Abstract
Circular RNAs (circRNAs) are a class of non-coding RNAs could affect expression of specific genes which may induce tumor occurrence and progression. In this study, we identified 32 differentially expressed circRNAs between five pairs of lung adenocarcinoma and paracancerous tissues using circRNA microarray. And circ_0007142 expression was the most upregulated in five lung adenocarcinoma tissues. Meanwhile, circ_0007142 expression was remarkably over-expressed in lung adenocarcinoma tissues and cells. In addition, knockdown of circ_0007142 inhibited proliferation, migration, invasion and induced apoptosis of lung adenocarcinoma cells. Furthermore, knockdown of circ_0007142 inhibited the biological behavior of lung adenocarcinoma through miR-186/FOXK1 axis and inactivated the Wnt/β-catenin signaling pathway. Altogether, our study suggests Circ_0007142/miR-186/FOXK1 axis may play as an important role in progression of lung adenocarcinoma, which provided a novel potential mechanism about this disease.
Keywords: Lung adenocarcinoma, circ_0007142, miR-186, FOXK1, Wnt/β-catenin signaling pathway
Introduction
Lung cancer is one of the most prevalent malignancies and the 5-year survival rate is only approximately 20% [1]. Lung cancers usually are grouped into two main types, namely, small cell lung cancer and non-small cell lung cancer. Lung adenocarcinoma is known as the most common histological subtypes and accounts for about 80% of non-small cell lung cancer [2]. Despite great advances in lung cancer diagnostic and therapeutic strategies, the clinical prognosis of patients remain unsatisfactory due to its recurrence and metastasis [3-5]. Thus, a detailed understanding of the molecular mechanisms underlying lung adenocarcinoma progression is essential.
As a class of non-coding RNAs, circular RNAs (circRNAs) have a covalently closed loop structure with neither a 5’ cap nor a 3’ polyadenylated tail [6]. Recent studies indicate circRNAs modulate diverse biological processes, including “miRNA sponge”, transcriptional regulation, protein-binding and translation [7,8]. However, aberrant circRNAs levels could affect expression of specific genes which may induce tumor occurrence or progression [9,10]. For instance, high circ_0023404 expression was correlated with poor prognosis of patients with lung cancer [11]. Circ-RAD23B could facilitate cell growth and metastasis in non-samll cell lung cancer [12]. Hence, exploring the functions of circRNAs in lung cancer development may lead to new strategies for lung cancer therapies.
MicroRNAs are short endogenous non-coding, single stranded RNAs (around 19-23 nucleotides in length) that could regulate gene expression by binding to the 3’-untranslated regions (3’-UTRs) of target mRNAs [13]. Competing endogenous RNA (ceRNA) hypothesis demonstrated that miRNAs sponging function of circRNAs [7,14]. However, the molecular function of circ_0007142 has yet to be fully explored. In the current study, we aimed to investigate the expression level and biological function of circ_0007142 in lung adenocarcinoma, and further study its potential mechanism.
Materials and methods
Participants and clinical specimens
Seventy clinical cancer tissues and paired adjacent normal tissues were acquired from patients who had received surgical resection between January 2016 and December 2017 in Peking Union Medical College Hospital. The patients had not yet received preoperative radiotherapy or chemotherapy. And the characteristics of participants enrolled in the study are shown in Table 1. All the specimens were immediately snap-frozen after isolation for further analysis. The study was approved by Peking Union Medical College Hospital, written informed consent was obtained from all participants.
Table 1.
The correlation of circ_0007142 expression with clinical parameters in patients with lung adenocarcinoma
| Variables | Clinical parameters | circ_0007142 expression | P value | |
|---|---|---|---|---|
|
| ||||
| High (n=38) | Low (n=32) | |||
| Gender | Male | 17 | 13 | 0.576 |
| Female | 21 | 21 | ||
| Age (years) | <65 | 18 | 14 | 0.762 |
| ≥65 | 20 | 18 | ||
| Size (cm) | <5 | 21 | 17 | 0.858 |
| ≥5 | 17 | 15 | ||
| Differentiation | Well/Moderate | 21 | 14 | 0.337 |
| Poor | 17 | 18 | ||
| TNM stage | I/II | 21 | 10 | 0.044* |
| III | 17 | 22 | ||
| Lymphatic metastasis | Yes | 26 | 13 | 0.020* |
| No | 12 | 19 | ||
P<0.05.
Circular RNA microarrays
The circular RNAs were enriched from total RNA of samples, amplified, transcribed into fluorescent cRNA, and then hybridized onto the Arraystar circRNA Array (8 × 15 K, Arraystar) (Rockville, MD, USA). After washing, the arrays were scanned by the Agilent Scanner G2505C. Agilent Feature Extraction software (version 11.0.1.1) was used to analyze acquired array images. A heat map of differentially expressed circRNAs/genes in cancer tissues and matched normal tissues was then constructed.
Cell culture and transfection
The human lung adenocarcinoma cell lines (HCC827, A549, H125, and NCI-23) and one human normal lung epithelial cell line BEAS-2B were purchased from the Chinese Academy of Sciences Cell Bank (Shanghai, China). Cells were cultured in dulbecco’s modified eagle medium (DMEM, Gibco, USA) supplemented with 10% fetal bovine serum (FBS; Solarbio) and 1% penicillin/streptomycin (Solarbio) at 37°C in a humidified atmosphere containing 5% CO2. Circ_0007142 small interfering RNA (siRNA) against circ_0007142, miR-186 mimics, miR-186 inhibitors and negative controls were all designed and synthesized by GenePharma (Shanghai, China). For cell transfection, A549 and H125 cells were seeded in six-well plates at a density of 5 × 105/well and cultured to a confluency of 50-60%. Then circ_0007142 expression vector (Invitrogen, CA, USA), miR-186 mimics, miR-186 inhibitors and circ_0007142 small interfering RNA (Gene-Pharma, Shanghai, China) were, respectively, alone or in combination transfected into A549 and H125 cells using Lipofectamine 2000 (Invitrogen, MA, USA) in accordance with manufacturer’s protocol. At 48 h after transfection, cells were harvested for RT-PCR analysis or western blotting.
CCK-8 cell proliferation assay
2 × 103 cells A549 and H125 cells were planted in 96-well plate and cultured in a humid atmosphere with 5% CO2 at 37°C for 96 h. Next, 10 μl CCK-8 solution (Beyotime, Shanghai, China) was added into each well for 3 hours incubation. The absorbance of each well at 490 nm was measured at 24 h, 48 h, 72 h and 96 h using FLx800 Fluorescence Microplate Reader (Biotek, USA).
Cell apoptosis assay
Cultured cells were harvested after transient transfection for 24 hours and washed with PBS, followed by cell resuspension in binding buffer. And then cells were fixed in ice-cold 70% ethanol, stained with Annexin V-FITC and propidium iodide (PI). Cell apoptosis was then detected by flow cytometry (FCM) FACS Calibur (BD Biosciences, CA, USA) and analyzed by software FACS Diva.
Scratch assay
Cells were collected and resuspended in RPMI-1640 medium. Each well of a six-well plate was seeded with 5 × 105 cells and cultured for 24 h to 100% confluence. The cells were scratched with the head of a 200 μl tip and washed with serum-free medium. These cells were further cultured for 24 h in serum-free medium. After that, serum-free medium were replaced with RPMI-1640 medium containing 3% FBS and continued to culture for 72 h; these cells were then photographed for analysis.
Cell migration and invasion assays
For cell migration, cells were resuspended in serum-free medium and added into the top chamber. Medium containing 10% FBS was added to the lower chamber. For cell invasion, 100 μl matrigel (BD, USA) was firstly added onto the bottom chamber. After 24 hours of incubation, cells on the upper surface of the transwell membrane were gently removed using a cotton swab, and cells on the lower surface of the transwell membrane were fixed in 4% paraformaldehyde, stained with 0.1% crystal violet, and counted under a microscope at 20 × magnification in random fields in each well.
Colony formation assay
Cells were plated into 6-well plates at a density of 1000 each well and cultured for two weeks. And then cells were washed with PBS three times, fixed with 4% paraformaldehyde, and stained with methyl violet. The number of colonies containing ≥50 cells was counted under an inverted microscope.
Real time quantitative PCR analysis
Total RNAs were extracted from tissues and cells using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer’s protocol. The concentrations of total RNAs were measured by the NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). For miRNA analysis, total RNAs were converted into cDNAs using a First-Strand cDNA Synthesis Kit (Takara, Dalian, China), followed by RT-qPCR analysis using SYBR Premix Ex TaqTM kit (Takara). For the quantification of circ_0007142, FOXK1, β-catenin, c-myc and cyclinD1, total RNAs were used to measure the expression by One Step TB Green™ PrimeScript™ RT-PCR Kit (Takara). All primers were designed and synthesized by Shanghai GenePharma Co., Ltd. The primers were as follows: circ_0007142 forward, 5’ GAACTCTGCCTCAGGATGAA 3’ and reverse, 5’ AACGTGTAACCTCGGTACCA 3’; miR186 forward, 5’ ACGAGGACGACAGAC 3’ and reverse, 5’ CTTGGGGATAGGTCATTGGGGG GT 3’; sicirc_0007142 5’ GATGGCCTCCTGGCGGT TA 3’; siNC 5’ UGGGGGG ACAACAUGGGGGGGCUCU 3’; miR186 mimic: 5’ ACGAGGUCUGUCUGAC 3’; miRNC 5’ AGUGCGUAUACGAGCUGU 3’; FOXK1 forward, 5’ ATGGGAA TACCCTGGGCCGGGGTGT 3’ and reverse, 5’ GGGACATCATGGGTCAGGGT 3’; U6 forward, 5’ CTCGCTTCGGCAGCACA 3’ and reverse, 5’ AACGCTTCACGAATTTGCGT 3’; GAPDH forward, 5’ GGGAGCCAAAAGGGTCAT 3’ and reverse, 5’ GAGTCCTTCCACGATACCAA 3’. The gene expression levels were calculated by 2-ΔΔCT method with GADPH or U6 as internal controls.
Luciferase reporter assay
Circ_0007142 sequences and FOXK1 3’-UTR with or without miR-186 binding sites were generated and inserted into luciferase reporter vectors psiCHECK-2, (Promega, Madison, WI, USA). Subsequently, reporter plasmid and miR-186 or control mimics were transduced into A549 and H125 cells by Lipofectamine 2000 (Invitrogen). Two days after transfection, renilla luciferase activities were detected using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) following the manufacturer’s instructions.
Western blotting analysis
Proteins were isolated from H125 cells using RIPA buffer containing protease inhibitors, and then separated by 10% sodium dodecyl sulfatepolyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes. After blocked with 5% skimmed milk, the membrane was incubated with the primary antibodies anti-FOXK1, anti-β-catenin, anti-c-myc, anti-cyclinD1 and anti-GAPDH at 4°C overnight, followed by incubating with anti-rabbit horseradish peroxidase-conjugated secondary antibody (ZSGB-BIO, Beijing, China), and detected using enhanced chemiluminescence detection system.
Statistical analysis
All statistical analyses were performed using the SPSS 17.0 software and GraphPad Prism 6. All data were expressed as the mean ± standard deviation. Differences were calcuated with Student’s t-test or one-way ANOVA. Pearson’s correlation analysis was used to analyze the expression correlation. Kaplan-Meier method and logrank test were utilized to analyze the overall survival rate of patients. The value of P less than 0.05 indicated a statistically significant difference.
Results
Circ_0007142 was enhanced in lung adenocarcinoma tissues and correlated with poor prognosis
To assess the expression of circRNAs in lung adenocarcinoma, we initially analyzed altered expression of circRNAs between five lung adenocarcinoma tissues and five paracancerous tissues using circRNA microarray. We observed 32 circRNAs were upregulated in lung adenocarcinoma tissues (Figure 1A). And circ_0007142 expression was the most upregulated in lung adenocarcinoma tissues (Figure 1B). Moreover, we measured the expression of circ_0007142 in 70 pairs of lung adenocarcinoma tissues and adjacent normal tissues. The results confirmed that circ_0007142 expression was obviously enhanced in cancer tissues (Figure 1C). In addition, we analyzed the association of circ_0007142 expression with lung adenocarcinoma clinical features. The results demonstrated that increased circ_0007142 expression was associated with TNM stage, lymphatic and distant metastases (Figure 1D; Table 1). The Kaplan-Meier curves and Log-rank test indicated that patients with high circ_0007142 expression had significantly shorter overall survival time than patients with low circ_0007142 expression (Figure 1E).
Figure 1.
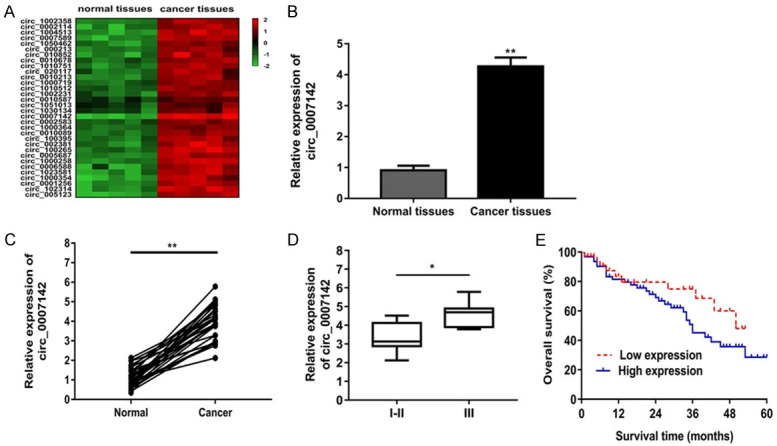
Circ_0007142 was enhanced in lung adenocarcinoma tissues and correlated with poor prognosis. A. 32 differentially expressed lncRNAs were upregulated in lung adenocarcinoma tissues and paracancerous tissues. B. The expression of circ_0007142 was most up-regulated in lung adenocarcinoma tissues. C. The expression of circ_0007142 in 60 pairs of lung adenocarcinoma tissues and adjacent normal tissues. D. Circ_0007142 expression was obviously higher in patients with advanced clinical stage (III phase) than that with early clinical stage (I-II phase). E. Kaplan-Meier curves of overall survivals and log-rank test showed that patients with high circ_0007142 expression had poor overall survivals. *P<0.05 compared to normal, I-II group; **P<0.01 compared to normal group.
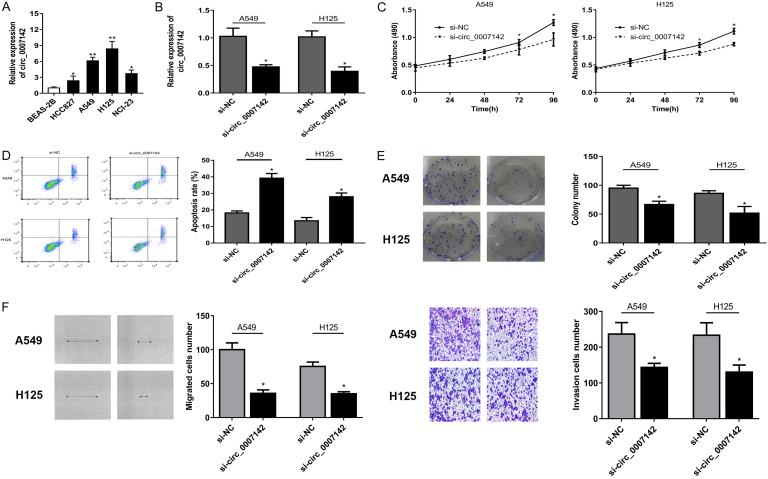
Knockdown of circ_0007142 suppressed proliferation, migration and invasion, induced apoptosis in lung adenocarcinoma cells
To identify the potential role of circ_0007142 in lung adenocarcinoma cells, we first measured the circ_0007142 expression in 4 lung adenocarcinoma cell lines. The results showed circ_0007142 expression was remarkably enhanced in cancer cell lines than that in normal cell line. Among all lung cancer cell lines, circ_0007142 expression was the highest in A549 and H125 cells (Figure 2A). Thus we constructed stable circ_0007142 knockdown in A549 and H125 cells via siRNA transfection (Figure 2B). CCK-8 assay revealed circ_0007142 knockdown inhibited the proliferation of A549 and H125 cells significantly (Figure 2C). The cell apoptosis assay manifested si-circ_0007142 induced a higher cell apoptosis percentage than the si-NC group (Figure 2D). In addition, colony formation assay showed circ_0007142 silencing decreased the colony forming ability of two cell lines (Figure 2E). In contrast to control si-NC group, knockdown of circ_0007142 inhibited the migration and invasion ability of cells (Figure 2F).
Figure 2.
Knockdown of circ_0007142 suppressed proliferation, migration and invasion, induced apoptosis in lung adenocarcinoma cells. A. Circ_0007142 expression level was remarkably enhanced in cancer cell lines than that in normal cell line. B. Circ_0007142 knockdown was established via siRNA transfection in A549 and H125 cell. C. CCK-8 assay revealed circ_0007142 knockdown inhibited the proliferation of A549 and H125 cells significantly. D. Flow cytometry revealed that silencing of circ_0007142 markedly augmented apoptosis in cells compared with the non-transfected cells. E. Colony formation assay showed circ_0007142 silencing decreased the colony forming ability of two cell lines. F. Transwell assay showed circ_0007142 silencing inhibited the migration and invasion ability of cells. *P<0.05 compared to BEAS-2B, si-NC group; **P<0.01 compared to BEAS-2B.
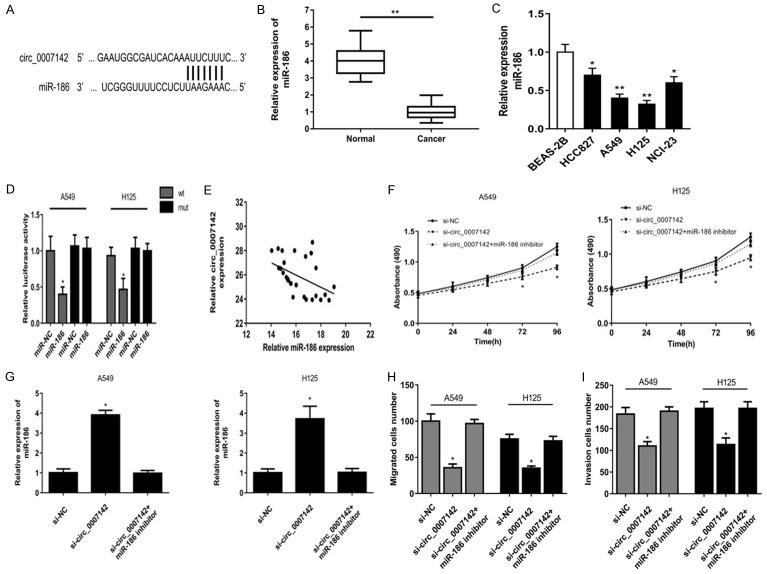
Circ_0007142 serves as a sponge for miR-186
Growing evidences had illustrated that circRNAs function as miRNA sponges to regulate gene expression post-transcriptionally. Thus, we investigated the potential candidate targets of circ_0007142 using circinteractome (https://circinteractome.nia.nih.gov/). And the bioinformatics analysis predicted that miR-186 was a potential target of circ_0007142 (Figure 3A). Besides, miR-186 level was significantly down-regulated in lung adenocarcinoma cancer tissues compared to adjacent normal tissues (Figure 3B). Similarly, miR-186 expression level was obviously lower in 4 cancer cell lines than that in normal cell line (Figure 3C). Luciferase reporter assay revealed miR-186 overexpression repressed the luciferase activity of only circ_0007142 wild type in A549 and H125 cells (Figure 3D). As shown, circ_0007142 expression was negatively correlated with miR-186 expression in cancer tissues (Figure 3E). CCK-8 assay revealed that miR-186 inhibitor could rescue the suppression by si-circ_0007142 in the proliferation of two cells (Figure 3F). After cells were transfected with si-circ_0007142, miR-186 expression was significantly up-regulated which was enhanced by miR-186 inhibitor (Figure 3G). Colony formation assay and transwell assay showed that miR-186 inhibitor could rescue the suppression by si-circ_0007142 in the migration and invasion number of two cells (Figure 3H, 3I).
Figure 3.
Circ_0007142 serves as a sponge for miR-186. A. Bioinformatics analysis showed that miR-186 was a potential target of circ_0007142. B. miR-186 level was significantly down-regulated in lung adenocarcinoma cancer tissues compared to adjacent normal tissues. C. miR-186 expression level was obviously lower in 4 cancer cell lines than that in normal cell line. D. The dual-luciferase reporter assay verified the binding with the decreasing fluorescence within miR-186 mimic and circ_0007142 wild type. E. Circ_0007142 expression was negatively correlated with miR-186 expression in cancer tissues. F. CCK-8 assay revealed that miR-186 inhibitor could rescue the suppression by si-circ_0007142 in the proliferation of two cells. G. After cells were transfected with si-circ_0007142, miR-186 expression was significantly up-regulated which was enhanced by miR-186 inhibitor. H, I. miR-186 inhibitor could rescue the suppression by si-circ_0007142 in the migration and invasion number of two cells. *P<0.05 compared to BEAS-2B, miR-NC, si-NC group; **P<0.01 compared to BEAS-2B, normal group.
FOXK1 is a direct target of miR-186
Increasing reports demonstrate miRNAs may regulate gene expression via binding to 3’-UTR of target mRNAs. Thus we explored potential target genes of miR-186 using TargetScan (http://www.targetscan.org/) and identified FOXK1 as a potential target candidate (Figure 4A). Luciferase reporter assay validated miR-186 significantly decreased the luciferase activity of wild type (WT) but not the mutant (Mut) 3’-UTR of FXOK1 in lung cancer cells (Figure 4B). Besides, we observed the FOXK1 expression was significantly increased in cancer tissues and cells (Figure 4C, 4D). Moreover, we confirmed a reverse correlation between miR-186 and FOXK1 in lung cancer tissues (Figure 4E). Furthermore, miR-186 overexpression obviously suppressed FOXK1 expression at both mRNA and protein level (Figure 4F).
Figure 4.
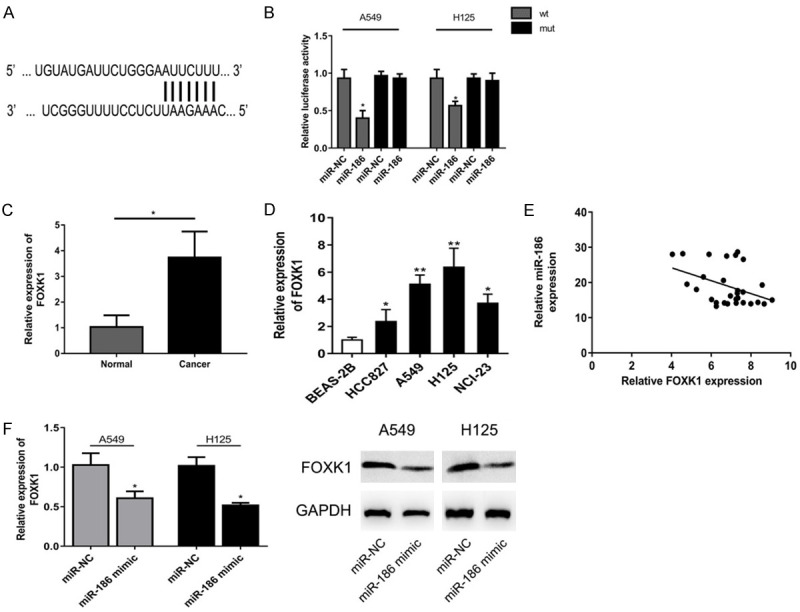
FOXK1 is a direct target of miR-186. A. TargetScan predicted FOXK1 as a potential target candidate of miR-186. B. Luciferase reporter assay validated miR-186 significantly decreased the luciferase activity of wild type (WT) but not the mutant (Mut) 3’-UTR of FXOK1 in lung cancer cells. C, D. FOXK1 expression was significantly increased in cancer tissues and cells. E. A reverse correlation was found between miR-186 and FOXK1 in lung cancer tissues. F. miR-186 overexpression obviously suppressed FOXK1 expression at both mRNA and protein level. *P<0.05 compared to miR-NC, normal group; **P<0.01 compared to BEAS-2B.
Circ_0007142 silencing alleviated the activation of Wnt/β-catenin signaling pathway
The Wnt/β-catenin signaling pathway has been documented to serve as an important role in tumor progression [15]. Thus, we explored the effect of circ_0007142 on this signaling pathway in H125 cells transfected with si-circ_0007142 using qRT-PCR and western blot. The results showed circ_0007142 knockdown downregulated the expression level of FOXK1 in H125 cells, which was reversed by miR-186 inhibitor (Figure 5A). Furthermore, miR-186 inhibition could rescue suppressed abilities caused by circ_0007142 knockdown in the expression of β-catenin, c-myc and cyclinD1 at both mRNA and protein levels (Figure 5B). These data suggested circ_0007142 silencing may suppress through the inactivation of Wnt/β-catenin pathway.
Figure 5.
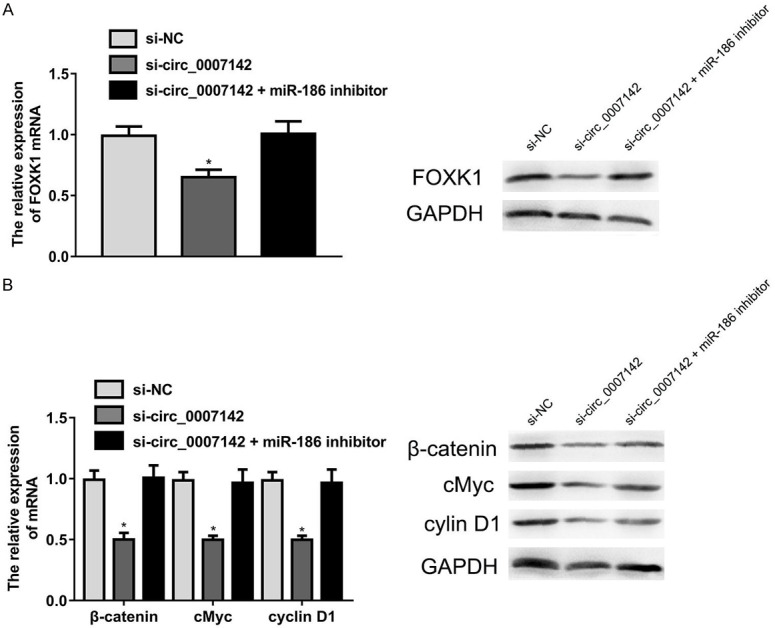
Circ_0007142 silencing alleviated the activation of Wnt/β-catenin signaling pathway. A. miR-186 inhibitor could rescue the suppression of FOXK1 expression by si-circ_0007142 in H125 cells. B. miR-186 inhibition could rescue suppressed abilities caused by circ_0007142 knockdown in the expression of β-catenin, c-myc and cyclinD1 at both mRNA and protein levels. *P<0.05 compared to si-NC group.
Discussion
Lung cancer is the world’s leading cause of cancer death (Available from: http://www.who.int/mediacentre/factsheets/fs297/en/). Approximately 40% of all the diagnosed cases are lung adenocarcinomas (LUADs). As one of pathological subtypes, lung adenocarcinoma comprises approximately 40% of all diagnosed lung cancer cases [16]. To date, circRNAs have been identified as important regulators in tumorigenesis and progression. In this present study, we first explored the functions and regulatory mechanism of circ_0007142 in lung adenocarcinoma progression. We found circ_0007142 expression was dramatically increased in lung adenocarcinoma tissues and cell lines. In addition, high expression of circ_0007142 was closely associated with advanced TNM stage and metastasis. Moreover, circ_0007142 level may act as a potential factor to predict overall survival. Furthermore, we observed circ_0007142 knockdown exhibited inhibitory effect on lung adenocarcinoma cell proliferation, migration and invasion. These data suggested that circ_0007142 may act as an important role in regulating cell proliferation and motility of lung adenocarcinoma cancer.
Growing evidence suggested that circRNA may execute its regulatory function in gene expression as miRNA sponge. Similarly, we predicted miR-186 was a potential target of circ_0007142 using bioinformatics analysis. Subsequently, we found miR-186 expression level was lower in cancer tissues and cell lines. And miR-186 inhibitor could rescue the suppression by si-circ_0007142 in the proliferation, migration and invasion of cancer cells. These results imply circ_0007142 may act as a sponge for miR-186 in lung adenocarcinoma. To further investigate the downstream effector molecule of circ_0007142/miR-186 axis, we identified FOXK1 as a potential target candidate of miR-186 target using TargetScan. FOXK1 belongs to FOX transcription factor family and regulates gene expression in cell proliferation, metastasis and metabolism [17,18]. Accumulating evidence has demonstrated FOXK1 level was enhanced in several malignant tumors, including esophageal cancer [19], gastric cancer [20] and colorectal cancer [21]. Similarly, we found FOXK1 was overexpression in lung adenocarcinoma tissue and cells. Meanwhile, miR-186 overexpression obviously suppressed FOXK1 expression at both mRNA and protein level. The Wnt/β-catenin signaling pathway plays an important role in the development of human cancers [22]. Previous studies have revealed aberrant activation of the Wnt/β-catenin pathway was involved in lung cancer progression [23]. Given that FOXK1 could regulate Wnt/β-catenin pathway in cancers [24-26], we further investigated the effect of circ_0007142 on this signaling. The results demonstrated knockdown of circ_0007142 could downregulate the expression level of FOXK1 in cells. Besides, circ_0007142 silencing resulted in inhibition of β-catenin, c-myc and cyclinD1 expression, indicating that circ_0007142 knockdown suppressed the activation of Wnt/β-catenin signaling pathway through miR-186/FOXK1 axis.
Conclusion
In summary, we first identified that circ_0007142 was upregulated in lung adenocarcinoma tissues and cells. Knockdown of circ_0007142 inhibited the proliferation and metastasis of lung adenocarcinoma through miR-186/FOXK1 axis and inactivated the Wnt/β-catenin signaling pathway, which provided a novel potential mechanism about lung adenocarcinoma development.
Acknowledgements
This work was supported by Peking Union Medical College Hospital.
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Devarakonda S, Morgensztern D, Govindan R. Genomic alterations in lung adenocarcinoma. Lancet Oncol. 2015;16:e342–351. doi: 10.1016/S1470-2045(15)00077-7. [DOI] [PubMed] [Google Scholar]
- 3.Calvayrac O, Pradines A, Pons E, Mazieres J, Guibert N. Molecular biomarkers for lung adenocarcinoma. Eur Respir J. 2017;49:1601734. doi: 10.1183/13993003.01734-2016. [DOI] [PubMed] [Google Scholar]
- 4.Greenhalgh J, Dwan K, Boland A, Bates V, Vecchio F, Dundar Y, Jain P, Green JA. First-line treatment of advanced epidermal growth factor receptor (EGFR) mutation positive non-squamous non-small cell lung cancer. Cochrane Database Syst Rev. 2016:CD010383. doi: 10.1002/14651858.CD010383.pub2. [DOI] [PubMed] [Google Scholar]
- 5.Gridelli C, Rossi A, Carbone DP, Guarize J, Karachaliou N, Mok T, Petrella F, Spaggiari L, Rosell R. Non-small-cell lung cancer. Nat Rev Dis Primers. 2015;1:15009. doi: 10.1038/nrdp.2015.9. [DOI] [PubMed] [Google Scholar]
- 6.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 8.Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20:1829–1842. doi: 10.1261/rna.047126.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kristensen LS, Hansen TB, Veno MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555–565. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang M, Xin Y. Circular RNAs: a new frontier for cancer diagnosis and therapy. J Hematol Oncol. 2018;11:21. doi: 10.1186/s13045-018-0569-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu C, Zhang Z, Qi D. Circular RNA hsa_circ_0023404 promotes proliferation, migration and invasion in non-small cell lung cancer by regulating miR-217/ZEB1 axis. Onco Targets Ther. 2019;12:6181–6189. doi: 10.2147/OTT.S201834. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Han W, Wang L, Zhang L, Wang Y, Li Y. Circular RNA circ-RAD23B promotes cell growth and invasion by miR-593-3p/CCND2 and miR-653-5p/TIAM1 pathways in non-small cell lung cancer. Biochem Biophys Res Commun. 2019;510:462–466. doi: 10.1016/j.bbrc.2019.01.131. [DOI] [PubMed] [Google Scholar]
- 13.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 14.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sokol SY. Spatial and temporal aspects of Wnt signaling and planar cell polarity during vertebrate embryonic development. Semin Cell Dev Biol. 2015;42:78–85. doi: 10.1016/j.semcdb.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kerdidani D, Chouvardas P, Arjo AR, Giopanou I, Ntaliarda G, Guo YA, Tsikitis M, Kazamias G, Potaris K, Stathopoulos GT, Zakynthinos S, Kalomenidis I, Soumelis V, Kollias G, Tsoumakidou M. Wnt1 silences chemokine genes in dendritic cells and induces adaptive immune resistance in lung adenocarcinoma. Nat Commun. 2019;10:1405. doi: 10.1038/s41467-019-09370-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katoh M. Human FOX gene family (Review) Int J Oncol. 2004;25:1495–1500. [PubMed] [Google Scholar]
- 18.Shi X, Wallis AM, Gerard RD, Voelker KA, Grange RW, DePinho RA, Garry MG, Garry DJ. Foxk1 promotes cell proliferation and represses myogenic differentiation by regulating Foxo4 and Mef2. J Cell Sci. 2012;125:5329–5337. doi: 10.1242/jcs.105239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen D, Wang K, Li X, Jiang M, Ni L, Xu B, Chu Y, Wang W, Wang H, Kang H, Wu K, Liang J, Ren G. FOXK1 plays an oncogenic role in the development of esophageal cancer. Biochem Biophys Res Commun. 2017;494:88–94. doi: 10.1016/j.bbrc.2017.10.080. [DOI] [PubMed] [Google Scholar]
- 20.Zhang P, Tang WM, Zhang H, Li YQ, Peng Y, Wang J, Liu GN, Huang XT, Zhao JJ, Li G, Li AM, Bai Y, Chen Y, Ren YX, Li GX, Wang YD, Liu SD, Wang JD. MiR-646 inhibited cell proliferation and EMT-induced metastasis by targeting FOXK1 in gastric cancer. Br J Cancer. 2017;117:525–534. doi: 10.1038/bjc.2017.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu M, Wang J, Tang W, Zhan X, Li Y, Peng Y, Huang X, Bai Y, Zhao J, Li A, Chen C, Chen Y, Peng H, Ren Y, Li G, Liu S. FOXK1 interaction with FHL2 promotes proliferation, invasion and metastasis in colorectal cancer. Oncogenesis. 2016;5:e271. doi: 10.1038/oncsis.2016.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene. 2017;36:1461–1473. doi: 10.1038/onc.2016.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Serman L, Nikuseva Martic T, Serman A, Vranic S. Epigenetic alterations of the Wnt signaling pathway in cancer: a mini review. Bosn J Basic Med Sci. 2014;14:191–194. doi: 10.17305/bjbms.2014.4.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen F, Xiong W, Dou K, Ran Q. Knockdown of FOXK1 suppresses proliferation, migration, and invasion in prostate cancer cells. Oncol Res. 2017;25:1261–1267. doi: 10.3727/096504017X14871164924588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao L, Chen B, Li J, Yang F, Cen X, Liao Z, Long X. Wnt/beta-catenin signaling pathway inhibits the proliferation and apoptosis of U87 glioma cells via different mechanisms. PLoS One. 2017;12:e0181346. doi: 10.1371/journal.pone.0181346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li P, Yu Z, He L, Zhou D, Xie S, Hou H, Geng X. Knockdown of FOXK1 inhibited the proliferation, migration and invasion in hepatocellular carcinoma cells. Biomed Pharmacother. 2017;92:270–276. doi: 10.1016/j.biopha.2017.05.087. [DOI] [PubMed] [Google Scholar]