can produce significant differences in batch-to-batch efficacy (Schiestl et al. 2011). An alternative strategy that has emerged is the chemoenzymatic synthesis of antibodies with predetermined homogenous glycosylation, although industrial application of this strategy has thus far been cost-prohibitive. Central to this strategy is the use of enzymes with activity on the Asn297 carbohydrate of IgG. The bacteria Streptococcus pyogenes secretes two endoglycosidases, EndoS and EndoS2, which are remarkably specific for IgG. They are “V”-shaped, multi-domain enzymes that rely on a glycoside hydrolase (GH) domain and carbohydrate binding module (CBM) for activity (Dixon et al. 2014; Trastoy et al. 2014, 2018; Klontz et al. 2019). EndoS has a narrower specificity, recognizing only non-bisected complex biantennary glycans, while EndoS2 recognizes complex biantennary, hybrid and high-mannose glycans. Together, they can remove more than 20 glycoforms from Asn297, helping the bacteria to evade the immune system. Since their discovery, EndoS, EndoS2 and specific point mutants thereof have been described in pathways to chemoenzymatically synthesize antibodies with homogenous glycosylation (Figure 1b). This review provides a brief overview of the mechanism of action of these enzymes, then delves into the structural basis by which they specifically recognize glycosylated IgG antibodies.
Mechanism of action of GHs and glycosynthases
GHs, which can be separated into 161 families based on amino acid sequence similarity, as in the Carbohydrate-Active Enzymes Database, (CAZy; www.cazy.org), are enzymes that cleave glycosidic bonds. GH family 18 (GH18) is predominantly composed of chitinases (EC 3.2.1.14) and endo-β-N-acetylglucosaminidases (ENGases) (EC 3.2.1.96), the latter of which contains EndoS, EndoS2 and other IgG-active endoglycosidases. Several other ENGases reside in GH85, but are not the focus of this review. Chitinases break down chitin, a linear polymer of β-1,4-linked-N-acetylglucosamine, while ENGases hydrolyze the chitobiose (GlcNAc2) core of N-linked glycans, such as those present on antibodies. Although their substrates differ, both chitinases and ENGases cleave between two β-1,4-linked-N-acetylglucosamine residues, and their catalytic mechanisms are thought to be conserved. The mechanism of chitinases has been studied extensively (White and Rose 1997; van Aalten et al. 2001; Williams et al. 2002; Jitonnom et al. 2011). Unlike other retaining GHs that form a covalent glycosyl-enzyme intermediate, chitinases hydrolyze the glycosidic bond through a mechanism that involves the participation of the 2-acetamide group of the substrate GlcNAc (−1) (van Aalten et al. 2001; Williams et al. 2002; Synstad et al. 2004). This mechanism involves a highly conserved D1XD2XE catalytic motif in the enzyme that performs a double displacement reaction (Figure 1c).
In the initial step, the binding of the substrate to specific residues of the binding pocket generates a distortion of GlcNAc (−1), and a general acid/base residue (e.g., E235 in EndoS, E186 in EndoS2) transfers a proton to the anomeric oxygen, while the carbonyl oxygen of the N-acetamido group of GlcNAc (−1) attacks the anomeric center to form an oxazoline intermediate (van Aalten et al. 2001, Williams et al. 2002; Jitonnom et al. 2011). Recent quantum mechanics/molecular mechanics (QM/MM) metadynamics simulations on chitinase B from Serratia marcescens (SmChiB) suggest that this reaction intermediate is a neutral oxazoline with an oxazolinium ion formed on the pathway toward the reaction products (Coines et al. 2018). A second carboxylate residue (D2: e.g., D233 in EndoS, D184 in EndoS2) assists the oxazoline intermediate through a hydrogen bond, orienting it and enhancing the nucleophilicity of the acetamido group that attacks the anomeric center (Williams et al. 2002). In the second step of the reaction, the same general acid/base residue from the first step now deprotonates an incoming water. This water molecule attacks the anomeric carbon, breaking the oxazoline ring and regenerating the sugar hemiacetal product with overall retention of stereochemistry (Figure 1c) (van Aalten et al. 2001). Before the product is released, GlcNAc (−1) can often be found in a skew-boat conformation, suggesting that this is a normal part of the catalytic cycle (Hsieh et al. 2010; Malecki et al. 2013; Speciale et al. 2014; Fadel et al. 2015; Ranok et al. 2015; Itoh et al. 2016, Klontz et al. 2019). In addition, other conserved residues in the GH18 ENGases contribute to stabilize the reaction intermediates (e.g., Q250 and Y252 in EndoS2), while Y70 and T138 stabilize the charge on D182 (D1), and D182 keeps D184-E235 protonated in EndoS2 (Figure 1c) (Synstad et al. 2004).
If, during the second step of the reaction, a sugar molecule replaces the role of water, a glycosidic linkage is created (Figure 1d). In this case, the reaction is referred to as transglycosylation. The GlcNAc (+1) in the active site is referred to as the acceptor, while the incoming sugar is the donor. Most ENGases are capable of performing transglycosylation in addition to hydrolysis; however, transglycosylation is usually very inefficient because the product remains an excellent substrate for hydrolysis. To get appreciable accumulation of transglycosylation product, a large excess of donor is usually required. Transglycosylation efficiency is determined by the ratio between transglycosylation and hydrolysis rates for the enzyme. Increasing transglycosylation or decreasing hydrolysis both serve to increase the amount of product produced. To circumvent the necessity for large excesses of donor, Mackenzie et al. (1998) introduced an alternative approach in which they mutated a catalytic residue (in their case, the nucleophile). Another key breakthrough in the field was the identification of N-linked oxazolines as glycosyl donors for these enzymes (Fujita et al. 2001; Noguchi et al. 2009; Wang 2011; Fairbanks 2013). The oxazoline simulates the high-energy intermediate in the ENGase catalytic site and can be efficiently attacked by the deprotonated O4 of the GlcNAc (+1) attached to a peptide or a protein (Figure 1b). When the enzyme is provided a suitably activated donor (oxazolines), the enzyme can still catalyze transglycosylation without the full catalytic machinery. However, it cannot catalyze hydrolysis, so product accumulation is very efficient. Such mutants are called glycosynthases, and although this term traditionally applied to enzymes in which the nucleophile has been mutated, the terminology has been expanded to ENGases that lack a proteinaceous nucleophile, so an assisting residue is mutated. In this case, glycosynthase versions of ENGases are usually created by mutating the assisting aspartic acid (D2).
Chemoenzymatic synthesis of IgG
In the past decade, a chemoenzymatic approach was developed in order to obtain homogenous glycoproteins, including therapeutic monoclonal antibodies (Fairbanks 2017; Li and Wang 2018). A general approach can be described in two steps (Figure 1b). In the first step, an ENGase from GH18 or GH85 hydrolyzes a heterogeneous mixture of glycoforms obtained by any expression system. In a second step, an ENGase with deficient hydrolytic activity (i.e., a glycosynthase mutant) or a wild-type ENGase transfers a synthetic glycan donor bearing a reactive oxazoline moiety to the deglycosylated protein that retains a GlcNAc (+1) from the first step. GH85 ENGases show the same substrate-mediated catalytic mechanism as GH18 ENGases, with the exception that the assisting catalytic residue (D2) is an asparagine instead of an aspartic acid. The glycosynthase mutants are obtained by mutating this assisting residue in order to abolish the hydrolytic activity of the enzymes. It has been described that mutation of this residue in a wide range of endoglycosidases has similar effects, creating a path to leverage the inherent substrate specificity and catalytic machinery of endoglycosidases to create glycosynthases for myriad applications. These endoglycosidases/glycosynthases exhibit a wide range of specificities for different glycans, allowing their use to be tailored to the desired glycoform.
The first example of remodeling the N-glycan of the Fc domain without denaturing the protein was described by Wei et al. (2008). The IgG1 Fc domain was expressed in yeast Pichia pastoris, and oligomannose N-glycans produced in this expression system were deglycosylated using EndoH to yield the pure GlcNAc-Fc glycoform. Then, EndoA was used to transfer homogenous biantennary glycans to the GlcNAc-Fc domain.
After the identification of enzymes that are specific for IgG Fc, EndoS and EndoS2 became important tools for glycoengineering monoclonal antibodies, as their reactions are efficient and can be used to specifically remodel the Fc glycan even when glycans are present on the Fab (Huang et al. 2012). In 2012, Goodfellow et al. (2012) showed that EndoSWT has transglycosylation activity, and is able to use Man3GlcNAc oxazoline to glycosylate GlcNAc-IgG, albeit with a very low yield because of the remaining hydrolytic activity of the enzyme. The same year, Huang et al. (2012) described two glycosynthase mutants of EndoS (EndoSD233A and EndoSD233Q; Figure 1f). They used wild-type EndoS to remove the glycans while the glycosynthase mutants were used to add synthetic glycans back onto the same site in order to generate G0F, G1F and G2F glycoforms, including fully sialylated (S2G2F), non-fucosylated (G2) and azido-tagged glycoforms, the last of which can be further modified into drug conjugates (Huang et al. 2012; Iwamoto et al. 2018b; Li and Wang 2018), imaging probes and epitope tags (Lopez Aguilar et al. 2017). The D233A mutation targets the assisting residue (D2), discussed above, which forms interactions with the N-acetyl group and helps to orient the substrate for hydrolysis. Mutation of this residue reduces hydrolysis while maintaining transglycosylation capabilities, although the precise mechanism is uncertain. Presumably, the mutant enzyme still allows oxazoline derivatives to enter the binding pocket while the acid/base deprotonates GlcNAc (+1) and catalyzes the formation of a glycosidic linkage. These studies also examined the ability of EndoS and glycosynthase mutants to accommodate core fucosylation. It was found that the glycosynthases could recognize both fucosylated and non-fucosylated rituximab (making it the first glycosynthase to do so), however, the transglycosylation rate was much higher with the fucosylated substrate. (Goodfellow et al. 2012; Huang et al. 2012). Analysis of the crystal structures of EndoS and EndoS2 in complex with glycans reveals there is sufficient space in the binding cavity to accommodate GlcNAc (+1) and its accompanying fucose. The region contains aromatic residues which may form interactions with fucose to increase the binding affinity of the acceptor glycan and increase transglycosylation rates; however, binding studies with fucosylated vs. afucosylated antibodies have not been reported.
EndoS2 shows broader N-glycan specificity than EndoS. Li et al. (2016) performed a systematic study of the hydrolytic and transglycosylation activity of 19 mutants of D184 (equivalent to D233 of EndoS) and identified several mutants, including EndoS2D184M, that are much more efficient than previously described mutants, with yields exceeding 90%. These EndoS2 glycosynthase mutants are able to transfer high-mannose (HM), complex type (CT) and hybrid type glycans to deglycosylated IgG. More recently, Tong et al. (2018) applied this knowledge to glycosynthase mutants of EndoS, and found that the same methionine mutation (EndoSD233M), produces the highest transglycosylation/hydrolysis ratio among any EndoS glycosynthase mutant, but is less efficient than EndoS2D184M. They performed a comparative kinetic analysis of the glycosynthase mutants generated from EndoS and EndoS2, and revealed that the catalytic efficiency of EndoSD233M over previously described EndoSD233A was contributed mainly by two factors: an increased turnover number (i.e., increased kcat) for the glycan oxazoline donor substrate, and an enhanced affinity (i.e., reduced KM) for the antibody substrate. This pattern also applied to the equivalent EndoS2 mutants. Recently, Shivatare et al. (2018) described other glycosynthase mutants, such as EndoS2T138Q, that show favorable transglycosylation/hydrolysis activity ratios, albeit slightly lower than EndoS2D184M. It is unclear why this mutation works, but it has been noted that the equivalent residue to T138 in SmChiB (S93) stabilizes D1 of the catalytic machinery (Synstad et al. 2004). Therefore, mutation to glutamine may disrupt the catalytic machinery in a way that promotes transglycosylation.
Others introduced additional mutations to the described glycosynthase mutant EndoSD233Q, including EndoSD233Q/Q303L, EndoSD233Q/E350Q and EndoSD233Q/Y402F/D405Q (Figure 1f), which resulted in transglycosylation efficiencies of 90% using reduced concentrations of oxazoline (Iwamoto et al. 2018). In addition to the assisting residue D233, Q303 is also involved in the catalytic cycle, stabilizing the reaction intermediate. Y402 is part of the largely hydrophobic floor of the binding pocket, forming a hydrogen bond with O4 of GlcNAc (−1). The role of the E350 in this glycosynthase mutant is more intriguing since this residue makes contacts with the α(1,3) antenna at some distance from the catalytic site. Mutations of D405 are even less explicable, as this residue does not interact directly with the glycan in the crystal structures of EndoS and EndoS2 (Trastoy et al. 2018; Klontz et al. 2019) (Figure 1f). It is worth noting that these mutations are not used individually, but when used in conjunction with D233Q, decrease hydrolysis and increase transglycosylation. Giddens et al. (2016) have identified EndoF3 glycosynthase mutants by targeting the assisting residue, D165. EndoF3D165A and EndoF3D165Q are the only known glycosynthases able to transfer tri-antennary CT glycans to IgG, but its use is limited to core-fucosylated glycoproteins for reasons unknown (Giddens et al. 2016).
Additional alternative enzymatic methods have been developed that avoid using sugar oxazoline derivatives, simplifying the chemical synthesis and preventing side reactions between these highly reactive donors and amino acids on the protein scaffold. These secondary products are produced in reactions that are not well controlled or when the enzymatic activity of the enzymes is not very efficient (Iwamoto et al. 2018; Liu et al. 2018). Liu et al. (2018) use EndoS2WT and an excess (over 1000 molar equivalents) of sialylglycopeptide (SGP) as the donor substrate instead of glycan oxazoline, producing a glycosylation yield of up to 80% with respect to the acceptor. Iwamoto et al. (2018) also described a one-pot reaction to transfer the glycan of SGP to the Fc of antibodies without purification of the deglycosylated antibody and generation of oxazoline donor derivatives. EndoMN175Q was used to target SGP to release the target glycan and EndoSD233Q/E350Q was then used to catalyze the attachment of the glycan to the Fc region of the antibody (Iwamoto et al. 2018).
Glycosynthase mutants active on antibodies have also been created from GH85 enzymes. For example, Eshima et al. (2015) discovered that the N180H mutation of EndoCC, obtained from Coprinopsis cinerea (EndoCCN180H), exhibited transglycosylation activity. Similar to design strategies applied to EndoD (Fan et al. 2012) and EndoM (Umekawa et al. 2008), this mutation targets the residue responsible for assisting oxazoline complex formation. Here, as well, transglycosylation can be performed using high concentrations of SGP as a donor substrate (Manabe et al. 2018).
Structural basis of glycan specificity by IgG processing enzymes
EndoS, encoded by the ndoS gene, was first reported in 2001 from S. pyogenes serotype M1 (Collin and Olsén 2001). EndoS2, encoded by the ndoS2 gene, was discovered over a decade later in a serotype M49 strain (Sjögren et al. 2013). X-ray crystal structures of EndoS and EndoS2 both alone and in complex with their respective glycan substrates have now been reported, providing a structural basis for glycan specificity by these enzymes (Trastoy et al. 2014; Trastoy et al. 2018; Klontz et al. 2019).
The enzymes share ~ 37% amino acid sequence identity, and form the same overall “V”-shape structure, which exists in both crystal structures and in solution (Figure 2a). The GH domain resides on one tip of the “V”, while a CBM (discussed later) is located on the other tip. Separating these two domains is a leucine rich repeat (LRR) domain and hybrid-Ig domain, which together form the characteristic “V”-shape scaffold. EndoS contains an additional 3-helix bundle domain on each terminus, which is likely involved in stabilizing the GH and CBM domains to which it is attached.
Fig. 2.

(a) Overall structure of EndoS and EndoS2 highlighting the GH domain (left) and CBM (right). GH domain loops are annotated, and CBM residues that have been studied are labeled. (b) In the upper panel: surface representation of the residues studied by alanine scanning mutagenesis, colored by loop number. Hydrolytic activity retained by alanine mutants is in parentheses, with the substrate glycan tested superimposed onto its crystal structure: EndoS with CT glycan (left), EndoS2 with CT glycan (center) and EndoS2 with HM glycan (right). In the lower panel: enzyme residues that form glycan contacts. Residues that mediate conserved interactions in the three crystal structures are labeled in black, while residues that mediate a unique contact are in orange.
Although their overall structures are quite similar, EndoS has a stricter glycan specificity than EndoS2, recognizing only non-bisected CT biantennary glycans. In contrast, EndoS2 recognizes CT biantennary glycans (bisected and non-bisected), HM and hybrid glycans, although it shows a preference for CT glycans over HM glycans (Figure 1a). Both enzymes are capable of recognizing CT glycans regardless of their galactose, sialic acid and fucose content (the major variability seen in IgG Asn297-linked glycosylation). The differences in glycan specificity between EndoS and EndoS2 can be attributed to certain structural differences in their GH domains. Both enzymes construct a (β/α)8 barrel, common to all GH18 enzymes. Connecting the β-strands and α-helices are a series of loops that form the binding site in which the glycan sits. The binding site has excellent shape complementarity for a biantennary carbohydrate, forming two distinct grooves to accommodate each of the antennae; Groove 1 is formed mainly by loops 2, 3 and 4, and accommodates the α(1,6) antenna. Groove 2 is formed mainly by loops 1, 2, 7 and 8, and accommodates the α(1,3) antenna (Figure 2b and c). Loop 2 bisects the two grooves with an aromatic residue (W153 in EndoS; H109 in EndoS2; Figure 2b). A well-conserved hydrophobic floor lies below the glycan pentasaccharide core in both enzymes, and is formed mostly by the distal ends of the β-strands. The catalytic residues lie more proximal to the β-barrel, on the β4-strand and loop. Loop 6 contributes a conserved glutamine (Q303 in EndoS; Q250 in EndoS2) and tyrosine (Y305 in EndoS; Y252 in EndoS2) to the active site, which form contacts with GlcNAc (−1) and may be involved in stabilizing the reaction intermediate. In both enzymes, ~ 75% of enzyme-glycan contacts are made to the pentasaccharide core, with the remaining contacts split between the α(1,3) and α(1,6) branches. Alanine scanning mutagenesis revealed a conserved mechanism of CT glycan recognition between both enzymes. More specifically, residues on loops 6 and 7, which form contacts with the glycan core, are critical to activity. Residues on loop 1, which form contacts with the glycan core and α(1,3) antenna, are also critical. The experimental data support a model in which CT glycan binding is driven by the core plus the α(1,3) antenna. In contrast, residues on loops 2, 3 and 4, which form contacts with the α(1,6) antenna, are unimportant for recognition of the CT glycan.
The main differences between EndoS and EndoS2 lie in loops 3 and 4 (Figures 2a and 3a), which accommodate the α(1,6) antenna. These loops are several residues shorter in EndoS2, forming a more open groove. This is important in the recognition of HM glycans, which have an additional branch on the α(1,6) antenna relative to biantennary CT glycans. Crystal structures with an HM glycan bound reveal that this space accommodates the extra branch, although the glycan is mostly disordered in this region. Alanine scanning mutagenesis suggests that the recognition of HM glycans by EndoS2 is predominantly driven by contacts with the glycan core and α(1,3) antenna, as residues that contact these areas cannot be mutated without nearly complete loss of hydrolytic activity. However, mutating loops 3 and 4 reduces activity on HM glycans, but not CT glycans. Together, these results suggest that EndoS2 accommodates HM glycans mainly through contacts with the glycan core and α(1,3) antenna, while creating extra space and fine-tuning binding to the α(1,6) antenna. EndoS cannot accommodate HM glycans because loops 3 and 4 sterically exclude it. Accordingly, swapping loops 3 and 4 from EndoS into EndoS2 produces an enzyme with dramatically reduced activity on HM glycans, and very little loss of activity on CT glycans. Although the structural basis for EndoS2 recognition of hybrid and bisected CT glycans has not yet been shown, one can speculate. Hybrid glycans are identical to CT glycans in the core and α(1,3) antenna, while resembling HM glycans in the α(1,6) antenna. Therefore, EndoS2 likely recognizes hybrid glycans by using a combination of the mechanisms by which it recognizes CT and HM glycans: by specifically recognizing the core and CT-like α(1,3) antenna while creating space and fine-tuning binding to the HM-like α(1,6) antenna. One can also speculate that having a smaller residue at the position that bisects the glycans (histidine versus tryptophan) allows EndoS2 to bind bisected CT glycans, which possess an additional GlcNAc at this location; however, this remains unproven.
Fig. 3.
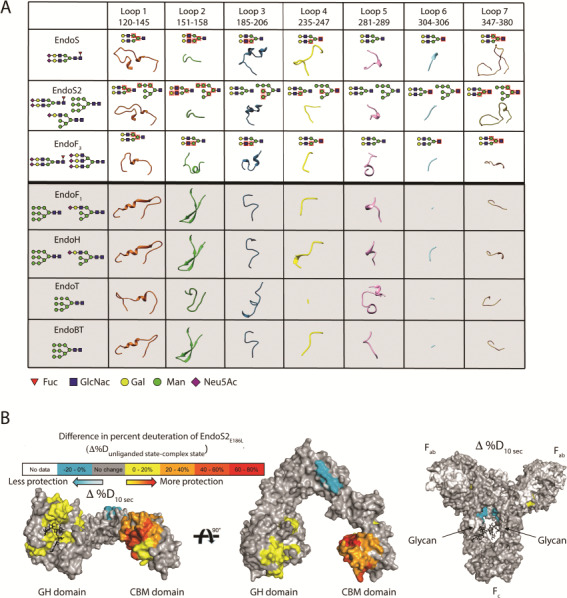
(a) Structural comparison of the loops (using EndoS numbering) surrounding the active site of GH18 family enzymes with ENGase activity. Oligosaccharide moieties that interact with each loop in the crystal structure of EndoS-CT, EndoF3-CT, EndoS2-CT and EndoS2-HM are marked with red squares. GH18 ENGases that hydrolyze CT glycans have a white background, while those that hydrolyze HM glycans have a gray background. (b) Hydrogen-deuterium exchange mass spectrometry of EndoS2 and IgG1. Difference in deuteration between unliganded- and IgG1-bound EndoS2E186L (left) and IgG1 and EndoS2E186L-bound IgG1 (right), mapped onto a surface representation of EndoS2 (PDB 6MDS) and IgG1 (PDB 1HZH) at the earliest time point tested (10 s). IgG1 glycosylation sites annotated, and Fc, Fab, GH and CBM domains are labeled for orientation.
Although EndoS and EndoS2 are the only known enzymes that are specific for IgG, many other enzymes that are agnostic to the protein component of their substrates can also deglycosylate IgG, including EndoF1 (Elder and Alexander 1982), EndoF3 (Tarentino and Plummer Jr 1994), EndoA and EndoH (Wei et al. 2008), EndoD (Fan et al. 2012), EndoM (Yamamoto et al. 1994), EndoBT (Trastoy et al. 2018) and EndoCC (Manabe et al. 2018). In fact, it is likely that most ENGases that can process glycoproteins can process IgG, as long as the IgG molecule possesses a glycan that the ENGase can normally recognize. Although EndoS and EndoS2 can process the vast majority of glycans naturally present on antibodies, other enzymes are sometimes needed in chemoenzymatic approaches to add unusual glycans to antibodies. For example, EndoF3 is uniquely useful in this context, as it is capable of processing tri-antennary CT glycans. Although a crystal structure of EndoF3 bound to a tri-antennary CT glycan does not exist, molecular docking suggests that the third antenna of this glycan would lie flat on the EndoF3 surface, in a location that would clash with EndoS/EndoS2 loop 7 (Figure 3a). It has been shown that EndoF3 recognizes bi- and tri-antennary CT glycans by forming contacts with the glycan core, while sterically excluding tetra-antennary CT glycans and HM glycans, in a mechanism that shows many similarities to EndoS (Waddling et al. 2000). An analysis of GH18 active site loops has revealed a pattern that is predictive of the glycan specificities of some enzymes (Figure 3a). For example, enzymes that are specific for HM glycoproteins (e.g., EndoF1, EndoH, EndoT and EndoBT) form a β-hairpin in loop 2, which might be a major determinant of their specificity. In contrast, enzymes specific for CT glycoproteins display a short loop in the equivalent location. This structural difference as observed solely in the unliganded crystal structure of EndoBT proved predictive for its ability to process IgG with HM glycans but not CT glycans. Whether this strategy can be extended to predicting ENGase specificity based on primary amino acid sequence alone (e.g., in the above mentioned key loops) remains to be determined. It is worth noting that EndoS2 is the only ENGase described to date that can process both CT and HM glycans on antibodies. In loop 2, it more closely resembles CT-processing ENGases than HM-processing ENGases. This is consistent with the observation that EndoS2 processes CT glycans ~20× more efficiently than HM glycans, and further consistent with the fact that CT glycans make up the vast majority of naturally occurring IgG glycoforms. Why EndoS2 evolved to process HM glycans when these are nearly absent on IgG remains unknown.
Structural basis for carbohydrate binding module contributions to enzymatic activity
CBMs are catalytically inactive domains frequently found on enzymes that modify carbohydrates. They have been grouped by CAZy into 84 families based on sequence similarities. They can alternatively be classified into seven families based on their 3D folds: β-sandwich, β-trefoil, cysteine knot, OB, hevein, hevein-like and unique fold (Boraston et al. 2004). CBMs have four main roles: glycan proximity, targeting, disruption and adhesion. The proximity effect occurs when CBMs bind carbohydrates, increasing their location concentration near the GH domain. Targeting occurs by binding a substrate in a distinct location so that the GH domain is properly oriented to the correct location of the glycan. Disruption occurs on tightly packed polysaccharides, such as starch, where CBM binding relaxes the packing to allow the GH domain to access its substrate. Adhesion pins enzymes via their CBM to locations where they are needed, such as the surface of a cell (Boraston et al. 2004; Guillén et al. 2010).
The β-sandwich CBMs of EndoS and EndoS2 have not been formally classified by CAZy (Figure 2A). However, structural homology suggests they belong to the CBM32 family. This assignment is consistent with the family’s description as diverse, promiscuous and frequently found appended to bacterial enzymes that interact with human N-glycans (Abbott et al. 2007). The binding site of CBMs is generally defined by the location of aromatic, solvent-exposed side chains, composed of tryptophan, tyrosine and occasionally phenylalanine residues. Computational alanine scanning mutagenesis on the CBM of EndoS revealed a potential role for W803 (Figure 2a) in binding. Mutational studies revealed that this residue is essential for EndoS activity, with a single alanine mutation at this location producing undetectable IgG binding by SPR, and barely detectible hydrolytic activity as monitored by mass-spectrometry. The same residue is conserved in EndoS2 (W712), and mutation in this enzyme results in a similar loss of activity. Together, these results implicate this conserved tryptophan residue as part of the binding surface of EndoS and EndoS2 CBMs. Domain swap experiments, described below, indicated that the EndoS and EndoS2 CBMs are not interchangeable. The region surrounding W712 in EndoS2 is packed with aromatic residues (Figure 2a), including two tyrosines and a phenylalanine. The same region in EndoS has three serines and a tyrosine (Figure 2a). While EndoS2 cannot be made more EndoS-like by simply mutating these residues to their corresponding counterparts, it is evident that EndoS2 Y820 plays an important role in hydrolysis for both CT and HM glycans. Interestingly, these residues are in the interior of the V-shape of EndoS and EndoS2, facing the GH domain (Figure 2a). It therefore remains unclear why EndoS and EndoS2 CBMs are not interchangeable in chimeric enzymes. It is possible that EndoS and EndoS2 CBMs have subtle differences in their binding sites that require them to be oriented by the scaffold in different ways relative to the GH domain. What is certain is that these domains are indispensable to EndoS and EndoS2 activity, and should not be overlooked when studying and engineering these enzymes.
It seems apparent that the main determinant of ENGase activity lies in the structure of the GH domain. In support of the concept that the specificity of EndoS and EndoS2 is determined by their GH domains, chimeric enzymes in which EndoS possessed the GH domain from EndoS2 instead of its own gained the ability to process HM glycans for the first time. However, activity toward both HM and CT substrates was very low, suggesting that one or more domains is/are working in conjunction with the GH domain to produce optimal activity. However, when the GH and CBM from EndoS2 onto EndoS were simultaneously swapped, an enzyme with activity nearing wild-type EndoS2 for both substrates resulted. Importantly, swapping the CBM alone had no effect on EndoS activity or specificity. These results suggest that (1) the EndoS2 GH domain and CBM coevolved to optimally recognize its substrate repertoire and (2) the LRR and hybrid-Ig domain “scaffold” impacts catalysis as well, since the chimeric enzyme with an EndoS2 GH and CBM did not return fully to wild-type levels of activity on an EndoS scaffold.
Structural basis of antibody specificity by EndoS and EndoS2
EndoS and EndoS2 are the only ENGases that have been reported to exhibit specificity to a narrow range of glycoproteins. Neither enzyme has general ENGase activity, processing glycans only from all human and mouse IgG subclasses, but not IgA, IgM, α2-macroglobulin, ovalbumin, lactoferrin, RNase B or fetuin. It has been reported that EndoS2 processes α1-acid glycoprotein (AGP), but further experiments demonstrated that this occurs with ~ 105-fold reduced efficiency compared to IgG. EndoS has also been reported to free glycans from gp120, however, it was not reported how much enzyme was needed to detect this (Goodfellow et al. 2012). With enough enzyme and a long enough time scale, activity on many other glycoproteins may also be detectable, raising the question as to what counts as having “specificity” for a single protein vs. a “strong preference” for one. It is our opinion that EndoS and EndoS2 can be described as “specific” for IgG. It is provocative that EndoS2 has been shown to lose activity on heat-denatured IgG and AGP. This suggests that EndoS2 specifically recognizes the native conformation of the IgG backbone and/or glycan. However, as heat denaturation tends to cause aggregation, it cannot be ruled out that this is not an artifact of the glycoproteins aggregating and becoming inaccessible to any enzymatic cleavage. Therefore, experiments using non-specific ENGases should be tested on heat-denatured IgG before concluding that this phenomenon is unique to EndoS/EndoS2.
The structural basis for the antibody specificity of EndoS and EndoS2 has not been fully elucidated. This has been hampered by the difficulty in obtaining an IgG-endoglycosidase co-crystal structure. EndoS and EndoS2 bind glycosylated IgG with ~ 1–10 μM affinity, depending on the point mutation used to reduce hydrolytic activity for binding studies. Neither enzyme binds deglycosylated IgG, which is frequently used as a negative control. While trace hydrolytic activity in “catalytically inactive” mutants is not detectable in most binding and kinetic studies, it is problematic in structural biology experiments such as protein crystallization, where enzyme concentrations and experimental time scales are orders of magnitude greater.
In the absence of high-resolution IgG-endoglycosidase structures, one can only speculate as to why EndoS and EndoS2 are specific to IgG. The question of specificity is 2-fold: first, how do these endoglycosidases exclude non-IgG substrates? Second, how do they accommodate IgG? The answer to the first question may be explained by the deep grooves that line the GH domain active site. The glycan binding site resembles a canyon, with loops 4 and 8 (and additionally in EndoS, N-3HB and residues 312–323) forming walls ~ 15–20 Å high. This type of architecture is not seen in other GH18 ENGases that are not IgG-specific and for which structural data is available. Therefore, it may be possible that EndoS and EndoS2 evolved to specifically accommodate the protein fold of IgG surrounding the Asn297 glycosylation site. This is supported by the observation that EndoS2 does not process glycans on IgG when they are placed in other nearby loops directly adjacent to the normal Asn297 location. It is also supported by the observation that both endoglycosidases process free glycans and short glycopeptides, suggesting that steric hindrance of the protein backbone may be responsible for the exclusion of non-IgG glycoproteins.
The answer to how these endoglycosidases accommodate IgG is murkier. Simply docking the antibody onto EndoS/EndoS2 using the glycan co-crystal structures as a guide results in major steric clashes with the antibody backbone. A model of the EndoS/IgG1 Fc encounter complex was described by docking an IgG1 Fc homodimer structure onto the EndoS crystal structure (Trastoy et al. 2014). In this study, the final model showed that the IgG1 Fc is bound to the EndoS CBM, while loop residues 312–323 of the EndoS GH domain bisects the two Fc monomers. However, this model fails to place any glycan in the active site. Therefore, a conformational change in the enzyme and/or antibody is likely required for catalysis. Hydrogen-deuterium exchange mass-spectrometry (HDX-MS) experiments with IgG and EndoS2 have yielded important insights into this topic (Figure 3b). HDX-MS provides information on the solvent accessibility of proteins, and when two proteins are mixed together, it can map the binding interface that is formed between them. Differences seen at early time points correspond to solvent exclusion of surface exposed regions, while differences seen at later time points can be attributed to changes in protein dynamics. HDX-MS experiments suggest that a large interface is formed between IgG and the GH and CBM domains of EndoS2. While protection is extensive on the enzyme side, it is nearly absent on the antibody side. Notably, glycans are invisible to HDX-MS analysis, so the absence of protection on the antibody suggests that EndoS2 ignores the protein backbone, forming contacts almost exclusively with the IgG glycans, consistent with NMR analysis demonstrating that the CBM binds carbohydrates (Dixon et al. 2014). HDX-MS supports conformational changes occurring in the enzyme, as changes in dynamics are seen to extend through a majority of the enzyme. In contrast, very few conformational changes are seen on the antibody side, with the possible exception of the glycan being pulled away from the protein backbone. These results, combined with the findings that EndoS/EndoS2 do not detectably bind deglycosylated IgG (Allhorn et al. 2008), suggest that the endoglycosidases predominantly recognize the glycan portion of IgG. However, further experimental data is required to establish the mode of recognition of IgG by EndoS/EndoS2.
It is provocative that the GH and CBM domains are located at opposite tips of the “V”-shaped enzymes with their glycan-binding surfaces pointing inwards. One might speculate that EndoS/EndoS2 use these two domains to simultaneously recognize the two glycans on the IgG Fc, with the scaffold providing the correct separation to specifically recognize IgG. If true, the CBM would have a proximity and targeting effect for the enzyme. However, this hypothesis has problems; foremost, if the CBM is holding onto one glycan while the other is in the active site, how would the enzyme work once the first glycan is removed? In such a case, one might expect bimodal kinetics, with the first glycan removed rapidly, and the second glycan removed slowly. However, this has not been reported. It may be possible that conformational changes in the enzyme allow the glycan to move from CBM to GH without the antibody dissociating, but this too remains unsupported. Thus, the precise nature of the IgG-endoglycosidase interaction remains a mystery.
Future directions
The discussion above highlights the substantial progress that has been made in understanding the mechanism of IgG-specific endoglycosidases. However, several areas necessitate further exploration. With respect to the GH domain, more studies are required to determine how similar the mechanism is between ENGases and chitinases. Although the main catalytic residues are conserved, there is some divergence in surrounding residues that may play important supporting roles. There is also relatively little known about the precise mechanism by which mutations in supporting catalytic residues create glycosynthases. Crystal structures of glycosynthase mutants with oxazoline mimetics would provide valuable insight into why some mutations are more effective than others.
In the CBM, future efforts should be directed toward understanding what sugar residues are recognized, and how. Although the general glycan-binding surface has been identified, precise details of molecular recognition are unknown. This is due in part to the relatively low (~millimolar) affinities that CBMs typically have for their substrates, making structural studies more difficult. It is also worth investigating if CBMs with different specificities can be swapped or subjected to directed evolution to alter the substrate preference of the overall enzyme. Efforts could also be directed toward engineering point mutations in the CBM to increase the efficiency of glycosynthases (e.g., by reducing residual hydrolytic activity).
More studies are also needed to determine how some ENGases are specific to antibodies, with a structure of an ENGase-IgG complex tantamount to success. Such studies may also reveal what conformational changes are needed to support catalysis. They may also reveal if there is a single mechanism to remove each glycan, or if the two glycans are removed by separate mechanisms. If the latter is the case, it may be possible to engineer these enzymes for asymmetric transglycosylation. IgG-specific enzymes, including EndoS, have already been studied with some success for treating various autoimmune diseases (Nandakumar et al. 2007; Marth and Grewal 2008; Collin et al. 2008; Allhorn et al. 2010; van Timmeren et al. 2010; Hirose et al. 2012; Mihai et al. 2017; Nandakumar et al. 2018). EndoS has also been explored as a supplement given with glycoengineered antibodies resistant to hydrolysis (e.g., bearing high-mannose glycans) in order to deplete the serum of other FcγR-binding antibodies and increase the efficacy of the delivered therapeutic antibody (Baruah et al. 2012). An IgG-degrading endopeptidase, IdeS, also produced by S. pyogenes, has recently been studied for its therapeutic application in the treatment of thrombotic thrombocytopenic purpura and graft rejection following kidney transplantation, and has passed safety and efficacy trials in humans (Winstedt et al. 2015; Jordan et al. 2017; Lorant et al. 2018; Stubbs et al. 2018). These results suggest that EndoS and EndoS2 might share similar successes in a therapeutic setting. By understanding the precise nature of their specificity for antibodies, it may also be possible to engineer variants that are specific for a single subclass of IgG, which could reduce the immunosuppressive side effects of using these enzymes. It may even be possible to target these enzymes toward specific autoantibodies by fusing them to specific self-epitopes, but these areas remain unexplored.
The homogenously glycoengineered IgG products created by these enzymes could be useful as therapeutics, as already discussed. However, they could also prove invaluable in probing the immune system to understand how it works. For example, what degree of sialylation is needed for an antibody to have anti-inflammatory effects? A systematic analysis of IgG glycoforms and their resulting properties would allow researchers to create antibodies with precisely tuned glycosylation for optimal effects.
References
- Abbott DW, Eirín-López JM, Boraston AB. 2007. Insight into ligand diversity and novel biological roles for family 32 carbohydrate-binding modules. Mol Biol Evol. 25:155–167. [DOI] [PubMed] [Google Scholar]
- Allhorn M, Briceño JG, Baudino L, Lood C, Olsson ML, Izui S, Collin M. 2010. The IgG-specific endoglycosidase EndoS inhibits both cellular and complement-mediated autoimmune hemolysis. Blood. 115:5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allhorn M, Olin AI, Nimmerjahn F, Collin M. 2008. Human IgG/FcγR interactions are modulated by streptococcal IgG glycan hydrolysis. PLoS One. 3:e1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony RM, Nimmerjahn F, Ashline DJ, Reinhold VN, Paulson JC, Ravetch JV. 2008a. Recapitulation of IVIG anti-inflammatory activity with a recombinant IgG fc. Science. 320:373–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony RM, Wermeling F, Karlsson MC, Ravetch JV. 2008b. Identification of a receptor required for the anti-inflammatory activity of IVIG. Proc Natl Acad Sci. 105:19571–19578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruah K, Bowden TA, Krishna BA, Dwek RA, Crispin M, Scanlan CN. 2012. Selective deactivation of serum IgG: A general strategy for the enhancement of monoclonal antibody receptor interactions. J Mol Biol. 420:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boraston AB, Bolam DN, Gilbert HJ, Davies GJ. 2004. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J. 382:769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell IK, Miescher S, Branch DR, Mott PJ, Lazarus AH, Han D, Maraskovsky E, Zuercher AW, Neschadim A, Leontyev D. 2014. Therapeutic effect of IVIG on inflammatory arthritis in mice is dependent on the fc portion and independent of sialylation or basophils. J Immun. 192:5031–5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coines J, Alfonso-Prieto M, Biarnés X, Planas A, Rovira C. 2018. Oxazoline or oxazolinium ion? The protonation state and conformation of the reaction intermediate of chitinase enzymes revisited. Chem Eur J. 24:19258–19265. [DOI] [PubMed] [Google Scholar]
- Collin M, Olsén A. 2001. EndoS, a novel secreted protein from Streptococcus pyogenes with endoglycosidase activity on human IgG. EMBO J. 20:3046–3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin M, Shannon O, Björck L. 2008. IgG glycan hydrolysis by a bacterial enzyme as a therapy against autoimmune conditions. Proc Natl Acad Sci. 105:4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalziel M, Crispin M, Scanlan CN, Zitzmann N, Dwek RA. 2014. Emerging principles for the therapeutic exploitation of glycosylation. Science. 343:1235681. [DOI] [PubMed] [Google Scholar]
- Dixon EV, Claridge JK, Harvey DJ, Baruah K, Yu X, Vesiljevic S, Mattick S, Pritchard LK, Krishna B, Scanlan CN. 2014. Fragments of bacterial endoglycosidase S and immunoglobulin G reveal subdomains of each that contribute to deglycosylation. J Biol Chem. 289:13876–13889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder JH, Alexander S. 1982. Endo-beta-N-acetylglucosaminidase F: endoglycosidase from Flavobacterium meningosepticum that cleaves both high-mannose and complex glycoproteins. Proc Natl Acad Sci. 79:4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshima Y, Higuchi Y, Kinoshita T, Nakakita S-i, Takegawa K. 2015. Transglycosylation activity of glycosynthase mutants of endo-β-N-acetylglucosaminidase from Coprinopsis cinerea. PLoS One. 10:e0132859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadel F, Zhao Y, Cachau R, Cousido-Siah A, Ruiz FX, Harlos K, Howard E, Mitschler A, Podjarny A. 2015. New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM. Acta Crystallogr D Biol Crystallogr. 71:1455–1470. [DOI] [PubMed] [Google Scholar]
- Fairbanks AJ. 2013. Endohexosaminidase-catalyzed synthesis of glycopeptides and proteins. Pure Appl Chem. 85:1847–1863. [Google Scholar]
- Fairbanks AJ. 2017. The ENGases: versatile biocatalysts for the production of homogeneous N-linked glycopeptides and glycoproteins. Chem Soc Rev. 46:5128–5146. [DOI] [PubMed] [Google Scholar]
- Fan S-Q, Huang W, Wang L-X. 2012. Remarkable transglycosylation activity of glycosynthase mutants of endo-D, an endo-β-N-acetylglucosaminidase from Streptococcus pneumoniae. J Biol Chem. 287:11272–11281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrara C, Grau S, Jäger C, Sondermann P, Brünker P, Waldhauer I, Hennig M, Ruf A, Rufer AC, Stihle M et al. 2011. Unique carbohydrate–carbohydrate interactions are required for high affinity binding between FcγRIII and antibodies lacking core fucose. Proc Natl Acad Sci. 108:12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Shoda S-i, Haneda K, Inazu T, Takegawa K, Yamamoto K. 2001. A novel disaccharide substrate having 1,2-oxazoline moiety for detection of transglycosylating activity of endoglycosidases. Biochim Biophys Acta. 1528:9–14. [DOI] [PubMed] [Google Scholar]
- Giddens JP, Lomino JV, Amin MN, Wang L-X. 2016. Endo-F3 glycosynthase mutants enable chemoenzymatic synthesis of core-fucosylated triantennary complex type glycopeptides and glycoproteins. J Biol Chem. 291:9356–9370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow JJ, Baruah K, Yamamoto K, Bonomelli C, Krishna B, Harvey DJ, Crispin M, Scanlan CN, Davis BG. 2012. An endoglycosidase with alternative glycan specificity allows broadened glycoprotein remodelling. J Am Chem Soc. 134:8030–8033. [DOI] [PubMed] [Google Scholar]
- Guillén D, Sánchez S, Rodríguez-Sanoja R. 2010. Carbohydrate-binding domains: multiplicity of biological roles. Appl Microbiol Biotechnol. 85:1241–1249. [DOI] [PubMed] [Google Scholar]
- Haltiwanger RS, Lowe JB. 2004. Role of glycosylation in development. Annu Rev Biochem. 73:491–537. [DOI] [PubMed] [Google Scholar]
- Hart GW, Copeland RJ. 2010. Glycomics hits the big time. Cell. 143:672–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A, Aebi M. 2001. Intracellular functions of N-linked glycans. Science. 291:2364–2369. [DOI] [PubMed] [Google Scholar]
- Hirose M, Vafia K, Kalies K, Groth S, Westermann J, Zillikens D, Ludwig RJ, Collin M, Schmidt E. 2012. Enzymatic autoantibody glycan hydrolysis alleviates autoimmunity against type VII collagen. J Autoimmun. 39:304–314. [DOI] [PubMed] [Google Scholar]
- Hsieh Y-C, Wu Y-J, Chiang T-Y, Kuo C-Y, Shrestha KL, Chao C-F, Huang Y-C, Chuankhayan P, W-g W, Li Y-K. 2010. Crystal structures of Bacillus cereus NCTU2 Chitinase complexes with Chitooligomers reveal novel substrate binding for catalysis a chitinase without chitin binding and insertion domains. J Biol Chem. 285:31603–31615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Giddens J, Fan S-Q, Toonstra C, Wang L-X. 2012. Chemoenzymatic glycoengineering of intact IgG antibodies for gain of functions. J Am Chem Soc. 134:12308–12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Hibi T, Suzuki F, Sugimoto I, Fujiwara A, Inaka K, Tanaka H, Ohta K, Fujii Y, Taketo A. 2016. Crystal structure of chitinase ChiW from Paenibacillus sp. str. FPU-7 reveals a novel type of bacterial cell-surface-expressed multi-modular enzyme machinery. PLoS One. 11:e0167310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto M, Sekiguchi Y, Nakamura K, Kawaguchi Y, Honda T, Hasegawa J. 2018a. Generation of efficient mutants of endoglycosidase from Streptococcus pyogenes and their application in a novel one-pot transglycosylation reaction for antibody modification. PLoS One. 13:e0193534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto M, Yamaguchi T, Sekiguchi Y, Oishi S, Shiiki T, Soma M, Nakamura K, Yoshida M, Chaya H, Mori Y. 2018b. Pharmacokinetic and pharmacodynamic profiles of glyco-modified atrial natriuretic peptide derivatives synthesized using chemo-enzymatic synthesis approaches. Bioconjug Chem. 29:2829–2837. [DOI] [PubMed] [Google Scholar]
- Jefferis R. 2009. Glycosylation as a strategy to improve antibody-based therapeutics. Nat Rev Drug Discov. 8:226. [DOI] [PubMed] [Google Scholar]
- Jennewein MF, Alter G. 2017. The Immunoregulatory roles of antibody glycosylation. Trends Immunol. 38:358–372. [DOI] [PubMed] [Google Scholar]
- Jitonnom J, Lee VS, Nimmanpipug P, Rowlands HA, Mulholland AJ. 2011. Quantum mechanics/molecular mechanics modeling of substrate-assisted catalysis in family 18 chitinases: Conformational changes and the role of Asp142 in catalysis in ChiB. Biochemistry. 50:4697–4711. [DOI] [PubMed] [Google Scholar]
- Jordan SC, Lorant T, Choi J, Kjellman C, Winstedt L, Bengtsson M, Zhang X, Eich T, Toyoda M, Eriksson B-M. 2017. IgG endopeptidase in highly sensitized patients undergoing transplantation. N Engl J Med. 377:442–453. [DOI] [PubMed] [Google Scholar]
- Klontz EH, Trastoy B, Deredge D, Fields JK, Li C, Orwenyo J, Marina A, Beadenkopf R, Günther S, Flores J. 2019. Molecular basis of broad Spectrum N-glycan specificity and processing of therapeutic IgG monoclonal antibodies by Endoglycosidase S2. ACS Cent Sci. 5:524–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wang L-X. 2018. Chemoenzymatic methods for the synthesis of glycoproteins. Chem Rev. 118:8359–8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Tong X, Yang Q, Giddens JP, Wang L-X. 2016. Glycosynthase mutants of endoglycosidase S2 show potent transglycosylation activity and remarkably relaxed substrate specificity for antibody glycosylation remodeling. J Biol Chem. 291:16508–16518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C-P, Tsai T-I, Cheng T, Shivatare VS, Wu C-Y, Wong C-H. 2018. Glycoengineering of antibody (Herceptin) through yeast expression and in vitro enzymatic glycosylation. Proc Natl Acad Sci. 115:720–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez Aguilar A, Briard JG, Yang L, Ovryn B, Macauley MS, Wu P. 2017. Tools for studying glycans: recent advances in chemoenzymatic glycan labeling. ACS Chem Biol. 12:611–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorant T, Bengtsson M, Eich T, Eriksson BM, Winstedt L, Järnum S, Stenberg Y, Robertson AK, Mosén K, Björck L. 2018. Safety, immunogenicity, pharmacokinetics, and efficacy of degradation of anti-HLA antibodies by IdeS (imlifidase) in chronic kidney disease patients. Am J Transplant. 18:2752–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Chu J, Zou Z, Hamacher NB, Rixon MW, Sun PD. 2015. Structure of FcγRI in complex with Fc reveals the importance of glycan recognition for high-affinity IgG binding. Proc Natl Acad Sci. 112:833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macauley MS, Crocker PR, Paulson JC. 2014. Siglec-mediated regulation of immune cell function in disease. Nat Rev Immunol. 14:653–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie LF, Wang Q, Warren RAJ, Withers SG. 1998. Glycosynthases: Mutant glycosidases for oligosaccharide synthesis. J Am Chem Soc. 120:5583–5584. [Google Scholar]
- Malecki PH, Raczynska JE, Vorgias CE, Rypniewski W. 2013. Structure of a complete four-domain chitinase from Moritella marina, a marine psychrophilic bacterium. Acta Crystallogr D Biol Crystallogr. 69:821–829. [DOI] [PubMed] [Google Scholar]
- Manabe S, Yamaguchi Y, Abe J, Matsumoto K, Ito Y. 2018. Acceptor range of endo-β-N-acetylglucosaminidase mutant endo-CC N180H: From monosaccharide to antibody. R Soc Open Sci. 5:171521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marth JD, Grewal PK. 2008. Mammalian glycosylation in immunity. Nat Rev Immunol. 8:874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihai S, Albert H, Ludwig RJ, Iwata H, Björck L, Collin M, Nimmerjahn F. 2017. In vivo enzymatic modulation of IgG antibodies prevents immune complex-dependent skin injury. Exp Dermatol. 26:691–696. [DOI] [PubMed] [Google Scholar]
- Nandakumar KS, Collin M, Happonen KE, Lundström SL, Croxford AM, Xu B, Zubarev RA, Rowley MJ, Blom AM, Kjellman C et al. 2018. Streptococcal Endo-β-N-Acetylglucosaminidase suppresses antibody-mediated inflammation in vivo. Front Immunol. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar KS, Collin M, Olsén A, Nimmerjahn F, Blom AM, Ravetch JV, Holmdahl R. 2007. Endoglycosidase treatment abrogates IgG arthritogenicity: Importance of IgG glycosylation in arthritis. Eur J Immunol. 37:2973–2982. [DOI] [PubMed] [Google Scholar]
- Nimmerjahn F, Anthony RM, Ravetch JV. 2007. Agalactosylated IgG antibodies depend on cellular fc receptors for in vivo activity. Proc Natl Acad Sci. 104:8433–8437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn F, Ravetch JV. 2008. Fcγ receptors as regulators of immune responses. Nat Rev Immunol. 8:34. [DOI] [PubMed] [Google Scholar]
- Noguchi M, Tanaka T, Gyakushi H, Kobayashi A, Shoda S-i. 2009. Efficient synthesis of sugar oxazolines from unprotected N-acetyl-2-amino sugars by using chloroformamidinium reagent in water. J Org Chem. 74:2210–2212. [DOI] [PubMed] [Google Scholar]
- Pereira NA, Chan KF, Lin PC, Song Z. 2018. The “less-is-more” in therapeutic antibodies: afucosylated anti-cancer antibodies with enhanced antibody-dependent cellular cytotoxicity. MAbs. 10:693–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrescu A-J, Wormald MR, Dwek RA. 2006. Structural aspects of glycomes with a focus on N-glycosylation and glycoprotein folding. Curr Opin Struct Biol. 16:600–607. [DOI] [PubMed] [Google Scholar]
- Ranok A, Wongsantichon J, Robinson RC, Suginta W. 2015. Structural and thermodynamic insights into chitooligosaccharide binding to human cartilage chitinase 3-like protein 2 (CHI3L2 or YKL-39). J Biol Chem. 290:2617–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl M, Stangler T, Torella C, Čepeljnik T, Toll H, Grau R. 2011. Acceptable changes in quality attributes of glycosylated biopharmaceuticals. Nat Biotechnol. 29(4):310–312. [DOI] [PubMed] [Google Scholar]
- Sharma M, Schoindre Y, Hegde P, Saha C, Maddur MS, Stephen-Victor E, Gilardin L, Lecerf M, Bruneval P, Mouthon L. 2014. Intravenous immunoglobulin-induced IL-33 is insufficient to mediate basophil expansion in autoimmune patients. Sci Rep. 4:5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields RL, Lai J, Keck R, O'Connell LY, Hong K, Meng YG, Weikert SH, Presta LG. 2002. Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human FcγRIII and antibody-dependent cellular toxicity. J Biol Chem. 277:26733–26740. [DOI] [PubMed] [Google Scholar]
- Shivatare SS, Huang L-Y, Zeng Y-F, Liao J-Y, You T-H, Wang S-Y, Cheng T, Chiu C-W, Chao P, Chen L-T. 2018. Development of glycosynthases with broad glycan specificity for the efficient glyco-remodeling of antibodies. Chem Commun. 54:6161–6164. [DOI] [PubMed] [Google Scholar]
- Sjögren J, Struwe WB, Cosgrave EF, Rudd PM, Stervander M, Allhorn M, Hollands A, Nizet V, Collin M. 2013. EndoS2 is a unique and conserved enzyme of serotype M49 group a streptococcus that hydrolyses N-linked glycans on IgG and α1-acid glycoprotein. Biochem J. 455:107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speciale G, Thompson AJ, Davies GJ, Williams SJ. 2014. Dissecting conformational contributions to glycosidase catalysis and inhibition. Curr Opin Struct Biol. 28:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs MJ, Thomas M, Vendramin C, Sonesson E, Kjellman C, Järnum S, Stenberg Y, Elfving C, Scully M. 2018. Administration of immunoglobulin G-degrading enzyme of Streptococcus pyogenes (IdeS) for persistent anti-ADAMTS 13 antibodies in patients with thrombotic thrombocytopenic purpura in clinical remission. Br J Haematol 186:137–140. [DOI] [PubMed] [Google Scholar]
- Subedi GP, Barb AW. 2015. The structural role of antibody N-glycosylation in receptor interactions. Structure. 23:1573–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synstad B, Gåseidnes S, Van Aalten DM, Vriend G, Nielsen JE, Eijsink VG. 2004. Mutational and computational analysis of the role of conserved residues in the active site of a family 18 chitinase. Eur J Biochem. 271:253–262. [DOI] [PubMed] [Google Scholar]
- Tarentino AL, Plummer TH Jr. 1994. [4] enzymatic deglycosylation of asparagine-linked glycans: Purification, properties, and specificity of oligosaccharide-cleaving enzymes from Flavobacterium meningosepticum. Methods Enzymol. 230:44–57. [DOI] [PubMed] [Google Scholar]
- Thomann M, Reckermann K, Reusch D, Prasser J, Tejada ML. 2016. Fc-galactosylation modulates antibody-dependent cellular cytotoxicity of therapeutic antibodies. Mol Immunol. 73:69–75. [DOI] [PubMed] [Google Scholar]
- Tong X, Li T, Li C, Wang L-X. 2018. Generation and comparative kinetic analysis of new Glycosynthase mutants from Streptococcus pyogenes Endoglycosidases for antibody glycoengineering. Biochemistry. 57:5239–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trastoy B, Klontz E, Orwenyo J, Marina A, Wang L-X, Sundberg EJ, Guerin ME. 2018. Structural basis for the recognition of complex-type N-glycans by Endoglycosidase S. Nat Commun. 9:1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trastoy B, Lomino JV, Pierce BG, Carter LG, Günther S, Giddens JP, Snyder GA, Weiss TM, Weng Z, Wang L-X. 2014. Crystal structure of Streptococcus pyogenes EndoS, an immunomodulatory endoglycosidase specific for human IgG antibodies. Proc Natl Acad Sci. 111:6714–6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umekawa M, Huang W, Li B, Fujita K, Ashida H, Wang L-X, Yamamoto K. 2008. Mutants of Mucor hiemalis endo-β-N-acetylglucosaminidase show enhanced transglycosylation and glycosynthase-like activities. J Biol Chem. 283:4469–4479. [DOI] [PubMed] [Google Scholar]
- van Aalten DM, Komander D, Synstad B, Gåseidnes S, Peter MG, Eijsink VG. 2001. Structural insights into the catalytic mechanism of a family 18 exo-chitinase. Proc Natl Acad Sci. 98:8979–8984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Timmeren MM, van der Veen BS, Stegeman CA, Petersen AH, Hellmark T, Collin M, Heeringa P. 2010. IgG glycan hydrolysis attenuates ANCA-mediated glomerulonephritis. J Am Soc Nephrol. 21:1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A. 1993. Biological roles of oligosaccharides: all of the theories are correct. Glycobiology. 3:97–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddling CA, Plummer TH, Tarentino AL, Van Roey P. 2000. Structural basis for the substrate specificity of endo-β-N-acetylglucosaminidase F3. Biochemistry. 39:7878–7885. [DOI] [PubMed] [Google Scholar]
- Wang L-X. 2011. The amazing transglycosylation activity of Endo-BETA-N-acetylglucosaminidases. Trends Glycosci Glyc. 23:33–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Li C, Huang W, Li B, Strome S, Wang L-X. 2008. Glycoengineering of human IgG1-fc through combined yeast expression and in vitro chemoenzymatic glycosylation. Biochemistry. 47:10294–10304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White A, Rose DR. 1997. Mechanism of catalysis by retaining β-glycosyl hydrolases. Curr Opin Struct Biol. 7:645–651. [DOI] [PubMed] [Google Scholar]
- Williams SJ, Mark BL, Vocadlo DJ, James MN, Withers SG. 2002. Aspartate 313 in the Streptomyces plicatushexosaminidase plays a critical role in substrate-assisted catalysis by orienting the 2-acetamido group and stabilizing the transition state. J Biol Chem. 277:40055–40065. [DOI] [PubMed] [Google Scholar]
- Winstedt L, Järnum S, Nordahl EA, Olsson A, Runström A, Bockermann R, Karlsson C, Malmström J, Palmgren GS, Malmqvist U. 2015. Complete removal of extracellular IgG antibodies in a randomized dose-escalation phase I study with the bacterial enzyme IdeS–a novel therapeutic opportunity. PLoS One. 10:e0132011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto KJ, Kadowaki S, Watanabe J, Kumagai H. 1994. Transglycosylation activity of Mucor hiemalis endo-β-N-acetylglucosaminidase which transfers complex oligosaccharides to the N-acetylglucosamine moieties of peptides. Biochem Biophys Res Commun. 203:244–252. [DOI] [PubMed] [Google Scholar]
- Yu X, Vasiljevic S, Mitchell DA, Crispin M, Scanlan CN. 2013. Dissecting the molecular mechanism of IVIg therapy: The interaction between serum IgG and DC-SIGN is independent of antibody glycoform or fc domain. J Mol Biol. 425:1253–1258. [DOI] [PubMed] [Google Scholar]


