Abstract
KLF14 belongs to the Krüppel-like factor (KLF) family of transcription factors. The KLF family activate and/or repress transcription in a promoter- and cell-dependent manner by interacting with co-suppressors or co-activators. However, the function and mechanism of KLF14 in osteogenic differentiation of human bone marrow mesenchymal stem cells (hMSCs) is unknown. This study explores the impact and molecular mechanism of KLF14 in hMSC osteogenic differentiation in vitro. We found that KLF14 was highly expressed in hMSCs, and KLF14 expression gradually decreased after inducing osteogenic differentiation. Inhibiting KLF14 expression promoted osteogenic differentiation of hMSCs. We also found that KLF14 interacted with the WNT3A promoter. This interaction decreased expression of WNT3A and downstream osteogenesis-related target genes in the WNT signaling pathway, and resulted in cell cycle arrest. In conclusion, we describe a new mechanism for KLF14 in differentiation of hMSCs into osteoblasts and suggest a new target for clinical therapeutics related to human bone development.
Keywords: KLF14, WNT3A, hMSC, osteogenic differentiation
Introduction
Krüppel-like factors (KLFs) are a family of DNA-binding transcription factors. The evolutionarily conserved KLF family of transcriptional regulators includes 17 identified members in the mammalian genome. These factors regulate gene expression for key biological processes, including proliferation, differentiation, apoptosis, and development. KLF regulators influence gene expression in both normal and pathological conditions, including cardiovascular disease, metabolic disease, and cancer [1-3]. Though many KLF genes are broadly expressed, some have tissue-specific expression patterns. Several organs and cells express a distinct profile of KLF proteins, and some interact in functional cooperation or antagonism depending on context [1,2,4].
Structurally, KLF proteins are characterized by a conserved DNA-binding domain with three Cys2/His2 zinc fingers, similar to a specificity protein (Sp)1, at the C terminus. This DNA-binding domain identifies and attaches to the GT/GC-rich regulatory sites within gene promoters and enhancers. KLF family members also have a nuclear localization signal sequence and transcriptional regulatory domain in the N-terminus. The N-terminal domain is highly flexible and specifically interacts with distinct nuclear proteins to repress and/or activate genes. Therefore, KLF transcription factors participate in a protein network that generates disease phenotypes in organisms ranging from flies to humans [2].
KLF14 is a relatively newly discovered member of the KLF family and is characterized by three highly conserved C-terminal Cys2His2 zinc finger domains that regulate gene transcription via DNA binding activity [5-7]. KLF14 is encoded by an intronless gene and has monoallelic maternal expression in all human and mouse tissues that have been examined to date [4]. Recently, large-scale genome-wide association studies (GWAS) reported that KLF14 is closely related to both high-density lipoprotein cholesterol (HDL-C) and type 2 diabetes mellitus in different populations, as well as being implicated in the etiology and susceptibility of atherosclerosis phenotypes [8-10]. Interestingly, maternal KLF14 expression is as a master trans-regulator of adipose gene expression in humans [4]. KLF14 is also reportedly associated with regulating T-regulatory cell differentiation [11] and tumorigenesis [12].
Although KLF14 has been widely biochemically characterized, the physiological roles of KLF14 remain largely unknown. To gain deep insight into how KLF14 functions in human bone marrow mesenchymal stem cell (hMSC) osteogenic differentiation, we investigated KLF14 expression of mRNA and protein in hMSCs. Additionally, we used both gain-of-function and loss-of-function studies to explore the role of KLF14 in osteogenic differentiation of hMSCs. We found that inhibiting KLF14 expression activated WNT3A expression, which increased expression of osteogenesis-related target genes in the WNT signaling pathway and promoted osteogenic differentiation of hMSCs.
Materials and methods
Compounds and reagents
We purchased RNA kits from RiboBio Co., Ltd. (Guangzhou, China), TRIzol reagents from Invitrogen (Rockville, MD, USA), and Cell Lysis Buffer from Cell Signaling Technology (Danvers, MA, USA). The antibodies against KLF14, GAPDH, cyclinD1, cyclin E, pRb, p-pRb, β-catenin, p84, and fluorochrome-conjugated β-catenin were purchased from Abcam (Cambridge, MA, USA). Nuclear/Cytosol Fractionation Kits were purchased from BioVision, Inc. (San Francisco, CA, USA) and Cell Cycle Detection kits from KeyGEN Biotech (Nanjing, China). OriCell Human Mesenchymal Stem Cell Osteogenic Differentiation Medium was purchased from Lonza (Alpharetta, GA, USA) and the Alizarin Red S Staining Quantification Assay (ARed-Q) from Sciencell Research Laboratories (Carlsbad, CA, USA).
Ethical considerations
The experimental protocol was approved by the Human Subjects Institutional Review Board and the Medical Ethics Committee of the Shenzhen People’s Hospital, Second Clinical Medical School of Jinan University.
Surgical procedures
We isolated hMSCs from 10 mL of bone marrow harvested from patients who underwent iliac bone graft harvesting for spine fusion. Bone marrow samples were aspirated and collected in a heparin-rinsed syringe.
hMSCs isolation and culture
We rinsed each bone marrow sample with Dulbecco’s phosphate buffered saline (DPBS) and added up to 2×108 nucleated cells in 5 mL of DPBS to 25 mL of Percoll cushion (Pharmacia Biotech). To remove unwanted cells in the marrow aspirate, we applied a density gradient and isolated a minor fraction of cells from the density interface. The cells were re-suspended and seeded in T-75 flask with 2×105 cells and maintained in DMEM-LG with 20% fetal bovine serum (FBS) and antibiotics in a 5% CO2 humidified incubator at 37°C. We removed non-adherent cells by changing the medium after 7 days of primary culture. Colonies of hMSCs were subcultured in fresh flasks at 10 to 14 days after trypsin.
Osteogenic differentiation of hMSCs
We cultured hMSCs at 37°C in a humidified atmosphere of 5% CO2 to approximately 80-90% confluency and then replated in 6-well cell culture plates with 2 mL growth medium/well at 3×103 cells/cm2. After 24 hours of incubation, we carefully aspirated the growth medium from each well and added 2 mL Human Mesenchymal Stem Cell Osteogenic Differentiation Medium. Cell media was refreshed every 3 days for 2-3 weeks by completely replacing the medium with fresh Human Mesenchymal Stem Cell Osteogenic Differentiation Medium. After 2-4 weeks of differentiation, cells were fixed and stained with Alizarin Red S (ARS).
ARS staiing
As previously reported [13], we stained differentiated hMSCs with 40 mM ARS for 10-15 minutes with gentle agitation. Cells were rinsed for 15 minutes with 1× phosphate-buffered saline with gentle agitation.
RNA preparation and reverse transcription quantitative-PCR assay
We extracted total RNA from cells using TRIzol following manufacturer instructions. cDNA was reverse transcribed using 2 μg of each sample RNA. The primer sequences used were: KLF14-forward, 5’-ACAGCCCCCAGGAACTGATA-3’; KLF14-reverse, 5’-GGATGGGTGAGACACCAGAG-3’; FASL-forward, 5’-GCACACAGCATCATCTTTGG-3’; FASL-reverse, 5’-GGACCTTGAGTTGGACTTGC-3’; P27-forward, 5’-AAGAAGCCTGGCCTCAGAAG-3’; P27-reverse, 5’-TTCATCAAGCAGTGATGTATCTGA-3’; Bim-forward, 5’-CCTCCCTACAGACAGAGCCA-3’; Bim-reverse, 5’-GATAGTGGTTGAAGGCCTGG-3’; P21-forward, 5’-AGTCAGTTCCTTGTGGAGCC-3’; P21-reverse, 5’-CATGGGTTCTGACGGACAT-3’; Caspase-9-forward, 5’-AGGCCCCATATGATCGAGGA-3’; Caspase-9-reverse, 5’-TCGACAACTTTGCTGCTTGC-3’; ALP-forward, 5’-CAGACGTTCCATACCCCCAC-3’; ALP-reverse, 5’-GGGACCTTTGGCTCTCGAC-3’; OCN-forward, 5’-CTCACACTCCTCGCCCTATT-3’; OCN-reverse, 5’-TTGGACACAAAGGCTGCAC-3’; SOST-forward, 5’-AATCACATCCGCCCCAACTT-3’; SOST-reverse, 5’-GGCCAGTGTCCTTGAACCTT-3’; DMP1-forward, 5’-TCTTTGTGAACTACGGAGGGT-3’; DMP1-reverse, 5’-GTTGGTGCCTGAGCCAAATG-3’; OPN-forward, 5’-CTGAACGCGCCTTCTGATTG-3’; OPN-reverse, 5’-GGGTTTCAGCACTCTGGTCA-3’. We normalized expression data to the geometric mean of housekeeping gene GAPDH (forward, 5’-GACTCATGACCACAGTCCATGC-3’, and reverse, 5’-AGAGGCAGGGATGATGTTCTG-3’) to control the variability in expression levels. We calculated expression using the equation 2-[(Ctof target) - (Ctof GAPDH)], where Ct represents the threshold cycle for each transcript.
Western blotting analysis
We collected wild type, KLF14 overexpression, or KLF14 silenced hMSCs and rinsed them using cold PBS. After adding the Cell Lysis Buffer, we incubated samples on ice for 30 min to separate the total proteins. We measured protein concentrations using the bicinchoninic acid assay. Equal amounts of proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a polyvinylidene fluoride (PVDF) membrane to determine protein expression levels after culturing with primary antibodies, and then further developed with enhanced chemiluminescence (ECL) reagents.
Cytosolic/nuclear protein extraction
We followed the instructions from a Nuclear/Cytosol Fractionation Kit to separate nuclear and cytosolic proteins from wild type, prepared KLF14 overexpression or U87 silenced hMSCs. Protein expression in each fraction was evaluated by western blotting using GAPDH and nuclear protein p84 as cytosolic and nuclear markers, respectively.
Cell cycle analysis
We incubated wild type, KLF14 overexpression, or KLF14 silenced hMSCs in 12-well tissue culture plates for 24 h (1×105 cells/well). Cell cycle phase was investigated using the Cell Cycle Detection kit according manufacturer instructions. Briefly, we fixed cultured cells overnight in 70% cold ethanol and washed using cold PBS, then treated cells with RNaseA (40 μl) at 37°C for 30 min, followed by another 30 min incubation using 160 μl of propidium iodide at 4°C in dark. We analyzed stained cells with a flow cytometer (Accuri C6, BD Biosciences, USA).
Cell viability assay
We assessed cell viability by MTT assay. In brief, we plated wild type, KLF14 overexpression, or KLF14 silenced hMSCs (1×105 cells/ml) in 96-well plates and incubated overnight to allow cells to attach. At the indicated time, we measured optical density (O.D.) values at the 570 nm wavelength.
TOP/FOP flash reporter examination
We co-transfected the pRL-TK plasmid (10 ng/well) together with the FOP flash plasmid (200 ng/well), or with the TOP flash plasmid, into wild type, KLF14 overexpression, or KLF14 silenced hMSCs. After 36-48 h, we evaluated luminescence intensity using a Dual-Luciferase Reporter Assay System and calculated ratios of TOP Flash and FOP Flash activity.
Chromatin immunoprecipitation (ChIP) assays
We performed ChIP assays as follows: briefly, wild type, KLF14 overexpression, or KLF14 silenced hMSCs were cross linked in 1% formaldehyde for 10 min followed by quenching with 125 mM glycine. We dissolved the nuclei pellet in lysis buffer with 0.1% SDS to obtain 200-600 bp fragments by sonication. Protein G magnetic beads and ChIP grade β-catenin antibody were used for analysis. We used IgG antibody as the negative control and conducted qPCR to confirm enrichment.
Immunofluorescence assay
We fixed cells by covering with 2-3 mm 4% formaldehyde diluted in warm PBS for 15 min at RT, rinsed 3 times in 1× PBS for 5 min. Cells were blocked by incubating in blocking buffer for 60 min and then incubated in diluted fluorochrome-conjugated β-catenin overnight at 4°C. After, we rinsed cells three times in 1× PBS for 5 min each wash, and cured with mountant overnight at RT. We observed treated samples under fluorescence microscope.
Statistical analysis
We used SPSS statistical software (SPSS Inc., Chicago, IL, USA) to analyze all data and presented data as mean ± standard deviation. To determine statistical significance of data differences between two groups, we applied a two-tailed Student’s t-test and considered P<0.05 to indicate statistical significance.
Results
KLF14 expression was downregulated in hMSCs after inducing osteogenic differentiation
To identify whether KLF14 contributes to hMSC osteogenic differentiation as a prospective biomarker or therapeutic target, we isolated hMSCs from patients and induced osteogenic differentiation. Then we compared KLF14 expression between hMSCs and hMSCs with induced osteogenic differentiation, as well as with human calvarial osteoblasts (HCOs), at both the protein (Figure 1A) and mRNA (Figure 1B) levels. We found that gradual downregulation of KLF14 expression in hMSCs at 15 and 25 days after inducing osteogenic differentiation. This result was further confirmed in HCOs, suggesting that KLF14 potentially contributes to osteogenic differentiation of hMSCs.
Figure 1.

KLF14 expression was higher in human bone marrow mesenchymal stem cells (hMSCs) than in osteogenic differentiated hMSCs and human calvarial osteoblasts (HCOs). KLF14 protein expression in hMSCs, osteogenic differentiated hMSCs, and HCOs was determined by western blot. A. Western blot analysis (left panel), Quantification of fold changes (right panel); B. KLF14 mRNA expression in hMSCs, osteogenic differentiated hMSCs, and HCOs determined by RT-qPCR assays.
KLF14 overexpression constrained proliferation, arrested the cell cycle, and inhibited osteogenic differentiation of hMSCs
Many factors mediate osteogenic differentiation of MSCs, which is vital for maintaining healthy bone tissue. To gain further insight into how KLF14 contributes to the osteogenic differentiation process in hMSCs, we used both gain-of-function and loss-of-function studies. We found that KLF14 expression levels were downregulated during osteogenic differentiation of hMSCs (Figure 1A), leading to the thought that KLF14 may be associated with the osteogenic differentiation of hMSCs. We performed lentivirus infection to overexpress or knockdown KLF14 expression in hMSCs and confirmed that KLF14 expression was either successfully overexpressed or knocked down in hMSCs using real-time qPCR (Figure 2A).
Figure 2.

KLF14 overexpression inhibited growth and osteogenic differentiation in hMSCs. A. RT-qPCR analysis to confirm KLF14 overexpression or knock-down in hMSCs with KLF14 or KLF14-siRNA from lentivirus infection. B. Flow cytometry analysis of the cell cycle distribution in KLF14 overexpression hMSCs. C. MTT assay showing the effect of KLF14 manipulation on hMSC viability. D. Alizarin red staining (ARS) showing effect of KLF14 manipulation on osteogenic differentiation of hMSCs. The images (left panel) and numbers (right panel) of the ARS positive cells were obtained by fluorescence microscopy.
The percentage of cells in S phase decreased from 32.62% to 20.92% and those in G1/G0 phase increased from 60.22% to 71.87% in a flow cytometry assay for cell cycle distribution in KLF14 overexpressed hMSCs (Figure 2B, middle panel). However, KLF14 silencing in hMSCs increased the percentage of cells in S phase from 32.62% to 55.23% and the cells in G1/G0 phase decreased from 60.22% to 37.79% (Figure 2B, right panel). We used an MTT assay to confirm that KLF14 upregulation inhibited cell viability, whereas KLF14 silencing promoted the viability of hMSCs viability (Figure 2C). Similarly, ARS showed that KLF14 overexpression inhibited osteogenic differentiation of hMSCs (approximately 50% less), whereas KLF14 silencing promoted osteogenic differentiation of hMSCs (approximately 5-fold more) (Figure 2D) of ARS positive cells viewed by florescent microscopy. These data suggest that KLF14 negatively regulates hMSC proliferation and differentiation.
KLF14 regulated key factors involved in cell proliferation, osteogenesis, and apoptosis in hMSCs
To explore if the main factors promoting cell proliferation were also regulated by KLF14, cyclinD1, cyclin E, pRb, and p-pRb, we determined protein expression levels in hMSCs by western blot assay. We determined mRNA expression of CCND1, LGRS, Nanog, CD133, Snail, FGF4, Frizzled7, SOX2, and Twist by real-time qPCR analysis.
Our results showed that KLF14 overexpression in hMSCs decreased protein expression for cyclinD1, cyclin E, and p-pRb, whereas KLF14 silencing promoted cyclinD1, cyclin E, and p-pRb protein expression (Figure 3A). KLF14 overexpression in hMSCs decreased mRNA expression for CCND1, LGRS, Nanog, CD133, Snail, FGF4, Frizzled7, SOX2, and Twist, whereas KLF14 silencing promoted expression of these genes (Figure 3B).
Figure 3.

KLF14 regulated key factors involved in cell proliferation, osteogenesis, and apoptosis. A. Protein bands in western blots (left panel) and fold changes after band quantification (right panel) for proliferation-associated proteins in KLF14 overexpression or KLF14 silenced hMSCs. B. RT-qPCR analysis to determine effect of KLF14 manipulation on proliferation-related gene expression, including CCND1, LGR5, Nanog, CD133, Snail, FGF4, Frizzled7, SOX2, and Twist. C. RT-qPCR analysis to determine effect of KLF14 manipulation on expression levels of osteogenic related genes, including ALP, OCN, SOST, DMP1, and OPN. D. RT-qPCR analysis to determine effect of KLF14 manipulation on expression levels of different apoptosis-related genes, including FASL, p27, Bim, p21, and Caspase-9.
We also explored expression of key factors regulated by KLF14 that promote the hMSC osteogenesis. Our real-time qPCR analysis for expression levels of ALP, OCN, SOST, DMP1, and OPN showed KLF14 overexpression decreased expression of these genes, whereas KLF14 silencing increased their expression (Figure 3C). When we measured gene expression for pro-apoptotic genes regulated by KLF14, we found that KLF14 overexpression increased expression of multiple pro-apoptotic genes, including FASL, p27, Bim, p21, and caspase-9, whereas KLF14 silencing decreased their expression (Figure 3D). These results further indicate that KLF14 negatively regulates hMSC growth and differentiation.
KLF14 inhibited the Wnt/β-catenin signaling pathway during hMSC osteogenic differentiation
Canonical Wnt/β-catenin signaling is crucial for MSC osteogenic differentiation [14-19]. β-catenin interacts with TCF/LEF transcription factors and upregulates WNT3A target genes in the nucleus. We investigated whether Wnt/β-catenin signaling related to hMSC osteogenic differentiation by measuring Wnt signaling pathway activity that was regulated by KLF14. To further assess activation of the β-catenin-TCF/LEF complex, we measured the activity of both TOP flash (containing wild type TCF binding sites) and FOP flash (mutant TOP flash) in KLF14 overexpression or KLF14 silenced hMSCs. Our results showed that KLF14 overexpression inhibited TOP/FOP activity, whereas KLF14 silencing promoted TOP/FOP activity in hMSCs (Figure 4A).
Figure 4.

KLF14 upregulation inactivated Wnt/β-catenin signaling. A. Wnt/β-catenin signaling pathway activity in KLF14 or KLF14-siRNA transfected hMSCs determined by TOP/FOP ratio. B. Nuclear β-catenin protein expression in KLF14 or KLF14-siRNA transfected hMSCs detected by western blot analysis. C. TOP/FOP ratio determining activity of Wnt/β-catenin signaling pathway in hMSCs transfected by KLF14-siRNA together with β-catenin siRNA or TCF4-dn. D. Immunofluorescence assay to determine effect of KLF14 manipulation on nuclear β-catenin staining. E. Chromatin immunoprecipitation (ChIP) assays in KLF14 over-expressed and KLF14 silenced hMSCs with anti-β-catenin antibody to recognize β-catenin enrollment in the promoters of Nanog, RUNX2, and OCN. IgG was served as a negative control.
We used western blots to detect nuclear expression of β-catenin in nuclear proteins isolated from KLF14 overexpression or KLF14 silenced hMSCs. These nuclear expression samples represented activated β-catenin, which has been translocated from the cytosol into the nucleus. Our data show that KLF14 overexpression inhibited β-catenin nuclear translocation in hMSCs, whereas KLF14 silencing promoted β-catenin nuclear translocation from the cytosol (Figure 4B).
To investigate the functional significance of KLF14 in Wnt/β-catenin signaling, we expressed β-catenin-siRNA and TCF4-dn in hMSC cells with silenced KLF14. As expected, KLF14 downregulation induced TOP/FOP luciferase reporter activity (Figure 4C), suggesting that KLF14 mediates Wnt/β-catenin signaling in hMSCs (Figure 4C). We further verified β-catenin translocation from cytosol to the nucleus by immunofluorescently staining β-catenin, and found that KLF14 overexpression in hMSCs weakened nuclear β-catenin staining, whereas KLF14 silencing in hMSCs increased the amount of nuclear β-catenin staining (Figure 4D). We performed ChIP assays with an anti-β-catenin antibody in KLF14 overexpression and KLF14 silenced hMSCs to identify β-catenin recruitment to promoters for its downstream genes, Nanog, RUNX2, and OCN, with IgG serving as a negative control. We found that β-catenin recruitment to the promoters of these genes was inhibited by KLF14 overexpression (Figure 4E). These results suggest that KLF14 inhibited the Wnt/β-catenin signaling pathway during hMSC osteogenic differentiation.
KLF14 regulated Wnt/β-catenin signaling pathway by targeting WNT3A
Our data above indicate that KLF14 inhibited the Wnt/β-catenin signaling pathway in MSC osteogenic differentiation. To further characterize the specific mechanism of this activity, we studied the target molecule. Based on predictions by JASPAR software, we found that the transcriptional factor KLF14 existed at two binding sites (BTE1 and BTE2) in the BTE region of the WNT3A gene promoter, suggesting that WNT3A is a KLF14 target (Figure 5A). We detected the effect of KLF14 on WNT3A activity using a dual-luciferase reporter assay in hMSCs transfected with KLF14 or KLF14-siRNA and wild type or mutant WNT3A reporters. Our results showed that KLF14 had a regulatory effect on the BTE2 locus, but no regulatory effect on the BTE1 locus, indicated by binding at the BTE2 locus in the BTE region of WNT3A gene promoter. KLF14 inactivated the Wnt/β-catenin signaling pathway during hMSC osteogenic differentiation (Figure 5B). Our ChIP assay for both the BTE1 and BTE2 locus in the BTE region of WNT3A gene promoter further suggested that KLF14 specifically binds to the BTE2 locus of the WNT3A gene promoter (Figure 5C). Additionally, analysis of luciferase activity showed that the TOP/FOP ratio was not changed in hMSCs co-transfected with WNT3A and KLF14 (Figure 5D, *P<0.05, vs. the control). This result suggests that KLF14 abolished WNT3A mediated Wnt/β-catenin pathway activity. Correlation analysis showed that WNT3A expression negatively correlated with KLF14 expression in hMSCs (Figure 5E), indicating a close relationship between KLF14 and WNT3A during osteogenic differentiation.
Figure 5.
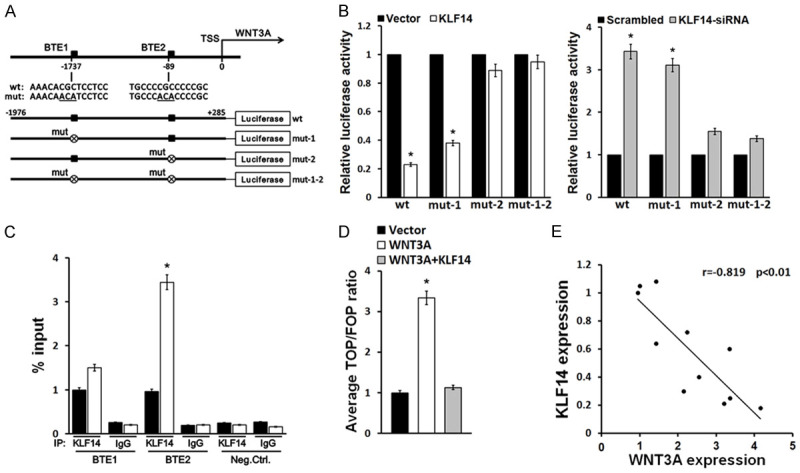
KLF14 downregulated Wnt/β-catenin signaling pathway by targeting WNT3A. A. JASPAR software prediction showing two KLF14 binding sites (BTE1and BTE2) in the BTE region of WNT3A gene promoter. B. Effect of KLF14 on WNT3A activity detected by dual-luciferase reporter assay in hMSCs transfected with KLF14 or KLF14-siRNA and the wild type or mutant WNT3A reporters. C. ChIP assay showing binding of KLF14 to the BTE2 locus. D. TOP/FOP ratio determining activity of Wnt/β-catenin signaling pathway in hMSCs transfected by KLF14 together with WNT3A. E. Correlation analysis of KLF14 and WNT3A expression. **P<0.01, vs. the control.
Discussion
MSCs are multipotent precursors with the potential to differentiate into osteogenic, chondrogenic, and adipogenic lineages, depending on both extracellular and intracellular signaling [14,15]. Wnt signaling, which leads to β-catenin stabilization and nuclear translocation, where it forms a complex with TCF/LEF to regulate target gene transcription, plays an important role in skeletal development [16-20]. Important for this study, Wnt signaling can regulate the self-renewal and proliferation of MSCs [21,22]. Furthermore, Wnt signaling also contributes to osteogenic differentiation in hMSCs [23]. To date, Wnt3a, Wnt5a, Wnt6, Wnt10a, and Wnt10b are recognized to control the stem cell properties of MSCs via Wnt signaling [21,24-26]. However, little is known about the interaction between KLF14 and Wnt signaling, or how this relationship functions in regulating MSC proliferation and differentiation.
Our results here show that KLF14 was downregulated during hMSC osteogenic differentiation. We also found that that KLF14 overexpression suppressed cell viability and osteogenic differentiation of hMSCs, and also induced cell cycle arrest of hMSCs. Therefore, our study shows that KLF14 negatively regulated hMSC proliferation and osteogenic differentiation. However, the specific regulatory mechanism of KLF14 in hMSC proliferation and osteogenic differentiation remains unknown. Here, we found that WNT3A expression negatively correlated with the expression of KLF14. KLF14 regulated WNT3A by binding to the BTE2 locus in the WNT3A gene promoter. Moreover, KLF14 negatively regulated hMSC proliferation and osteogenic differentiation and abolished WNT3A mediated Wnt/β-catenin pathway activity.
It will be important for future studies to investigate the specific mechanism of KLF14 regulatory activity in animal models and patients with bone defects. Additionally, we only investigated the relationship between KLF14 and Wnt/β-catenin signaling pathway in hMSCs in vitro. Therefore, our work must be verified by in vivo studies that support a role for KLF14 and WNT3A in osteogenic differentiation of hMSCs.
Our study provides reasonable evidences that KLF14 expression was downregulated in osteogenically differentiated hMSCs and HCO. We found that cell viability and osteogenic differentiation in hMSCs were inhibited, and the percentage of cells in the G1/G0 phase of the cell cycle increased KLF14 overexpression. Moreover, we found that WNT3A was a target of KLF14. Therefore, we identified a role for KLF14 in regulating Wnt signaling in hMSCs, thus impacting how these cells proliferate and differentiate into osteoblasts of hMSCs. These findings provide new information about the function and molecular mechanisms of KLF14 in regulating the osteogenic differentiation of hMSCs.
Acknowledgements
This study was funded by the National Natural Science Foundation of China (81900972) and Natural Science Foundation of Guangdong Province (2017A030310624).
Disclosure of conflict of interest
None.
References
- 1.Bureau C, Hanoun N, Torrisani J, Vinel JP, Buscail L, Cordelier P. Expression and function of Kruppel like-factors (KLF) in carcinogenesis. Curr Genomics. 2009;10:353–360. doi: 10.2174/138920209788921010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lomberk G, Urrutia R. The family feud: turning off Sp1 by Sp1-like KLF proteins. Biochem J. 2005;392:1–11. doi: 10.1042/BJ20051234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fan Y, Lu H, Liang W, Hu W, Zhang J, Chen YE. Kruppel-like factors and vascular wall homeostasis. J Mol Cell Biol. 2017;9:352–363. doi: 10.1093/jmcb/mjx037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parker-Katiraee L, Carson AR, Yamada T, Arnaud P, Feil R, Abu-Amero SN, Moore GE, Kaneda M, Perry GH, Stone AC, Lee C, Meguro-Horike M, Sasaki H, Kobayashi K, Nakabayashi K, Scherer SW. Identification of the imprinted KLF14 transcription factor undergoing human-specific accelerated evolution. PLoS Genet. 2007;3:e65. doi: 10.1371/journal.pgen.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, Balkau B, Heude B, Charpentier G, Hudson TJ, Montpetit A, Pshezhetsky AV, Prentki M, Posner BI, Balding DJ, Meyre D, Polychronakos C, Froguel P. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 6.Zhao C, Meng A. Sp1-like transcription factors are regulators of embryonic development in vertebrates. Dev Growth Differ. 2005;47:201–211. doi: 10.1111/j.1440-169X.2005.00797.x. [DOI] [PubMed] [Google Scholar]
- 7.Fernandez-Zapico ME, Lomberk GA, Tsuji S, DeMars CJ, Bardsley MR, Lin YH, Almada LL, Han JJ, Mukhopadhyay D, Ordog T, Buttar NS, Urrutia R. A functional family-wide screening of SP/KLF proteins identifies a subset of suppressors of KRAS-mediated cell growth. Biochem J. 2011;435:529–537. doi: 10.1042/BJ20100773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, Johansen CT, Fouchier SW, Isaacs A, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Aulchenko YS, Thorleifsson G, Feitosa MF, Chambers J, Orho-Melander M, Melander O, Johnson T, Li X, Guo X, Li M, Shin Cho Y, Jin Go M, Jin Kim Y, Lee JY, Park T, Kim K, Sim X, Twee-Hee Ong R, Croteau-Chonka DC, Lange LA, Smith JD, Song K, Hua Zhao J, Yuan X, Luan J, Lamina C, Ziegler A, Zhang W, Zee RY, Wright AF, Witteman JC, Wilson JF, Willemsen G, Wichmann HE, Whitfield JB, Waterworth DM, Wareham NJ, Waeber G, Vollenweider P, Voight BF, Vitart V, Uitterlinden AG, Uda M, Tuomilehto J, Thompson JR, Tanaka T, Surakka I, Stringham HM, Spector TD, Soranzo N, Smit JH, Sinisalo J, Silander K, Sijbrands EJ, Scuteri A, Scott J, Schlessinger D, Sanna S, Salomaa V, Saharinen J, Sabatti C, Ruokonen A, Rudan I, Rose LM, Roberts R, Rieder M, Psaty BM, Pramstaller PP, Pichler I, Perola M, Penninx BW, Pedersen NL, Pattaro C, Parker AN, Pare G, Oostra BA, O’Donnell CJ, Nieminen MS, Nickerson DA, Montgomery GW, Meitinger T, McPherson R, McCarthy MI, McArdle W, Masson D, Martin NG, Marroni F, Mangino M, Magnusson PK, Lucas G, Luben R, Loos RJ, Lokki ML, Lettre G, Langenberg C, Launer LJ, Lakatta EG, Laaksonen R, Kyvik KO, Kronenberg F, König IR, Khaw KT, Kaprio J, Kaplan LM, Johansson A, Jarvelin MR, Janssens AC, Ingelsson E, Igl W, Kees Hovingh G, Hottenga JJ, Hofman A, Hicks AA, Hengstenberg C, Heid IM, Hayward C, Havulinna AS, Hastie ND, Harris TB, Haritunians T, Hall AS, Gyllensten U, Guiducci C, Groop LC, Gonzalez E, Gieger C, Freimer NB, Ferrucci L, Erdmann J, Elliott P, Ejebe KG, Döring A, Dominiczak AF, Demissie S, Deloukas P, de Geus EJ, de Faire U, Crawford G, Collins FS, Chen YD, Caulfield MJ, Campbell H, Burtt NP, Bonnycastle LL, Boomsma DI, Boekholdt SM, Bergman RN, Barroso I, Bandinelli S, Ballantyne CM, Assimes TL, Quertermous T, Altshuler D, Seielstad M, Wong TY, Tai ES, Feranil AB, Kuzawa CW, Adair LS, Taylor HA Jr, Borecki IB, Gabriel SB, Wilson JG, Holm H, Thorsteinsdottir U, Gudnason V, Krauss RM, Mohlke KL, Ordovas JM, Munroe PB, Kooner JS, Tall AR, Hegele RA, Kastelein JJ, Schadt EE, Rotter JI, Boerwinkle E, Strachan DP, Mooser V, Stefansson K, Reilly MP, Samani NJ, Schunkert H, Cupples LA, Sandhu MS, Ridker PM, Rader DJ, van Duijn CM, Peltonen L, Abecasis GR, Boehnke M, Kathiresan S. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X, Li S, Yang Y, Yang X, Liu Y, Hu W, Jin L, Wang X. Genome-wide association study validation identifies novel loci for atherosclerotic cardiovascular disease. J Thromb Haemost. 2012;10:1508–1514. doi: 10.1111/j.1538-7836.2012.04815.x. [DOI] [PubMed] [Google Scholar]
- 10.Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, Mora S, Beckmann JS, Bragg-Gresham JL, Chang HY, Demirkan A, Den Hertog HM, Do R, Donnelly LA, Ehret GB, Esko T, Feitosa MF, Ferreira T, Fischer K, Fontanillas P, Fraser RM, Freitag DF, Gurdasani D, Heikkilä K, Hyppönen E, Isaacs A, Jackson AU, Johansson Å, Johnson T, Kaakinen M, Kettunen J, Kleber ME, Li X, Luan J, Lyytikäinen LP, Magnusson PKE, Mangino M, Mihailov E, Montasser ME, Müller-Nurasyid M, Nolte IM, O’Connell JR, Palmer CD, Perola M, Petersen AK, Sanna S, Saxena R, Service SK, Shah S, Shungin D, Sidore C, Song C, Strawbridge RJ, Surakka I, Tanaka T, Teslovich TM, Thorleifsson G, Van den Herik EG, Voight BF, Volcik KA, Waite LL, Wong A, Wu Y, Zhang W, Absher D, Asiki G, Barroso I, Been LF, Bolton JL, Bonnycastle LL, Brambilla P, Burnett MS, Cesana G, Dimitriou M, Doney ASF, Döring A, Elliott P, Epstein SE, Ingi Eyjolfsson G, Gigante B, Goodarzi MO, Grallert H, Gravito ML, Groves CJ, Hallmans G, Hartikainen AL, Hayward C, Hernandez D, Hicks AA, Holm H, Hung YJ, Illig T, Jones MR, Kaleebu P, Kastelein JJP, Khaw KT, Kim E, Klopp N, Komulainen P, Kumari M, Langenberg C, Lehtimäki T, Lin SY, Lindström J, Loos RJF, Mach F, McArdle WL, Meisinger C, Mitchell BD, Müller G, Nagaraja R, Narisu N, Nieminen TVM, Nsubuga RN, Olafsson I, Ong KK, Palotie A, Papamarkou T, Pomilla C, Pouta A, Rader DJ, Reilly MP, Ridker PM, Rivadeneira F, Rudan I, Ruokonen A, Samani N, Scharnagl H, Seeley J, Silander K, Stančáková A, Stirrups K, Swift AJ, Tiret L, Uitterlinden AG, van Pelt LJ, Vedantam S, Wainwright N, Wijmenga C, Wild SH, Willemsen G, Wilsgaard T, Wilson JF, Young EH, Zhao JH, Adair LS, Arveiler D, Assimes TL, Bandinelli S, Bennett F, Bochud M, Boehm BO, Boomsma DI, Borecki IB, Bornstein SR, Bovet P, Burnier M, Campbell H, Chakravarti A, Chambers JC, Chen YI, Collins FS, Cooper RS, Danesh J, Dedoussis G, de Faire U, Feranil AB, Ferrières J, Ferrucci L, Freimer NB, Gieger C, Groop LC, Gudnason V, Gyllensten U, Hamsten A, Harris TB, Hingorani A, Hirschhorn JN, Hofman A, Hovingh GK, Hsiung CA, Humphries SE, Hunt SC, Hveem K, Iribarren C, Järvelin MR, Jula A, Kähönen M, Kaprio J, Kesäniemi A, Kivimaki M, Kooner JS, Koudstaal PJ, Krauss RM, Kuh D, Kuusisto J, Kyvik KO, Laakso M, Lakka TA, Lind L, Lindgren CM, Martin NG, März W, McCarthy MI, McKenzie CA, Meneton P, Metspalu A, Moilanen L, Morris AD, Munroe PB, Njølstad I, Pedersen NL, Power C, Pramstaller PP, Price JF, Psaty BM, Quertermous T, Rauramaa R, Saleheen D, Salomaa V, Sanghera DK, Saramies J, Schwarz PEH, Sheu WH, Shuldiner AR, Siegbahn A, Spector TD, Stefansson K, Strachan DP, Tayo BO, Tremoli E, Tuomilehto J, Uusitupa M, van Duijn CM, Vollenweider P, Wallentin L, Wareham NJ, Whitfield JB, Wolffenbuttel BHR, Ordovas JM, Boerwinkle E, Palmer CNA, Thorsteinsdottir U, Chasman DI, Rotter JI, Franks PW, Ripatti S, Cupples LA, Sandhu MS, Rich SS, Boehnke M, Deloukas P, Kathiresan S, Mohlke KL, Ingelsson E, Abecasis GR Global Lipids Genetics Consortium. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei X, Yang R, Wang C, Jian X, Li L, Liu H, Yang G, Li Z. A novel role for the Kruppel-like factor 14 on macrophage inflammatory response and atherosclerosis development. Cardiovasc Pathol. 2017;27:1–8. doi: 10.1016/j.carpath.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 12.Fan G, Sun L, Shan P, Zhang X, Huan J, Li D, Wang T, Wei T, Gu X, Yao L, Xuan Y, Hou Z, Cui Y, Cao L, Li X, Zhang S, Wang C. Loss of KLF14 triggers centrosome amplification and tumorigenesis. Nat Commun. 2015;6:8450. doi: 10.1038/ncomms9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weng J, Peng W, Zhu S, Chen S. Long noncoding RNA sponges miR-454 to promote osteogenic differentiation in maxillary sinus membrane stem cells. Implant Dent. 2017;26:178–186. doi: 10.1097/ID.0000000000000569. [DOI] [PubMed] [Google Scholar]
- 14.Moon RT, Kohn AD, De Ferrari GV, Kaykas A. WNT and beta-catenin signalling: diseases and therapies. Nat Rev Genet. 2004;5:691–701. doi: 10.1038/nrg1427. [DOI] [PubMed] [Google Scholar]
- 15.Yu JM, Kim JH, Song GS, Jung JS. Increase in proliferation and differentiation of neural progenitor cells isolated from postnatal and adult mice brain by Wnt-3a and Wnt-5a. Mol Cell Biochem. 2006;288:17–28. doi: 10.1007/s11010-005-9113-3. [DOI] [PubMed] [Google Scholar]
- 16.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 17.Luu HH, Song WX, Luo X, Manning D, Luo J, Deng ZL, Sharff KA, Montag AG, Haydon RC, He TC. Distinct roles of bone morphogenetic proteins in osteogenic differentiation of mesenchymal stem cells. J Orthop Res. 2007;25:665–677. doi: 10.1002/jor.20359. [DOI] [PubMed] [Google Scholar]
- 18.Glass DA 2nd, Karsenty G. In vivo analysis of Wnt signaling in bone. Endocrinology. 2007;148:2630–2634. doi: 10.1210/en.2006-1372. [DOI] [PubMed] [Google Scholar]
- 19.Kim JH, Liu X, Wang J, Chen X, Zhang H, Kim SH, Cui J, Li R, Zhang W, Kong Y, Zhang J, Shui W, Lamplot J, Rogers MR, Zhao C, Wang N, Rajan P, Tomal J, Statz J, Wu N, Luu HH, Haydon RC, He TC. Wnt signaling in bone formation and its therapeutic potential for bone diseases. Ther Adv Musculoskelet Dis. 2013;5:13–31. doi: 10.1177/1759720X12466608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Otero JJ, Fu W, Kan L, Cuadra AE, Kessler JA. Beta-catenin signaling is required for neural differentiation of embryonic stem cells. Development. 2004;131:3545–3557. doi: 10.1242/dev.01218. [DOI] [PubMed] [Google Scholar]
- 21.Boland GM, Perkins G, Hall DJ, Tuan RS. Wnt 3a promotes proliferation and suppresses osteogenic differentiation of adult human mesenchymal stem cells. J Cell Biochem. 2004;93:1210–1230. doi: 10.1002/jcb.20284. [DOI] [PubMed] [Google Scholar]
- 22.Shang YC, Wang SH, Xiong F, Zhao CP, Peng FN, Feng SW, Li MS, Li Y, Zhang C. Wnt3a signaling promotes proliferation, myogenic differentiation, and migration of rat bone marrow mesenchymal stem cells. Acta Pharmacol Sin. 2007;28:1761–1774. doi: 10.1111/j.1745-7254.2007.00671.x. [DOI] [PubMed] [Google Scholar]
- 23.He W, Wang Z, Zhou Z, Zhang Y, Zhu Q, Wei K, Lin Y, Cooper PR, Smith AJ, Yu Q. Lipopolysaccharide enhances Wnt5a expression through toll-like receptor 4, myeloid differentiating factor 88, phosphatidylinositol 3-OH kinase/AKT and nuclear factor kappa B pathways in human dental pulp stem cells. J Endod. 2014;40:69–75. doi: 10.1016/j.joen.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 24.Bilkovski R, Schulte DM, Oberhauser F, Gomolka M, Udelhoven M, Hettich MM, Roth B, Heidenreich A, Gutschow C, Krone W, Laudes M. Role of WNT-5a in the determination of human mesenchymal stem cells into preadipocytes. J Biol Chem. 2010;285:6170–6178. doi: 10.1074/jbc.M109.054338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brun J, Fromigue O, Dieudonne FX, Marty C, Chen J, Dahan J, Wei Y, Marie PJ. The LIM-only protein FHL2 controls mesenchymal cell osteogenic differentiation and bone formation through Wnt5a and Wnt10b. Bone. 2013;53:6–12. doi: 10.1016/j.bone.2012.11.020. [DOI] [PubMed] [Google Scholar]
- 26.Cawthorn WP, Bree AJ, Yao Y, Du B, Hemati N, Martinez-Santibanez G, MacDougald OA. Wnt6, Wnt10a and Wnt10b inhibit adipogenesis and stimulate osteoblastogenesis through a beta-catenin-dependent mechanism. Bone. 2012;50:477–489. doi: 10.1016/j.bone.2011.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]


