Abstract
Purpose
Diabetic retinopathy (DR) is a microvascular complication caused by prolonged hyperglycemia and characterized by leaky retinal vasculature and ischemia-induced angiogenesis. Vitreous humor is a gel-like biofluid in the posterior segment of the eye between the lens and the retina. Disease-related changes are observed in the biochemical constituents of the vitreous, including proteins and macromolecules. Recently, we found that IL-6 trans-signaling plays a significant role in the vascular leakage and retinal pathology associated with DR. Therefore, in this study, comprehensive proteomic profiling of the murine vitreous was performed to identify diabetes-induced alterations and to determine effects of IL-6 trans-signaling inhibition on these changes.
Methods
Vitreous samples from mice were collected by evisceration, and proteomic analyses were performed using liquid chromatography–tandem mass spectrometry (LC-MS/MS).
Results
A total of 154 proteins were identified with high confidence in control mice and were considered to be characteristic of healthy murine vitreous fluid. The levels of 72 vitreous proteins were significantly altered in diabetic mice, including several members of heat shock proteins, 14-3-3 proteins, and tubulins. Alterations in 52 out of 72 proteins in diabetic mice were mitigated by IL-6 trans-signaling inhibition.
Conclusions
Proteomic analysis of murine vitreous fluid performed in this study provides important information about the changes caused by diabetes in the ocular microenvironment. These diabetes-induced alterations in the murine vitreous proteome were mitigated by IL-6 trans-signaling inhibition. These findings further support that IL-6 trans-signaling may be an important therapeutic target for the treatment of DR.
Keywords: diabetic retinopathy, IL-6 trans-signaling, vitreous fluid, proteomics, LC-MS/MS
Increased retinal vascular permeability in diabetic retinopathy (DR) has been linked to inflammatory processes and the release of proinflammatory cytokines, including members of the interleukin family.1–4 IL-6 is a pleiotropic cytokine known to be elevated in patients with DR and associated with DR pathology.5–7 Importantly, IL-6 signaling occurs via two different mechanisms: the classical and trans-signaling pathways.5,8,9 Classical signaling is mediated by the membrane-bound IL-6 receptor; in contrast, IL-6 trans-signaling uses a soluble form of the IL-6 receptor (sIL-6R) and is primarily proinflammatory. Existing anti–IL-6 therapeutics inhibit both classical and trans-signaling together. However, IL-6 trans-signaling can be selectively inhibited using sgp130Fc, a fusion protein consisting of the endogenous IL-6 trans-signaling inhibitor soluble gp130 (sgp130) and the Fc region of IgG1.10 Our lab has previously demonstrated that sgp130Fc can decrease inflammation, restore oxidative balance, and prevent endothelial barrier dysfunction in retinal endothelial cells.5,11
Vitreous humor is a gel-like biofluid in the posterior segment of the eye between the lens and the retina. Composed primarily of water, vitreous also contains a variety of macromolecules, both native to the vitreous and from neighboring tissues.12–14 Because of the proximity of the vitreous to the retina and the contribution of retinal vasculature to the maintenance of the vitreous body, disease-related retinal changes can alter the protein composition of the vitreous.15–18 Several proteomic studies have been conducted on human vitreous fluid,12,13,15,18–22 providing important and clinically relevant insight into DR pathogenesis. However, controlled murine studies play a crucial role in the preclinical evaluation of novel therapies, and studies examining murine vitreous are very limited.23,24 This is largely due to the small vitreous volume, and murine samples are often pooled for analysis. Recent technological advancements in mass spectrometry (MS) have improved proteomic profiling in small sample volumes, providing a new method for evaluating the effects of novel DR therapies on the ocular microenvironment, including the murine vitreous proteome.
The purpose of this study was to identify the protective effects of IL-6 trans-signaling inhibition on the diabetic mouse vitreous proteome using advanced MS technology. The vitreous proteomes of healthy, diabetic, and diabetic mice treated with sgp130Fc were characterized. Murine vitreous samples were processed and analyzed individually, without pooling, resulting in increased power to detect the variation in biological replicates. Our analyses identified vitreous proteomic changes associated with diabetes, as well as a large set of proteins that normalized in response to sgp130Fc treatment, providing novel insight into the role of IL-6 trans-signaling in the pathogenesis of DR.
Methods
Animal Studies
All animal studies were conducted in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. Diabetes was induced in 8-week-old male C57BL/6 mice (Jackson Laboratory, Bar Harbor, ME, USA) by intraperitoneal injection of 65 mg/kg streptozotocin (Sigma, St. Louis, MO, USA) in sodium citrate buffer (0.05 M, pH 4.5) once daily for 5 days. Mice were considered diabetic when glucose levels were consistently above 250 mg/dL, and both blood glucose and body weight were assessed continually for the duration of the study. After 8 weeks of hyperglycemia, diabetic mice were randomly assigned to untreated or sgp130Fc treatment groups. Treated mice received 5 mg/kg sgp130Fc (Mouse sgp130Fc Chimera Protein; R&D Systems, Minneapolis, MN, USA) by intraperitoneal injection twice weekly for the last 2 weeks of study. At the end of the treatment period, all mice (control, diabetic, and diabetic treated with sgp130Fc) were euthanized for sample collection. Eyes were dissected and vitreous bodies extracted as previously described by Skeie et al.23 Briefly, a linear incision was made in the cornea, and lens-vitreous tissue was extracted by gently applying pressure to the external surface of the sclera. To isolate vitreous fluid, tissue was washed in 20 μL PBS and centrifuged in Costar Spin-X 0.22-mm centrifuge tube filters (Corning, Inc., Corning, NY, USA) at 14,000 × g for 15 minutes at 4°C. The vitreous was collected as filtrate and stored at –80°C until analysis.
Proteomic Profiling by Liquid Chromatography–Tandem Mass Spectrometry (LC-MS/MS)
Following protein digestion, vitreous samples were analyzed using an Ultimate 3000 RSLC nano system (Thermo Scientific, Waltham, MA, USA) coupled to an Orbitrap Fusion Tribrid mass spectrometer (Thermo Scientific). Raw MS peptide data were analyzed using Proteome Discoverer (v1.4; Thermo Scientific) and searched against the Uniprot protein database using TurboSequest. Parallel reaction monitoring (PRM) was used to confirm the findings of discovery proteomic analyses by targeting and quantifying specific peptides of interest.25,26 Additional details for LC-MS/MS methodology are provided in the supplemental materials and methods section.
Statistical and Bioinformatics Analyses
All statistical analyses were conducted using the R Project for Statistical Computing (v3.2.5, R Foundation for Statistical Computing, Vienna, Austria; www.r-project.org). Peptide spectral match (PSM) count data for the identified proteins were log2 transformed and differential expression analyses were conducted using the “edgeR” package.27 False discovery rate (FDR)-adjusted P values <0.05 were considered significant. In order to identify biological processes, cellular components, and molecular functions, a functional annotation analysis was conducted using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) Bioinformatics Resources v6.8.28 Similarly, Ingenuity Pathway Analysis (IPA) software was used for network analysis to display the interactions between proteins of interest.
Results
Proteomic Constituents of Healthy Murine Vitreous
A total of 443 unique proteins were identified in all mouse vitreous samples, which is comparable to the number of proteins identified in other studies.29 Of these, 154 proteins were identified with high confidence in healthy control mice (present in at least three of six control animals, a distinction not possible with pooled samples) and were considered to be characteristic of healthy murine vitreous fluid. The top 50 proteins identified in the healthy murine vitreous are listed in Table 1, and a complete list of all the 154 proteins is available in Supplementary Table S1. The most abundant proteins found in murine vitreous are several crystallin isoforms, including γ-crystallin C (average PSM = 1561), γ-crystallin B (average PSM = 1431), γ-crystallin D (average PSM = 797), β-crystallin S (average PSM = 613), γ-crystallin E (average PSM = 608), β-crystallin B2 (average PSM = 584), β-crystallin A4 (average PSM = 342), β-crystallin A1 (average PSM = 262), β-crystallin B3 (average PSM = 189), γ-crystallin A (average PSM = 179), β-crystallin A2 (average PSM = 183), α-crystallin A (average PSM = 155), and β-crystallin B1 (average PSM = 131). Other abundant vitreous proteins include albumin (average PSM = 359), hemoglobin subunit β1 (average PSM = 141), phosphatidylethanolamine-binding protein 1 (average PSM = 88), and phosphoglycerate kinase 1 (average PSM = 64). Functional annotation analysis revealed that two highly enriched molecular functions in the vitreous proteins are structural constituents of the eye and cadherin binding involved in cell-cell junctions (Fig. 1A). The major biological processes associated with vitreous proteins include visual perception, eye development, lens development, glycolytic processes, and response to peptide hormones (Fig. 1B). The major cellular components include cytoplasm, extracellular exosomes, extracellular space, myelin sheath, axons, cell-cell adherens junctions, and neuronal projections (Fig. 1C).
Table 1.
Top 50 Proteins Identified in the Healthy Murine Vitreous Humor using Mass Spectrometry (MS)
| Accession | Symbol | Description | Average PSM |
|---|---|---|---|
| Q61597 | Crygc | Gamma-crystallin C | 1561.13 |
| P04344 | Crygb | Gamma-crystallin B | 1430.51 |
| P04342 | Crygd | Gamma-crystallin D | 797.49 |
| O35486 | Crygs | Beta-crystallin S | 613.42 |
| Q03740 | Cryge | Gamma-crystallin E | 607.54 |
| P62696 | Crybb2 | Beta-crystallin B2 | 583.62 |
| P07724 | Alb | Albumin | 359.00 |
| Q9JJV0 | Cryba4 | Beta-crystallin A4 | 342.11 |
| P02525 | Cryba1 | Beta-crystallin A1 | 262.33 |
| Q9JJU9 | Crybb3 | Beta-crystallin B3 | 188.71 |
| Q9JJV1 | Cryba2 | Beta-crystallin A2 | 182.59 |
| P04345 | Cryga | Gamma-crystallin A | 179.41 |
| P24622 | Cryaa | Alpha-crystallin A chain | 155.10 |
| P02088 | Hbb-b1 | Hemoglobin subunit beta-1 | 140.66 |
| Q9WVJ5 | Crybb1 | Beta-crystallin B1 | 131.34 |
| P70296 | Pebp1 | Phosphatidylethanolamine-binding protein 1 | 87.74 |
| P09411 | Pgk1 | Phosphoglycerate kinase 1 | 63.82 |
| Q5DTX6 | Jcad | Junctional protein associated with coronary artery disease | 57.60 |
| Q9R0P9 | Uchl1 | Ubiquitin carboxyl-terminal hydrolase isozyme L1 | 45.38 |
| P10649 | Gstm1 | Glutathione S-transferase Mu 1 | 43.79 |
| P00920 | Ca2 | Carbonic anhydrase 2 | 41.42 |
| P17182 | Eno1 | Alpha-enolase | 39.81 |
| P60710 | Actb | Actin, cytoplasmic 1 | 37.63 |
| P17742 | Ppia | Peptidyl-prolyl cis-trans isomerase A | 36.18 |
| Q9CQI6 | Cotl1 | Coactosin-like protein | 34.37 |
| P17751 | Tpi1 | Triosephosphate isomerase | 30.81 |
| Q05816 | Fabp5 | Fatty acid binding protein, epidermal | 29.45 |
| P19157 | Gstp1 | Glutathione S-transferase P 1 | 28.56 |
| Q99LX0 | Park7 | Protein deglycase DJ-1 | 26.67 |
| O08709 | Prdx6 | Peroxiredoxin-6 | 26.54 |
| Q921I1 | Tf | Serotransferrin | 25.48 |
| P11590 | Mup4 | Major urinary protein 4 | 25.35 |
| Q9CPU0 | Glo1 | Lactoylglutathione lyase | 25.27 |
| O09131 | Gsto1 | Glutathione S-transferase omega-1 | 23.60 |
| P62962 | Pfn1 | Profilin-1 | 21.17 |
| Q9DBJ1 | Pgam1 | Phosphoglycerate mutase 1 | 17.95 |
| Q8BGZ7 | Krt75 | Keratin, type II cytoskeletal 75 | 17.58 |
| Q61598 | Gdi2 | Rab GDP dissociation inhibitor beta | 17.39 |
| P31786 | Dbi | Acyl-CoA-binding protein | 17.26 |
| P16015 | Ca3 | Carbonic anhydrase 3 | 15.94 |
| Q9R0P5 | Dstn | Destrin | 11.81 |
| Q04447 | Ckb | Creatine kinase B-type | 11.74 |
| Q6UGQ3 | Scgb2b2 | Secretoglobin family 2B member 2 | 11.55 |
| P0CG49 | Ubb | Polyubiquitin-B | 11.24 |
| P99029 | Prdx5 | Peroxiredoxin-5, mitochondrial | 11.05 |
| P27005 | S100a8 | Protein S100-A8 | 10.58 |
| Q6XVG2 | Cyp2c54 | Cytochrome P450 2C54 | 10.23 |
| P01942 | Hba | Hemoglobin subunit alpha | 10.00 |
| P62774 | Mtpn | Myotrophin | 9.62 |
Figure 1.
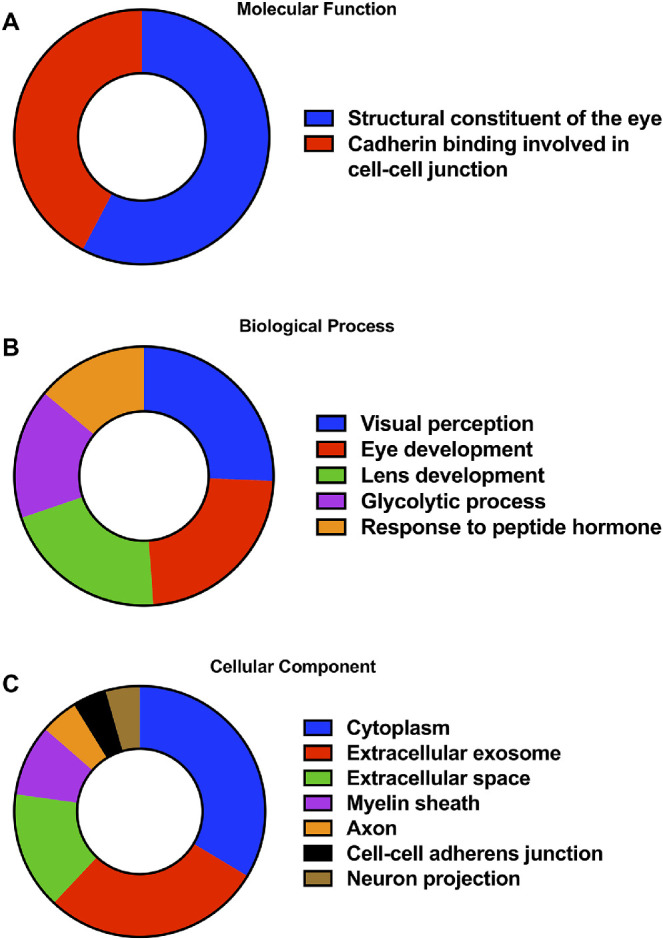
Characterization of the healthy murine vitreous humor proteome. The Gene Ontology (GO) terms, including molecular functions (A), biological processes (B), and cellular components (C), significantly associated with the 154 proteins characteristic of the healthy murine vitreous humor. Associations with Benjamini-corrected P value <0.05 were considered significant.
Vitreous Proteomic Changes in Diabetic Mice
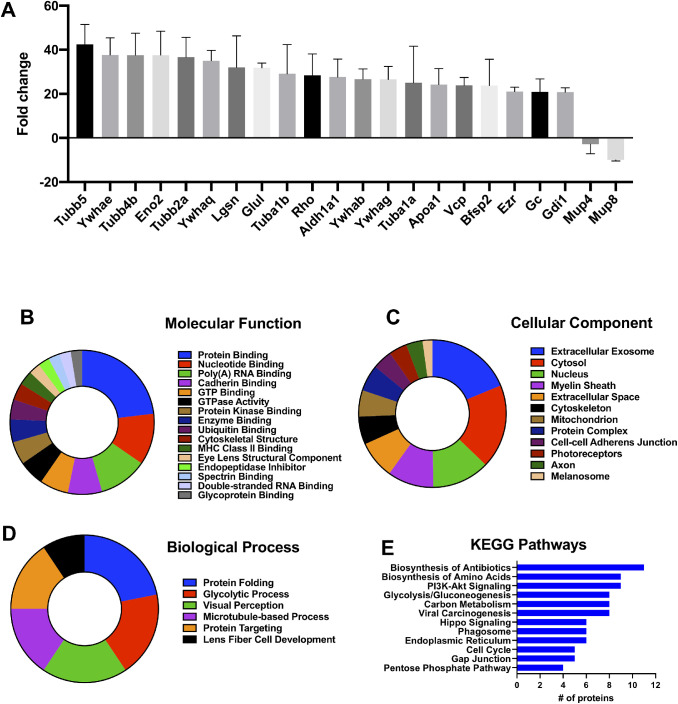
Vitreous protein levels in diabetic mice were compared to healthy mice. A total of 72 proteins were significantly altered in diabetic mice as compared to healthy controls (Table 2). The proteins with the largest change in vitreous from diabetic mice are tubulin β5 (Tubb5: 42-fold), 14-3-3 protein-ε (Ywhae: 38-fold), tubulin β4B (Tubb4b: 38-fold), γ-enolase (Eno2: 38-fold), tubulin β2A (Tubb2a: 37-fold), 14-3-3 protein-θ (Ywhaq: 35-fold), lengsin (Lgsn: 32-fold), glutamine synthetase (Glul: 32-fold), tubulin α1B (Tuba1b: 29-fold), and rhodopsin (Rho: 28-fold) (Fig. 2A). Molecular functions associated with the altered proteins include protein binding, nucleotide binding, poly(A) RNA binding, cadherin binding, GTP binding, GTPase activity, and protein kinase binding (Fig. 2B). Enriched biological processes are protein folding, glycolytic processes, visual perception, microtubule-based processes, protein targeting, and lens fiber cell development (Fig. 2C). The major cellular components of these proteins are extracellular exosomes, cytosol, nucleus, myelin sheath, extracellular space, cytoskeleton, mitochondrion, and protein complexes (Fig. 2D). The major Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways associated with the proteins altered in diabetic vitreous are biosynthesis of antibiotics, biosynthesis of amino acids, PI3K-Akt signaling, glycolysis/gluconeogenesis, and carbon metabolism (Fig. 2E).
Table 2.
Changes in the Vitreous Humor Proteome of Diabetic Mice with and without sgp130Fc Treatment as Compared to Healthy Controls
| Accession | Symbol | Description | Fold Change Diabetic * | FDR | Fold Change Diab + Sgp130Fc † | FDR |
|---|---|---|---|---|---|---|
| P99024 | Tubb5 | Tubulin beta-5 chain | 42.45 | 4.13E-07 | 28.89 | 1.18E-04 |
| P62259 | Ywhae | 14-3-3 protein epsilon | 37.66 | 8.25E-07 | 28.65 | 1.18E-04 |
| P68372 | Tubb4b | Tubulin beta-4B chain | 37.58 | 8.25E-07 | 9.44 | 0.2751 |
| P17183 | Eno2 | Gamma-enolase | 37.58 | 8.25E-07 | 8.36 | 0.4094 |
| Q7TMM9 | Tubb2a | Tubulin beta-2A chain | 36.66 | 1.32E-06 | 21.16 | 0.0026 |
| P68254 | Ywhaq | 14-3-3 protein theta | 35.03 | 2.20E-06 | 26.02 | 3.08E-04 |
| Q8CIX8 | Lgsn | Lengsin | 32.02 | 6.79E-06 | 30.28 | 1.18E-04 |
| P15105 | Glul | Glutamine synthetase | 31.83 | 1.06E-05 | 12.53 | 0.1065 |
| P05213 | Tuba1b | Tubulin alpha-1B chain | 29.11 | 3.63E-05 | 1.00 | 1.0000 |
| P15409 | Rho | Rhodopsin | 28.36 | 3.63E-05 | 11.33 | 0.1065 |
| P24549 | Aldh1a1 | Retinal dehydrogenase 1 | 27.65 | 6.68E-05 | 21.01 | 0.0026 |
| Q9CQV8 | Ywhab | 14-3-3 protein beta/alpha | 26.68 | 9.39E-05 | 12.34 | 0.0767 |
| P61982 | Ywhag | 14-3-3 protein gamma | 26.59 | 9.74E-05 | 19.27 | 0.0087 |
| P68369 | Tuba1a | Tubulin alpha-1A chain | 25.02 | 1.75E-04 | 23.20 | 0.0016 |
| Q00623 | Apoa1 | Apolipoprotein A-I | 24.21 | 3.03E-04 | 3.68 | 1.0000 |
| Q01853 | Vcp | Transitional endoplasmic reticulum ATPase | 23.82 | 3.03E-04 | 19.20 | 0.0087 |
| Q6NVD9 | Bfsp2 | Phakinin | 23.76 | 3.03E-04 | 23.51 | 9.08E-04 |
| P26040 | Ezr | Ezrin | 21.04 | 8.72E-04 | 10.48 | 0.1628 |
| P21614 | Gc | Vitamin D binding protein | 20.93 | 8.94E-04 | 6.10 | 0.5569 |
| P50396 | Gdi1 | Rab GDP dissociation inhibitor alpha | 20.76 | 8.72E-04 | 10.25 | 0.1628 |
| O08553 | Dpysl2 | Dihydropyrimidinase-related protein 2 | 20.06 | 8.72E-04 | 5.45 | 0.7703 |
| P16125 | Ldhb | L-lactate dehydrogenase B chain | 19.13 | 0.0016 | 12.02 | 0.1065 |
| P22599 | Serpina1b | Alpha-1-antitrypsin 1-2 | 18.59 | 0.0027 | 1.00 | 1.0000 |
| P16546 | Sptan1 | Spectrin alpha chain, nonerythrocytic 1 | 18.20 | 0.0027 | 11.67 | 0.1065 |
| P16460 | Ass1 | Argininosuccinate synthase | 17.52 | 0.0027 | 15.24 | 0.0443 |
| Q78ZA7 | Nap1l4 | Nucleosome assembly protein 1–like 4 | 17.43 | 0.0027 | 13.73 | 0.0767 |
| O35945 | Aldh1a7 | Aldehyde dehydrogenase, cytosolic 1 | 16.92 | 0.0050 | 1.00 | 1.0000 |
| P01027 | C3 | Complement C3 | 16.38 | 0.0082 | 1.00 | 1.0000 |
| P48774 | Gstm5 | Glutathione S-transferase Mu 5 | 16.33 | 0.0049 | 3.68 | 1.0000 |
| Q61838 | A2m | Alpha-2-macroglobulin | 16.28 | 0.0082 | 2.34 | 1.0000 |
| P13020 | Gsn | Gelsolin | 15.55 | 0.0082 | 14.01 | 0.0443 |
| Q61753 | Phgdh | D-3-phosphoglycerate dehydrogenase | 15.22 | 0.0082 | 11.04 | 0.1628 |
| P58252 | Eef2 | Elongation factor 2 | 14.51 | 0.0153 | 7.14 | 0.4094 |
| Q62433 | Ndrg1 | Protein NDRG1 | 14.05 | 0.0153 | 9.27 | 0.2751 |
| P15499 | Prph2 | Peripherin-2 | 13.93 | 0.0153 | 4.11 | 1.0000 |
| Q02053 | Uba1 | Ubiquitin-like modifier-activating enzyme 1 | 13.22 | 0.0271 | 4.11 | 1.0000 |
| P06745 | Gpi | Glucose-6-phosphate isomerase | 13.18 | 8.80E-05 | 6.65 | 0.1065 |
| P29699 | Ahsg | Alpha-2-HS-glycoprotein | 12.69 | 1.27E-04 | 4.15 | 0.5569 |
| Q9WU62 | Incenp | Inner centromere protein | 12.56 | 0.0271 | 13.49 | 0.0767 |
| P70168 | Kpnb1 | Importin subunit beta-1 | 12.42 | 0.0271 | 5.80 | 0.5569 |
| Q6P1F6 | Ppp2r2a | Serine/threonine-protein phosphatase 2A 55 Bα | 11.74 | 0.0488 | 9.27 | 0.2751 |
| Q80ZQ5 | Jazf1 | Juxtaposed with another zinc finger protein 1 | 11.72 | 0.0488 | 7.70 | 0.4094 |
| P62874 | Gnb1 | Guanine nucleotide-binding protein subunit β-1 | 11.40 | 3.72E-04 | 4.82 | 0.3183 |
| Q9ES00 | Ube4b | Ubiquitin conjugation factor E4 B | 11.18 | 0.0488 | 9.34 | 0.2751 |
| P14602 | Hspb1 | Heat shock protein beta-1 | 9.23 | 9.15E-05 | 13.12 | 4.54E-06 |
| A2AMT1 | Bfsp1 | Filensin | 8.75 | 9.71E-04 | 7.97 | 0.0136 |
| Q91XV3 | Basp1 | Brain acid soluble protein 1 | 8.67 | 0.0050 | 6.05 | 0.1471 |
| P52480 | Pkm | Pyruvate kinase PKM | 8.29 | 3.58E-06 | 7.23 | 1.72E-04 |
| P05063 | Aldoc | Fructose-bisphosphate aldolase C | 8.20 | 3.09E-05 | 7.09 | 9.08E-04 |
| P20152 | Vim | Vimentin | 7.94 | 5.68E-04 | 5.28 | 0.0784 |
| P63017 | Hspa8 | Heat shock cognate 71-kDa protein | 7.05 | 5.68E-04 | 3.41 | 0.3183 |
| Q61171 | Prdx2 | Peroxiredoxin-2 | 6.20 | 0.0026 | 3.40 | 0.4131 |
| P49194 | Rbp3 | Retinol-binding protein 3 | 6.10 | 5.59E-06 | 3.71 | 0.0405 |
| P11499 | Hsp90ab1 | Heat shock protein HSP 90-beta | 6.03 | 0.0066 | 3.72 | 0.4131 |
| P34884 | Mif | Macrophage migration inhibitory factor | 5.96 | 0.0106 | 2.98 | 0.7414 |
| P63268 | Actg2 | Actin, gamma-enteric smooth muscle | 5.13 | 5.68E-04 | 4.11 | 0.0406 |
| Q9D1U0 | Grifin | Grifin | 4.65 | 0.0081 | 3.18 | 0.2751 |
| P20443 | Sag | S-arrestin | 4.55 | 9.15E-05 | 2.69 | 0.1709 |
| P20612 | Gnat1 | Guanine nucleotide-binding protein subunit α-1 | 4.54 | 0.0291 | –1.31 | 1.0000 |
| P07901 | Hsp90aa1 | Heat shock protein HSP 90-alpha | 4.50 | 0.0488 | 2.19 | 1.0000 |
| P05064 | Aldoa | Fructose-bisphosphate aldolase A | 4.38 | 0.0024 | 2.68 | 0.3359 |
| P10126 | Eef1a1 | Elongation factor 1–alpha 1 | 4.25 | 0.0033 | 3.14 | 0.1726 |
| Q9JI02 | Scgb2b20 | Secretoglobin family 2B member 20 | 4.02 | 0.0051 | –1.39 | 1.0000 |
| Q03734 | Serpina3m | Serine protease inhibitor A3M | 3.47 | 0.0184 | –1.23 | 1.0000 |
| P40142 | Tkt | Transketolase | 3.34 | 0.0020 | 2.64 | 0.1065 |
| Q8BFZ3 | Actbl2 | Beta-actin-like protein 2 | 3.11 | 0.0056 | 1.14 | 1.0000 |
| P06151 | Ldha | L-lactate dehydrogenase A chain | 2.87 | 0.0167 | 1.81 | 0.7448 |
| P63101 | Ywhaz | 14-3-3 protein zeta/delta | 2.43 | 0.0352 | 2.46 | 0.1065 |
| P16858 | Gapdh | Glyceraldehyde-3-phosphate dehydrogenase | 2.33 | 0.0233 | 2.13 | 0.1389 |
| P23927 | Cryab | Alpha-crystallin B chain | 2.21 | 0.0294 | 2.42 | 0.0360 |
| P11590 | Mup4 | Major urinary protein 4 | –2.83 | 0.0241 | –3.18 | 0.0352 |
| P04938 | Mup8 | Major urinary proteins 11 and 8 (fragment) | –9.94 | 0.0029 | –4.01 | 0.0784 |
FDR, false discovery rate.
Diabetic versus controls.
Diabetic with sgp130Fc treatment versus controls.
Figure 2.
Proteomic alterations in the vitreous of diabetic mice. A total of 72 proteins were significantly altered in diabetic mice vitreous as compared to healthy controls. (A) The fold change of the top 20 upregulated and two downregulated vitreous proteins. The molecular functions (B), cellular components (C), biological processes (D), and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (E) associated with proteins altered in vitreous of diabetic mice are shown. Associations with Benjamini-corrected P value <0.05 were considered significant.
Network Analyses of Vitreous Proteins Altered in Diabetic Mice
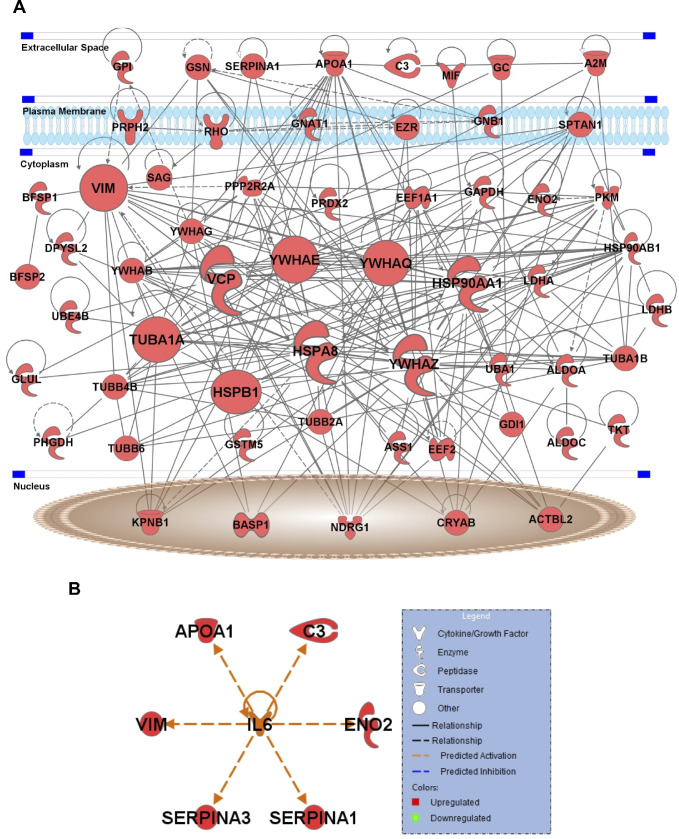
To understand the possible regulatory interactions between the 72 proteins altered in the vitreous of diabetic mice, IPA was performed. This analysis revealed extensive connections between these proteins, including nine protein hubs with more than 15 connections. These key hubs include three heat shock proteins (Hsp90α, Hsp90β, Hsp70-member 8), tubulin α1A (Tuba1a), transitional endoplasmic reticulum ATPase (Vcp), vimentin (Vim), and three members of 14-3-3 protein family (Ywhae, Ywhaq, Ywhaz) (Fig. 3A). Furthermore, upstream regulator analysis revealed known connections between IL-6 and six other upregulated proteins, including apolipoprotein A1 (Apoa1), complement 3 (C3), γ-enolase (Eno2), vimentin (Vim), and two serine protease inhibitors (Serpina1, Serpina3) (Fig. 3B).
Figure 3.
Network analysis of vitreous proteins altered in diabetic mice. IPA software was used to identify interactions among the vitreous proteins altered in diabetic mice (A). Nodes in the network represent proteins, and edges represent known protein-protein interaction. Nine proteins, including Hsp90aa1, Hsp90ab1, Hspa8, Tuba1a, Vcp, Vim, Ywhae, Ywhaq, and Ywhaz, are the key hubs of this network having more than 15 interactions. (B) Upstream regulator analysis revealed known connections between IL-6 and other proteins, including Apoa1, C3, Eno2, Serpina1, Serpina3, and Vim.
Inhibition of IL-6 Trans-Signaling Prevents Vitreous Proteomic Changes in Diabetic Mice
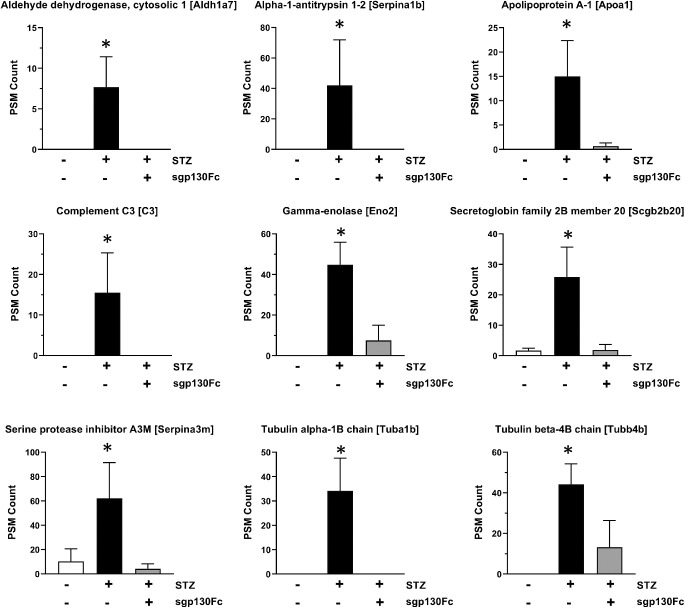
Sgp130Fc treatment was used for selective inhibition of IL-6 trans-signaling in diabetic mice. Inhibition of IL-6 trans-signaling prevented the changes in 52 out of the 72 vitreous proteins that were significantly altered in diabetic mice (Table 2), including all of those identified by upstream regulator analysis. The bar plots of nine highly upregulated proteins in diabetic mice that were normalized to control levels by sgp130Fc treatment are shown in Figure 4. These proteins include aldehyde dehydrogenase cytosolic 1 (Aldh1a7), alpha-1-antitrypsin 1-2 (Serpina1b), Apoa1, C3, Eno2, secretoglobin family 2B member 20 (Scgb2b20), serine protease inhibitor A3M (Serpina3m), Tuba1b, and Tubb4b.
Figure 4.
Relative PSM counts of proteins upregulated in vitreous of diabetic mice. The relative PSM counts of nine candidate proteins, including Aldh1a7, Serpina1b, Apoa1, C3, Eno2, Scgb2b20, Serpina3m, Tuba1b, and Tubb4b, are shown in three different groups (control, diabetes, diabetes with sgp130Fc treatment). There was a significant upregulation in the levels of these proteins in the vitreous of diabetic mice, and inhibition of IL-6 trans-signaling mitigated these changes.
Confirmation of Vitreous Proteomic Alterations Using PRM Analysis
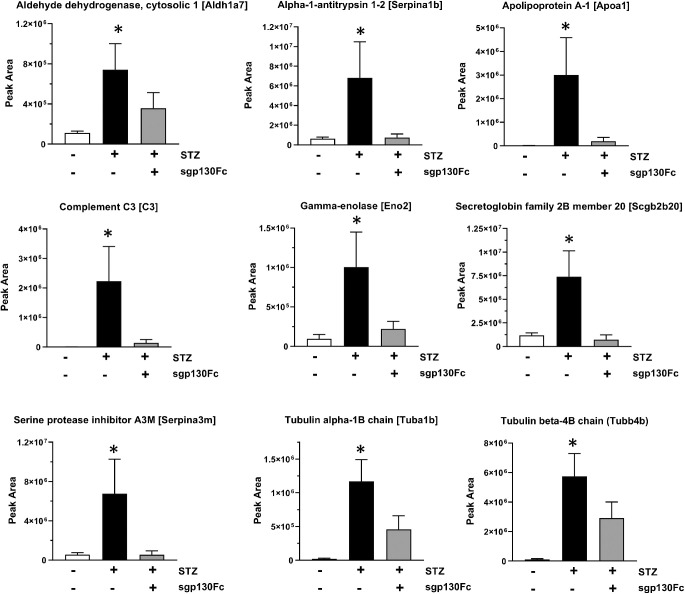
PRM analysis was performed to confirm the increased abundance of these nine proteins in diabetic mice vitreous (Fig. 5). Aldh1a7 (6.71-fold), Serpina1b (10.78-fold), Apoa1 (515.5-fold), C3 (209.3-fold), Eno2 (10.51-fold), Scgb2b20 (6.27-fold), Serpina3m (12.05-fold), Tuba1b (59-fold), and Tubb4b (54.47-fold) were all upregulated in diabetic mice as compared to healthy control animals (Table 3). PRM analysis also confirmed that all these proteins were not significantly upregulated in diabetic animals treated with sgp130Fc.
Figure 5.
Confirmation of vitreous proteomic alterations using PRM analysis. Differential abundance of nine candidate proteins (Aldh1a7, Serpina1b, Apoa1, C3, Eno2, Scgb2b20, Serpina3m, Tuba1b, and Tubb4b) in three groups (control, diabetes, diabetes with sgp130Fc treatment) was confirmed using PRM high-precision mass spectrometry analysis.
Table 3.
Confirmation of Changes in the Vitreous Humor Proteome Using PRM Analyses
| Fold Change | Fold Change | |||||
|---|---|---|---|---|---|---|
| Accession | Symbol | Description | Diabetic * | P Value | Diab + Sgp130Fc † | P Value |
| O35945 | Aldh1a7 | Aldehyde dehydrogenase, cytosolic 1 | 6.71 | 0.0116 | 3.24 | 0.8255 |
| P22599 | Serpina1b | Alpha-1-antitrypsin 1-2 | 10.78 | 0.0461 | 1.18 | 0.3521 |
| Q00623 | Apoa1 | Apolipoprotein A-1 | 515.5 | 0.0140 | 33.15 | 0.9006 |
| P01027 | C3 | Complement C3 | 209.3 | 0.0004 | 12.77 | 0.5185 |
| P17183 | Eno2 | Gamma-enolase | 10.51 | 0.0181 | 2.30 | 0.2210 |
| Q9JI02 | Scgb2b20 | Secretoglobin family 2B member 20 | 6.27 | 0.0176 | 0.61 | 0.0688 |
| Q03734 | Serpina3m | Serine protease inhibitor A3M | 12.05 | 0.0382 | 0.99 | 0.4340 |
| P05213 | Tuba1b | Tubulin alpha-1B chain | 59.0 | 0.0360 | 23.13 | 0.0790 |
| P68372 | Tubb4b | Tubulin beta-4B chain | 54.47 | 0.0481 | 27.67 | 0.3364 |
Diabetic versus controls.
Diabetic with sgp130Fc treatment versus controls.
Discussion
Diabetic retinopathy is a microvascular complication that is a leading cause of visual disability and blindness in diabetic patients.30 Increasing evidence suggests that the IL-6 pathway plays a prominent role in DR pathogenesis,31,32 and interactions between IL-6 and endothelial cells regulate recruitment of leukocytes and expression of inflammatory proteins. A soluble form of the IL-6R is generated by limited proteolysis and alternative splicing. In cells lacking membrane bound IL-6 receptor, the IL-6/sIL-6R complex associates with the ubiquitously expressed signal transducing protein gp130, initiating dimerization and intracellular signaling (IL-6 trans-signaling).
Studies have shown that IL-6 is upregulated in the vitreous of patients with type 2 diabetes.33 Also, levels of sIL-6R have been found to be increased in the vitreous of patients with proliferative DR as compared to nondiabetics.34 Using experimental models, we and others have shown that IL-6 trans-signaling plays an important role in endothelial barrier dysfunction, inflammation, and oxidative stress in DR and other diabetic complications.5,8,11,35,36 In this study, we sought to gain further insight into the role of IL-6 trans-signaling in the ocular microenvironment in a murine model of DR through proteomic analysis of vitreous fluid following systemic inhibition of IL-6 trans-signaling with sgp130Fc.
The vitreous is a transparent, highly aqueous extracellular matrix overlaying the retina. Due to close proximity to the retina and the breakdown of the blood-retinal barrier, the proteomic composition of the vitreous can be altered by pathologic retinal changes. Understanding these changes in vitreal biochemical composition due to the progression of retinal disease may be beneficial in the development of preventative measures and therapies. While murine studies are a critical part of preclinical therapeutic evaluation, vitreous sample collection and analysis from mouse models are difficult due to a very small vitreous body. However, the technological revolution during the past decade has helped in overcoming this limitation. In this study, we performed comprehensive proteomic profiling of the mouse vitreous humor using the latest LC-MS/MS technology to evaluate the effects of IL-6 trans-signaling inhibition using sgp130Fc. To the best of our knowledge, this is the first study to evaluate the effects of IL-6 trans-signaling inhibition on the diabetic vitreous proteome.
Our analysis identified 443 unique proteins, of which 154 were characteristic of healthy murine vitreous. These findings are similar to a previous murine vitreous proteomic profiling study that identified 675 unique vitreous proteins in samples pooled from eight eyes.29 The most abundant proteins identified in our analysis are crystallins, which are structural proteins primarily expressed in the lens, cornea, and retina.37 Previous proteomic profiling studies have also shown that crystallins are the most abundant proteins in the vitreous humor.14–16,19,22,38 Like Skeie et al.,24,29,39 we also identified fatty acid binding protein 5 (Fabp5), acyl-CoA-binding protein (diazepam binding inhibitor, Dbi), phosphoglycerate kinase 1 (Pgk1), phosphatidylethanolamine binding protein 1 (Pebp1), carbonic anhydrase 2 (Ca2), peroxiredoxin 5 (Prdx5), and hemoglobin subunit beta 1 (Hbb-b1) among the most abundant proteins in healthy murine vitreous fluid. Our analysis also revealed a large number of intracellular proteins and proteins associated with extracellular exosomes, as previously reported in both mouse and human vitreous studies.
Comparison of the vitreous proteome of healthy mice to that of diabetic mice revealed a higher abundance of proteins in diabetic vitreous than in control. This could possibly be due to increased vascular leakage and breakdown of the blood-retinal barrier commonly seen in DR. Furthermore, it is also possible that diabetes-induced retinal damage released retinal proteins into the vitreous, thereby increasing protein content. We confirmed that vitreous samples were free of cellular contaminants by hematoxylin and eosin (H&E) staining (data not shown). Interestingly, protein abundance was decreased in the vitreous of diabetic mice after treatment with sgp130Fc. While this offers support to the vascular leakage hypothesis, as sgp130Fc has been shown to have protective effects on endothelial barrier function,5 it is also possible that any changes to vascular permeability are instead concordant with an overall decrease in retinal inflammation. Our data demonstrate the protective effects of IL-6 trans-signaling inhibition on the diabetic vitreous proteome, although further studies are needed to delineate the exact mechanism(s) underlying this response.
The most upregulated proteins in diabetic vitreous humor are tubulin beta-5 chain (Tubb5), 14-3-3 protein epsilon (Ywhae), tubulin beta-4B (Tubb4b), gamma-enolase (Eno2), and tubulin beta-2A chain (Tubb2a). Both beta-tubulins40 and Eno241 have been previously identified in proteomic analyses of diabetic animal models. Tubb5 is a major component of microtubules known to be upregulated in retinopathy.42 Ywhae plays a role in the cellular response to heat stress and regulation of apoptotic signaling and is known to be expressed in murine photoreceptors.43 Two proteins downregulated in the vitreous of diabetic mice as compared to controls are major urinary protein 4 (Mup4) and 8 (Mup8). MUPs are part of the lipocalin family, which are involved in communication and regulate glucose and lipid metabolism. Others have reported decreased Mup4 expression in diabetes,44 while Mup8 levels are reportedly decreased45 or unchanged.46 Despite significant changes in the abundance of these proteins in diabetic mouse vitreous, their significance to DR pathology remains unclear.
Inhibition of IL-6 trans-signaling with sgp130Fc prevented diabetes-induced proteomic changes in 52 out of the 72 vitreous proteins. Since the effects of IL-6 trans-signaling in vitreous fluid have not previously been characterized, we performed IPA analyses to identify any known interactions between proteins in this subset. Network and upstream regulator analyses revealed a highly interconnected network of regulatory relationships among these proteins. Vimentin, an intermediate filament protein identified as one of the key regulatory hubs in the network, plays a role in retinal response to injury and is upregulated in DR.47,48 IL-6 promotes epithelial-mesenchymal transition through vimentin upregulation in a STAT3-dependent manner.49 Vimentin is also known to play a role in the regulation of Notch signaling and angiogenesis.50
Other protein hubs identified by network analysis include several heat shock proteins (HSPs), including Hsp90α, Hsp90β, and Hspa8. Hsp90α and β are members of the Hsp90 family of proteins and serve as critical molecular chaperones during protein synthesis.51,52 Hspa8 is a member of the Hsp70 family of proteins and plays roles in protein folding, clathrin-mediated endocytosis, and ubiquitin-mediated protein degradation.53 HSPs are important for the cellular response to damage or injury,54 and Hsp70 specifically has been implicated in both DR and the retinal damage response.55,56 While IL-6 has been associated with several HSPs,57,58 this interaction in the context of DR has not yet been described.
We also found several 14-3-3 family proteins that were upregulated in the vitreous of diabetic mice, including Ywhab, Ywhae, Ywhag, Ywhaq, and Ywhaz. These proteins constitute five out of the seven members of a highly conserved protein family responsible for regulating the activity of phosphoproteins and modulate several signaling pathways, including those related to metabolism, cell proliferation, and stress responses.59,60 The 14-3-3 proteins have been shown to be crucial for the cellular response to insulin signaling.61 While members of this protein family have been reported to be overexpressed in DR,41 their significance to disease pathology and relationship to IL-6 trans-signaling remains unclear.
The five other proteins identified by upstream regulator analysis (Apoa1, C3, Eno2, Serpina1, and Serpina3) are increased in vitreous from diabetic mice and have predicted activation by IL-6. These proteins were normalized to control levels in diabetic mice after sgp130Fc treatment. Apoa1 is a well-known biomarker of DR and is elevated in the serum, retina, vitreous, and tears of patients with DR,62,63 and its expression is known to be regulated by IL-6.64 Similarly, complement C3 and the acute phase response protein α-1-antitrypsin 1-2 (Serpina1b) are known to be elevated in diabetic vitreous15,65 and regulated by IL-6.66–68 Eno2 has also been associated with DR,41 and as a neuron-specific isoform, the presence of this protein in vitreous could indicate neuronal damage. Regulation of serpina3m (alpha-1-antichymotrypsin) by IL-6 has been well characterized in some cell types,69 but to our knowledge, this protein has not previously been associated with DR.
Several tubulin proteins, including Tuba1a, Tuba1b, Tubb2a, Tubb4b, and Tubb5, were highly upregulated in diabetic mice, and this change was prevented by inhibition of IL-6 trans-signaling. Tubulin is a major cytoskeletal component regulated by inflammatory signaling,70 and tubulin dysfunction can disrupt cell proliferation and differentiation.71 Tubulin α1B (Tuba1b) has been shown to be increased in the vitreous of patients with DR,16 and this increase may be due to increased endothelial cell proliferation during angiogenesis.16
While the significance of many specific dysregulated proteins remains unclear, the restoration of normal protein abundance in vitreous fluid following sgp130Fc treatment emphasizes the importance of IL-6 trans-signaling to DR pathogenesis. As previously mentioned, IL-6 is known to regulate several of the proteins identified in our study, but it is also possible that the observed proteomic changes are the indirect consequence of retinal tissue damage due to inflammation or oxidative stress, both of which are known to be associated with IL-6 trans-signaling.5,11 Corticosteroids and nonsteroidal anti-inflammatory drugs have shown benefits in patients with DR by reducing leukocyte recruitment into the retinal vasculature,72–74 and as leukocyte recruitment is often linked to IL-6 trans-signaling,75–77 it is likely that this mechanism is a major contributor to the effectiveness of sgp130Fc.
The use of unpooled samples offers an advantage in being able to distinguish between biological replicates for statistical purposes, but it appears to be less sensitive for the detection of low-abundance proteins. Qualitative comparison of our proteomics data from control mice with that of a similar study by Skeie and Mahajan29 showed similar vitreous protein composition for highly abundant proteins but greater disparities in low-abundance proteins. To confirm that the elevated protein in diabetic vitreous was not due to contamination from retinal tissue, we verified the acellularity of vitreous samples through H&E staining as previously described.29 The absence of the retina-specific protein rhodopsin in control samples further supports the efficacy of our vitreous isolation method, meaning its consistent detection in only diabetic samples is likely caused by increased retinal damage and cell death during disease progression, thus releasing protein contents into the adjacent vitreous fluid. This hypothesis is also supported by the decreased detection of rhodopsin in diabetic animals treated with sgp130Fc.
In conclusion, this study identified the effects of IL-6 trans-signaling inhibition on diabetes-induced alterations to the mouse vitreous proteome. Several members of heat shock proteins, 14-3-3 proteins, and tubulins were highly dysregulated in diabetic mice. Inhibition of IL-6 trans-signaling using sgp130Fc prevented the alteration of the majority of these proteins, including those with both well-known and novel associations with IL-6 signaling. Restoration of protein levels following sgp130Fc treatment emphasizes the importance of IL-6 trans-signaling in DR pathophysiology. These findings further our understanding of the role of IL-6 trans-signaling within the ocular microenvironment and provide additional insight into the pathogenesis of DR.
Supplementary Material
Acknowledgments
The authors thank Lara Churchwell, BS, for conducting the Ingenuity Pathway Analysis (IPA).
Supported by the National Institutes of Health, National Eye Institute (Bethesda, MD, USA) grant R01-EY026936 awarded to SS.
Disclosure: R. Robinson, None; H. Youngblood, None; H. Iyer, None; J. Bloom, None; T.J. Lee, None; L. Chang, None; Z. Lukowski, None; W. Zhi, None; A. Sharma, None; S. Sharma, None
References
- 1. Joussen AM, Poulaki V, Le ML, et al.. A central role for inflammation in the pathogenesis of diabetic retinopathy. FASEB J. 2004; 18: 1450–1452. [DOI] [PubMed] [Google Scholar]
- 2. Gerhardinger C, Costa MB, Coulombe MC, Toth I, Hoehn T, Grosu P. Expression of acute-phase response proteins in retinal Muller cells in diabetes. Invest Ophthalmol Vis Sci. 2005; 46: 349–357. [DOI] [PubMed] [Google Scholar]
- 3. Krady JK, Basu A, Allen CM, et al.. Minocycline reduces proinflammatory cytokine expression, microglial activation, and caspase-3 activation in a rodent model of diabetic retinopathy. Diabetes. 2005; 54: 1559–1565. [DOI] [PubMed] [Google Scholar]
- 4. Antonetti DA, Klein R, Gardner TW. Diabetic retinopathy. N Engl J Med. 2012; 366: 1227–1239. [DOI] [PubMed] [Google Scholar]
- 5. Valle ML, Dworshak J, Sharma A, Ibrahim AS, Al-Shabrawey M, Sharma S. Inhibition of interleukin-6 trans-signaling prevents inflammation and endothelial barrier disruption in retinal endothelial cells. Exp Eye Res. 2019; 178: 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kauffmann D, Van Meurs J, Mertens D, Peperkamp E, Master C, Gerritsen M. Cytokines in vitreous humor: interleukin-6 is elevated in proliferative vitreoretinopathy. Invest Ophthalmol Vis Sci. 1994; 35: 900–906. [PubMed] [Google Scholar]
- 7. Koleva-Georgieva DN, Sivkova NP, Terzieva D. Serum inflammatory cytokines IL-1β, IL-6, TNF-α and VEGF have influence on the development of diabetic retinopathy. Folia Med. 2011; 53: 44–50. [DOI] [PubMed] [Google Scholar]
- 8. Rose-John S. IL-6 trans-signaling via the soluble IL-6 receptor: importance for the pro-inflammatory activities of IL-6. Int J Biol Sci. 2012; 8: 1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen H, Zhang X, Liao N, Wen F. Increased levels of IL-6, sIL-6R, and sgp130 in the aqueous humor and serum of patients with diabetic retinopathy. Mol Vis. 2016; 22: 1005. [PMC free article] [PubMed] [Google Scholar]
- 10. Scheller J, Garbers C, Rose-John S. Interleukin-6: from basic biology to selective blockade of pro-inflammatory activities. Semin Immunol. 2014; 26: 2–12. [DOI] [PubMed] [Google Scholar]
- 11. Robinson R, Srinivasan M, Shanmugam A, et al.. Interleukin-6 trans-signaling inhibition prevents oxidative stress in a mouse model of early diabetic retinopathy. Redox Biology. 2020; 34: 101574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schori C, Trachsel C, Grossmann J, Zygoula I, Barthelmes D, Grimm C. The proteomic landscape in the vitreous of patients with age-related and diabetic retinal disease. Invest Ophthalmol Vis Sci. 2018; 59: AMD31–AMD40. [DOI] [PubMed] [Google Scholar]
- 13. Li J, Lu Q, Lu P. Quantitative proteomics analysis of vitreous body from type 2 diabetic patients with proliferative diabetic retinopathy. BMC Ophthalmol. 2018; 18: 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mahajan VB, Skeie JM. Translational vitreous proteomics. Proteomics Clin Appl. 2014; 8: 204–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gao B-B, Chen X, Timothy N, Aiello LP, EPJJopr Feener. Characterization of the vitreous proteome in diabetes without diabetic retinopathy and diabetes with proliferative diabetic retinopathy. J Proteome Res 2008; 7: 2516–2525. [DOI] [PubMed] [Google Scholar]
- 16. Wang H, Feng L, Hu J, Xie C, Wang F. Differentiating vitreous proteomes in proliferative diabetic retinopathy using high-performance liquid chromatography coupled to tandem mass spectrometry. Exp Eye Res. 2013; 108: 110–119. [DOI] [PubMed] [Google Scholar]
- 17. Murthy KR, Goel R, Subbannayya Y, et al.. Proteomic analysis of human vitreous humor. Clin Proteomics. 2014; 11: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Balaiya S, Zhou Z, Chalam KV. Characterization of vitreous and aqueous proteome in humans with proliferative diabetic retinopathy and its clinical correlation. Proteomics Insights. 2017; 8: 1178641816686078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Angi M, Kalirai H, Coupland SE, Damato BE, Semeraro F, Romano MR. Proteomic analyses of the vitreous humour. Mediators Inflamm. 2012; 2012: 148039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chiang SY, Tsai ML, Wang CY, et al.. Proteomic analysis and identification of aqueous humor proteins with a pathophysiological role in diabetic retinopathy. J Proteomics. 2012; 75: 2950–2959. [DOI] [PubMed] [Google Scholar]
- 21. Csosz E, Boross P, Csutak A, et al.. Quantitative analysis of proteins in the tear fluid of patients with diabetic retinopathy. J Proteomics. 2012; 75: 2196–2204. [DOI] [PubMed] [Google Scholar]
- 22. Loukovaara S, Nurkkala H, Tamene F, et al.. Quantitative proteomics analysis of vitreous humor from diabetic retinopathy patients. J Proteome Res. 2015; 14: 5131–5143. [DOI] [PubMed] [Google Scholar]
- 23. Skeie JM, Tsang SH, Mahajan VB. Evisceration of mouse vitreous and retina for proteomic analyses. J Vis Exp. 2011; 50: 2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Skeie JM, Roybal CN, Mahajan VB. Proteomic insight into the molecular function of the vitreous. PLoS One. 2015; 10: e0127567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rauniyar N. Parallel reaction monitoring: a targeted experiment performed using high resolution and high mass accuracy mass spectrometry. Int J Mol Sci. 2015; 16: 28566–28581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Peterson AC, Russell JD, Bailey DJ, Westphall MS, Coon JJ. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol Cell Proteomics. 2012; 11: 1475–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010; 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009; 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 29. Skeie JM, Mahajan VB. Proteomic interactions in the mouse vitreous-retina complex. PLoS One. 2013; 8: e82140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang X, Saaddine JB, Chou CF, et al.. Prevalence of diabetic retinopathy in the United States, 2005–2008. JAMA. 2010; 304: 649–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gabay C. Interleukin-6 and chronic inflammation. Arthritis Res Ther. 2006; 8(suppl 2): S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rincon M. Interleukin-6: from an inflammatory marker to a target for inflammatory diseases. Trends Immunol. 2012; 33: 571–577. [DOI] [PubMed] [Google Scholar]
- 33. Tsai T, Kuehn S, Tsiampalis N, et al.. Anti-inflammatory cytokine and angiogenic factors levels in vitreous samples of diabetic retinopathy patients. PLoS One. 2018; 13: e0194613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kawashima M, Shoji J, Nakajima M, Kamura Y, Sato Y. Soluble IL-6 receptor in vitreous fluid of patients with proliferative diabetic retinopathy. Jpn J Ophthalmol. 2007; 51: 100–104. [DOI] [PubMed] [Google Scholar]
- 35. Feigerlová E, Battaglia-Hsu S-F. IL-6 signaling in diabetic nephropathy: from pathophysiology to therapeutic perspectives. Cytokine Growth Factor Rev. 2017; 37: 57–65. [DOI] [PubMed] [Google Scholar]
- 36. Wei LH, Chou CH, Chen MW, et al.. The role of IL-6 trans-signaling in vascular leakage: implications for ovarian hyperstimulation syndrome in a murine model. J Clin Endocrinol Metab. 2013; 98: E472–E484. [DOI] [PubMed] [Google Scholar]
- 37. Andley UP. Crystallins in the eye: function and pathology. Progress Retinal Eye Res. 2007; 26: 78–98. [DOI] [PubMed] [Google Scholar]
- 38. Aretz S, Krohne TU, Kammerer K, et al.. In-depth mass spectrometric mapping of the human vitreous proteome. Proteome Sci. 2013; 11: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhao Y, Weber SR, Lease J, et al.. Liquid biopsy of vitreous reveals an abundant vesicle population consistent with the size and morphology of exosomes. Transl Vis Sci Technol. 2018; 7: 6–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Semba RD, Huang H, Lutty GA, Van Eyk JE, Hart GW. The role of O‐GlcNAc signaling in the pathogenesis of diabetic retinopathy. Proteomics Clin Appl. 2014; 8: 218–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Safaei A, Tavirani MR, Azodi MZ, et al.. Diabetic retinopathy and laser therapy in rats: a protein-protein interaction network analysis. J Lasers Med Sci. 2017; 8: S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang X, Dong X, Jia C, Wang Y.. Profiling of genes associated with the murine model of oxygen-induced retinopathy. Mol Vis. 2013; 19: 775. [PMC free article] [PubMed] [Google Scholar]
- 43. Santos FM, Gaspar LM, Ciordia S, et al.. iTRAQ quantitative proteomic analysis of vitreous from patients with retinal detachment. Int J Mol Sci. 2018; 19: 1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Charkoftaki G, Wang Y, McAndrews M, et al.. Update on the human and mouse lipocalin (LCN) gene family, including evidence the mouse Mup cluster is result of an “evolutionary bloom.” Hum Genomics. 2019; 13: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schönke M, Björnholm M, Chibalin AV, Zierath JR, Deshmukh AS. Proteomics analysis of skeletal muscle from leptin‐deficient ob/ob mice reveals adaptive remodeling of metabolic characteristics and fiber type composition. Proteomics. 2018; 18: 1700375. [DOI] [PubMed] [Google Scholar]
- 46. Tsai F-J, Chen S-Y, Liu Y-C, Liao H-Y, Chen C-J. The comparison of CHCA solvent compositions for improving LC-MALDI performance and its application to study the impact of aflatoxin B1 on the liver proteome of diabetes mellitus type 1 mice. PLoS One. 2017; 12: e0181423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wunderlich KA, Tanimoto N, Grosche A, et al.. Retinal functional alterations in mice lacking intermediate filament proteins glial fibrillary acidic protein and vimentin. FASEB J. 2015; 29: 4815–4828. [DOI] [PubMed] [Google Scholar]
- 48. Zhou T, Di Che YL, Fang Z, et al.. Mesenchymal marker expression is elevated in Müller cells exposed to high glucose and in animal models of diabetic retinopathy. Oncotarget. 2017; 8: 4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sullivan N, Sasser A, Axel AE, et al.. Interleukin-6 induces an epithelial–mesenchymal transition phenotype in human breast cancer cells. Oncogene. 2009; 28: 2940–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Antfolk D, Sjöqvist M, Cheng F, et al.. Selective regulation of Notch ligands during angiogenesis is mediated by vimentin. Proc Natl Acad Sci USA. 2017; 114: E4574–E4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zuehlke AD, Beebe K, Neckers L, Prince T. Regulation and function of the human HSP90AA1 gene. Gene. 2015; 570: 8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Haase M, Fitze G. HSP90AB1: Helping the good and the bad. Gene. 2016; 575: 171–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stricher F, Macri C, Ruff M, Muller S. HSPA8/HSC70 chaperone protein: structure, function, and chemical targeting. Autophagy. 2013; 9: 1937–1954. [DOI] [PubMed] [Google Scholar]
- 54. Atalay M, Oksala NK, Laaksonen DE, et al.. Exercise training modulates heat shock protein response in diabetic rats. J Appl Physiol. 2004; 97: 605–611. [DOI] [PubMed] [Google Scholar]
- 55. Sayed KM, Mahmoud AA. Heat shock protein‐70 and hypoxia inducible factor‐1α in type 2 diabetes mellitus patients complicated with retinopathy. Acta Ophthalmol. 2016; 94: e361–e366. [DOI] [PubMed] [Google Scholar]
- 56. Kayama M, Nakazawa T, Thanos A, et al.. Heat shock protein 70 (HSP70) is critical for the photoreceptor stress response after retinal detachment via modulating anti-apoptotic Akt kinase. Am J Pathol. 2011; 178: 1080–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Stephanou A, Amin V, Isenberg D, Akira S, Kishimoto T, Latchman DS. Interleukin 6 activates heat-shock protein 90 β gene expression. Biochem J. 1997; 321: 103–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Parikh AA, Moon MR, Kane CD, Salzman AL, Fischer JE, Hasselgren P-O. Interleukin-6 production in human intestinal epithelial cells increases in association with the heat shock response. J Surg Res. 1998; 77: 40–44. [DOI] [PubMed] [Google Scholar]
- 59. Yang X, Lee WH, Sobott F, et al.. Structural basis for protein–protein interactions in the 14-3-3 protein family. Proc Natl Acad Sci USA. 2006; 103: 17237–17242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Paul G, van Heusden H. 14-3‐3 proteins: regulators of numerous eukaryotic proteins. IUBMB Life. 2005; 57: 623–629. [DOI] [PubMed] [Google Scholar]
- 61. Chen S, Synowsky S, Tinti M, MacKintosh C. The capture of phosphoproteins by 14-3-3 proteins mediates actions of insulin. Trends Endocrinol Metab. 2011; 22: 429–436. [DOI] [PubMed] [Google Scholar]
- 62. Sasongko MB, Wong TY, Nguyen TT, et al.. Serum apolipoprotein AI and B are stronger biomarkers of diabetic retinopathy than traditional lipids. Diabetes Care. 2011; 34: 474–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Simó R, García-Ramírez M, Higuera M, Hernández C. Apolipoprotein A1 is overexpressed in the retina of diabetic patients. Am J Ophthalmol. 2009; 147: 319–325. e311. [DOI] [PubMed] [Google Scholar]
- 64. Müller N, Schulte DM, Türk K, et al.. IL-6 blockade by monoclonal antibodies inhibits apolipoprotein expression and lipoprotein synthesis in humans. J Lipid Res. 2015; 56: 1034–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nakanishi T, Koyama R, Ikeda T, Shimizu A. Catalogue of soluble proteins in the human vitreous humor: comparison between diabetic retinopathy and macular hole. J Chromatogr B Analyt Technol Biomed Life Sci. 2002; 776: 89–100. [DOI] [PubMed] [Google Scholar]
- 66. Falus A, Rokita H, Walcz E, Brozik M, Hidvégi T, Meretey K. Hormonal regulation of complement biosynthesis in human cell lines—II. Upregulation of the biosynthesis of complement components C3, factor B and Cl inhibitor by interleukin-6 and interleukin-1 in human hepatoma cell line. Mol Immunol. 1990; 27: 197–201. [DOI] [PubMed] [Google Scholar]
- 67. Stapp JM, Sjoelund V, Lassiter HA, Feldhoff RC, Feldhoff PW. Recombinant rat IL-1β and IL-6 synergistically enhance C3 mRNA levels and complement component C3 secretion by H-35 rat hepatoma cells. Cytokine. 2005; 30: 78–85. [DOI] [PubMed] [Google Scholar]
- 68. Perlmutter DH, May LT, Sehgal PB. Interferon beta 2/interleukin 6 modulates synthesis of alpha 1-antitrypsin in human mononuclear phagocytes and in human hepatoma cells. J Clin Invest. 1989; 84: 138–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Baker C, Belbin O, Kalsheker N, Morgan K. SERPINA3 (aka alpha-1-antichymotrypsin). Front Biosci. 2007; 12: 2821–2835. [DOI] [PubMed] [Google Scholar]
- 70. Chu CC, Paul WE.. Expressed genes in interleukin-4 treated B cells identified by cDNA representational difference analysis. Mol Immunol. 1998; 35: 487–502. [DOI] [PubMed] [Google Scholar]
- 71. Lu C, Zhang J, He S, et al.. Increased α-tubulin1b expression indicates poor prognosis and resistance to chemotherapy in hepatocellular carcinoma. Dig Dis Sci. 2013; 58: 2713–2720. [DOI] [PubMed] [Google Scholar]
- 72. Stewart MW. Corticosteroid use for diabetic macular edema: old fad or new trend? Curr Diabetes Rep. 2012; 12: 364–375. [DOI] [PubMed] [Google Scholar]
- 73. Tamura H, Miyamoto K, Kiryu J, et al.. Intravitreal injection of corticosteroid attenuates leukostasis and vascular leakage in experimental diabetic retina. Invest Ophthalmol Vis Sci. 2005; 46: 1440–1444. [DOI] [PubMed] [Google Scholar]
- 74. Joussen AM, Poulaki V, Mitsiades N, et al.. Nonsteroidal anti-inflammatory drugs prevent early diabetic retinopathy via TNF-α suppression. FASEB J. 2002; 16: 438–440. [DOI] [PubMed] [Google Scholar]
- 75. Hurst SM, Wilkinson TS, McLoughlin RM, et al.. Il-6 and its soluble receptor orchestrate a temporal switch in the pattern of leukocyte recruitment seen during acute inflammation. Immunity. 2001; 14: 705–714. [DOI] [PubMed] [Google Scholar]
- 76. Noda K, Nakao S, Ishida S, Ishibashi T. Leukocyte adhesion molecules in diabetic retinopathy. J Ophthalmol. 2012; 2012: 279037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Romano M, Sironi M, Toniatti C, et al.. Role of IL-6 and its soluble receptor in induction of chemokines and leukocyte recruitment. Immunity. 1997; 6: 315–325. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.