Abstract
Although cutaneous squamous cell carcinoma (cSCC) is treatable in the majority of cases, deadly invasive and metastatic cases do occur. To date there are neither reliable predictive biomarkers of disease progression nor FDA-approved targeted therapies as standard of care. To address these issues, we screened patient-derived primary cultured cells from invasive/metastatic cSCC with 107 small-molecule inhibitors. In-house bioinformatics tools were used to cross-analyze drug responses and DNA mutations in tumors detected by whole-exome sequencing (WES). Aberrations in molecular pathways with evidence of potential drug targets were identified, including the Eph-ephrin and neutrophil degranulation signaling pathways. Using a screening panel of siRNAs, we identified EPHA6 and EPHA7 as targets within the Eph-ephrin pathway responsible for mitigating decreased cell viability. These studies form a plausible foundation for detecting biomarkers of high-risk progressive disease applicable in dermatopathology and for patient-specific therapeutic options for invasive/metastatic cSCC.
Keywords: squamous cell carcinoma of the skin
INTRODUCTION
Cutaneous squamous cell carcinoma (cSCC) accounts for up to one-third of nonmelanoma skin cancer, with 200,000 –300,000 new cases in the United States per year (Maubec et al. 2011) and 3000–4500 deaths (Karia et al. 2013). Initial treatment for cSCC is primarily surgical, with the option of radiation therapy for cases not amenable to surgery (Stratigos et al. 2015). Despite the surgical cure rate of >90%, ∼8% of patients diagnosed with cSCC experience recurrence within 5 years of initial treatment, and 5% progress to metastatic disease (Lewis et al. 2012). Furthermore, 10-year survival rates are <20% for patients with local lymph node metastases and <10% for those with distant metastases (Alam and Ratner 2001). Several risk factors for aggressive cSCC have been identified. These include involvement of subcutaneous tissue, size >2 cm, perineural invasion, lymphovascular invasion, recurrence, poorly differentiated tumors, location on the head or neck, immunosuppression, and invasion deeper than 7 mm (Alam and Ratner 2001; O'Hara et al. 2011; Warren et al. 2016; Garrett et al. 2017; Skulsky et al. 2017). However, identification of these aggressive cSCCs remains challenging, with significant variability in the criteria used for high-risk classification (Parikh et al. 2014).
Currently, there are no standard treatment options for cSCC with distant metastases. Previous studies have shown a high frequency of overexpression of the epidermal growth factor receptor (EGFR) in cutaneous and oral SCC (Jalili et al. 2008; Taoudi Benchekroun et al. 2010). Targeted EGFR inhibitor therapy with cetuximab, a chimeric antibody, has been utilized as a treatment strategy for cSCC with some promising results in recurrent cases and as frontline therapy (Bauman et al. 2007; Maubec et al. 2011). More recently cemiplimab, an immunotherapy targeting PD-1, has been approved for the treatment of advanced cSCC (Migden et al. 2018). However, the use of immune checkpoint blockade in individuals who develop cSCC post–organ transplant is not advised because of the risk of transplant rejection, and it is often these patients who develop the most severe cSCC, emphasizing the need for therapeutic strategies targeting the malignant cell (Fisher et al. 2020). Additionally, there are no standard protocols guiding treatment (Stratigos et al. 2015) and targeted therapy options remain limited (Fung and Grandis 2010; Blasco et al. 2017; Migden et al. 2018), thus presenting vast opportunities for drug repurposing and discovery.
We present our molecular case study to address the diagnostic difficulties, provide deeper molecular characterization, and offer priority targets and pathways in an effort to reduce the gap in treatment options for advanced cSCC. We unite both genomic and functional analyses to identify potential druggable targets and shed light on shared molecular abnormalities that may lead to aggressive disease. To date, only small sequencing studies have been conducted with a focus on cSCC (Pickering et al. 2014; Chitsazzadeh et al. 2016). Thus, we seek to add to the published literature on the mutational landscape of cSCC while also testing the susceptibility of patient-derived tumor cells from three deeply invasive/metastatic cases to our panel of 107 small-molecule inhibitors. We then use the combined genomic and functional data to identify prioritized potential targets for molecularly guided therapy for aggressive cSCC utilizing our in-house algorithm, HitWalker2 (Bottomly et al. 2016), which prioritizes targets according to patient-specific mutation information. Last, we confirm EPHA6 and EPHA7 within the Eph-ephrin signaling pathway as responsible for mediating decreased cell viability in two of the patient cell lines using small interfering RNA (siRNA) screening panels.
RESULTS
Clinical Presentation and Treatment Outcomes
We evaluated three immunocompetent patients with invasive cSCC associated with metastatic disease. All three patients were male, Caucasian, and older than 60 years of age. Two of the patients had a smoking history of >20 pack years—a known risk factor for development of cSCC (Dusingize et al. 2017)—and one patient had concurrent oral SCC (Patient 3). Clinical information, including tumor grade, primary location, and presence of metastasis, is provided in the narratives below and in Supplemental Table 1.
Patient 1 is a >90-yr-old male with a history of smoking (20 pack years), significant sun exposure, and prior keratinocytic carcinomas. He presented with progressively worsening left-sided facial swelling and intermittent pain that was unresponsive to antibiotics. A computed tomography (CT) scan of the neck showed a left parotid superficial mass, and fine needle aspirate (FNA) revealed keratinizing SCC, thought to be metastasis from his prior skin cancer. Upon left parotidectomy and neck dissection, a specimen from the parotid was procured for this study. This specimen was assessed in pathology to be moderately differentiated SCC. Subsequent lymph node dissection identified one of six parotid nodes and zero of 15 neck nodes positive for disease. The patient passed away 45 months after initial diagnosis and 43 months after surgical removal and collection for our study.
Patient 2 is a 61-yr-old male with a family history of leukemia and breast, prostate, and bladder cancer and a prior right parietal scalp SCC (T2N0M0 (II) G2) that was treated with Mohs two-stage excision followed by 60 Gy of adjuvant radiation. Twenty-four months after his scalp SCC diagnosis and 12 months post-Mohs, the patient was seen at the ear, nose, and throat (ENT) clinic for a progressively enlarging right posterior neck mass. FNA demonstrated p16 negative metastatic cSCC. Subsequent neck dissection confirmed five of 25 positive lymph nodes from which a specimen was procured for our study. The patient presented 3 months postexcision with recurrent, right neck cSCC. Positron emission tomography (PET)/CT revealed multiple regional nodules with pulmonary and lymph node lesions compatible with metastatic disease. The patient then underwent 66 Gy radiation therapy and systemic treatment with cetuximab; however, he was unable to tolerate the chemotherapy. The patient passed away ∼22 months after resection of his neck cSCC.
Patient 3 is a 68-yr-old male with a history of smoking (50 pack years) and alcohol use. He initially presented with a 7-month history of poorly differentiated p16 positive cSCC of the left ear and left mastoid process. The patient underwent surgical resection, during which a tumor specimen from the ear lesion was procured for our study. Pathology findings of left ear specimen showed invasive cSCC, T2N3M0. Additionally, a floor of mouth specimen showed invasive SCC, T1N0M0. The patient declined adjuvant chemotherapy and radiation and elected surveillance. At 4 months post–initial surgery, the patient developed lung metastases, and biopsy of a left hilar node confirmed metastatic, p16-positive SCC. The patient was started on nivolumab but was taken off therapy because of development of bullous pemphigoid. Two years post–primary resection and 1 year after discontinuing treatment, the patient developed a superficially invasive, moderately differentiated SCC on the right floor of the mouth. The patient was last seen at the time of floor of mouth lesion removal, 34 months after the initial diagnosis of left ear SCC.
Patient-Derived Tumor Cell Models
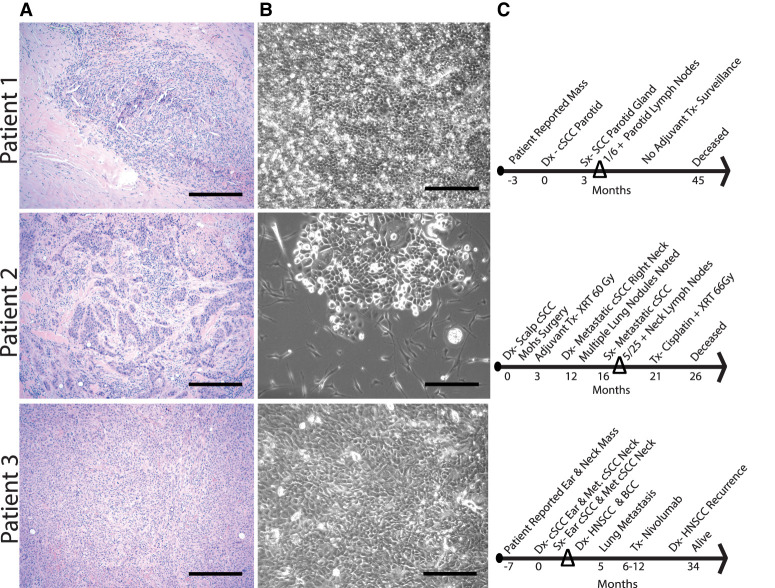
Tissue collected from participants included a suspected metastatic cSCC deeply invasive to the parotid gland, metastatic cSCC from a lymph node, and primary deeply invasive cSCC from the ear, from Patients 1, 2, and 3, respectively. Tumor morphology, percentage of sample composed of tumor, and differentiation status were evaluated by a pathologist. Micrographs of tumor and tumor-derived cells in culture are shown in Figure 1A,B. Patient-derived tumor cell cultures were subjected to differential trypsinization to remove fibroblasts, and morphology was >90% epithelial cells when cells were subjected to small-molecule inhibitor panels. The time line of each patient's cSCC progression and survival relative to specimen collection is shown in Figure 1C.
Figure 1.
Original cutaneous squamous cell carcinoma (cSCC) tumor and primary cultured tumor cell morphology, cultured patient-derived tumor cells, and clinical time line. For each of the three deeply invasive/metastatic cSCCs indicated, micrographs are shown to include hematoxylin and eosin (H&E)-stained sections (A), phase contrast images of primary cultures (B), and time lines of key cSCC progression features from patient chart review (C). The percent tumors determined by pathologist review of entire original tumor sections were 50%, 10%, and 80% for Patient 1, Patient 2, and Patient 3, respectively. Magnification bars, 300 µm. (Dx) Diagnosis, (Tx) treatment, (Sx) surgery.
Genomic Analysis
Whole-exome sequencing was performed on both the tumor cells and patient-matched blood cells from each of the three cSCC cases with the goal of identifying mutations that could potentially be responsible for the observed response to drugs on the panel. The median coverage was 209×–213×.
Single-Nucleotide Variants
The observed mutational load in the tumor samples relative to patient-matched blood cell controls averaged 3470 mutations per case (range 458–6226) and 52 mutations per Mb (range 6.9–93). Patient 1 had 8022 mutation calls, Patient 2 had 990 mutation calls, and Patient 3 had 13,495 mutation calls. A full list of the DNA mutation calls for each patient can be accessed via the database of Genotypes and Phenotypes (dbGaP; https://www.ncbi.nlm.nih.gov/gap/) and the controlled access Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra/docs/).
Integrative Analysis: HitWalker2 Prioritization
Using the network-based prioritization tool, HitWalker2 (Bottomly et al. 2016), each patient's single-nucleotide variation data were incorporated into a protein–protein interaction (PPI) network in order to determine high-priority genes by virtue of their network relationships to areas of increased mutational burden (Supplemental Table 2). These prioritized genes were then overlaid onto curated pathways using the pathway knowledgebase, Reactome (Fabregat et al. 2016). We filtered the pathways to emphasize pathways that do not contain highly overlapping gene members (Supplemental Table 3) given the hierarchical/nested nature of the pathways that can lead to bias in pathway enrichment. Table 1 highlights mutations within the neutrophil degranulation and Eph-ephrin pathways, identified via the HitWalker2 prioritization analysis, that were predicted to have functional consequences at the protein level and that also matched the following criteria: There must be multiple novel mutations within a single sample, novel mutations spanning multiple samples, or mutations matching known health-related phenotypes, including cancer (see Methods). In our three patients, we identified 10 such mutations across four genes: BPI, HRAS, EPHA7, and TP53. Finally, by integrating drug target annotation from the Cancer Targetome (Blucher et al. 2017) with the prioritized pathways, we identified four highly ranked molecular pathways that had targets across all three patients (Table 2). Given the enrichment of aberrations in the Eph-ephrin signaling pathway across our three patients, this pathway was further analyzed to show the network properties of its ranked aberrations and drug response data (Fig. 2). This showed involvement by the Eph family of proteins and revealed RAC1 to be a significant hub within the network with 14 network connections.
Table 1.
Highlighted damaging and novel variants in cutaneous squamous cell carcinoma (cSCC)
| Gene | Chromosome | HGVS DNA reference | HGVS protein reference | Variant type | Predicted effect | dbSNP | COSMIC | ExAC | ClinVar | Novel | Genotype | Patient 1 | Patient 2 | Patient 3 | Reactome pathway |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BPI | 20 | g.36938975G > A | ENSENSP00000262865: p.Gly157Ser | Single AA change | Deleterious/damaging | None | None | None | No match | Yes | Heterozygous | X | - | - | R-HSA-6798695 |
| BPI | 20 | g.36954682C > T | ENSENSP00000262865: p.Pro341Ser | Single AA change | Deleterious/damaging | None | None | None | No match | Yes | Heterozygous | - | - | X | R-HSA-6798696 |
| HRAS | 11 | g.534286C > G | ENSENSP00000309845: p.Gly13Arg | Single AA change | Deleterious/damaging | rs104894228 | None | None | HRAS13GR pathogenic/likely pathogenic | No | Heterozygous | - | X | - | R-HSA-2682334 |
| HRAS | 11 | g.534285C > T | ENSENSP00000309845: p.Gly13Asp | Single AA change | Deleterious/damaging | rs104894226 | CDS = c.38G > A; AA = p.G13D, | None | HRAS13GD pathogenic | No | Heterozygous | - | - | X | R-HSA-2682335 |
| EPHA7 | 6 | g.93956676G > A | ENSENSP00000358309: p.Arg854Cys | Single AA change | Deleterious/Damaging | None | None | None | No match | Yes | Heterozygous | X | - | - | R-HSA- 2682336 |
| EPHA7 | 6 | g.93953241C > T | ENSENSP00000358309: p.Gly967Glu | Single AA change | Deleterious/damaging | None | None | None | No match | Yes | Heterozygous | - | X | - | R-HSA- 2682337 |
| TP53 | 17 | g.7577085C > T | ENSENSP00000269305: p.Glu285Lys | Single AA change | Deleterious/damaging | rs112431538 | CDS = c.853G > A; AA = p.E285K, | None | TP53285EK pathogenic/likely pathogenic, drug response | No | Heterozygous | X | - | - | NA |
| TP53 | 17 | g.7577099C > T | ENSENSP00000269305: p.Arg280Lys | Single AA change | Deleterious/damaging | None | CDS = c.839G > A; AA = p.R280K, | None | TP53280RK conflicting interpretations of pathogenicity | No | Heterozygous | - | X | - | NA |
| TP53 | 17 | g.7577538C > T | ENSENSP00000269305: p.Arg248Gln | Single AA change | Deleterious/damaging | rs11540652 | CDS = c.743G > A; AA = p.R248Q, CDS = c.464G > A; AA = p.R155Q, | 5.77E-05 | TP53248RQ pathogenic/likely pathogenic | No | Heterozygous | - | - | X | NA |
| TP53 | 17 | g.7578526C > T | ENSENSP00000269305: p.Cys135Tyr | Single AA change | Deleterious/damaging | None | CDS = c.125G > A; AA = p.C42Y, CDS = c.404G > A; AA = p.C135Y, CDS = c.8G>A; AA = p.C3Y, | None | TP53135CY likely pathogenic | No | Heterozygous | - | - | X | NA |
Whole-exome sequencing (Illumina Hi-Seq) was performed on the three cases of collected samples with a median coverage of 209×–213×. DNA mutations were called relative to matched patient blood. Genes within the neutrophil degranulation and Eph-ephrin pathways were selected for this table if they met any of the following criteria: multiple novel mutations within a single sample, novel mutations spanning multiple samples, or mutations matching known health related phenotypes, including cancer.
Table 2.
Highly aberrational pathways identified in our cutaneous squamous cell carcinoma (cSCC) cohort
| Pathway | Pathway ID | Pathlength | % Pathway with SNVs | % Pathway with CNVs |
|---|---|---|---|---|
| Inactivation, recovery, and regulation of the phototransduction cascade | R-HSA-2514859 | 33 | 30.30 | 18.18 |
| Eph-ephrin signaling | R-HSA-2682334 | 92 | 26.09 | 39.13 |
| Eph-ephrin mediated repulsion of cellsa | R-HSA-3928665 | 49 | 47.06 | 36.73 |
| EPHA-mediated growth cone collapse | R-HSA-3928663 | 15 | 37.93 | 46.67 |
Using HitWalker2, we identified high-priority genes based on their mutational burden in pathways that do not contain highly overlapping gene members. Pathway Names, IDs, and pathway size are provided from Reactome annotation. Percentage of the pathway with SNVs and CNVs were calculated by dividing the total number of proteins in the pathway by the number of unique genes with SNVs and CNVs respectively.
(SNV) Single-nucleotide variant, (CNV) copy-number variant. aIndicates daughter pathways within the parent Eph-ephrin signaling pathway.
Figure 2.

RAC1, a significant hub within the Eph-ephrin signaling pathway. The 92 proteins of the Eph-ephrin signaling pathway were each annotated with an aberrational score, based on the type of aberration seen in the patients: single-nucleotide variation (SNV) only (1-light blue), copy-number variation (CNV) only (2-medium blue), or both SNV and CNV (3-dark blue). Asterisks (*) within the circles indicate proteins that are targeted by drugs present on our inhibitor assay screen at IC50 values <100 nM as determined by targetome. Fifty percent of the pathway had a nonzero aberrational score. A protein–protein interaction network using high-confidence (0.9) edges from String was generated, and we focused only on the subnetwork within the aberrational nodes (zero-order interaction network). RAC1 was the largest hub (degree = 14; node with brown outline).
Copy-Number Variants
We identified 2009, 3064, and 2733 genes in segments of copy-number gain in Patients 1, 2, and 3, respectively. We identified 5582, 1891, and 535 genes in segments of copy-number loss in Patients 1, 2, and 3, respectively. Across these patients, we identified 330 genes with copy-number gain and 29 genes with copy-number loss that had been previously reported (Li et al. 2015b). Given the tendency of whole-exome sequencing to have false-positive calls, we filtered our genes list to only those present in two or more samples that did not contain genes that had been called as both copy-number increase and copy-number decrease across multiple samples. We also filtered out a curated list of known problematic variants/genes (Tyner et al. 2018) and excluded any genes on Chromosomes 8 or 9, as almost the entire lengths of these chromosomes were copy-number-variable. We identified seven genes in segments of copy-number gain and 259 genes in segments of copy-number loss (Supplemental Table 4). Finally, we identified 36 genes from the Eph-ephrin signaling pathway that were copy-number variable in one or more of our samples (Supplemental Table 5).
Functional Analysis
Tumor-derived cell cultures from Patients 1, 2, and 3 were exposed to 107 small-molecule inhibitors. As an approach to placing our assays in the context of U.S. Food and Drug Administration (FDA)-approved EGFR targeted treatments, and as an initial approach to testing combinations of agents that include a drug targeting EGFR, we tested each of the drugs on our panel without and with the addition of an EGFR inhibitor, gefitinib. The use of cetuximab, which targets EGFR and is approved for head and neck squamous cell carcinoma (HNSCC), was considered; however, because of the storage of our inhibitor assays, it was not suitably stable for inclusion. Further, cetuximab activity may be through extrinsic antitumor immunity beyond the intrinsic effects approached in our study (Ferris 2015; Miyauchi et al. 2019). The drug panels were originally created for leukemia (Tyner et al. 2013) and include a breadth of kinase inhibitors (tyrosine kinases as well as select families of non–tyrosine kinases, including PI3K/AKT, PKC, PKA, IκK, RAF/MEK/ERK, JNK, p38, AMPK, aurora kinases, and cyclin-dependent kinases), histone deacetylase inhibitors, bromodomain inhibitors, BH3 mimetics, proteosome inhibitors, and HSP90 inhibitors. (Supplemental Table 6). These panels have been used to identify kinase dependence in models of gallbladder cancer, nasopharyngeal carcinoma, and by us in leukemia and HNSCC (Weber et al. 2017; Yuan et al. 2017; Tyner et al. 2018).
Sensitivity and resistance to each drug on the panel was determined on a patient-specific basis by calculating the area under the dose–response curve (AUC) for each drug and subsequently classifying the lowest 20% of AUCs as sensitive and the highest 20% of AUCs as resistant. These data are shown in Figure 3.
Figure 3.
Efficacy of small-molecule inhibitors on cutaneous squamous cell carcinoma (cSCC) patient–derived cells. Patient-derived tumor cells were exposed to 107 drugs with and without the addition of an EGFR inhibitor, gefitinib (50 nM). Sensitivity (blue) and resistance (red) to each drug on the panel was determined on a patient-specific basis by calculating the lowest and highest 20% of the area under the dose–response curve (AUC) values for each drug. Drugs with AUC values between the highest and lowest 20% across all three patients were excluded from the table. *Indicates drugs with varied starting concentrations; see Supplemental Table 6 for dosing information.
All three patients’ cultured cells were sensitive to dasatinib, ponatinib, and sunitinib, both with and without the addition of gefitinib. Utilizing drug–target information classified according to levels of evidence as described in the Cancer Targetome (Blucher et al. 2017), overlapping targets for these three drugs include ABL1 and RET tyrosine kinases. However, multiple patient samples show resistance to both bosutinib and imatinib, making these targets alone less likely to be those responsible for the observed drug sensitivity. There is significant overlap between targets for all three agents, which include the tyrosine kinases CSF1R (dasatininb and sunitinib), FLT3 (dasatininb and sunitinib), and KDR (ponatinib and sunitinib). None of these targets overlap with targets of the resistant agents (bosutinib, imatinib, and ibrutinib). Of note, using this same level of evidence for drug–target interactions, there was significant resistance to agents targeting EGFR. Additionally, tankyrase inhibitor, XAV-939, in combination with gefitinib appeared to drive resistance in all three patient-derived cell lines.
Drugs included on the inhibitor assay that target members of the Eph-ephrin pathway with varying degrees of sensitivity include axitinib, bosutinib, dasatinib, imatinib, ponatinib, and sunitinib. Although dasatinib, ponatinib, and sunitinib show sensitivity in all three patient-derived cell lines, it is important to recognize that these drugs have a wide distribution of targets, including many outside of the Eph-ephrin pathway, making it difficult to interpret whether their effect is due to targeting Eph-ephrin pathway members specifically. In an attempt to identify specific genes responsible for mitigating cell viability, we utilized the siRNA screening panel described below.
For a subset of drugs on our panel, we do not have extensive drug–target interaction data available through the Cancer Targetome, which currently covers only FDA-approved cancer drugs. Therefore, the response to these agents must be analyzed carefully. The pansensitivity seen with A-674563, AT-7519, and flavopiridol shares significant target overlap with GSK3B, CDK5, CDK4, and CDK6, without sharing targets of any of the panresistant drugs. Patient 1, however, showed resistance to targeting of GSK3B, making it less likely that this target was solely responsible for the sensitivity pattern. The pansensitivity seen with PI-103 and PP242 shares overlap with MTOR inhibitors. Furthermore, there were strong responses to the combined PI3K/MTOR inhibitor, dactilisib, across all three cases, independent of gefitinib treatment.
siRNA RAPID Assay
We utilized a siRNA RNAi-assisted protein target identification (RAPID) screening assay (Tyner et al. 2009) to identify potential single targets responsible for mediating cell viability. Two Eph-ephrin pathway members, EPHA7 and EPHA6, were among the top targets in Patient 1 and Patient 2, respectively (Fig. 4). A reduction in cell viability in response to siRNA-mediated knockdown of Eph-ephrin signaling genes was not enhanced by the addition of gefitinib. It is important to note that EPHA1-8, EPHB1-6, LYN, and Yes1 were screened Eph-ephrin signaling pathway members; however, only EPHA6 and EPHA7 showed >30% reductions in cell viability. EPHA6 is a particularly interesting target as it has been implicated in metastasis, angiogenesis, and cancer progression (Li et al. 2015a). Many of the inhibitors on the small-molecule inhibitor screen targeted EPHA6 with varying degrees of sensitivity, likely because of the promiscuous nature of these inhibitors and their target binding affinities.
Figure 4.
Eph-ephrin signaling pathway members, EPHA6 and EPHA7, among top siRNA targets. Patient-derived tumor cells from Patient 1 and Patient 2 were transfected with small interfering RNA (siRNA) pools cotreated with vehicle or gefitinib (50 nM). Cell viability was measured at 490 nM (as determined by the MTS assay) after 96 h of treatment and was normalized to the median plate value. Dotted lines indicate 2 S.D. from the mean. Mean individual siRNA targets are represented with error bars representing ± S.E.M., each containing three replicates (n = 3). Targets highlighted are outside 2 S.D. from the mean (EPHA7, JAK2, LYN, STYK1) and/or are statistically different from nonspecific siRNA (EPHA6, JAK2, LYN, STYK1).
DISCUSSION
The acquisition and application of genomics data for risk stratification, identification of disease drivers, and development of targeted therapies for cSCC has been hampered by multiple factors. As nonmelanoma skin cancer is often cured by surgery alone, the same efforts to characterize more aggressive cancer types by large-scale projects such as the NCI's The Cancer Genome Atlas or the Leukemia and Lymphoma Society's BeatAML have not been applied to cSCC. Yet, total mortality from cSCC remains significant because of the large overall incidence (Maubec et al. 2011; Karia et al. 2013). Furthermore, the skin, as the body's main protective barrier, has many carcinogenic stressors, including UV sunlight, viral agents (principally human papillomavirus [HPV]), and environmental exposures. This results in cells that contain a higher baseline mutational burden, even prior to development of cSCC (Martincorena et al. 2015). Indeed, well-differentiated primary cSCCs have a mutational burden of ∼50 mutations/Mb (Chitsazzadeh et al. 2016). Given the high mutational burden, identifying which mutations are drivers in metastatic disease is important, including by functional annotation.
Application of a functional genomics approach to characterize deeply invasive/metastatic cSCC is unprecedented to our knowledge. Potential drug targets identified in our patient samples include the tyrosine kinase FGR and the serine/threonine kinase STK10. As part of a highly aberrant neutrophil degranulation pathway, FGR, STK10, and PYGL were identified as potential targets in nonoverlapping patient samples. Previous studies have shown that FGR expression is associated with histone deacetylase (HDAC) inhibitor resistance in lymphoma (Joosten et al. 2016). Mutations in the STK10 gene have been associated with a decrease in its apoptotic function in peripheral T-cell lymphoma, and PYGL has been shown to be a target of p53 and is part of a group of genes that show dysregulation under the influence of mutated p53 (Garritano et al. 2013).
Gefitinib in combination with other agents was variable in effect across the patient samples. While gefitinib has largely lacked benefit over other treatments in clinical trials in head and neck SCC (Sundvall et al. 2010; Tang et al. 2019), it showed promise in vitro in Fanconi anemia–associated HNSCC when used in combination with afatinib (Montanuy et al. 2020). In our study, tumor-derived cells from Patient 2, which were insensitive to an Alk inhibitor NVP-TAE-684, showed increased sensitivity when tested in combination with gefitinib. Synergistic activity of Alk inhibitors in combination with gefitinib was previously reported by us in HNSCC patient–derived cell lines (Ouyang et al. 2018).
Of the highly ranked molecular pathways identified in our samples, the Eph-ephrin signaling pathway showed a particularly heavy aberrational burden relative to its size. Eph-ephrin signaling is important in the signaling of cancer because of its roles in cell adhesion, migration, anoikis, and cell cycle regulation (Lisabeth et al. 2013). In particular, Eph-ephrin signaling can promote or repress tumorigenesis and invasion by influencing many different signaling networks, each of which is cell- and context-dependent (Xu et al. 1992; Surawska et al. 2004).
Many of the Eph-ephrin pathway member genes carry a high mutational rate, including EPHA7 with mutations rates up to 34% in cSCC (cBioPortal). There is also a high cooccurrence of mutations in multiple gene members of the Ephrin signaling pathway, suggesting that members of this pathway contribute collectively to tumor formation and development (cBioPortal) (Cui 2010). RAC1, a significant hub within the Eph-ephrin pathway, is highly expressed in skin and is a purported driver of angiogenesis and metastases and is also implicated in the epithelial-to-mesenchymal transition of cancer cells (Patel et al. 2007; Bid et al. 2013; Yoon et al. 2017). More specifically, HNSCC cells have been found to exhibit high levels of constitutively activated RAC1, further highlighting its likely significance in cSCC (Patel et al. 2007). Outside of the Eph-ephrin pathway, RAC1 plays a significant role in the initiation and progression of cancer by acting through multiple commonly aberrant signaling pathways including cell growth and survival, angiogenesis, and cell migration (Bid et al. 2013). Additionally, RAC1 deregulation has been linked to cardiovascular and neurodegenerative disease, making it an attractive therapeutic target with a wide variety of potential applications (Marei and Malliri 2017).
Other well-characterized oncogenes also present in the EPHA-ephrin signaling pathway include TIAM, YES1, and HRAS (Yang et al. 2015; Dotto and Rustgi 2016; Garmendia et al. 2019). MYH10, the only pathway member in our cohort with alterations in both single-nucleotide variation (SNV) and copy-number variation (CNV), has been identified as an important hub in other quantitative analyses and plays an important role in cancer migration and invasion (Wang et al. 2018; Guo et al. 2019; Li et al. 2019). These data combined with our HitWalker2 analyses suggest great therapeutic potential in targeting RAC1 and other members of the Eph-ephrin pathway in cSCC. Further studies examining the role of RAC1 inhibition on cSCC cell viability and cSCC development and metastasis in animal models are needed.
Progress in the treatment of cSCC will be aided by standardized annotation of clinical and pathological features, integrated with genomic and functional analyses. Our work in the evaluation of patient-derived tumor samples offers a window into the missing dimension of functional association of genetic profiling and has implications in response to therapy, guidance in repurposing current targeted agents, and development of new drug therapies. Moving forward, we will utilize the Targetome evidence for drug activity and our functional genomic findings in order to develop a cSCC-specific screening panel that will include a broader distribution of drugs under development, including those that have not been previously approved for cancer as well as selected natural products. The identifications of pathways and candidate gene targets in advanced cSCC in this study need to be validated mechanistically and in a larger set of cSCC cases. These findings have potential to open opportunities in development of diagnostics or predictive signature biomarkers for cSCC at high risk for recurrence, invasion, or metastasis and further precision medical treatments for aggressive cSCC.
METHODS
Patient Sample Collection and Cell Culture
We collected fresh tissues, including tumor, uninvolved skin, and matched blood samples, from three consenting patients who were being treated surgically in Oregon Health and Science University (OHSU) Otolaryngology for advanced cSCC (Dermatology Molecular Profiling Resource Repository, IRB #10071). Tissue was de-identified and processed for cell culture and DNA sequencing. Tissue for cell culture was minced and cultured in DMEM/F12 Media (Gibco 11320082), supplemented with 5% supplemented BCS (Hyclone), 1× antibiotic/antimitotic (Invitrogen), 1.8 × 10−4 M adenine (Sigma-Aldrich), 0.4 µg/mL hydrocortisone (Sigma-Aldrich), 1 × 10−10 M cholera enterotoxin (Sigma-Aldrich), 2 × 10−11 M triiodothyronine (Sigma-Aldrich), 5 µg/mL insulin (Sigma-Aldrich), and 10 µg/mL epidermal growth factor (Invitrogen). The cells were allowed to grow to 90% confluence, with differential 1-min trypsinization (0.25% and EDTA, Gibco) being used to remove fibroblasts as needed. Successive passages were made and cells used for experiments were of passage 3 or lower. Cell culture methods developed by James Rheinwald of the Harvard Skin Disease Research Center, Boston, were used as described (Rasmussen et al. 2013).
DNA Exome Sequencing
Using the QIAGEN kit DNeasy for Blood and Tissue, DNA was isolated for Patient 1 from primary tissue culture, and for Patient 2 and Patient 3 from tumor tissue previously snap-frozen in liquid nitrogen. Blood DNA was used as the normal control for all three subjects and was stored in blood collection vacutainers containing EDTA at −80°C. Whole-exome sequencing on both the tumor cells and patient-matched blood cells from each of the three cSCC cases were performed at the Massively Parallel Sequencing Shared Resource at OHSU on an Illumina HiSeq 2500. Samples were sequenced at a median coverage of ∼210×.
Bioinformatics Analysis
DNA Exome Sequencing
Whole-exome sequencing (WES) was the basis of mutation calls from original tumor tissue compared to patient-matched normal DNA taken from blood samples. After alignment of the reads to human genome using the Burrows–Wheeler Aligner (Li and Durbin 2010), the aligned reads were separated into read groups and any duplicate aligned reads were marked (Institute B 2015b; Cibulskis et al. 2013). To increase accuracy, misalignment of reads that span indels were subjected to local multiple sequence realignment and systematically inaccurate base quality scores were recalibrated (Institute 2015a, 2017b). Finally, MuTect2 was used to identify mutations in the tumor sample relative to the matched normal sample (Cibulskis et al. 2013; Institute 2016). The called mutants can be subsequently analyzed with confidence that they are not the result of artifact (DePristo et al. 2011).
HitWalker2
HitWalker2 (Bottomly et al. 2016) utilizes a random walk with restarts algorithm for each individual patient sample. It can prioritize patient variants relative to their weighted proximity to other data types in a protein–protein interaction network. It is highly extensible, allowing incorporation of diverse data types to refine prioritization. Further, HitWalker2 was extended to allow cohort-level identification of subset of cohort with the same hit results in a given set of genes, as well as the pathway context for results (individual, subset, and full cohort). This will allow the three cases to be compared to larger cohort studies as they become available.
Copy-Number Variation
We analyzed WES from the three cSCC samples matched with normal controls from the same patient in order to identify copy-number alterations in the tumor relative to normal (Li and Durbin 2010). After alignment of the reads to the human genome, the aligned reads were separated into read groups and any duplicates were marked (Institute, 2015b; Cibulskis et al. 2013). To decrease erroneous mutation calls, misalignment of reads that span indels were subject to local multiple sequence realignment and systematically inaccurate base quality scores were recalibrated (Institute 2015a, 2017b). These aligned reads were then used to determine the depth-of-coverage across each exonic segment in both the tumor and normal samples (Institute 2017a). To identify areas of CNV, we used a tool called ExomeCNV, which utilizes the ratio—tumor to normal—of normalized depth-of-coverage in order to call CNVs at the exonic level (Sathirapongsasuti et al. 2011). The benefit of ExomeCNV is that the user can define cutoff values for the sensitivity, specificity, and false discovery rate for called CNV exons. This allows for a higher level of confidence in the outputted CNV regions. Once the exonic CNVs are called, they are combined with neighboring exonic regions by circular binary segmentation in order to determine CNV segments in the tumor samples (Olshen et al. 2004).
Inhibitor Assays
Inhibitor assays were completed using 107 drugs that target specific cancer-related pathways. The drugs were prealiquoted onto custom 384-well plates with seven concentrations per drug, resulting in three plates per complete panel (Supplemental Table 6). Patient-derived cSCC cells were plated at a concentration of 2500 cells per well with and without the presence of 50 nM gefitinib (Selleck S1025) and then assessed for viability after 72 hours of incubation at 37°C using a tetrazolium-based cell viability assay (MTS, Promega PR-G3581) by measuring absorption at 490 nM using a Biotek Synergy H1 Microplate Reader. Values were then normalized to wells that received no drug. Dose response curves were created for AUC analysis.
siRNA RAPID Assay
Patient-derived cells were plated in RNAi-assisted protein target identification (RAPID) functional screens as previously described (Tyner et al. 2009). Cells were plated with and without the presence of 50 nM gefitinib (Selleck S1025) into three replicate 96 wells at a density of 8000 cells per well containing Lipofectamine RNAiMAX (Life Technologies 13778-150), Opti-Mem (Gibco 31985-062), and the siRNA. All siRNAs were SMARTpool: siGENOME siRNA pools (GE Dharmacon). Cell viability was assessed after 96 hours of incubation using a tetrazolium-based cell viability assay (MTS, Promega PR-G3581) by measuring absorption at 490 nM using a Biotek Synergy H1 Microplate Reader. Values were then normalized to the median plate value.
ADDITIONAL INFORMATION
Data Deposition and Access
Data is available via dbGAP (https://www.ncbi.nlm.nih.gov/gap/) and controlled-access SRA (https://www.ncbi.nlm.nih.gov/sra).
Ethics Statement
Oral and written patient consent was obtained under OHSU IRB protocol 10071 and work was performed under OHSU IRB protocol 809.
Acknowledgments
We thank the OHSU Gene Profiling Shared Resource for whole-exome sequencing and Clara Stemwedel for her editorial assistance. D.M. is grateful to her medical student advisor, John Kirkwood, MD, University of Pittsburgh, for discussion, and the University of Pittsburgh Medical School Grant for travel to present preliminary results at the Society of Investigative Dermatology Annual Meeting, May 2017, Portland, OR.
Author Contributions
A.N.A. obtained patient consent and collected specimens, performed drug inhibitor and siRNA assays and data analysis, and prepared the manuscript. D.M. performed drug inhibitor assays and DNA sequencing and prepared the manuscript. J.J. performed mutation and CNV analysis and prepared the manuscript. S.J. performed the drug response AUC analysis, HitWalker2 prioritization, and manuscript preparation. M.V. prepared the manuscript. A.S.B. performed pathway analysis and Targetome. C.Z. performed DNA sequencing analysis and prepared the manuscript. Y.J.Y. and C.H. performed patient clinical chart review and prepared the data presentation. X.O. was responsible for inhibitor assay, data analysis, and DNA preparation consultation. D.C. consulted on the manuscript and, with P.A., prepared surgical specimens. J.W.T. was consulted for drug inhibitor assays and study design. A.B., O.M.L., and J.J.L. consulted on the manuscript. S.M. was responsible for study design, bioinformatics analysis, HitWalker2, data accessibility, and manuscript preparation. M.K.-M. was responsible for study design, patient consent and specimen collection consultation, data analysis, and manuscript preparation.
Funding
This work was supported by the OHSU Dermatology Research Star Fund, by the National Institutes of Health, National Cancer Institute R01 CA192405 (M.K.-M. and S.M.) and by the Knight Cancer Institute P30 CA069533. J.W.T. received grants from the V Foundation for Cancer Research, the Gabrielle's Angel Foundation for Cancer Research, and the National Cancer Institute (1R01CA183947, 1U01CA217862, 1U54CA224019).
Competing Interest Statement
J.W.T. has received research support from Agios, Aptose, Array, AstraZeneca, Constellation, Genentech, Gilead, Incyte, Janssen, Petra, Seattle Genetics, Syros, and Takeda. All other authors have declared no competing interest.
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Alam M, Ratner D. 2001. Cutaneous squamous-cell carcinoma. N Engl J Med 344: 975–983. 10.1056/NEJM200103293441306 [DOI] [PubMed] [Google Scholar]
- Bauman JE, Eaton KD, Martins RG. 2007. Treatment of recurrent squamous cell carcinoma of the skin with cetuximab. Arch Dermatol 143: 889–892. 10.1001/archderm.143.7.889 [DOI] [PubMed] [Google Scholar]
- Bid HK, Roberts RD, Manchanda PK, Houghton PJ. 2013. RAC1: an emerging therapeutic option for targeting cancer angiogenesis and metastasis. Mol Cancer Ther 12: 1925–1934. 10.1158/1535-7163.MCT-13-0164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco MA, Svider PF, Raza SN, Jacobs JR, Folbe AJ, Saraf P, Eloy JA, Baredes S, Fribley AM. 2017. Systemic therapy for head and neck squamous cell carcinoma: historical perspectives and recent breakthroughs. Laryngoscope 127: 2565–2569. 10.1002/lary.26629 [DOI] [PubMed] [Google Scholar]
- Blucher AS, Choonoo G, Kulesz-Martin M, Wu G, McWeeney SK. 2017. Evidence-based precision oncology with the cancer targetome. Trends Pharmacol Sci 38: 1085–1099. 10.1016/j.tips.2017.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottomly D, McWeeney SK, Wilmot B. 2016. HitWalker2: visual analytics for precision medicine and beyond. Bioinformatics 32: 1253–1255. 10.1093/bioinformatics/btv739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitsazzadeh V, Coarfa C, Drummond JA, Nguyen T, Joseph A, Chilukuri S, Charpiot E, Adelmann CH, Ching G, Nguyen TN, et al. 2016. Cross-species identification of genomic drivers of squamous cell carcinoma development across preneoplastic intermediates. Nat Commun 7: 12601 10.1038/ncomms12601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. 2013. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 31: 213–219. 10.1038/nbt.2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Q. 2010. A network of cancer genes with co-occurring and anti-co-occurring mutations. PLoS ONE 5: e13180 10.1371/journal.pone.0013180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43: 491–498. 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotto GP, Rustgi AK. 2016. Squamous cell cancers: a unified perspective on biology and genetics. Cancer Cell 29: 622–637. 10.1016/j.ccell.2016.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusingize JC, Olsen CM, Pandeya NP, Subramaniam P, Thompson BS, Neale RE, Green AC, Whiteman DC, Study QS. 2017. Cigarette smoking and the risks of basal cell carcinoma and squamous cell carcinoma. J Invest Dermatol 137: 1700–1708. 10.1016/j.jid.2017.03.027 [DOI] [PubMed] [Google Scholar]
- Fabregat A, Sidiropoulos K, Garapati P, Gillespie M, Hausmann K, Haw R, Jassal B, Jupe S, Korninger F, McKay S, et al. 2016. The Reactome Pathway Knowledgebase. Nucleic Acids Res 44: D481–D487. 10.1093/nar/gkv1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris RL. 2015. Immunology and immunotherapy of head and neck cancer. J Clin Oncol 33: 3293–3304. 10.1200/JCO.2015.61.1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher J, Zeitouni N, Fan W, Samie FH. 2020. Immune checkpoint inhibitor therapy in solid organ transplant recipients: a patient-centered systematic review. J Am Acad Dermatol 82: 1490–1500. 10.1016/j.jaad.2019.07.005 [DOI] [PubMed] [Google Scholar]
- Fung C, Grandis JR. 2010. Emerging drugs to treat squamous cell carcinomas of the head and neck. Expert Opin Emerg Drugs 15: 355–373. 10.1517/14728214.2010.497754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmendia I, Pajares MJ, Hermida-Prado F, Ajona D, Bertolo C, Sainz C, Lavin A, Remirez AB, Valencia K, Moreno H, et al. 2019. YES1 drives lung cancer growth and progression and predicts sensitivity to dasatinib. Am J Respir Crit Care Med 200: 888–899. 10.1164/rccm.201807-1292OC [DOI] [PubMed] [Google Scholar]
- Garrett GL, Blanc PD, Boscardin J, Lloyd AA, Ahmed RL, Anthony T, Bibee K, Breithaupt A, Cannon J, Chen A, et al. 2017. Incidence of and risk factors for skin cancer in organ transplant recipients in the United States. JAMA Dermatol 153: 296–303. 10.1001/jamadermatol.2016.4920 [DOI] [PubMed] [Google Scholar]
- Garritano S, Inga A, Gemignani F, Landi S. 2013. More targets, more pathways and more clues for mutant p53. Oncogenesis 2: e54 10.1038/oncsis.2013.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T, Ma H, Zhou Y. 2019. Bioinformatics analysis of microarray data to identify the candidate biomarkers of lung adenocarcinoma. PeerJ 7: e7313 10.7717/peerj.7313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Institute B. 2015a. Base Quality Score Recalibration. Vol 2017. Broad Institute.
- Institute B. 2015b. Mapping, processing, and duplicate marking with Picard tools. Vol 2017. Broad Institute.
- Institute B. 2016. MuTect2: Call somatic SNPs and indels via local re-assembly of haplotypes. Vol 2017. Broad Institute.
- Institute B. 2017a. DepthOfCoverage. Vol 2018. Broad Institute.
- Institute B. 2017b. (howto) Recalibrate base quality scores=run BQSR. Vol 2017. Broad Institute.
- Jalili A, Pinc A, Pieczkowski F, Karlhofer FM, Stingl G, Wagner SN. 2008. Combination of an EGFR blocker and a COX-2 inhibitor for the treatment of advanced cutaneous squamous cell carcinoma. J Dtsch Dermatol Ges 6: 1066–1069. 10.1111/j.1610-0387.2008.06861.x [DOI] [PubMed] [Google Scholar]
- Joosten M, Ginzel S, Blex C, Schmidt D, Gombert M, Chen C, Linka RM, Grabner O, Hain A, Hirsch B, et al. 2016. A novel approach to detect resistance mechanisms reveals FGR as a factor mediating HDAC inhibitor SAHA resistance in B-cell lymphoma. Mol Oncol 10: 1232–1244. 10.1016/j.molonc.2016.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karia PS, Han J, Schmults CD. 2013. Cutaneous squamous cell carcinoma: estimated incidence of disease, nodal metastasis, and deaths from disease in the United States, 2012. J Am Acad Dermatol 68: 957–966. 10.1016/j.jaad.2012.11.037 [DOI] [PubMed] [Google Scholar]
- Lewis CM, Glisson BS, Feng L, Wan F, Tang X, Wistuba II, El-Naggar AK, Rosenthal DI, Chambers MS, Lustig RA, et al. 2012. A phase II study of gefitinib for aggressive cutaneous squamous cell carcinoma of the head and neck. Clin Cancer Res 18: 1435–1446. 10.1158/1078-0432.CCR-11-1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26: 589–595. 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Ma Y, Xie C, Wu Z, Kang Z, Fang Z, Su B, Guan M. 2015a. EphA6 promotes angiogenesis and prostate cancer metastasis and is associated with human prostate cancer progression. Oncotarget 6: 22587–22597. 10.18632/oncotarget.4088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YY, Hanna GJ, Laga AC, Haddad RI, Lorch JH, Hammerman PS. 2015b. Genomic analysis of metastatic cutaneous squamous cell carcinoma. Clin Cancer Res 21: 1447–1456. 10.1158/1078-0432.CCR-14-1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Qian S, Li B, Zhan X. 2019. Quantitative analysis of the human ovarian carcinoma mitochondrial phosphoproteome. Aging (Albany NY) 11: 6449–6468. 10.18632/aging.102199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisabeth EM, Falivelli G, Pasquale EB. 2013. Eph receptor signaling and ephrins. Cold Spring Harb Perspect Biol 5: a009159 10.1101/cshperspect.a009159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marei H, Malliri A. 2017. Rac1 in human diseases: the therapeutic potential of targeting Rac1 signaling regulatory mechanisms. Small GTPases 8: 139–163. 10.1080/21541248.2016.1211398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincorena I, Roshan A, Gerstung M, Ellis P, Van Loo P, McLaren S, Wedge DC, Fullam A, Alexandrov LB, Tubio JM, et al. 2015. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348: 880–886. 10.1126/science.aaa6806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maubec E, Petrow P, Scheer-Senyarich I, Duvillard P, Lacroix L, Gelly J, Certain A, Duval X, Crickx B, Buffard V, et al. 2011. Phase II study of cetuximab as first-line single-drug therapy in patients with unresectable squamous cell carcinoma of the skin. J Clin Oncol 29: 3419–3426. 10.1200/JCO.2010.34.1735 [DOI] [PubMed] [Google Scholar]
- Migden MR, Rischin D, Schmults CD, Guminski A, Hauschild A, Lewis KD, Chung CH, Hernandez-Aya L, Lim AM, Chang ALS, et al. 2018. PD-1 blockade with cemiplimab in advanced cutaneous squamous-cell carcinoma. N Engl J Med 379: 341–351. 10.1056/NEJMoa1805131 [DOI] [PubMed] [Google Scholar]
- Miyauchi S, Kim SS, Pang J, Gold KA, Gutkind JS, Califano JA, Mell LK, Cohen EEW, Sharabi AB. 2019. Immune modulation of head and neck squamous cell carcinoma and the tumor microenvironment by conventional therapeutics. Clin Cancer Res 25: 4211–4223. 10.1158/1078-0432.CCR-18-0871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montanuy H, Martínez-Barriocanal A, Antonio Casado J, Rovirosa L, Ramírez MJ, Nieto R, Carrascoso-Rubio C, Riera P, González A, Lerma E, et al. 2020. Gefitinib and afatinib show potential efficacy for Fanconi anemia–related head and neck cancer. Clin Cancer Res 26: 3044–3057. 10.1158/1078-0432.CCR-19-1625 [DOI] [PubMed] [Google Scholar]
- O'Hara J, Ferlito A, Takes RP, Rinaldo A, Strojan P, Shaha AR, Rodrigo JP, Paleri V. 2011. Cutaneous squamous cell carcinoma of the head and neck metastasizing to the parotid gland—a review of current recommendations. Head Neck 33: 1789–1795. 10.1002/hed.21583 [DOI] [PubMed] [Google Scholar]
- Olshen AB, Venkatraman ES, Lucito R, Wigler M. 2004. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics 5: 557–572. 10.1093/biostatistics/kxh008 [DOI] [PubMed] [Google Scholar]
- Ouyang X, Barling A, Lesch A, Tyner JW, Choonoo G, Zheng C, Jeng S, West TM, Clayburgh D, Courtneidge SA, et al. 2018. Induction of anaplastic lymphoma kinase (ALK) as a novel mechanism of EGFR inhibitor resistance in head and neck squamous cell carcinoma patient-derived models. Cancer Biol Ther 19: 921–933. 10.1080/15384047.2018.1451285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh SA, Patel VA, Ratner D. 2014. Advances in the management of cutaneous squamous cell carcinoma. F1000Prime Rep 6: 70 10.12703/P6-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel V, Rosenfeldt HM, Lyons R, Servitja JM, Bustelo XR, Siroff M, Gutkind JS. 2007. Persistent activation of Rac1 in squamous carcinomas of the head and neck: evidence for an EGFR/Vav2 signaling axis involved in cell invasion. Carcinogenesis 28: 1145–1152. 10.1093/carcin/bgm008 [DOI] [PubMed] [Google Scholar]
- Pickering CR, Zhou JH, Lee JJ, Drummond JA, Peng SA, Saade RE, Tsai KY, Curry JL, Tetzlaff MT, Lai SY, et al. 2014. Mutational landscape of aggressive cutaneous squamous cell carcinoma. Clin Cancer Res 20: 6582–6592. 10.1158/1078-0432.CCR-14-1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen C, Thomas-Virnig C, Allen-Hoffmann BL. 2013. Classical human epidermal keratinocyte cell culture. Methods Mol Biol 945: 161–175. 10.1007/978-1-62703-125-7_11 [DOI] [PubMed] [Google Scholar]
- Sathirapongsasuti JF, Lee H, Horst BA, Brunner G, Cochran AJ, Binder S, Quackenbush J, Nelson SF. 2011. Exome sequencing-based copy-number variation and loss of heterozygosity detection: ExomeCNV. Bioinformatics 27: 2648–2654. 10.1093/bioinformatics/btr462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skulsky SL, O'Sullivan B, McArdle O, Leader M, Roche M, Conlon PJ, O'Neill JP. 2017. Review of high-risk features of cutaneous squamous cell carcinoma and discrepancies between the American Joint Committee on Cancer and NCCN Clinical Practice Guidelines in Oncology. Head Neck 39: 578–594. 10.1002/hed.24580 [DOI] [PubMed] [Google Scholar]
- Stratigos A, Garbe C, Lebbe C, Malvehy J, del Marmol V, Pehamberger H, Peris K, Becker JC, Zalaudek I, Saiag P, et al. 2015. Diagnosis and treatment of invasive squamous cell carcinoma of the skin: European consensus-based interdisciplinary guideline. Eur J Cancer 51: 1989–2007. 10.1016/j.ejca.2015.06.110 [DOI] [PubMed] [Google Scholar]
- Sundvall M, Karrila A, Nordberg J, Grenman R, Elenius K. 2010. EGFR targeting drugs in the treatment of head and neck squamous cell carcinoma. Expert Opin Emerg Drugs 15: 185–201. 10.1517/14728211003716442 [DOI] [PubMed] [Google Scholar]
- Surawska H, Ma PC, Salgia R. 2004. The role of ephrins and Eph receptors in cancer. Cytokine Growth Factor Rev 15: 419–433. 10.1016/j.cytogfr.2004.09.002 [DOI] [PubMed] [Google Scholar]
- Tang X, He J, Li B, Zheng Y, Li K, Zou S, Chen L. 2019. Efficacy and safety of gefitinib in patients with advanced head and neck squamous cell carcinoma: a meta-analysis of randomized controlled trials. J Oncol 2019: 6273438 10.1155/2019/6273438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoudi Benchekroun M, Saintigny P, Thomas SM, El-Naggar AK, Papadimitrakopoulou V, Ren H, Lang W, Fan YH, Huang J, Feng L, et al. 2010. Epidermal growth factor receptor expression and gene copy number in the risk of oral cancer. Cancer Prev Res (Phila) 3: 800–809. 10.1158/1940-6207.CAPR-09-0163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyner JW, Deininger MW, Loriaux MM, Chang BH, Gotlib JR, Willis SG, Erickson H, Kovacsovics T, O'Hare T, Heinrich MC, et al. 2009. RNAi screen for rapid therapeutic target identification in leukemia patients. Proc Natl Acad Sci 106: 8695–8700. 10.1073/pnas.0903233106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyner JW, Yang WF, Bankhead A III, Fan G, Fletcher LB, Bryant J, Glover JM, Chang BH, Spurgeon SE, Fleming WH, et al. 2013. Kinase pathway dependence in primary human leukemias determined by rapid inhibitor screening. Cancer Res 73: 285–296. 10.1158/0008-5472.CAN-12-1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyner JW, Tognon CE, Bottomly D, Wilmot B, Kurtz SE, Savage SL, Long N, Schultz AR, Traer E, Abel M, et al. 2018. Functional genomic landscape of acute myeloid leukaemia. Nature 562: 526–531. 10.1038/s41586-018-0623-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yang Q, Cheng Y, Gao M, Kuang L, Wang C. 2018. Myosin heavy chain 10 (MYH10) gene silencing reduces cell migration and invasion in the glioma cell lines U251, T98G, and SHG44 by inhibiting the Wnt/β-catenin pathway. Med Sci Monit 24: 9110–9119. 10.12659/MSM.911523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren TA, Broit N, Simmons JL, Pierce CJ, Chawla S, Lambie DL, Quagliotto G, Brown IS, Parsons PG, Panizza BJ, et al. 2016. Expression profiling of cutaneous squamous cell carcinoma with perineural invasion implicates the p53 pathway in the process. Sci Rep 6: 34081 10.1038/srep34081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Valbuena JR, Barbhuiya MA, Stein S, Kunkel H, Garcia P, Bizama C, Riquelme I, Espinoza JA, Kurtz SE, et al. 2017. Small molecule inhibitor screening identified HSP90 inhibitor 17-AAG as potential therapeutic agent for gallbladder cancer. Oncotarget 8: 26169–26184. 10.18632/oncotarget.15410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YY, Pettersson K, Blomberg K, Hemmila I, Mikola H, Lovgren T. 1992. Simultaneous quadruple-label fluorometric immunoassay of thyroid-stimulating hormone, 17 α-hydroxyprogesterone, immunoreactive trypsin, and creatine kinase MM isoenzyme in dried blood spots. Clin Chem 38: 2038–2043. 10.1093/clinchem/38.10.2038 [DOI] [PubMed] [Google Scholar]
- Yang H, Cai YC, Cao Y, Song M, An X, Xia Y, Wei J, Jiang WQ, Shi YX. 2015. The prognostic value of Tiam1 protein expression in head and neck squamous cell carcinoma: a retrospective study. Chin J Cancer 34: 614–621. 10.1186/s40880-015-0053-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon C, Cho SJ, Chang KK, Park DJ, Ryeom SW, Yoon SS. 2017. Role of Rac1 pathway in epithelial-to-mesenchymal transition and cancer stem-like cell phenotypes in gastric adenocarcinoma. Mol Cancer Res 15: 1106–1116. 10.1158/1541-7786.MCR-17-0053 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Yuan J, Jiang YY, Mayakonda A, Huang M, Ding LW, Lin H, Yu F, Lu Y, Loh TKS, Chow M, et al. 2017. Super-enhancers promote transcriptional dysregulation in nasopharyngeal carcinoma. Cancer Res 77: 6614–6626. 10.1158/0008-5472.CAN-17-1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is available via dbGAP (https://www.ncbi.nlm.nih.gov/gap/) and controlled-access SRA (https://www.ncbi.nlm.nih.gov/sra).