Abstract
Colon cancer is the third leading cause of cancer-related deaths all around the world. LncRNA methylation has been verified to participate in some kinds of malignancies. The aim of this study was to investigate the function of NEAT1 in colon cancer and further explore the potential mechanism between NEAT1 and ALKBH5. Differential expression of lncRNAs between colon cancer tissues and normal tissues was identified using ArrayStar lncRNA microarrays. The levels of NEAT1 and ALKBH5 expression in colon cancer tissues and cells were measured using qRT-PCR. MTT, transwell migration assays and qRT-PCR were performed to detect cell proliferation and migration. Flow cytometry and qRT-PCR was used for apoptosis analysis. Then, m6A RNA immunoprecipitation was performed to detect methylated NEAT1 in colon cancer cells. The results showed NEAT1 was remarkably enhanced in colon cancer tissues and correlated with poor prognosis. Knockdown of NEAT1 inhibited cell proliferation and migration, induced cell apoptosis in colon cancer cell lines. Besides, ALKBH5 could upregulate NEAT1 expression by demethylation. In addition, ALKBH5 knockdown suppressed malignant behavior of colon cancer partially through NEAT1 in vitro and vivo. In a word, we observed NEAT1 expression level was up-regulated in colon cancer tissues and cells. ALKBH5 knockdown suppressed malignant behavior of colon cancer partially through NEAT1 by demethylation in vitro and vivo, suggesting that ALKBH5-NEAT1 axis maybe potential therapeutic target for colon cancer treatment.
Keywords: Colon cancer, NEAT1, ALKBH5
Introduction
Colon cancer is the third leading cause of cancer-related deaths all around the world with approximately 1 million new cases each year [1]. Though great progress has been made in the field of colon cancer treatment in recent years, the 5-year survival rate remains around 30% due to recurrence and metastasis [2,3]. Thus, a clear understanding of genetic changes and signaling pathways related to development of colon cancer is essential.
Long noncoding RNAs (lncRNAs) belong to a group of noncoding RNAs that are over 200 nucleotides in length and have no protein-coding potential [4]. Emerging evidence has revealed lncRNAs modulate diverse cellular processes and tumorigenesis, including transcription regulation, posttranscriptional regulation, protein modification [5,6]. As a newly identified nuclear-restricted lncRNA, nuclear paraspeckle assembly transcript 1 (NEAT1) is located on chromosome 11 (11q13.1) and has been confirmed to facilitate tumorigenesis in various cancers, including prostate [7], lung [8], esophageal [9] and gastric [10] cancers. However, the role of lncRNA NEAT1 in colon cancer is still poorly understood.
RNA modification is one of the most important ways to regulate gene expression and is involved in many pathophysiological processes in eukaryotes [11]. N6-methyladenosine (m6A) is a very common RNA methylation modification, which is involved in eukaryotic mRNA transportation, shearing, translation and posttranslational processing [12]. Therefore, the abnormal changes of this modification may cause the expression disorder of the downstream genes, affecting cell biological behaviors such as cell proliferation and apoptosis, finally leading to the occurrence of cancer. As a member of the AlkB family, ALKBH5 is a demethylated enzyme of m6A and has been shown to be associated with proliferation, invasion and metastasis of breast cancer [13], glioma [14] and pancreatic cancer [15]. Nevertheless, there are few studies about the role of ALKBH5 in colon cancer progression. The aim of this study was to investigate the function of NEAT1 in colon cancer and further explore the potential mechanism between NEAT1 and ALKBH5.
Materials and methods
Patients
Each patient signed an informed consent prior to surgery which was approved by the Ethics Committee of The Fourth Affiliated Hospital of Anhui Medical University. From January 2018 to December 2019, seventy colon cancer tissues and adjacent normal tissues were collected from patients who visited The Fourth Affiliated Hospital of Anhui Medical University. And the patients’ data in the study are shown in Table 1. All the specimens were immediately snap-frozen and preserved in liquid nitrogen for further use.
Table 1.
The correlation of NEAT1 expression with clinical parameters in patients with colon cancer
| Variables | Clinical parameters | NEAT1 expression | P value | |
|---|---|---|---|---|
|
| ||||
| High (n = 38) | Low (n = 32) | |||
| Gender | Male | 20 | 20 | 0.406 |
| Female | 18 | 12 | ||
| Age (years) | <65 | 16 | 14 | 0.890 |
| ≥65 | 22 | 18 | ||
| Size (cm) | <5 | 18 | 17 | 0.631 |
| ≥5 | 20 | 15 | ||
| Differentiation | Well/Moderate | 21 | 14 | 0.337 |
| Poor | 17 | 18 | ||
| TNM stage | I/II | 10 | 21 | 0.001* |
| III/IV | 28 | 11 | ||
| Lymphatic metastasis | Yes | 26 | 13 | 0.020* |
| No | 12 | 19 | ||
| Distant metastasis | Yes | 26 | 14 | 0.038* |
| No | 12 | 18 | ||
P<0.05.
Cell culture and transfection
The human colon cancer cell lines (Lovo, RKO, SW620 and HCT8) and the normal colon epithelial cell line HIEC cells were cultivated in DMEM with 10% FBS at 37°C with 5% CO2. For transfection, colon cancer cells were firstly seeded into 24-well plates before transfection, and then transiently transfected with pcDNA-NEAT1 vector, si-NEAT1, si-ALKBH5, NEAT1 mimic and corresponding specific controls (50 nM, Ribobio Company, Guangzhou, China) using Lipofectamine 2000 reagent (Invitrogen, USA) based on manufacturer’s protocols for potential mechanism research.
MTT assay
Cells (3 × 103) were cultured in 96-well plates and incubated for 24 h and stained with 0.5 mg/ml MTT for 4 h. And then 200 μl of dimethylsulfoxide (DMSO) was added to dissolve precipitates. At indicated time points, cells were detected according the manufacturer’s instructions.
Cell migration assay
The transfected colon cancer cells were firstly planted to approximately 60~80% confluence in 12-well plates, and then added into upper chambers of transwell inserts (8 μm pore size; Millipore, USA) with the lower chambers containing DMEM with 10% FBS. After 12 h of incubation, the migrated cells were fixed in 4% paraformaldehyde and stained with crystal violet, and then measured using a light microscope (Olympus BX51, Olympus Corporation, Japan).
Cell apoptosis assay
5 × 105 cells were harvested and washed twice with ice-cold PBS, incubated with 5 mL Annexin V-FITC and 5 mL propidium iodide (PI) in the dark at 37°C. Then, the flow cytometry analysis was employed for detecting apoptotic events.
Real-time quantitative polymerase chain reaction (qRT-PCR)
Total RNA from tissues or cells was isolated using Trizol reagent (Invitrogen). And cDNA synthesis was conducted using the using the PrimeScript RT reagent Kit (Promega). SYBR green Premix DimerEraser (TaKaRa) was used to calculate the relative levels of associated parameters. The relative expression of each mRNA expression was analyzed using 2-ΔΔCT method. The primers for qRT-PCR were shown as follow: ALKBH5, forward 5’-GCTTCAGGGTATGGGAGTTG-3’ and reverse 5’-TTCCAGGATCTGAGTGGATAGA-3’; NEAT1, forward 5’-CTTCCTCCCTTTAACTTATCCATTCAC-3’ and reverse 5’-CTCTTCCTCCACCATTACCAACAATAC-3’; PCNA, forward 5’-GCCGAGATCTCAGCCATATT-3’ and reverse 5’-ATGTACTTAGAGGTACAAAT-3’; Bcl-2, forward 5’-TTCGCCGAGATGTCCAGGC-3’ and reverse 5’-TCACTTGTGGCCCAGATAGG-3’; GAPDH, forward 5’-TGGTGAAGGTCGGTGTGAAC-3’ and reverse 5’-TTCCCATTCTCAGCCTTGAC-3’.
Western blotting analysis
Proteins were extracted from cultured cells by RIPA buffer containing a mixture of protease inhibitors, and then transferred to polyvinylidene fluoride (PVDF) membrane blocked with non-fat milk by SDS-PAGE. After the protein samples were incubated with primary antibodies overnight, the secondary antibodies were employed to incubate the PVDF membranes for 1 h at room temperature. Bands were quantified using ImageJ software.
Methylated RNA immuneprecipitation assay (MeRIP)
The total m6A content of Total RNA was determined using an m6A methylation quantification kit (EpiGentek, USA). After total RNA was isolated and purified, the bind RNA was cultured with the capture antibody, subsequently the detection antibody and enhancer solution were added. Finally, the m6A level was measured according to the fluorescence.
RNA immunoprecipitation assay (RIP)
The Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore Corporation) was applied to perform RIP assay. 3 μg ALKBH5, METTL3, METTL14, WTAP and FTO and IgG control antibodies were used for RIP assay. Co-precipitated RNAs were then detected by qRT-PCR.
Tumor xenografts
The animal experiments were approved by the Animal Care and Use Committee of Anhui Medical University and were performed in accordance with the institutional guide for the care and use of laboratory animals. To establish xenograft model, RKO cells (1 × 107) tansduced with si-control (si-NC) or si-ALKBH5 or si-ALKBH5+NEAT1 were injected subcutaneously into the right flank of the nude mice (n = 6 each group) every 5 days for 4 times. The tumor volumes were calculated by the formula: Volume = 0.5 × length × width × width. The tumor weights were measured after the mice were sacrificed.
Statistical analysis
All data were presented as mean ± SD. SPSS 17.0 software (IBM Software, Chicago, IL, USA) were applied for the statistical analysis. Data were analyzed using one-way analysis of variance followed by Dunnett’s post hoc test for multiple comparisons or a Student’s t test for comparisons between two groups. Kaplan-Meier method and logrank test were utilized to analyze the overall survival rate of patients. P<0.05 was considered as statically significant.
Results
NEAT1 was remarkably enhanced in colon cancer tissues and correlated with poor prognosis
To acquire the role of NEAT1 in colon cancer progression, we analyzed the colon cancer tissues and normal tissues (n = 4) using ArrayStar lncRNA microarrays. We identified sixteen candidate lncRNAs were markedly up-regulated in cancer tissues (Figure 1A). And we found NEAT1 expression level was the biggest increase among these lncRNAs (Figure 1A). Besides, NEAT1 expression level was elevated significantly in colon cancer tissues of patients (Figure 1B). Subsequently, we divided the 70 patients into a high and a low NEAT1 expression group based on its median value (Figure 1C; Table 1). The analysis results revealed that NEAT1 mRNA level was significantly higher in patients with TNM stage, lymphatic or distant metastases (Figure 1D; Table 1). In addition, the Kaplan-Meier curves manifested patients with high NEAT1 expression had a worse prognosis than those with low NEAT1 expression (Figure 1E).
Figure 1.
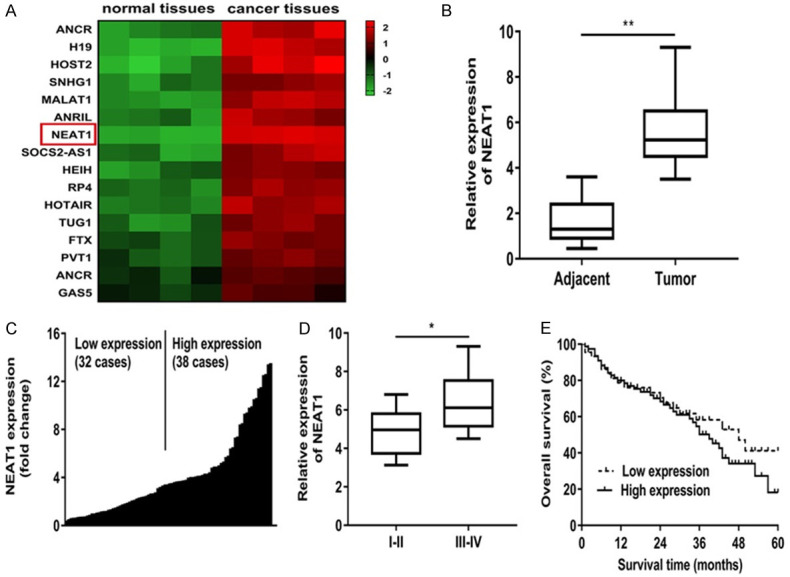
NEAT1 was remarkably enhanced in colon cancer tissues and correlated with poor prognosis. A: Differently expressed lncRNAs in colon cancer tissues and normal tissues using ArrayStar lncRNA microarrays. B: NEAT1 expression level was elevated significantly in colon cancer tissues of patients. C: 70 patients were divided into a high and a low NEAT1 expression group based on its median value. D: NEAT1 expression was obviously higher in patients with advanced clinical stage (III-IV phase) than that with early clinical stage (I-II phase). E: Kaplan-Meier curves of overall survivals and log-rank test showed that patients with high NEAT1 levels had poor overall survivals. *P<0.05 compared to I-II group, **P<0.01 compared to adjacent group.
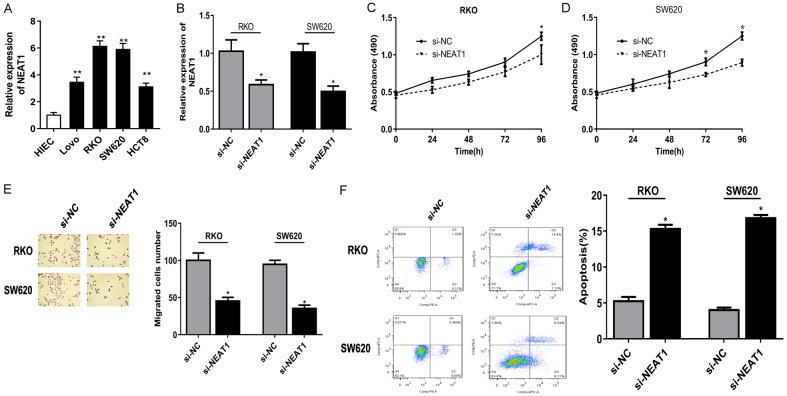
Knockdown of NEAT1 inhibited cell proliferation and migration, induced cell apoptosis in colon cancer cell lines
Subsequently, we assessed the oncogenic role of NEAT1 in the biological behavior of colon cancer cell lines, and observed that NEAT1 expression was remarkably up-regulated in RKO and SW620 cell lines among 4 colon cancer lines (Figure 2A). Thus, we transiently established NEAT1 knockdown in RKO and SW620 cell lines by RNA interference for the following study (Figure 2B). And NEAT1 knockdown suppressed cell growth of RKO and SW620 cells (Figure 2C, 2D). Besides, knockdown of NEAT1 inhibited the migration of RKO and SW620 cells (Figure 2E). Flow cytometry revealed NEAT1 knockdown markedly augmented apoptosis in cells compared with the non-transfected cells (Figure 2F).
Figure 2.
Knockdown of NEAT1 inhibited cell proliferation and migration, induced cell apoptosis in colon cancer cell lines. A: NEAT1 expression was remarkably up-regulated in RKO and SW620 cell lines among 4 colon cancer lines. B: NEAT1 knockdown was established in RKO and SW620 cell lines. C, D: The growth rate of cells transfected with si-NEAT1 was significantly down-regulated compared to that with si-NC in RKO and SW620 cell lines. E: Transwell assay showed si-NEAT1 inhibited the migration ability of colon cancer cells. F: Flow cytometry revealed that silencing of NEAT1 markedly augmented apoptosis in cells compared with the non-transfected cells. *P<0.05 compared to si-NC group, **P<0.01 compared to HIEC.
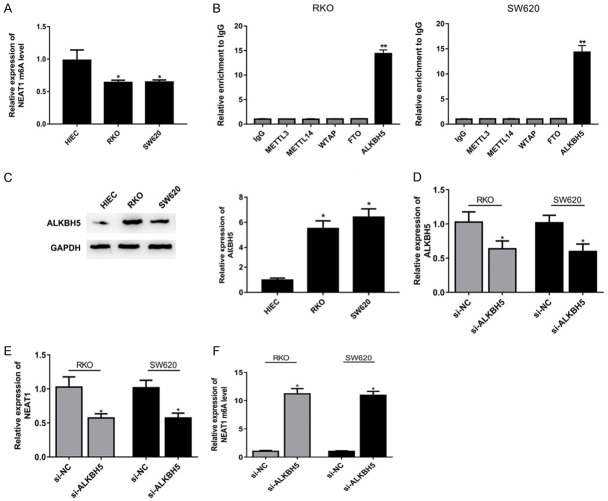
ALKBH5 upregulated NEAT1 expression by demethylation
Given that NEAT1 is verified to be enriched in the nucleus [16], thus we measured the m6A level of NEAT1 in RKO and SW620 cells. The results showed NEAT1 m6A enrichment was lower in RKO and SW620 cells than in HIEC cell (Figure 3A). As predicted by StarBase v2.0, NEAT1 is a target biomarker of ALKBH5. And we observed NEAT1 could be significantly enriched by ALKBH5 compared to IgG, METTL3, METTL14, WTAP and FTO (Figure 3B). Besides, the overexpression of ALKBH5 at mRNA and western blot was detected in RKO and SW620 cells (Figure 3C). Then we knocked down the expression of ALKBH5 by RNA interference (Figure 3D). And si-ALKBH5 could downregulate the NEAT1 expression level (Figure 3E), and upregulate NEAT1 m6A enrichment (Figure 3F).
Figure 3.
ALKBH5 upregulated NEAT1 expression by demethylation. A: NEAT1 m6A enrichment was lower in RKO and SW620 cells than in HIEC cell. B: NEAT1 could be significantly enriched by ALKBH5 compared to IgG, METTL3, METTL14, WTAP and FTO in RKO and SW620 cells. C: The overexpression of ALKBH5 at mRNA and western blot was detected in RKO and SW620 cells. D: ALKBH5 knockdown was established in RKO and SW620 cell lines. E: si-ALKBH5 could downregulate the NEAT1 expression level. F: si-ALKBH5 could upregulate NEAT1 m6A enrichment. *P<0.05 compared to HIEC, si-NC group, **P<0.01 compared to IgG group.
ALKBH5 knockdown suppressed malignant behavior of colon cancer partially through NEAT1 in vitro and vivo
Finally, we investigated the functional role of ALKBH5-NEAT1 axis in biological behavior of colon cancer in vitro and vivo. MTT assay showed ALKBH5 knockdown significantly attenuated colon cancer cells proliferation, which was partially rescued by NEAT1 at 96 h (Figure 4A). And ALKBH5 knockdown significantly inhibited the expression of PCNA, which was partially rescued by NEAT1 (Figure 4A). Besides, NEAT1 could abolish the suppression of ALKBH5 knockdown in cell migration (Figure 4B). In addition, ALKBH5 knockdown could obviously enhance cell apoptosis rates in colon cancer cells, which was reversed by NEAT1 obviously (Figure 4C). Meanwhile, NEAT1 could reverse the suppression of ALKBH5 knockdown in Bcl-2 expression (Figure 4C). Furthermore, we verified the functional role of ALKBH5-NEAT1 axis in biological behavior of colon cancer in vivo. Compared with control group, si-ALKBH5 could decrease tumor growth including tumor volume and weight (Figure 4D, 4E). Besides, NEAT1 could was capable to partly counteract the suppression of si-ALKBH5 in tumor growth (Figure 4D, 4E).
Figure 4.
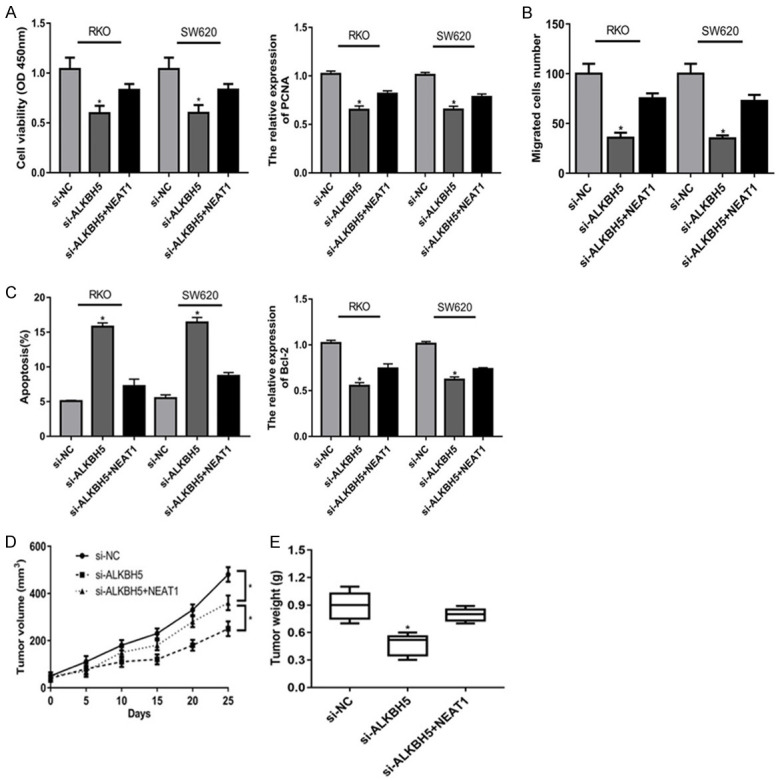
ALKBH5 knockdown suppressed malignant behavior of colon cancer partially through NEAT1 in vitro and vivo. A: ALKBH5 knockdown significantly attenuated colon cancer cells proliferation, which was partially rescued by NEAT1 at 96 h; ALKBH5 knockdown significantly inhibited the expression of PCNA, which was partially rescued by NEAT1. B: NEAT1 could abolish the suppression of ALKBH5 knockdown in cell migration. C: ALKBH5 knockdown could obviously enhance cell apoptosis rates in colon cancer cells, which was reversed by NEAT1 obviously; NEAT1 could reverse the suppression of ALKBH5 knockdown in Bcl-2 expression. D, E: NEAT1 could was capable to partly counteract the suppression of si-ALKBH5 in tumor growth. *P<0.05 compared to si-NC, si-ALKBH5+NEAT1 group.
Discussion
Recently, emerging evidences indicate lincRNAs act as important regulators for cancer progression [17,18]. Previous researches have investigated the role of lncRNAs in colon cancer progression [19-21]. Although dysregulation of NEAT1 is involved in various cancers, the NEAT1 in colon cancer have not been fully elucidated. In this study, we observed NEAT1 expression was remarkably in colon cancer tissue and cell lines. In addition, we noticed that NEAT1 is closely associated with cancer phenotypes and as a potential factor to predict overall survival. Furthermore, NEAT1 silencing suppressed proliferation and migration of colon cancer cells significantly. These data indicated NEAT1 may serve as an oncogene in the development of colon cancer and targeting NEAT1 may represent a favorable therapeutic strategy for colon cancer treatment.
RNA epigenetic modification is the regulation of post-transcriptional level, which could occur in transport RNA, ribosomal RNA, mRNA, non-coding small RNA and long non-coding RNA. Among them, m6A is the most important methylation modification of mRNA in eukaryotes [11,22]. Some research has focused on the importance of lncRNA methylation in tumorigenesis. For instance, ALKBH5 promoted gastric cancer progression by demethylating the lncRNA NEAT1 [12]. ALKBH5 could inhibit pancreatic cancer motility by demethylating lncRNA KCNK15-AS1 [13]. ALKBH5-mediated m6A modification of PVT1 contributes to OS tumorigenesis [14]. Here, we first investigated the role of ALKBH5 in biological behavior of colon cancer cells. And we observed ALKBH5 expression level was significantly upregulated. Besides, NEAT1 could be significantly enriched by ALKBH5. In addition, ALKBH5 knockdown inhibited cell proliferation and migration, induced cell apoptosis in colon cancer cells, and facilitated tumor growth in vivo. The above results suggested that ALKBH5 may act as a tumor promoter to participate in the development of colon cancer as a tumor promoter. In a word, we observed NEAT1 expression level was up-regulated in colon cancer tissues and cells. ALKBH5 knockdown suppressed malignant behavior of colon cancer partially through NEAT1 by demethylation in vitro and vivo, suggesting that ALKBH5-NEAT1 axis maybe potential therapeutic target for colon cancer treatment.
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Azvolinsky A. Colorectal cancer: to stack or sequence therapy? J Natl Cancer Inst. 2015;107:djv138. doi: 10.1093/jnci/djv138. [DOI] [PubMed] [Google Scholar]
- 3.Lee YC, Lee YL, Chuang JP, Lee JC. Differences in survival between colon and rectal cancer from SEER data. PLoS One. 2013;8:e78709. doi: 10.1371/journal.pone.0078709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 5.Deng G, Sui G. Noncoding RNA in oncogenesis: a new era of identifying key players. Int J Mol Sci. 2013;14:18319–18349. doi: 10.3390/ijms140918319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 7.Chakravarty D, Sboner A, Nair SS, Giannopoulou E, Li R, Hennig S, Mosquera JM, Pauwels J, Park K, Kossai M, MacDonald TY, Fontugne J, Erho N, Vergara IA, Ghadessi M, Davicioni E, Jenkins RB, Palanisamy N, Chen Z, Nakagawa S, Hirose T, Bander NH, Beltran H, Fox AH, Elemento O, Rubin MA. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun. 2014;5:5383. doi: 10.1038/ncomms6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang P, Wu X, Wang X, Huang W, Feng Q. NEAT1 upregulates EGCG-induced CTR1 to enhance cisplatin sensitivity in lung cancer cells. Oncotarget. 2016;7:43337–43351. doi: 10.18632/oncotarget.9712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X, Kong J, Ma Z, Gao S, Feng X. Up regulation of the long non-coding RNA NEAT1 promotes esophageal squamous cell carcinoma cell progression and correlates with poor prognosis. Am J Cancer Res. 2015;5:2808–2815. [PMC free article] [PubMed] [Google Scholar]
- 10.Fu JW, Kong Y, Sun X. Long noncoding RNA NEAT1 is an unfavorable prognostic factor and regulates migration and invasion in gastric cancer. J Cancer Res Clin Oncol. 2016;142:1571–1579. doi: 10.1007/s00432-016-2152-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee M, Kim B, Kim VN. Emerging roles of RNA modification: m(6)A and U-tail. Cell. 2014;158:980–987. doi: 10.1016/j.cell.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J, Guo S, Piao HY, Wang Y, Wu Y, Meng XY, Yang D, Zheng ZC, Zhao Y. ALKBH5 promotes invasion and metastasis of gastric cancer by decreasing methylation of the lncRNA NEAT1. J Physiol Biochem. 2019;75:379–389. doi: 10.1007/s13105-019-00690-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He Y, Hu H, Wang Y, Yuan H, Lu Z, Wu P, Liu D, Tian L, Yin J, Jiang K, Miao Y. ALKBH5 inhibits pancreatic cancer motility by decreasing long non-coding RNA KCNK15-AS1 methylation. Cell Physiol Biochem. 2018;48:838–846. doi: 10.1159/000491915. [DOI] [PubMed] [Google Scholar]
- 14.Chen S, Zhou L, Wang Y. ALKBH5-mediated m(6)A demethylation of lncRNA PVT1 plays an oncogenic role in osteosarcoma. Cancer Cell Int. 2020;20:34. doi: 10.1186/s12935-020-1105-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, Chen Y, Sulman EP, Xie K, Bogler O, Majumder S, He C, Huang S. m(6)A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31:591–606. e596. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holoch D, Moazed D. RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet. 2015;16:71–84. doi: 10.1038/nrg3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 19.He X, Tan X, Wang X, Jin H, Liu L, Ma L, Yu H, Fan Z. C-Myc-activated long noncoding RNA CCAT1 promotes colon cancer cell proliferation and invasion. Tumour Biol. 2014;35:12181–12188. doi: 10.1007/s13277-014-2526-4. [DOI] [PubMed] [Google Scholar]
- 20.Zhai H, Fesler A, Schee K, Fodstad O, Flatmark K, Ju J. Clinical significance of long intergenic noncoding RNA-p21 in colorectal cancer. Clin Colorectal Cancer. 2013;12:261–266. doi: 10.1016/j.clcc.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 21.Zhou X, Li X, Cheng Y, Wu W, Xie Z, Xi Q, Han J, Wu G, Fang J, Feng Y. BCLAF1 and its splicing regulator SRSF10 regulate the tumorigenic potential of colon cancer cells. Nat Commun. 2014;5:4581. doi: 10.1038/ncomms5581. [DOI] [PubMed] [Google Scholar]
- 22.Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat Rev Genet. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]