Abstract
Methylation profiling is widely used to study tumor biology and perform cluster analysis, particularly in brain cancer research where tissue biopsies are scarce. We have recently reported on the development of novel mouse models for germ line mutations in pineoblastoma (Nature Communications, 2020). Here, we present unpublished methylation profiling of 8 Rb-deleted/p53-deleted pineoblastoma from our mouse model as well as 3 normal cerebellum tissues as control. The primary dataset can be accessed via SRA (PRJNA638504). These methylation data can be used to perform inter- and intra-species comparisons with other brain cancers as well as with specific subtypes of pineoblastoma, and to investigate potential epigenetic mechanisms and pathways underlying Rb-deficient pineoblastoma-genesis..
Keywords: Pineoblastoma, Methylation sequencing, Mouse models, Rb, p53
Specifications Table
| Subject | Cancer Research |
| Specific subject area | Pediatric brain tumor, mouse pineoblastoma |
| Type of data | Methylation sequencing read (raw) and Figures |
| How data were acquired | Reduced Representation Bisulfite Sequencing using Illumina HiSeq 2500 |
| Data format | Raw |
| Parameters for data collection | Methylation sequencing was performed on genomic DNA collected from mouse Rb/p53-deficient pineoblastoma and normal age-matched mouse cerebellum |
| Description of data collection | Rb-deficient pineoblastoma were collected from WAP-Cre:Rbf/f:p53f/f mice and normal cerebellum samples.. Genomic DNA was extracted using DNeasy Blood & Tissue Kit and was subjected to Methyl-MiniSeq methylation sequencing (Zymo Research, California, USA) |
| Data source location | University Health Network Toronto, Ontario Canada |
| Data accessibility | Repository name: Sequence Read Archive Data identification number: PRJNA638504 Direct URL to data: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA638504 |
| Related research article | Philip E. D. Chung, Deena M. A. Gendoo, Ronak Ghanbari-Azarnier, Jeff C. Liu, Zhe Jiang, Jennifer Tsui, Dong-Yu Wang, Xiao, Bryan Li, Adrian Dubuc, David Shih, Marc Remke, Ben Ho, Livia Garzia, Yaacov Ben-David, Seok-Gu Kang, Sidney Croul, Benjamin Haibe-Kains, Annie Huang, Michael D. Taylor & Eldad Zacksenhaus. Modeling germline mutations in pineoblastoma uncovers lysosome disruption-based therapy. Nature Communications. 11, 1825 (2020).https://doi.org/10.1038/s41467–020–15585–2 |
Value of the Data
-
•
Pineoblastoma is a rare but lethal disease. We have generated mouse models for germ line mutations that drive pineoblastoma and performed RNA-based cluster analysis. Here we provide unpublished DNA methylation data from a mouse model of RB-deficient pineoblastoma.
-
•
This methylation dataset can benefit investigators of brain cancer, pineoblastoma, Rb and p53 tumor suppressors, and metastatic dissemination.
-
•
The methylation dataset can be further used for clustering analysis, discovering de novo epigenetic changes, investigating known methylation sites, pathway analysis and therapeutic development.
1. Data Description
1.1. Methylation profiling of a mouse model of RB-deficient pineoblastoma
Pineoblastoma (PB) is a pediatric brain cancer of the pineal gland with several distinct subtypes, including germline mutations in the tumor suppressors RB or Dicer [1]. We have recently established mouse models for germline PB by disrupting Rb (or Dicer1) together with p53 via the WAP-Cre delete line [2]. Through transcriptomic profiling of brain tumors, we clustered the Rb/p53-deficient mouse model of PB close to human PB as well as to Group 3 and Group 4 MBs, but relatively far from GBM [2], as is the case in human brain tumors [3,4]. We have also generated methylation profiling by Methyl-MiniSeq (Zymo Research) sequencing of Rb/p53-deficient mouse pineoblastoma and normal cerebellum. The raw dataset was deposited in the NCBI Sequence Read Archive under PRJNA638504.
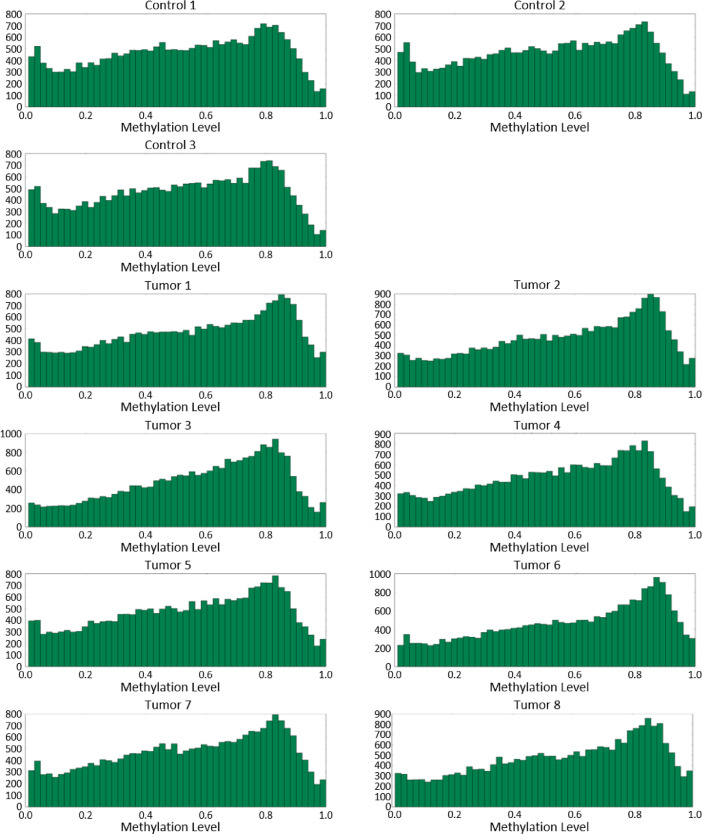
Fig. 1 shows overall methylation levels of 8 Rb-deleted/p53-deleted pineoblastoma and 3 controls.
Fig. 1.
Overall methylation level profiles per chromosome in 8 Rb-deficient pineoblastomas and 3 cerebellum controls.
Figs. 2, Fig. 3, Fig. 4 show CpG island methylation levels, CpG gene methylation levels, and CpG promoter methylation levels, respectively.
Fig. 2.
A histogram summarizing the number of sites of CpG at each methylation ratio level in the context of CpG islands.
Fig. 3.
A histogram summarizing the number of sites of CpG at each methylation ratio level in the context of gene bodies.
Fig. 4.
A histogram summarizing the number of sites of CpG at each methylation ratio level in the context of gene promoter.
2. Experimental design, materials and methods
2.1. Methyl-MiniSeq methylation sequencing
Tumor and tissue samples were collected from 8 Rb-deleted/p53-deleted pineoblastoma and 3 normal cerebellum as control. Genomic DNA was extracted using DNeasy Blood & Tissue Kit (Qiagen) according to manufacturer's instruction. DNA samples (50 – 200 ng/µl in 40 µl) were subjected to Methyl-MiniSeq methylation sequencing (Zymo Research (California, USA)). 200 – 500 ng of genomic DNA was used for library construction and sequencing. Genomic DNA was fragmented using TaqαI and MspI, ligated to adapters containing 5′-methyl-cytosine, bisulfite-treated using EZ DNA Methylation-Lightning Kit (Zymo Research), and PCR amplified for sequencing on Illumina HiSeq 2500. Sequence reads were identified using Illumina base-calling software and aligned using Zymo Research proprietary analysis pipeline.
Ethics statement
All mouse experiments were performed in accordance with the current Canadian Animal Care Council guide for the care and use of laboratory animals and were approved by the Toronto General Research Institute Animal Research Committee, UHN.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships, which have, or could be perceived to have influenced the work reported in this article.
Acknowledgments
This work was supported by a grant from the Canadian Institute of Health Research (CIHR) to EZ.
Contributor Information
Philip E.D. Chung, Email: eu.chung@mail.utoronto.ca.
Eldad Zacksenhaus, Email: eldad.zacksenhaus@utoronto.ca.
References
- 1.Li B.K. Pineoblastoma segregates into molecular sub-groups with distinct clinico-pathologic features: a rare brain tumor consortium registry study. Acta Neuropathol. 2019;139:223–241. doi: 10.1007/s00401-019-02111-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chung P.E.D. Modeling germline mutations in pineoblastoma uncovers lysosome disruption-based therapy. Nat Commun. 2020;11:1825. doi: 10.1038/s41467-020-15585-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sturm D. New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell. 2016;164:1060–1072. doi: 10.1016/j.cell.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Capper D. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555:469–474. doi: 10.1038/nature26000. [DOI] [PMC free article] [PubMed] [Google Scholar]