Abstract
SLC6A1 is associated with an autosomal dominant early-onset seizure and epileptic encephalopathy associated with intellectual disability. We present a 2-yr-old girl with developmental delay and epilepsy, using a new computational filtering impact score to show the patient's variant ranks with other pathogenic variants. Genomic studies within the patient revealed a G443D variant of uncertain significance. Structural and evolutionary assessments establish this variant as a loss of function to the protein. Compiled metrics through our custom tools on sequence, structure, and protein dynamics combined with PolyPhen-2, PROVEAN, SIFT, and Align-GVGD reveal this variant to rank in the top functional outcome changes relative to gnomAD, TOPMed, and ClinVar variants known to date. The patient was resistant to multiple epileptic drugs, finally finding that valproic acid controls the seizures. This is consistent with additional groups studying SLC6A1 variants within patients.
Keywords: absence seizures, autism, moderate global developmental delay
CASE PRESENTATION
The patient is a 2-yr-old Hispanic child with developmental delay and epilepsy. She did not walk until 20 mo of age. She has speech delays and has been diagnosed with autism spectrum disorder. She had previously been evaluated by physical therapy and speech therapy and was measured to be delayed by ∼3 mo. At ∼19 mo of age, her mother noted recurrent episodes of staring and unresponsiveness. During these events, she would display sudden behavioral arrest and staring, lasting 10–15 sec at a time. Upon resolution, she would immediately return to her baseline. An electroencephalogram (EEG) was obtained and confirmed the presence of absence seizures involving generalized 3-Hz spike-and-wave complexes lasting <10 sec. Clinically, she displayed brief behavioral arrest with upward eye deviation and eyelid flutter.
In light of the clinical presentation and EEG findings, the patient was started on ethosuximide. Her seizures continued despite escalating doses of ethosuximide. Over the next 3 mo, her mother also witnessed multiple falls with her seizures. The patient was transitioned from ethosuximide to zonisamide. Her seizures continued on zonisamide despite escalating doses. A 24-h EEG was obtained at 24 mo of age. This displayed generalized background slowing, rhythmic posterior δ activity, loosely organized generalized 3-Hz spike-and-wave discharges, and numerous absence seizures. This EEG was remarkable for worsening encephalopathy when compared to the tracing at 19 mo of age. Valproic acid was added to the patient's regimen after her long-term video-EEG monitoring study. Her seizure burden became significantly improved after starting valproic acid; however, she had occasional breakthrough seizures in the setting of fatigue and illness. Clobazam was added to her regimen, which provided further seizure control. She was diagnosed to be on the autism spectrum by 30 mo of age, after undergoing a full physical and psychological evaluation at the Autism Spectrum Disorders Assessment Clinic.
TECHNICAL ANALYSIS
The patient was referred to Medical Genetics at 22 mo of age. Neither the maternal nor paternal family history shared her clinical findings (Table 1). A chromosomal microarray was obtained and found to be normal. A comprehensive epilepsy gene panel evaluating 181 known epilepsy-causing genes was obtained (Invitae), with follow-up performed on both parents. Variants were assessed through our previously published sequence-to-structure-to-function workflow (Prokop et al. 2017), comparing the patient variant to all gnomAD, TOPMed, and ClinVar missense variants for SLC6A1. These tools develop a high density conservation and evolutionary selection map of each amino acid and the additive conservation of motifs while also developing a quantitative map of biophysical insights of the protein structure. All variants were assessed with PolyPhen-2, PROVEAN, SIFT, and Align-GVGD. A total of 18 nsec of molecular dynamics simulations (mds) were run on a lipid membrane-embedded SLC6A1 protein model using the AMBER03 force field (Duan et al. 2003). Using mds is a way to track atomic movement based on biophysical parameters, allowing for taking 3D structure qualitative insights and translating them into quantitative assessments of protein folding.
Table 1.
Clinical findings
| Feature | Proband | Mother | Father |
|---|---|---|---|
| Developmental delay | Yes | No | No |
| Epilepsy | Yes | No | No |
| Episodes of staring and unresponsiveness | Yes | No | No |
| Autism | Yes | No | No |
VARIANT INTERPRETATION
Genetic testing revealed six gene mutations as noted on Table 2. One de novo heterozygous variant of uncertain significance (VUS) was present in SLC6A1. Heterozygous variants for GRIN2B, KANSL1, and MBD5 were inherited from the unaffected father, changing their classification from VUS to likely benign. CERS1 and SLC25A22 also had one heterozygous variant each, but these are both autosomal recessive linked disorders, thus classifying the variants as noncausal.
Table 2.
Genomic findings
| Gene | Chromosome | HGVS DNA reference | HGVS protein reference | Variant type | Predicted effect (substitution, deletion, etc.) | dbSNP/dbVar ID | Genotype (heterozygous/ homozygous) | ClinVar ID | Parent of origin | Comments |
|---|---|---|---|---|---|---|---|---|---|---|
| SLC6A1 | 3: 11031181 (GRCh38) | c.1328G > A | p.Gly443Asp | SNV | Missense | N/A | Heterozygous | VCV000845407.1 | De novo | VUS |
| GRIN2B | 12: 13865884 (GRCh38) | c.325G > T | p.Ala109Ser | SNV | Missense | rs772078838 | Heterozygous | VCV000246440.3 | Paternal | Likely benign |
| KANSL1 | 17: 46039111 (GRCh38) | c.2308C > T | p.Arg770Cys | SNV | Missense | N/A | Heterozygous | VCV000833548.1 | Paternal | Likely benign |
| MBD5 | 2: 148469427-148469429 (GRCh38) | c.1484_1486CAA | p.Thr496del | 3-bp microsatellite | deletion | N/A | Heterozygous | VCV000833549.1 | Paternal | Likely benign |
| CERS1 | 19: 18884102 (GRCh38) | c.575C > T | p.Ser192Phe | SNV | Missense | rs1009080328 | Heterozygous | VCV000656194.2 | Not evaluated | Noncausal (recessive gene) |
| SLC25A22 | 11: 792588 (GRCh38) | c.552C > A | p.Ala184= | SNV | Synonymous | rs368807589 | Heterozygous | VCV000506576.2 | Not evaluated | Noncausal (recessive gene) |
(HGVS) Human Genome Variation Society, (SNV) single-nucleotide variation, (N/A) not applicable, (VUS) variant of uncertain significance.
Our top VUS was found within the SLC6A1 gene that codes for the sodium- and chloride-dependent GABA transporter one protein (GAT-1). GAT-1 is important in GABA concentration management in synapsis and has been associated with myoclonic–atonic epilepsy with some variability in epilepsy types, whereas most patients have an intellectual disability with autistic features similar to this patient (Carvill et al. 2015; Johannesen et al. 2018; Mattison et al. 2018). Johannesen et al. also report that the genetic etiology of childhood absence epilepsy can include SLC6A1. This is consistent with our patient's epilepsy phenotype. Similar to the findings from Johannesen et al., we find that valproic acid is an effective drug in seizure control, whereas clobazam has also provided some benefit. This may be related in part to the positive effects that both of these medications have on the GABA system. GAT-1 acts as a voltage-regulated transmembrane transporter that reuptakes GABA from the synaptic cleft by process of a Na+ and Cl− exchange for GABA, with mouse knockouts showing spike-wave discharges (Jin et al. 2011; Carvill et al. 2015). Modulation of GABA by various means has been shown to minimize the myoclonic–atonic seizures (Palmer et al. 2016).
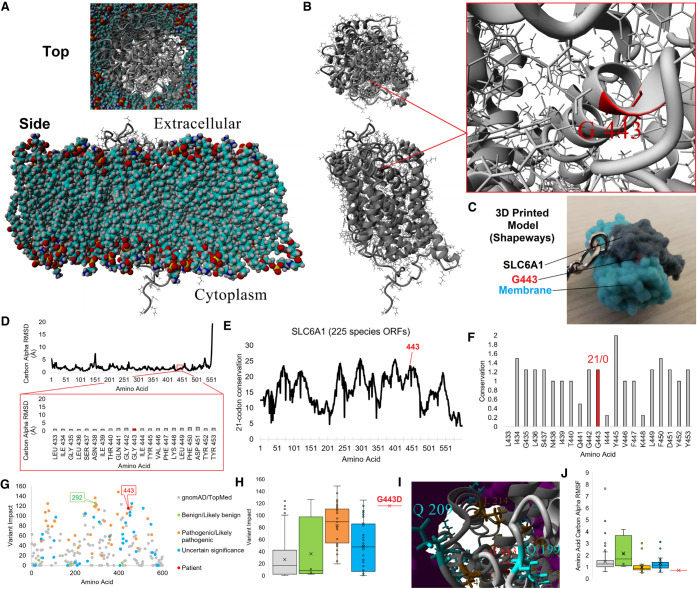
As of early 2020, ClinGen annotates “sufficient” evidence for haploinsufficiency-linked disorder for SLC6A1 with 229 deposited cases of SLC6A1 variants in ClinVar. Of these, 29 missense variants are annotated as pathogenic or likely pathogenic. A total of 67 (29.3%) have been annotated as VUSs similar to the patient presented here, highlighting the need for additional insights of variant linked pathology outside of phenotype matching, linking VUSs to pathogenic variants using bioinformatics. Evolution and structural analysis of the SLC6A1 G443D variant suggests a loss of function to the protein. A protein model for SLC6A1 was generated and embedded into a lipid membrane (Fig. 1A), revealing the G413 site to be found at the base of an α-helix (Fig. 1B,C) that is well-packed within the structure based on mds (Fig. 1D). Alterations of the flexible Gly to a polar acidic residue at this position will disrupt helix formation, disrupting the transmembrane helix packing. A deep evolutionary analysis of 225 species’ open reading frames (ORFs) for SLC6A1 reveals amino acid 443 to fall in a highly conserved region (Fig. 1E) under very high selection (Fig. 1F). The conservation and selection confirm the critical need for the flexible Gly residue at this position. Integrating the conservation for each amino acid with a sliding window of conservation to map important motifs and functional predictions (PolyPhen-2, PROVEAN, SIFT, and Align-GVGD), we developed a numerical value for all known SLC6A1 variants. Variant impact for all gnomAD/TOPMed, ClinVar, and patient variants reveals the G443D to fall near several other ClinVar variants with high variant impact scores including several pathogenic variants (Fig. 1G). The G443D combined impact is one of the highest categorized annotations, ranking 14th out of 254 variants assessed (Fig. 1H) and falls three-dimensionally near L214 and S437 previously associated with disease (Fig. 1I). The mds data supports G443 to be critical in the folding of the protein, with movement far below the average of all other variant groups (Fig. 1J). The structural, evolutionary, and functional bioinformatics can confirm clustering of impactful variants between VUS and pathogenic. This data supports the G443D variant to be a loss-of-function site for SLC6A1, ranking as one of the most impactful VUSs within ClinVar annotations, clustering in score to pathogenic variants. We also noted that one variant annotated as likely benign for intellectual disability, I292T, scores very high with conservation in all species assessed and functional damaging call in all tools used. A second ClinVar inclusion for I292T linked to myoclonic–atonic epilepsy has the variant entered as a VUS.
Figure 1.
SLC6A1 variant analysis. (A,B) Structural model of SLC6A1 (gray) in a lipid membrane (A, multicolored) or alone (B). The top represents a view of the protein from the extracellular surface, whereas the bottom is from a cross section of the membrane. On B, the G443 position is marked and the zoom-in view is provided on the right. (C) 3D printed model of G443 (red) on SLC6A1 (gray) with a cutout of the membrane (cyan) available at www.shapeways.com/product/AAUK2K5JU/slc6a1?li=marketplace&optionId=150795805. (D) Molecular dynamic simulation of the SLC6A1 protein model in A shown for the carbon α root mean squared deviation (RMSD) for each amino acid averaged throughout 18 nsec of simulation. These values provide quantitative values for amino acids well packed (low RMSD) and those found within loops with no atomic hindrance in movement, with most values of SLC6A1 here low, suggestive of a well-folded protein. The bottom red box is a zoom-in view of amino acid 443 region showing high stability of the structure. (E) Deep evolutionary analysis using 225 species open reading frame sequences for SLC6A1. The plot shows a sliding window calculation for each site (plus 10 upstream and downstream), identifying the most selected and conserved linear motifs within the gene. Amino acid 443 is identified in red. (F) Zoom-in view of conservation for amino acid 443 (red) linear motif. The numbers above represent the percent of sequences with synonymous/nonsynonymous variants throughout evolution. (G) Variant impact scoring for all TOPMed/gnomAD (gray), ClinVar (benign or likely benign in green, pathogenic or likely pathogenic in orange, VUS in cyan), and patient (red) variants for SLC6A1. (H) Box and whisker plot for each group plotted in G, with colors consistent within G–J. This shows that the value clustering for each ClinVar annotation was likely pathogenic, pathogenic values are seen elevated over gnomAD/TOPMed and benign annotation, and only a few VUSs score high including G443D in red. (I) Clustered 3D variants on SLC6A1 relative to the patients with the same colors as those labeled in H. The lipid membrane is shown in magenta. (J) Root mean squared fluctuation (RMSF) of variant groups. The values show the average movement of each amino acid throughout biochemical simulations in which benign and gnomAD/TOPMed variants have higher movement and all other groups (excluding a few VUSs and pathogenic variants) are low, suggestive of well-packed amino acids including G443D.
In combination we show the variant is de novo in proband with confirmed lack of presence in parents and paternity confirmed through the presence of other rare variants (PS2) and is absent in controls of gnomAD/TOPMed (PM2), and we have multiple lines of computational evidence (PP3) ranking the variant as likely pathogenic using American College of Medical Genetics and Genomics (ACMG) standards (Richards et al. 2015). In addition, we show that pharmacogenomics aligns our likely pathogenic variant in SLC6A1 with known successful drug response.
SUMMARY
In conclusion, we identify a variant in SLC6A1, G443D, that associates with seizures and developmental delay. Based on assessments to all known variants in SLC6A1, we show that G443 is a far outlier for functional impact and structural fold contribution, which indicates its critical loss-of-function role in SLC6A1. This represents a novel strategy for SLC6A1 assessments for VUSs that can be used in future screening projects.
ADDITIONAL INFORMATION
Data Deposition and Access
All variants have been submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) with accession numbers VCV000845407.1, VCV000246440.3, VXV000833548.1, VCV000833549.1, VCV000656194.2, and VCV000506576.2.
Ethics Statement
Signed consent was obtained for research publication from the patient's parents. Spectrum Health IRB approval is not needed for a single care report.
Acknowledgments
We thank the patient and family for consenting to share this story and providing all clinical samples.
Author Contributions
S.D. was lead clinician and oversaw all clinical care, M.M. managed patient care and extracted clinical information, J.G.C. and J.W.P. performed all bioinformatic variant interpretation work, and P.R.M. was lead geneticist on the project. All authors contributed to the writing of the manuscript.
Funding
Funding for variant characterization came from the National Institutes of Health (NIH) Office of the Director and National Institute of Environmental Health Sciences (NIEHS) grant K01ES025435 (J.W.P.).
Competing Interest Statement
The authors have declared no competing interest.
REFERENCES
- Carvill GL, McMahon JM, Schneider A, Zemel M, Myers CT, Saykally J, Nguyen J, Robbiano A, Zara F, Specchio N, et al. 2015. Mutations in the GABA transporter SLC6A1 cause epilepsy with myoclonic–atonic seizures. Am J Hum Genet 96: 808–815. 10.1016/j.ajhg.2015.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, et al. 2003. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem 24: 1999–2012. 10.1002/jcc.10349 [DOI] [PubMed] [Google Scholar]
- Jin X-T, Galvan A, Wichmann T, Smith Y. 2011. Localization and function of GABA transporters GAT-1 and GAT-3 in the basal ganglia. Front Syst Neurosci 5: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannesen KM, Gardella E, Linnankivi T, Courage C, de Saint Martin A, Lehesjoki A-E, Mignot C, Afenjar A, Lesca G, Abi-Warde M-T, et al. 2018. Defining the phenotypic spectrum of SLC6A1 mutations. Epilepsia 59: 389–402. 10.1111/epi.13986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattison KA, Butler KM, Inglis GAS, Dayan O, Boussidan H, Bhambhani V, Philbrook B, da Silva C, Alexander JJ, Kanner BI, et al. 2018. SLC6A1 variants identified in epilepsy patients reduce γ-aminobutyric acid transport. Epilepsia 59: e135–e141. 10.1111/epi.14531 [DOI] [PubMed] [Google Scholar]
- Palmer S, Towne MC, Pearl PL, Pelletier RC, Genetti CA, Shi J, Beggs AH, Agrawal PB, Brownstein CA. 2016. SLC6A1 mutation and ketogenic diet in epilepsy with myoclonic–atonic seizures. Pediatr Neurol 64: 77–79. 10.1016/j.pediatrneurol.2016.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokop JW, Lazar J, Crapitto G, Smith DC, Worthey EA, Jacob HJ. 2017. Molecular modeling in the age of clinical genomics, the enterprise of the next generation. J Mol Model 23: 75 10.1007/s00894-017-3258-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. 2015. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17: 405–424. 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All variants have been submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) with accession numbers VCV000845407.1, VCV000246440.3, VXV000833548.1, VCV000833549.1, VCV000656194.2, and VCV000506576.2.