Abstract
Hemangioblastomas (HBs) histologically overlap with TFE3 rearrangement-associated tumors, which present as alveolar architecture and clear or eosinophilic granular cytoplasm. However, whether TFE3 is expressed in HBs remains unexplored. Herein, we analyzed the clinicopathologic features of 42 HBs emphasizing studies of TFE3 expression. Of 42 cases, 38 were sporadic and 4 were regarded as a part of von Hippel-Lindau (VHL) syndrome according to clinical presentation. Nineteen patients were male and 23 were female. Patient age ranged from 17 to 70 years (median 43). Tumor size ranged from 0.4 to 4.8 cm (mean 2.2 cm). Follow-up ranged from 1 to 60 months and 6 patients developed recurrence. Immunohistochemistry staining showed that 36 (86%) of 42 HBs expressed TFE3 in nuclei of tumor cells, of which 21 were evaluated as high TFE3 expression levels. Increased TFE3 expression was significantly associated with older ages (P=0.018) and larger tumor size (P=0.001). Seventeen HBs with high TFE3 expression were negative for rearrangement and amplification of TFE3 by FISH analysis, 3 of which including 2 sporadic and 1 VHL-related HBs demonstrated trisomies or tetrasomies of X-chromosome in 7%~18% of tumor cells. All 3 cases occurred in female, presented with a larger tumor size and displayed a similar morphologic appearance with high cellularity and hyperchromatic nuclei. Our study first reports TFE3 expression and its clinicopathological relevance in HBs. We hypothesize that TFE3 might be involved in the pathogenesis of non-VHL-related HBs. Furthermore, HBs with strong TFE3 expression should be differentiated from brain-metastatic TFE3-rearranged tumors.
Keywords: Hemangioblastoma, TFE3, immunohistochemistry staining, clinicopatholgical features
Introduction
Hemangioblastomas (HBs) are benign neoplasms of undetermined origin and account for up to 2.5% of all intracranial tumors [1,2]. HBs can occur in the cerebellum (16-69%), brainstem (5-22%), spinal cord (13-53%), cauda equina (11%), supratentorial region (1-7%) or other locations outside of central nervous system (CNS) [2,3]. On gross appearance, HBs are often described as red vascular masses with a thin layer of capsule. Microscopically, the tumor is composed of two main components: polygonal-shaped stromal cells with clear or granular eosinophilic cytoplasm and vascular cells comprising of endothelial cells and pericytes [2,4]. The stromal cells are considered as the neoplastic cells harboring the genetic alterations [5]. HBs can occur sporadically (75%) or as a part of von Hippel-Lindau (VHL) syndrome (25%) [6]. Germline mutations or aberrations in the VHL gene have been identified as the cause of VHL-related HBs [7-10]. However, the pathogenesis of sporadic HBs remains unclear. Recent studies have explored additional genetic alterations that may contribute to the tumorigenesis of sporadic HBs [11,12].
Microphthalmia-associated transcription factor/transcription factor E (MiTF/TFE) family members include TFE3, MITF, TFEB, and TFEC. The TFE3 (transcription factor binding to IGHM enhancer 3) gene is located on chromosome Xp11.2 and can fuse to multiple partners owing to chromosomal translocations. Known TFE3 fusion partners include ASPSCR1 (ASPL), PRCC, SFPQ1 (PSF), NONO, CLTC, PARP14, LUC7L3, DVL2, KHSRP, RBM10, NEAT1, and KAT6A [13-22]. These TFE3 gene fusion partners contribute more robust promoters that cause overexpression of TFE3 fusion proteins and transactivation of other genes [23,24]. An increasing number of TFE3 rearrangement-associated tumors, such as Xp11 translocation perivascular epithelioid cell neoplasm (PEComas), Xp11 translocation renal cell carcinoma, melanotic Xp11 translocation renal cancers, and a subset of epithelioid hemangioendothelioma have been reported [13,14,25-27]. It has been suggested that TFE3-associated tumors have distinctive histological features presented as solid nests or alveolar architecture, epithelioid tumor cells with abundant clear to finely granular eosinophilic cytoplasm and delicate vascular septa.
It is noted that, according to a review of literature, TFE3 expression in CNS has not hitherto been reported with any frequency. Due to the recognition that there are morphological similarity between HBs and TFE3-rearranged neoplasms while TFE3 expression in HBs remains unclear, herein we analyzed TFE3 expression by immunohistochemistry staining and its clinicopathologic features in HBs, then further performed fluorescence in situ hybridization (FISH) analysis in HBs with high TFE3 expression.
Materials and methods
Patients
Forty-two patients of HB who were operated between 2014 and 2018 in our Hospital, Sun Yat-sen University were eligible. The surgical specimens were available in the tissue bank of the Department of Pathology. A retrospective review was performed in these cases, including clinicopathologic data, pathomorphology, immunohistochemistry analyses, treatments and follow-up information (all patients were followed until June 2019). Of the 42 patients, 4 with a clinical manifestation of VHL syndrome were classified as having VHL-related HBs. All the patients received magnetic resonance imaging (MRI) at 1- to 3-month intervals to evaluate extent of preoperative disease progression and postoperative recovery. Lesion size were acquired based on pre-operative MRI scan and on written surgery reports. HBs are frequently cystic & solid lesion, but just the solid nodule size calculated by the largest diameter rather than the cyst size was defined as tumor size. For multiple lesions, the size of the largest tumor was counted. For the use of clinical materials for analysis, prior patient consent and approval from the Institutional Research Ethics Committee had been obtained.
Immunohistochemistry
As previously described [28], immunohistochemistry staining for TFE3 was performed on formalin-fixed, paraffin-embedded sections (4 μm thick) using manual protocols. Briefly, tissue slides were deparaffinized in xylene (10 min, thrice) and rehydrated using graded ethanol concentrations. Antigen retrieval was performed with steaming treatment in 1 mmol/L EDTA (PH 9.0) buffer. Following blocking endogenous peroxidase activity with 3% hydrogen peroxidase for 10 min, tissue sections were incubated overnight at 4°C with anti-TFE3 antibody (Cell Marque; clone MRQ-37; prediluted). Then the sections were incubated with the ChemMateTm EnvisionTm/HRP, Rabbit/Mouse secondary antibody (Gene Tech, GK500705)for an hour at 37°C. Diaminobenzidine (3,3’-Diaminobenzidine) was used as the chromogen, and Mayer’s hematoxylin was used as a counterstain.
The immunohistochemistry staining was independently evaluated by two experienced pathologists blinded to the clinicopathologic information. Only distinct nuclear staining for TFE3 was considered positive. As previously described [29,30], immunoreactivity was scored in a semiquantitative manner based on both labeling intensity and the percentage of immunopositive tumor cells among total tumor cells. The staining intensity was defined as 0 (negative), 1 (weak staining), 2 (moderate staining) and 3 (strong staining). The score was calculated by multiplying the staining intensity by the percentage of tumor cells showing immunoreactivity in the nucleus (0-100). The immunostaining results were considered to be 0 when the score was <25; 1+ when the score was 26-100; 2+ when the score was 101-200; or 3+ when the score was 201-300. For statistical analysis, 2+ and 3+ stainings were defined as high-TFE3 level, whereas 0 and 1+ as low-TFE3 level.
Fluorescence in situ hybridization (FISH)
To identify TFE3 rearrangements, FISH was performed on interphase nuclei using commercial dual-color break-apart probes (Guangzhou LBP medical Technology Co., Ltd., China) specific to the TFE3 gene locus at Xp11.2. The TFE3 break-apart probe set is composed of clones flanking TFE3 including those centromeric to 5’TFE3 (red) and telomeric to 3’TFE3 (green).
As previously described [30,31], 3-µM formalin-fixed, paraffin-embedded tissue sections were mounted on positively charged slides. A serial of matched H&E-stained slides were used to identify the neoplastic cells. The unstained slides were deparaffinized at 90°C, immersed twice in xylene (15 minutes each) and twice in 100% ethanol (10 minutes each), air dried, and then microwaved in 0.1 mM citric acid (PH 6.0) for 10 minutes. The tissue sections were subsequently pretreated with 2× standard saline citrate (SSC) for 5 minutes, digested with 0.4 ml of pepsin (5 mg/ml in 0.1 N HCl/0.9 NaCl) (Sigma, St Louis, MO, USA) at 37°C for 40 minutes, and immersed in graded ethanol solutions for a second dehydration step. Following drying, the probes were applied to the marked tumoral region on each slide, covered with a cover slip, sealed with rubber cement, co-denatured with the target DNA at 80°C for 5 minutes and then hybridized overnight at 37°C. After stringent washing with 0.3% NP40 in 2× SSC, the sections were counterstained with DAPI, and mounted with anti-fade solution.
All samples were analyzed in a blind manner by two technologists. For each case, a minimum of 100 tumor nuclei were scored using an Olympus BX51 fluorescent microscope (Tokyo, Japan), controlled by IMSTAR software (Paris, France). Only non-overlapping tumor nuclei were evaluated. A fused or closely approximated red-green signal pattern was interpreted as a normal result, whereas splitting of the probes indicated the presence of a translocation/rearrangement. The criterion used for calling two signals as ‘separate’ was the presence of at least one signal width of space between the red and green signals. According to the generally accepted criteria, gene rearrangement was reported as present if ≥ 10% of the tumor nuclei showed split signals defined as separation of signals.
Since TFE3 is located in X chromosome, the positive signal pattern differs between males and females. For balanced translocation, males should show one red and one green signal (1R1G), whereas females show one red, one green and one fusion signal (1R1G1F). For unbalanced translocation, males should show one red and one fusion signal (1R1F), while females show one red and two fusion signals (1R2F). For negative results, interphase nuclei from a normal male should show one fusion signal (1F), while a normal female should exhibit two fusion signals (2F) because of an intact copy of TFE3 on each of the two X chromosomes.
Scoring of polyploid of X chromosome also differs between males and females. For male, cells which present to have more than one pair of red and green signals in the nuclei are considered to be positive, while for female, cells which contain more than two pairs of red-green signals are regarded as positive.
Statistical methods
Statistical analyses were performed using SPSS 22.0 statistical software (IBM, USA). The relationship between qualitative variables were analyzed by Pearson Chi-square test or Fisher’s exact test. For comparison of continuous variables, Independent-Sample T test or Mann-Whitney U test was used. P-values <0.05 were considered significant.
Results
Clinicopathological characteristics of HBs
The main clinical characteristics of the patients with HBs were summarized in Table 1. Of the 42 patients, 19 were male and 23 were female. Patient ages at the time of diagnosis ranged from 17 to 70 years (mean, 44 years; median, 43 years). Eight cases were multiple tumors and the others were solitary. Twenty-nine cases arose in cerebellum, of which 4 had multiple nodules, 5 in medulla oblongata and 4 in spinal canal. Additionally, one case occurred simultaneously in cerebellum and medulla oblongata, 2 in cerebellum and spinal canal, and 1 in cerebellum and cerebellopontine angle. Tumor size ranged from 0.4 to 4.8 cm (mean, 2.2 cm) in diameter.
Table 1.
Clinical characteristics of HBs patients
| Characteristics | All samples/Patients (N=42) | Sporadic HBs (N=38) | VHL-related HBs (N=4) | P |
|---|---|---|---|---|
| Age, years | ||||
| Mean ± SD | 44±14 | 46±14 | 29±13 | 0.026a |
| Sex | ||||
| Male | 19 | 18 | 1 | 0.613b |
| Female | 23 | 20 | 3 | |
| Lesions number | ||||
| Multiple | 8 | 5 | 3 | 0.018b |
| Solitary | 34 | 33 | 1 | |
| Tumor size, cm | ||||
| Mean ± SD | 2.2±1.3 | 2.2±1.3 | 2.4±0.7 | 0.813a |
| Tumor location | ||||
| Cerebellum | 33 | 30 | 3 | 0.289b |
| Oblongata | 6 | 4 | 2 | |
| Spinal canal | 6 | 5 | 1 | |
| CPA | 1 | 1 | 0 | |
| Recurrence | ||||
| Present | 6 | 6 | 0 | 1.000b |
| Absent | 36 | 32 | 4 |
Mann-Whitney U test.
Fisher exact test.
All cases presented with clinical symptoms related to tumor compression. The majority of patients were treated by tumor resection whereas 2 patients with multiple tumors underwent incomplete excision. Follow up information (range, 1 to 60 months) was available for all cases. Thirty-six patients were alive without recurrence or metastasis. However, 6 patients experienced tumor recurrence.
There were 38 sporadic HBs and 4 VHL syndrome-related HBs. In the 4 VHL-related HBs, one patient with a solitary HB had a family history with his father suffering from adrenal tumor, and his grandfather and uncle both having kidney benign tumors (specific information unavailable). Meanwhile, this patient had also adrenal pheochromocytoma and multiple pancreatic cysts. All of the other 3 patients without family history of tumors had multiple HBs, of which one had renal angiomyolipoma and polycystic pancreas. The sporadic HBs patients were elderly (mean, 46±14 years) while the VHL-related HBs mostly occurred in young patients (mean, 29±13 years) (P=0.026). Compared with sporadic HBs, VHL-related HBs were more prone to multiple lesions. In the sporadic HBs patients, the tumors were solitary in 33 cases and multiple in 5 cases, while out of 4 VHL-related HBs, 3 were multifocal lesions (P=0.018).
VHL-related and sporadic HBs had identical histological features. Microscopically, the tumors were composed of nests of stromal cells which presented as oval to polygonal shape with a palely eosinophilic to clear and often multi-vacuolated cytoplasm, irregularly contoured nuclei and small nucleoli (Figure 1A-D). Some tumor cell nuclei was hyperchromatic or vesicular with inconspicuous nucleoli. Mitotic figures were rare. No necrosis was observed in all tumors. Immunostaining showed that Inhibin α (Figure 1E, 1F), S-100 protein (Figure 1G, 1H), CD56, and NSE were expressed in tumor cells, as well as CD31 and CD34 were positive in vascular endothelial cells (Figure 1I, 1J). Ki-67 proliferation index was about 1% to 15% (median, 2%).
Figure 1.
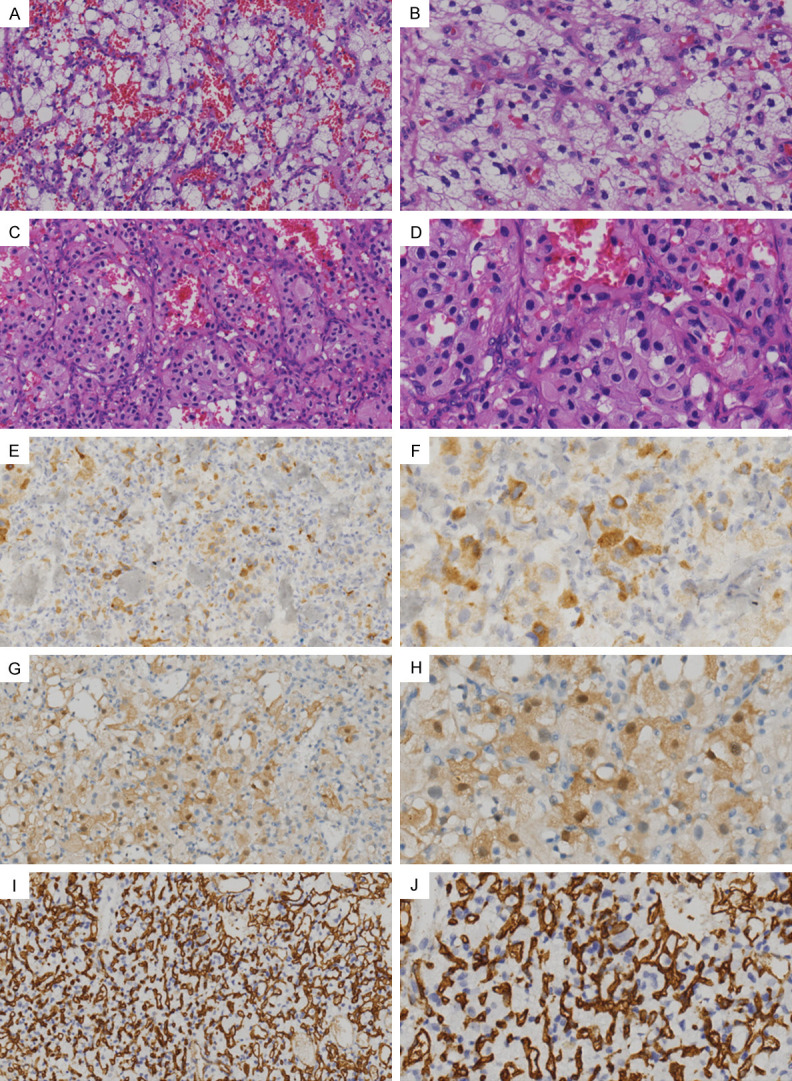
Histological and immunophenotypic features of HBs. (A-D) Microscopically, stromal tumor cells of HBs displayed nested or alveolar growth pattern, accompanied by a predominant delicate vasculature. The tumor cell cytoplasm varied from abundant clear vacuolated (A and B; H&E) to eosinophilic granular (C and D; H&E), and the nuclei were irregularly contoured with small nucleoli. (E-H) The tumor cells were positive for the expression of inhibin α (E and F; IHC) and S-100 protein (G and H; IHC). (I and J) Vascular endothelial cells expressed CD34 (IHC). Original magnification, left column, 200×; right column, 400×.
TFE3 expression in HBs by immunohistochemical staining
In our series, 36 of 42 (85.7%) HBs including 32 cases of sporadic HBs and 4 cases of VHL-related HBs showed TFE3 nuclear staining, while 6 HBs (14.3%) completely lacked TFE3 expression in the nucleus. Among the 36 HBs labeled positively for TFE3, 23 cases (54.8%) exhibited strong TFE3 staining in a mean of 53% of tumor cells (range, 5%-90%), 11 cases (26.2%) demonstrated moderate immunostaining for TFE3 in a mean of 56% of tumor cells (range, 5%-90%), while weak TFE3 labeling was observed in only 2 cases (60% and 90%, respectively) (Figure 2A-D). Following the scoring criteria described in Materials and methods, half of all cases exhibiting nuclear TFE3 labeling index more than 100 were regarded as high TFE3 expression level and the other half were regarded as low TFE3 expression level.
Figure 2.
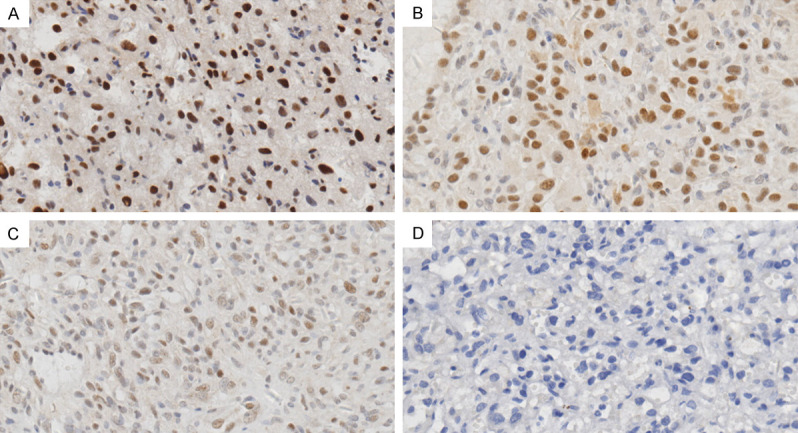
Nuclear TFE3 expression in HBs by immunohistochemistry staining. Representative images for IHC staining of HB tissues with variable nuclear TFE3 expression, including strong (A), intermediate (B) and weak (C) intensity as well as negative TFE3 expression (D). (A-D) Original magnification, 400×.
The relationship between TFE3 expression and the clinicopathologic features in HBs
The relationship between TFE3 expression and the clinicopathologic features of HBs was analyzed (Table 2). Our results showed that high TFE3 expression level was significantly associated with older age (P=0.018) and larger tumor size (P=0.001). No significant relationship between TFE3 expression and any other clinicopathologic variable was found.
Table 2.
Relationships between nuclear TFE3 expression levels and clinicopathologic features in HBs
| Characteristics | No. | TFE3 levels | P | |
|---|---|---|---|---|
|
| ||||
| High | Low | |||
| Age, years | ||||
| Mean ± SD | 49±14 | 39±13 | 0.018a | |
| Sex | ||||
| Male | 19 | 10 | 9 | 0.757b |
| Female | 23 | 11 | 12 | |
| Lesions number | ||||
| Multiple | 8 | 5 | 3 | 0.697c |
| Solitary | 34 | 16 | 18 | |
| Tumor size, cm | ||||
| Mean ± SD | 2.9±1.0 | 1.5±1.1 | 0.001d | |
| Tumor location | ||||
| Cerebellum | 33 | 21 | 12 | 0.061c |
| Oblongata | 6 | 1 | 5 | |
| Spinal canal | 6 | 2 | 4 | |
| CPAe | 1 | 0 | 1 | |
| Recurrence | ||||
| Present | 6 | 3 | 3 | 1.000c |
| Absent | 36 | 18 | 18 | |
| HBs subtype | ||||
| Sporadic | 38 | 19 | 19 | 1.000c |
| VHL-related | 4 | 2 | 2 | |
Independent-Sample T Test.
Pearson Chi-square test.
Fisher’s exact test.
Mann-Whitney U test.
‘CPA’ denotes cerebellopontine angle.
FISH findings
This pattern of TFE3 over-expression prompted us to perform break-apart FISH assay to detect whether it is related to TFE3 rearrangement. Seventeen HBs with high nuclear TFE3 levels including 15 cases of sporadic HBs and 2 VHL-related HBs were analyzed by FISH. The FISH findings were summarized in Table 3. All 17 cases were negative for TFE3 break-apart by FISH. A normal male showed one fusion signal (Figure 3A) whereas a normal female showed two fusion signals (Figure 3B). However, our data showed that polyploid X-chromosome with increased TFE3 copy numbers occurred in 3 HBs including 2 sporadic HBs and 1 VHL-related HB. Of the 2 sporadic cases, one had trisomies and tetrasomies of X-chromosome (7%) and the other had tetraploid X-chromosome (7%). While in the VHL-related case, trisomies and tetrasomies of X-chromosome was found in about 18% tumor cells.
Table 3.
Summary of FISH findings
| Case | Age/Gender | Tumor type | Diameter (cm) | Nuclear TFE3 labeling (intensity, %) | TFE3 break-apart FISH | X-chromosome |
|---|---|---|---|---|---|---|
| 1 | 48/F | VHL | 3.0 | 3+, 80% | 1R1G2F, 2%; 1R1F, 1% | trisomies and tetrasomies, 18% |
| 2 | 48/M | Sporadic | 3.0 | 3+, 70% | 1R1G, 3% | Normal |
| 3 | 29/M | Sporadic | 0.4 | 3+, 50% | 1R1G, 3% | Normal |
| 4 | 31/F | Sporadic | 2.5 | 2+, 80% | 1R1G1F, 3% | tetrasomies, 7% |
| 5 | 50/M | Sporadic | 3.0 | 3+, 50% | 1R1G, 2% | Normal |
| 6 | 34/M | Sporadic | 4.8 | 3+, 70% | 1R1G, 2% | Normal |
| 7 | 38/F | Sporadic | 2.5 | 3+, 70% | 1G1F, 2%; 1R1F, 1% | trisomies and tetrasomies, 7% |
| 8 | 54/F | Sporadic | 3.5 | 3+, 70% | 1R1G1F, 2% | Normal |
| 9 | 50/M | Sporadic | 3.5 | 3+, 50% | 1R1F, 1%; 1G1F, 2% | Normal |
| 10 | 17/F | VHL | 3.0 | 2+, 70% | 1R1G, 2% | Normal |
| 11 | 68/M | Sporadic | 3.5 | 3+, 90% | 1R1G1F, 3% | Normal |
| 12 | 63/F | Sporadic | 1.0 | 3+, 90% | NS | Normal |
| 13 | 60/F | Sporadic | 2.0 | 3+, 50% | 1R1G1F, 2%; 1G1F, 2% | Normal |
| 14 | 57/F | Sporadic | 3.0 | 3+, 50% | NS | Normal |
| 15 | 36/F | Sporadic | 2.4 | 3+, 80% | 1R1G1F, 2%; 1G1F, 2% | Normal |
| 16 | 56/M | Sporadic | 3.0 | 2+, 80% | 1R1G, 1% | Normal |
| 17 | 55/M | Sporadic | 4.0 | 3+, 80% | 1R1G, 2% | Normal |
‘NS’ denotes no signal that can be detected.
Figure 3.
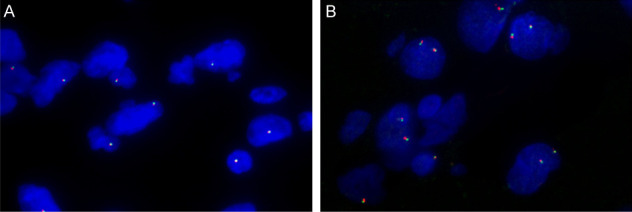
No TFE3 rearrangements detected in HBs. Break-apart FISH analyses using probes flanking TFE3 gene are negative. Normal combination of red-green signals differs between a male and a female (red, centromeric; green, telomeric). A. Representative FISH image of a normal male showing one fusion signal (1F). B. Representative FISH image of a normal female exhibiting two fusion signals (2F). A and B. Original magnification, 1000×.
All of the 3 cases were female ranging in age from 31 to 48 years (mean, 39 years). The size of each tumor was larger than average (2.2 cm) of the overall tumor population, ranging from 2.5 to 3.0 cm. Two sporadic HBs were solitary tumors and were both located in cerebellar hemisphere, while the VHL-related HB had multifocal lesions respectively located in cerebellar vermis and Lumbar vertebral canal. Microscopically, the 3 tumors exhibited a similar morphological features including high cellularity (Figure 4A) and hyperchromatic nuclei (Figure 4B). Represent images of high TFE3 expression examined by IHC and of triploid as well as tetraploid tumor cells detected by FISH analysis were shown in Figure 4C-E, respectively. All the 3 cases had no recurrence during the follow-up period.
Figure 4.
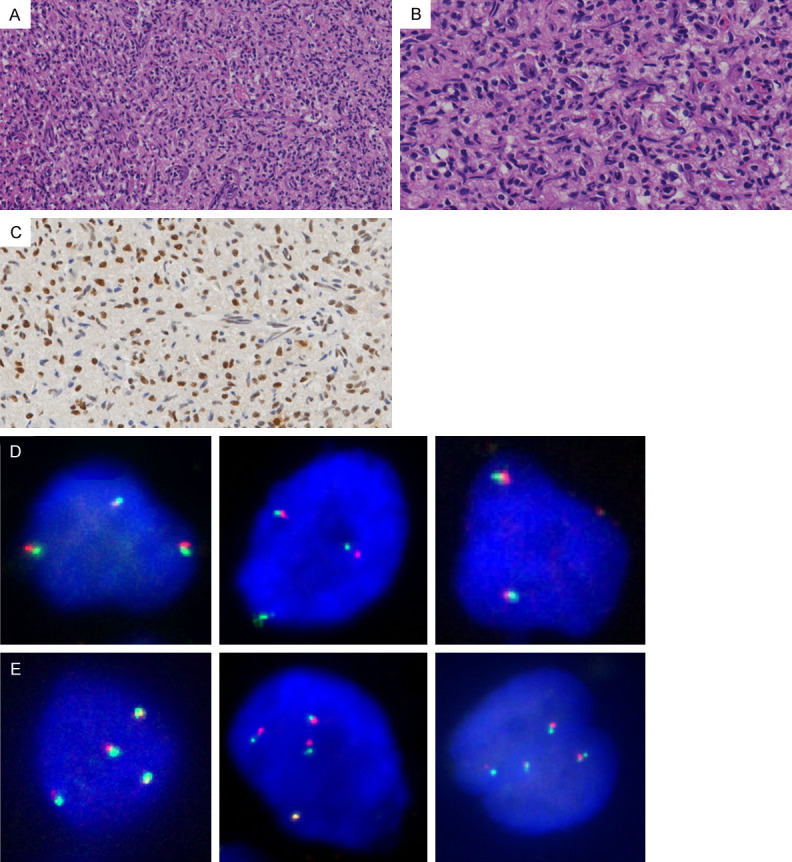
Histopathological features of HBs harboring polyploid X-chromosome. Representative images from one case of HB. A. Microscopically, the tumor showed high cellularity (H&E; original magnification, 200×). B. The stromal tumor cells contained hyperchromatic nuclei (H&E; original magnification, 400×). C. The stromal tumor cells demonstrated high TFE3 expression (IHC; original magnification, 400×). D. Representative FISH images showing triploid tumor cells (original magnification, 1000×). E. Representative FISH images showing tetraploid tumor cells (original magnification, 1000×).
Discussion
Most of HBs occur sporadically except a minority of cases are manifestation of VHL syndrome. VHL syndrome is an autosomal-dominant, multi-organ, and familiar neoplastic disease, including HBs in CNS and retina, clear cell renal cell carcinoma (ccRCC), pheochromocytoma, paragangliomas, and pancreatic neuroendocrine tumors [2,32-36]. Among these VHL-associated tumors, HBs in CNS are the most common symptom and affect 60-80% of VHL patients [3,36]. The VHL gene that is a tumor suppressor gene located on chromosome 3p25 has been recognized as the cause of VHL syndrome [35]. Due to VHL mutation or deletion, normal VHL protein has been lost and thus results in VHL disease. Somatic mutations and biallelic inactivation of VHL gene have been also found in a subset of sporadic HBs [9,37-39] and the rates of VHL mutation and biallelic VHL inactivation were 56% and 46% in sporadic HBs by next-generation sequencing, respectively [39]. This observation suggests that there might be undetermined alternative mechanism contributing to the tumorigenesis of HBs without loss of function of VHL gene.
The TFE3 gene is a member of the MiTF/TFE family of transcription factors. A number of studies have determined TFE3 as an oncogene. Oncogenic activation of TFE3 is driven by the fusion of TFE3 with a number of different gene partners. Chromosomal translocation results in gene fusion involving TFE3. The most commonly identified fusion partner was ASPSCR1 (ASPL) resulting from a t(X;17)(p11.2;q25.3) [13]. Other known partners of TFE3 include PRCC, SFPQ1 (PSF), NONO or CLTC respectively resulting from t(X;1)(p11.2;q21), t(X;1)(p11.2;p34), inv(X) (p11.2;q12), or t(X;17)(p11.2;q23) [14-16]. These chimeric transcription factors are involved in sustaining proliferation, driving metabolism and overcoming stress in the related tumors [40]. Besides, recent reports have also highlighted the role of TFE3 in promoting Wnt signaling pathway [41-43]. Overexpression of the fusion proteins can be demonstrated by strong nuclear TFE3 immunoreactivity [28], which has been accepted as a highly sensitive immunohistochemical marker for initial screening of TFE3 translocations. Immunohistochemistry coupled with break-apart FISH is now the optimized method for the clinical diagnosis of TFE3 rearrangement-associated tumors [25,28,44-46].
TFE3 immunoreactivity, as observed by Argani P et al., was closely associated with morphologic features and its positivity could be predicted by morphologic appearance [28]. Obviously, HBs share some morphological overlap with known TFE3-rearranged tumors that is typically described as acinar or nested architecture with a delicate vasculature and clear or eosinophilic cytomorphology, therefore prompting our focus on the status of TFE3 expression in HBs.
In this study, we retrospectively collected a total of 42 cases of HBs from our institution, of which 38 cases were sporadic type and 4 were VHL syndrome-related type. Although there might be great differences between the two subtypes of HBs in terms of genetics and clinic process, they share identical histopathology and are both composed of polygonal-shaped stromal tumor cells and a prominent delicate vasculature. Further diagnostic evidence was conferred by a characteristic immunohistochemical profile presented as expression of inhibin α and S-100 protein in stromal cells. We compared the clinicopathological features of the two types of HBs. The results displayed that the mean age of patients and the proportion of multiple tumors were significantly different between 38 sporadic HBs and 4 syndromic HBs. Compared with sporadic HBs, VHL-related HBs demonstrated a predilection for young adults (P<0.05) and frequent multifocal presentation (P<0.05), which was in keeping with the prior reports [11].
By immunohistochemistry staining, our results first showed that TFE3 protein was expressed in tumor cells of HBs. In the samples we tested, the majority (36 out of 42 cases; about 85.7%) of HBs, including 32 cases of sporadic HBs and 4 cases of VHL-related HBs, exhibited variable degree of nuclear TFE3 reactivity. Increased TFE3 expression levels were significantly related to larger tumor size (P=0.001), indicating that there might be an effect of nuclear TFE3 expression on tumor growth and proliferation in HBs. Additionally, our data showed that HBs with high TFE3 expression had a predilection for older patients (P=0.018). Further investigation is necessary to clarify the biological significance of nuclear TFE3 localization.
To identify whether TFE3 expression in HBs is related to TFE3 rearrangements, we further performed TFE3 break-apart FISH analysis in 17 HB cases haboring high nuclear TFE3 expression, including 15 sporadic and 2 VHL-related HBs. The result showed that all cases were negative for TFE3 gene rearrangement. In addition, no obvious TFE3 amplification was detected. However, recent advances have prompted us to be aware that the false-negative rate of TFE3 break-apart FISH has been increasing, because the currently designed TFE3 break-apart probes are difficult to efficiently detect the TFE3 gene rearrangements caused by an inversion of chromosome X which can produce the NONO-TFE3, RBM10-TFE3, and GRIPAP1-TFE3 gene fusions [15,19,47-49]. Thus, it still cannot be entirely ruled out the possibility that TFE3 rearrangements and gene fusions may exist in some HBs. Perhaps reverse transcriptase polymerase chain reaction (RT-PCR) and next generation sequencing are the preferred methods for identification of TFE3 gene fusions.
An additional finding by FISH detection was that three cases of HBs displayed polysomies (trisomies or tetrasomies) of chromosome X with increased TFE3 copy numbers, occupying about 7% to 18% of tumor cells. DNA polyploidy can confer genomic instability and is often linked to tumor occurrences. Moreover, it has already been reported that high chromosomal copy number alterations are also associated with aggressive behavior in Xp11.2 translocation renal cell carcinoma [50]. In our cohort, 3 HBs harboring X-chromosome polyploid all exhibited larger tumor size on gross examination, and higher cellularity as well as hyperchromatic nuclei seen under microscopy, suggesting that these tumors might have an increased proliferative potential. Additionally, all 3 cases were female, indicating that these neoplasms might have an predisposition to female. However, X-chromosome polyploid is not a very common event accounting for about 17.6% (3/17) in the cases we evaluated and its actual frequency in HBs needs to be investigated in a larger patients population.
Several studies have reported that by immunohistochemistry, TFE3 overexpression were also observed in solid-pseudopapillary neoplasms of the pancreas [51,52] and granular cell tumors [53,54], but FISH assay indicated that TFE3 break-apart was negative in these neoplasms. In addition, Yang et al. [29] have found that the strong positive immunostaining of TFE3 was not unique of Xp11 translocation renal cell carcinomas, and also occurred in a small minority (0.4%) of clear cell renal cell carcinomas with neither translocation nor amplification of TFE3 detected by FISH. In particular, the possibility of TFE3 rearrangement caused by an inversion of chromosome X had also been excluded by further RNA-sequencing in their study. These findings suggest that there might be some additional genetic, epigenetic or physiologic factors leading to increased TFE3 protein levels in HBs.
Diagnosis of HBs is typically straightforward but our findings suggest a potential pitfall that HBs could be misdiagnosed as some other brain metastatic tumors sharing common histology and aberrant TFE3 immunoreactivity, especially alveolar soft-part sarcoma (ASPS), PEComa, and Xp11 translocation renal cell carcinoma. Pathologists should be aware of the possibility of intracranial metastasis of these entities and distinguish them from HBs based on their distinct clinicopathological features, together with a judicious immunohistochemistry panel as well as molecular cytogenetic testing.
In summary, we first report that TFE3 expression has been found in the vast majority of HBs. Our findings expand the currently known list of tumors with TFE3 expression, and highlight a potential diagnostic pitfall caused by strong expression of TFE3 in some HBs. Future studies evaluating the possible role of TFE3 protein in pathogenesis of HBs would be interesting as only limited studies to date have explored specific molecular alterations associated with sporadic HBs.
Acknowledgements
This work was supported by 2 grants from the National Natural Science Foundation of China (81772862 and 81402413 to Y.Y).
Disclosure of conflict of interest
None.
References
- 1.Conway JE, Chou D, Clatterbuck RE, Brem H, Long DM, Rigamonti D. Hemangioblastomas of the central nervous system in von Hippel-Lindau syndrome and sporadic disease. Neurosurgery. 2001;48:55–62. doi: 10.1097/00006123-200101000-00009. [DOI] [PubMed] [Google Scholar]
- 2.Lonser RR, Glenn GM, Walther M, Chew EY, Libutti SK, Linehan WM, Oldfield EH. von Hippel-Lindau disease. Lancet. 2003;361:2059–2067. doi: 10.1016/S0140-6736(03)13643-4. [DOI] [PubMed] [Google Scholar]
- 3.Chittiboina P, Lonser RR. Von Hippel-Lindau disease. Handb Clin Neurol. 2015;132:139–156. doi: 10.1016/B978-0-444-62702-5.00010-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, editors. WHO Classification of Tumours of the Central Nervous System. 4th edition. Lyon: International Agency for Research on Cancer; 2007. [Google Scholar]
- 5.Vortmeyer AO, Gnarra JR, Emmert-Buck MR, Katz D, Linehan WM, Oldfield EH, Zhuang Z. von Hippel-Lindau gene deletion detected in the stromal cell component of a cerebellar hemangioblastoma associated with von Hippel-Lindau disease. Hum Pathol. 1997;28:540–543. doi: 10.1016/s0046-8177(97)90075-7. [DOI] [PubMed] [Google Scholar]
- 6.Neumann HP, Bender BU. Genotype-phenotype correlations in von Hippel-Lindau disease. J Intern Med. 1998;243:541–545. doi: 10.1046/j.1365-2796.1998.00336.x. [DOI] [PubMed] [Google Scholar]
- 7.Iliopoulos O, Levy AP, Jiang C, Kaelin WG Jr, Goldberg MA. Negative regulation of hypoxia-inducible genes by the von Hippel-Lindau protein. Proc Natl Acad Sci U S A. 1996;93:10595–10599. doi: 10.1073/pnas.93.20.10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliffe PJ. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 9.Gläsker S, Bender BU, Apel TW, van Velthoven V, Mulligan LM, Zentner J, Neumann HP. Reconsideration of biallelic inactivation of the VHL tumour suppressor gene in hemangioblastomas of the central nervous system. J Neurol Neurosurg Psychiatry. 2001;70:644–648. doi: 10.1136/jnnp.70.5.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nordstrom-O’Brien M, van der Luijt RB, van Rooijen E, van den Ouweland AM, Majoor-Krakauer DF, Lolkema MP, van Brussel A, Voest EE, Giles RH. Genetic analysis of von Hippel-Lindau disease. Hum Mutat. 2010;31:521–537. doi: 10.1002/humu.21219. [DOI] [PubMed] [Google Scholar]
- 11.Takayanagi S, Mukasa A, Tanaka S, Nomura M, Omata M, Yanagisawa S, Yamamoto S, Ichimura K, Nakatomi H, Ueki K, Aburatani H, Saito N. Differences in genetic and epigenetic alterations between von Hippel-Lindau disease-related and sporadic hemangioblastomas of the central nervous system. Neuro Oncol. 2017;19:1228–1236. doi: 10.1093/neuonc/nox034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma D, Yang J, Wang Y, Huang X, Du G, Zhou L. Whole exome sequencing identified genetic variations in Chinese hemangioblastoma patients. Am J Med Genet A. 2017;173:2605–2613. doi: 10.1002/ajmg.a.38350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Argani P, Antonescu CR, Illei PB, Lui MY, Timmons CF, Newbury R, Reuter VE, Garvin AJ, Perez-Atayde AR, Fletcher JA, Beckwith JB, Bridge JA, Ladanyi M. Primary renal neoplasms with the ASPL-TFE3 gene fusion of alveolar soft part sarcoma: a distinctive tumor entity previously included among renal cell carcinomas of children and adolescents. Am J Pathol. 2001;159:179–192. doi: 10.1016/S0002-9440(10)61684-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Argani P, Antonescu CR, Couturier J, Fournet JC, Sciot R, Debiec-Rychter M, Hutchinson B, Reuter VE, Boccon-Gibod L, Timmons C, Hafez N, Ladanyi M. PRCC-TFE3 renal carcinomas: morphologic, immunohistochemical, ultrastructural, and molecular analysis of an entity associated with the t(X;1)(p11.2;q21) Am J Surg Pathol. 2002;26:1553–1566. doi: 10.1097/00000478-200212000-00003. [DOI] [PubMed] [Google Scholar]
- 15.Clark J, Lu YJ, Sidhar SK, Parker C, Gill S, Smedley D, Hamoudi R, Linehan WM, Shipley J, Cooper CS. Fusion of splicing factor genes PSF and NonO (p54nrb) to the TFE3 gene in papillary renal cell carcinoma. Oncogene. 1997;15:2233–2239. doi: 10.1038/sj.onc.1201394. [DOI] [PubMed] [Google Scholar]
- 16.Argani P, Lui MY, Couturier J, Bouvier R, Fournet JC, Ladanyi M. A novel CLTC-TFE3 gene fusion in pediatric renal adenocarcinoma with t(X;17)(p11.2;q23) Oncogene. 2003;22:5374–5378. doi: 10.1038/sj.onc.1206686. [DOI] [PubMed] [Google Scholar]
- 17.Huang W, Goldfischer M, Babayeva S, Mao Y, Volyanskyy K, Dimitrova N, Fallon JT, Zhong M. Identification of a novel PARP14-TFE3 gene fusion from 10-year old FFPE tissue by RNA-seq. Genes Chromosomes Cancer. 2015;54:500–505. doi: 10.1002/gcc.22261. [DOI] [PubMed] [Google Scholar]
- 18.Argani P, Zhong M, Reuter VE, Fallon JT, Epstein JI, Netto GJ, Antonescu CR. TFE3-fusion variant analysis defines specific clinicopathologic associations among Xp11 translocation cancers. Am J Surg Pathol. 2016;40:723–737. doi: 10.1097/PAS.0000000000000631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xia QY, Wang XT, Zhan XM, Tan X, Chen H, Liu Y, Shi SS, Wang X, Wei X, Ye SB, Li R, Ma HH, Lu ZF, Zhou XJ, Rao Q. Xp11 translocation renal cell carcinomas (RCCs) with RBM10-TFE3 gene fusion demonstrating melanotic features and overlapping morphology with t(6;11) RCC: interest and diagnostic pitfall in detecting a paracentric inversion of TFE3. Am J Surg Pathol. 2017;41:663–676. doi: 10.1097/PAS.0000000000000837. [DOI] [PubMed] [Google Scholar]
- 20.Malouf GG, Monzon FA, Couturier J, Molinié V, Escudier B, Camparo P, Su X, Yao H, Tamboli P, Lopez-Terrada D, Picken M, Garcia M, Multani AS, Pathak S, Wood CG, Tannir NM. Genomic heterogeneity of translocation renal cell carcinoma. Clin Cancer Res. 2013;19:4673–4684. doi: 10.1158/1078-0432.CCR-12-3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Classe M, Malouf GG, Su X, Yao H, Thompson EJ, Doss DJ, Grégoire V, Lenobin J, Fantoni JC, Sudour-Bonnange H, Khayat D, Aubert S, Tannir NM, Leroy X. Incidence, clinicopathological features and fusion transcript landscape of translocation renal cell carcinomas. Histopathology. 2017;70:1089–1097. doi: 10.1111/his.13167. [DOI] [PubMed] [Google Scholar]
- 22.Pei J, Cooper H, Flieder DB, Talarchek JN, Al-Saleem T, Uzzo RG, Dulaimi E, Patchefsky AS, Testa JR, Wei S. NEAT1-TFE3 and KAT6A-TFE3 renal cell carcinomas, new members of MiT family translocation renal cell carcinoma. Mod Pathol. 2019;32:710–716. doi: 10.1038/s41379-018-0191-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weterman MJ, van Groningen JJ, Jansen A, van Kessel AG. Nuclear localization and transactivating capacities of the papillary renal cell carcinoma-associated TFE3 and PRCC (fusion) proteins. Oncogene. 2000;19:69–74. doi: 10.1038/sj.onc.1203255. [DOI] [PubMed] [Google Scholar]
- 24.Tsuda M, Davis IJ, Argani P, Shukla N, McGill GG, Nagai M, Saito T, Laé M, Fisher DE, Ladanyi M. TFE3 fusions activate MET signaling by transcriptional up-regulation, defining another class of tumors as candidates for therapeutic MET inhibition. Cancer Res. 2007;67:919–929. doi: 10.1158/0008-5472.CAN-06-2855. [DOI] [PubMed] [Google Scholar]
- 25.Argani P, Aulmann S, Illei PB, Netto GJ, Ro J, Cho HY, Dogan S, Ladanyi M, Martignoni G, Goldblum JR, Weiss SW. A distinctive subset of PEComas harbors TFE3 gene fusions. Am J Surg Pathol. 2010;34:1395–1406. doi: 10.1097/PAS.0b013e3181f17ac0. [DOI] [PubMed] [Google Scholar]
- 26.Argani P, Aulmann S, Karanjawala Z, Fraser RB, Ladanyi M, Rodriguez MM. Melanotic Xp11 translocation renal cancers: a distinctive neoplasm with overlapping features of PEComa, carcinoma, and melanoma. Am J Surg Pathol. 2009;33:609–619. doi: 10.1097/PAS.0b013e31818fbdff. [DOI] [PubMed] [Google Scholar]
- 27.Antonescu CR, Le Loarer F, Mosquera JM, Sboner A, Zhang L, Chen CL, Chen HW, Pathan N, Krausz T, Dickson BC, Weinreb I, Rubin MA, Hameed M, Fletcher CD. Novel YAP1-TFE3 fusion defines a distinct subset of epithelioid hemangioendothelioma. Genes Chromosomes Cancer. 2013;52:775–784. doi: 10.1002/gcc.22073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Argani P, Lal P, Hutchinson B, Lui MY, Reuter VE, Ladanyi M. Aberrant nuclear immunoreactivity for TFE3 in neoplasms with TFE3 gene fusions: a sensitive and specific immunohistochemical assay. Am J Surg Pathol. 2003;27:750–761. doi: 10.1097/00000478-200306000-00005. [DOI] [PubMed] [Google Scholar]
- 29.Yang B, Duan H, Cao W, Guo Y, Liu Y, Sun L, Zhang J, Sun Y, Ma Y. Xp11 translocation renal cell carcinoma and clear cell renal cell carcinoma with TFE3 strong positive immunostaining: morphology, immunohistochemistry, and FISH analysis. Mod Pathol. 2019;32:1521–1535. doi: 10.1038/s41379-019-0283-z. [DOI] [PubMed] [Google Scholar]
- 30.Wang XT, Xia QY, Ye SB, Wang X, Li R, Fang R, Shi SS, Zhang RS, Tan X, Chen JY, Sun K, Teng XD, Ma HH, Lu ZF, Zhou XJ, Rao Q. RNA sequencing of Xp11 translocation-associated cancers reveals novel gene fusions and distinctive clinicopathologic correlations. Mod Pathol. 2018;3:1346–1360. doi: 10.1038/s41379-018-0051-5. [DOI] [PubMed] [Google Scholar]
- 31.Hodge JC, Pearce KE, Wang X, Wiktor AE, Oliveira AM, Greipp PT. Molecular cytogenetic analysis for TFE3 rearrangement in Xp11.2 renal cell carcinoma and alveolar soft part sarcoma: validation and clinical experience with 75 cases. Mod Pathol. 2014;27:113–127. doi: 10.1038/modpathol.2013.83. [DOI] [PubMed] [Google Scholar]
- 32.Maher ER. Von Hippel-Lindau disease. Eur J Cancer. 1994;30:1987–1990. doi: 10.1016/0959-8049(94)00391-h. [DOI] [PubMed] [Google Scholar]
- 33.Gossage L, Eisen T, Maher ER. VHL, the story of a tumour suppressor gene. Nat Rev Cancer. 2015;15:55–64. doi: 10.1038/nrc3844. [DOI] [PubMed] [Google Scholar]
- 34.Butman JA, Linehan WM, Lonser RR. Neurologic manifestations of von Hippel-Lindau disease. JAMA. 2008;300:1334–1342. doi: 10.1001/jama.300.11.1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Latif F, Tory K, Gnarra J, Yao M, Duh FM, Orcutt ML, Stackhouse T, Kuzmin I, Modi W, Geil L. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science. 1993;260:1317–1320. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- 36.Cassol C, Mete O. Endocrine manifestations of von Hippel Lindau disease. Arch Pathol Lab Med. 2015;139:263–268. doi: 10.5858/arpa.2013-0520-RS. [DOI] [PubMed] [Google Scholar]
- 37.Kanno H, Kondo K, Ito S, Yamamoto I, Fujii S, Torigoe S, Sakai N, Hosaka M, Shuin T, Yao M. Somatic mutations of the von Hippel-Lindau tumor suppressor gene in sporadic central nervous system hemangioblastomas. Cancer Res. 1994;54:4845–4847. [PubMed] [Google Scholar]
- 38.Muscarella LA, la Torre A, Faienza A, Catapano D, Bisceglia M, D’Angelo V, Parrella P, Coco M, Fini G, Tancredi A, Zelante L, Fazio VM, D’Agruma L. Molecular dissection of the VHL gene in solitary capillary hemangioblastoma of the central nervous system. J Neuropathol Exp Neurol. 2014;73:50–58. doi: 10.1097/NEN.0000000000000024. [DOI] [PubMed] [Google Scholar]
- 39.Shankar GM, Taylor-Weiner A, Lelic N, Jones RT, Kim JC, Francis JM, Abedalthagafi M, Borges LF, Coumans JV, Curry WT, Nahed BV, Shin JH, Paek SH, Park SH, Stewart C, Lawrence MS, Cibulskis K, Thorner AR, Van Hummelen P, Stemmer-Rachamimov AO, Batchelor TT, Carter SL, Hoang MP, Santagata S, Louis DN, Barker FG, Meyerson M, Getz G, Brastianos PK, Cahill DP. Sporadic hemangioblastomas are characterized by cryptic VHL inactivation. Acta Neuropathol Commun. 2014;2:167. doi: 10.1186/s40478-014-0167-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slade L, Pulinilkunnil T. The MiTF/TFE family of transcription factors: master regulators of organelle signaling, metabolism, and stress adaptation. Mol Cancer Res. 2017;15:1637–1643. doi: 10.1158/1541-7786.MCR-17-0320. [DOI] [PubMed] [Google Scholar]
- 41.Calcagnì A, Kors L, Verschuren E, De Cegli R, Zampelli N, Nusco E, Confalonieri S, Bertalot G, Pece S, Settembre C, Malouf GG, Leemans JC, de Heer E, Salvatore M, Peters DJ, Di Fiore PP, Ballabio A. Modelling TFE renal cell carcinoma in mice reveals a critical role of WNT signaling. Elife. 2016;5 doi: 10.7554/eLife.17047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mathieu J, Detraux D, Kuppers D, Wang Y, Cavanaugh C, Sidhu S, Levy S, Robitaille AM, Ferreccio A, Bottorff T, McAlister A, Somasundaram L, Artoni F, Battle S, Hawkins RD, Moon RT, Ware CB, Paddison PJ, Ruohola-Baker H. Folliculin regulates mTORC1/2 and WNT pathways in early human pluripotency. Nat Commun. 2019;10:632. doi: 10.1038/s41467-018-08020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ploper D, De Robertis EM. The MITF family of transcription factors: role in endolysosomal biogenesis, Wnt signaling, and oncogenesis. Pharmacol Res. 2015;99:36–43. doi: 10.1016/j.phrs.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 44.Camparo P, Vasiliu V, Molinie V, Couturier J, Dykema KJ, Petillo D, Furge KA, Comperat EM, Lae M, Bouvier R, Boccon-Gibod L, Denoux Y, Ferlicot S, Forest E, Fromont G, Hintzy MC, Laghouati M, Sibony M, Tucker ML, Weber N, Teh BT, Vieillefond A. Renal translocation carcinomas: clinicopathologic, immunohistochemical, and gene expression profiling analysis of 31 cases with a review of theliterature. Am J Surg Pathol. 2008;32:656–670. doi: 10.1097/PAS.0b013e3181609914. [DOI] [PubMed] [Google Scholar]
- 45.Mosquera JM, Dal Cin P, Mertz KD, Perner S, Davis IJ, Fisher DE, Rubin MA, Hirsch MS. Validation of a TFE3 break-apart FISH assay for Xp11.2 translocation renal cell carcinomas. Diagn Mol Pathol. 2011;20:129–137. doi: 10.1097/PDM.0b013e31820e9c67. [DOI] [PubMed] [Google Scholar]
- 46.Green WM, Yonescu R, Morsberger L, Morris K, Netto GJ, Epstein JI, Illei PB, Allaf M, Ladanyi M, Griffin CA, Argani P. Utilization of a TFE3 break-apart FISH assay in a renal tumor consultation service. Am J Surg Pathol. 2013;37:1150–1163. doi: 10.1097/PAS.0b013e31828a69ae. [DOI] [PubMed] [Google Scholar]
- 47.Argani P, Zhang L, Reuter VE, Tickoo SK, Antonescu CR. RBM10-TFE3 renal cell carcinoma: a potential diagnostic pitfall due to cryptic intrachromosomal Xp11.2 inversion resulting in false-negative TFE3 FISH. Am J Surg Pathol. 2017;41:655–662. doi: 10.1097/PAS.0000000000000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Just PA, Letourneur F, Pouliquen C, Dome F, Audebourg A, Biquet P, Vidaud M, Terris B, Sibony M, Pasmant E. Identification by FFPE RNA-Seq of a new recurrent inversion leading to RBM10-TFE3 fusion in renal cell carcinoma with subtle TFE3 break-apart FISH pattern. Genes Chromosomes Cancer. 2016;55:541–548. doi: 10.1002/gcc.22356. [DOI] [PubMed] [Google Scholar]
- 49.Wang XT, Xia QY, Zhou XJ, Rao Q. Incidence, clinicopathological features and fusion transcript landscape of translocation renal cell carcinomas. Histopathology. 2017;71:835–836. doi: 10.1111/his.13256. [DOI] [PubMed] [Google Scholar]
- 50.Pan CC, Sung MT, Huang HY, Yeh KT. High chromosomal copy number alterations in Xp11 translocation renal cell carcinomas detected by array comparative genomic hybridization are associated with aggressive behavior. Am J Surg Pathol. 2013;37:1116–1119. doi: 10.1097/PAS.0b013e318293d872. [DOI] [PubMed] [Google Scholar]
- 51.Harrison G, Hemmerich A, Guy C, Perkinson K, Fleming D, McCall S, Cardona D, Zhang X. Overexpression of SOX11 and TFE3 in solid-pseudopapillary neoplasms of the pancreas. Am J Clin Pathol. 2017;149:67–75. doi: 10.1093/ajcp/aqx142. [DOI] [PubMed] [Google Scholar]
- 52.Jiang Y, Xie J, Wang B, Mu Y, Liu P. TFE3 is a diagnostic marker for solid pseudopapillary neoplasms of the pancreas. Hum Pathol. 2018;81:166–175. doi: 10.1016/j.humpath.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 53.Liu Y, Zheng Q, Wang C, Wang J, Ming J, Zhang Y, Li X, Cho WC, Wang L, Li QC, Qiu XS, Wang EH. Granular cell tumors overexpress TFE3 without gene rearrangement: evaluation of immunohistochemistry and break-apart FISH in 45 cases. Oncol Lett. 2019;18:6355–6360. doi: 10.3892/ol.2019.10995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chamberlain BK, McClain CM, Gonzalez RS, Coffin CM, Cates JM. Alveolar soft part sarcoma and granular cell tumor: an immunohistochemical comparison study. Hum Pathol. 2014;45:1039–1044. doi: 10.1016/j.humpath.2013.12.021. [DOI] [PubMed] [Google Scholar]


