Abstract
Over the past 15 years, downy mildew became the most destructive foliar disease in cultivated Impatiens species (Balsaminaceae) worldwide. A previous study had revealed that the causal agent was not Plasmopara obducens (Oomycota, Peronosporales) but Plasmopara destructor on Impatiens walleriana, and Plasmopara velutina on Impatiens balsamina. This hints to a relatively high degree of specialization of Plasmopara on Balsaminaceae. Therefore, it was the aim of the present study to perform multigene phylogenetic analysis and detailed morphological investigation for several Korean downy mildew samples parasitic to cultivated I. walleriana, and I. balsamina, but also to a northeast Asian wild plant, Impatiens textori. It was revealed that I. textori harbors a new species, which is introduced and described here as Plasmopara elegantissima.
Keywords: cox2 mtDNA, multi-locus phylogeny, Peronosporaceae, taxonomy, allopatric speciation
1. Introduction
The phylum Oomycota comprises ecologically diverse organisms of various degrees of complexity and diverse life histories [1,2]. The highest diversity of the Oomycota is in the family Peronosporaceae, containing ca. 900 species [3]. Within the Peronosporaceae, the vast majority of species is obligate biotrophic, causing downy mildew disease on numerous flowering plants, including economically relevant crops, such as aromatic herbs, beets, berries, cabbages, oilseed rape, onions, quinoa, radishes, roses, soybean, spinach, tobacco, and various ornamentals [4]. Most of the recent molecular phylogenetic and taxonomic studies paid attention to downy mildews affecting crops of economic value, leading to significant progress in their identification, taxonomy, and nomenclature [4]. For example, for 50 years the suggestion of Yerkes and Shaw [5] that the downy mildew pathogens affecting Chenopodiaceae plants, including beet, spinach, and quinoa, should all be included in “Peronospora farinosa (Botrytis farinosa)”, which has been proposed for rejection [6], has been widely followed by applied plant pathologists. However, recent multi-locus phylogenetic analyses have shown that the complex includes a variety of highly specialized and distinct species, such as P. effusa on spinach [7], P. schachtii on beet [8], and P. variabilis on quinoa [9]. But also in other genera, such as Hyaloperonospora, new species were described for pathogens infecting economically important crops, for example, Hyaloperonospora erucae on arugula [10] and Perofascia macaicola on maca [11].
Increasing global trade raises the risk of introduction of new downy mildew diseases, as exemplified by P. belbahrii on basil [12]. But also on sages [13] and balsamines [14,15] newly occurring downy mildew diseases were reported. Impatiens (Balsaminaceae) is one of the largest genera of flowering plants, covering about 900 species, which are distributed mainly in highlands and mountains of the Paleotropics, but extends with various species into parts of temperate Asia, Europe, and North America [16]. On two balsams, Impatiens walleriana and I. balsamina, which are popular ornamental plants grown worldwide, emerging downy mildew diseases have been reported. Since 2003 when the infection on cultivated I. walleriana appeared in the UK [17], this disease speedily spread worldwide over the past 15 years [14,15,18]. Except for a highly divergent species, Plasmopara constantinescui [19], all downy mildew agents parasitic on different species of Impatiens had been identified as Plasmopara obducens until this species complex was reexamined and it was revealed that the downy mildew pathogen of European Impatiens, P. obducens s.str., was not conspecific with the downy mildew pathogens of cultivated Impatiens, which, consequently, were described as new species – P. destructor on I. walleriana and Plasmopara velutina on I. balsamina [14].
In Korea, downy mildew infections have been recorded from four species of Impatiens: I. textori [20], I. noli-tangere [21], I. walleriana, and I. balsamina [22]. In line with various studies (see references in [14]), the diseases were attributed to P. obducens [20–22], because among the different pathogen/host accessions there was no sequence variation in LSU rDNA region [22] and morphological differences were subtle [21]. However, the recent finding that each of the latter three hosts harbors a specialized species of Plasmopara [14], but also the potential overlap in host ranges reported recently [15] prompts to the necessity of a careful reexamination of the pathogens. Given the high host specificity known for Plasmopara species [14,23–26] and the high phylogenetic distance between I. textori and other Impatiens species that are host to downy mildew pathogens [27], it seemed likely that the downy mildew pathogen of I. textori is an undescribed species. Thus, the aim of the current study was to resolve the identity of the pathogens causing impatiens downy mildew in Korea, using detailed morphological and multi-locus phylogenetic investigations.
2. Materials and methods
2.1. Herbarium specimens
In total, 30 downy mildew specimens originating from I. balsamina, I. textori, and I. walleriana were morphologically examined, and 10 specimens were selected for molecular phylogenetic analysis based on their morphology and host species. For comparison, the reference sequences of Plasmopara species originated from Impatiens plants were retrieved from GenBank. Information on the specimens sequenced in this study is shown in Table 1.
Table 1.
List of herbarium specimens sequenced in this study.
| DNA no. | Herb. no. | Pathogen | Host plant | Geographic origin (Year) | GenBank Acc. No. ITS1/cox2/cox2-1 spacera |
|---|---|---|---|---|---|
| YC22731 | KUS-F22731 | Plasmopara velutina | Impatiens balsamina | Korea; Gangneung (2007) | MK067070/MK067058/MK067082 |
| YC23751 | KUS-F23751 | Plasmopara velutina | Impatiens balsamina | Korea; Yongin (2008) | MK067074/MK067062/MK067086 |
| YC25940 | KUS-F25940 | Plasmopara velutina | Impatiens balsamina | Korea; Osan (2011) | MK067076/MK067064/MK067088 |
| YC22732 | KUS-F22732 | Plasmopara destructor | Impatiens walleriana | Korea; Gangneung (2007) | MK067071/MK067059/MK067083 |
| YC21824 | KUS-F21824 | Plasmopara sp. | Impatiens textori | Korea; Chuncheon (2006) | MK067072/MK067060/MK067084 |
| YC23341 | KUS-F23341 | Plasmopara sp. | Impatiens textori | Korea; Hongcheon (2008) | MK067073/MK067061/MK067085 |
| YC24170 | KUS-F24170 | Plasmopara sp. | Impatiens textori | Korea; Dongducheon (2009) | MK067075/MK067063/MK067087 |
| D178 | KUS-F19509 | Plasmopara sp. | Impatiens textori | Korea; Hongcheon (2003) | MK067067/MK067055/MK067079 |
| D179 | KUS-F19566 | Plasmopara sp. | Impatiens textori | Korea; Pyeongchang (2003) | MK067068/MK067056/MK067080 |
| D201 | KUS-F20250 | Plasmopara sp. | Impatiens textori | Korea; Yangpyeong (2004) | MK067069/MK067057/MK067081 |
The spacer region between cox2 and cox1 genes.
2.2. DNA extraction, PCR amplification, and sequencing
From the herbarium specimens, 5–20 mg of infected plant tissue was taken and ground in a mixer mill (MM2, Retsch, Germany), using 170 mg of glass beads (BioSpec Products, Bartlesville, OK, USA) of 1 mm diameter per sample. Genomic DNA was extracted using the MagListo 5 M plant Genomic DNA Extraction Kit (Bioneer, Daejeon, Korea). Three highly variable regions, ITS1 rDNA, cytochrome c oxidase subunit II (cox2), and the spacer region between cox2 and cox1 genes (coxS) were amplified using oomycete-specific primers, as outlined previously [8,14,28]. Amplicons were visualized on 1.2% agarose gel, purified using an AccuPrep PCR Purification Kit (Bioneer), and sequenced by a DNA sequencing service (Macrogen, Seoul, Korea), with the primers used for amplification.
2.3. Molecular phylogenetic analysis
Sequences were edited with the DNAStar software package (DNAStar, Madison, WI, USA), version 5.05. Alignment of each locus was performed using MAFFT 7 [29] employing the Q-INS-i algorithm [30]. SequenceMatrix 1.7.8 [31] was used for concatenating individual gene sequences and for checking unusually similar or divergent sequences. Phylogenetic trees were constructed by two different methods, minimum evolution (ME) and maximum likelihood (ML) inference. ME analysis was done using MEGA 7.0 [32], with the default settings of the program, except for using the Tamura-Nei model instead of the maximum composite likelihood model. For ML analyses, 1000 rounds of random addition of sequences as well as 1000 fast bootstrap replicates were performed using RAxML 7.0.3 [33] as implemented in raxmlGUI 1.3 [34] using the GTRCAT variant.
2.4. Morphological analysis
Morphological characteristics of sporangiophores and sporangia were investigated using dried herbarium specimens. Photographs were taken at 100× or 200× for sporangiophores and 400× for sporangia and ultimate branchlets, using an AxioCam MRc5 digital camera mounted to a Zeiss Imager M2 AX10 microscope (Carl Zeiss, Jena, Germany). Measurements were done with the software AxioVision LE (Carl Zeiss Imaging Solutions, Munich, Germany) after calibration using a stage micrometer, and reported as follows; (minimum–) standard deviation toward the minimum – mean – standard deviation toward the maximum (–maximum) (n = 50).
3. Results
3.1. Molecular phylogenetic analysis
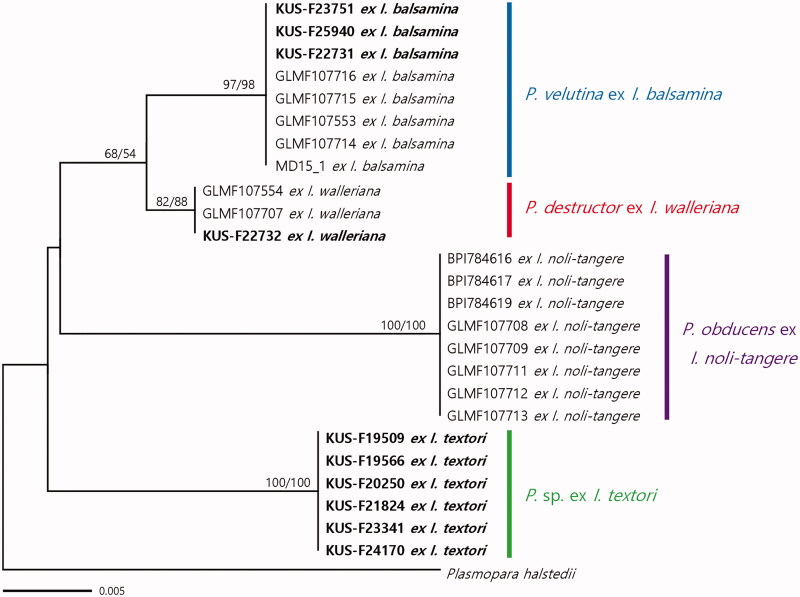
A phylogeny based on the concatenated alignment showed neither conflicting support nor a markedly different topology as compared to individual gene tree inferred from ITS1, cox2, and coxS. The final concatenated alignment had 625 total characters, including 29 variable characters, 23 of which were parsimony-informative. Since the dataset revealed no significant conflicts in the topologies derived from ML and ME analyses, only the tree from the ME inference is shown in Figure 1. In all phylogenetic trees, the six Plasmopara samples from I. textori formed a well-supported group, which was related to P. obducens, P. destructor, and P. velutina. Even in the ITS1 rDNA sequences that cannot be used to discriminate among P. destructor, P. obducens, and P. velutina, Plasmopara sp. ex I. textori exhibited a single nucleotide substitution. In addition, the sequences of Plasmopara sp. ex I. textori were different at 7, 5, and 7 characters out of 203 characters of the cox2 and at 11, 10, and 11 out of 227 in the cox2-1 spacer, as compared to P. destructor, P. obducens, and P. velutina, respectively. The Korean samples ex I. balsamina (n = 3) and I. walleriana (n = 1), clustered with the reference sequences of P. velutina and P. destructor, respectively, with high support values in both ME and ML analyses.
Figure 1.
Minimum evolution tree based on a concatenated alignment of ITS1 rDNA, partial cox2 mtDNA, and a spacer region between cox2 and cox1 genes. Bootstrap support values for minimum evolution and maximum likelihood analyses higher than 50% are given above the branches. The Korean specimens investigated in this study are highlighted in bold. The scale bar represents the number of nucleotide substitutions per site.
3.2. Morphological analysis
Measurements of morphological characteristics of Plasmopara species from four different species of Impatiens are summarized in Table 2. The main morphological characters differentiating Plasmopara sp. affecting I. textori was the height of sporangiophores, the length of the ultimate branchlets, and the breadth of sporangia. In Plasmopara sp., the height of sporangiophores was 541 µm on average, in contrast to those of P. obducens s.str., P. destructor, and P. velutina, which were less than 400 µm on average. The sporangial breadth was 15.3 µm on average in Plasmopara sp., wider than 13.0, 13.8, and 14.6 µm on average as measured for the other three respective species. This also resulted in a lower length to breadth ratio of 1.15 on average, as compared to the respective averages of 1.20, 1.21, and 1.20 in the other three species. The ultimate branchlets of were 10.0 µm long on average in Plasmopara sp., longer than those of P. obducens (av. 7.8 µm), P. destructor (av. 8.4 µm), and P. velutina (av. 8.0 µm). Thus, Plasmopara sp. ex I. textori can be easily distinguished from the other three species known to affect balsamines in Eurasia.
Table 2.
Morphological characteristics of Plasmopara species parasitic on Impatiens species.
| Plasmopara species | P. sp. | P. obducensa | P. velutinaa | P. destructora |
|---|---|---|---|---|
| Host species | Impatiens textori | Impatiens noli-tangere | Impatiens balsamina | Impatiens walleriana |
| Sporangia | ||||
| Length (µm) | (10.5–)15.4–17.6–19.8 (–23.9) | (9.6–)13.6–15.9–18.1 (–23.5) | (10.6–)14.9–17.3–19.6(–31.2) | (10.7–)13.7–15.9–17.9(–23.3) |
| Width (µm) | (9.7–)13.7–15.3–16.8(–19.8) | (8.5–)11.3–13.3–15.4–(–15.5) | (9.1–)12.8–14.6–16.3(–22.9) | (8.5–)11.9–13.7–15.4 (–18.9) |
| Ratio of length/width | (1.00)1.07–1.15–1.24(–1.65) | (1.00)1.08–1.20–1.32(–1.76) | (1.01)1.09–1.20–1.31(–2.00) | (1.00)1.09–1.21–1.33(–1.69) |
| Ultimate branchlets | ||||
| Length (µm) | (4.2–)7.6–10.0–12.4(–17.9) | (3.7–)5.7–7.8–9.9(–14.2) | (3.9–)5.5–7.4–9.35(–16.6) | (3.2–)6.5–9.0–11.5(–16.7) |
| Sporangiophores | ||||
| Length (µm) | (250–)410–540–670(–90) | (90–)261–373–485(–659) | (209–)302–393–483(–650) | (189–)325–429–532(–728) |
| Height of first branching (µm) | (110–)230–340–450(–710) | (54–)124–197–271(–417) | (60–)127–202–277(–422) | (60–)142–217–292(–433) |
| Ratio of first branching/length | (0.30–)0.50–0.63–0.76(–0.85) | (0.19–)0.43–0.53–0.63(–0.80) | (0.16–)0.38–0.51–0.64(–0.79) | (0.24–)0.39–0.51–0.62(–0.71) |
Measurements of Görg et al. [14].
Numbers with the underlines indicate the average of the measurements.
4. Discussion
The phylogenetic and morphological data of the present study demonstrated that Plasmopara sp. affecting I. textori is not conspecific with the three known species of Plasmopara parasitic on Impatiens species in Eurasia, P. obducens s.str., P. destructor, and P. velutina. These results support a previous study that Plasmopara species parasitic on Impatiens spp. exhibits a high degree of differentiation according to the host species [14]. In addition, it adds evidence to the recently re-appraised view that a narrow species concept reflects the evolutionary history of downy mildews much better than a broad species concept, as exemplified on other species complexes, such as the ones around Bremia lactucae [35–41], Hyaloperonospora parasitica [42–48], Peronospora farinosa [7–9,49], Peronospora lamii [12,13,50], and Plasmopara halstedii [24,26,51–53]. The downy mildew of I. textori is one of the most common downy mildew diseases throughout South Korea [21] and is common in China [54,55] and Japan [56] where the host plant is also widely distributed. Notably, the present result that the downy mildew pathogens of I. textori and I. noli-tangere are distinct reflects the occurrence patterns of the disease on the host plants well. I. textori often grows together with I. noli-tangere, the host plant of P. obducens, and both prefer to moist to wet habitats, but in most of the cases, only I. textori exhibits downy mildew infection, while I. noli-tangere remains healthy and downy mildew incidences are still comparatively rare.
In Korea, the downy mildews of I. balsamina and I. walleriana have recently been reported [22]. In the present study, the downy mildew on I. walleriana and I. balsamina, which have previously attributed to P. obducens, were re-identified as P. destructor and P. velutina, respectively. Interestingly, Salgado-Salazar et al. [15] in a population genetics approach limited to a small region of the ribosomal cistron came to the conclusion that the three closely related species on Impatiens, P. obducens, P. destructor, and P. velutina were probably conspecific, as in the sequence stretch analyzed there were no clear-cut differences between P. destructor and P. velutina and they reported that there is some overlap in the morphology. However, this result is ambiguous, because (i) morphological characters can be significantly different even if there is some overlap, (ii) most parts of the ITS were considered in the population study, which has the issue of repeat-driven instability of ITS in downy mildews with pyriform haustoria [57], (iii) cloned sequences from a non-proofreading enzyme were used to determine sequence types, (iv) no dedicated measures have been taken to make sure that herbarium specimens used were not contaminated with spores of other, more recent collections. Considering also that downy mildew on cultivated I. walleriana has not been reported prior to this millennium, while the native pathogens, P. obducens (in Europe and probably North America), Plasmopara sp. in North-East Asia and P. velutina (in Asia) were present for decades before the epidemics, an invasion by a previously unrecognized species, such as in the case of Pe. belbahrii [12] or P. muralis [25] seems more plausible. While given a dynamic host shift ability of obligate biotrophic pathogens [58], including downy mildews, it is conceivable that outside the native range the newly occurring pathogen, P. destructor is able to infect additional species of Impatiens, lumping the species on impatiens together would be a huge step backwards. The influential publication of Yerkes and Shaw [5] who favored a very broad species, hampered appropriate recognition and quarantine regulations for newly occurring pathogens, resulting in the too late recognition that newly occurring species were the cause of disease [4]. Thus, even in the event that population genetics studies should in the future reveal that P. destructor and P. velutina should rather be treated as subspecies because of significant admixture, it seems better to treat them as separate species, also as the evidence reported in this study rather supports recognition as independent species. As I. walleriana was not cultivated in Korea before the early 2000s when it has been imported for commercial purposes, it is conceivable that I. walleriana might have been accompanied by the pathogen when being introduced to Korea. In the case of I. balsamina, it is unclear from where the downy mildew species responsible for the present epidemic in Korea has originated. However, it seems plausible that the pathogen has been introduced rather recently, as this plant had been widely distributed throughout Korea for a long time, but there was no report of downy mildew incidence until 2007 [22]. Since the two cultivated balsamines are popular in outdoor gardens and indoor flowerpots in Korea, their diseases have the potential to cause significant economic losses in nurseries and landscape businesses, like in many other countries [14,15]. However, the causal agent of downy mildew on I. textori is deeply separated from the other downy mildew species on balsamines and unlikely to be of concern for the growing of ornamental balsamines, even though infection trials should probably be done to clarify the risk of a potential host jump.
5. Taxonomy
Based on the results of both the phylogenetic and morphological analyses, the downy mildew pathogen affecting I. textori is described below as a new species in the genus Plasmopara. The downy mildew specimens from I. walleriana and I. balsamina were confirmed as P. destructor and P. velutina, respectively, both of which are new to Korea.
Plasmopara elegantissima Y.J. Choi, Görg & Thines, sp. nov. MB 820725 (Figure 2)
Figure 2.
Plasmopara sp. on Impatiens textori. (A & B) Downy mildew symptoms on the lower and upper surface on Impatiens textori. (C & D) Sporangiophore; (E–H) Ultimate branchlets; (I–L) Sporangium. Scale bars: 100 µm for sporangiophores; 10 µm for ultimate branchlets and sporangia.
Etymology: Referring to the elegant appearance of the sporangiophores of this species.
Description: Lesions commonly present mainly on leaves, but rarely on stems, vein-limited, poly-angular, frequently covering larger areas by coalescing; infected tissues become reddish, later turning necrotic. Down developing when infected leaf tissue is still alive, hypophyllous, whitish, consisting of agglomerated to scattered sporangiophores, often felt-like, dense. Haustoria intracellular, not branched, flask-shaped, symmetrical, 11–20 μm diam., with a stalk at the part of entry into the host cell, surrounded by sheaths of 0.5–2 µm thickness. Sporangiophores emerging through stomata, straight to substraight, (250–)410–540–670(–900) µm long; trunk straight, (110–)230–340–450(–710) µm long, 4–9 µm wide, a ratio of the length of sporangiophores to the length of trunk (1.23–)1.37–1.63–1.89(–2.91), base not or somewhat swollen, up to 13 µm wide, callose plugs commonly present; branches arising at right angle to the main axis, monopodial, branching 3 to 5(–6) orders; callose plugs frequently present. Ultimate branchlets in pairs or three, straight to slightly curved, (4.2–)7.6–10.0–12.4(–17.9) µm long, 1.5–2.5 µm wide at the base; tip truncate or cup-like, rarely swollen. Sporangia oblong, broadly ellipsoidal or ovoid, but sometimes nearly globose, (10.5–)15.4–17.6–19.8(–23.9) µm long, (9.7–)13.7–15.3–16.8(–19.8) µm wide, a ratio of length to width (1.00–)1.07–1.15–1.24(–1.65), greatest width sub-median or median, tip round, base broadly round or subtruncate; pedicel absent or only a minute protuberance visible at the point of attachment to the sporangiophores; pore 2–5 µm wide, with a plug of 0.5–1.5 µm thickness, dissolving during germination. Zoospores releasing through an apical pore; encysted zoospores 8–10 µm diam., with thick wall, germinating by a germ tube, up to 750 µm long, sometimes branched 1–3 times. Resting organ not seen.
Diagnosis: Sporangiophores and ultimate branchlets longer and sporangia wider, as compared to P. obducens, P. destructor, and P. velutina.
Habitat: On living leaves of I. textori (Balsaminaceae).
Typus: KOREA; Dongduchoen-si; Sangbongam-dong; Mt. Soyo (37°56'40"N 127°05'11"E), on leaves of Impatiens textori, 22 June 2009, leg. Y.J. Choi & H.D. Shin, ZEVCFG0000000020 (holotypus), KUS-F24170 (isotypus).
Additional specimens for morphological investigation: KUS-F15905 (29 May 1999, Seochon-ri, Chuncheon), 19204 (11 Oct. 2002, Seochon-ri, Chuncheon), 19547 (4 Jun. 2003, Experimental Forest of Korea University, Yangpyeong), 19790 (7 Oct. 2003, Goeun-ri, Chuncheon), 19821 (7 Oct. 2003, Seochon-ri, Chuncheon), 20228 (26 May 2004, near Yeonhwa Temple, Bukbang-myon, Hongcheon), 20223 (24 May 2004, Sangnam-myon, Inje), 20255 (1 Jun. 2004, Seosang-ri, Chuncheon), 20270 (1 Jun. 2004, Seochon-ri, Chuncheon), 20291 (4 Jun. 2004, near Sangdang fortress, Cheongju), 20310 (18 Jun. 2004, Experimental Forest of Kangwon National University, Hongcheon), 20343 (23 Jun. 2004, Mt. Cheongtae, Hoengseong), 20691 (10 Sept. 2004, Goeun-ri, Chuncheon), 20719 (18 Sept. 2004, near Yeonhwa Temple, Bukbang-myon, Hongcheon), 21144 (27 May 2005, Goeun-ri, Chuncheon), 21215 (6 Jun. 2005, Bukbang-myon, Hongcheon), 21267 (12 Jun. 2005, Ecological Park, Hongcheon), 21902 (22 Jun. 2006, Hoengseong Natural Recreation Forest, Hoengseong).
Plasmopara destructor Görg & Thines, Mycological Progress 16 (8): 797 (2017) [MB#820725]
Description: Sporangiophores emerging through stomata, hyaline, 200–700 µm long, base slightly swollen, branched monopodially 4–6 orders, ultimate branchlets were straight but often slightly curved, 3–15 µm long, sporangia globose to ovoid, hyaline, 10–25 × 10–20 µm.
Specimen examined: KOREA; Gangneung-si; Gyeongpo-dong; near Gyeongpo provincial park (37°47′20″N 128°53′28″E), on leaves of I. walleriana, 26 July 2007, leg. Y.J. Choi & H.D. Shin, ZEVCFG0000000021 (KUS-F22732).
Habitat: On living leaves of I. walleriana (Balsaminaceae)
Plasmopara velutina Görg & Thines, Mycological Progress 16 (8): 797 (2017) [MB#820726]
Description: Sporangiophores emerging through stomata, hyaline, 200–700 µm long, bases not or somewhat swollen; branched monopodially 4–6 orders; ultimate branchlets slightly curved, 5–14 µm long with truncate tip. Sporangia globose to ovoid, hyaline, 10–30 × 10–22 µm.
Specimen examined: KOREA; Gangneung-si; Gyeongpo-dong; near Gyeongpo Provincial Park (37°47′20″N 128°53′28″E), on leaves of I. walleriana, 26 July 2007, leg. Y.J. Choi & H.D. Shin, ZEVCFG0000000022 (KUS-F22731).
Habitat: On living leaves of I. balsamina (Balsaminaceae).
Acknowledgements
YJC and HDS collected all downy mildew samples included in the present study.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- 1.Beakes GW, Honda D, Thines M.. Systematics of the Straminipila: Labyrinthulomycota, Hyphochytriomycota, and Oomycota In: McLaughlin DJ, Spatafora J, editors. Systematics and evolution. New York: Springer; 2014. p. 39–97. [Google Scholar]
- 2.Thines M. Phylogeny and evolution of plant pathogenic oomycetes – a global overview. Eur J Plant Pathol. 2014;138(3):431–447. [Google Scholar]
- 3.Beakes GW, Thines M.. Hyphochytriomycota and Oomycota In: Archibald JM, Simpson AGB, Slamovits CH, Margulis L, Melkonian M, Chapman DJ, Corliss JO, editors. Handbook of the Protists. Cham: Springer International Publishing; 2016. p. 1–71. [Google Scholar]
- 4.Thines M, Choi YJ.. Evolution, diversity, and taxonomy of the Peronosporaceae, with focus on the genus Peronospora. Phytopathology. 2016;106(1):6–18. [DOI] [PubMed] [Google Scholar]
- 5.Yerkes WD, Shaw CG.. Taxonomy of the Peronospora species on Cruciferae and Chenopodiaceae. Phytopathology. 1959;49:499–507. [Google Scholar]
- 6.Choi YJ, Thines M.. (2288) Proposal to reject the name Botrytis farinosa (Peronospora farinosa) (Peronosporaceae: Oomycetes). Taxon. 2014;63(3):675–676. [Google Scholar]
- 7.Choi YJ, Hong SB, Shin HD.. Re-consideration of Peronospora farinosa infecting Spinacia oleracea as distinct species, Peronospora effusa. Mycol Res. 2007;111(4):381–391. [DOI] [PubMed] [Google Scholar]
- 8.Choi YJ, Klosterman SJ, Kummer V, et al. . Multi-locus tree and species tree approaches toward resolving a complex clade of downy mildews (Straminipila, Oomycota), including pathogens of beet and spinach. Mol Phylogenet Evol. 2015;86:24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi YJ, Danielsen S, Lübeck M, et al. . Morphological and molecular characterization of the causal agent of downy mildew on quinoa (Chenopodium quinoa). Mycopathologia. 2010;169(5):403–412. [DOI] [PubMed] [Google Scholar]
- 10.Choi YJ, Kruse J, Thines M.. Hyaloperonospora erucae sp. nov. (Peronosporaceae; Oomycota), the downy mildew pathogen of arugula (Eruca sativa). Eur J Plant Pathol. 2018;151(2):549–555. [Google Scholar]
- 11.Choi YJ, Thines M, Choi IY, et al. . Perofascia is not monotypic – the description of the second taxon affecting the South American crop maca (Lepidium meyenii). Mycol Prog. 2017;16(9):857–864. [Google Scholar]
- 12.Thines M, Telle S, Ploch S, et al. . Identity of the downy mildew pathogens of basil, coleus, and sage with implications for quarantine measures. Mycol Res. 2009;113(5):532–540. [DOI] [PubMed] [Google Scholar]
- 13.Choi YJ, Shin HD, Thines M.. Two novel Peronospora species are associated with recent reports of downy mildew on sages. Mycol Res. 2009;113(12):1340–1350. [DOI] [PubMed] [Google Scholar]
- 14.Görg M, Ploch S, Kruse J, et al. . Revision of Plasmopara (Oomycota, Peronosporales) parasitic to Impatiens. Mycol Prog. 2017;16(8):791–799. [Google Scholar]
- 15.Salgado-Salazar C, LeBlanc N, Ismaiel A, et al. . Genetic variation of the pathogen causing impatiens downy mildew predating and including twenty-first century epidemics on Impatiens walleriana. Plant Dis. 2018;102(12):2411–2420. [DOI] [PubMed] [Google Scholar]
- 16.Hattori M, Nagano Y, Shinohara Y, et al. . Pattern of flower size variation along an altitudinal gradient differs between Impatiens textori and Impatiens noli-tangere. J Plant Interact. 2016;11(1):152–157. [Google Scholar]
- 17.Lane CR, Beales PA, O'Neill TM, et al. . First report of Impatiens downy mildew (Plasmopara obducens) in the UK. Plant Pathol. 2005;54(2):243–243. [Google Scholar]
- 18.Farr DF, Rossman AY.. Fungal databases. ARS, USDA: Systematic Mycology and Microbiology Laboratory; 2020. [Google Scholar]
- 19.Voglmayr H, Thines M.. Phylogenetic relationships and nomenclature of Bremiella sphaerosperma (Chromista, Peronosporales). Mycotaxon. 2007;100:11–20. [Google Scholar]
- 20.Shin HD, Choi YJ.. A first check-list of Peronosporaceae from Korea. Mycotaxon. 2003;86:249–267. [Google Scholar]
- 21.Shin HD, Choi YJ.. Peronosporaceae of Korea. Suwon: National Institute of Agricultural Science and Technology; 2006. [Google Scholar]
- 22.Choi YJ, Han JG, Park MJ, et al. . Downy mildew of Impatiens balsamina and I. walleriana in Korea. Plant Pathol J. 2009;25(4):433–433. [Google Scholar]
- 23.Voglmayr H, Fatehi J, Constantinescu O.. Revision of Plasmopara (Chromista, Peronosporales) parasitic on Geraniaceae. Mycol Res. 2006;110(6):633–645. [DOI] [PubMed] [Google Scholar]
- 24.Constantinescu O, Thines M.. Plasmopara halstedii is absent from Australia and New Zealand. Pol Bot J. 2010;55:293–298. [Google Scholar]
- 25.Thines M. Recent outbreaks of downy mildew on grape ivy (Parthenocissus tricuspidata, Vitaceae) in Germany are caused by a new species of Plasmopara. Mycol Prog. 2011;10(4):415–422. [Google Scholar]
- 26.Choi YJ, Kiss L, Vajna L, et al. . Characterization of a Plasmopara species on Ambrosia artemisiifolia, and notes on P. halstedii, based on morphology and multiple gene phylogenies. Mycol Res. 2009;113(10):1127–1136. [DOI] [PubMed] [Google Scholar]
- 27.Yuan YM, Song Y, Geuten K, et al. . Phylogeny and biogeography of Balsaminaceae inferred from ITS sequences. Taxon. 2004;53(2):391–403. [Google Scholar]
- 28.Choi YJ, Beakes G, Glockling S, et al. . Towards a universal barcode of oomycetes – a comparison of the cox1 and cox2 loci. Mol Ecol Resour. 2015;15(6):1275–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katoh K, Standley DM.. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katoh K, Toh H.. Improved accuracy of multiple ncRNA alignment by incorporating structural information into a MAFFT-based framework. BMC Bioinformatics. 2008;9:212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaidya G, Lohman DJ, Meier R.. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics. 2011;27(2):171–180. [DOI] [PubMed] [Google Scholar]
- 32.Kumar S, Stecher G, Tamura K.. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. [DOI] [PubMed] [Google Scholar]
- 34.Silvestro D, Michalak I.. raxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 2012;12(4):335–337. [Google Scholar]
- 35.Voglmayr H, Riethmüller A, Göker M, et al. . Phylogenetic relationships of Plasmopara, Bremia and other genera of downy mildew pathogens with pyriform haustoria based on Bayesian analysis of partial LSU rDNA sequence data. Mycol Res. 2004;108(9):1011–1024. [DOI] [PubMed] [Google Scholar]
- 36.Choi YJ, Thines M, Runge F, et al. . Evidence for high degrees of specialisation, evolutionary diversity, and morphological distinctiveness in the genus Bremia. Fungal Biol. 2011;115(2):102–111. [DOI] [PubMed] [Google Scholar]
- 37.Choi YJ, Hong SB, Shin HD.. Extreme size and sequence variation in the ITS rDNA of Bremia lactucae. Mycopathologia. 2007;163(2):91–95. [DOI] [PubMed] [Google Scholar]
- 38.Park JH, Thines M, Lee HB, et al. . Bremia polycephala and Bremia sawadae spp. nov. (Peronosporaceae; Oomycota), parasitic to Northeast Asian Asteraceae. Nova Hedw. 2018;107(3):303–314. [Google Scholar]
- 39.Choi YJ, Thines M.. Host jumps and radiation, not co-divergence drives diversification of obligate pathogens. A case study in downy mildews and Asteraceae. PLoS One. 2015;10(7):e0133655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi YJ, Wong J, Runge F, et al. . BrRxLR11 – a new phylogenetic marker with high resolution in the downy mildew genus Bremia and related genera. Mycol Prog. 2017;16(2):185–190. [Google Scholar]
- 41.Choi YJ, Park JH, Lee J, et al. . Bremia itoana (Oomycota, Peronosporales), a specialized downy mildew pathogen on an East Asian plant, Crepidiastrum sonchifolium (Asteraceae). Mycobiology. 2018;46(4):416–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi YJ, Hong SB, Shin HD.. Diversity of the Hyaloperonospora parasitica complex from core brassicaceous hosts based on ITS rDNA sequences. Mycol Res. 2003;107(11):1314–1322. [DOI] [PubMed] [Google Scholar]
- 43.Voglmayr H, Choi YJ, Shin HD.. Multigene phylogeny, taxonomy and reclassification of Hyaloperonospora on Cardamine. Mycol Prog. 2014;13(1):131–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Voglmayr H, Göker M.. Morphology and phylogeny of Hyaloperonospora erophilae and H. praecox sp. nov., two downy mildew species co-occurring on Draba verna sensu lato. Mycol Progress. 2011;10(3):283–292. [Google Scholar]
- 45.Choi YJ, Shin HD, Voglmayr H.. Reclassification of two Peronospora species parasitic on Draba in Hyaloperonospora based on morphological and molecular phylogenetic data. Mycopathologia. 2011;171(2):151–159. [DOI] [PubMed] [Google Scholar]
- 46.Göker M, Voglmayr H, García-Blázquez G, et al. . Species delimitation in downy mildews: the case of Hyaloperonospora in the light of nuclear ribosomal ITS and LSU sequences. Mycol Res. 2009;113(3):308–325. [DOI] [PubMed] [Google Scholar]
- 47.Göker M, Riethmüller A, Voglmayr H, et al. . Phylogeny of Hyaloperonospora based on nuclear ribosomal internal transcribed spacer sequences. Mycol Prog. 2004;3(2):83–94. [Google Scholar]
- 48.Lee JS, Lee HB, Shin HD, et al. . Diversity, phylogeny, and host-specialization of Hyaloperonospora species in Korea. Mycobiology. 2017;45(3):139–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choi YJ, Denchev CM, Shin HD.. Morphological and molecular analyses support the existence of host-specific Peronospora species infecting Chenopodium. Mycopathologia. 2008;165(3):155–164. [DOI] [PubMed] [Google Scholar]
- 50.Belbahri L, Calmin G, Pawlowski J, et al. . Phylogenetic analysis and real time PCR detection of a presumbably undescribed Peronospora species on sweet basil and sage. Mycol Res. 2005;109(11):1276–1287. [DOI] [PubMed] [Google Scholar]
- 51.Spring O, Voglmayr H, Riethmüller A, et al. . Characterization of a Plasmopara isolate from Helianthus × laetiflorus based on cross infection, morphological, fatty acids and molecular phylogenetic data. Mycol Prog. 2003;2(3):163–170. [Google Scholar]
- 52.Komjáti H, Walcz I, Virányi F, et al. . Characteristics of a Plasmopara angustiterminalis isolate from Xanthium strumarium. Eur J Plant Pathol. 2007;119(4):421–428. [Google Scholar]
- 53.Duarte LL, Choi YJ, Soares DJ, et al. . Plasmopara invertifolia sp. nov. causing downy mildew on Helichrysum bracteatum (Asteraceae). Mycol Prog. 2014;13(2):285–289. [Google Scholar]
- 54.Ling L. Host index of the parasitic fungi of Szechwan, China. 1948. [Google Scholar]
- 55.Tai FL. Sylloge fungorum sinicorum. Peking: Science Press; 1979. [Google Scholar]
- 56.Ito S. Mycological Flora of Japan. Tokyo: Yokendo; 1936. [Google Scholar]
- 57.Thines M. Characterisation and phylogeny of repeated elements giving rise to exceptional length of ITS2 in several downy mildew genera (Peronosporaceae). Fungal Genet Biol. 2007;44(3):199–207. [DOI] [PubMed] [Google Scholar]
- 58.Thines M. An evolutionary framework for host shifts-jumping ships for survival. New Phytol. 2019;224(2):605–617. [DOI] [PubMed] [Google Scholar]