SUMMARY
Complex ethological behaviors could be constructed from finite modules that are reproducible functional units of behavior. Here, we test this idea for foraging and develop methods to dissect rich behavior patterns in mice. We uncover discrete modules of foraging behavior reproducible across different strains and ages, as well as nonmodular behavioral sequences. Modules differ in terms of form, expression frequency, and expression timing and are expressed in a probabilistically determined order. Modules shape economic patterns of feeding, exposure, activity, and perseveration responses. The modular architecture of foraging changes developmentally, and different developmental, genetic, and parental effects are found to shape the expression of specific modules. Dissecting modules from complex patterns is powerful for phenotype analysis. We discover that both parental alleles of the imprinted Prader-Willi syndrome gene Magel2 are functional in mice but regulate different modules. Our study found that complex economic patterns are built from finite, genetically controlled modules.
Graphical Abstract
In Brief
The principles and mechanisms involved in constructing complex behavior patterns are not well defined. Stacher Hörndli et al. find that complex foraging patterns in mice are constructed from finite modules, defined as significantly reproducible behavioral sequences. Modules are expressed in a probabilistically defined order to construct complex patterns and controlled by genetic mechanisms.
INTRODUCTION
Behavioral studies often focus on the analysis of specific behavioral responses. Consequently, the principles and mechanisms involved in constructing complex behavior patterns are not well defined, and most genomic elements influencing brain development and function shape instinctive and learned behavior patterns in ways that are unknown (Bakken et al., 2016; Kang et al., 2011; Miller et al., 2014; Silbereis et al., 2016; Thompson et al., 2014). Behavior could be constructed from finite “modules” that are reproducible units of behavior (Vogelstein et al., 2014; Wiltschko et al., 2015; Yang and Shah, 2014). From this perspective, genomic elements evolved to shape behavioral modules, which in turn are building blocks for more complex patterns. Changes to the genome, and brain structure and function, would create new behavior patterns by altering the expression of existing modules and/or by creating novel modules. However, for most complex ethological behaviors, methods are lacking to test this hypothesis. It is generally unclear which behavioral parameters delineate modules and what level of repetition and standardization characterize modules in a population of individuals versus nonmodular, stochastic behavioral sequences. Solving these problems is important to understand the architecture of complex, ethological patterns and determine whether changes to the expression frequency, timing, and sequential order of specific modules underlies different patterns. Developing methods to resolve modules will also facilitate studies of the mechanistic basis of behavior, allowing us to study how module form and expression changes in response to targeted genetic and/or environmental perturbations to better understand the basis of normal and abnormal patterns.
Our study investigates the existence of modules in foraging. Foraging encompasses a rich and deeply conserved repertoire of behavioral responses (Stephens et al., 2007). Many neural systems are involved in foraging, including novelty seeking, anxiety, reward, perseveration, sensorimotor integration, hunger, satiety, attention, activity, navigation, and learning and memory (Stephens et al., 2007). Natural selection shapes foraging patterns through these systems to minimize predation risk and energy expenditure while maximizing caloric intake (Brown and Laundré, 1999; Pulliam and Charnov, 1977; Stephens et al., 2007). Understanding different foraging patterns and tradeoffs is important for understanding the basis of economic behavior (Brown and Laundré, 1999; Pulliam and Charnov, 1977; Stephens et al., 2007), which in turn shapes human obesity, addiction, fear, anxiety, and psychiatric disorders (Monterosso et al., 2012; Rowland et al., 2008; Sharp et al., 2012). In addition, selective pressures on foraging shaped mammalian life histories, including brain and behavioral developmental processes (Barton, 2012; Jones, 2011; Melin et al., 2014; Reader and Laland, 2002; Sol et al., 2005; Walker et al., 2006). Indeed, major mammalian developmental milestones are linked to foraging, such as the elimination of suckling during nursing and emergence of independent foraging after weaning (Galef, 1981; Hewlett and Lamb, 2010). Therefore, uncovering modules of foraging at different ages could deepen our understanding of the basis of complex economic patterns and mammalian behavioral development.
In general, behavioral sequences during foraging could be infinitely variable or involve finite, genetically controlled modules. Here, we test the hypothesis that foraging is composed of finite modules of behavior that underlie different economic patterns of food intake, exposure, activity, and perseveration in mice (Figure 1A). We introduce a rich foraging behavior paradigm in which freely behaving mice express complex, ethological patterns. The behavioral measures that best delineate which behavioral sequences may be modules are investigated and a methodology to identify their reproducibility is introduced. We test whether different modules have different forms, expression frequencies, temporal properties, and orders of expression. Our study then determines whether modules are building blocks for different economic behavior patterns and have a stereotyped developmental progression across different strains of mice. Finally, we test whether modules are genetically regulated and if different genetic and parental effects can be mapped onto discrete modules. In a proof-of-principle experiment, we investigate foraging modules in mutant mice for the imprinted Prader-Willi syndrome (PWS) and autism-linked gene Magel2 (Bervini and Herzog, 2013; Cassidy et al., 2012; Kozlov et al., 2007; Mercer et al., 2009; Schaller et al., 2010). We interrogate the functions of the expressed paternal and imprinted maternal Magel2 alleles, uncovering significant but distinct behavioral effects for each parental allele. Our data show foraging involves finite, genetically controlled modules of behavior and provide a deeper understanding of how the genome shapes complex economic behavior patterns, behavioral development, and abnormal behavior in mouse models of human brain disorders.
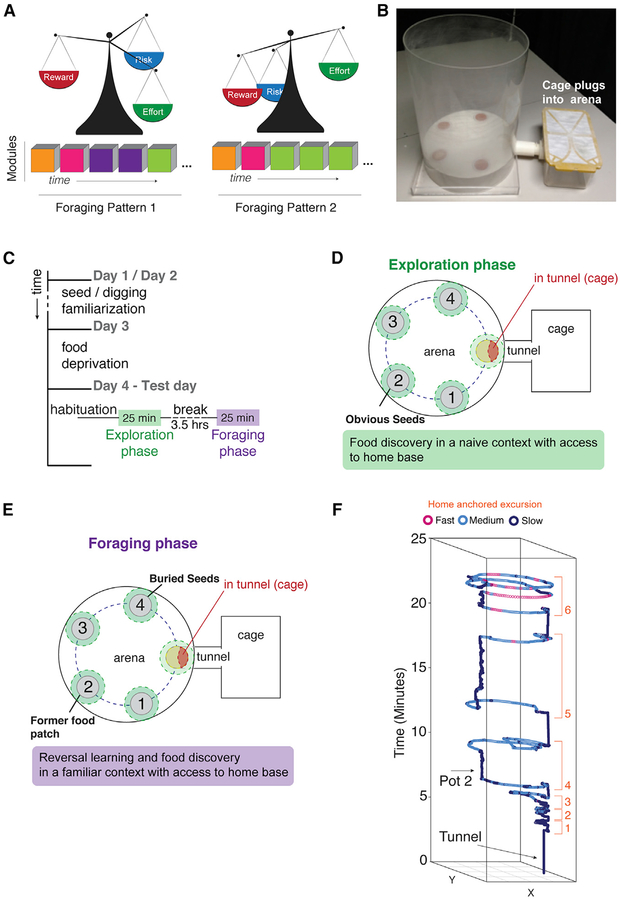
Figure 1. A Paradigm to Dissect Rich Foraging Patterns in Mice.
(A) Schematic depiction of the study hypothesis. Modules are finite, reproducible behavioral sequences that are building blocks for different patterns over time (pattern 1 versus 2). Different patterns are created by changing the expression frequency, timing, and sequential order of modules of behavior (colored blocks). Changes to foraging patterns shape the relative economic balance of caloric intake (reward), predation risk (risk), and energy expenditure (effort).
(B–E) Foraging assay paradigm.
(B) The foraging arena is constructed with a platform that holds pots containing sand and one with sand and seeds. The mouse home base cage plugs into the arena through a tunnel on the side.
(C) A summary of the foraging testing workflow.
(D) The naive exploration phase arena configuration.
(E) The familiar foraging phase arena configuration in which the seeds are hidden in the sand in pot 4 and pot 2 is the formerly learned food patch.
(F) An example of x-y tracking of the body center point over time during the foraging assay. The mice perform repeated round trip excursions from the home base cage (orange numbered track sections). Different excursions have different behavioral and locomotor features (gait speed shown by colors in legend). Modules are uncovered from excursions using DeepFeats.
RESULTS
A Paradigm to Study the Architecture of Spontaneous Mouse Foraging Patterns
We define modules as functionally and mechanistically distinct behavioral sequences that are reproducible across different individuals. To test for modules in complex, spontaneous mouse foraging patterns, we devised a conceptual and experimental workflow that guides the structure of this paper (Figure S1A). To begin, we required an assay in which mice exhibit rich, spontaneous, and ethologically relevant foraging patterns. Our foraging paradigm integrates concepts from behavioral ecology, including free access to a home base (Figure 1B; STAR Methods) and food patches of seeds in sand pots (Brown and Kotler, 1994; Stephens et al., 2007). Although most lab tests do not incorporate a home, it could be critical for natural, modular structure in complex behavior (Eilam and Golani, 1989). For the home base, standard mouse cages are “plugged” into the foraging arena via a connecting tunnel (Figure 1B). The foraging arena contains four pots filled with sand; one has seeds sprinkled atop to create a food patch.
Behavior testing encompasses two phases: a naive exploration phase and a more familiar foraging phase (Figures 1C–1E). During the 25-min exploration phase (Figure 1D), naive mice express behavioral sequences related to the exploration of a novel environment and the discovery and consumption of food in a novel food patch (pot 2; Video S1). 4 h later, the mice are again given 25 min of access to the arena in the foraging phase, in which the seeds are now buried in the sand in pot 4 (Figure 1E). Therefore, mice express behavioral sequences related to their expectation of the former food patch in the now-familiar environment and the discovery of a new, hidden food source (pot 4) (Video S2).
Our study tests whether foraging modules exist and are controlled by different developmental, genetic, and parental effects. Since independent foraging emerges at weaning, we profile developmental changes in early-weaning age (postnatal day 15 [P15]), weaning age (P20), post-weaning age (P25), and adult mice. To test for genetic and parental effects, our design uses reciprocal crosses of the distant subspecies, Mus musculus castaneous (Cast) and C57BL6/J (B6) (Zheng et al., 2014). By comparing the F1cb (Cast mother × B6 father) and F1bc (B6 mother × Cast father) hybrid offspring from these crosses, we test how parental effects influence foraging (Figures 1 and S1B). Parental effects in this design encompass environmental parental effects related to the in utero environment or parental behaviors and genetic parental effects involving changes to genes preferentially inherited from one parent (e.g., mitochondrial DNA) (Lacey, 1998). Genetic effects due to differences between Cast and B6 alleles are uncovered by comparing F1bc offspring to B6 offspring or F1cb offspring to Cast offspring (Figure S1B). This allows us to enrich for genetic effects because the mice share maternal effects but are genetically different (Figure S1B); however, possible paternal influences are not eliminated.
At P15, only ~20% of B6 offspring entered the arena from the home base, while over 80% of F1cb, F1bc, and Cast offspring entered. Of the P15 F1cb, F1bc, and Cast pups tested, only ~20% of the animals consumed seeds. Thus, while the full repertoire of foraging behavior has not developed at P15, early-stage patterns are emerging. By P20, all mice enter the arena, forage, and consume seeds. We profiled foraging for 191 mice across the different ages (P15, P20, P25, and adult), genetic backgrounds, and crosses (B6, F1cb, F1bc, and Cast).
Identification of Behavioral Measures to Resolve Modularity from Foraging
We hypothesized that home base foraging excursions constitute discrete modules of behavior (Figure 1F). However, the best approach to detect potential modules was not known. In the process of solving this problem, we developed a data analysis pipeline we call “DeepFeats” (Figure S1A). DeepFeats takes x-y tracking data from commercial tracking software as an input and extracts discrete behavioral sequences. We extract data for round trip foraging excursions from the home. Each excursion is assigned a concise idiosyncratic module alignment report (CIMAR) string to record the position of the excursion in the data, the subject (mouse), time of expression, and other details. CIMAR string keys allow data retrieval, parsing, quantification, and analyses of the expression of behavioral excursions. For each excursion, DeepFeats extracts a vector of 57 different behavioral measures describing what the animal did during the excursion (Table S1; STAR Methods). Redundant measures could dominate informative measures for module detection, while a measure set that is too sparse may fail to resolve important modules. We used a multi-step approach to ascertain the best measures to resolve modules.
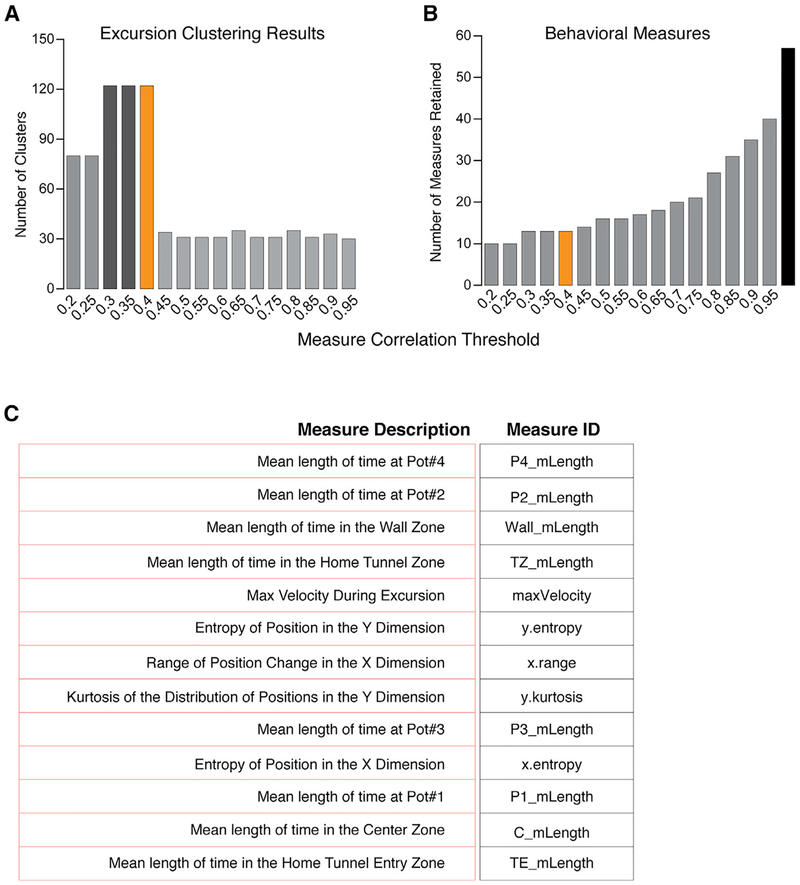
Data were collected for the 57 behavioral measures for each of the 5,687 excursions performed by the 191 mice profiled. Next, we calculated the Pearson correlation for each measure pair and discarded one measure if the pair was correlated above a set threshold ranging from r < 0.1 to 0.95 (STAR Methods). At each threshold, unsupervised hierarchical clustering was performed on the retained measures and excursion clusters were defined using the dynamic tree cut algorithm (Langfelder et al., 2008) (STAR Methods). This approach statistically identifies clusters throughout a dendrogram (Langfelder et al., 2008). We tallied the number of clusters at each threshold (Figure 2A). The r = 0.3 to 0.4 thresholds yield the largest number of clusters and therefore the finest resolution of candidate modules (Figure 2A). At these thresholds, 13 of the 57 measures are retained (Figures 2B and 2C; Table S1).
Figure 2. Identification of Behavioral Measures Best Resolving Candidate Modules from Foraging Patterns.
(A) The bar plot shows the number of excursion clusters detected by Dynamic Tree Cut from unsupervised hierarchical clustering (Ward) of the foraging excursions based on the retained measures at different correlation thresholds. A Pearson correlation threshold of r < 0.3 to 0.4 yields the best resolution of distinct clusters (candidate modules; r < 0.4 was selected for our study [orange bar]). More relaxed thresholds (r > 0.4) may reduce cluster detection due to redundant measures that mask the effect of other measures. Thresholds that are too stringent (r < 0.3) may reduce cluster detection by pruning informative measures.
(B and C) The bar plot shows the number of behavioral measures retained at different Pearson correlation thresholds (B). A total of 57 (black bar) measures are tested and the number retained at the selected threshold (r < 0.4) is 13 (orange bar).
(C) The identity of the 13 measures that best resolve excursion clusters (candidate modules) are shown.
Based on the above results, we chose a threshold of r < 0.4, identifying 122 clusters in total (Figure 2A). We tested whether this number of clusters could be found by chance (Figure S2A; STAR Methods) and determined the observed number of clusters is significantly fewer than the number found from randomly sampled data (Figure S2B; p < 1 × 10−4, lower-tailed test). Therefore, bona fide clusters of excursions exist and our results uncover 13 behavioral measures that effectively resolve discrete subtypes of foraging excursions.
Discovery of 71 Foraging Modules, as well as Nonmodular Behavior Sequences
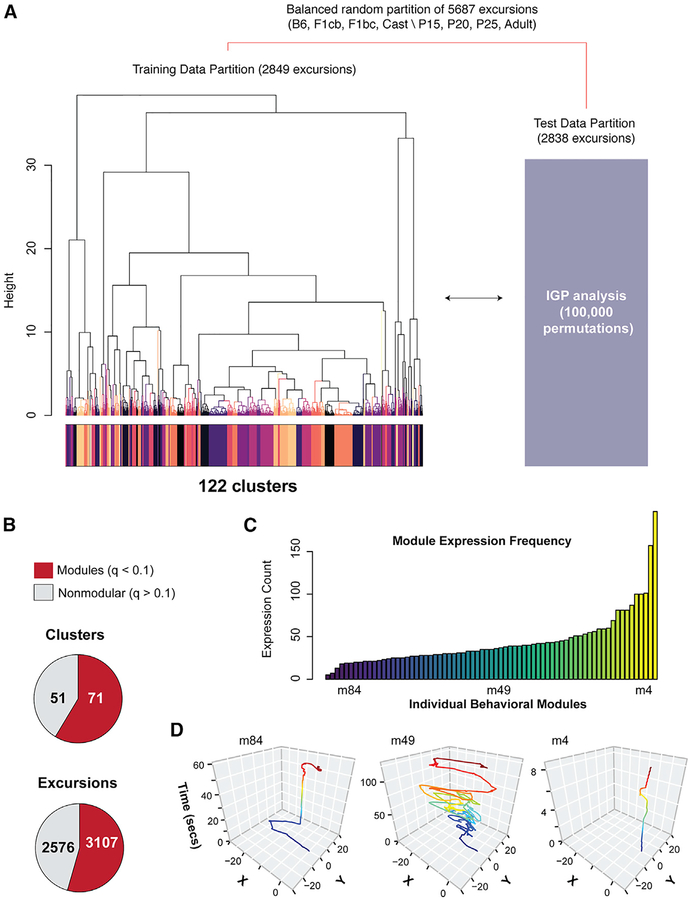
Foraging modules are behavioral sequences reproducible across different mice. Although we identified subtypes of excursions above, our null hypothesis is that the excursion clusters are not reproducible modules and instead reflect stochastic behavioral sequences that we call “nonmodular.” To test this null, we used the in-group proportion (IGP) statistical method for evaluating cluster reproducibility (Kapp and Tibshirani, 2007). The data for the 5,687 excursions performed by the 191 mice profiled were randomly partitioned into training and test datasets balanced by genotype, cross, and age (Figure 3A). Clustering and dynamic tree cut identified 122 clusters in the training data (Figure 3A). We then investigated their reproducibility in the test data. We used the training set centroids (mean values for the 13 behavioral measures) to compute the IGP statistic for each cluster based on the test dataset (Figure 3A; STAR Methods). Permutation testing determined if the observed IGP for a cluster was greater than chance, indicating a cohesive, isolated and reproducible cluster across the two datasets (Kapp and Tibshirani, 2007). p values were corrected for multiple testing using the q-value method (Storey, 2003) (Figures S3A and S3B).
Figure 3. Discovery of 71 Significant Modules of Foraging Behavior across Different Ages and Strains of Mice.
(A) Details of the training and testing data partitions and approach to test for significant cluster reproducibility using the IGP permutation test. Unsupervised hierarchical clustering (Ward) and Dynamic Tree Cut analysis identified 122 clusters (colored track) in the training data. The centroids for each training data cluster are used in the IGP permutation test with the test data (100,000 permutations yielded over 5,000 permutations per cluster).
(B) Pie charts show the number of significantly reproducible training data clusters found by the IGP test. The data reveal 71 modules (q < 0.1, red), as well as nonmodular excursions (q > 0.1, gray). The number of excursions in each class is also shown.
(C) A histogram showing the frequency at which each module is expressed across all 191 mice tested. Some are relatively infrequently (e.g., module 84 [m84]), moderately frequently (e.g., module 49), or frequently (e.g., module 4) expressed.
(D) The plots show representative x-y traces of mouse movement over time (seconds) in the arena for examples of infrequently (module 84), moderately (module 49), and frequently (module 4) expressed modules. The traces track the center of the mouse’s body during the round-trip excursions, which start and end at the tunnel to the home (0,0). They are colored according to relative time. Different modules have different movement and temporal characters.
We found 71 significantly reproducible (q < 0.1) excursion clusters. These constitute modules of foraging behavior (Figure 3B). The remaining 51 clusters are not significant (q > 0.1), and we refer to them as nonmodular. The mean IGP was higher for modular clusters than nonmodular clusters (IGPmod/IGPnonmod = 1.6). Overall, 55% of all excursions are revealed to be modular, and 45% are nonmodular (Figure 3B). Some modules are frequently expressed, such as module 4, while others are less frequent (e.g., modules 49 and 84) (Figures 3C and 3D). Modules have different forms (Figure 3D), and the distinguishing characteristics of different modules are apparent from a scaled heatmap of the centroids for each module type (Figure S3C). Our results show that complex foraging patterns from different ages and strains of mice are constructed from finite, reproducible modules, as well as nonmodular behavioral sequences.
The Sequential Order of Module Expression Is Probabilistically Determined and Not Random
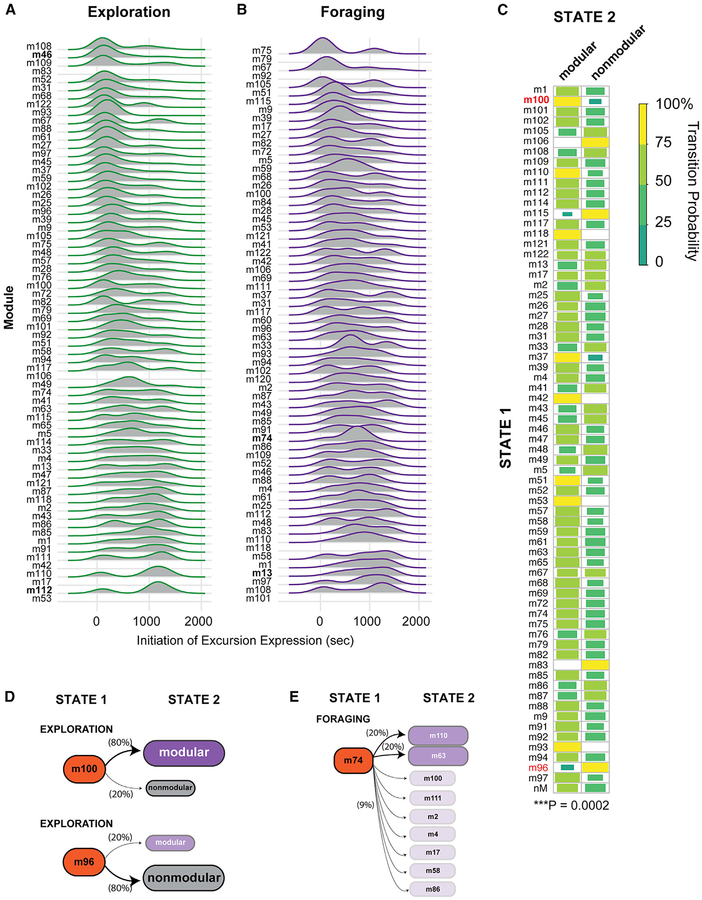
We hypothesized that different modules are characterized by differences in not only form and expression frequency but also expression timing and order. To begin to test this, we determined the expression start time for each excursion relative to the start of the exploration and foraging phase trials. We visualized expression timing patterns for excursions in different modules with ridge-line plots (Figures 4A and 4B). Some are preferentially expressed early, such as module 46 during the exploration phase (Figure 4A). Others are preferentially expressed later, such as module 112 in the exploration phase or modules 74 and 13 in the foraging phase (Figures 4A and 4B). A linear regression analysis of the excursion start times revealed a significant effect of module type on excursion timing in the exploration (p < 2.2 × 10−16) and foraging phases (p = 2.3 × 10−8). Therefore, different modules have significantly different temporal expression properties.
Figure 4. The Expression Order of Modular and Nonmodular Excursions Is Probabilistically Determined.
(A and B) Ridgeplots reveal the distribution of start times for excursions of different module types in the exploration (A) and foraging (B) phases relative to the beginning of the 25-min testing period (data are from all 191 study mice). Modules detailed in the main text are highlighted in bold.
(C) The chart shows the transition probabilities between specific modules (y axis, state 1) and modular versus nonmodular excursions (x axis, state 2) in the exploration phase. The plot is computed from a transition matrix of the number of times each transition type occurred across all 191 study mice. The transition to a modular versus nonmodular excursion (state 2) depends significantly on the type of module expressed (state 1) (Fisher’s exact test, p = 0.0002, two-sided test, Monte Carlo simulated p value). The strength of the transition probability is indicated by the size and shade of the box (see legend). If a box is not shown, then no association exists. Main text examples are highlighted in red.
(D) Transition probabilities for modular versus nonmodular excursions following expression of module 100 or module 96. Probabilities are shown in brackets.
(E) Transition probabilities of different modules following expression of module 74 in the foraging phase. Transitions to specific modules occur in probabilistically determined manner (Data S1 and S2). Dark purple modules, 20% probability; light purple modules, 9% probability.
Module expression order could be probabilistically determined, such that the expression of one type significantly influences the nature of the next excursion type. To test this, we ordered the modular and nonmodular excursions for each mouse by increasing expression start time to study the sequence of events. From the event sequence data for all 191 mice, we constructed a transition matrix and first asked whether the expression of a modular versus nonmodular excursion depends significantly on the nature of the previously expressed excursion. A Fisher’s exact test found the transition matrix is statistically significant for the exploration phase (p = 0.0002, two-sided test, Monte Carlo simulated p value), but not the foraging phase (p = 0.58). By plotting the exploration phase transition probabilities, we identified modules with biased transitions to modular versus nonmodular excursions (Figure 4C). For example, if a mouse expresses module 100, then the probability of expressing a modular excursion next is 83% (Figure 4D). On the other hand, if a mouse expresses module 96, then the probability of expressing another modular excursion is only 19% and the transition probability to a nonmodular excursion is 81% (Figure 4E). Thus, transitions between modular and nonmodular excursion types are probabilistically determined in some contexts.
We next investigated transitions between specific modules. A Fisher’s exact test of the transition matrix revealed that the expression of a given module depends significantly on the nature of the previously expressed module in both the exploration (p = 0.009; Data S1) and foraging (p = 1 × 10−5; Data S2) phases. Plots of the transition probabilities reveal the major transition patterns between different modules in each assay phase (exploration, Data S1; foraging, Data S2). During the foraging phase, module 74 expression is associated with a 40% probability of transitioning to the expression of module 110 or 63 and a 63% probability of transitioning to one of seven other modules (Figure 4E). Therefore, foraging patterns are constructed not from randomly ordered behavioral sequences but from probabilistically determined transitions from one module type to another.
Different Modules Link to Different Economic Patterns of Food Intake, Exposure, Activity, and Perseveration Responses
Although we do not know the functions of the 71 modules we uncovered, foraging patterns are proposed to be under selective pressure to minimize predation risk and energy expenditure while maximizing caloric intake (Brown and Laundré, 1999; Pulliam and Charnov, 1977; Stephens et al., 2007). Therefore, one hypothesis is that modules are building blocks for different feeding, exposure (predation risk), activity, and perseverative response patterns. To test this, we defined keystone features in our assay that are measures of food intake, exposure, activity, and memory and perseverative responses (Table S2; Figure S4). For food intake, we measure the total seeds eaten by the mice in each phase (TFC.0; Table S2), and the total time spent at the food pot (TTP2, exploration phase; TTP4, foraging phase), and the total visits to the food pot (TVP2, exploration phase; TVP4, foraging phase). We capture time and visit data over a whole trial (−0) and in 5-min time bins (−1, −2, −3, −4, and −5) to resolve potential changes over time. To measure increases and decreases in exposure, we collect the percentage of time and visits in the exposed center versus more sheltered wall zones (Figures S4C and S4D) and home base tunnel zones (Figures S4C and S4E) in the arena. To measure activity, we captured the total distance traveled (e.g., Figure S4F), total sand dug from the pots, and total visits to different pots in the arena (Table S2; Videos S1 and S2). Finally, mice exhibit memory and perseverative responses in the foraging phase due to the changed food location (Videos S1 and S2). During the foraging phase, mice repeatedly investigate the former food pot (pot 2). These perseverative responses are captured from measures of the latency to visit pot 2 (LVP2), total time spent at pot 2 (TTP2; Figures 5 and S1G), total visits to pot 2 (TVP2; Figure S4H), and the total sand dug from pot 2 (TSDP2; Figure S4I). Overall, our keystone features not only detect the expected responses in the mice but also show variability that indicates a diversity of economic patterns among the animals tested (Figures S4D–S4I). We capture much of this diversity across 106 measures of the keystone features (Table S2).
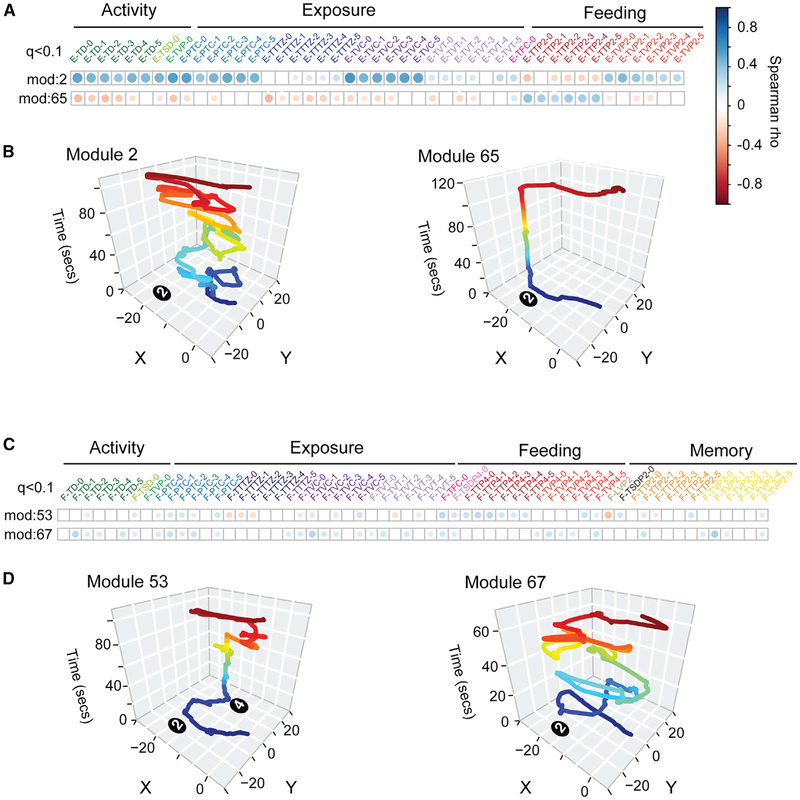
Figure 5. Individual Modules Are Associated with Different Feeding, Exposure, Activity, and Perseveration Response Patterns.
(A) The plot shows statistically significant correlations between module 2 or 65 (rows) and keystone measures of feeding, exposure, and activity in the exploration phase (columns). Correlations are calculated using the Spearman test on data from all 191 study mice (q < 0.1). Empty white squares indicate no significant correlation (q > 0.1). The magnitude of significant positive (blue) and negative (red) correlations between modules and keystone measures are indicated by dot size and shade (see legend). Both modules are significantly associated with the total food consumed (E-TFC-0) but are linked to different patterns of activity, exposure, and feeding in the exploration phase based on time spent and visits in different zones of the arena at different times during the trial (five 5-min time bins shown: −1, −2, −3, −4, and −5 and overall [0]). Thus, the different modules link to different economic patterns.
(B) Representative traces of the x-y movement patterns for modules 2 and 65 are shown. Each module involves a different movement and temporal pattern.
(C) The plot shows correlations between modules 53 and 67 with keystone measures of activity, exposure, feeding, and perseverative responses in the foraging phase. Both modules are significantly associated with total time at the former food pot (pot 2) at the beginning of the trial (F-TTP2–1), a measure of memory of the former food location. However, the two modules differ in links to other features describing activity, exposure, feeding, and memory and perseveration responses.
(D) Representative foraging phase traces of the x-y movement patterns of excursions for modules 53 and 67 are shown. The different module forms and links to economic patterns are consistent with different functions.
Different economic patterns could be constructed from the expression of particular modules, such that specific modules correlate with specific feeding, activity, exposure, and/or perseveration patterns. Alternatively, modules are functionally interchangeable and do not shape different economic patterns. To test these alternatives, we computed a pairwise correlation matrix (Spearman rank) between the expression frequencies of each module and the magnitude of the values for each keystone feature measure from the data for the 191 mice tested (exploration phase, Data S3; foraging phase, Data S4). p values for the correlations between modules and measures were corrected for multiple testing using the q-value. We found distinct groups of modules with significant positive and/or negative correlations to different measures of feeding, activity, exposure, and perseveration (Data S3 and S4). Therefore, modules are not functionally interchangeable and associate with distinct economic patterns.
In the exploration phase, modules 2 and 65 are both significantly positively associated with the total food consumed (E-TFC-0) (Figure 5A) but have important differences. Module 2 expression is correlated with increased activity and exposure throughout the trial and is negatively correlated with the total time spent at the food pot (E-TTP2). In contrast, module 65 is positively associated with increased total time spent at the food pot but negatively correlated with measures of activity, exposure, and visits to pot 2 (Figure 5A). The form of module 2 indicates substantial movement and repeated visits to pot 2, suggesting a “grazing” pattern of food consumption, whereas module 65 shows a direct round trip from the home to the food and back (Figure 5B). Thus, while both modules are associated with increased food consumption, they are distinct behavioral sequences that appear to have different functions.
The foraging phase includes memory and preservation responses, which further resolve links between modules and economic patterns (Data S4). For example, modules 53 and 67 are significantly associated with increased total time at the former food pot (pot 2) during the first 5 min of the trial (Figure 5C; F-TTP2–1). However, module 53 is significantly correlated with a shorter latency to visit pot 2 and sand displaced from pot 2, but module 67 is not (Figure 5C). On the other hand, module 67 is associated with more total visits to pot 2 at the beginning of the trial (F-TVP2–1; Figure 5C). The two modules have different forms (Figure 5D) and differ in how they shape memory and perseverative responses. These data support the conclusion that the modules are building blocks for constructing different feeding, exposure, activity, and perseverative patterns.
Modules Have a Stereotyped Developmental Progression across Different Genetic Backgrounds of Mice
The emergence of independent foraging at weaning is a major developmental milestone in mammals involving changes to behavior, metabolism, and brain function (Galef, 1981; Hewlett and Lamb, 2010). We hypothesized that changes to foraging modules are an integral part of behavioral development. To begin, we investigated the relative expression frequency of modular versus nonmodular excursions for the P15, P20, P25, and adult mice from the different strains profiled. We found that the relative expression of modular versus nonmodular excursions depends significantly on age (Figure 6A; p = 1.7 × 10−14). Younger mice express more modular excursions and fewer nonmodular excursions (Figure 6A). Nonmodular excursions become increasingly prevalent at later ages and are associated with adulthood (Figure 6A). Thus, modules play the predominant role in constructing early economic behavior patterns and more stochastic, nonmodular behavioral sequences emerge as development progresses.
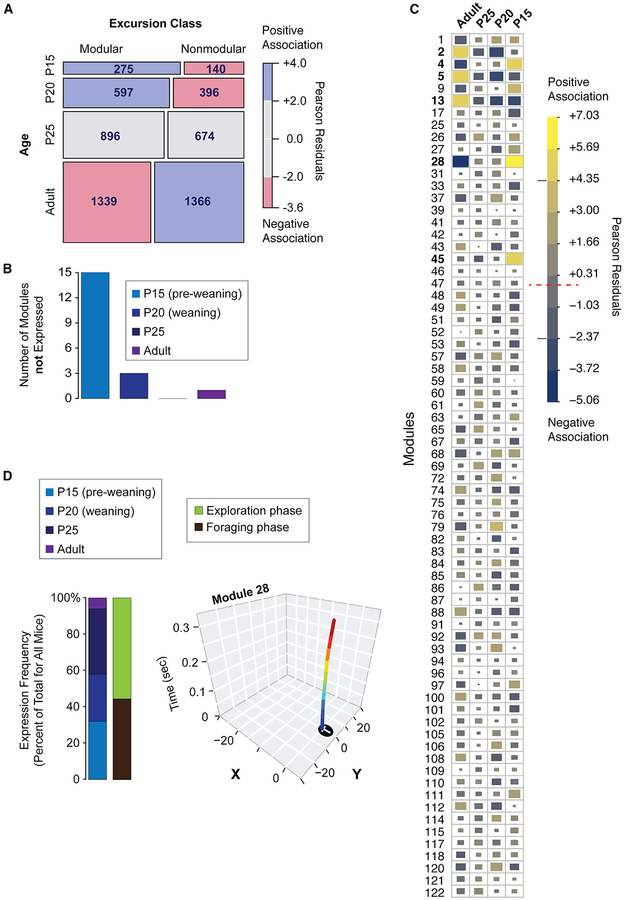
Figure 6. Module Expression Develops in a Stereotyped Manner in Offspring.
(A) A mosaic plot depicting a table of the numbers of modular and nonmodular excursions expressed at different ages. A chi-square test indicates that the relative expression of modular versus nonmodular excursions is dependent on age (p = 1.7 × 10−14). The mosaic plot boxes are scaled according to the relative numbers of excursions and colored according to positive (blue) and negative (red) associations between the rows and columns based on the Pearson residuals. P15 and P20 juveniles are positively associated with modular excursions, while adulthood is associated with relatively more nonmodular excursions. n = 20 adult, n = 16 P25, n = 15 P20, n = 16 P15.
(B) Bar plot shows the number of modules that are not detected at different ages across all of the mouse strains tested. Some modules emerge at specific ages.
(C) The chart shows the top modules impacted by age differences. Pearson residuals computed from a chi-square test are shown for module expression frequency data. The data show modules with expression frequencies that are positively (yellow) or negatively (blue) associated with each age. Relative effect size is depicted by color (see legend) and block size. The red dashed line in the legend shows the threshold at which the observed and expected module expression counts are the same (Pearson residuals equal zero). The gray lines show the threshold for the top age affected modules (bolded modules).
(D) Stacked bar plots show the relative expression frequency of module 28 by age and phase (exploration, green; Foraging, brown). Representative x-y tracks for excursions for module 28 are shown. Module 28 is expressed predominantly by juveniles, not adults, and in both phases.
We investigated the appearance of specific modules at different ages and found 15 modules are not expressed at P15 and three are not expressed at P20 (Figure 6B). All module types were expressed by P25 offspring. One module is not expressed by adults and is preferentially expression by juveniles (Figure 6B). To more generally test whether developmental effects significantly affect module expression, we fit a generalized linear model testing for an interaction between module type and age in the expression frequency data (STAR Methods). We found the module-age interaction effect is significant (p < 2.2 × 10−16, likelihood ratio test). Therefore, the expression of specific modules changes in an age-dependent manner across different strains of mice.
To help unravel the developmental progression of module expression changes, we tallied the expression frequency of each module for each age and computed Pearson residuals from a chi-square test of independence (STAR Methods). Some modules that are more positively (yellow) or negatively (blue) associated with each age than expected (Figure 6C). For example, module 28 expression is positively associated with juveniles, especially P15 pups, and negatively associated with adults (Figure 6C). This module is rarely expressed by adults, involves sub-second darts in-and-out of the arena, and is expressed in both the exploration (55% of cases) and foraging (45% of cases) phases (Figure 6D). The keystone feature analysis shows module 28 is linked to increased time in the tunnel zone during the exploration phase (TTTZ), shapes the total visits to the home cage tunnel (TVT), and reduces time at the food pot during later time bins (TTP2–4 and TTP2–5) (Data S3). In the foraging phase, expression of this module reduces the distance traveled during the first 5 min (TD-1) and during later time bins is linked to increased time (TTP2–2, TTP2–3, TTP2–4, and TTP2–5) and visits (TVP2–3 and TVP2–4) to the former food pot (Data S4). Based on these results, we speculate that this module functions in juveniles to reduce risk taking and exposure to predators. Overall, behavioral development involves age-dependent changes to the expression of modules shaping economic patterns.
Module Expression Is Controlled by Genetic Mechanisms
We hypothesized that foraging modules are discrete functional units of behavior shaped by evolution and therefore have a genetic basis. Potential genetic effects could be uncovered by comparing module expression in B6 and F1bc mice (Figure S1B). Beginning with adults, we performed generalized linear modeling of the B6 and F1bc module expression data and found the interaction between module and mouse genotype (B6 versus F1bc) is significant (p < 9.5 × 10−8, likelihood ratio test; STAR Methods). We performed a similar analysis for P20 and P25 juveniles (P15s were omitted due to lack of B6 data). In this case, the full model and nested models included age as an explanatory variable to absorb variance due to the P20 and P25 age difference. As in adults, the results show the module-genotype interaction is significant (p = 1.7 × 10−10, likelihood ratio test). Therefore, module expression is regulated by genetic mechanisms in juveniles and adults, which lays foundations to map specific mechanisms onto specific modules.
Different Genetic and Parental Effects Can Be Mapped onto Specific Modules at Different Ages
We investigated genetic and parental effects on module expression at different ages. Parental effects were tested by comparing F1cb and F1bc mice and genetic effects from comparisons of F1bc and B6 mice (Figure S1B). Above, we found significant genetic effects exist, which are dissected below. Using a similar generalized modeling approach, we tested for parental effects by testing for an interaction between module and cross (F1cb versus F1bc). The results revealed a significant interaction effect in adults (p = 0.01, likelihood ratio test). Unexpectedly, however, this was not the case for the juveniles. In our analysis of parental effects in juveniles, which includes P15, P20, and P25 offspring, the interaction was not significant (p = 0.7, likelihood ratio test). Thus, parental effects are undetectable in juveniles. This observation is intriguing, because weaning is the stage when the bond between mother and offspring changes and one might have predicted the opposite result (Lee, 1996; Trivers, 1974). Instead, while genetic effects are significant in juveniles and adults, significant parental effects only emerge at later ages. These data show that module expression is shaped by different factors at different stages of life and reveal that parental effects on foraging manifest in adulthood.
We sought to identify the specific modules most sensitive to genetic differences between B6 and F1bc mice and those that are sensitive to differences in parental effects in F1cb and F1bc mice. We analyzed the module expression counts for each strain and cross by computing the Pearson residuals from a chi-square test of independence. The results reveal modules more positively (yellow) or negatively (dark blue) associated with the B6, F1cb, or F1bc mice than expected (Figures S5A and S5B). In the juveniles (P20 and P25), five modules are strongly impacted by the genetic effects (red text, Figure S5A). Strong differences between F1cb and F1bc mice are not apparent, consistent with the absence of significant parental effects in juveniles. In adults, however, eight modules are impacted by genetic effects and five are impacted by parental effects (Figure S5B; genetic effects, red text; parental effects, orange text). Our keystone feature analysis shows that the five modules impacted by parental effects (modules 2, 5, 9, 86, and 112) are linked to a similar economic pattern and promote increased activity and exposure and total food pot visits (pot 2) in the exploration phase while reducing the total time at the food pot (Data S3, orange highlighted modules). The modules differ in terms of their temporal properties and link to specific measures in specific time bins (Data S3). Similar results were found in the foraging phase (Data S4, orange modules). Therefore, parental effects impact a discrete set of modules shaping economic patterns. Overall, these data show that different mechanisms can be mapped onto different modules.
The PWS and Autism Gene Magel2 Regulates the Expression of Specific Foraging Modules
Our findings lay foundations for targeted mechanistic studies to map specific genomic elements onto specific modules and learn how different mechanisms function to shape complex economic patterns. This opens the possibility of using our approach to help dissect the basis of abnormal behavior patterns in mouse models of human disease. In a proof-of-principle study, we focused on PWS, which involves complex cognitive, feeding, and motivated behavior abnormalities that change developmentally and result in obesity during childhood (Cassidy and Driscoll, 2009). The paternally expressed, imprinted gene Magel2 is mutated in PWS and contributes to some phenotypes (Bervini and Herzog, 2013; Cassidy et al., 2012; Kozlov et al., 2007; Mercer et al., 2009; Schaller et al., 2010). Magel2 mutant mice exhibit altered circadian rhythm, feeding, anxiety, arousal, and serotonin signaling compared to controls (Kozlov et al., 2007; Mercer et al., 2009). However, the underlying basis of these phenotypes is not well understood, and it is unknown whether Magel2 functions to regulate specific modules of economic behavior. Since Magel2 is imprinted and expressed from the paternal allele, Magel2+/− offspring are considered loss-of-function mutants and reciprocal Magel2−/+ offspring are typically considered unaffected. We bred available Magel2 mutant mice to generate female Magel2+/− mutants, as well as Magel2−/+ and Magel2+/+ littermate controls (n = 15–28). Foraging patterns were profiled at weaning (P20) and in adults (9–12 weeks of age). DeepFeats found that the 173 mice profiled performed 5,301 excursions, enabling us to investigate the existence of Magel2 regulated modules.
With these data, we first determined which of the “reference modules” identified in the F1cb, F1bc, B6, and Cast mice are expressed by the Magel2 mice. The 57 excursion behavioral measures for Magel2 mice were pruned to the same 13 measures as for the reference modules. Then, an IGP permutation test was performed using the centroids for the 71 reference module centroids (Figure S3C) and the excursion data from Magel2 mice. We determined the proportion of reference modules not expressed by Magel2 mice (true nulls) by computing the pi0 statistic from a q-value analysis of the IGP p values (STAR Methods) (Storey, 2003). We found the majority of the reference modules (61%; 43 reference modules) are expressed by Magel2 mice. These results demonstrate that the reference modules are well preserved, as expected, but do not reveal the full landscape of potential modules expressed by the Magel2+/−, Magel2−/+, and Magel2+/+ mice, the identity of nonmodular excursions, or the full profile of possible phenotypic effects. To achieve this, we needed to discover modules directly from the Magel2 mouse cohort.
To directly analyze modules in the Magel2 cohort, we ran our DeepFeats pipeline to first determine the set of behavioral measures that best reveal clusters of excursions in the Magel2 data. Behavioral measures were assessed with different Pearson correlations, as above, and again we found that a threshold of r < 0.4 yielded the best resolution of clusters (Data S5A). This threshold retains 14 out of the 57 behavioral measures (Data S5B; Table S1), of which two differ from those that delineate the reference modules, namely, MeanGear2Length and TotalGearChanges (Table S1). Our resampling test found significant clustering based on these measures (p < 1 × 10−3), supporting the existence of bona fide clusters (Data S5C). IGP permutation analysis uncovered modular and nonmodular excursions using training and test partition datasets balanced by genotype, age, and assay phase. The results identified 59 modules (q < 0.1) and reveal 67% of the 5301 Magel2 excursions are modular, while 33% are nonmodular (q > 0.1) (Figure S6A). Our data reveal the full modular architecture of foraging in the P20 and adult Magel2+/−, Magel2−/+, and Magel2+/+ mice, allowing us to investigate the functional effects of the paternal and maternal Magel2 alleles.
We next tested whether loss of the paternal Magel2 allele affects specific modules. The expression frequency of each module was determined for the Magel2+/− paternal mutants and the paternal allele wild-type controls (Magel2−/+ and Magel2+/+). Generalized linear modeling was performed to test the interaction between module type and paternal allele genotype (mutant versus wild-type; STAR Methods). Our results found a statistically significant interaction in P20 juveniles (p = 0.039) and adults (p = 1.7 × 10−5, likelihood ratio test). Therefore, loss of the paternal Magel2 allele significantly changes the expression of specific modules in juvenile and adult offspring.
To identify the modules affected, a chi-square test of independence was performed to compute Pearson residuals that show modules with positive or negative associations with the Magel2+/− versus control genotypes (Figure S6B). In P20 juveniles, seven modules (8%) are positively associated with the Magel2+/− genotype (bright yellow; modules 20, 49, 50, 56, 64, 73, and 84), and four (5%) are negatively associated (dark blue; modules 16, 32, and 80) (Figure S6B; P20). Module 20 excursions, for example, are almost exclusively expressed by Magel2+/− mice and not control P20 juveniles (Figure S6C). In the controls, this module is instead expressed by adults (Figure S6D). In adults, we found eight affected modules: five positively associated (6%; modules 5, 39, 76, 81, and 82) and three negatively associated (4%; modules 7, 20, and 50) with the Magel2+/− mutants (Figure S6B, adult). These modules are distinct from those most strongly affected in juveniles. For example, module 81 is preferentially expressed by adult, but not P20, Magel2+/− mutants (Figures S6B and S6C). The module involves several seconds of hovering near the tunnel zone to the home and is predominantly expressed by adults in the exploration and foraging phases (Figure S6E). The paternal Magel2 allele therefore regulates a discrete set of foraging modules in juvenile and adult offspring.
Foraging Module Analysis Uncovers Functional Effects for the Imprinted Maternal Magel2 Allele
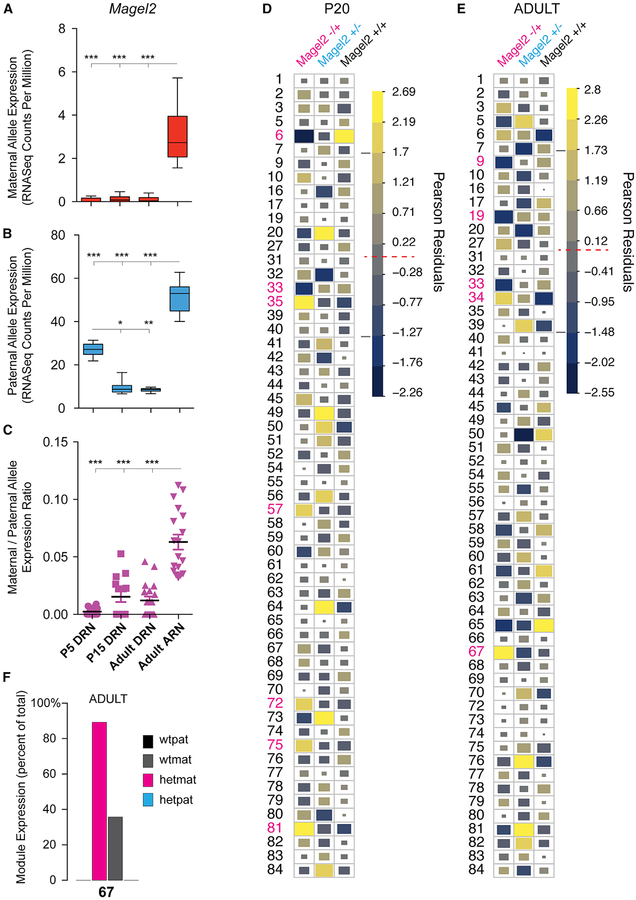
The maternal allele for Magel2 is typically considered to be silenced (imprinted) and therefore nonfunctional. However, a recent study found the maternal Magel2 allele is expressed at low levels in some brain regions in Magel2+/− paternal allele knockout mice (Matarazzo and Muscatelli, 2013), raising the questions of whether it is expressed in wild-type mice and has any functional effects. Analyzing behavior at the module level could enhance the detection of phenotypic effects otherwise difficult to identify. We first tested whether the maternal Magel2 allele is expressed in the normal mouse brain by reanalyzing RNA sequencing (RNA-seq) datasets of imprinting and allelic expression in the mouse brain using reciprocal F1 hybrid Cast × B6 mice (Bonthuis et al., 2015; Huang et al., 2017). Magel2 maternal and paternal allele expression was examined in the dorsal raphe nucleus (DRN) and arcuate nucleus (ARN) of the adult female brain and in the DRN for P5 pups and P15 juveniles (Figures 7A–7C). The maternal Magel2 allele is expressed and the highest expression occurs in the adult ARN compared to P5, P15, and adult DRN (Figure 7A). The expression pattern of the paternal allele differs from the maternal allele (Figure 7B). By normalizing maternal allele expression to the paternal allele, we determined that the maternal allele is significantly activated in the ARN (Figure 7C).
Figure 7. The Imprinted Maternal Magel2 Allele Is Expressed in the Brain and Has Significant but Distinct Effects on Module Expression Compared to the Paternal Allele.
(A and B) Box plots of allele-specific F1cb and F1bc RNA-seq data show expression of the maternal (A, red) and paternal (B, blue) Magel2 alleles in the P5 (n = 14), P15 (n = 14), and adult (n = 18) female mouse dorsal raphe nucleus (DRN) and the adult female arcuate nucleus (ARN) (n = 18). Allele RNA-seq data are presented as read counts per million reads. The main effect of brain region is significant for the maternal (A) and paternal (B) allele data (p < 1 × 10−4, nonparametric Kruskal-Wallis test with Dunn’s post-test). Asterisks indicate significant post-test results and brain regions and/or ages with different expression levels. Plots indicate mean ± minimum maximum values. ***p < 0.001, **p < 0.01, and *p < 0.05,
(C) The plot shows the expression of the maternal Magel2 allele normalized to the expression level of the paternal allele for each P5, P15, and adult DRN and ARN sample. Maternal allele expression is significantly increased in the ARN after normalizing for paternal allele activity (p < 1 × 10−4, nonparametric Kruskal-Wallis test with Dunn’s post-test). Plot indicates mean ± SEM.
(D and E) The charts show the identity of the top modules impacted by loss of the maternal Magel2 allele (pink text) in P20 (D) and adult (E) offspring. The charts plot the Pearson residuals computed from a chi-square test of independence between modules and different Magel2 genotypes, including Magel2−/+, Magel2−/+, and Magel2+/+ controls for P20 juveniles (D) and adults (E). The data show modules with expression frequencies that are positively (yellow) and negatively (blue) associated with Magel2+/− mutants versus controls. Relative effect size is depicted by color (see legend) and block size and the most affected modules are highlighted (pink numbers). The data show specific modules with expression frequencies changed by loss of the maternal Magel2 allele. The modules differ from those impacted by loss of the paternal allele. For P20 and adult Magel2 mice, n = 22/18 Magel2m+’/p+, n = 28/23 Magel2m−/p+, n = 22/20 Magel2m+/p+’ and n = 22/20 Magel2m+/p−.
(F) The bar plots show the percentage of excursions that are expressed by the different Magel2 genotypes for module 67 in adults. This module is preferentially expressed by Magel2−/+ maternal allele mutants (hetmat) compared to paternal allele mutants (Magel2+/−, hetpat) and controls allele (Magel2+/+’, wtpat; Magel2+’/+, wtmat).
We tested whether loss of the maternal allele in Magel2−/+ mice has effects on foraging module expression frequency by testing for an interaction between module and maternal allele genotype. The fit of the full model was compared to a nested model of the main effects only and absorbs variance due to the paternal allele genotype (STAR Methods). In support of functional effects for the maternal allele, we found a significant interaction in both the P20 (p = 0.023) and adult offspring (p = 0.024, likelihood ratio test). Plots of Pearson residuals show the specific modules that are affected by loss of the maternal allele (Figures 7D and 7E). Modules affected by loss of the maternal allele differ from those affected by loss of the paternal allele (Figures 7D and 7E). In adults, for example, module 67 is preferentially expressed by Magel2−/+ maternal allele mutants, but not by Magel2+/− paternal allele mutants (Figures 7E and 7F).
We used keystone feature analysis to investigate the relationships between the affected modules and economic patterns in the Magel2+/− and −/+ offspring. In adults in the exploration phase, we found both parental alleles affect modules associated with increased activity and exposure and reduced time at the food pot (pot 2) (Figure S7). However, loss of the paternal allele decreased expression of some modules significantly correlated with increased food intake (E-TFC-0), which did not occur for the maternal allele (arrow, Figure S7). Moreover, loss of the paternal allele increased module 76 expression, which is negatively correlated with food intake, but loss of the maternal allele increased module 34 expression, which is positively correlated with food intake, and module 67 expression, which is linked to increased time at the food pot (Figure S7). Thus, the paternal and maternal Magel2 alleles regulate distinct modules linked to distinct economic patterns.
DISCUSSION
Since most behavioral studies evaluate specific behavioral responses, the principles and mechanisms involved in constructing complex behavior patterns are ripe for discovery. Here, we found that complex foraging patterns are constructed from finite, genetically controlled modules of home base excursions, as well as nonmodular behavioral sequences. The modules are distinct in form, expression frequency, and timing and are expressed in a probabilistically determined order. Different modules link to different economic patterns of food intake, exposure, activity, and perseveration. These data support the discovery of functionally distinct modules of economic behavior. Modules are revealed to be integral to behavioral development and regulated by specific age, genetic, and parental effects. By analyzing modules in a mouse model of PWS with a mutation in the imprinted gene Magel2, we found that both the paternal and maternal Magel2 alleles are functional but regulate the expression of distinct modules. Our study establishes a framework for behavior analysis that will help elucidate how evolution and specific molecular and neural mechanisms shape complex economic behavior patterns in health and disease.
Foraging Modules Elucidate How the Genome Shapes Complex Economic Behavior Patterns
The importance of deeply analyzing relatively spontaneous behavioral sequences in rodents was emphasized decades ago (Kavanau, 1967). Since then, invertebrate model organisms have shown how computational dissections of behavioral sequences can ultimately help resolve links between specific mechanisms and behavior patterns (Anderson and Perona, 2014; Robie et al., 2017; Vogelstein et al., 2014), and advances are being made in mammals (Hong et al., 2015; Kabra et al., 2013; Markowitz et al., 2018; Weissbrod et al., 2013). Recently, sub-second syllables of body language were uncovered using machine learning in mice (Wiltschko et al., 2015), revealing an important new dimension of behavior. Here, we also identified modules that are 1–2 s or less in duration, as well as modules that are hundreds of seconds long, which underscores the importance of searching for modules across broad timescales. The study of modularity in free patterns of complex ethological behaviors presents many challenges in terms of how to design the assay, select features for analysis, and analyze the data to uncover biologically important modules. Our study makes advances on these fronts for economic patterns involved in foraging, but our general methodology could be applied to other behavior tests involving animal tracking and ecological landmarks.
Economic behavior patterns that regulate the balance among reward, risk, and effort during foraging are under strong natural selection. Theoretical work has modeled the selective pressures and tradeoffs shaping different foraging decisions (Pulliam and Charnov, 1977). However, our data in mice show that behavioral sequences during foraging are not infinitely variable and instead involve a finite number of reproducible and genetically controlled modules. The total number of different foraging modules encoded in the genome and brain remains to be determined; additional modules are likely to be revealed in more complex, natural environments. Another future direction is to define the regulatory logic controlling module expression frequency, timing, and order under different circumstances and contexts. Different patterns will shape gains versus costs differently in different contexts, such as a naive versus familiar context, and the general principles controlling pattern construction are now ripe for discovery.
An important finding from our study is that the home base is an anchor point naturally breaking foraging behavior into many distinct modules. The home base is typically omitted in lab behavior studies, with the exception of some pioneering work (Eilam and Golani, 1989; Gorny et al., 2002). Indeed, for most species in the wild, including humans, behaviors are structured within a defined home range (Börger et al., 2008). Neural mechanisms for navigating and dead reckoning relative to a landmark, such as the home, have been identified (Wallace et al., 2008). However, we know little about the role of the home in structuring economic patterns. Given our data showing foraging modules have a genetic basis in mice and the conservation of home range behavior across different species, the modules we observe were likely selected for and have analogs in other species.
Our study focused on food-deprived mice, but new modules may be uncovered from mice in other internal states. Using the methodology we introduced here, future studies could construct a reference atlas of modules for different internal states that can be used to help interpret module expression phenotypes detected in mice following targeted genetic, epigenetic, neural, or environmental perturbations. Future studies could also assess whether different modules have antagonistic functional and mechanistic relationships that shape different economic tradeoffs (Stephens et al., 2007). These different lines of investigation will deepen our understanding of the basis of complex patterns of behavior.
Nonmodular Excursions Are an Important Feature of Foraging Patterns
The nonmodular excursions uncovered in our study are relatively more stochastic behavioral sequences. One enticing possibility is that nonmodular excursions reflect states of increased behavioral flexibility and the expression of idiosyncratic impulses to explore and exploit the environment. Future studies of nonmodular excursions might uncover mechanisms shaping creative and cognitive processes. Alternatively, some nonmodular excursions could be rarely expressed modules in the current assay that are expressed more frequently in another context.
Uncovering the Modular Architecture of Behavioral Development
Much remains to be understood about how complex behavior patterns develop and change with age. This is an important area for study, since most forms of mental illness involve early-life antecedents (Rutter et al., 2006) and arise at characteristic ages (Silbereis et al., 2016). Our data suggest that behavioral development for foraging involves creating and changing the expression of modules with age, as well as expressing more nonmodular sequences. Currently, we do not know the functions or the mechanisms involved. While the genomics field has described developmental gene expression changes in the human, non-human primate, and rodent brain (Bakken et al., 2016; Kang et al., 2011; Miller et al., 2014; Silbereis et al., 2016; Thompson et al., 2014), less is known about how molecular and neural changes map to specific behavioral changes. The majority of the cellular gene expression programs in the cortex of rodents and humans are highly conserved (Zeng et al., 2012), as are the trajectories of most neurodevelopmental gene expression programs in the developing brain (Bakken et al., 2016) and the temporal sequence of neurodevelopmental processes (Workman et al., 2013). Thus, the trajectory of module development that we uncovered in mice could be rooted in conserved neurodevelopmental programs, help stage mouse behavioral development, and reveal abnormal patterns.
Foraging Modules and the Basis of Abnormal Economic Behavior Patterns
Obesity, addiction, depression, and anxiety disorders have been framed as maladaptive neuroeconomic processes (Camerer, 2013; Hartley and Phelps, 2012; Monterosso et al., 2012; Rowland et al., 2008; Sharp et al., 2012). The behaviors that lead to these diagnoses are often highly heterogenous; for instance, many different feeding and activity patterns might lead to obesity. Analyzing the form and expression of modules and nonmodular behavioral sequences in mouse models could deepen our understanding of the basis of different phenotypes, enhance the detection of phenotypic effects, and thereby reveal strategies to help normalize behavior. Our analysis of Magel2 mutant mice supports this idea. In humans, paternally inherited MAGEL2 mutations contribute to PWS and autism (Bervini and Herzog, 2013; Schaaf et al., 2013), which involve major phenotypic changes across development (Cassidy et al., 2012; Lord et al., 2015). Our data show that both the paternal and maternal Magel2 alleles are expressed and have functional effects that change the expression of specific modules in an age-dependent manner. Thus, foraging module analysis is sufficiently sensitive to detect effects from loss of the imprinted maternal Magel2 allele and reveal the identity of the different modules affected by each Magel2 allele. A recent study also observed functional expression of the imprinted maternal allele for Ndn (Rieusset et al., 2013), which is a paternally expressed imprinted gene neighboring Magel2. Thus, there is growing evidence for functional effects of lowly expressed imprinted alleles in the PWS imprinted gene cluster. Overall, our methodology and findings lay foundations for investigations into the mechanistic basis of complex economic behavior patterns.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Christopher Gregg (chris.gregg@neuro.utah.edu).
EXPERIMENT MODEL AND SUBJECT DETAILS
Animal experiments were performed in accordance with protocols approved by the University of Utah Institutional Animal Care and Use Committee (#14–12012). C57BL/6J (B6), CastEiJ (CAST), Magel2 mice were obtained from Jackson Laboratory. Mice were maintained on reverse 12 h light/dark cycle (lights off at 11:00) for foraging behavior experiments, and given water and food ad libitum (Harlan Tecklad rodent diet 2920X; Madison, WI.). All studies were performed on females. Cage bedding was Paperchip bedding (Shepherd Specialty Paper) and animals were housed with sex-matched littermates (2–5 mice per cage). Breeding pairs were housed together continuously for breeding. Maximum litter number was capped at 6 for each breeding pair and large litters were culled to a maximum of 6 pups. We tested female P15, P20, P25 and adult (8–12 week old) mice. For P15 behavior testing, offspring were returned to the parental cage after testing and weaned at P21. For P20 behavior testing, mice were weaned after testing was completed. For P25 behavior testing, mice had been weaned at P21.
METHOD DETAILS
Mouse Behavior Experiments
All B6, Cast, F1cb and F1bc mice were only tested once in the foraging assay and therefore mice at all ages were naive to the test. Adult mice for all groups were at ~3 months of age when tested. Magel2 mice were tested twice in the foraging assay, once at P20 and again as adults. For B6 females: N = 20 adult, N = 16 P25, N = 15 P20 and N = 16 P15 mice; For F1cb: N = 17 adult, N = 15 P25, N = 15 P20 and N = 15 P15 mice; For F1bc: N = 18 adult, N = 15 P25, N = 18 P20 and N = 15 P15; For Cast: N = 15 P15 mice. For Magel2 at P20, N = 22 maternal wt (Magel2m+’/p+), N = 28 maternal het (Magel2m−/p+), N = 22 paternal wt (Magel2m+/p+’) and N = 22 paternal het (Magel2m+/p−). For Magel2 adults, N = 18 maternal wt, N = 23 maternal het, N = 20 paternal wt and N = 20 paternal het. Since each litter yielded ~3 females, each group involved testing pups derived from 5–9 different litters and at least five different breeding pairs. A detailed protocol for the behavior analysis is provided in the Supplemental Information.
Foraging Behavior Testing
In preparation for the foraging assay, mice were first habituated with sand (Jurassic play sand, Jurassic Sand) and seeds (Whole millet, Living Whole Foods) for two days in their home cage. On day one, seeds are spread on top of sand in the bottom of a Petri dish and the dish is placed on the bedding in the home cage for the mice and pups to explore. On day two, seeds are covered with sand in the bottom of the Petri dish in the home cage for the mice and pups to dig in and explore. To motivate animals to feed, mice were food deprived prior to testing to achieve 8%–10% weight loss at the time of testing. For P15, P20 and P25 mice, weight gain normally occurring during the period of food deprivation was taken into account to achieve the intended weight loss. We selected this weight loss target after several pilot studies with the goal of achieving some consistency in the motivational states of the animals at different ages and not compromising health or activity. To achieve the intended weight loss and motivational state, adult mice were food deprived for 24 hours, P25 mice were food deprived for 17 hours, P20 mice were food deprived for 17 hours and housed with the mother, P15 mice were food deprived for 4 hours prior to testing in the absence of the mother. For food deprivation, mice are moved into a fresh cage with some soiled bedding to ensure familiar smell, but no leftover food in the cage. Water is available ad libitum at all times except when mice are in testing cage (2 × 1 hour).
Mice are housed in a room with an 11:00 – 23:00 dark cycle, so that testing is performed during the dark cycle. For testing, mice are moved into the behavior room prior to the start of testing for at least 1 hour for habituation to the new room. All testing is performed in the dark and video recording is done using infrared illumination and all manual procedures are done in the dark using a headlamp with red light. The mouse to be tested, and their home cage soiled bedding, are moved to the testing-cage attached to the arena 30 minutes prior to testing and allowed to habituate. At the start of testing, the testing-cage is attached to the arena via the tunnel, the mouse now has access to the arena and video recording starts for the Exploration phase. Mouse behavior is recorded continuously during the 30 min Exploration phase trial under infrared lights. Noldus Ethovision software v8 and v9 were used for video tracking. After completing the Exploration phase, the mouse is transferred to a holding cage with water but no food until the Foraging phase four hours later. For the Foraging phase, mice are again placed in the testing cage and habituated for 30 min. The testing cage is then gently attached to the tunnel and access to the arena is possible and the Foraging trial begins. Video recording of the Foraging phase is performed for 30 minutes. After testing, mice are placed in a new cage with food and are returned to the mouse colony room. Between each Exploration and Foraging phase trial, the entire arena, including walls, platform, tunnel and steel pots, are wiped clean with 70% ethanol.
Foraging Assay Design
The initial prototype foraging assay design and protocol, as performed in this study, is provided below. In a separate supplemental Protocol file, we provide a detailed protocol for an updated arena design, assay methodology and computer code for the data analysis.
Preparation of Sand and Seed Pots for the Foraging Assay
Three stainless steel pots (Resco, diameter 5.5cm, depth 4cm) were filled with 95 g of sand. For the Exploration phase, 1 pot is filled with 80 g of sand covered with 2.5g of seeds. On top of seeds, a layer of 12 g of sand is added to cover seeds. This sand is then covered with 0.5g of seeds. This pot is placed in position 2 in the arena. For the Foraging phase, 1 pot is filled with 80 g of sand, 3g of seeds on top of sand and additional 12 g of sand to cover all seeds. This pot is placed in position 4 in the arena. All pots are weighed before and after the trial to measure the sand displaced from each pot. Remaining seeds and hulls left in the pot and on the platform are measured after each Exploration or Foraging trial to determine the amount of seeds consumed by the mouse during the trial. Used sand is collected after every trial and set aside. At the completion of all testing, the used sand is autoclaved before reuse in future trials.
Foraging Arena Construction
The foraging arena, tunnel and testing cage were custom built with acrylic plastic (Delvies Plastics, Salt Lake City, UT, USA). The 14 cm long tunnel enters the platform from underneath through one of the five 5.5 cm diameter holes in the arena platform. The platform is 8 cm above stage level and is made from 0.5 cm thick white Plexiglas. The arena is made from a transparent Plexiglas tube and the 0.4 cm thick walls raise 42 cm above the platform. The arena has a diameter of 35 cm. The walls of the arena were roughened with sand paper to limit glaring and recording artifacts.
Arena Tracking Zones
The arena is organized into zones that are used to breakdown the behavior and foraging strategies used by each animal in the assay as detailed in Figure S1. Here, we provide additional details regarding the definition of these zones. The arena is divided into five sectors and the boundary of each sector is the mid point between two pots (see Figure S1). The arena is further divided into three concentric circles, including the middle center zone, the intermediate zone and the outer wall zone. The outer radius of the Intermediate zone intersects with the center of the pots and tunnel entry. The radius of the Center zone is half the radius of the Intermediate zone. A zone is also created around each pot in the arena. Pot zones have a radius of 1.7x the radius of the pot itself. Finally, to learn about the behavior of the animal related to entries to and exits from the arena, we define zones around the tunnel entry. The Tunnel Entry zone aligns with the entry hole of the actual tunnel. The In Tunnel zone is covering the most peripheral area of the Tunnel Entry and tracks the mouse just before leaving the arena completely. Whenever the mouse is in the cage, the tracking system is recording the mouse as being in the In Tunnel zone. The Tunnel Zone area has the same radius as the Pot zones.
Automated Tracking
At the start of the trial, the tracking begins with a 10 s delay to allow time for the connection of the testing-cage to the arena. The mouse is first tracked when it appears in the In Tunnel zone (Figures 6 and S1C) and position and movement is continuously recorded after this time point until the end of the 30 min Exploration or Foraging Phase. The XY position of the center of the mouse is video tracked with at a rate of 30 frames per second. For the data analysis, tracking data from 0–25 minutes are used. Time spent in each zone, latency to visit a zone and number of visits to each zone, as well as the distance traveled, are calculated using the Ethovision software. The data is exported as results for the total 25-minute trial duration, as well as in 5 minute time bins. Sand displacement and food consummation measures are collected and calculated manually.
Behavioral Measures
All measures captured during Exploration or Foraging phases of the assay are presented in Table S2. Each measure is assigned a custom ID, as indicated in the table. To calculate the time spent in the Tunnel Entry zone, time in the In Tunnel zone is subtracted from the raw time spent in the Tunnel Entry zone. To calculate the time spent in Tunnel Zone, the raw time in the Tunnel Entry zone is subtracted from the raw time spent in Tunnel zone. To calculate the total time spent in the arena, the time spent in the In Tunnel zone is subtracted the raw time in the arena. For calculation of these measures for specific time bins, the corresponding measures for each time 5 min time bin output by Noldus Ethovision are used instead of the total time measures. For the time spent in all other zones in the arena, the Noldus Ethovision output was used directly for both total time and time binned results. The latency to appear on the platform is scored manually, by selecting the last video frame when the center of the mouse is tracked in the Tunnel Entry zone before climbing onto the arena and standing with all four paws on the platform.
Behavioral Measure Data Normalization:
As indicated in Table S2, all ‘time spent in zone’, sand displaced, food consumed and zone visit measures were normalized to the total time spent in the arena (TTA) by dividing each value by TTA (x/TTA). For time bin values, the TTA for the corresponding time bin is used for normalization (i.e., x-1/TTA-1). All latencies to visit pots were normalized to the latency to enter the platform (LEP) by subtracting the LEP (x-LEP). Latency to the center zone after arena entry (LCAE) is already normalized to LEP and does not need any further normalization. All percentage measures do not need any normalization. Time on the platform itself and time in the tunnel (cage) are not included in the normalized dataset because they are closely related and redundant to TTA.
Locomotor Measures
The raw data files generated by Noldus Ethovision listing all XY coordinated and all zones as well as distance traveled and velocity for each frame were used to extract data describing excursions and locomotor patterns using custom code, including the duration and number of bouts at different velocities. An excursion is defined as beginning when the mouse leaves the tunnel (In Tunnel zone) and ending when the mouse returns to the tunnel (one round trip). Continuous velocity values in the data were categorized into three velocity classes (gears in Table S2), slow: velocity ≤ 5 equals; medium: velocity > 5 ≤ 15; fast: velocity > 15. The length of the bout is calculated using the number of frames in the sequence and all sequences of the same velocity class longer than 3 frames are counted as a single velocity bout.
QUANTIFICATION AND STATISTICAL ANALYSIS
Excursion Data Capture
Our DeepFeats approach for analyzing modularity in XY behavior tracking data can be adapted for most commercial tracking software. In our study, mice were tracked with Noldus Ethovision software. Noldus settings were used to define regions of interest in the foraging arena and indicated when the mouse was in each area. To ensure the tracking is equivalent across different mice, a Procrustes transformation of the XY coordinates was performed to put every tracking file in the same coordinate space. The track coordinates were zero’d to the center of the tunnel to the home cage. We then generated custom code in R to parse the raw Noldus tracking files into discrete, round trip home base excursions from the home cage tunnel. Each excursion is assigned a unique ID key that we call the Concise Idiosyncratic Module Alignment Report (CIMAR) string key. It stores the coordinates of the excursion in the data and the CIMAR string includes metadata regarding the mouse number, excursion number, sex, age, genotype and phase. Next, custom code compares the CIMAR coordinates to the raw Noldus data files and constructs a new dataset that extracts 57 measures from the Noldus output, which we use to initially statistically describe each excursion. The 57 measures are presented in Table S1 and are designed to capture a relatively comprehensive array of different behavioral and locomotor parameters, as well as describe interactions with food and non-food containing patches and exposed regions in the environment. These measures consist of shape, frequency, order and location statistics of an animal’s X and Y movements, numbers of visits and time spent at different features in the arena, including food patches (Pots#2 and 4), non-food containing patches (Pots#1 and 3), the tunnel zone, wall zone and center zone of the arena and data describing locomotor patterns, including velocity, gait and distance traveled. The 57 measures for each excursion are normalized and centered because they are in different units.
Identification of a Set of Behavioral Measures to Resolve Modularity in Excursions
This section details the methods to define the set of behavioral measures that best resolve candidate modules. A data matrix was constructed in which the rows are excursions performed by the mice, labeled by CIMAR keys, and the columns are the 57 behavioral measures. A correlation matrix was constructed from the data using the Pearson correlation statistic. The measures were then systematically filtered from the data as detailed in the main text based on different correlation thresholds using the “findCorrelation” function in the caret package in R. With this approach, the absolute values of pairwise correlations are considered. If two variables have a high correlation, the function looks at the mean absolute correlation of each variable and removes the variable with the largest mean absolute correlation. We systematically threshold the data in r = 0.5+ value increments as shown in the main text to identify the best set of measures to resolve clusters of excursions. At each threshold, the retained measures are used in an unsupervised clustering analysis to define clusters of excursions. We used the Ward. D2 minimum variance method implemented using the “hclust” function in R to perform the clustering and define compact, spherical clusters. We then statistically define discrete excursion clusters from the results using the Dynamic Tree Cut algorithm (Langfelder et al., 2008). This is a powerful approach because it is adaptive to the shape of the dendrogram compared to typical constant height cutoff methods and offers the following advantages: (1) identification of nested clusters; (2) suitable for automation; and (3) can combine the advantages of hierarchical clustering and partitioning around medoids, giving better detection of outliers. We detect clusters using the “hybrid” method and use the DeepSplit parameter set to 4 and the minimum cluster size set to 2 to maximize nested cluster detection in the dendrogram. The total number of clusters detected is quantified at each correlation threshold. Conceptually, more relaxed correlation threshold cutoffs could reduce cluster detection by retaining redundant measures that mask important effects from other measures. On the other hand, thresholds that are too stringent could reduce cluster detection by pruning informative measures. Our objective is to identify the threshold that uncovers the most informative and sensitive set of measures for resolving different clusters of excursions, setting the stage for the discovery of potential modules.
Statistical Validation of Significant Clusters of Excursions Based on Retained Behavioral Measures
In our study, Dynamic Tree Cut will deeply cut branches in a dendrogram generating large numbers of small clusters if there are few bona fide relationships in the data. Thus, to test whether bona fide clusters of excursions exist in the data we implemented a random sampling procedure in R in which we randomly sample from the matrix of the retained behavioral measure data to break the relationships between the excursions and the measures. The sampled null data matrix is then subjected to the same clustering and quantification procedure to determine the number of clusters found by Dynamic Tree Cut. A null distribution is created from 10,000 iterations and compared to the observed number of clusters, which is expected to be significantly less than the null due to bona fide biological relationships between the excursions and set of retained measures. A lower tailed p value was computed to test this outcome.
IGP Permutation Test to Identify Significant Modules of Behavior
To test whether reproducible modules of behavior exist in the data for the foraging excursions, we use the in-group proportion (IGP) statistical method for testing for reproducible clusters between two datasets, which is implemented in the clusterRepro package in R (Kapp and Tibshirani, 2007). We built a modified version of this function for parallelized computing to speed the analysis for large number numbers of permutations. The excursion data for the mice is separated into a training data and test data partition for reproducibility testing. A balanced partition is generated by genotype, age, animal and phase factors using the “createDataPartition” function in the caret package in R. Unsupervised hierarchical clustering was performed on the Training data partition excursions and clusters were defined using Dynamic Tree Cut. Next, the centroids for each training data cluster were computed as the mean values of the behavioral data for the excursions in the cluster. The training data centroids were then used to compute the IGP statistic for each training data cluster based on the test partition data, thereby evaluating the reproducibility of each cluster. The permutation testing approach implemented in the clusterRepro R package was used to compute p values for each cluster to determine whether the IGP value is greater than chance. False positives due to multiple testing errors were controlled using the q-value method (Storey, 2002). Modules are thus defined as significantly reproducible training partition excursion clusters (q < 0.1). Each module detected was assigned an ID number and individual excursions in the data were annotated based on the module they match to. This approach facilitated quantifications of module expression frequency by the mice.
Generalized Linear Modeling and Likelihood Ratio Testing of Module Expression Frequency
To statistically evaluate the factors that significantly affect module expression frequency, we used generalized linear modeling functions implemented in R. The full and nested models tested are detailed in the text for each analysis. The generalized linear modeling is performed using a Poisson distribution. We tested the goodness-of-fit of the full model with a chi-square test of the residual deviance and degrees of freedom in R. If the test result was not significant (p > 0.05), we concluded the model fit the data and proceeded based on Poisson-distributed errors – which was the case for all analyses in the paper. The likelihood ratio test comparing the full and nested models was performed using the anova() function in R with the additional option test = “Chisq.”
Testing Age Effects on Module Expression (Figure 6):
The generalized linear model was fit based on a Poisson distribution chosen from a goodness-of-fit test (p > 0.05). The full model (expression ~module + mouse strain + age + module:age) was compared to a nested model of the main effects (expression ~module + mouse strain + age) and absorbs variance related to the mouse strain.
Testing Genetic Effects on Module Expression (main text):
We compared the full model (expression ~module + genotype + module:genotype) to a nested model of the main effects (expression ~module + genotype) to test the B6 and F1bc module expression data for an interaction effect between module and mouse genotype. For juveniles, we included a variable for the main effect of age to absorb variance between P20 and P25 offspring.
Testing Parental Effects on Module Expression (main text):
For adults, we compared the full model (expression ~module + cross + module:cross) to a nested model of the main effects (expression ~module + genotype) to test the F1cb and F1bc module expression data for an interaction effect between module and parental cross. For juveniles, we included a variable for the main effect of age to absorb variance between P15, P20 and P25 offspring.
Testing Loss of the Paternal Magel2 Allele on Module Expression (main text):
The full model (expression ~module + paternal allele genotype + module:paternal allele genotype) was compared to a nested model of the main effects alone (expression ~module + paternal allele genotype).
Testing Loss of the Maternal Magel2 Allele on Module Expression (main text):
The fit of the full model (expression ~module + paternal allele genotype + maternal allele genotype + module:maternal allele genotype) was compared to a nested model of the main effects only (expression ~module + paternal allele genotype + maternal allele genotype) and absorbs variance due to the paternal allele genotype.
Module Expression Timing Analysis
To evaluate the temporal characteristics of module expression, we record the start time of each excursion relative to the start times of the Exploration or Foraging phase trial. Using custom code in R, this data was formatted as an event sequence object in which the temporal properties (start time, end time, duration, expression order) of each excursion for each animal are stored along with the identity of the module assignments and metadata for the animal (ID, genotype, age, sex, phase). To visualize the temporal expression patterns of each module type in the Exploration or Foraging phases as density ridgeplots, we used the “ggplot2” package in R. To determine whether modules differ significantly in their timing characteristics, a linear regression analysis was performed on the data using the “lm” function in R to test whether variance in excursion start times (response variable) is significantly explained by the factor “module”: (excursion start time ~module). The effect of the explanatory variable was determined using the anova() function in R on the fitted linear model. This same general approach was used to test other explanatory variables in the study, such as age, genotype or the interaction between module and genotype.
Module Expression Sequential Order Analysis
To test whether modules are expressed in a random versus probabilistically determined order, the event sequences of the excursions were sorted in order according to start time for each animal. Using the “createSequenceMatrix” function in the markovchain package in R, we used the event sequence data object to create a transition matrix that counts the number of transitions from one module type to another, or from one module type to a modular or nonmodular excursion class, across all of the mice tested. The same function was used to create a transition probability matrix, which is displayed in the main text using the “corrplot” and “viridis” packages in R. To test whether transitions between modules are significantly dependent on module type, we performed a Fisher’s Exact Test on the transition matrix count table. The Fisher’s Exact Test was performed in R using the “fisher.test” function and simulate.p.value option and 100,000 Monte Carlo replicates.
Developmental Changes to Modular Architecture
To test whether changes to the relative expression frequency of modules versus nonmodular behavioral sequences occur developmentally, we quantified the modular and nonmodular excursions for P20, P25 and adult mice. A Chi-Square Test of Independence was performed on the count matrix using the “vcd” package in R, which also displays mosaic plots of the results. The mosaic plot shows the Pearson residuals from the Chi-Square test, which are defined as: (observed – expected) / sqrt(expected).
Identification of Individual Modules Positively or Negatively Associated with Specific Ages or Genotypes
To define individual modules that are most strongly positively or negatively associated with a given age, cross or genotype, we quantified module expression frequencies according to the factor of interest (age, cross or genotype). The count table was then analyzed using a Chi-Square Test of Independence to compute the Pearson residuals, defined as: (observed – expected) / sqrt(expected). The results are displayed in a chart visualized using the “corrplot” package in R and show how strong the positive (positive residual value) or negative (negative residual value) association is between individual modules and a given factor of interest. We identify the top modules impacted by specific genetic, parental or developmental effects from these results.
Associating Modules with Economic Behavior Patterns Using Keystone Features of Foraging
Measures of Keystone features of foraging, including feeding, activity, exposure and preservation responses were collected for each mouse (see Table S2), along with the expression frequency counts of each module. From this data, a pairwise nonparametric Spearman rank correlation matrix was computed that relates the module expression frequencies to the measures of the different keystone features using the “corr.test” function in the psych package in R. The p values for each correlation event in the Exploration and Foraging phase data were computed using the “corr.test” function. False positives due to multiple testing were corrected using the q-value approach and significant correlations between specific modules and measures of keystone features were defined (q < 0.1). The results were displayed using the “corrplot” function in the corrplot package in R.
Testing Preservation of Reference Modules in the Magel2 Mouse Cohort
Behavioral measures for the Magel2 mouse excursions were captured and pruned to the 13 measures used to identify the reference modules (ie. Cast, B6, F1bc and F1bc modules). The excursion data for the Magel2 mice was compared to the centroids for the 71 significant reference modules using the IGP permutation testing method. Each of the 71 modules received a p value for reproducibility in the Magel2 mouse data. The p values were analyzed using the qvalue package in R to compute the proportion of true positives and true nulls in the data (ie. the pi0 statistic).
DATA AND CODE AVAILABILITY
Raw Noldus Ethovision Mouse tracking data from GREGG Lab is available upon request. Keystone Feature and Module Expression Data deposited in DRYAD (https://doi.org/10.5061/dryad.1cr4qd3).
Supplementary Material
KEY RESOURCES TABLE
Highlights.
A methodology to dissect the architecture of complex behavior patterns
Foraging patterns are built from finite, genetically controlled modules of behavior
Different modules are linked to different economic behavior patterns
Parental alleles of the Prader-Willi syndrome gene Magel2 regulate distinct modules
ACKNOWLEDGMENTS
The methodology in this study was reviewed by the University of Utah Biostatistics Core Facility. The authors wish to thank Drs. Monica Vetter, Adam Douglass, Matt Wachowiak, Jan Christian, and Jared Rutter and the Gregg lab for comments on earlier versions of the manuscript. C.N.S.H. was supported by a Swiss National Science Foundation postdoctoral fellowship and the Gottfried & Julia Bangerter-Rhyner Stiftung. E.W. and P.T.F. are supported by NSF grant IIS-1251049. The study was funded by NIH grant R01MH109577 (C.G.). C.G. is a New York Stem Cell Foundation Robertson Investigator. This research was supported by the New York Stem Cell Foundation.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.07.038.
DECLARATION OF INTERESTS
The authors declare that a patent on this work is pending.
REFERENCES
- Anderson DJ, and Perona P (2014). Toward a science of computational ethology. Neuron 84, 18–31. [DOI] [PubMed] [Google Scholar]
- Bakken TE, Miller JA, Ding S-L, Sunkin SM, Smith KA, Ng L, Szafer A, Dalley RA, Royall JJ, Lemon T, et al. (2016). A comprehensive transcriptional map of primate brain development. Nature 535, 367–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton RA (2012). Embodied cognitive evolution and the cerebellum. Philos. Trans. R. Soc. Lond. B Biol. Sci 367, 2097–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bervini S, and Herzog H (2013). Mouse models of Prader-Willi syndrome: a systematic review. Front. Neuroendocrinol 34, 107–119. [DOI] [PubMed] [Google Scholar]
- Bonthuis PJ, Huang W-C, Stacher Hörndli CN, Ferris E, Cheng T, and Gregg C (2015). Noncanonical Genomic Imprinting Effects in Offspring. Cell Rep. 12, 979–991. [DOI] [PubMed] [Google Scholar]
- Börger L, Dalziel BD, and Fryxell JM (2008). Are there general mechanisms of animal home range behaviour? A review and prospects for future research. Ecol. Lett 11, 637–650. [DOI] [PubMed] [Google Scholar]
- Brown J, and Kotler B (1994). Foraging theory, patch use, and the structure of a Negev Desert granivore community. Ecology 75, 2286–2300. [Google Scholar]
- Brown J, and Laundré J (1999). The ecology of fear: optimal foraging, game theory, and trophic interactions. J. Mammal 80, 385–399. [Google Scholar]
- Camerer CF (2013). Goals, methods, and progress in neuroeconomics. Annu. Rev. Econ 5, 425–455. [Google Scholar]
- Cassidy SB, and Driscoll DJ (2009). Prader-Willi syndrome. Eur. J. Hum. Genet 17, 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy SB, Schwartz S, Miller JL, and Driscoll DJ (2012). Prader-Willi syndrome. Genet. Med 14, 10–26. [DOI] [PubMed] [Google Scholar]
- Eilam D, and Golani I (1989). Home base behavior of rats (Rattus norvegicus) exploring a novel environment. Behav. Brain Res 34, 199–211. [DOI] [PubMed] [Google Scholar]
- Galef BG Jr. (1981). The ecology of weaning. Parental Care in Mammals (Springer US), pp. 211–241. [Google Scholar]
- Gorny JH, Gorny B, Wallace DG, and Whishaw IQ (2002). Fimbria-fornix lesions disrupt the dead reckoning (homing) component of exploratory behavior in mice. Learn. Mem 9, 387–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley CA, and Phelps EA (2012). Anxiety and decision-making. Biol. Psychiatry 72, 113–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewlett I, and Lamb ME (2010). Hunter-Gatherer Childhoods: Evolutionary, Developmental, and Cultural Perspectives (Transaction Publishers). [Google Scholar]
- Hong W, Kennedy A, Burgos-Artizzu XP, Zelikowsky M, Navonne SG, Perona P, and Anderson DJ (2015). Automated measurement of mouse social behaviors using depth sensing, video tracking, and machine learning. Proc. Natl. Acad. Sci. USA 112, E5351–E5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W-C, Ferris E, Cheng T, Hörndli CS, Gleason K, Tamminga C, Wagner JD, Boucher KM, Christian JL, and Gregg C (2017). Diverse non-genetic, allele-specific expression effects shape genetic architecture at the cellular level in the mammalian brain. Neuron 93, 1094–1109.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JH (2011). Primates and the evolution of long, slow life histories. Curr. Biol 21, R708–R717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabra M, Robie AA, Rivera-Alba M, Branson S, and Branson K (2013). JAABA: interactive machine learning for automatic annotation of animal behavior. Nat. Methods 10, 64–67. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AMM, Pletikos M, Meyer KA, Sedmak G, et al. (2011). Spatio-temporal transcriptome of the human brain. Nature 478, 483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapp AV, and Tibshirani R (2007). Are clusters found in one dataset present in another dataset? Biostatistics 8, 9–31. [DOI] [PubMed] [Google Scholar]
- Kavanau JL (1967). Behavior of captive white-footed mice. Science 155, 1623–1639. [DOI] [PubMed] [Google Scholar]
- Kozlov SV, Bogenpohl JW, Howell MP, Wevrick R, Panda S, Hogenesch JB, Muglia LJ, Van Gelder RN, Herzog ED, and Stewart CL (2007). The imprinted gene Magel2 regulates normal circadian output. Nat. Genet 39, 1266–1272. [DOI] [PubMed] [Google Scholar]
- Lacey ER (1998). What is an adaptive environmentally induced parental effect? In Maternal Effects as Adaptations, Mousseau TA and Fox CW, eds. (Oxford University Press; ), pp. 54–66. [Google Scholar]
- Langfelder P, Zhang B, and Horvath S (2008). Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics 24, 719–720. [DOI] [PubMed] [Google Scholar]
- Lee PC (1996). The meanings of weaning: growth, lactation, and life history. Evol. Anthropol 5, 87–98. [Google Scholar]
- Lord C, Bishop S, and Anderson D (2015). Developmental trajectories as autism phenotypes. Am. J. Med. Genet. C. Semin. Med. Genet 169, 198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz JE, Gillis WF, Beron CC, Neufeld SQ, Robertson K, Bhagat ND, Peterson RE, Peterson E, Hyun M, Linderman SW, et al. (2018). The striatum organizes 3D behavior via moment-to-moment action selection. Cell 174, 44–58.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matarazzo V, and Muscatelli F (2013). Natural breaking of the maternal silence at the mouse and human imprinted Prader-Willi locus: a whisper with functional consequences. Rare Dis 1, e27228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melin AD, Young HC, Mosdossy KN, and Fedigan LM (2014). Seasonality, extractive foraging and the evolution of primate sensorimotor intelligence. J. Hum. Evol 71, 77–86. [DOI] [PubMed] [Google Scholar]
- Mercer RE, Kwolek EM, Bischof JM, van Eede M, Henkelman RM, and Wevrick R (2009). Regionally reduced brain volume, altered serotonin neurochemistry, and abnormal behavior in mice null for the circadian rhythm output gene Magel2. Am. J. Med. Genet. B. Neuropsychiatr. Genet 150B, 1085–1099. [DOI] [PubMed] [Google Scholar]
- Miller JA, Ding S-L, Sunkin SM, Smith KA, Ng L, Szafer A, Ebbert A, Riley ZL, Royall JJ, Aiona K, et al. (2014). Transcriptional landscape of the prenatal human brain. Nature 508, 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monterosso J, Piray P, and Luo S (2012). Neuroeconomics and the study of addiction. Biol. Psychiatry 72, 107–112. [DOI] [PubMed] [Google Scholar]
- Pulliam H, and Charnov E (1977). Optimal foraging: a selective review of theory and tests. Q. Rev. Biol 52, 137–154. [Google Scholar]
- Reader SM, and Laland KN (2002). Social intelligence, innovation, and enhanced brain size in primates. Proc. Natl. Acad. Sci. USA 99, 4436–4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieusset A, Schaller F, Unmehopa U, Matarazzo V, Watrin F, Linke M, Georges B, Bischof J, Dijkstra F, Bloemsma M, et al. (2013). Stochastic loss of silencing of the imprinted Ndn/NDN allele, in a mouse model and humans with Prader-Willi syndrome, has functional consequences. PLoS Genet. 9, e1003752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robie AA, Hirokawa J, Edwards AW, Umayam LA, Lee A, Phillips ML, Card GM, Korff W, Rubin GM, Simpson JH, et al. (2017). Mapping the neural substrates of behavior. Cell 170, 393–406.e28. [DOI] [PubMed] [Google Scholar]
- Rowland NE, Vaughan CH, Mathes CM, and Mitra A (2008). Feeding behavior, obesity, and neuroeconomics. Physiol. Behav 93, 97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter M, Kim-Cohen J, and Maughan B (2006). Continuities and discontinuities in psychopathology between childhood and adult life. J. Child Psychol. Psychiatry 47, 276–295. [DOI] [PubMed] [Google Scholar]
- Schaaf CP, Gonzalez-Garay ML, Xia F, Potocki L, Gripp KW, Zhang B, Peters BA, McElwain MA, Drmanac R, Beaudet AL, et al. (2013). Truncating mutations of MAGEL2 cause Prader-Willi phenotypes and autism. Nat. Genet 45, 1405–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller F, Watrin F, Sturny R, Massacrier A, Szepetowski P, and Muscatelli F (2010). A single postnatal injection of oxytocin rescues the lethal feeding behaviour in mouse newborns deficient for the imprinted Magel2 gene. Hum. Mol. Genet 19, 4895–4905. [DOI] [PubMed] [Google Scholar]
- Sharp C, Monterosso J, and Montague PR (2012). Neuroeconomics: a bridge for translational research. Biol. Psychiatry 72, 87–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silbereis JC, Pochareddy S, Zhu Y, Li M, and Sestan N (2016). The cellular and molecular landscapes of the developing human central nervous system. Neuron 89, 248–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sol D, Duncan RP, Blackburn TM, Cassey P, and Lefebvre L (2005). Big brains, enhanced cognition, and response of birds to novel environments. Proc. Natl. Acad. Sci. USA 102, 5460–5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens DW, Brown JS, and Ydenberg RC (2007). Foraging: Behavior and Ecology (University of Chicago Press). [Google Scholar]
- Storey JD (2002). A direct approach to false discovery rates. J. R. Statist. Soc. B 64, 479–498. [Google Scholar]
- Storey J (2003). The positive false discovery rate: a Bayesian interpretation and the q-value. Ann. Stat 31, 2013–2035. [Google Scholar]
- Thompson CL, Ng L, Menon V, Martinez S, Lee C-K, Glattfelder K, Sunkin SM, Henry A, Lau C, Dang C, et al. (2014). A high-resolution spatiotemporal atlas of gene expression of the developing mouse brain. Neuron 83, 309–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivers RL (1974). Parent-offspring conflict. Integr. Comp. Biol 14, 249–264. [Google Scholar]
- Vogelstein JT, Park Y, Ohyama T, Kerr RA, Truman JW, Priebe CE, and Zlatic M (2014). Discovery of brainwide neural-behavioral maps via multi-scale unsupervised structure learning. Science 344, 386–392. [DOI] [PubMed] [Google Scholar]
- Walker R, Burger O, Wagner J, and Von Rueden CR (2006). Evolution of brain size and juvenile periods in primates. J. Hum. Evol 51, 480–489. [DOI] [PubMed] [Google Scholar]
- Wallace DG, Martin MM, and Winter SS (2008). Fractionating dead reckoning: role of the compass, odometer, logbook, and home base establishment in spatial orientation. Naturwissenschaften 95, 1011–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissbrod A, Shapiro A, Vasserman G, Edry L, Dayan M, Yitzhaky A, Hertzberg L, Feinerman O, and Kimchi T (2013). Automated long-term tracking and social behavioural phenotyping of animal colonies within a semi-natural environment. Nat. Commun 4, 2018. [DOI] [PubMed] [Google Scholar]
- Wiltschko AB, Johnson MJ, Iurilli G, Peterson RE, Katon JM, Pashkovski SL, Abraira VE, Adams RP, and Datta SR (2015). Mapping sub-second structure in mouse behavior. Neuron 88, 1121–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman AD, Charvet CJ, Clancy B, Darlington RB, and Finlay BL (2013). Modeling transformations of neurodevelopmental sequences across mammalian species. J. Neurosci 33, 7368–7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CF, and Shah NM (2014). Representing sex in the brain, one module at a time. Neuron 82, 261–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Shen EH, Hohmann JG, Oh SW, Bernard A, Royall JJ, Glattfelder KJ, Sunkin SM, Morris JA, Guillozet-Bongaarts AL, et al. (2012). Large-scale cellular-resolution gene profiling in human neocortex reveals species-specific molecular signatures. Cell 149, 483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Chen Y, Deng F, Huang R, Petersen F, Ibrahim S, and Yu X (2014). mtDNA sequence, phylogeny and evolution of laboratory mice. Mitochondrion 17, 126–131. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw Noldus Ethovision Mouse tracking data from GREGG Lab is available upon request. Keystone Feature and Module Expression Data deposited in DRYAD (https://doi.org/10.5061/dryad.1cr4qd3).