Abstract
Objective: To study the expression of IL13RA2 in gliomas and to analyze its correlation with clinicopathological/molecular features, immune cell infiltration and prognostic significance. Methods: mRNA expression data for IL13RA2 were downloaded and analyzed from two open access datasets (TCGA & CGGA). IL13RA2 protein expression was examined by immunohistochemistry. The association between IL13RA2 and important clinicopathological/molecular markers was examined using χ2 and Spearman correlation tests. The TIMER tool was used to evaluate the correlation of IL13RA2 with multiple intra-tumoral immune cell types in glioma. Kaplan-Meier test and multivariate Cox analyses were applied to evaluate the prognosis. Results: Out of the 297 glioma tissues and 20 normal brain tissues in our cohort, IL13RA2 protein was highly expressed in 115 glioma tissues (115/297, 38.7%), but no expression was detected in normal brain tissues (0/20, 0%). The expression of IL13RA2 was significantly higher in GBMs (P<0.001). More than half of GBMs (68/132, 51.5%) were high expression of IL13RA2 protein, especially GBM patients with IDH wild-type and TERT promoter mutated (60/78, 76.9%). Moreover, 11/13 (84.6%) diffuse midline gliomas and 31/51 (64.7%) IDH wild-type LGGs also highly expressed IL13RA2 in our cohorts. Chi-square test showed that the expression of IL13RA2 was correlated with patient age, WHO grade, Ki67 index, IDH status, TERT promoter status and immune cell infiltration. Additionally, IL13RA2 was strongly associated with patients’ OS and served as a negative prognostic marker in infiltrating gliomas. Conclusion: IL13RA2 was high expression in some glioma subtypes, and significantly correlated with poor prognosis. Based on its role in CAR-T therapy, it might act as an extremely important and specific therapeutic target for human malignant gliomas, especially in IDH wild-type LGG, “IDH wild-type and TERT promoter mutated” GBM and H3K27M-mutated diffuse midline glioma, and improve the clinical outcomes of these patients.
Keywords: IL13RA2, glioma, glioblastoma, prognosis, IDH, TERT, H3K27M
Introduction
Malignant glioma is one of the most aggressive human tumor that arises from the neuroectoderm [1,2]. The multiformity and variability of these glioma tumors are present at the histomorphological and genetic molecular level; moreover, the heterogeneous morphology is also exhibited both between different patients and between different tumor areas [3,4]. The most outstanding characteristic of malignant glioma is its infiltrative and invasive nature, leading to a high recurrence rate and a dismal prognosis. At present, although several kinds of treatment means, such as neurosurgery, radiotherapy, and chemotherapy, have been used for this disease, the treatment of patients with glioma is still a major challenge, and the median survival of GBM patients remains approximately 1 year [5,6]. Thus, new therapy and genetic treatments are urgently needed for patients with GBM [7-9]. IL13RA2, an immunotherapeutic target, is considered a promising direction for the treatment of glioma. Interleukin-13 receptor subunit alpha-2 (IL13RA2) is a high-affinity membrane receptor of IL-13 and is abundantly expressed in a variety of tumors, such as pancreatic cancer [10], ovarian cancer [11], breast cancer [12], colon cancer [13] and melanoma [14]. Recently, several researchers found that the IL-13 receptor could serve as a brand-new target in immunotherapy trials of malignant glioma [15-17]. In glioma cells, IL13RA2 is highly expressed and affects glioma proliferation and invasion, but the mechanism is controversial [18,19]. In addition, its correlation with glioma molecular subtypes, its effects on glioma immunity and tumor microenvironment, and its prognostic significance have yet to be further studied.
Materials and methods
Patients and specimens
Paraffin-embedded tissues were obtained from the archives of the Department of Pathology, Cancer Center, Sun Yat-sen University, Guangzhou, China, between 2008 and 2016; these tissues were from 297 patients with gliomas (WHO II-IV) and 20 patients who received surgery for reasons other than gliomas. Written informed consent was acquired in all cases before this study and the protocols of this research were approved by the Scientific Ethics Committee of the Sun Yat-sen University Cancer Center (SYSUCC). The choice of samples was based on the following factors: the availability of resected tissue, the accuracy of follow-up data, and lack of radiotherapy or chemotherapy before surgery. The final samples included 83 cases of WHO II (oligodendroglioma and astrocytoma), 82 cases of WHO III (anaplastic oligodendroglioma and astrocytoma) and 132 cases of WHO IV (glioblastoma and diffuse midline glioma) with complete molecular information (IDH, 1p/19q, MGMT, TERT and H3K27M). Patients who died of non-glioma causes were also excluded from the research. Overall survival represents the time interval between the date of diagnosis and the date of death or the last known follow-up.
TCGA and CGGA dataset
A total of 1006 glioma cases from the CGGA and TCGA datasets were included in our research. A total of 325 glioma cases with detailed clinical information were obtained from the CGGA database [20] (http://www.cgga.org.cn). The TCGA cohort consisted of 681 patients with detailed clinical information and was downloaded from the public database (https://tcga-data.nci.nih.gov/tcga/tcgaDownload.jsp) and analysed with the GEPIA tool [21] (http://gepia.cancer-pku.cn/).
Oncomine database analysis
IL13RA2 mRNA expression was explored by using the Oncomine database [22] (https://www.oncomine.org/resource/login.html).
TIMER (tumor immune estimation resource) database analysis
The TIMER database is an integrated resource to systematically analyze the abundance of immune infiltrates in diverse cancer types [23] (https://cistrome.shinyapps.io/timer/). We used the “correlation” module plots to evaluate the correlation of IL13RA2 and immune cells including neutrophils, dendritic cells, B cells, CD4+ T cells, CD8+ T cells and macrophages in the LGG and GBM sample sets.
Protein-protein interaction (PPI) network, gene ontology, and pathway analysis
Differentially expressed genes were further analyzed at the protein level using protein-protein interaction network functional enrichment analysis through STRING (Protein-Protein Interaction Network Functional Enrichment Analysis) (https://string-db.org/). Results from STRING were further analyzed using Cytoscape to visualize molecular interaction networks and integrating gene expression profiles, in order to identify clusters of protein interaction that are highly related to IL13RA2. The list of genes generated in the cluster was then analyzed again in DAVID for significant functional annotation enrichment.
Tissue microarray (TMA) construction, immunohistochemistry (IHC) staining of tissue specimens and molecular tests
TMA, IHC, and molecular tests were constructed as the method described previously [24]. A mouse monoclonal human anti-IL13RA2 antibody was used for IHC and WB (Abcam, Cambridge, UK, clone number: ab55275). Details are provided in Supplementary Materials.
Scoring of staining
IL13RA2 IHC scoring criteria were established by combining the positive proportion (1 for 0-25%, 2 for 26%-50%, 3 for 51%-75%,4 for >75%) and staining intensity (0 for no staining,1 for light yellow, 2 for yellowish brown, 3 for brown) of the stained tumor cells. Therefore, we achieved the IL13RA2 expression score (0-12) by positive ratio × staining intensity. ROC curves were used to find the best cut-off values of IL13RA2 with respect to OS. A two-level grade system of IL13RA2 staining was quantified as follows: 0-3 (no/low expression) indicates negative and 4-12 (high expression) indicates positive. All the samples were scored separately by two independent neuropathologists, who were blinded to the patient data, and the final scores were averaged for further comparative evaluation of IL13RA2 expression.
Statistical analysis
The association between IL13RA2 expression and clinicopathological characteristics and variables were assessed using 2 × 2 contingency tables and the chi-square (χ2) test. Kaplan-Meier survival curves were generated to estimate overall survival. Survival differences according to IL13RA2 expression were analyzed by the log-rank test. The influence of variables on survival was assessed using univariate and/or multivariate Cox regression analyses. The TIMER “correlation” module used the Spearman correlation coefficient. All statistical analyses were performed with SPSS (version 16.0, Chicago, USA), and significance was defined as P<0.05.
Results
IL13RA2 is highly expressed in malignant gliomas, particularly in LGG with IDH wild-type, GBM with IDH wild-type & TERT promoter mutation and diffuse midline glioma with H3K27M mutation
Out of all 297 glioma cases and 20 normal brain tissues in our cohort, IL13RA2 protein was highly expressed in 115 glioma cases (115/309, 38.7%), but no expression was found in normal brain tissues (0/20, 0%). Typical IHC figures were presented in Figure 1A-D. High expression of IL13RA2 was correlated with the WHO grade of the tumor (P<0.001), with 21.7% (23/83) in WHO II, 32.9% (26/82) in WHO III and 51.5% (68/132) in WHO IV (Figure 1E). Notably, we found IL13RA2 was significantly expressed in IDH wild-type LGG (31/51, 64.7%), IDH wild-type & TERTp-mutated GBM (60/78, 76.9%) and H3K27M-mutated diffuse midline glioma (11/13, 84.6%) (Figure 1F). The expression of IL13RA2 mRNA was also significantly higher (P<0.001) in glioma than in normal brain tissues and in GBM versus LGG (P<0.001). However, no significant difference was found between the LGG groups and normal brain tissues (Figure 1G).
Figure 1.
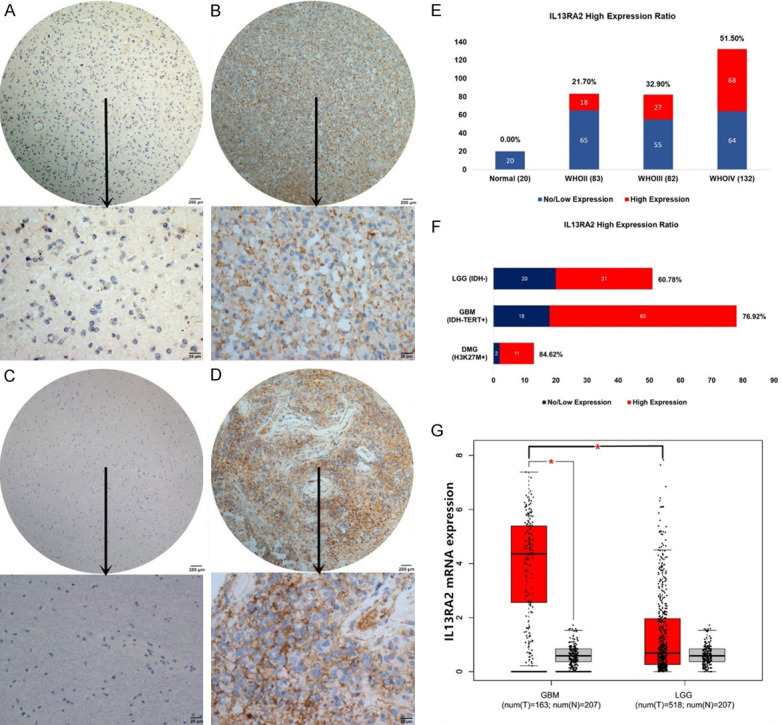
The IL13RA2 protein was primarily expressed in the membrane. Representative sections of IL13RA2 in gliomas and normal brain tissues for (A) a WHO II case of diffuse astrocytoma, (B) a WHO III case of anaplastic astrocytoma, (C) a normal brain sample and (D) a GBM sample. (E) IL13RA2 was positively correlated with histological grades in gliomas. WHO IV patients had the highest IL13RA2 expression ratio of 51.5%. (F) IL13RA2 was significantly highly expressed in IDH wild-type LGG (31/51, 64.7%), IDH wild-type & TERTp mutated GBM (60/78, 76.9%) and diffuse midline glioma with H3K27M mutation (11/13, 84.6%). (G) IL13RA2 mRNA expression in TCGA datasets. IL13RA2 was upregulated in high-grade gliomas (especially in GBM) compared to low-grade gliomas (*P<0.001) and normal brain tissues (*P<0.001).
To further confirm the results, we queried IL13RA2 expression in the Oncomine database. In addition to TCGA datasets, 4 other datasets also showed that IL13RA2 expression was significantly higher in GBM patient tissues than in normal brain tissues (Table 1).
Table 1.
IL13RA2 expression in Oncomine glioma database
| Upregulation of IL13RA2 in glioblastoma | P-value | Database |
|---|---|---|
| Glioblastoma(542) vs Normal(10) | 4.50E-06 | TCGA Brain |
| Glioblastoma(80) vs Normal(4) | 2.09E-09 | Murat Brain |
| Glioblastoma(22) vs Normal(3) | 1.85E-06 | Lee Brain |
| Glioblastoma(81) vs Normal(23) | 0.003 | Sun Brain |
| Glioblastoma(27) vs Normal(7) | 0.011 | Shai Brain |
IL13RA2 correlates with patient age, WHO grade, Ki67 index, IDH, TERT promoter and H3K27M status, and predicts unfavorable prognosis in gliomas
The correlation between IL13RA2 expression and the clinicopathological characteristics of glioma patients is shown in Table 2. High expression of IL13RA2 was significantly associated with patient age (P<0.001), WHO grade (P<0.001), Ki67 index (P<0.001), IDH (P<0.001), TERTp (P<0.001) and H3K27M (P<0.001). No significant correlation of IL13RA2 with sex (P=0.802), tumor location (P=0.081), ATRX status (P=0.067), 1p/19q status (P=0.159), MGMT promoter status (P=0.757) or P53 expression (P=0.617) was observed. IL13RA2 is a significantly unfavorable prognostic marker in all malignant gliomas (WHO grade II-IV) in both TCGA/CGGA and our SYSUCC cohorts. Further stratification analysis revealed that it was a clear indicator of poor prognosis in LGG but inconsistent in GBM (Figure 2). Univariate analysis revealed that old patient age (P<0.005), high WHO grade (P<0.001), high Ki67 index (P<0.001) and high expression of IL13RA2 (P<0.001) were negatively correlated with the prognosis of 297 malignant glioma patients (WHO II-IV), while IDH mutation (P<0.001) and 1p/19q codeletion (P=0.002) were positively correlated with prognosis. Multivariate Cox analysis confirmed that IL13RA2 expression was an independent risk factor in all malignant glioma patients, with an adjusted HR of 1.528 (95% CI: 1.054-2.216, P=0.025, Table 3).
Table 2.
Clinical-pathological Characteristics of the patients and IL13RA2 expression
| No. of cases | IL13RA2 | P | ||
|---|---|---|---|---|
|
| ||||
| Low | High | |||
| Age | <0.001* | |||
| ≤45 | 178 | 128 | 50 | |
| >45 | 131 | 65 | 66 | |
| Sex | 0.802 | |||
| Male | 181 | 112 | 69 | |
| Female | 128 | 81 | 47 | |
| WHO grade | <0.001* | |||
| Low (I+II) | 95 | 74 | 21 | |
| High (III+IV) | 214 | 119 | 95 | |
| Location | 0.081 | |||
| Supratentorial | 292 | 179 | 113 | |
| Subtentorial | 17 | 14 | 3 | |
| IDH | <0.001* | |||
| Wildtype | 177 | 87 | 90 | |
| Mutated | 132 | 106 | 26 | |
| ATRX | 0.067 | |||
| Wildtype | 182 | 106 | 76 | |
| Mutated | 127 | 87 | 40 | |
| 1p/19q | 0.159 | |||
| Co-deleted | 46 | 33 | 13 | |
| Intact | 263 | 160 | 103 | |
| TERT | <0.001* | |||
| Wildtype | 184 | 130 | 54 | |
| Mutated | 125 | 63 | 62 | |
| MGMT | 0.757 | |||
| Promoter methylated | 151 | 93 | 58 | |
| Promoter Unmethylated | 158 | 100 | 58 | |
| H3K27M | <0.001* | |||
| Mutated | 13 | 2 | 11 | |
| Wildtype | 296 | 191 | 105 | |
| P53 | 0.617 | |||
| ≤10% | 112 | 72 | 40 | |
| >10% | 197 | 121 | 76 | |
| Ki67 | <0.001* | |||
| ≤10% | 74 | 59 | 15 | |
| >10% | 235 | 134 | 101 | |
P<0.05.
Figure 2.
The survival curves of overall survival in different datasets based on the expression level of IL13RA2. Patients with high IL13RA2 expression had significantly shorter OS than those with low IL13RA2 expression in all gliomas (A-C) and LGG (D-F) (P<0.001) in TCGA, CGGA and SYSUCC cohort. But only CGGA dataset (H) showed prognostic significance in GBM (P=0.003). Both TCGA and our SYSUCC GBM dataset (G, I) had no statistical significance (P=0.16 and 0.419). TPM = Transcripts Per Kilobase Million. TCGA = The Cancer Genome Atlas, CGGA = Chinese Glioma Genome Atlas, SYSUCC = Sun Yat-sen University Cancer Center.
Table 3.
Univariate and Multivariate analysis for overall survivals
| Variable | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
|
|
|
|||
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | |
| Sex: (Male) | 1.144 (0.821~1.595) | 0.425 | ||
| Age: (years >45) | 1.579 (1.145~2.178) | 0.005* | 1.010 (0.687-1.485) | 0.959 |
| Location: (Supratentorial) | 1.218 (0.597~2.486) | 0.587 | ||
| WHO Grade: (High) | 2.617 (1.733~3.953) | <0.001* | 1.472 (0.754-2.874) | 0.258 |
| IDH1/2: (Mutated) | 0.362 (0.247~0.529) | <0.001* | 0.609 (0.385-0.961) | 0.033* |
| 1p/19q: (Co-deleted) | 0.319 (0.156~0.653) | 0.002* | 0.427 (0.200-0.915) | 0.029* |
| ATRX: (Mutated) | 0.736 (0.531~1.020) | 0.065 | ||
| TERTp: (Mutated) | 0.965 (0.688~1.355) | 0.839 | ||
| MGMTp: (Methylated) | 1.010 (0.616~1.657) | 0.967 | ||
| P53: (>10%) | 1.249 (0.884~1.764) | 0.208 | ||
| Ki67: (>10%) | 2.292 (1.444~3.638) | <0.001* | 1.470 (0.700-3.089) | 0.309 |
| IL13RA2: (High) | 2.036 (1.464~2.832) | <0.001* | 1.528 (1.054-2.216) | 0.025* |
P<0.05.
IL13RA2 expression is correlated with immune cell infiltration in glioma
We used the TIMER [23] tool to evaluate the correlation of IL13RA2 and multiple intra-tumoral immune cell types in gliomas. This analysis included the 6 most common immune cell types (B cells, CD4+ T cells, CD8+ T cells, macrophages, neutrophils and dendritic cells). Although a weak correlation was found, the IL13RA2 expression level was positively correlated with the levels of infiltrating macrophages (R=0.108, P=0.0273), neutrophils (R=0.157, P=0.00125) and dendritic cells (R=0.104, P=0.0334) in GBM. Similarly, positive correlations were also found in the levels of infiltrating macrophages (R=0.103, P=0.0255), neutrophils (R=0.164, P=0.000337) and dendritic cells (R=0.149, P=0.00108) in LGG. In addition, B cells (R=0.123, P=0.00702) and CD8+ T cells (R=0.325, P<0.001) were also positively correlated with IL13RA2 in LGG (Figure 3B). Additionally, we also found significant correlations of IL13RA2 with 28 types of TILs across heterogeneous human cancers, including GBM and LGG (Figure 3A). These findings suggested that IL13RA2 might a specific role in immune infiltration in glioma, especially in macrophages, neutrophils and dendritic cells. The same results were found in TCGA by GEPIA correlation analysis (Figure 4).
Figure 3.
A. Relationships between the expression of IL13RA2 and 28 types of TILs across human heterogeneous cancers. IL13RA2 is significantly correlated with abundant TILs in both GBM and LGG. B. Scatterplot showing a significant positive correlation between immune score and IL13RA2 expression in TCGA data sets. A significant positive correlation was found between the infiltration of macrophages, neutrophils, dendritic cells and IL13RA2 expression in GBM. The same results were also found in LGG. In addition to macrophages, neutrophils and dendritic cells, IL13RA2 was also positively correlated with B cells and CD8+ T cells in LGG. The calculated tumour purity was negatively correlated with IL13RA2 expression in LGG, but not in GBM. CD4+ T cells were not correlated with IL13RA2 expression in either GBM or LGG.
Figure 4.

B cell markers (CD19, CD22 and PAX5), CD4+ T cell markers (CD4), CD8+ T cell maker (CD8), Macrophage markers (ITGAM/CD11b, CD68 and CD163), Neutrophil markers (FUT4/CD15) and Dendritic cell markers (CD1a, CD83 and ITGAE/CD103) were selected in the GEPIA TCGA glioma database. IL13RA2 mRNA expression level is positively correlated with macrophage [(A) ITGAM/CD11b (R=0.21, P<0.001), (B) CD68 (R=0.19, P<0.001), (C) CD163 (R=0.21, P<0.001)]; CD4+ T cell [(D) CD4 (R=0.21, P<0.001)]; CD8+ T cell [(E) CD8A (R=0.24, P<0.001)]; neutrophil [(F) FUT4/CD15 (R=0.26, P<0.001)] and dendritic cell [(G) CD1A (R=0.079, P=0.04), (H) CD83 (R=0.15, P<0.001), (I) ITGAE/CD103 (R=0.31, P<0.001)] infiltration, but no correlation was found between B cell markers [(J) CD19 (P=0.16), (K) CD79A (P=0.43), (L) PAX5 (P=0.39)] and IL13RA2 in the GEPIA TCGA glioma data sets. TPM = Transcripts Per Kilobase Million.
Identification of IL13RA2 differentially expressed genes and functional enrichment analysis
325 CGGA glioma cases and 681 TCGA glioma cases were divided into IL13RA2 Low and IL13RA2 High groups based on the median IL13RA2 expression value, respectively. Then the DESeq R package was used to screen the Differentially Expressed Genes (DEGs), cut-off criteria was set as |logFC| >1, and the p-value <0.05. A total of 1577 DEGs were obtained in TCGA, including 1209 upregulated genes and 368 downregulated genes (Figure 5A). A total of 1649 DEGs were obtained in CGGA, including 897 upregulated genes and 752 downregulated genes (Figure 5B). The intersection of these two arrays was shown in Figure 5C, and 818 overlapping genes were found. The 818 genes were further analyzed using STRING and Cytoscape MCODE for their protein-protein interactions, and obtained 147 hub genes (Figure 5D). To explore the biological processes and pathways in which the hub genes were involved, we uploaded the hub gene lists into the DAVID software, and obtained the results. A p-value <0.05 was considered significant. Biological-process-enrichment analysis suggested that overlapping genes were significantly enriched such as inflammatory response, immune response, regulation of cell growth and angiogenesis (Figure 5E). As for KEGG pathways, the overlapping genes were enriched such as the Jak-STAT signaling pathway, Toll-like receptor signaling pathway, ECM-receptor interaction, PI3K-Akt signaling pathway and cytokine-cytokine receptor interaction (Figure 5F).
Figure 5.
(A, B) The results of Volcano Plot of differentially expressed genes (DEGs) in TCGA (A) and CGGA (B). The vertical lines demark the fold-change values. The right vertical line corresponds to log2FC>1 change, while the left vertical line corresponds to log2FC<-1 change. The horizontal line marks a -log10 p-value of 0.05. (C) 818 overlapping DEGs were identified in both TCGA and CGGA. (D) Protein-protein interaction (PPI) network analysis by means of running Cytoscape software via the STRING database. The network constructed from 147 genes with combined score >0.4 out of 818 overlapping genes. (E) The top 12 significantly enriched biological processes were ranked including regulation of cell growth and angiogenesis. (F) The top 12 significantly enriched KEGG pathways are shown including Jak-STAT, Toll-like receptor and PI3K-AKT signaling pathway. A p-value <0.05 was regarded as significant.
Discussion
With the development of immunotherapy, increasing attention has been paid to the immune status and biomarkers in glioma. As an extremely malignant tumor of the central nervous system, conventional therapy does not significantly prolong patient survival due to the aggressive nature of glioma, particularly in GBM. Immunotherapy is considered to be the fifth most common cancer treatment technology after surgery, radiotherapy, chemotherapy and targeted therapy, and the use of immunotherapy has contributed to an increasing number of breakthroughs in clinical research for a variety of cancers. However, for a long time, due to the lack of immune organs in the central nervous system and the presence of the blood-brain barrier, the brain is considered to be one of the “immunity-privileged” organs in the body, making glioma a notoriously capable immune evader of solid tumors, suggesting that immunotherapy for glioma must confront many challenges [25,26]. However, breakthrough report was published in 2016; Brown et al. [27] succeeded in eliminating the tumor foci of a recurrent multifocal GBM patient for 7.5 months by using CAR-T cells targeting IL13RA2. This was the first case around the world to confirm that CAR-T technology may be effective against GBM. Moreover, this research showed that the IL13RA2 target is worthy of attention. IL13RA2, a new and potential immunotherapy target, is a membrane-bound protein that is encoded by the IL13RA2 gene in humans.
In this study, we revealed that the expression of IL13RA2 significantly increased in GBM compared to LGG and normal brain tissues, indicating its oncogenic role. In addition, we found that IL13RA2 was highly expressed in IDH wild-type and TERT promoter mutated GBM, diffuse midline glioma and in a considerable proportion of IDH wild-type LGG (WHO II and III) gliomas, suggesting that these patients could benefit from CAR-T therapy targeting IL13RA2 with high precision and specificity.
IDH mutation, 1p/19q codeletion, ATRX mutation, TERT promoter mutation and MGMT promoter methylation are the five key molecular markers in the routine assessment of gliomas due to their diagnostic and therapeutic value at present. Jing Han et al. [28] found no correlation between IL13RA2 mRNA expression and previously reported biomarkers of GBM subtypes such as IDH1, EGFR, MGMT, and PDGFRA. However, the association between important molecular biomarkers and IL13RA2 protein expression in gliomas has not been studied before. In contrast to Jing Han’s study, we found that IL13RA2 was correlated with IDH status and TERT promoter status, and IL13RA2 protein was highly expressed in IDH wild-type LGG and IDH wild-type & TERTp-mutated GBM cases, suggesting its important role not only in immunotherapy, but also in the development and progression of glioma. The functional enrichment analysis in TCGA and CGGA databases also confirmed this hypothesis, for the biological process analysis found IL13RA2 involved in regulation of cell growth and angiogenesis and KEGG pathway analysis found IL13RA2 might promote the progression of glioma through PI3K-AKT signaling pathway. Further experimental studies are still needed to confirm our results.
Tumor infiltrating immune cells are part of the complex microenvironment of tumors and promote and regulate tumor pathogenesis and development. Depending on the type of cells and their functional interactions, immune cells might play an important role in tumors [29]. However, the general role of IL13RA2 and local immune response status in glioma remains unknown. Our results demonstrated that the infiltration of some immune cells was correlated with IL13RA2 expression and that there was a positive relationship between the IL13RA2 expression level and the infiltration level of macrophages, neutrophils and dendritic cells in both GBM and LGG. Several studies [28,30-32] exhibit possible mechanisms that explain why IL13RA2 expression correlates with immune infiltration. IL13RA2 is supposed to be an inhibitory subunit of the type II receptor and acts as a “decoy” receptor for IL-13 and IL-4 which are secreted by Th2-polarized T cells, granulocytes and monocytes/macrophages. These cytokines play a key immune balancing role in the brain and efficiently induce STAT6 activation in normal tissues. However, GBMs with high IL13RA2 expression fail to respond to either IL-4 or IL-13. This might explain why IL13RA2 has an impact on immune cell infiltration. Fully understanding the role of IL13RA2 in systemic immunity is helpful for improving treatment, monitoring the negative effects of treatment and improving the prognosis of patients.
Regarding the prognostic significance of IL13RA2 in gliomas, IL13RA2 was found to be closely related to prognosis, especially in LGG, with high expression indicating poor prognosis in our study. A possible explanation is that IL13RA2 is mainly expressed in LGG with wild-type IDH, and the biological behavior of this subtype was similar to that of GBM. In the multivariate Cox analysis, IL13RA2 was an independent prognostic factor, accompanied by IDH and 1p/19q status, also indicating its important prognostic significance. However, in the stratification analysis, we did not find a significant prognostic difference in GBM in either the TCGA or our own cohort, but only found a difference in the CGGA datasets. Jing Han et al. [28] also analyzed the TCGA database but they identified an inverse relationship between IL13RA2 expression and overall survival and potential temozolomide resistance in GBM. However, despite P value <0.05, K-M survival curves were not markedly different, with a median survival time of 335 days in IL13RA2 high expression patients compared to 384 days in IL13RA2-negative patients. It is possible that the sample size is too small, and further large sample confirmation is required.
Another phenomenon worth noting is that 11/13 diffuse midline gliomas, the most fatal glioma type with H3K27M mutation (WHO IV), have high IL13RA2 expression in our cohort. Deng, H et al. [33] also found the same result in a cell model; they performed microarray analysis in a midline glioma cell model harboring H3K27M, which caused the upregulation of IL13RA2. These results strongly indicate that IL13RA2 may be a potential therapeutic target in diffuse midline glioma. However, due to the limited cases in our study, further studies will be needed to confirm the prediction in larger cohorts.
In conclusion, IL13RA2 is overexpressed in malignant gliomas and serves as an important poor prognostic marker in predicting overall survival independent of the traditional clinicopathological characteristics in malignant glioma. Based on its role in CAR-T therapy, it may act as an extremely important and specific therapeutic target for human malignant gliomas, especially in IDH wild-type LGG, “IDH wild-type and TERT promoter-mutated” GBM and H3K27M-mutated diffuse midline glioma, and improve the clinical outcomes of these patients.
Disclosure of conflict of interest
None.
Abbreviations
- GBM
Glioblastoma
- LGG
lower grade glioma (WHO II&III)
- IL13RA2
Interleukin-13 receptor subunit alpha-2
- TCGA
The Cancer Genome Atlas
- CGGA
Chinese Glioma Genome Atlas
- SYSUCC
Sun Yat-sen University cancer center
- IL-4
Interleukin 4
- IL-13
Interleukin 13
- IHC
Immunohistochemistry
- TMA
Tissue microarray
- TPM
Transcripts Per Kilobase Million
- DEGs
Differentially Expressed Genes
Supporting Information
References
- 1.Jungk C, Chatziaslanidou D, Ahmadi R, Capper D, Bermejo J, Exner J, Deimling A, Herold-Mende C, Unterberg A. Chemotherapy with BCNU in recurrent glioma: analysis of clinical outcome and side effects in chemotherapy-naive patients. BMC Cancer. 2016;16:81. doi: 10.1186/s12885-016-2131-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu C, Lin G, Lin Z, Zhang J, Chen L, Liu S, Tang W, Qiu X, Zhou C. Peritumoral edema on magnetic resonance imaging predicts a poor clinical outcome in malignant glioma. Oncol Lett. 2015;10:2769–2776. doi: 10.3892/ol.2015.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Natsume A, Kinjo S, Yuki K, Kato T, Ohno M, Motomura K, Iwami K, Wakabayashi T. Glioma-initiating cells and molecular pathology: implications for therapy. Brain Tumor Pathol. 2011;28:1–12. doi: 10.1007/s10014-010-0011-3. [DOI] [PubMed] [Google Scholar]
- 4.McLendon R, Rich J. Glioblastoma stem cells: a neuropathologist’s view. J Oncol. 2011;2011:397195. doi: 10.1155/2011/397195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li B, Meng C, Zhang X, Cong D, Gao X, Gao W, Ju D, Hu S. Effect of photodynamic therapy combined with torasemide on the expression of matrix metalloproteinase 2 and sodium-potassium-chloride cotransporter 1 in rat peritumoral edema and glioma. Oncol Lett. 2016;11:2084–2090. doi: 10.3892/ol.2016.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lou M, Zhao Y. Satisfactory therapy results of combining nimustine with nicardipine against glioma at advanced stage. J Cancer Res Ther. 2015;11:1030. doi: 10.4103/0973-1482.154033. [DOI] [PubMed] [Google Scholar]
- 7.Chaudhuri S, Singh M, Bhattacharya D, Acharya S, Chatterjee S, Kumar P, Bhattacharjee P, Basu A, Sa G, Das T, Ghosh T, Chaudhuri S. The novel immunotherapeutic molecule T11TS modulates glioma-induced changes of key components of the immunological synapse in favor of T cell activation and glioma abrogation. J Neurooncol. 2014;120:19–31. doi: 10.1007/s11060-014-1528-9. [DOI] [PubMed] [Google Scholar]
- 8.Eid R, Demattei M, Episkopou H, Auge-Gouillou C, Decottignies A, Grandin N, Charbonneau M. Genetic Inactivation of ATRX leads to a decrease in the amount of telomeric cohesin and level of telomere transcription in human glioma cells. Mol Cell Biol. 2015;35:2818–2830. doi: 10.1128/MCB.01317-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Claus E, Walsh K, Wiencke J, Molinaro A, Wiemels J, Schildkraut J, Bondy M, Berger M, Jenkins R, Wrensch M. Survival and low-grade glioma: the emergence of genetic information. Neurosurg Focus. 2015;38:E6. doi: 10.3171/2014.10.FOCUS12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujisawa T, Joshi B, Nakajima A, Puri R. A novel role of interleukin-13 receptor alpha2 in pancreatic cancer invasion and metastasis. Cancer Res. 2009;69:8678–8685. doi: 10.1158/0008-5472.CAN-09-2100. [DOI] [PubMed] [Google Scholar]
- 11.Kioi M, Kawakami M, Shimamura T, Husain S, Puri R. Interleukin-13 receptor alpha2 chain: a potential biomarker and molecular target for ovarian cancer therapy. Cancer. 2006;107:1407–1418. doi: 10.1002/cncr.22134. [DOI] [PubMed] [Google Scholar]
- 12.Kawakami K, Kawakami M, Puri R. Specifically targeted killing of interleukin-13 (IL-13) receptor-expressing breast cancer by IL-13 fusion cytotoxin in animal model of human disease. Mol Cancer Ther. 2004;3:137–147. [PubMed] [Google Scholar]
- 13.Valverde A, Povedano E, Montiel V, Yanez-Sedeno P, Garranzo-Asensio M, Barderas R, Campuzano S, Pingarron J. Electrochemical immunosensor for IL-13 Receptor alpha2 determination and discrimination of metastatic colon cancer cells. Biosens Bioelectron. 2018;117:766–772. doi: 10.1016/j.bios.2018.07.017. [DOI] [PubMed] [Google Scholar]
- 14.Okamoto H, Yoshimatsu Y, Tomizawa T, Kunita A, Takayama R, Morikawa T, Komura D, Takahashi K, Oshima T, Sato M, Komai M, Podyma-Inoue K, Uchida H, Hamada H, Fujiu K, Ishikawa S, Fukayama M, Fukuhara T, Watabe T. Interleukin-13 receptor alpha2 is a novel marker and potential therapeutic target for human melanoma. Sci Rep. 2019;9:1281. doi: 10.1038/s41598-019-39018-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Husain S, Puri R. Interleukin-13 receptor-directed cytotoxin for malignant glioma therapy: from bench to bedside. J Neurooncol. 2003;65:37–48. doi: 10.1023/a:1026242432647. [DOI] [PubMed] [Google Scholar]
- 16.Kunwar S, Prados M, Chang S, Berger M, Lang F, Piepmeier J, Sampson J, Ram Z, Gutin P, Gibbons R, Aldape K, Croteau D, Sherman J, Puri R. Direct intracerebral delivery of cintredekin besudotox (IL13-PE38QQR) in recurrent malignant glioma: a report by the Cintredekin Besudotox Intraparenchymal Study Group. J. Clin. Oncol. 2007;25:837–844. doi: 10.1200/JCO.2006.08.1117. [DOI] [PubMed] [Google Scholar]
- 17.Kioi M, Husain S, Croteau D, Kunwar S, Puri R. Convection-enhanced delivery of interleukin-13 receptor-directed cytotoxin for malignant glioma therapy. Technol Cancer Res Treat. 2006;5:239–250. doi: 10.1177/153303460600500307. [DOI] [PubMed] [Google Scholar]
- 18.Tu M, Wange W, Cai L, Zhu P, Gao Z, Zheng W. IL-13 receptor alpha2 stimulates human glioma cell growth and metastasis through the Src/PI3K/Akt/mTOR signaling pathway. Tumour Biol. 2016;37:14701–14709. doi: 10.1007/s13277-016-5346-x. [DOI] [PubMed] [Google Scholar]
- 19.Brown C, Warden C, Starr R, Deng X, Badie B, Yuan Y, Forman S, Barish M. Glioma IL13Ralpha2 is associated with mesenchymal signature gene expression and poor patient prognosis. PLoS One. 2013;8:e77769. doi: 10.1371/journal.pone.0077769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bao Z, Chen H, Yang M, Zhang C, Yu K, Ye W, Hu B, Yan W, Zhang W, Akers J, Ramakrishnan V, Li J, Carter B, Liu Y, Hu H, Wang Z, Li M, Yao K, Qiu X, Kang C, You Y, Fan X, Song W, Li R, Su X, Chen C, Jiang T. RNA-seq of 272 gliomas revealed a novel, recurrent PTPRZ1-MET fusion transcript in secondary glioblastomas. Genome Res. 2014;24:1765–1773. doi: 10.1101/gr.165126.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhodes D, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs B, Barrette T, Anstet M, Kincead-Beal C, Kulkarni P, Varambally S, Ghosh D, Chinnaiyan A. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li T, Fan J, Wang B, Traugh N, Chen Q, Liu J, Li B, Liu X. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu W, Yang Y, Xi S, Sai K, Su D, Zhang X, Lin S, Zeng J. Expression of CPEB4 in human glioma and its correlations with prognosis. Medicine (Baltimore) 2015;94:e979. doi: 10.1097/MD.0000000000000979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engelhardt B, Vajkoczy P, Weller R. The movers and shapers in immune privilege of the CNS. Nat Immunol. 2017;18:123–131. doi: 10.1038/ni.3666. [DOI] [PubMed] [Google Scholar]
- 26.MEDAWAR PB. Immunity to homologous grafted skin; the fate of skin homografts transplanted to the brain, to subcutaneous tissue, and to the anterior chamber of the eye. Br J Exp Pathol. 1948;29:58–69. [PMC free article] [PubMed] [Google Scholar]
- 27.Brown C, Alizadeh D, Starr R, Weng L, Wagner J, Naranjo A, Ostberg J, Blanchard M, Kilpatrick J, Simpson J, Kurien A, Priceman S, Wang X, Harshbarger T, D’Apuzzo M, Ressler J, Jensen M, Barish M, Chen M, Portnow J, Forman S, Badie B. Regression of glioblastoma after chimeric antigen receptor T-Cell therapy. N Engl J Med. 2016;375:2561–2569. doi: 10.1056/NEJMoa1610497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han J, Puri R. Analysis of the cancer genome atlas (TCGA) database identifies an inverse relationship between interleukin-13 receptor alpha1 and alpha2 gene expression and poor prognosis and drug resistance in subjects with glioblastoma multiforme. J Neurooncol. 2018;136:463–474. doi: 10.1007/s11060-017-2680-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Domingues P, Gonzalez-Tablas M, Otero A, Pascual D, Miranda D, Ruiz L, Sousa P, Ciudad J, Goncalves J, Lopes M, Orfaoand A, Tabernero M. Tumor infiltrating immune cells in gliomas and meningiomas. Brain Behav Immun. 2016;53:1–15. doi: 10.1016/j.bbi.2015.07.019. [DOI] [PubMed] [Google Scholar]
- 30.McCormick SM, Heller NM. Commentary: IL-4 and IL-13 receptors and signaling. Cytokine. 2015;75:38–50. doi: 10.1016/j.cyto.2015.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rahaman S, Sharma P, Harbor P, Aman M, Vogelbaum M, Haque S. IL-13R(alpha)2, a decoy receptor for IL-13 acts as an inhibitor of IL-4-dependent signal transduction in glioblastoma cells. Cancer Res. 2002;62:1103–1109. [PubMed] [Google Scholar]
- 32.Suzuki A, Leland P, Joshiand B, Puri R. Targeting of IL-4 and IL-13 receptors for cancer therapy. Cytokine. 2015;75:79–88. doi: 10.1016/j.cyto.2015.05.026. [DOI] [PubMed] [Google Scholar]
- 33.Deng H, Zeng J, Zhang T, Gong L, Zhang H, Cheung E, Jones C, Li G. Histone H3.3K27M Mobilizes Multiple Cancer/Testis (CT) antigens in pediatric glioma. Mol Cancer Res. 2018;16:623–633. doi: 10.1158/1541-7786.MCR-17-0460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.





