Abstract
There is a desperate need for safe and effective vaccines, therapies, and diagnostics for SARS– coronavirus 2 (CoV-2), the development of which will be aided by the discovery of potent and selective antibodies against relevant viral epitopes. Human phage display technology has revolutionized the process of identifying and optimizing antibodies, providing facile entry points for further applications. Herein, we use this technology to search for antibodies targeting the receptor-binding domain (RBD) of CoV-2. Specifically, we screened a naïve human semisynthetic phage library against RBD, leading to the identification of a high-affinity single-chain fragment variable region (scFv). The scFv was further engineered into two other antibody formats (scFv-Fc and IgG1). All three antibody formats showed high binding specificity to CoV-2 RBD and the spike antigens in different assay systems. Flow cytometry analysis demonstrated specific binding of the IgG1 format to cells expressing membrane-bound CoV-2 spike protein. Docking studies revealed that the scFv recognizes an epitope that partially overlaps with angiotensin-converting enzyme 2 (ACE2)–interacting sites on the CoV-2 RBD. Given its high specificity and affinity, we anticipate that these anti-CoV-2 antibodies will be useful as valuable reagents for accessing the antigenicity of vaccine candidates, as well as developing antibody-based therapeutics and diagnostics for CoV-2.
Keywords: RBD, scFv, ACE2, mAb, IgG1, monoclonal antibody, virus, phage display, infectious disease, antigen
The recently identified novel human coronavirus, referred to as severe acute respiratory syndrome (SARS)–coronavirus 2 (CoV-2), is the causative agent of the ongoing pandemic of COVID-19. After the report of first case of the CoV-2 infection in December 2019, it has spread all over the globe. To slow down the spread of COVID-19, many countries have introduced lockdown measures. However, these measures will not be enough to eradicate the coronavirus pandemic from the world, and there is an urgent requirement for some medical intervention to control the spread of infection either in the form of a vaccine or other therapeutic options such as small molecule or therapeutic antibodies. So far, no clinically approved therapeutics are available for CoV-2, and only symptomatic treatment is offered (1). One of the approaches is to look for neutralizing antibodies (NAbs) for CoV-2 either from convalescent patient samples or from synthetic antibody library sources. The antibodies can work via two different mechanisms, i.e. by direct neutralization of target viral antigen and also by indirect effector mechanisms such as antibody-dependent cell-mediated cytotoxicity and complement-dependent cytotoxicity, wherein antibody binds to infected cells and potentially clears the viral reservoirs (2, 3).
The spike (S) protein on the surface of CoV2 plays the primary role in viral attachment to the host cell receptor followed by fusion and entry. The S protein comprises two components: S1, which contains the distinct RBD (residues 318–510); and S2, which contains the fusion peptide. The virus gains entry into permissive host cells through interactions of the RBD with the cell surface receptor ACE2. Therefore, the RBD of CoV-2 S protein is the most likely target for development of virus attachment inhibitors, NAbs, and vaccines (4). An antibody against RBD is expected to block the attachment of the surface spike of virions to the ACE2 receptors on the host-cell surface and is thereby supposed to neutralize virus entry. Recent success with plasma therapy from convalescent patient samples against CoV-2 is a good indicator of possible successful antibody-based therapy to avoid further fatalities (5, 6). However, plasma therapy is a crude method of treatment, and a mAb, the main ingredient of plasma therapy, is needed for wide and safe application (7, 8).
Thus, novel tools and reagents for therapy and diagnosis are urgently needed. Keeping this need in mind, we here used a scFv phage display library to identify a novel scFv, II62, against the RBD of CoV-2 that targets an epitope immediately adjacent to and slightly overlapping with the ACE2-binding region on RBD. We further characterized the binding properties of II62–scFv and its two other formats, scFv-Fc and IgG1, against both the RBD and S protein of CoV-2.
Results
Identification of CoV-2 RBD reactive clones from phage display library
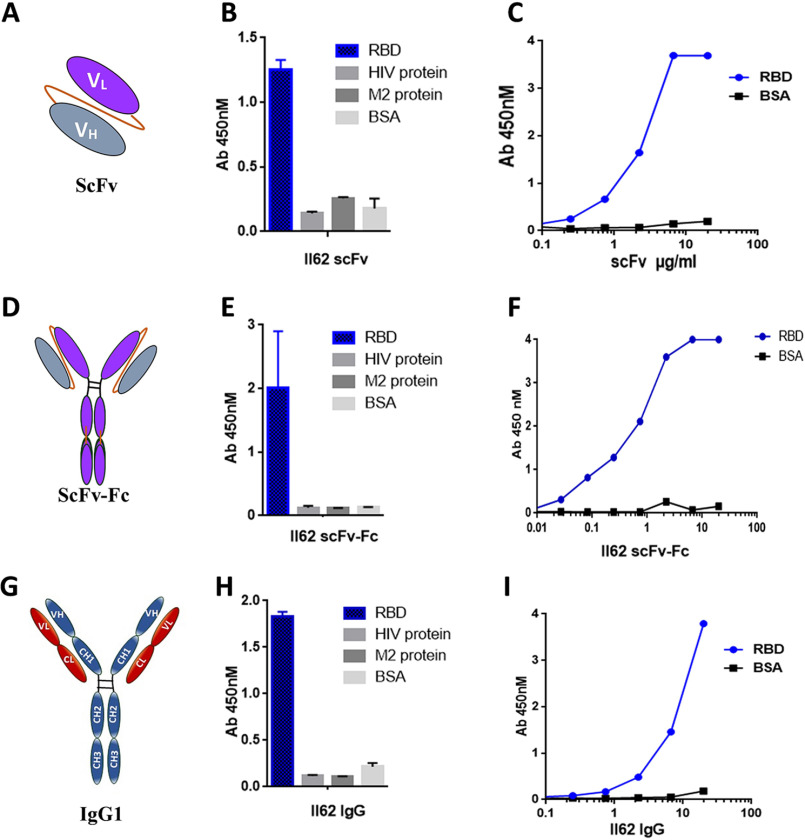
The human synthetic Tomlinson I single fold phage library was used for screening of scFvs against the purified (>95% pure) RBD protein from mammalian expression system. Three rounds of panning were done, and the titers of input and output phage were calculated at the end of each round to monitor the efficiency of enrichment. After three rounds of selection, ∼1100-fold enrichment of antigen-specific clones was observed (Fig. 1A). After the third round of panning, 50 clones were randomly picked and assessed for binding to RBD protein by monoclonal scFv-phage ELISA (Fig. 1B). Twenty clones of the 50 showed five times better binding to RBD in ELISA as compared with the negative controls (BSA and helper phage) and hence were considered as RBD-specific binders. One clone of scFv, II62, was dominantly selected via panning (50% of the selected binders) and was selected for further characterization (Fig. 1C).
Figure 1.
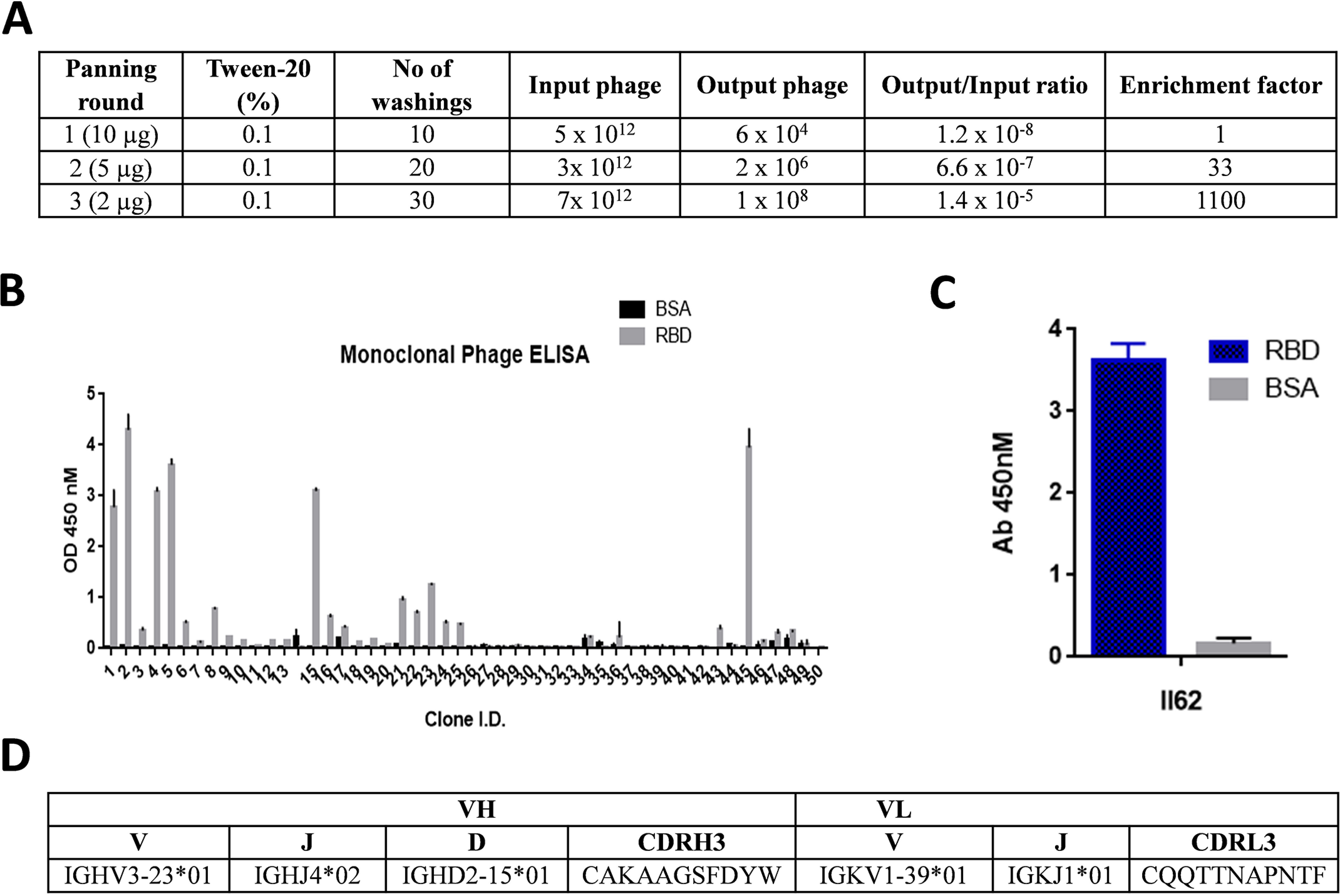
A, table showing enrichment of phage library with each round of panning. In each round the stringency of selection was increased by reducing the amount of antigen and increasing the number of washings. B, phage ELISA of clones screened for binding to RBD after the third round of selection. C, the best binding clone, II62, selected for further characterization. D, table showing the antibody gene locus as determined by blasting against the IMGT database.
Sequence analysis
The complete nucleotide sequence of the heavy and light chain variable region was determined using the immunoglobulin BLAST homology search. The result indicated that the coding sequences are composed of the VH gene (351 bp) and the VL gene (327 bp). The closest germline sequence for the II62 scFv-antibody gene was identified by comparison with the database. The VH gene use the VH3 family-derived germline V3-23, D2-15, and JH4 genes, whereas the VL gene uses the VL1 (Vκ subgroup I, V1–39) family-derived germline and Jk1. Also, a high degree of mutation appeared in VH complementarity-determining region 2. The II62 mAb sequence showed 70.8% (bases) homology to the closest germline match (Fig. 1D and Fig. S1).
Expression, purification, and characterization of II62–scFv
The His-tagged II62–scFv was overexpressed in a bacterial system and purified as soluble protein using the Ni–NTA affinity column. Overall yield of the II62–scFv was 3 mg/liter with >90% purity. The purified II62–scFv migrated as a single band on SDS-PAGE with an estimated molecular mass of 28 kDa (Fig. S2D) and same was confirmed by Western blotting using HRP-conjugated anti-His antibody (data not shown). The functional activity and specificity of the purified II62–scFv was assessed by its binding to the RBD protein in ELISA. The II62–scFv showed specific binding to the RBD and did not bind to unrelated proteins like BSA and the envelope proteins from HIV and Chikungunya (Fig. 2, B and C).
Figure 2.
A, representative image of scFv. The VH and VL chains are connected by a flexible linker. B, binding specificity of purified II62–scFv against the target antigen, RBD. Unrelated antigens used as control showed no binding. C, purified II62–scFv used in ELISA for titration with increasing concentration of RBD. D, representative image of scFv-Fc. The scFv is fused to the Fc region of the heavy chain. Through dimerization of Fc parts, this format forms a bivalent antibody. E, binding specificity of purified II62–scFv-Fc against the target antigen, RBD. Unrelated antigens used as control showed no binding. F, purified II62–scFv-Fc used in ELISA for titration with increasing concentration of RBD. G, representative image of full-length antibody constructed by fusing the light and heavy chain of II62–scFv to respective constant regions of IgG1 framework. H, binding specificity of purified II62–IgG1 against the target antigen, RBD. Unrelated antigens used as control showed no binding. I, purified II62–IgG1 used in ELISA for titration with increasing concentrations of RBD.
Expression, purification, and characterization of II62–scFv–Fc format
The scFvs are monovalent in nature and lack the Fc-mediated functions. The Fc domain of human IgG1 was introduced to produce a homodimeric II62–scFv-Fc chimeric protein that is capable of bivalent binding and that retains Fc functions (Fig. 2D). The II62–scFv-Fc construct was transiently expressed in Expi293F cells and subsequently purified by using protein G affinity column, with >95% purity and yields of 40–60 mg/liter. The purified II62–scFv-Fc migrated as a single protein band on SDS–PAGE with an estimated molecular mass of 55 kDa (Fig. S2E). The scFv-Fc showed specific binding to RBD in ELISA (Fig. 2, E and F).
Expression and purification of full-length II62–IgG1 mAb
The full-length version of the II62–scFv (II62–IgG1) was purified from the supernatant of transiently transfected Expi293F cells using protein A/G affinity columns (Fig. 2G). The purified IgG1 migrated as two protein bands on SDS–PAGE with estimated molecular mass of 50 kDa for heavy chain and 25 kDa for light chain (Fig. S2F). Overall yield of the II62–IgG1 was 80–100 mg/liter with >98% purity. The purified II62–IgG1 also showed specific binding to CoV-2 RBD protein (Fig. 2, H and I).
Affinity determination of II62 mAb formats
To evaluate the affinity of scFv-Fc and IgG1 format of II62 against the RBD, the antibodies were captured on the anti-human Fc biosensor and RBD was used as analyte in concentrations ranging from 10.0 to 0.6 μm with half-fold serial dilution. The scFv-Fc format showed an affinity of ∼700 nm with both slow on (1.5 × 102 (1/Ms)) and off (1.1 × 10−4 (1/s)) rate, suggesting that the scFv-Fc format binds slowly to the RBD, but once bound it also dissociates slowly (Fig. 3A). The full-length format, II62–IgG1, showed an affinity of ∼160 nm, with an on rate of (kon) of 3.3 × 102 (1/Ms) and off rate (koff) of 5.3 × 10−5 (1/s) (Fig. 3B).
Figure 3.
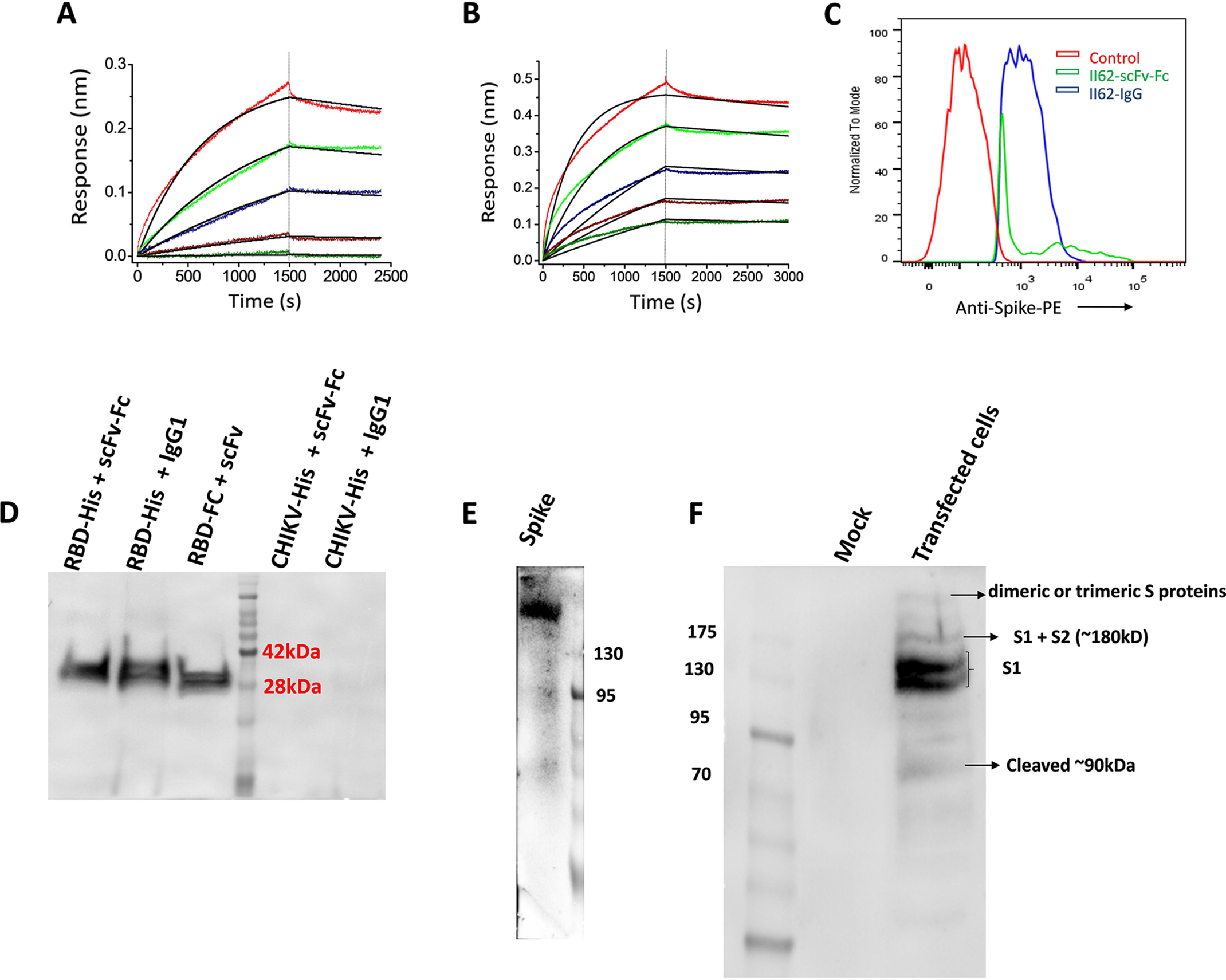
A and B, binding kinetics of II62–scFv-Fc (A) and II62–IgG1 (B) against RBD. In both cases the antibody was immobilized on anti-human Fc sensors and RBD used as analyte. C, FACS analysis showing shift compared with control when II62–IgG1 and II62–scFv-Fc used in staining of cell expressing membrane-bound full-length spike protein. D, IP followed by Western blotting analysis using HRP-conjugated anti-His antibody. In the first and second lanes, RBD-His was pulled down with Fc-bearing antibodies (scFv-Fc and IgG1) immobilized to protein A–agarose resin. In the third lane, RBD–Fc was immobilized to protein A–agarose resin, and scFv-His was pulled down. Nonspecific antigens (His-tagged CHIKV envelope protein) used as control. E, His-tagged full-length spike is detected on Western blotting by using II62–scFv-Fc antibody using HRP-conjugated anti-Fc antibody. F, cell lysate of pseudovirus transfected 293T cells was run on SDS-PAGE and probed with II62–IgG1 mAb.
Binding of II62 mAb to cell surface–expressed CoV–2 S protein
We investigated the binding of II62–scFv-Fc and IgG1 with the cell surface–expressed CoV-2 S protein in native conformation. For this, HEK293T cells were transiently transfected with plasmid containing the full-length S protein gene to express the same on the cell surface. The cells were then incubated with II62 scFv or IgG1 and analyzed by flow cytometry. The CoV-2 S proteins expressed on the 293T cell surface were readily detected by II62–scFv-Fc and IgG1. The results suggest that both formats of II62 recognizes the CoV-2 S protein on the cell surface (Fig. 3C and Figs. S3 and S4).
Specificity of II62 antibody formats to purified RBD and spike protein
Specificity of different antibody formats of II62 (scFv, scFv-Fc, and IgG1) were further individually tested for their specific binding to CoV-2 RBD by immunoprecipitation (IP) followed by Western blotting analysis. All the three antibody formats showed specific binding to RBD protein in co-IP (Fig. 3D). Binding specificity of II62 mAb was also tested toward purified soluble spike protein through Western blotting analysis and ELISA. Antibody II62 specifically recognizes the band of ∼180-kDa (uncleaved soluble spike) (Fig. 3E and Fig. S5).
Binding specificity of II62 mAbs toward cleaved and uncleaved spike S protein
Like other coronaviruses, the S protein of CoV-2 contains cleavage sites, and the precursor S protein is cleaved into the S1 and S2 subunits by host-cell proteases (9), an important step for initializing of infection. To further assess the effect of the binding of II62–IgG1 to cleaved and uncleaved forms of the S protein, 293T cells were transiently transfected with pseudo virus expressing CoV-2 S protein plasmid. Cell lysates of these transfected cells were immunoblotted with II62–IgG1. Immunoblotting of the cell lysate revealed two bands for the CoV-2 S protein at ∼110–130 kDa, which might be the uncleaved S1 protein (Fig. 3F and Fig. S6). The molecular mass of each monomer of trimeric S protein is ∼180 kDa and contains two subunits: S1 (110 kDa) and S2 (70 kDa) (10). The Western blotting showed another band at the position of 90 kDa reflecting the presence of cleaved S1 protein (Fig. 3F and Fig. S6). The band size above 200 kDa likely shows the presence of dimeric or trimeric S proteins as reported previously (11).
Molecular modeling and docking to explore the interaction site and key residues between RBD and mAb
The hot-spot residues involved in mAb–RBD interactions have been identified using protein–protein docking approaches. Through multiple quantitative analysis, we found 24 residues of RBD interact with II62–scFv. Of these residues Ser344, Arg341, Phe481, Tyr484, Tyr346, Tyr446, and Tyr488 of RBD were found to contribute significantly (Fig. 4A). While comparing the interacting interfaces of the complex of RBD–scFv with RBD–ACE2, 11 residues were found in common between RBD–scFv and RBD–ACE2. The sequence-wise comparisons among CoV/CoV-2/MERS revealed the structurally and sequentially conserved residues i.e. Tyr346, Tyr446, and Leu487, indicating that this mAb is possibly occupying the RBD–ACE2–interacting site (Fig. 4 and supporting information).
Figure 4.
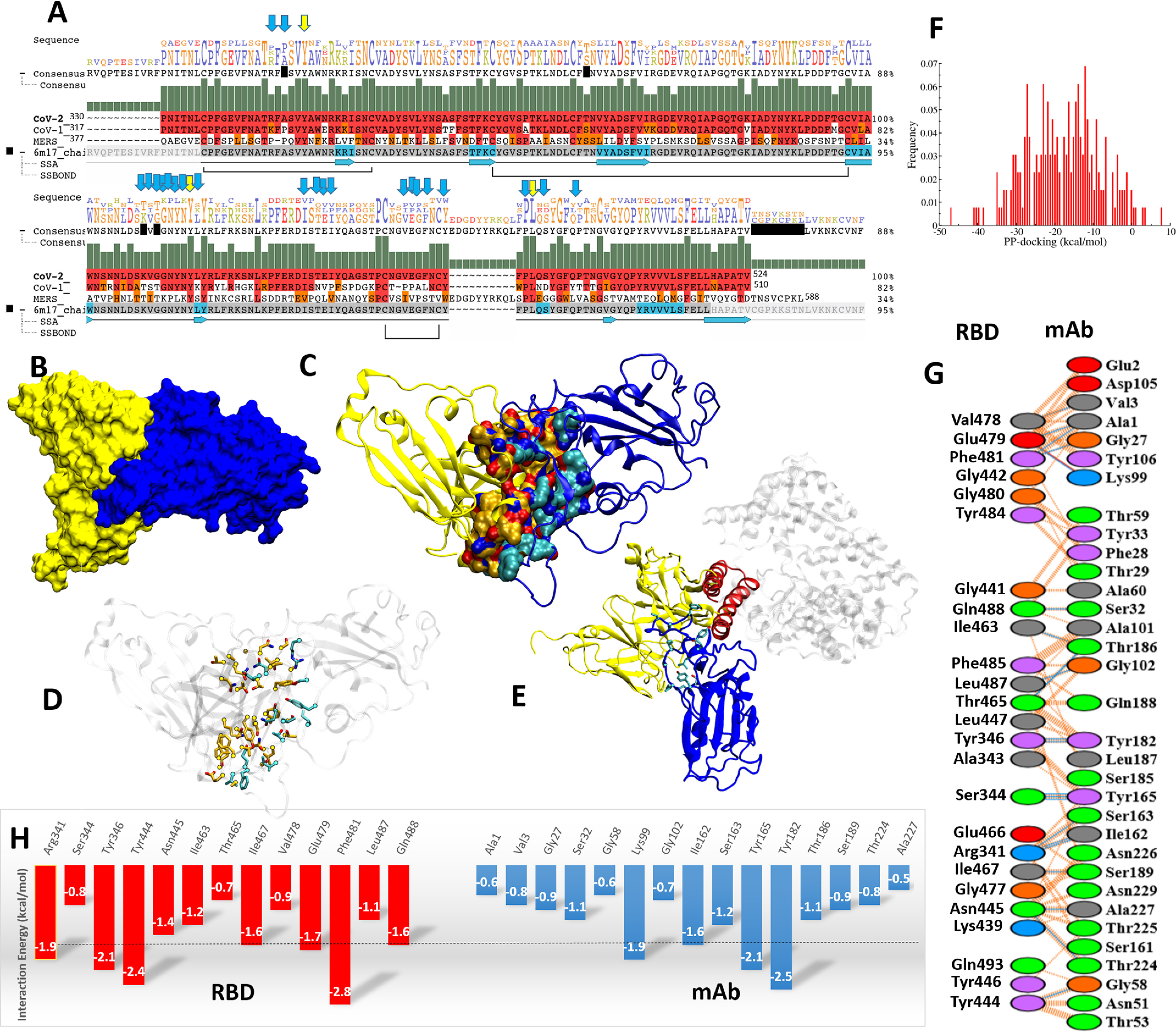
Protein–protein interaction and interface characterization. A, sequence alignment of CoV-2, CoV, and MERS viruses. Conserved residues are shown with yellow and blue arrows. B, the complex of II62–scFv and RBD in surface view (yellow and blue, respectively). C, the interacting interface residues in yellow and blue cartoons. D, the interacting residues are shown in atom-wise coloring: orange (II62) and cyan (RBD). E, the overlay of RBD-mAb and RBD–ACE2. The ACE2-interacting secondary structure, α-helix, is shown with a red color cartoon. F, histogram of protein–protein docking score in kcal/mol. G, the quantification of interacting residues such as hydrogen bonds (blue line) and VdW (dotted orange line). The residues are mentioned: basic (blue), acidic (red), polar (green), aromatic (purple), and hydrophobic contacts (gray). H, the interaction energetics quantification (less than −1.5 is cut off with a dotted black line).
Discussion
The RBD of SARS–CoV and CoV-2 S protein uses ACE2 on the host cell surface as receptor. The NAbs can block this interaction and prevent virus entry into the host cell (12). Phage display technique is a powerful tool that has been used for years for the discovery of therapeutic mAbs against infectious diseases (13). The advantage of usage of such libraries is that it avoids the direct use of patients' samples in a pandemic situation and related ethical, safety, and handling concerns (14). Biopanning of Tomlinson I library against CoV-2 RBD captured II62 as predominant clone that showed specific binding to CoV-2 RBD protein. We further engineered and characterized three different formats of the selected II62–scFv, i.e. scFv, scFv-Fc, and IgG1, respectively. We examined the binding of different formats to CoV-2 RBD protein by ELISA and by co-IP followed by Western blotting analysis. The results show that all three formats efficiently and specifically bind to the CoV-2 RBD protein. Next, we tested whether these formats bind with native-like cell surface–expressed S protein by flow cytometry analysis of transiently transfected HEK293T cells expressing membrane-bound CoV-2 S protein. Our data suggest that the II62 antibody formats (scFv-Fc and IgG1) bind to a conformational epitope on the full-length S protein as presented on the cell surface. Furthermore, we determined the binding affinity of these mAb formats using biolayer interferometry. Both scFv-Fc and IgG1 effectively bind with nanomolar affinity to CoV-2 RBD. The RBD in the S1 domain of CoV-2 S protein interacts with the ACE2 receptors expressed on the host cell that triggers a conformational change of the S2 domain, resulting in formation of six helix bundles and insertion of fusion peptide into the host cell. It is reported that most of the CoV-2 S proteins in pseudovirions are cleaved, and our results suggest that II62 mAb binds efficiently to various forms of S protein. This mAb could be potentially used to address various research questions on CoV-2 S protein biosynthesis, cleavage, intracellular processing, and expression and will help to identify non-neutralizing epitopes to facilitate design of conformationally stable vaccine candidates. Similar findings have been previously reported for MERS (10). Because of sequence divergence among MERS, CoV, and CoV-2, most of the polyclonal and mAbs raised against MERS and CoV poorly cross-react with CoV-2 (15). In future it will be important to determine whether this mAb can also cross-react with MERS and CoV S proteins. Interestingly, the gene sequence of II62 reveals only few somatic mutations compared with the closest germline genes.
The II62 mAb did not show significant reduction in neutralization potential in cytopathic effect–based and plaque reduction assay when tested up to 100 µg/ml against CoV-2 live virus (Figs. S7 and S8). Docking study suggests that the II62–scFv epitope on RBD partially overlaps with the ACE2-binding site. It was further supported by the ACE2–RBD competition assay (Fig. S9). The II62–scFv-Fc antibody showed a concentration-dependent inhibition of RBD binding to ACE2 (starting from 5 to 0.008 μm). The other possible explanation is that the epitope is inaccessible in the closed prefusion form of S protein and accessible only in open post-fusion conformation. Pinto et al. (16) also reported similar phenomenon with mAbs S306 and S310 that recognize post-fusion conformation of CoV-2 S glycoprotein and were found to be poorly neutralizing. Similar findings have been recently reported for non-neutralizing behavior of CR3022 to CoV-2 (17). Human and animal studies for various viral infection model and vaccine studies clearly suggests that non-NAb in vitro may play an important role to block cell surface infection in vivo without disturbing virus entry (18). The high affinity and specificity of II62 mAb makes it an attractive target in mucosal antibody–based prevention strategies to trap the virus particles even with non-NAb mAbs before they initiate infection. In the past it has been shown that topical application of mAb was ∼100 times more effective than systemic delivery (19).
Our study also provides a way to compare different engineered versions of a mAb and their suitability for different applications. The one advantage of scFv format is that it is devoid of Fc-mediated functions that sometimes exaggerate the infection through ADE (20). Wan et al. (21) suggested that ADE events could also happen with anti-CoV-2 Abs. To overcome the ADE-related issues, one approach is to develop either scFv or llamas antibodies that are devoid of Fc-related functions (22). Our study opens a path forward toward isolation of scFv molecules with high affinity and specificity. To the best of our knowledge, this is the first report of isolation and characterization of scFv and its other formats for CoV-2 using the human semisynthetic Tomlinson I phagemid library.
The other interesting observation of our study is that II62 mAb showed more ELISA binding reactivity with baculovirus expressed RBD-His protein as compared with mammalian expressed RBD–His protein (Fig. S10). Similar findings have also been recently reported in a preprint from Bertoglio et al. (23), in which they have compared the binding of mammalian cell– and high five cell–expressed S1 protein with ACE2. One probable reason for this is the different protein glycosylation pathways of the heterologous hosts.
Experimental procedures
Library amplification and panning
The stock of Tomlinson I library was expanded and amplified in 2× TY (tryptone and yeast extract) medium containing 100 μg/ml ampicillin and 2% glucose. The panning procedure was performed according to the Tomlinson manual (28, 29). A detailed methodology for this has been provided in the supporting information.
Screening of clones (monoclonal phage ELISA)
After the third round of biopanning, individual colonies were picked and tested for specificity by phage ELISA. Monoclonal phage ELISA was carried out as described by Kumar et al. (13) and Spriestersbach et al. (24) and detailed in the supporting information. Briefly, individual colonies from third round of selection were grown in 2× TY medium containing 100 μg/ml ampicillin and 2% glucose and infected with helper phage in 96-well plate format. The plate was then incubated at 30 °C with shaking overnight. 50 μl of the supernatant from each well was used for phage ELISA.
Cloning, expression, and purification of proteins and antibody formats
His-tagged codon optimized genes for mammalian expression were used for transient transfection of CoV-2 RBD and S protein ectodomain in Expi293F cells to produce the recombinant proteins and purified by Ni–NTA affinity chromatography following standard protocol (25). For scFv-Fc format, the II62–scFv gene was amplified using gene-specific primers and cloned into pCMX2.5–hIgG1–XP vector using NcoI and NotI restriction enzymes. For II62–IgG1 the variable regions of the heavy and light chains were cloned into respective expression plasmids containing the constant regions of human IgG1 heavy chain and Igκ light chain (InvivoGen). In each case proper cloning was confirmed by gene sequencing.
For expression of soluble II62–scFv HB2151 strain of Escherichia coli bearing scFv plasmid was grown in 1 liter of 2× TY medium at 37 °C with shaking at 200 rpm. The culture was induced with 1 mm isopropyl β-d-thiogalactopyranoside at A600 0.4–0.6 and incubated at 18 °C for 16–20 h. The II62–scFv antibody was purified from the periplasmic fraction using Ni–NTA affinity chromatography. The scFv-Fc and IgG1 formats were expressed by transient transfection of respective plasmids in Expi293F cells, and both the antibody formats were purified using protein G affinity column.
ELISA
For phage ELISA, NUNC Maxisorp plates (Thermo Scientific) were coated with 100 μl of RBD protein or soluble S protein (200 ng per well), 50 μl of phage-rescued supernatants diluted in 50 μl of 2% milk PBS were added to each well and incubated for 1 h. HRP-conjugated anti-M13 (Sigma) was used for developing ELISA with tetramethylbenzidine substrate.
For CoV-2 antigen-specific ELISA, the plates were coated with equimolar amounts of antigens. Different formats of antibody were added in serial dilutions starting from 20 μg/ml. In case of II62–scFv antibody, HRP-conjugated protein L was used as secondary antibody. For Fc-bearing antibody formats, HRP-conjugated goat anti-human secondary antibody (Jackson Immunoresearch) was used. Standard protocols of blocking and washing of ELISA plates were followed.
IP and Western blotting analysis
For IP, 1 µg of purified protein was incubated overnight at 4 °C with 2 µg of mAbs of choice in the presence of protein G–agarose (100 µl of 50% slurry; G Biosciences) followed by washing of beads with PBS before SDS-PAGE analysis. In the case of scFv, the Fc-tagged RBD protein was immobilized to protein G beads, and His-tagged scFv was pulled down. For scFv-Fc and IgG1, the antibodies were immobilized to protein G beads, and RBD-His was pulled down. For Western blotting analysis, the proteins were transferred from PAGE to polyvinylidene difluoride membrane. The membrane was blocked with 5% skim milk and developed with HRP-conjugated anti-His antibody in 1:3000 dilutions for 2 h at room temperature.
Binding kinetics using biolayer interferometry
For measuring binding kinetics, anti-human Fc sensors (ForteBio Inc.) were used to capture the scFv-Fc or the IgG1 antibody formats, and the RBD-His was used as analyte. The analytes were used in various concentrations with half-fold serial dilution in the PBS buffer background supplemented with 0.1% BSA. The ligands were used at a concentration of 10 µg/ml. Associations and dissociations were recorded for 1500 s. The data were analyzed using the ForteBio data analysis software, 10.0 (Forte-Bio Inc.). The kinetic parameters were calculated using a global fit 1:1 model.
Molecular modeling and protein–protein docking study
For RBD the crystal structure 6M17 was used. Based on sequence identity/similarity homology model was generated and validated. Protein–protein docking tools were used to identify the most likely binding interface (26, 27) (see supporting information).
Data availability
The data supporting the findings of this study are available within the article and in its supporting information.
Supplementary Material
Acknowledgments
We thank Dr. Gagandeep Kang (Translational Health Science & Technology Institute) for development of the project and Dr. Anna George for critical inputs. We thank the Medical Research Council of the United Kingdom for allowing us to use the Tomlinson libraries and Prof. S. Pöhlmann (Infection Biology Unit, Göttingen, Germany) for the kind gift of ACE2-Fc plasmids. SARS–CoV-2-S–RBD–Fc was a gift from Erik Procko (Addgene Plasmid 141183). We also thank Prof. M. Hust for providing pCMX2.5-hIgG1-XP vector. The RBD-His is a proprietary reagent with IP No. 202011018845. We thank Dr. B. Graham (Vaccine Research Center, NIAID, National Institutes of Health) for providing us with spike construct (SARS-2-CoV S 2P). The following reagent was deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, National Institutes of Health: SARS-related coronavirus 2, Isolate USA-WA1/2020, NR-52281. Part of the work was carried out at the Advanced Technology Platform Centre (ATPC) of Regional Centre for Biotechnology (RCB), and is funded by the Department of Biotechnology, Govt. of India (Grant No. BT.MED-II/ATPC/BSC/01/2010).
This article contains supporting information.
Author contributions—H. A. P., A. K. C., S. Asthana, N. Y., S. M., C. S., K. P., and R. K. methodology; S. Asthana, N. Y., T. S., P. V., S. D., S. Samal, S. Ahmed, and R. K. investigation; S. Sinha, and R. K. supervision; S. Sinha, S. Ahmed, and R. K. writing-review and editing; S. Ahmed validation; S. Ahmed and R. K. writing-original draft; R. K. conceptualization; R. K. project administration; H. A. P. experiment design and performed.
Funding and additional information—This work was supported by the Department of Biotechnology and by a Translational Health Science & Technology Institute core grant.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- CoV-2
- coronavirus 2
- SARS
- severe acute respiratory syndrome
- RBD
- receptor-binding domain
- scFv
- single-chain fragment variable region
- ACE
- angiotensin-converting enzyme
- NAb
- neutralizing antibody
- HRP
- horseradish peroxidase
- IP
- immunoprecipitation
- MERS
- Middle East respiratory syndrome
- Ni–NTA
- nickel–nitrilotriacetic acid
- ADE
- antibody dependent enhancement.
References
- 1. Felsenstein S., Herbert J. A., McNamara P. S., and Hedrich C. M. (2020) COVID-19: immunology and treatment options. Clin. Immunol. 215, 108448 10.1016/j.clim.2020.108448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hey A. (2015) History and practice: antibodies in infectious diseases. Microbiol. Spectr. 3, AID-0026-2014 [DOI] [PubMed] [Google Scholar]
- 3. Kumar R., Shrivastava T., Samal S., Ahmed S., and Parray H. A. (2020) Antibody-based therapeutic interventions: possible strategy to counter chikungunya viral infection. Appl. Microbiol. Biotechnol. 104, 3209–3228 10.1007/s00253-020-10437-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Premkumar L., Segovia-Chumbez B., Jadi R., Martinez D. R., Raut R., Markmann A., Cornaby C., Bartelt L., Weiss S., Park Y., Edwards C. E., Weimer E., Scherer E. M., Rouphael N., Edupuganti S., et al. (2020) The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS–CoV-2 patients. Sci. Immunol. 5, eabc8413 10.1126/sciimmunol.abc8413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Casadevall A., and Pirofski L. A. (2020) The convalescent sera option for containing COVID-19. J. Clin. Invest. 130, 1545–1548 10.1172/JCI138003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brown B. L., and McCullough J. (2020) Treatment for emerging viruses: Convalescent plasma and COVID-19. Transfus. Apher. Sci. 59, 102790 10.1016/j.transci.2020.102790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Burnouf T., and Seghatchian J. (2014) Ebola virus convalescent blood products: where we are now and where we may need to go. Transfus. Apher. Sci. 51, 120–125 10.1016/j.transci.2014.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Marano G., Vaglio S., Pupella S., Facco G., Catalano L., Liumbruno G. M., and Grazzini G. (2016) Convalescent plasma: new evidence for an old therapeutic tool?. Blood Transfus. 14, 152–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., and Li F. (2020) Cell entry mechanisms of SARS–CoV-2. Proc. Natl. Acad. Sci. U.S.A. 26, 11727–11734 10.1073/pnas.2003138117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Qian Z., Dominguez S. R., and Holmes K. V. (2013) Role of the spike glycoprotein of human Middle East respiratory syndrome coronavirus (MERS-CoV) in virus entry and syncytia formation. PLoS One 8, e76469 10.1371/journal.pone.0076469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., et al. (2020) Characterization of spike glycoprotein of SARS–CoV-2 on virus entry and its immune cross-reactivity with SARS–CoV. Nat. Commun. 11, 1620 10.1038/s41467-020-15562-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., Gao G., Hu X., Zhang Y., Tong Z., Huang W., et al. (2020) A human neutralizing antibody targets the receptor binding site of SARS–CoV-2. Nature 10.1038/s41586-020-2381-y [DOI] [PubMed] [Google Scholar]
- 13. Kumar R., Andrabi R., Tiwari A., Prakash S. S., Wig N., Dutta D., Sankhyan A., Khan L., Sinha S., and Luthra K. (2012) A novel strategy for efficient production of anti-V3 human scFvs against HIV-1 clade C. BMC Biotechnol. 12, 87 10.1186/1472-6750-12-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kumar R., Parray H. A., Shrivastava T., Sinha S., and Luthra K. (2019) Phage display antibody libraries: A robust approach for generation of recombinant human monoclonal antibodies. Int. J. Biol. Macromol. 135, 907–918 10.1016/j.ijbiomac.2019.06.006 [DOI] [PubMed] [Google Scholar]
- 15. Haynes B. F., Ma B., Montefiori D. C., Wrin T., Petropoulos C. J., Sutherland L. L., Scearce R. M., Denton C., Xia S. M., Korber B. T., and Liao H. X. (2006) Analysis of HIV-1 subtype B third variable region peptide motifs for induction of neutralizing antibodies against HIV-1 primary isolates. Virology 345, 44–55 10.1016/j.virol.2005.08.042 [DOI] [PubMed] [Google Scholar]
- 16. Pinto D., Park Y.-J., Beltramello M., Walls A. C., Tortorici M. A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., Peter A., Guarino B., Spreafico R., Cameroni E., Case J. B., et al. (2020) Cross-neutralization of SARS–CoV-2 by a human monoclonal SARS–CoV antibody. Nature 583, 290–295 10.1038/s41586-020-2349-y [DOI] [PubMed] [Google Scholar]
- 17. Yuan M., Wu N. C., Zhu X., Lee C.-C. D., So R. T. Y., Lv H., Mok C. K. P., and Wilson I. A. (2020) A highly conserved cryptic epitope in the receptor binding domains of SARS–CoV-2 and SARS–CoV. Science 368, 630–633 10.1126/science.abb7269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Excler J. L., Ake J., Robb M. L., Kim J. H., and Plotkin S. A. (2014) Nonneutralizing functional antibodies: a new “old” paradigm for HIV vaccines. Clin. Vaccine Immunol. 21, 1023–1036 10.1128/CVI.00230-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Prince G. A., Hemming V. G., Horswood R. L., Baron P. A., and Chanock R. M. (1987) Effectiveness of topically administered neutralizing antibodies in experimental immunotherapy of respiratory syncytial virus infection in cotton rats. J. Virol. 61, 1851–1854 10.1128/JVI.61.6.1851-1854.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ngono A. E., and Shresta S. (2018) Immune response to dengue and Zika. Annu. Rev. Immunol. 36, 279–308 10.1146/annurev-immunol-042617-053142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wan Y., Shang J., Sun S., Tai W., Chen J., Geng Q., He L., Chen Y., Wu J., Shi Z., Zhou Y., Du L., and Li F. (2020) Molecular mechanism for antibody-dependent enhancement of coronavirus entry. J. Virol. 94, e02015–19 10.1128/JVI.02015-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Benjathummarak S., Pipattanaboon C., Boonha K., Wongwit W., Ramasoota P., and Pitaksajjakul P. (2018) Human single-chain variable fragment antibody expressed in E. coli with optimal in vitro cross-neutralizing and no enhancing activity. Biologicals. 56, 54–62 10.1016/j.biologicals.2018.10.003 [DOI] [PubMed] [Google Scholar]
- 23. Bertoglio F., Meier D., Langreder N., Steinke S., Rand U., Simonelli L., Heine P. A., Ballmann R., Schneider K.-T., Roth K. D. R., Ruschig M., Riese P., Eschke K., Kim Y., Schäckermann D., et al. (2020) SARS–CoV-2 neutralizing human recombinant antibodies selected from pre-pandemic healthy donors binding at RBD–ACE2 interface. bioRxiv 10.1101/2020.06.05.135921 [DOI] [PMC free article] [PubMed]
- 24. Sankhyan A., Sharma C., Dutta D., Sharma T., Chosdol K., Wakita T., Watashi K., Awasthi A., Acharya S. K., Khanna N., Tiwari A., and Sinha S. (2016) Inhibition of preS1-hepatocyte interaction by an array of recombinant human antibodies from naturally recovered individuals. Sci. Rep. 6, 21240 10.1038/srep21240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Spriestersbach A., Kubicek J., Schäfer F., Block H., and Maertens B. (2015) Purification of His-tagged proteins. Methods Enzymol. 559, 1–15 10.1016/bs.mie.2014.11.003 [DOI] [PubMed] [Google Scholar]
- 26. Kanwal A., Kasetti S., Putcha U. K., Asthana S., and Banerjee S. K. (2016) Protein kinase C–mediated sodium glucose transporter 1 activation in precondition-induced cardioprotection. Drug Des. Devel. Ther. 10, 2929–2938 10.2147/DDDT.S105482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mattapally S., Singh M., Murthy K. S., Asthana S., and Banerjee S. K. (2018) Computational modeling suggests impaired interactions between NKX2.5 and GATA4 in individuals carrying a novel pathogenic D16N NKX2.5 mutation. Oncotarget 9, 13713–13732 10.18632/oncotarget.24459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Abou El-Magd R. M., Vozza N. F., Tuszynski J. A., and Wishart D. S. (2016) Isolation of soluble scFv antibody fragments specific for small biomarker molecule, L-Carnitine, using phage display. J. Immunol. Methods 428, 9–19 10.1016/j.jim.2015.11.006 [DOI] [PubMed] [Google Scholar]
- 29. Bagheri S., Yousefi M., Safaie Qamsari E., Riazi-Rad F., Abolhassani M., Younesi V., Dorostkar R., Movassaghpour A. A., and Sharifzadeh Z. (2017) Selection of single chain antibody fragments binding to the extracellular domain of 4-1BB receptor by phage display technology. Tumour Biol. 39 10.1177/1010428317695924 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available within the article and in its supporting information.