Abstract
Objectives: Preeclampsia (PE), a pregnancy-specific disorder characterized by hypertension and a variety of organ failures, currently lacks effective treatments. Urate (hydroxyiso-) hydrolase, pseudogene (URAHP), which is also a long noncoding RNA (lncRNA), has higher expression in PE placentae than in normal controls and therefore acquires an investigation for the specific mechanism of regulation. Methods: Placentae were divided into two groups: those from patients with normal pregnancy (NP) (n = 3) and those from patients with PE (n = 3). Total RNA was extracted from the placentae and differentially expressed lncRNAs and mRNAs in PE and NP were identified by Arraystar Human LncRNA Expression Microarray V4.0 analysis. The microarray data were validated by profiling the noncoding RNA expression of URAHP in NP and PE placental tissues through quantitative real-time PCR (qRT-PCR). Then, we uncover the effect of URAHP on cell proliferation by CCK-8 assay and by 3D colony forming assay. Gene coexpression analysis was conducted to identify mRNAs coexpressed with URAHP. qRT-PCR and western blotting assays were used to measure the expression levels of URAHP and KISS1R in JAR and JET-3 cell lines. Results: A total of 675 differentially expressed lncRNAs (DELRs) [184 upregulated DELRs and 491 downregulated DELRs] and a total of 205 differently expressed genes (DEGs) [56 upregulated mRNAs and 149 downregulated mRNAs] were finally identified between PE and NP samples through high-throughput sequencing analysis. The expression of lncRNA URAHP was increased significantly in the placentae of women with preeclampsia when compared to those with normal pregnancies. The functional assay suggested that the downregulation of URAHP alters the proliferative capacity of JAR/JET-3 cells and that the overexpression of URAHP promotes the proliferation of HTR-8/SVneo cells. We also determined that URAHP and KISS1R are coexpressed. Conclusion: We demonstrated for the first time that the pseudogene URAHP may be associated with PE. The results of this study provide a new target for the comprehensive treatment of preeclampsia.
Keywords: URAHP, pseudogene, preeclampsia, KISS1R
Introduction
Preeclampsia (PE) is a prevalent disease characterized by hypertension (blood pressure ≥ 140/90 mmHg) in previously normotensive women and by proteinuria (≥ 300 mg/24-hour urine collection or random urine protein positive), and it affects approximately 5%-8% of pregnant women worldwide, seriously threatening the health of fetuses and pregnant women [1-3]. The pathogenesis of preeclampsia is complex and accumulating evidence has demonstrated that the abnormal expression of multiple genes, including those encoding miRNAs, circRNAs and lncRNAs, is involved in pregnancies complicated by preeclampsia [4-7].
Long noncoding RNAs (lncRNAs), which are longer than 200 nucleotides in length, are reported to be an important class of gene regulators, both at the transcription and posttranscription levels, and are involved in diverse biological functions, such as cell proliferation, development, motility and death [8-10]. In the past 5 years, many studies have proposed that lncRNAs are aberrantly expressed in many types of diseases and are closely associated with cell apoptosis, migration, and invasion [11,12].
Although preeclampsia is a pathological process that occurs during pregnancy, how the abnormal expression and biological functions of lncRNAs contribute to preeclampsia progression in the placenta remains unknown. In this research, we aimed to investigate the differentially expressed lncRNAs in preeclampsia placentas. Thus, we analyzed the lncRNA profiles in preeclampsia placentas. Our study showed for the first time that aberrant expression of the pseudogene URAHP might contribute to the abnormal condition of trophoblast cells and human choriocarcinoma cell lines.
Urate (hydroxyiso-) hydrolase, pseudogene (URAHP), which encodes a long non-coding RNA (full length 1379 bp), is a pseudogene with several inactivating mutations located on chromosome 16q24.3 based on an alignment of the URAHP sequence (GenBank BC044232) with the genomic sequence (GRCh37) in humans [13]. URAHP participates in uric acid degradation to inert allantoin in humans. First, uric acid is oxidized to 5-hydroxyisourate (HIU) by urate oxidase [14]; second, urate oxidase is hydrolyzed to generate 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline (OHCU) via HIU hydrolase (URAHP). Finally, OHCU is decarboxylated by OHCU decarboxylase to produce (S)-allantoin [15]. LncRNA URAHP is expressed in many normal tissues, including placentae, while little is known regarding whether lncRNA URAHP expression levels are abnormal in preeclampsia.
In this study, we determined that preeclamptic placentas exhibited higher levels of URAHP than controls. Moreover, we also detected the effects of URAHP on trophoblast proliferation and assessed the possible mechanisms that might be involved. Our data suggest that high levels of lncRNA URAHP might promote trophoblast cell proliferation. Therefore, we proposed URAHP as a novel lncRNA molecule that might be associated with the development of preeclampsia, which might provide a new molecular biomarker for the early diagnosis and treatment of preeclampsia.
Materials and methods
Study population and decidual sample collection
Three women with pregnancies complicated by preeclampsia (PE, blood pressure ≥ 160 mmHg or diastolic blood pressure ≥ 110 mmHg, systolic or and/or proteinuria > 3+ protein on dip stick) [16] and three women with normal pregnancies (NP) were recruited from the Department of Obstetrics, First Affiliated Hospital of China Medical University between July 2018 and October 2018. The study was approved by the First Affiliated Hospital of China Medical University Research and Ethics Committees. Informed consent was obtained from all participants before the collection of decidual tissues. All clinical investigations were conducted according to the principles expressed in the Declaration of Helsinki.
Microarray
Total RNA was extracted from placental tissue using TRIzol (Cat. No. 15596-018; Invitrogen; Thermo Fisher Scientific, Inc.). The sample preparation and microarray hybridization were performed according to the manufacturer’s standard protocols with minor modifications (Arraystar Human LncRNA V4.0 analysis) [17].
Cell culture
The human Choriocarcinoma JAR and JET-3 cell lines were purchased from American Type Culture Collection (ATCC), and the human EVT-derived cell line HTR-8/SVneo (obtained from the Department of Cell Biology, China Medical University, China) was seeded in RPMI 1640 medium (Cat. No. 31870-082, Gibco™) supplemented with 10% fetal bovine serum (FBS, Cat. No. 10100147, Gibco™, Thermo Fisher Scientific, Inc., Waltham, MA, USA), 100 U/mL penicillin and 100 µg/mL streptomycin (Cat. No. 15070063, Gibco™). Cells were cultured in a 37°C incubator with 5% CO2.
Small interfering RNA and plasmid construction
For the RNAi-mediated knockdown of URAHP, three different siRNAs against URAHP were provided by Invitrogen. Among these, si-URAHP-1 had the highest transfection efficiency. The target sequence of the si-URAHP was 5’-UUA GUG CCA GCA CCU GCA AAG CUG U-3’. To overexpress URAHP, a plasmid vector expressing the full-length URAHP (1379 bp) was constructed by using In-Fusion kits (Takara Bio. USA, Inc.) and it was named pCDNA-URAHP. Empty vector was used as the control. We constructed the pGM2196-3 × flag-URAHP overexpression lentiviral vector and the pLenti-GFP-si-URAHP knockdown lentiviral vector.
Lentiviral production
HEK-293T cells were transiently transfected with the pLenti-GFP-si-URAHP Lentiviral vector or a negative control vector using the calcium phosphate reagent. Then, the plasmid mixture was transfected into HEK-293T cells when cells reached ~80% confluency in 10 cm dishes. The supernatants were centrifuged to concentrate the viruses after 48 hrs. The virus was used to infect cells in a 12-well plate, and then the cells were selected with puromycin (1.5 μg/mL). Infected cells were identified by real-time PCR analysis.
RNA extraction and real-time PCR analysis
Total RNA from placental tissues was extracted by using TRIzol (Invitrogen, Carlsbad, CA, USA). Then, the human immortal line (HTR-8/SVneo) and human choriocarcinoma cell lines (JAR and JET-3) were grown in a 6-well culture plate to ~70% confluence before total RNA extraction with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Reverse transcriptions was performed with total RNA (500 ng) using a Reverse Transcription Kit (TaKaRa, Shiga, Japan). SYBR green dye (Takara, Shiga, Japan) was used for the amplification of cDNA. The mRNA levels of URAHP and the internal standard β-actin were measured by real-time quantitative PCR in triplicate on an Mx3000PTM Real-Time PCR System by Agilent (Stratagene, La Jolla, CA, USA). The specific primers used for these genes are listed in Table 1.
Table 1.
Gene primers
| Gene | Primers (F: Forward; R: Reverse) | Amplicon size (bp) |
|---|---|---|
| URAHP | F: 5’-AGGTGTCAATGAAGTTAGTGCCA-3’ | 287 |
| R: 5’-ATCTTCCCAGCGAGATGTCC-3’ | ||
| β-actin | F: 5’-GTGGCCGAGGACTTTGATTG-3’ | 73 |
| R: 5’-CCTGTAACAACGCATCTCATATT-3’ |
Western blotting
Cells or placental tissues were washed with cold PBS before being lysed in RIPA (radioimmunoprecipitation assay) lysis buffer containing PI (protease inhibitor) cocktail (Roche, Basel, Switzerland). SDS-PAGE was performed to separate cell proteins, and then the protein was transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). Membranes were probed with specific primary antibodies (KISS1R and an equal loading control β-actin) and then with peroxidase-conjugated secondary antibodies. The bands were visualized by chemiluminescence (ECL, Tanon, Shanghai, China). ImageJ software was used for densitometric analyses of western blots, and the quantification results were normalized to the loading control.
3D colony formation assay
The complete medium was removed from the culture dish, and the cells were washed with 1 × PBS (phosphate-buffered saline). Then, 1 ml of 0.25% trypsin was added to the cells for 1-2 min, and the cells were pipetted well to create a single-cell suspension by adding complete medium. The cells were spun at 200 × g for 5 min resuspended in complete medium; then, the cells were counted and the concentration of cells was adjusted to 30 cells/ml. A total of 100 µl (30 cells/ml) of complete cell medium was added to the ultralow cluster 96-well round bottom culture plate (7007, Costar, Corning Inc., USA) and mixed until homogeneous. After 1, 3 and 7 days of culture, cell morphology was observed by phase-contrast microscopy and fluorescence microscopy.
Statistical analysis
In this study, all statistical analyses were performed using SPSS version 19.0 software (SPSS, Chicago, IL, USA) and GraphPad Prism V6.0 (GraphPad Software, Inc. San Diego, CA, USA). All experiments included at least three independent biological assays (n ≥ 3) and the results are presented as mean ± standard deviation (SD); To assess the statistical significance of the gene expression of URAHP and KISSR in cell lines, Student’s t-test (two tailed) was performed. For clone formation and MTT assays statistical differences between the control groups or overexpression of URAHP groups by using analysis of variance (one-way ANOVA); The results were considered to be significant when the P value was * < 0.05, ** < 0.01 or *** < 0.001. Corresponding significance levels are indicated in the figures.
Results
Upregulation of URAHP in preeclampsia placentas
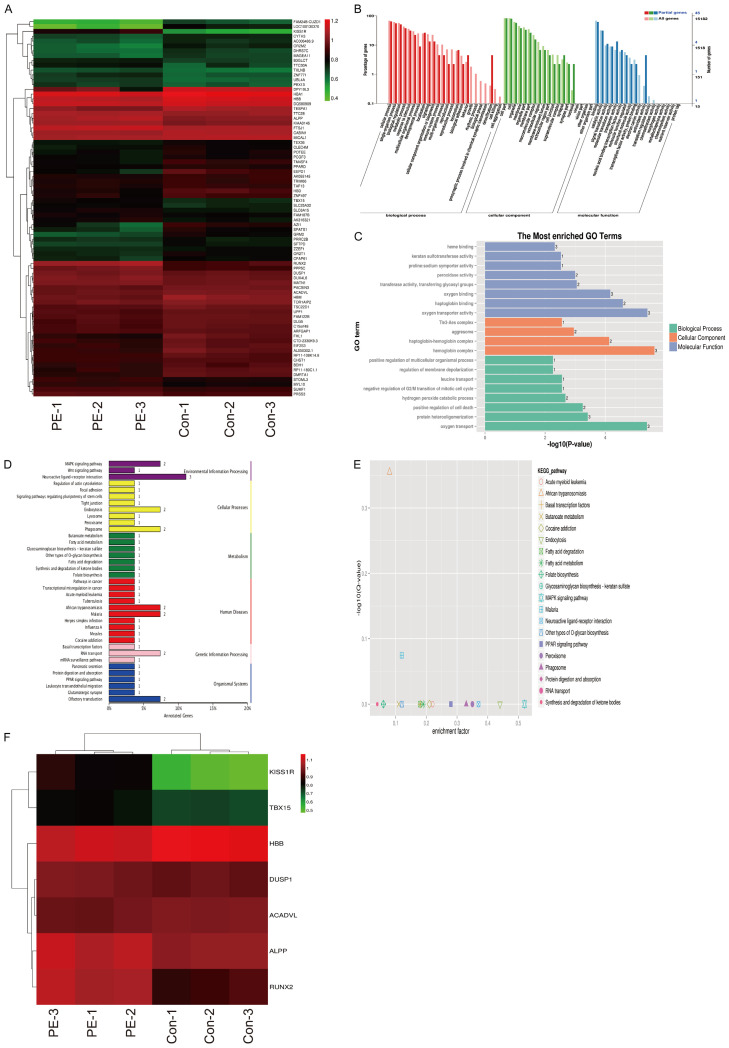
We profiled lncRNA and mRNA expression in placental samples from 3 normal pregnancies (Con) and 3 preeclampsia (PE) patients by Arraystar Human LncRNA V4.0 analysis (Figure 1A and 1B). LncRNA analysis from three pairs of Con and PE placentae samples detected a total of 585 lncRNAs differentially expressed (≥ 1.5-fold-change) in human placentae, with 184 upregulated lncRNAs and 391 downregulated lncRNAs. To recapitulate the different states of the placentae, we heuristically searched for the top 3 dysregulated lncRNAs in the comparisons of PE placentae with Con placentae. We examined the expression levels of URAHP (Figure 1C), CLSTN2-AS1 (Supplementary Figure 1A) and G030771 (Supplementary Figure 1B) in pairs of Con and PE placental samples by qRT-PCR analysis. As presented in Figure 1C, the level of URAHP was approximately 4 times greater in PE placentas than in normal controls (**P < 0.01, Con), whereas the levels of CLSTN2-AS1 and G030771 in PE placentas did not differ from those of normal controls (Con). The trophoblast cell line, HTR-8/SVneo, and two choriocarcinoma cell lines, JAR and JET-3, were surveyed for the presence of URAHP expression. Pseudogene URAHP was more highly expressed in JAR and JET-3 cell lines than in HTR-8/SVneo cell lines (Figure 1D).
Figure 1.
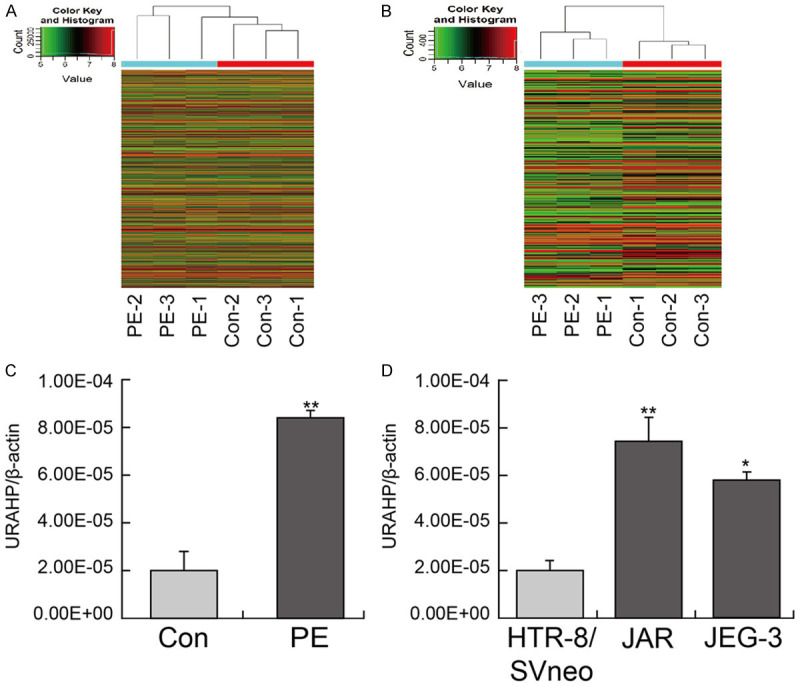
Elevated expression of lncRNA URAHP in preeclampsia placenta tissues. (A and B) Differentially expressed lncRNAs or mRNA in 3 pairs of placental tissues from women with PE and normal pregnancy (Con). (C) The Transcriptional expression of lncRNA URAHP in placental tissues from women with PE and normal pregnancy (Con) examined by real-time PCR, and (D) indicates lncRNA URAHP in HTR-8/SVneo, JAR and JEG-3 cell lines. All the experiments were carried out for three times. *P < 0.05; **P < 0.01. β-actin was used as a loading control.
Genes associated with URAHP identified by gene coexpression network analysis
Weighted gene coexpression network analysis (WGCNA) was used to construct coexpression networks to explore the associations between the URAHP gene and differentially expressed mRNA expression in PE/Con from our microarray data. In this study, genes were defined by module connectivity, measured by the absolute value of Pearson’s correlation (> 0.9) and differentially expressed mRNA (**P < 0.01). Seventy-eight genes were identified in the coexpression networks, and the expression of these genes was highly correlated with URAHP expression. The heatmap and data for the 78 differentially expressed genes are shown in Figure 2A and Table 2. To gain further insight into the function of genes in association with URAHP, we performed Gene Ontology (GO) enrichment analysis for modules by using the online Database for Annotation, Visualization and Integrated Discovery (DAVID; https://david.ncifcrf.gov/summary.jsp). The gene lists of modules were uploaded, and we obtained the results of biological process, cellular component, molecular function (Figure 2B and 2C), and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. A P-value ≤ 0.05 was regarded as significant (Figure 2D and 2E). Based on the current research articles (https://pubmed.ncbi.nlm.nih.gov), 7 genes (KISS1R, ALPP, DUSP1 [18,19], RUNX2 [20], TBX15 [21], HBB [22] and ACADVL [23]) were correlated with PE progression. The gene heatmap was generated (Figure 2F).
Figure 2.
Genes in association with URAHP identified by gene co-expression network analysis. A. Heat map visualized that the differentially expressed genes (DEGs) was associated with URAHP gene in PE/Con from our Microarray data. B. Web Gene Ontology Annotation Plotting (WEGO) analysis of the different comparisons for the transcriptome. The abscissa represents each GO term. The left Y-axis represents the percentage of each term, whereas the right Y-axis represents the genes/proteins corresponding to each GO term. Different colored bars represent different comparisons. C. The Gene Ontology (GO) terms in the DEGs associated with URAHP, the bar plot shows the enrichment scores of the significant enrichment GO terms. D. KEGG pathway analysis of the DEGs associated with URAHP. E. KEGG annotation DEGs associated with URAHP in the midnight blue module. F. Heat map showed that the hub DEGs associated with URAHP were correlated with PE progression.
Table 2.
URAHP related genes
| #ID | regulation | Fold change | Seqname | #ID | regulation | Fold change | Seqname |
|---|---|---|---|---|---|---|---|
| ALPP | up | 3.0989046 | NM_001632 | TEX36 | down | 1.5988173 | NM_001128202 |
| SUMF1 | up | 1.5663929 | NM_182760 | ZZEF1 | down | 1.5372339 | NM_015113 |
| STOML3 | up | 1.8681112 | NM_145286 | CTD-2330K9.3 | down | 2.585386 | ENST00000419183 |
| SLC6A15 | up | 2.0508868 | NM_182767 | TSC22D1 | down | 1.898143 | NM_183422 |
| B3GLCT | up | 1.6177645 | NM_194318 | PRRC2B | down | 1.5444275 | NM_013318 |
| KIAA0146 | up | 2.6028365 | uc010lxs.3 | TRIM66 | down | 1.5538624 | NM_014818 |
| FAM187B | up | 1.552513 | NM_152481 | MICAL1 | down | 1.6571165 | NM_022765 |
| KISS1R | up | 14.908618 | NM_032551 | AK055145 | down | 1.798836 | uc001vsc.2 |
| DUX4L6 | up | 2.037708 | uc031sie.1 | AZI1 | down | 4.9295394 | ENST00000269392 |
| UBL4A | up | 1.7236341 | NM_014235 | EEPD1 | down | 6.4123984 | NM_030636 |
| TTC30A | up | 2.1445221 | NM_152275 | RP11-108K14.8 | down | 2.0069334 | ENST00000468317 |
| AK316321 | up | 1.8507868 | uc021yhu.1 | FHL1 | down | 4.9840823 | NM_001159702 |
| PRSS3 | up | 1.5635353 | NM_002771 | FAM24B-CUZD1 | down | 3.1734195 | uc010qty.2 |
| DUSP1 | up | 1.614964 | NM_004417 | OR2T1 | down | 1.8854301 | NM_030904 |
| RUNX2 | up | 11.940299 | NM_001024630 | HBB | down | 3.1732027 | NM_000518 |
| MYL10 | up | 1.6647977 | NM_138403 | TAF13 | down | 1.5973 | NM_005645 |
| ZNF771 | up | 1.8384515 | NM_016643 | AL050302.1 | down | 3.5609417 | ENST00000540061 |
| TTC28 | up | 1.5004858 | NM_001145418 | ZNF497 | down | 2.2710302 | NM_198458 |
| PEX13 | up | 1.536914 | NM_002618 | OR2M2 | down | 2.9343417 | NM_001004688 |
| TBX15 | up | 2.1599624 | NM_152380 | CLEC4M | down | 1.7559813 | NM_014257 |
| TXLNB | up | 1.5952302 | NM_153235 | TESPA1 | down | 3.3768755 | NM_001098815 |
| FTSJ1 | up | 2.4551991 | NM_012280 | TM4SF4 | down | 3.0101494 | NM_004617 |
| PPP5C | up | 4.0733685 | NM_006247 | DQ580909 | down | 1.5887699 | uc003wuy.1 |
| SLC25A32 | up | 1.6817792 | NM_030780 | AC006486.9 | down | 1.8049138 | ENST00000594664 |
| MATN1 | up | 1.5542979 | NM_002379 | LOC100130370 | down | 9.1540149 | NM_001272086 |
| HBM | down | 2.9375454 | NM_001003938 | ACADVL | down | 1.6224552 | NM_000018 |
| BDH1 | down | 2.1354939 | NM_004051 | POTEE | down | 1.8094717 | NM_001083538 |
| EIF2S3 | down | 3.1844454 | NM_001415 | DLG5 | down | 2.0103653 | NM_004747 |
| RP11-180C1.1 | down | 2.7730187 | ENST00000507759 | CHST1 | down | 1.7205269 | NM_003654 |
| HBA1 | down | 2.2221738 | NM_000558 | FAM122B | down | 1.7496979 | NM_001166599 |
| PACSIN3 | down | 1.5214466 | NM_016223 | ARFGAP1 | down | 1.5236505 | NM_018209 |
| GRM2 | down | 3.5268663 | NM_000839 | PPARD | down | 1.6728892 | NM_006238 |
| C15orf48 | down | 1.704554 | NM_032413 | MAGEA11 | down | 1.5433937 | NM_005366 |
| DPY19L3 | down | 33.545205 | NM_207325 | DMRTA1 | down | 3.3841163 | NM_022160 |
| UPF1 | down | 1.6402329 | NM_001297549 | TOR1AIP2 | down | 1.8264003 | NM_022347 |
| CABIN1 | down | 1.7659698 | NM_012295 | CFAP61 | down | 1.7162132 | NM_015585 |
| CYTH3 | down | 1.733661 | NM_004227 | SPATS1 | down | 2.9702945 | NM_145026 |
| DHRS7C | down | 1.6933636 | NM_001220493 | PCGF3 | down | 1.7342107 | NM_006315 |
| HBD | down | 3.7968126 | NM_000519 | SFTPD | down | 1.9090047 | NM_003019 |
URAHP promotes trophoblast proliferation
To explore the functional role of URAHP in PE pathogenesis, we manipulated its expression by shRNA targeting URAHP (shRNA-URAHP) and the URAHP overexpression vector (OE-URAHP). Then, to further explore the biological function of URAHP in trophoblasts, we first transfected HTR-8/SVneo cells with OE-URAHP or empty vector (Con), and URAHP mRNA expression was measured by qRT-PCR. The results revealed that the expression of URAHP was remarkably elevated in cells transfected with OE-URAHP compared with the Con (Figure 3A). CCK-8 and 3D colony formation assay showed that URAHP overexpression promoted cell growth and increased the colony number in the OE-URAHP groups compared with the control groups (Figure 3B and 3C).
Figure 3.
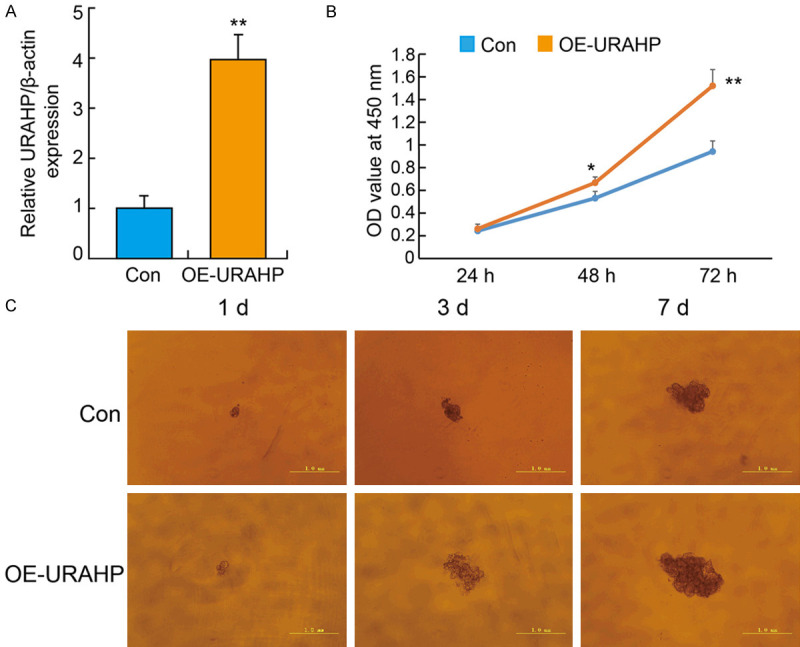
Overexpression of URAHP affect the proliferation of HTR8/SVneo cells. A. The HTR/SVneo cells were transfected with OE-URAHP or Con lentivirus, and qPCR results of URAHP expression in cells. Data are shown as means ± SD. Independent experiments were repeated in triplicate. **P < 0.01 (OE-URAHP versus Con group). B. Transfected cells were subjected to CCK-8 assays, and the changes in cell number at 24, 48, and 72 h were measured. C. Representative images of OE-URAHP or Control in HTR8/SVneo cells were detected by 3D culture at 1 d, 3 d and 7 d. Scale bar, 1.0 mm.
Knockdown of URAHP suppresses choriocarcinoma cell proliferation
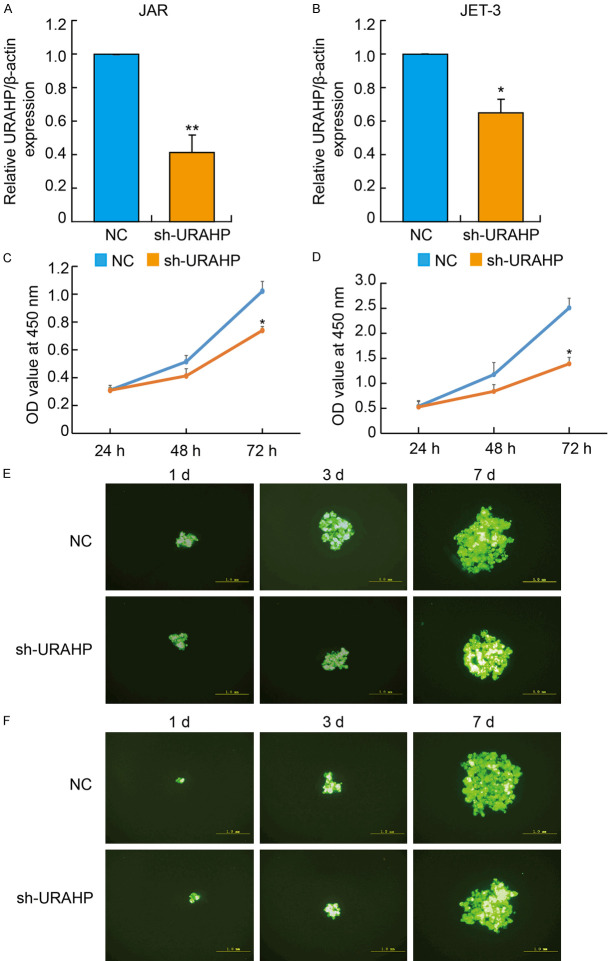
To further elucidate the function of URAHP in vitro, we used RNA interference technology to downregulate the level of URAHP. Then, we transfected interfering URAHP (shRNA-URAHP) into JAR and JET-3 cells to obtain URAHP knockdown cells. The qRT-PCR results showed that shRNA-URAHP significantly repressed URAHP expression compared with that in the control (NC) (Figure 4A and 4B). CCK-8 assays revealed that knockdown of URAHP markedly suppressed the cell proliferative capacity of JAR and JET-3 cells (Figure 4C and 4D). Meanwhile, the 3D colony size in JAR and JET-3 cells transfected with shRNA-URAHP was significantly smaller than that in the NC group, which is consistent with the results above (Figure 4E and 4F).
Figure 4.
Knockdown of URAHP suppressed trophoblasts to proliferate. Real-time PCR assay showed transfection efficacy of sh-URAHP in JAR (A) and JET-3 (B) cells. (C and D) CCK-8 assay showed the viability in JAR (C) and JET-3 (D) cells transfected sh-URAHP at 24 hr, 48 hr and 72 hr. *P < 0.05. 3D culture assay showed the colonies in JAR (E) and JET-3 (F) cells transfected sh-URAHP. NC, negative control.
KISS1R is a coexpressed gene of URAHP in preeclampsia placentas
A total of 205 differentially expressed genes (DEGs), 56 upregulated mRNAs and 149 downregulated mRNAs were finally identified. Among all of the genes in the coexpression network, only KISS1R, RUNX2, ALPP and PPP5C were upregulated (Fold Change > 3, P-value < 0.05). First, qPCR and western blotting revealed that KISS1R was highly expressed in preeclampsia placentas (Figure 5A and 5B). URAHP overexpression significantly increased KISS1R mRNA and protein expression in JAR and JET-3 cell lines (Figure 5C and 5F). All the original western blotting data are attached in Supplementary Figure 2.
Figure 5.
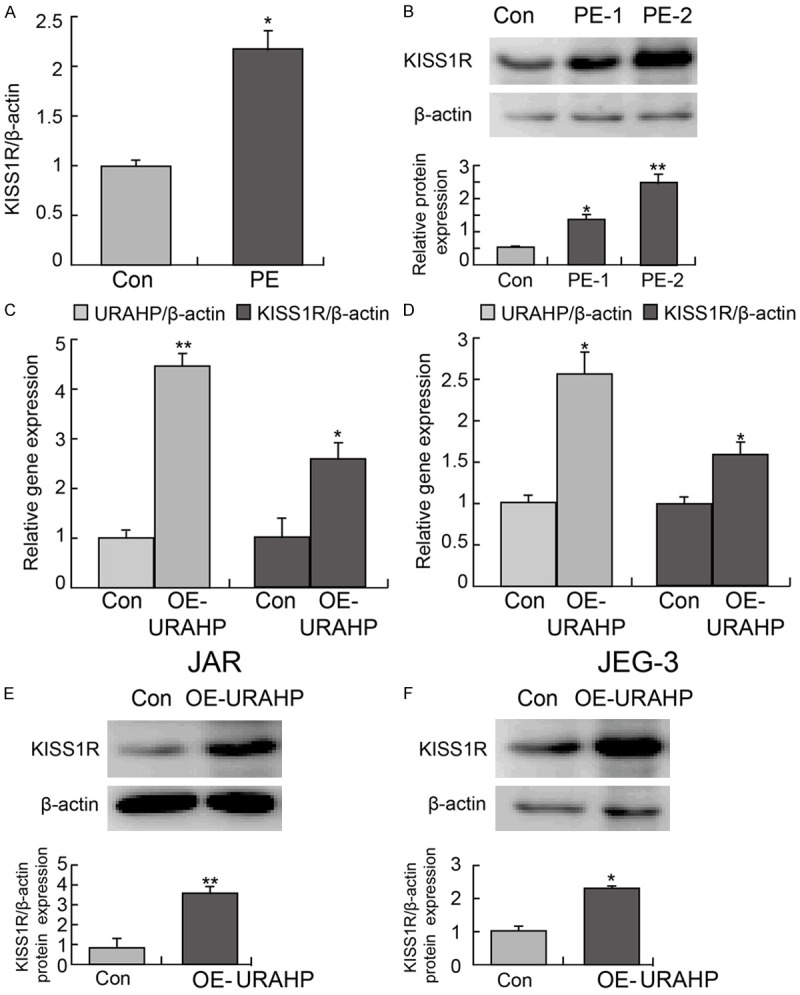
URAHP regulated KISS1R level. A. Relative level of KISS1R in PE pregnancies and normal pregnancies by real-time PCR assay. B. Protein levels of KISS1R in PE pregnancies and normal pregnancies by western blotting. C and D. Gene expression of URAHP and KISS1R in transfected JAR and JET-3 cells. *P < 0.05 and **P < 0.01. E and F. KISS1R expression in transfected OE-URAHP or Con JAR and JET-3 cells examined by western blotting assay. β-actin was used as a loading control.
Discussion
Pseudogenes, which are fairly common (~0.7% of DNA sequence) in the human genome, were regarded as nonfunctional genomic fossils for a long time [24]. However, recent experimental data indicated that many pseudogenes have important genetic functions [25]. Studies have found that pseudogenes also matter in the occurrence and development of PE. For instance, phosphoglycerate kinase 1, pseudogene 2 (PGK1P2) expression is correlated with abnormal decidualization and might lead to the occurrence of PE. However, the related mechanisms and the roles of abnormally expressed pseudogenes in PE have not been functionally characterized to date. Therefore, the identification of key pseudogenes associated with PE is critical to clarifying this disease and identifying novel therapeutic targets.
The focus of this study was to characterize differentially expressed lncRNA genes between normal pregnancy placentae and preeclampsia placentae by lncRNA expression microarray data analysis and to verify the function of differentially expressed genes in vitro, including URAHP by qRT-PCR analysis. We found that URAHP was abnormally highly expressed in preeclampsia placentae and human choriocarcinoma cell lines (JAR and JET-3). These results indicated that URAHP may play crucial roles in increasing the risk for preeclampsia occurrence or tumorigenesis. Subsequently, we predicted the biological roles of URAHP and showed that URAHP has a close connection with preeclampsia occurrence and cancer proliferation.
URAHP, as a pseudogene, had significantly increased expression in the placentae of women with preeclampsia. The functional assay suggests that the downregulation of URAHP alters the proliferative capacity of JAR/JET-3 cells and that the overexpression of URAHP promotes the proliferation of HTR-8/SVneo cells. Pseudogenes can interact with parental genes or other gene loci, leading to alterations in their sequences and/or transcriptional activities. Growth arrest specific 8 (Gas8), a microtubule-associated subunit of the dynein regulatory complex (DRC), is the parental gene of URAHP [26,27]. However, we did not reveal the regulatory relationship between URAHP and GAS8 in vitro (data not shown). The downstream target regulator of URAHP is still not clear in preeclampsia. Interestingly, the URAHP gene was consistently coexpressed with KISS1R [28,29], which is associated with the development of preeclampsia, upon URAHP knockdown or heterologous expression in vitro.
This study has several shortcomings. First, the downstream target genes of URAHP need to be screened and validated to better clarify the mechanism by which URAHP promotes cancer progression. Second, URAHP is coexpressed with KISS1R, so we should verify the regulatory relationship between URAHP and KISS1R in PE.
In conclusion, our results suggest that the upregulated expression of lncRNA URAHP might be a key factor in monitoring the biological function of trophoblast cells, which might provide a theoretical basis and therapeutic targets for the treatment of PE.
Acknowledgements
This work was supported by the Project of Natural Science Foundation of Liaoning Province (20180530037), and by grants from the National Natural Science Foundation of China (No. 81902701).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Zheng L, Huang J, Su Y, Wang F, Kong H, Xin H. Overexpression of tissue factor pathway inhibitor 2 attenuates trophoblast proliferation and invasion in preeclampsia. Hum Cell. 2020;33:512–520. doi: 10.1007/s13577-020-00322-0. [DOI] [PubMed] [Google Scholar]
- 2.Xu Y, Xia X, Jiang Y, Wu D, Wang S, Fu S, Yang N, Zhang Y, Sun L. Down-regulated lncRNA AGAP2-AS1 contributes to pre-eclampsia as a competing endogenous RNA for JDP2 by impairing trophoblastic phenotype. J Cell Mol Med. 2020;24:4557–4568. doi: 10.1111/jcmm.15113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang L, Cao Z, Feng F, Xu YN, Li L, Gao H. A maternal GOT1 novel variant associated with early-onset severe preeclampsia identified by whole-exome sequencing. BMC Med Genet. 2020;21:49. doi: 10.1186/s12881-020-0989-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xueya Z, Yamei L, Sha C, Dan C, Hong S, Xingyu Y, Weiwei C. Exosomal encapsulation of miR-125a-5p inhibited trophoblast cell migration and proliferation by regulating the expression of VEGFA in preeclampsia. Biochem Biophys Res Commun. 2020;525:646–653. doi: 10.1016/j.bbrc.2020.02.137. [DOI] [PubMed] [Google Scholar]
- 5.Jelena M, Sopić M, Joksić I, Zmrzljak UP, Karadžov-Orlić N, Košir R, Egić A, Miković Ž, Ninić A, Spasojević-Kalimanovska V. Placenta-specific plasma miR518b is a potential biomarker for preeclampsia. Clin Biochem. 2020;79:28–33. doi: 10.1016/j.clinbiochem.2020.02.012. [DOI] [PubMed] [Google Scholar]
- 6.Zhou B, Zhang X, Li T, Xie R, Zhou J, Luo Y, Yang C. CircZDHHC20 represses the proliferation, migration and invasion in trophoblast cells by miR-144/GRHL2 axis. Cancer Cell Int. 2020;20:19. doi: 10.1186/s12935-020-1097-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang X, Ning Q. Long noncoding RNAs as novel players in the pathogenesis of hypertension. Hypertens Res. 2020;43:597–608. doi: 10.1038/s41440-020-0408-2. [DOI] [PubMed] [Google Scholar]
- 8.Zhu H, Kong L. LncRNA CRNDE regulates trophoblast cell proliferation, invasion, and migration via modulating miR-1277. Am J Transl Res. 2019;11:5905–5918. [PMC free article] [PubMed] [Google Scholar]
- 9.Kamrani A, Alipourfard I, Ahmadi-Khiavi H, Yousefi M, Rostamzadeh D, Izadi M, Ahmadi M. The role of epigenetic changes in preeclampsia. Biofactors. 2019;45:712–724. doi: 10.1002/biof.1542. [DOI] [PubMed] [Google Scholar]
- 10.He T, Qiao Y, Lv Y, Wang J, Hu R, Cao Y. lncRNA FAM99A is downregulated in preeclampsia and exerts a regulatory effect on trophoblast cell invasion, migration and apoptosis. Mol Med Rep. 2019;20:1451–1458. doi: 10.3892/mmr.2019.10350. [DOI] [PubMed] [Google Scholar]
- 11.Feng Q, Zhang H, Yao D, Chen WD, Wang YD. Emerging role of non-coding RNAs in esophageal squamous cell carcinoma. Int J Mol Sci. 2019;21:258. doi: 10.3390/ijms21010258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei L, Wang X, Lv L, Liu J, Xing H, Song Y, Xie M, Lei T, Zhang N, Yang M. The emerging role of microRNAs and long noncoding RNAs in drug resistance of hepatocellular carcinoma. Mol Cancer. 2019;18:147. doi: 10.1186/s12943-019-1086-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramazzina I, Folli C, Secchi A, Berni R, Percudani R. Completing the uric acid degradation pathway through phylogenetic comparison of whole genomes. Nat Chem Biol. 2006;2:144–148. doi: 10.1038/nchembio768. [DOI] [PubMed] [Google Scholar]
- 14.Yamauchi K, Kasai K. Sequential molecular events of functional trade-offs in 5-hydroxyisourate hydrolase before and after gene duplication led to the evolution of transthyretin during chordate diversification. J Mol Evol. 2018;86:457–469. doi: 10.1007/s00239-018-9858-4. [DOI] [PubMed] [Google Scholar]
- 15.French JB, Ealick SE. Structural and mechanistic studies on Klebsiella pneumoniae 2-Oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase. J Biol Chem. 2010;285:35446–35454. doi: 10.1074/jbc.M110.156034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tong J, Yang J, Lv H, Lv S, Zhang C, Chen ZJ. Dysfunction of pseudogene PGK1P2 is involved in preeclampsia by acting as a competing endogenous RNA of PGK1. Pregnancy Hypertens. 2018;13:37–45. doi: 10.1016/j.preghy.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 17.Yang L, Li P, Yang W, Ruan X, Kiesewetter K, Zhu J, Cao H. Integrative transcriptome analyses of metabolic responses in mice define pivotal LncRNA metabolic regulators. Cell Metab. 2016;24:627–639. doi: 10.1016/j.cmet.2016.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo S, Cao N, Tang Y, Gu W. Identification of key microRNAs and genes in preeclampsia by bioinformatics analysis. PLoS One. 2017;12:e0178549. doi: 10.1371/journal.pone.0178549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bach MK, Brashler JR, White GJ, Galli SJ. Experiments on the mode of action of piriprost (U-60,257), an inhibitor of leukotriene formation in cloned mouse mast cells and in rat basophil leukemia cells. Biochem Pharmacol. 1987;36:1461–1466. doi: 10.1016/0006-2952(87)90111-0. [DOI] [PubMed] [Google Scholar]
- 20.Martinez-Fierro ML, Carrillo-Arriaga JG, Luevano M, Lugo-Trampe A, Delgado-Enciso I, Rodriguez-Sanchez IP, Garza-Veloz I. Serum levels of miR-628-3p and miR-628-5p during the early pregnancy are increased in women who subsequently develop preeclampsia. Pregnancy Hypertens. 2019;16:120–125. doi: 10.1016/j.preghy.2019.03.012. [DOI] [PubMed] [Google Scholar]
- 21.Chelbi ST, Doridot L, Mondon F, Dussour C, Rebourcet R, Busato F, Gascoin-Lachambre G, Barbaux S, Rigourd V, Mignot TM, Tost J, Vaiman D. Combination of promoter hypomethylation and PDX1 overexpression leads to TBX15 decrease in vascular IUGR placentas. Epigenetics. 2011;6:247–255. doi: 10.4161/epi.6.2.13791. [DOI] [PubMed] [Google Scholar]
- 22.AbdelHalim RM, Ramadan DI, Zeyada R, Nasr AS, Mandour IA. Circulating maternal total cell-free DNA, cell-free fetal DNA and soluble endoglin levels in preeclampsia: predictors of adverse fetal outcome? A cohort study. Mol Diagn Ther. 2016;20:135–149. doi: 10.1007/s40291-015-0184-x. [DOI] [PubMed] [Google Scholar]
- 23.Shin EK, Kang HY, Yang H, Jung EM, Jeung EB. The regulation of fatty acid oxidation in human preeclampsia. Reprod Sci. 2016;23:1422–1433. doi: 10.1177/1933719116641759. [DOI] [PubMed] [Google Scholar]
- 24.Zheng LL, Zhou KR, Liu S, Zhang DY, Wang ZL, Chen ZR, Yang JH, Qu LH. dreamBase: DNA modification, RNA regulation and protein binding of expressed pseudogenes in human health and disease. Nucleic Acids Res. 2018;46:D85–D91. doi: 10.1093/nar/gkx972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen X, Wan L, Wang W, Xi WJ, Yang AG, Wang T. Re-recognition of pseudogenes: from molecular to clinical applications. Theranostics. 2020;10:1479–1499. doi: 10.7150/thno.40659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patoughi M, Ghafouri-Fard S, Arsang-Jang S, Taheri M. GAS8 and its naturally occurring antisense RNA as biomarkers in multiple sclerosis. Immunobiology. 2019;224:560–564. doi: 10.1016/j.imbio.2019.04.005. [DOI] [PubMed] [Google Scholar]
- 27.Evron T, Philipp M, Lu J, Meloni AR, Burkhalter M, Chen W, Caron MG. Growth arrest specific 8 (Gas8) and G protein-coupled receptor kinase 2 (GRK2) cooperate in the control of Smoothened signaling. J Biol Chem. 2011;286:27676–27686. doi: 10.1074/jbc.M111.234666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kapustin RV, Drobintseva AO, Alekseenkova EN, Onopriychuk AR, Arzhanova ON, Polyakova VO, Kvetnoy IM. Placental protein expression of kisspeptin-1 (KISS1) and the kisspeptin-1 receptor (KISS1R) in pregnancy complicated by diabetes mellitus or preeclampsia. Arch Gynecol Obstet. 2020;301:437–445. doi: 10.1007/s00404-019-05408-1. [DOI] [PubMed] [Google Scholar]
- 29.Qiao C, Wang C, Zhao J, Liu C, Shang T. Elevated expression of KiSS-1 in placenta of Chinese women with early-onset preeclampsia. PLoS One. 2012;7:e48937. doi: 10.1371/journal.pone.0048937. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.