Abstract
Circular RNAs (circRNAs), formed from pre-messenger RNAs by back-splicing, are a novel class of evolutionarily-conserved endogenous non-coding RNAs. While circRNAs are involved in various diseases, the role of circRNAs in intracerebral hemorrhage (ICH) remains unknown. In the present study, we performed high-throughput sequencing to profile the expression of circRNAs in the rat brain at 24 and 48 hours after ICH onset, and utilized bioinformatics methods to make predictions about the function of dysregulated circRNAs. Compared with the sham group, 346 and 389 circRNAs changed significantly (|log2 (fold change)| > 1 and P < 0.05) at 24 and 48 hours after ICH, respectively. Bioinformatics analyses indicated that parent genes of dysregulated circRNAs were involved in biological processes, cellular component, and molecular function following ICH, and that they were enriched in the dopaminergic synapses, glutamatergic synapses, endocytosis, regulation of actin cytoskeleton, the mitogen-activated protein kinase signaling pathway, and the retrograde endocannabinoid signaling pathway. Enrichment analyses of target mRNAs showed that these mRNAs were enriched in synaptic plasticity, ion channel activity, and pathways including the phospholipase D signaling and the cGMP-PKG signaling. Our study indicates that the expression profile of circRNAs changes significantly after ICH in rat brains, and suggests that circRNAs may be crucial for the pathophysiological process following ICH.
Keywords: Intracerebral hemorrhage, rat, circular RNAs, high-throughput sequencing
Introduction
Stroke is one of the leading diseases with the highest mortality and disability rate in the world, especially in low- and middle-income countries. While ischemic stroke is the most common type of stroke, hemorrhagic stroke accounts for more deaths and disability-adjusted life-years lost [1-3]. Although research on mechanisms of brain injury after intracerebral hemorrhage (ICH) has progressed significantly, there is a lack of therapies that can effectively reduce the mortality and disability of ICH. The effectiveness of therapies such as acute phase decompression, hemostasis, minimally invasive surgery, anti-inflammatory therapy, and neuroprotection remains to be assessed [4,5]. Therefore, it is important to explore the pathogenesis of brain injury following ICH to find new treatment methods.
Over the past few years, non-coding RNAs (ncRNAs), such as long non-coding RNAs (lncRNAs) and microRNAs (miRNAs), are involved in various diseases and have attracted significant attention. Accumulating evidence indicates that mammalian brain expresses a wide variety of ncRNAs [6-15]. Circular RNAs (circRNAs), formed from pre-messenger RNAs by back-splicing, are a novel class of ncRNAs [16]. Despite the fact that circRNAs can regulate the transcription processes of parent genes and act as “miRNA sponges” [17], their functions need to be further explored. Altered circRNAs expression is associated with the pathophysiological processes of cancer, cardiac hypertrophy, atherosclerosis and central nervous system (CNS) diseases [18-23]. Mehta et al. [24] found significant changes in the expression of circRNAs in the ischemic penumbral cortex in mice with transient middle cerebral artery occlusion, indicating their possible implication in post-stroke pathophysiology. CircRNAs in the rat hippocampus were significantly altered after brain trauma, suggesting that changes in circRNAs are related to not only brain injury but also post-traumatic neural regeneration [25]. In this study, we utilized high-throughput sequencing to explore the circRNAs expression profile in the rat brain after ICH and bioinformatics methods to predict the potential function of those dysregulated circRNAs. The present findings would broaden our understanding of changes in genomic regulation following ICH and provide novel targets for the treatment of secondary brain injury after ICH.
Materials and methods
Ethics statement and animal preparation
All experiments received the approval of the Animal Care and Experiment Committee of Jinan University, Guangzhou, China, and were performed according to national and international guidelines. Male Sprague-Dawley rats (250-310 g) were housed for at least 7 days before the operation in the laboratory with well-controlled temperature (24-26°C) and humidity (50% relative) and a 12-hour light/dark cycle, and were free to food and water.
Animal groups and ICH model
Animals were randomly divided into sham and ICH groups. The operation was performed with a stereotaxic apparatus and ICH was induced using Type VII collagenase in adult male Sprague-Dawley rats. Under anesthesia with sodium pentobarbital (50 mg/kg, intraperitoneally), 2.0 µL of 0.9% sterile saline containing 0.4 U Type VII collagenase was steadily infused into the right caudate putamen (1.0 mm anterior, 3.1 mm lateral, and 6.0 mm ventral to the bregma) at an even speed for 2 minutes using a 26-gauge needle. After infusion, the needle was kept static for 10 minutes in the site before being slowly removed. Sham-operated rats underwent a similar surgical procedure without the addition of Type VII collagenase to the 0.9% sterile saline. During the operation, rectal temperature was maintained at 37.0 ± 0.5°C. Rats showing symptoms of neurological deficits were included in this study.
RNA extraction and quality control
Animals were euthanized 24 and 48 hours after the induction of ICH. The brain tissue ipsilateral to the hematoma was rapidly dissected and stored in liquid nitrogen. Total RNA was extracted from brain samples using TRIzol reagent (Invitrogen, Carlsbad, CA). The purity and concentration of total RNA were measured using NanoDrop ND-1000 (NanoDrop, Wilmington, DE). RNA integrity was evaluated by Agilent 2200 TapeStation (Agilent Technologies, USA).
High-throughput sequencing and analysis of circRNAs
Briefly, circRNAs were enriched by the removal of ribosomal RNA (rRNA) and linear RNAs using rRNA Removal kit and RNase R (Epicentre, USA). Then, enriched circRNAs were transcribed into fluorescent complementary deoxyribonucleic acid (cDNA) and purified. After PCR amplification, small RNA single-end sequencing was conducted on Illumina HiSeq 3000 (Illumina, San Diego, CA). Raw reads were processed with Trimmomatic tools (V0.36), reads quality was inspected using the FastQC software, and statistical result were then output. CIRI2 (Beijing Institutes of Life Science, Beijing, China) and CIRCexplorer2 (Shanghai Institutes for Biological Sciences, Shanghai, China) were used to detect circRNAs. Identified circRNAs were from the overlapped results of both methods. |log2 (fold change)| > 1 and P-value (P < 0.05) were used as criteria for identifying the statistical significance of differentially expressed circRNAs between ICH groups and sham.
Gene ontology (GO) and kyoto encyclopedia of genes and genomes (KEGG) pathway analyses
GO (http://www.geneontology.org/) and KEGG pathway analyses (http://www.genome.jp/) were performed to investigate potential roles of parent genes of the differentially expressed circRNAs (P < 0.05). |Fold change| > 1 and P < 0.05 were computed between the sham group and each ICH time point (24 and 48 hours after ICH onset) by cross-comparison analysis. GO and KEGG analyses were conducted for target mRNAs to further understand the function of significantly altered circRNAs.
Quantitative RT-PCR verification
Four significantly dysregulated circRNAs were randomly selected for qRT-PCR to validate the high-throughput sequencing data. In brief, total RNA was extracted from the brain samples of the two groups (48 h ICH group and sham, n = 6 each) using TRIzol reagent (Invitrogen), RNA was reverse transcribed into cDNA, and then qPCR was conducted using Bulge-LoopTM miRNA qRT-PCR Starter Kit on a C1000 Touch PCR system (Bio-Rad, Hercules, CA). Three independent experiments were performed, and GAPDH was used as the internal control. The 2-ΔΔCT method was used for comparative quantitation. The primers of four circRNAs and GAPDH are presented in Table 2.
Table 2.
The sequences of primers used for qRT-PCR verification of circRNAs
| circRNA ID | gene symbol | Primer sequences (5’-3’) |
|---|---|---|
| rno_circ:chr9:31604268-31660864 | Adgrb3 | F: CACTTGGCGAATGGCACTTT |
| R: GCATCTTCCCATTTCTCCTTGTT | ||
| rno_circ:chr18:17224244-17264715 | Fhod3 | F: GACTCACAAGAGGCTCTCACGG |
| R: GGACGATGCGGAAGATGGA | ||
| rno_circ:chr13:35769419-35784163 | Ptpn4 | F: CGGATGAATACTTTTCTGTGGTATC |
| R: CAAGGACAGGATAATCTTCCAGTAA | ||
| rno_circ:chr1:281093685-281095506 | Rab11fip2 | F: ACTATAATGCCATCTTCAAGGTGGT |
| R: CATGCTTGCTGTCATGTTGTTC | ||
| GAPDH | F: GAACGGGAAGCTCACTGG | |
| R: GCCTGCTTCACCACCTTCT |
Analysis of circRNA regulatory network
CircRNA regulatory networks were constructed to show the interactions among dysregulated circRNAs and their targets. Among the 86 differentially expressed circRNAs that were altered at both time points, 40 circRNAs (30 upregulated and 10 downregulated; ranked by |log2 (fold change)|) and their top five target miRNAs were selected to construct the circRNA-miRNA network. According to qRT-PCR results, the two confirmed circRNAs (rno_circ:chr18:17224244-17264715 and rno_circ:chr1:281093685-281095506) were selected for the annotation and function prediction. CircRNA-miRNA interactions and mRNAs targeted by the circRNA-miRNA network were predicted using proprietary software based on TargetScan and miRanda, and RNA regulatory networks were constructed utilizing Cytoscape software (San Diego, CA, USA). Target miRNAs intersected by miRanda and TargetScan prediction results were ranked by miRanda to identify the top five miRNAs for establishing the RNA interaction networks. Based on the intersection results of target mRNAs predicted by both software, 1962 mRNAs were selected for KEGG analysis according to “total score > 500” in miRanda, and 72 mRNAs that were enriched in the top 10 KEGG pathways were selected for constructing the circRNA-miRNA-mRNA network.
Statistical analysis
Student’s t-test was utilized to compare the statistical significance of data between two groups, data were presented as mean ± SD and analyzed by Statistical Program for Social Sciences (SPSS) 20.0 software (SPSS, Chicago, IL). A P value < 0.05 was considered statistically significant.
Results
CircRNA expression profiles in the perihematomal brain tissue after ICH
High-throughput sequencing was conducted to identify the circRNA expression profile in the brain tissue surrounding the hematoma of ICH rats. All samples showed similar distributions of circRNA expression patterns. A total of 31132 circRNAs were detected in all the ipsilateral brain samples (sham, 24 and 48 hours after ICH; n = 3 per group). Compared with the sham group, 346 circRNAs were differentially expressed (|log2 (fold change)| > 1 and P-value < 0.05) at 24 hours after ICH, of which 198 were upregulated and 148 were downregulated; and 389 circRNAs were differentially expressed at 48 hours after ICH, including 248 upregulated and 141 downregulated. Altered circRNAs data was delineated using hierarchical clustering analyses (Figure 1A, 1C) and volcano plots (Figure 1B, 1D). Eighty-six circRNAs showed persistent changes at 24 and 48 hours after ICH compared with the sham group. The 10 significantly upregulated and downregulated circRNAs among them are shown in Table 1. Of the differentially expressed circRNAs, most originated from exons (Figure 2B). The distribution of dysregulated circRNAs indicated that they were transcribed from all chromosomes (Figure 2A).
Figure 1.
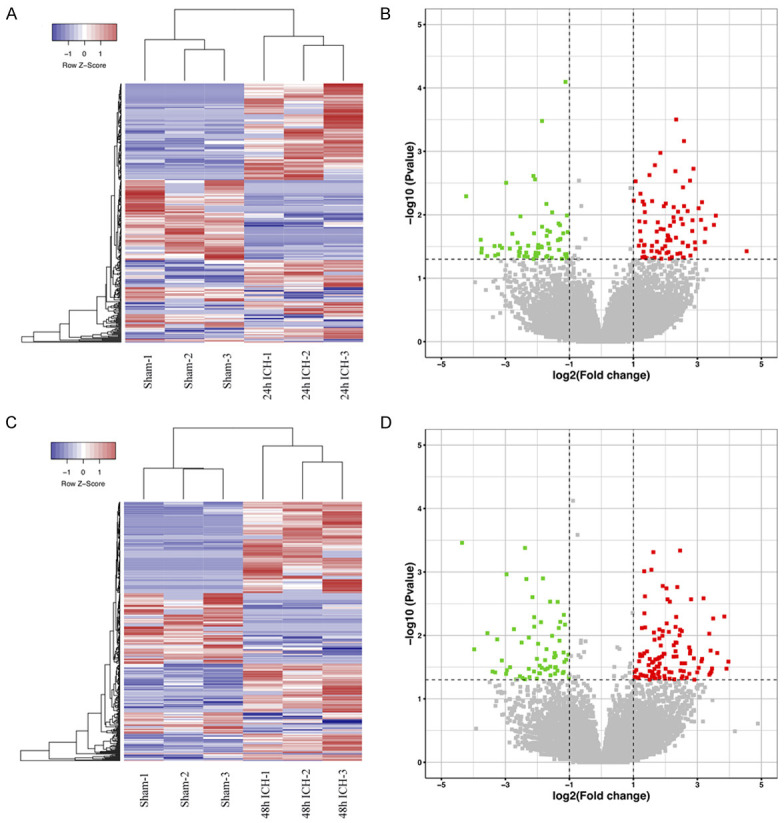
The expression profile of circRNAs in the perihematomal brain tissue after ICH. The hierarchical clustering analyses of circRNAs differentially expressed at 24 hours (A) and 48 hours (C) after ICH compared with the sham group, “Blue” represents low intensity, and “Red” represents strong intensity. Volcano plots show all the detected circRNAs (sham vs. 24 hours (B) and sham vs. 48 hours after ICH (D)), the red points represent the differentially upregulated circRNAs, while the green points represent the differentially downregulated circRNAs (|log2 (fold change)| > 1, P-value < 0.05 (-log10 scaled)).
Table 1.
The 10 differentially upregulated and downregulated circRNAs at 24 hours and 48 hours in ICH rats compared with the sham group
| CricRNA ID | |log2 (Fold change)| | P-Value | Regulation | Chromoso-me | type | Gensymbol | ||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| 24 h | 48 h | 24 h | 48 h | |||||
| rno_circ:chr12:37603986-37605288 | 11.9045 | 10.6433 | 0.010323969 | 0.017333116 | Up | chr12 | exonic | Sbno1 |
| rno_circ:chr20:50725400-50771244 | 11.4364 | 14.3141 | 0.014442869 | 0.005031039 | Up | chr20 | exonic | Hace1 |
| rno_circ:chr2:22264781-22267377 | 2.55942 | 2.57513 | 0.009064435 | 0.007492125 | Up | chr2 | exonic | Serinc5 |
| rno_circ:chr12:31504843-31537157 | 2.99192 | 3.15639 | 0.030914823 | 0.033726052 | Up | chr12 | splicing:exonic | Rimbp2 |
| rno_circ:chr13:35769419-35784163 | 3.39318 | 3.20916 | 0.013152745 | 0.023978029 | Up | chr13 | exonic | Ptpn4 |
| rno_circ:chr1:281093685-281095506 | 3.08086 | 2.43936 | 0.034259741 | 0.006119182 | Down | chr1 | exonic | Rab11fip2 |
| rno_circ:chr18:17224244-17264715 | 2.17911 | 3.15088 | 0.000080400 | 0.000000031 | Down | chr18 | exonic | Fhod3 |
| rno_circ:chr2:212471565-212487554 | 3.05132 | 3.28599 | 0.018498214 | 0.016463408 | Down | chr2 | exonic | Vav3 |
| rno_circ:chr4:152138047-152214576 | 2.94274 | 4.29274 | 0.047673343 | 0.005153764 | Down | chr4 | splicing:exonic | Erc1 |
| rno_circ:chr11:84575501-84577375 | 7.88282 | 4.81551 | 0.003129564 | 0.010837388 | Down | chr11 | exonic | Yeats2 |
Figure 2.
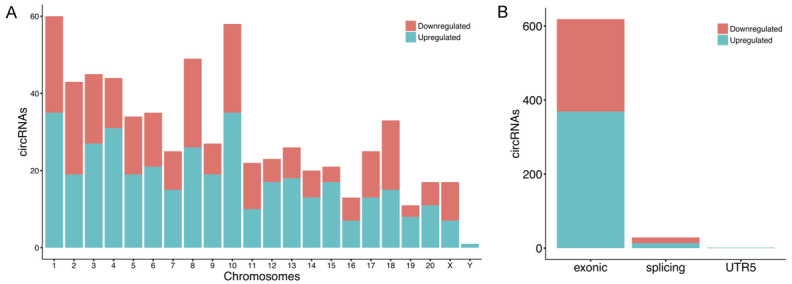
Features of differentially expressed circRNAs. Distribution of dysregulated circRNAs in rat chromosomes (A). Most circRNAs after ICH are from the exonic region, and few are from the splicing region (B).
GO and KEGG analyses for parent genes of significantly altered circRNAs
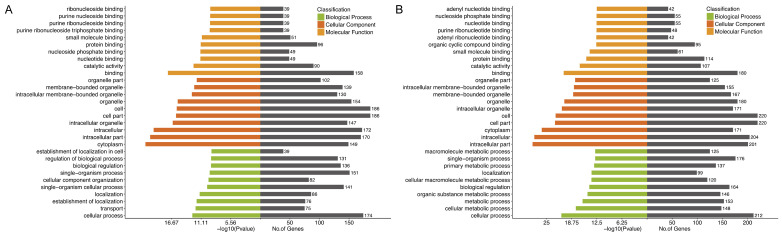
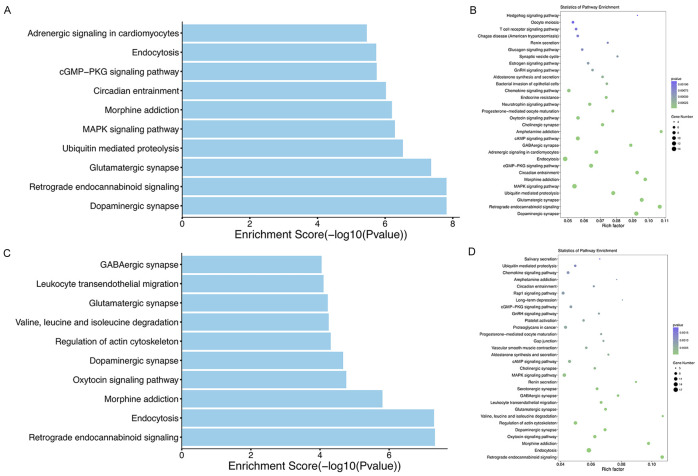
CircRNAs can regulate the expression of parent genes and affect the transcription of related mRNAs, thus have regulatory functions in the pathophysiological processes of various diseases [17,26,27]. To speculate about the pathophysiological significance of circRNAs dysregulated following ICH, we performed GO and KEGG analyses for parent genes of dysregulated circRNAs. GO analysis revealed that the major enriched GO terms for biological processes were “cellular process”, “transport”, and “establishment of localization” at 24 hours after ICH and “cellular process”, “cellular metabolic process”, and “metabolic process” at 48 hours after ICH. The majority of the enriched cellular component terms were “cytoplasm”, “intracellular part”, and “intracellular” at both time points after ICH. As for molecular function, the major GO terms were “binding”, “catalytic activity”, and “nucleotide binding” at 24 hours after ICH, and “binding”, “catalytic activity”, and “protein binding” at 48 hours after ICH (Figure 3A, 3B). Correspondingly, KEGG pathway analysis indicated that parent genes of the dysregulated circRNAs at 24 hours after ICH were significantly involved in the dopaminergic synapses, glutamatergic synapses, MAPK signaling pathway, and the retrograde endocannabinoid signaling pathway (Figure 4A, 4B), but those at 48 hours after ICH were significantly involved in regulation of actin cytoskeleton, endocytosis, and the retrograde endocannabinoid signaling pathway (Figure 4C, 4D).
Figure 3.
GO enrichment analyses for the parent genes of altered circRNAs. Major enriched GO terms for biological process, cellular component, and molecular function at 24 hours and 48 hours after ICH in rats (A, B).
Figure 4.
KEGG pathway analyses for the parent genes of altered circRNAs at 24 hours (A, B) and 48 hours (C, D) after ICH in rats. The top 10 (A, C) and top 30 (B, D) enriched pathways.
Verification of circRNA expression
To verify the high-throughput sequencing data, four differentially expressed circRNAs (rno_circ:chr9:31604268-31660864, rno_circ:chr18:17224244-17264715, rno_circ:chr13:35769419-35784163, and rno_circ:chr1:281093685-281095506) were selected for qRT-PCR. The validated results of rno_circ:chr9:31604268-31660864 and rno_circ:chr13:35769419-35784163 showed no statistical differences between ICH and the sham group (P > 0.05). The expression levels of rno_circ:chr18:17224244-17264715 (P < 0.05) and rno_circ:chr1:281093685-281095506 (P < 0.01) were similar to the sequencing data (Figure 5).
Figure 5.
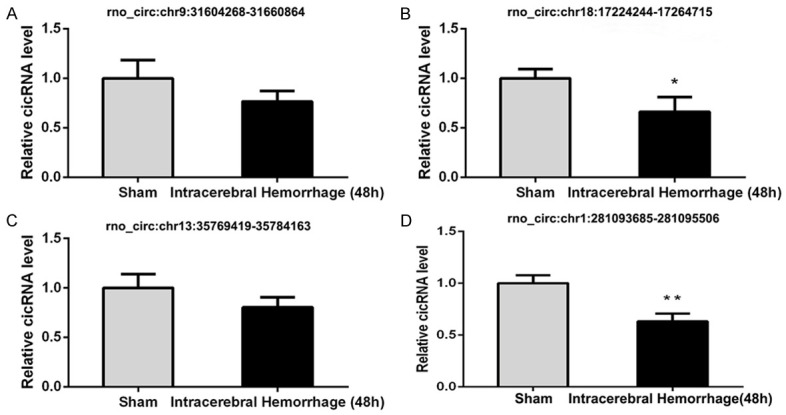
qRT-PCR validation results of four randomly selected circRNAs (rno_circ:chr9:31604268-31660864, rno_circ:chr18:17224244-17264715, rno_circ:chr13:35769419-35784163, and rno_circ:chr1:281093685-281095506). Data are shown as mean ± SD. *P < 0.05, **P < 0.01, compared with the sham group.
CircRNA-miRNA and circRNA-miRNA-mRNA network and enrichment analyses of target mRNAs
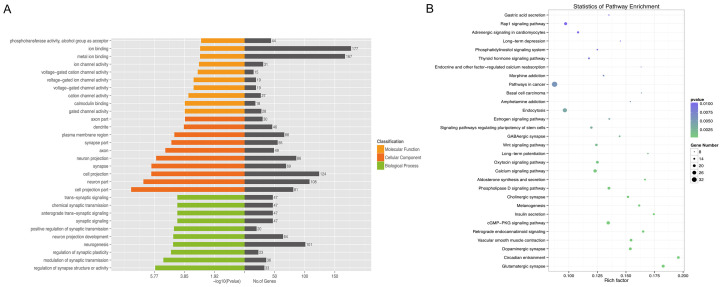
Currently, it is widely believed that miRNAs lead to the cleavage and translational repression of messenger RNA (mRNA) [28], and that circRNAs act as sponges for specific miRNAs to regulate miRNA-associated functions and to affect the activity of multiple genes [29]. The interaction between circRNAs and ICH-related miRNAs would indicate that circRNAs play a role in the pathophysiological processes after ICH. The circRNA-miRNA network for 40 differentially expressed circRNAs at both time points after ICH is shown in Figure 6, and the circRNA-miRNA-mRNA network for the two validated circRNAs is shown in Figure 7. GO and KEGG analyses were performed for target mRNAs to understand their potential functions after ICH and results are shown in Figure 8. As shown in Figure 7, rno_circ:chr18:17224244-17264715 has a binding site for miR-105, and rno_circ:chr1:281093685-281095506 can bind to miR-298-5p. MiR-105 and miR-298-5p are predicted to have binding sites for many mRNAs such as Wnt9a, Fzd7, Akt, Mapk1, and Trpm4. The enrichment analyses suggested that the target mRNAs might be associated with the regulation of synaptic plasticity and activity, ion channel activity and trans-synaptic signaling, the cGMP-PKG signaling pathway, the phospholipase D signaling pathway, and the retrograde endocannabinoid signaling.
Figure 6.
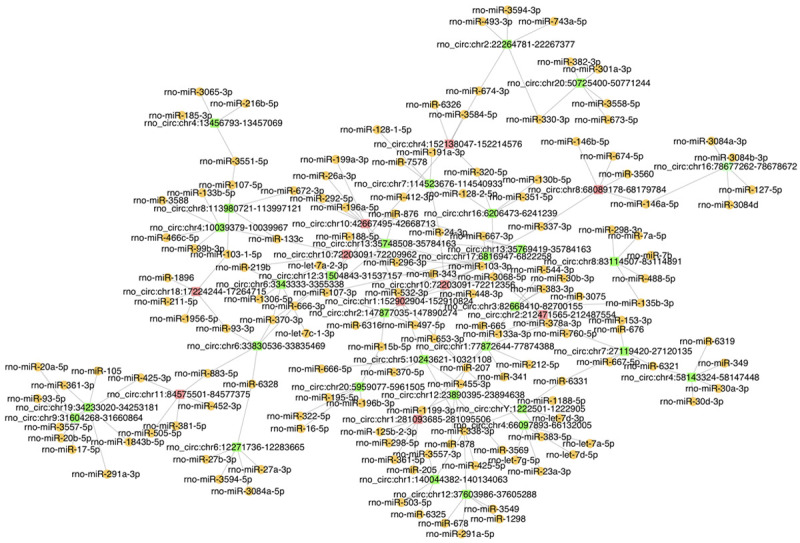
The circRNA-miRNA network. The network consists of 40 differentially expressed circRNAs and their top five miRNAs. Red squares, green squares, and orange spheres indicate downregulated circRNAs, upregulated circRNAs, and miRNAs, respectively.
Figure 7.
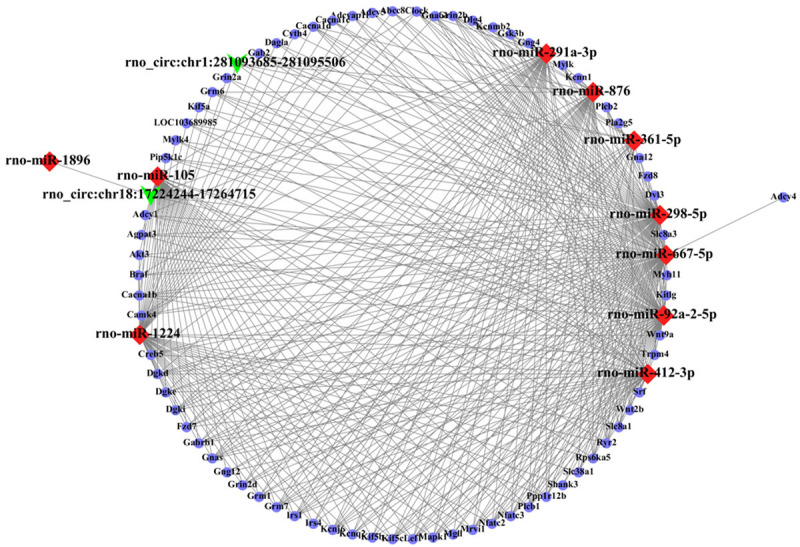
The circRNA-miRNA-mRNA interaction network. The network includes two validated circRNAs, 10 miRNAs, and 72 mRNAs. Green arrows, red rhombuses, and blue spheres indicate circRNAs, miRNAs, and mRNAs, respectively.
Figure 8.
GO and KEGG pathway analyses for mRNAs targeted by the circRNA (confirmed by qRT-PCR)-miRNA regulatory network. Major enriched GO terms for biological process, cellular component, and molecular function (A) and the 30 major enriched pathways (B) at 48 hours after ICH in rats.
Discussion
This study indicates that the expression profile of circRNAs in the rat brain changes significantly at 24 h and 48 h after ICH, and significantly dysregulated circRNAs may be involved in the pathophysiologic processes of secondary brain damage and the control of neurological dysfunction. The findings suggest that differentially expressed circRNAs might be novel molecular targets for protecting neuronal function and promoting neurological recovery after ICH.
ICH results in entry of glutamate from bloodstream into the extracellular space, and production of thrombin leads to phosphorylation of N-methyl-D-aspartate (NMDA) receptor and greater calcium influx into neurons. A large amount of intracellular calcium leads to mitochondrial dysfunction and the release of reactive oxygen species, resulting in apoptosis. In addition, brain injury activates glial cells in the CNS and results in infiltration of peripheral inflammatory cells that secrete inflammatory mediators, cytokines and chemokines, thereby exacerbating destruction of the blood-brain barrier and causing secondary brain injury [5]. Secondary brain injury after ICH involves various pathophysiological mechanisms, such as inflammation, oxidative stress, apoptosis, and autophagy. Accumulating evidence indicates that various classes of ncRNAs play important roles in brain injury and may have neuroprotective effects by modulating apoptosis, autophagy, neurogenesis, and angiogenesis after acute CNS injury [30,31]. The expression profile of circRNAs has been demonstrated to change significantly in the penumbral cortex after ischemic stroke and in the hippocampus after traumatic brain injury, suggesting that circRNAs might be involved in the regulation of secondary brain injury [24,25]. Cui et al. [32] found that lncRNA and mRNA were differentially expressed in the rat brain at 21 days following ICH, suggesting that lncRNA- and mRNA-related changes might regulate neuronal dysfunction. Dou et al. [33] explored the potential role of circRNAs at 6, 12 and 24 hours after ICH and predicted that the significantly altered circRNAs were associated with synaptic regulation, phosphorylation, and inflammation. Considering the drivers of acute and subacute neurological deterioration after ICH and the duration of cerebral edema [34], early intervention may promote neurological recovery and improve prognosis. In this study, we detected the differential expression of circRNAs at both 24 h and 48 h time points after ICH and used bioinformatics methods to explore their potential functions.
Based on the regulatory function of circRNAs on parent genes [17], we speculated on the possible role of ICH-related circRNAs by understanding the potential function of their parent genes. GO analysis for parent genes of dysregulated circRNAs showed the main enrichment of biological regulation, protein and nucleotide binding, localization, and metabolism process at both time points. Our findings suggest that these parent genes might be involved in the biological functions essential for the survival of neurons and play roles in the regulation of brain injury following ICH. KEGG analysis suggested that these parent genes were greatly enriched in dopaminergic and glutamatergic synapses, MAPK signaling pathway, regulation of actin cytoskeleton, endocytosis, and the retrograde endocannabinoid signaling pathway at 48 hours following ICH. Actin cytoskeleton is essential for junction function, and endocytosis is important for junction remodeling. After ICH, actin cytoskeleton may be a target of treatment for preventing blood-brain barrier (BBB) disruption [35], and cerebrovascular integrity plays a crucial role in disease pathology and neurologic deficits. MAPK families are strongly activated by environmental stresses and inflammatory cytokines and their activations contribute to inflammation, apoptosis, cytokine production, and metabolism [36]. Retrograde endocannabinoid signaling mediates biological functions after brain injury. Activation of the endocannabinoid system not only inhibits presynaptic GABAergic and glutamatergic synaptic transmission but also enhances post-synaptic responses at excitatory and inhibitory synapses [37,38]. Furthermore, inhibition of 2-arachidonoyl glycerol metabolism by inactivating monoacyl glycerol produces profound anti-inflammatory and neuroprotective effects [39,40]. Taken together, our data indicate that glutamatergic synapses, MAPK families, and the retrograde endocannabinoid signaling pathway may play crucial roles in the pathophysiological processes of secondary brain damage of ICH.
It is known that most circRNAs can act as “miRNA sponges” and affect the expression of downstream genes and their biological functions [29,41,42]. We used bioinformatics methods to predict miRNA targets and further circRNA-miRNA targets, constructed the circRNA-miRNA-mRNA network, and performed enrichment analyses for these target mRNAs. We found that the differentially expressed circRNAs had binding sites for different miRNAs. The two validated circRNAs, rno_circ:chr18:17224244-17264715 and rno_circ:chr1:281093685-281095506, can bind to miR-105 and miR-298-5p, respectively. Evidence shows that miR-105 participates in the regulation of necroptosis/apoptosis in myocardial infarction rat hearts [43] and miR-298-5p modulates signaling pathways of apoptosis [44]. Moreover, GO analysis indicated that target mRNAs were enriched in regulation of synaptic plasticity and activity, ion channel activity, and trans-synaptic signaling, and KEGG results suggested that the major enriched pathways were the cGMP-PKG signaling pathway, phospholipase D signaling pathway, retrograde endocannabinoid signaling, and glutamatergic and dopaminergic synapse. It is known that cGMP/PKG signaling pathways are involved in modulation of neuroinflammation, oxidative stress, apoptosis, and synaptic plasticity [45]. Notably, mRNAs shown in the circRNA-miRNA-mRNA network, such as those of Wnt, Gsk3b, Akt, Mapk1, and Trpm4, have many important functions in biological responses. Accumulating studies suggest that TRPM4 is related to secondary hemorrhage after spinal cord injury and contributes to neuronal injury under inflammatory conditions [46,47]. MAPK1 and MAPK14 and their upstream signaling pathways are essential for mitophagy [48]. These results indicate that the altered circRNAs after ICH possibly regulate functions of downstream mRNAs through signaling pathways mentioned above, especially the cGMP/PKG signaling pathway.
There are several limitations to this study. First, the significantly dysregulated circRNAs need to be verified in human samples, and the predicted target miRNAs and mRNAs require further identification. Second, we used bioinformatics methods to predict potential function of the differentially expressed circRNAs following ICH, and the biological function of the significantly altered circRNAs should be discovered in further studies. Third, it is not clear whether the differentially expressed circRNAs function in a cell-specific manner.
In conclusion, this study suggests that the significantly dysregulated circRNAs might be involved in the regulation of the pathophysiological processes after ICH, including blood-brain barrier permeability, inflammation, oxidative stress, apoptosis, and autophagy. The findings indicate that rno_circ:chr18:17224244-17264715 and rno_circ:chr1:281093685-281095506 might be key biological targets for the treatment of ICH. Further studies are needed to explore the specific regulatory mechanisms of circRNAs to delineate whether they can be used as novel biomarkers for treatment of ICH to promote neurological recovery and improve cerebral hemorrhage patients’ quality of life.
Acknowledgements
This study was supported by the grants from the Natural Science Foundation of Guangdong Province of China (2018A030313586), the Science and Technology Project of Shenzhen (JCYJ20190808123611319), Natural Science Foundation of Shenzhen University General Hospital (0000040559), Sanming Project of Medicine in Shenzhen (SZSM201812047), and Shenzhen Key Medical Discipline Construction Fund (SZXK074).
Disclosure of conflict of interest
None.
References
- 1.Johnson CO, Nguyen M, Roth GA, Nichols E, Alam T, Abate D, Abd-Allah F, Abdelalim A, Abraha HN, Abu-Rmeileh NME, Adebayo OM, Adeoye AM, Agarwal G, Agrawal S, Aichour AN, Aichour I, Aichour MTE, Alahdab F, Ali R, Alvis-Guzman N, Anber NH, Anjomshoa M, Arabloo J, Arauz A, Ärnlöv J, Arora A, Awasthi A, Banach M, Barboza MA, Barker-Collo SL, Bärnighausen TW, Basu S, Belachew AB, Belayneh YM, Bennett DA, Bensenor IM, Bhattacharyya K, Biadgo B, Bijani A, Bikbov B, Bin Sayeed MS, Butt ZA, Cahuana-Hurtado L, Carrero JJ, Carvalho F, Castañeda-Orjuela CA, Castro F, Catalá-López F, Chaiah Y, Chiang PPC, Choi JYJ, Christensen H, Chu DT, Cortinovis M, Damasceno AAM, Dandona L, Dandona R, Daryani A, Davletov K, de Courten B, De la Cruz-Góngora V, Degefa MG, Dharmaratne SD, Diaz D, Dubey M, Duken EE, Edessa D, Endres M, Faraon EJA, Farzadfar F, Fernandes E, Fischer F, Flor LS, Ganji M, Gebre AK, Gebremichael TG, Geta B, Gezae KE, Gill PS, Gnedovskaya EV, Gómez-Dantés H, Goulart AC, Grosso G, Guo Y, Gupta R, Haj-Mirzaian A, Haj-Mirzaian A, Hamidi S, Hankey GJ, Hassen HY, Hay SI, Hegazy MI, Heidari B, Herial NA, Hosseini MA, Hostiuc S, Irvani SSN, Islam SMS, Jahanmehr N, Javanbakht M, Jha RP, Jonas JB, Jozwiak JJ, Jürisson M, Kahsay A, Kalani R, Kalkonde Y, Kamil TA, Kanchan T, Karch A, Karimi N, Karimi-Sari H, Kasaeian A, Kassa TD, Kazemeini H, Kefale AT, Khader YS, Khalil IA, Khan EA, Khang YH, Khubchandani J, Kim D, Kim YJ, Kisa A, Kivimäki M, Koyanagi A, Krishnamurthi RK, Kumar GA, Lafranconi A, Lewington S, Li S, Lo WD, Lopez AD, Lorkowski S, Lotufo PA, Mackay MT, Majdan M, Majdzadeh R, Majeed A, Malekzadeh R, Manafi N, Mansournia MA, Mehndiratta MM, Mehta V, Mengistu G, Meretoja A, Meretoja TJ, Miazgowski B, Miazgowski T, Miller TR, Mirrakhimov EM, Mohajer B, Mohammad Y, Mohammadoo-khorasani M, Mohammed S, Mohebi F, Mokdad AH, Mokhayeri Y, Moradi G, Morawska L, Moreno Velásquez I, Mousavi SM, Muhammed OSS, Muruet W, Naderi M, Naghavi M, Naik G, Nascimento BR, Negoi RI, Nguyen CT, Nguyen LH, Nirayo YL, Norrving B, Noubiap JJ, Ofori-Asenso R, Ogbo FA, Olagunju AT, Olagunju TO, Owolabi MO, Pandian JD, Patel S, Perico N, Piradov MA, Polinder S, Postma MJ, Poustchi H, Prakash V, Qorbani M, Rafiei A, Rahim F, Rahimi K, Rahimi-Movaghar V, Rahman M, Rahman MA, Reis C, Remuzzi G, Renzaho AMN, Ricci S, Roberts NLS, Robinson SR, Roever L, Roshandel G, Sabbagh P, Safari H, Safari S, Safiri S, Sahebkar A, Salehi Zahabi S, Samy AM, Santalucia P, Santos IS, Santos JV, Santric Milicevic MM, Sartorius B, Sawant AR, Schutte AE, Sepanlou SG, Shafieesabet A, Shaikh MA, Shams-Beyranvand M, Sheikh A, Sheth KN, Shibuya K, Shigematsu M, Shin M-J, Shiue I, Siabani S, Sobaih BH, Sposato LA, Sutradhar I, Sylaja PN, Szoeke CEI, Te Ao BJ, Temsah MH, Temsah O, Thrift AG, Tonelli M, Topor-Madry R, Tran BX, Tran KB, Truelsen TC, Tsadik AG, Ullah I, Uthman OA, Vaduganathan M, Valdez PR, Vasankari TJ, Vasanthan R, Venketasubramanian N, Vosoughi K, Vu GT, Waheed Y, Weiderpass E, Weldegwergs KG, Westerman R, Wolfe CDA, Wondafrash DZ, Xu G, Yadollahpour A, Yamada T, Yatsuya H, Yimer EM, Yonemoto N, Yousefifard M, Yu C, Zaidi Z, Zamani M, Zarghi A, Zhang Y, Zodpey S, Feigin VL, Vos T, Murray CJL. Global, regional, and national burden of stroke, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019;18:439–458. doi: 10.1016/S1474-4422(19)30034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Katan M, Luft A. Global burden of stroke. Semin Neurol. 2018;38:208–211. doi: 10.1055/s-0038-1649503. [DOI] [PubMed] [Google Scholar]
- 3.Feigin VL, Norrving B, Mensah GA. Global burden of stroke. Circ Res. 2017;120:439–448. doi: 10.1161/CIRCRESAHA.116.308413. [DOI] [PubMed] [Google Scholar]
- 4.Hankey GJ. Stroke. Lancet. 2017;389:641–654. doi: 10.1016/S0140-6736(16)30962-X. [DOI] [PubMed] [Google Scholar]
- 5.Keep RF, Hua Y, Xi G. Intracerebral haemorrhage: mechanisms of injury and therapeutic targets. Lancet Neurol. 2012;11:720–731. doi: 10.1016/S1474-4422(12)70104-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, Herzog M, Schreyer L, Papavasileiou P, Ivanov A, Ohman M, Refojo D, Kadener S, Rajewsky N. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 7.Qureshi IA, Mehler MF. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat Rev Neurosci. 2012;13:528–541. doi: 10.1038/nrn3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X, Tang X, Liu K, Hamblin MH, Yin KJ. Long noncoding RNA Malat1 regulates cerebrovascular pathologies in ischemic stroke. J Neurosci. 2017;37:1797–1806. doi: 10.1523/JNEUROSCI.3389-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dykstra-Aiello C, Jickling GC, Ander BP, Shroff N, Zhan X, Liu D, Hull H, Orantia M, Stamova BS, Sharp FR. Altered expression of long noncoding RNAs in blood after ischemic stroke and proximity to putative stroke risk loci. Stroke. 2016;47:2896–2903. doi: 10.1161/STROKEAHA.116.013869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mehta SL, Kim T, Vemuganti R. Long noncoding RNA FosDT promotes ischemic brain injury by interacting with REST-associated chromatin-modifying proteins. J Neurosci. 2015;35:16443–16449. doi: 10.1523/JNEUROSCI.2943-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Selvamani A, Sathyan P, Miranda RC, Sohrabji F. An antagomir to microRNA Let7f promotes neuroprotection in an ischemic stroke model. PLoS One. 2012;7:e32662. doi: 10.1371/journal.pone.0032662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu DZ, Tian Y, Ander BP, Xu H, Stamova BS, Zhan X, Turner RJ, Jickling G, Sharp FR. Brain and blood microRNA expression profiling of ischemic stroke, intracerebral hemorrhage, and kainate seizures. J Cereb Blood Flow Metab. 2010;30:92–101. doi: 10.1038/jcbfm.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dharap A, Nakka VP, Vemuganti R. Effect of focal ischemia on long noncoding RNAs. Stroke. 2012;43:2800–2802. doi: 10.1161/STROKEAHA.112.669465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dharap A, Bowen K, Place R, Li LC, Vemuganti R. Transient focal ischemia induces extensive temporal changes in rat cerebral microRNAome. J Cereb Blood Flow Metab. 2009;29:675–687. doi: 10.1038/jcbfm.2008.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang H, Wang Y, Ai M, Wang H, Duan Z, Wang H, Zhao L, Yu J, Ding Y, Wang S. Long noncoding RNA CRNDE stabilized by hnRNPUL2 accelerates cell proliferation and migration in colorectal carcinoma via activating Ras/MAPK signaling pathways. Cell Death Dis. 2017;8:e2862. doi: 10.1038/cddis.2017.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J, Liu T, Wang X, He A. Circles reshaping the RNA world: from waste to treasure. Mol Cancer. 2017;16:58. doi: 10.1186/s12943-017-0630-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang S, Li X, Zheng H, Si X, Li B, Wei G, Li C, Chen Y, Chen Y, Liao W, Liao Y, Bin J. Loss of super-enhancer-regulated circRNA Nfix induces cardiac regeneration after myocardial infarction in adult mice. Circulation. 2019;139:2857–2876. doi: 10.1161/CIRCULATIONAHA.118.038361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J, Chen T, Zhu Y, Li Y, Zhang Y, Wang Y, Li X, Xie X, Wang J, Huang M, Sun X, Ke Y. circPTN sponges miR-145-5p/miR-330-5p to promote proliferation and stemness in glioma. J Exp Clin Cancer Res. 2019;38:398. doi: 10.1186/s13046-019-1376-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Y, Alexandrov PN, Jaber V, Lukiw WJ. Deficiency in the ubiquitin conjugating enzyme UBE2A in Alzheimer’s Disease (AD) is linked to deficits in a natural circular miRNA-7 Sponge (circRNA; ciRS-7) Genes (Basel) 2016;7:116. doi: 10.3390/genes7120116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, Kohlmaier A, Herbst A, Northoff BH, Nicolaou A, Gabel G, Beutner F, Scholz M, Thiery J, Musunuru K, Krohn K, Mann M, Teupser D. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 2016;7:12429. doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ouyang Q, Huang Q, Jiang Z, Zhao J, Shi GP, Yang M. Using plasma circRNA_002453 as a novel biomarker in the diagnosis of lupus nephritis. Mol Immunol. 2018;101:531–538. doi: 10.1016/j.molimm.2018.07.029. [DOI] [PubMed] [Google Scholar]
- 23.Li X, Wang J, Zhang C, Lin C, Zhang J, Zhang W, Zhang W, Lu Y, Zheng L, Li X. Circular RNA circITGA7 inhibits colorectal cancer growth and metastasis by modulating the Ras pathway and upregulating transcription of its host gene ITGA7. J Pathol. 2018;246:166–179. doi: 10.1002/path.5125. [DOI] [PubMed] [Google Scholar]
- 24.Mehta SL, Pandi G, Vemuganti R. Circular RNA expression profiles alter significantly in mouse brain after transient focal ischemia. Stroke. 2017;48:2541–2548. doi: 10.1161/STROKEAHA.117.017469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie BS, Wang YQ, Lin Y, Zhao CC, Mao Q, Feng JF, Cao JY, Gao GY, Jiang JY. Circular RNA expression profiles alter significantly after traumatic brain injury in rats. J Neurotrauma. 2018;35:1659–1666. doi: 10.1089/neu.2017.5468. [DOI] [PubMed] [Google Scholar]
- 26.Li J, Zhen L, Zhang Y, Zhao L, Liu H, Cai D, Chen H, Yu J, Qi X, Li G. Circ-104916 is downregulated in gastric cancer and suppresses migration and invasion of gastric cancer cells. Onco Targets Ther. 2017;10:3521–3529. doi: 10.2147/OTT.S136347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao S, Chen G, Yan L, Li L, Huang X. Contribution of dysregulated circRNA_100876 to proliferation and metastasis of esophageal squamous cell carcinoma. Onco Targets Ther. 2018;11:7385–7394. doi: 10.2147/OTT.S177524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 29.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 30.Chandran R, Mehta SL, Vemuganti R. Non-coding RNAs and neuroprotection after acute CNS injuries. Neurochem Int. 2017;111:12–22. doi: 10.1016/j.neuint.2017.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu Z, Wu P, Zuo X, Yu N, Qin Y, Xu Q, He S, Cen B, Liao W, Ji A. LncRNA-N1LR enhances neuroprotection against ischemic stroke probably by inhibiting p53 phosphorylation. Mol Neurobiol. 2016;54:7670–7685. doi: 10.1007/s12035-016-0246-z. [DOI] [PubMed] [Google Scholar]
- 32.Cui H, Liu T, Li P, Yang A, Zhou H, Luo J, Wang Y, Tang T. Altered long noncoding RNA and messenger RNA expression in experimental intracerebral hemorrhage - a preliminary study. Cell Physiol Biochem. 2018;45:1284–1301. doi: 10.1159/000487464. [DOI] [PubMed] [Google Scholar]
- 33.Dou Z, Yu Q, Wang G, Wu S, Reis C, Ruan W, Yan F, Chen G. Circular RNA expression profiles alter significantly after intracerebral hemorrhage in rats. Brain Res. 2020;1726:146490. doi: 10.1016/j.brainres.2019.146490. [DOI] [PubMed] [Google Scholar]
- 34.Lord AS, Gilmore E, Choi HA, Mayer SA VISTA-ICH Collaboration. Time course and predictors of neurological deterioration after intracerebral hemorrhage. Stroke. 2015;46:647–652. doi: 10.1161/STROKEAHA.114.007704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keep RF, Andjelkovic AV, Xiang J, Stamatovic SM, Antonetti DA, Hua Y, Xi G. Brain endothelial cell junctions after cerebral hemorrhage: changes, mechanisms and therapeutic targets. J Cereb Blood Flow Metab. 2018;38:1255–1275. doi: 10.1177/0271678X18774666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morrison DK. MAP kinase pathways. Cold Spring Harb Perspect Biol. 2012;4:a011254. doi: 10.1101/cshperspect.a011254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freund T, Katona I, Piomelli D. Role of endogenous cannabinoids in synaptic signaling. Physiol Rev. 2003;83:1017–1066. doi: 10.1152/physrev.00004.2003. [DOI] [PubMed] [Google Scholar]
- 38.Kano M, Ohno-Shosaku T, Hashimotodani Y, Uchigashima M, Watanabe M. Endocannabinoid-mediated control of synaptic transmission. Physiol Rev. 2009;89:309–380. doi: 10.1152/physrev.00019.2008. [DOI] [PubMed] [Google Scholar]
- 39.Panikashvili D, Simeonidou C, Ben-Shabat S, Hanus L, Breuer A, Mechoulam R, Shohami E. An endogenous cannabinoid (2-AG) is neuroprotective after brain injury. Nature. 2001;413:527–531. doi: 10.1038/35097089. [DOI] [PubMed] [Google Scholar]
- 40.Drew PD, Xu J, Storer PD, Chavis JA, Racke MK. Peroxisome proliferator-activated receptor agonist regulation of glial activation: relevance to CNS inflammatory disorders. Neurochem Int. 2006;49:183–189. doi: 10.1016/j.neuint.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 41.Jian X, He H, Zhu J, Zhang Q, Zheng Z, Liang X, Chen L, Yang M, Peng K, Zhang Z, Liu T, Ye Y, Jiao H, Wang S, Zhou W, Ding Y, Li T. Hsa_circ_001680 affects the proliferation and migration of CRC and mediates its chemoresistance by regulating BMI1 through miR-340. Mol Cancer. 2020;19:20. doi: 10.1186/s12943-020-1134-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin J, Chen A, Qiu W, Chen Y, Li Q, Zhou X, Jin D. Dysregulated circRNA_100876 suppresses proliferation of osteosarcoma cancer cells by targeting microRNA-136. J Cell Biochem. 2019;120:15678–15687. doi: 10.1002/jcb.28837. [DOI] [PubMed] [Google Scholar]
- 43.Shin S, Choi JW, Moon H, Lee CY, Park JH, Lee J, Seo HH, Han G, Lim S, Lee S, Kim SW, Hwang KC. Simultaneous suppression of multiple programmed cell death pathways by miRNA-105 in cardiac ischemic injury. Mol Ther Nucleic Acids. 2019;14:438–449. doi: 10.1016/j.omtn.2018.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barbagallo D, Piro S, Condorelli AG, Mascali LG, Urbano F, Parrinello N, Monello A, Statello L, Ragusa M, Rabuazzo AM, Pietro CD, Purrello F, Michele P. miR-296-3p, miR-298-5p and their downstream networks are causally involved in the higher resistance of mammalian pancreatic α cells to cytokine-induced apoptosis as compared to β cells. BMC Genomics. 2013;14:62. doi: 10.1186/1471-2164-14-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Franca MER, Peixoto CA. cGMP signaling pathway in hepatic encephalopathy neuroinflammation and cognition. Int Immunopharmacol. 2020;79:106082. doi: 10.1016/j.intimp.2019.106082. [DOI] [PubMed] [Google Scholar]
- 46.Gerzanich V, Woo SK, Vennekens R, Tsymbalyuk O, Ivanova S, Ivanov A, Geng Z, Chen Z, Nilius B, Flockerzi V, Freichel M, Simard JM. De novo expression of Trpm4 initiates secondary hemorrhage in spinal cord injury. Nat Med. 2009;15:185–191. doi: 10.1038/nm.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schattling B, Steinbach K, Thies E, Kruse M, Menigoz A, Ufer F, Flockerzi V, Bruck W, Pongs O, Vennekens R, Kneussel M, Freichel M, Merkler D, Friese MA. TRPM4 cation channel mediates axonal and neuronal degeneration in experimental autoimmune encephalomyelitis and multiple sclerosis. Nat Med. 2012;18:1805–1811. doi: 10.1038/nm.3015. [DOI] [PubMed] [Google Scholar]
- 48.Hirota Y, Yamashita S, Kurihara Y, Jin X, Aihara M, Saigusa T, Kang D, Kanki T. Mitophagy is primarily due to alternative autophagy and requires the MAPK1 and MAPK14 signaling pathways. Autophagy. 2015;11:332–343. doi: 10.1080/15548627.2015.1023047. [DOI] [PMC free article] [PubMed] [Google Scholar]