Abstract
Inhalation of the ambient air pollutant ozone causes lung inflammation and can suppress host defense mechanisms, including impairing macrophage phagocytosis. Ozone reacts with cholesterol in the lung to form oxysterols, like secosterol A and secosterol B (SecoA and SecoB), which can form covalent adducts on cellular proteins. How oxysterol-protein adduction modifies the function of lung macrophages is unknown. Herein, we used a proteomic screen to identify lung macrophage proteins that form adducts with ozone-derived oxysterols. Functional ontology analysis of the adductome indicated that protein binding was a major function of adducted proteins. Further analysis of specific proteins forming adducts with SecoA identified the phagocytic receptors CD206 and CD64. Adduction of these receptors with ozone-derived oxysterols impaired ligand binding and corresponded with reduced macrophage phagocytosis. This work suggests a novel mechanism for the suppression of macrophage phagocytosis following ozone exposure through the generation of oxysterols and the formation of oxysterol-protein adducts on phagocytic receptors.
Keywords: ozone, oxysterol, macrophage, phagocytosis, lung, Fc receptor, cholesterol, CD206
Ground-level ozone is a common air pollutant found in photochemical smog that is formed from the reaction of volatile organic compounds with oxides of nitrogen. Inhalation of ozone has been shown to cause pulmonary inflammation (1–3), exacerbate lung disease (4, 5), and increase susceptibility to pathogen infection (6, 7). Although many studies have examined the effects of ozone inhalation on the lung, the mechanism leading to inflammation and suppression of host response to infection is not well understood.
The first point of contact for inhaled ozone is the lung lining fluid which is made up of numerous components, including phospholipids, proteins, and cholesterol (8). Although the interaction of ozone with proteins and phospholipids has been explored, much less is known about the role of cholesterol in ozone-induced lung disease. Ozone reacts with cholesterol, a common component of the lung lining fluid and of cell membranes, to form oxysterols species, including secosterol A (SecoA) and its aldol condensation product secosterol B (SecoB) (9). Ozone-derived oxysterols, such as SecoA and SecoB, are electrophiles and form protein adducts with cellular proteins at nucleophilic lysine residues (10). Our previous work showed that SecoA can form adducts in lung epithelial cells with specific targets, namely the liver X receptor, which leads to alterations in cholesterol signaling and increased inflammation (9).
Pulmonary macrophages patrol the airways and play important roles in host defense by recognizing and internalizing foreign bodies and debris. Human and animal studies show that ozone exposure leads to impairment in macrophage phagocytosis (11–14), yet the mechanisms of this immune-suppressive effect are unclear.
Phagocytosis is mediated by a number of pattern recognition receptors which can recognize distinct target motifs. For example, different surface receptors are utilized to recognize terminal sugar residues, specific lipoproteins, or antibody-coated targets. The mannose receptor, also known as CD206, is one such receptor that recognizes terminal mannose moieties and is expressed highly on lung macrophages. CD206 has been shown to play an important role in the protection against certain bacterial and fungal infections (15–17). Expression of CD206 and other pathogen recognition receptors at the cell surface makes them particularly susceptible to interaction with extracellular electrophiles, like oxysterols. Yet, it is not known whether oxysterols can form adducts with proteins in lung macrophages, and the impact of oxysterol adduction on macrophage function has not been elucidated. Similar to our previous studies (9, 10), we utilized a “click” chemistry approach to identify protein-oxysterol adducts formed in human pulmonary macrophages obtained through bronchoalveolar lavage (BAL) which were exposed ex vivo to an alkynyl-tagged SecoA. We describe here that targets identified through this protein adductome screen include pattern recognition receptor, including CD206, and describe the functional effects of adduct formation on macrophage phagocytosis.
Results
Identification of oxysterol adducted proteins in macrophages
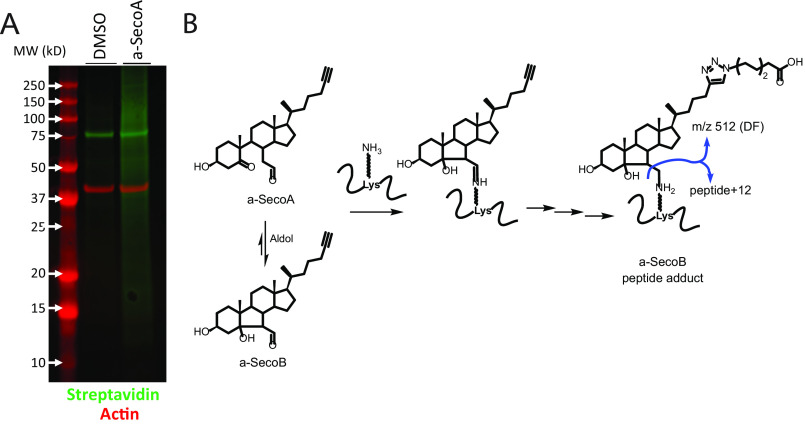
To determine the impact of oxysterols on macrophage function in the lung, we first carried out experiments to identify which macrophage proteins formed oxysterol-protein adducts using an unbiased proteomic approach. To do this, we treated primary human BAL macrophages, which represent the macrophages that would be exposed to ozone in the lung, with an alkynyl-tagged SecoA (a-SecoA), as described under “Materials and methods,” and lysed the cells after 4 h of treatment. We have shown previously that this exposure paradigm forms oxysterol-protein adducts in lung epithelial cells (9). To isolate oxysterol-protein adducts, the alkyne tag was reacted to allow the cycloaddition of azido biotin (Fig. 1). Biotinylated proteins were immobilized with streptavidin beads and then released using UV light (365 nm) after serial washes to remove unmodified proteins (Fig. 1). This pool of photoreleased proteins was used for subsequent in-gel trypsin digest and analysis by MS to provide an unbiased proteomic analysis of SecoA-protein adducts in human BAL macrophages. Streptavidin blot analysis of proteins following biotin addition revealed that many proteins in the a-SecoA–treated cells incorporated the biotin tag compared with the vehicle treatment, thus validating the efficiency of the click reaction (Fig. 2A).
Figure 1.
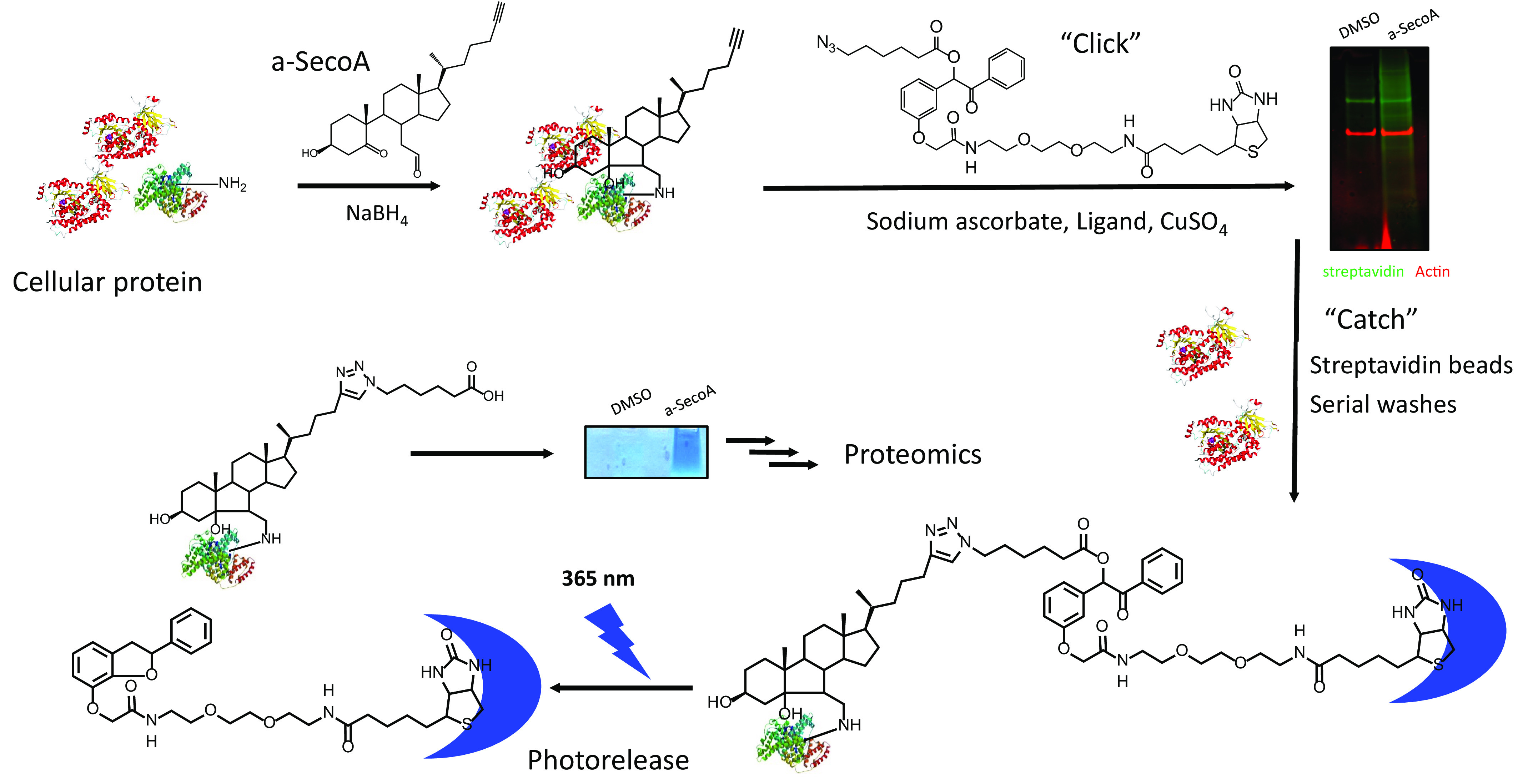
Oxysterol adduct identification using a-SecoA. Primary human BAL macrophages were treated with vehicle (DMSO) or a-SecoA (20 μm) for 4 h. Cells were harvested and NaBH4 added to stabilize adducts. Adducted proteins were isolated by reacting the alkynyl tag with cycloaddition of azido-biotin as described under “Materials and methods” and captured with streptavidin beads. Unbound proteins were washed away and captured proteins were photoreleased with UV light. Samples were concentrated on a stacking gel and trypsinized for MS analysis.
Figure 2.
a-SecoA forms adducts on macrophage proteins. A, biotin click mixture from vehicle and a-SecoA–treated cells was analyzed by Western blotting and probed with streptavidin fluorophore indicating a-SecoA–adducted proteins clicked with biotin. Actin is served as a loading control. B, oxysterol adducts can be formed through acid catalyzed immine formation of a-SecoA that then transforms to a-SecoB or through conversion of a-SecoA to a-SecoB and then formation of an a-SecoB adduct. The a-SecoB adduct can be identified by the formation of a diagnostic fragment (DF) with a signature 512 m/z.
Proteome-wide analysis after stringent filtration counted 756 spectra having a loss of a 512 m/z fragment diagnostic of a-SecoA/B adduction with b and y ions identifying the specific lysine site of adduction (Table S1). This diagnostic 512 m/z fragment served as a signature fragment of a SecoB adducted peptide shown in Fig. 2B. The conversion of SecoA to SecoB can occur by aldol condensation of free secosterol or at the adduct SecoA-lysine imine stage via an enamine condensation, meaning a protein exposure with either SecoA or SecoB typically yields a set of SecoB peptide adducts (Fig. 2B). The 756 spectra were assigned to 215 unique peptides of BAL macrophage proteins modified with SecoA/B. These adducted peptides were associated with 141 different proteins (Table S2).
Gene ontology analysis of the proteins adducted in BAL macrophages showed that most of the SecoA-adducted proteins were found on the vesicle and cell membrane (Fig. 3A). Similarly, functional analysis of adducted proteins showed that protein binding was a major function of adducted proteins (Fig. 3B). Together these data suggested that surface receptors and recognition of foreign material could be impacted by SecoA adduction.
Figure 3.
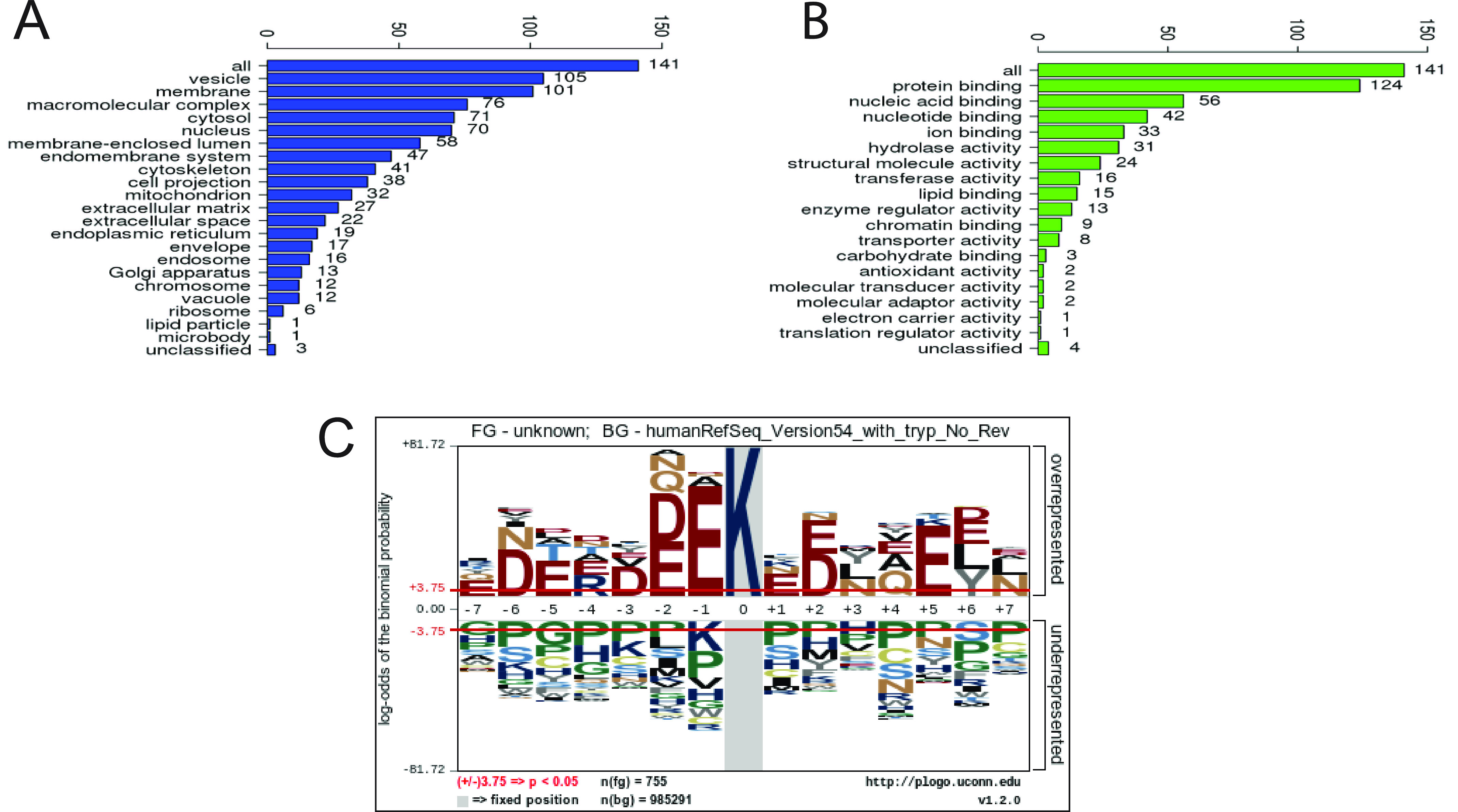
Oxysterol adduct cellular peptides. Adducted peptides were mapped to corresponding proteins. A and B, gene ontology analysis categorized proteins based on (A) cellular compartment and (B) protein function. C, sequence motif analysis using pLogo of adducted peptides was performed to determine amino acid sequence surrounding modified lysine residues. The red horizontal lines in the plot indicate p < 0.05 thresholds.
A sequence motif analysis plot was generated from comparison of the 756 modified lysine-containing sequences to the reference human database used for this study (33,487 proteins) giving the image shown in Fig. 3C. The motif shows unequivocally that acidic residues located one or two units away from the reactive lysine are significantly overrepresented in the adducted peptides. Indeed, glutamate and aspartate residues are overrepresented across the sequence, with sites at −6, −5, +5, and +6 being favored in addition to the nearest neighbor locations from −2 to +2, suggesting that acidic residues in these positions promote the formation of a-SecoA adducts.
Similar to our previous study in lung epithelial cells (9), heat-shock proteins and histone proteins were among the most highly adducted by SecoA in lung macrophages (Table 1 and Fig. S1). Interestingly, many proteins were identified in our macrophage analysis that were not found in our previous studies. One protein in particular, CD206 (also known as the mannose receptor C type 1, or MRC1), stood out as a protein of particular interest (Table 1 and Fig. 4A). Matching our gene ontology analysis, CD206 is expressed on the cell surface and is a pattern recognition receptor that binds to moieties on various respiratory pathogens (18).
Table 1.
Select proteins adducted by a-SecoA
| Gene ID | Description | Modified Peptides | |
|---|---|---|---|
| Abundant proteins adducted by lipid electrophiles highly in various cells | HSP90AA1 or AB1 | heat shock protein 90-a or b | ELISNSSDALD58K*IR, Q649K*AEADKNDK, YESLTDPS69K*LDSGK, YIDQEELN292K*TK, AEAD654 K* NDK, E283K*YIDQEELNK |
| CFL1 | cofilin 1 | YALYDATYET92K*ESK, NIILEEG53K*EILVGDVGQTVDDPYATFVK, HELQANCYEEV144K*DR | |
| HSP90B1 | endoplasmin precursor | EVEEDEY356K*AFYK, FDESE603K*TK | |
| HIST1H3A | histone H3.1 | EIAQDF80K*TDLR | |
| Unique adducts in BALMac | LTA4H | leukotriene A4 hydrolase | WITA480K*EDDLNSFNATDLK, DLAAFD580K*SHDQAVR, SLVLDT58K*DLTIEK |
| MRC1 | mannose receptor, C type 1, CD206 | SQGPEIVEVE502K*GCR | |
| APOE | apolipoprotein E | AY93K*S.E.LEEQLTPVAEETR | |
| ERLIN1 | ER lipid raft associated 1 | NFELMEAE192K*TK | |
| GLUD1 | glutamate dehydrogenase 1 | GASIVED84K*LVEDLR | |
| LDHA | lactate dehydrogenase A | DLADELALVDVIED57K*LK |
residue numberK* = Lys modified with a-SecoA. Identified MS/MS spectra are included in Fig. S1.
Figure 4.
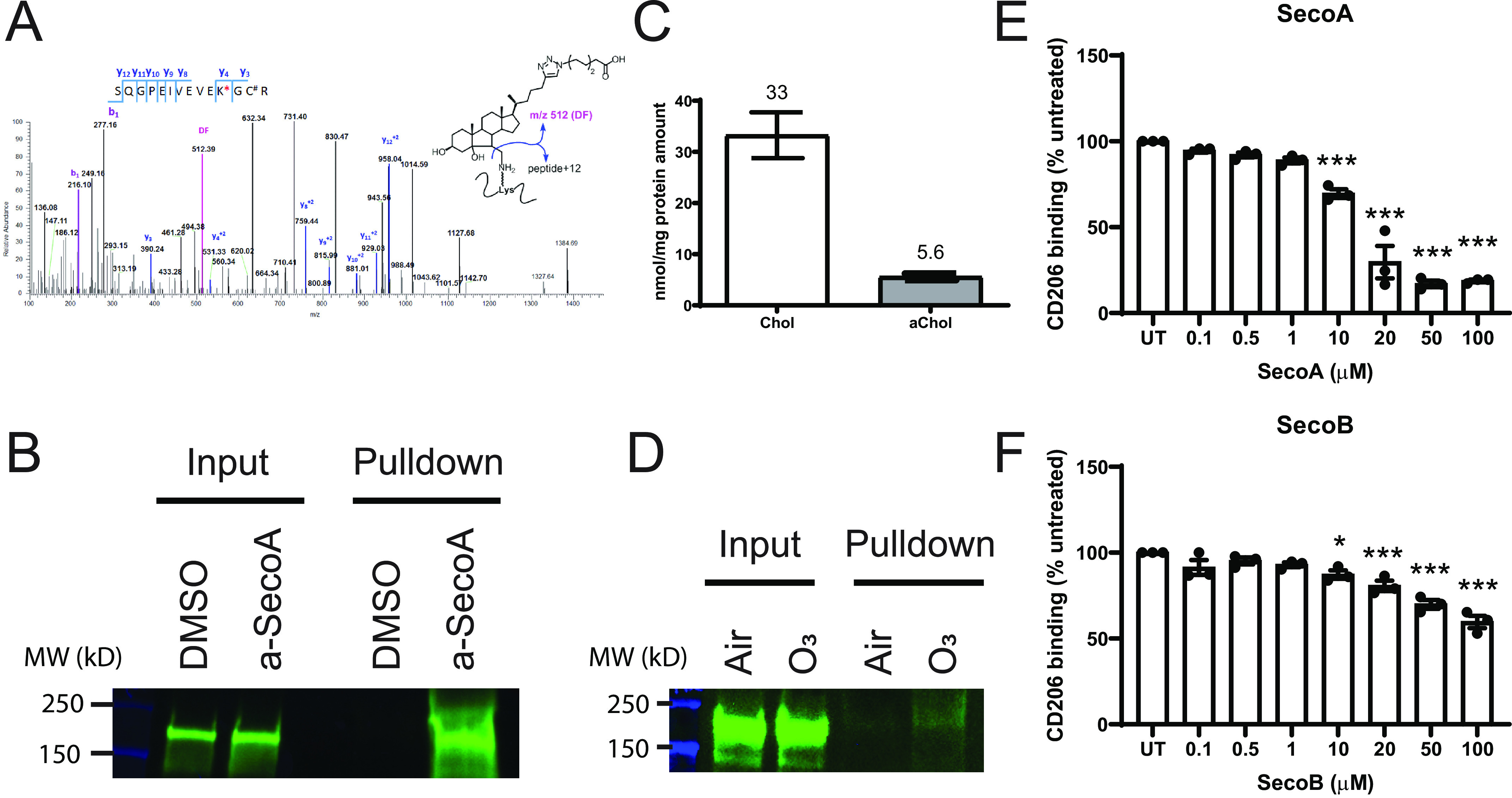
CD206 is adducted by oxysterols, which alters protein binding. A, shotgun proteomics identified a-SecoA adduction on the Lys-502 residue in the 493SQGEIVEVEK*GC#R (C# = carbamidomethyl-Cys) of MRC1 with 3.6 ppm precursor accuracy (calculated 2128.1621, observed 2128.1680). Diagnostic fragment (DF) m/z 512 is found shown in magenta. B, Western blot analysis of isolated adducted proteins for CD206. C, BAL macrophages were cultured in the presence a-Chol for 48 h and the levels of incorporated a-Chol is was determined by MS quantification. D, a-Chol–supplemented macrophages were exposed to air or 0.4 ppm ozone for 4 h to mimic formation of endogenous oxysterols. Following click and pulldown, lysate was probed for CD206. E and F, the effect of (E) SecoA or (F) SecoB on CD206 binding was determined utilizing an in-house ELISA as described under “Materials and methods.” Bars for cholesterol measurement represent the mean (±) S.D. of three replicate wells. Bars for binding assays represent the mean (±) S.E. of three independent experiments with quadruplicate wells per treatment. *, p < 0.05; ***, p < 0.001 by one-way ANOVA with Dunnett's multiple comparison test of treated groups to untreated (UT) control.
It is well known that ozone exposure impairs macrophage phagocytosis in vivo (11, 13, 19), thus, we hypothesized that ozone-derived adduction of CD206 played a role in the suppression of phagocytosis seen following ozone exposure. Analysis of the proteomic data identified CD206 modified by a-SecoA on Lys-502 in the peptide sequence SQGEIVEVE502K*GCR (Fig. 4A). We confirmed that a-SecoA forms protein adducts with CD206 by Western blotting using the proteins isolated from the click cycloaddition pulldown. Equal levels of CD206 were found in the lysates from vehicle and a-SecoA–treated macrophages prior to the isolation of adducted proteins (Fig. 4B, input). In contrast, CD206 was present in the purified pool of adducted proteins following a-SecoA treatment of BAL macrophages but absent in pulldown pool of proteins in vehicle-treated cells, thus confirming the presence of a-SecoA adduction to CD206 (Fig. 4B, pulldown).
Ozone exposure can replicate formation of oxysterols in vitro and adduct on phagocytic receptors
We next looked to confirm that the adduction of CD206 was relevant in the context of lung macrophage exposure to ozone. We have previously shown that alkynyl-tagged cholesterol (a-Chol) can be incorporated into cells and, in response to ozone, can form oxysterols, including a-SecoA (9). To confirm that the adduction of CD206 occurs following the formation of endogenous oxysterols during ozone exposure, we loaded BAL macrophages with a-Chol for 48 h to allow for incorporation of tagged cholesterol into the cell membrane. Cholesterol analysis of macrophages showed that a-Chol represented 14.5% of all cholesterol in the macrophage samples (Fig. 4C). Macrophages loaded with a-Chol were exposed to either filtered air or 0.4 ppm ozone for 4 h and the formation of adducted proteins was assessed 1 h after ozone exposure. Click chemistry isolation of adducted proteins revealed that ozone exposure of BAL macrophages formed adducts on CD206 (Fig. 4D).
Effect of oxysterols on CD206 binding
The peptides that were identified by MS identified a SecoA adduction site on lysine 502, which is located in between the second and third of eight tandem c-type lectin-binding domains of CD206. These c-type lectin-binding domains (CTLD) are responsible for receptor binding to sugar moieties (20, 21). We hypothesized that oxysterol adduction of CD206 would thus lead to impaired binding function and consequently affect phagocytosis, similar to what is seen in ozone-exposed macrophages in vivo (11–13). To test this, we used a cell-free binding assay to measure the effect of oxysterols on CD206 binding. ELISA plates were coated with yeast-derived mannan, a known CD206 ligand, to “capture” CD206. Recombinant, His-tagged, human CD206 was treated with oxysterols before incubation on the coated plates, unbound CD206 was washed away, and bound CD206 was detected with an HRP-linked secondary antibody against the His tag. We found that treatment of recombinant human CD206 with increasing concentrations of SecoA and SecoB resulted in a dose-dependent impairment of binding to yeast-derived mannan (Fig. 4, E and F). Interestingly, SecoA reduced binding of CD206 to a greater degree than treatment with SecoB.
To determine whether decreased binding of CD206 following adduction with oxysterols resulted in biologically relevant changes in macrophage phagocytosis, we used a THP-1 cell culture model. Although BAL macrophages represent a more relevant cell model for lung macrophages, they are not conducive to genetic silencing/overexpression techniques. Undifferentiated THP-1 monocytes show low expression of CD206; the expression of CD206 can be induced with PMA and subsequent treatment with IL-4 and IL-13 (22), thus providing a convenient model to examine the role of CD206 in the oxysterol-dependent impairment of phagocytosis. Analysis of mRNA transcripts (Fig. 5A) and protein expression (Fig. 5B) confirmed low abundance of CD206 mRNA and protein in undifferentiated THP-1 cells and a robust increase in the expression of CD206 with IL-4 and IL-13 treatment.
Figure 5.
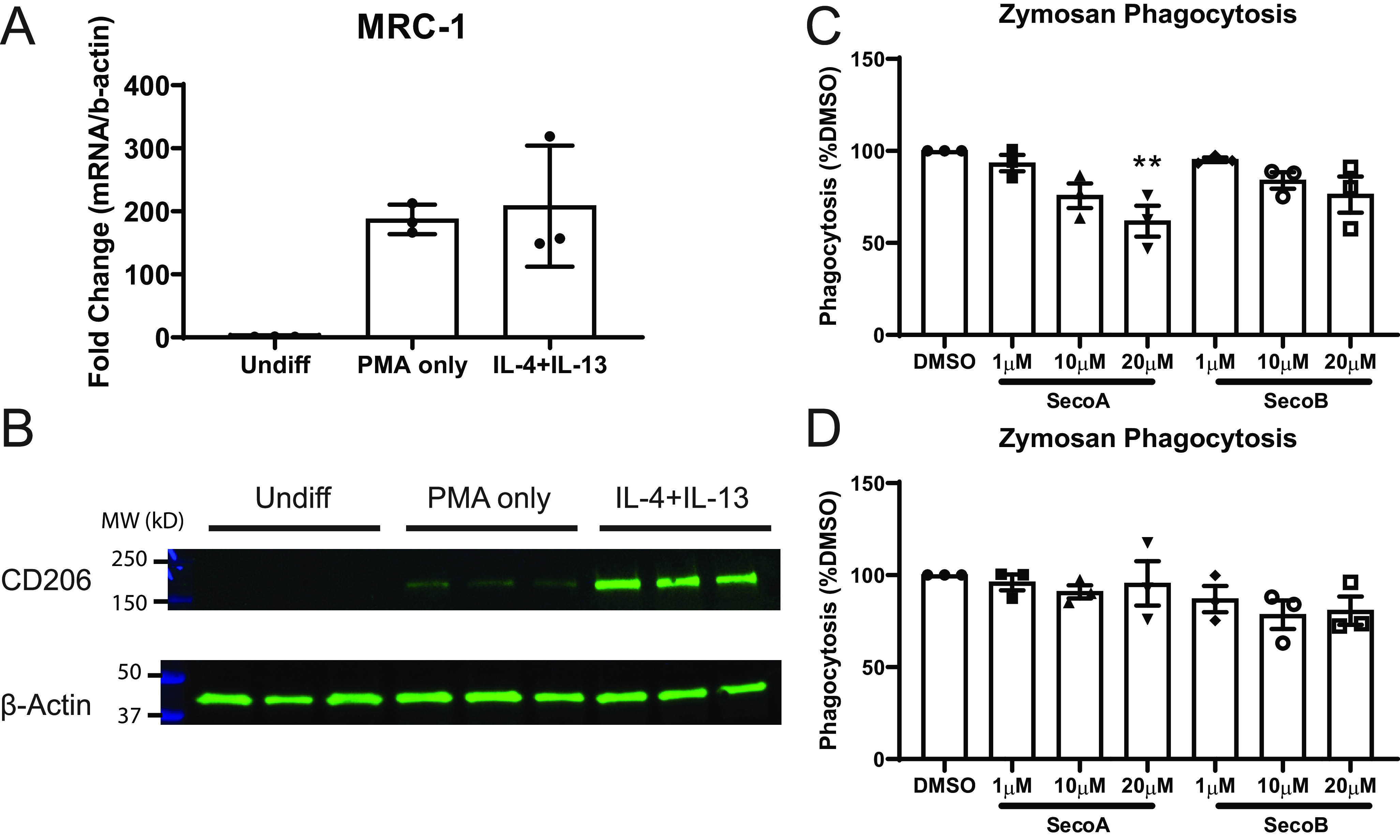
Oxysterols impair phagocytosis in CD206-expressing cells. THP-1 cells were differentiated into M2-like cells as described under “Materials and methods.” A and B, RNA (A) and protein (B) levels of CD206 were measured in undifferentiated, PMA-treated, and PMA+IL-4/IL-13–treated THP-1 cells. C and D, uptake of pH-sensitive zymosan bioparticles in (C) M2-like and (D) undifferentiated THP-1 cells. Bars for gene and protein expression represent the mean ± S.D. of three replicate wells. Bars for binding assays represent the mean ± S.E. of three independent experiments with quadruplicate wells per treatment. **, p < 0.01 by one-way ANOVA with Dunnett's multiple comparison test of treated groups to DMSO control.
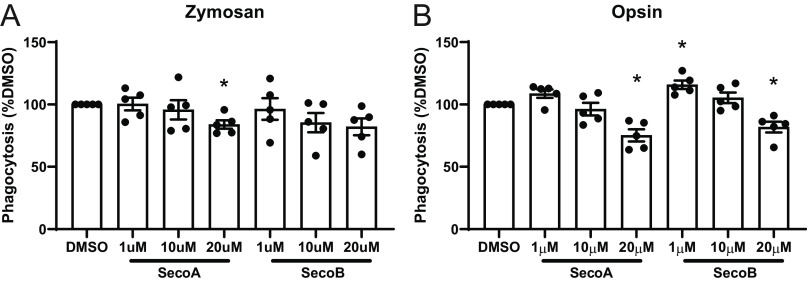
We used the IL-4/IL-13–differentiated THP-1 cells to investigate the effect of ozone-derived oxysterols on phagocytosis through CD206. Zymosan bioparticles contain mannose residues that can be recognized by CD206 (17). We cultured oxysterol-treated cells with pH-sensitive zymosan bioparticles which increase fluorescence following phagocytosis and the drop to acidic pH in the phagosome (23). We found that treatment of differentiated THP-1 cells with SecoA and SecoB led to decreases in uptake of zymosan bioparticles (Fig. 5C). Furthermore, oxysterol treatment of undifferentiated THP-1 cells did not affect uptake of zymosan particles suggesting the importance of CD206 adduction in the suppressed zymosan uptake (Fig. 5D).
Effect of oxysterol treatment on other phagocytic receptors
Macrophages express several different surface receptors that recognize different motifs of pathogens or dead and dying cells and aid in phagocytosis (24). We aimed to expand our findings to investigate whether the function of other phagocytic receptors is impaired by oxysterols. To do this, we investigated the effects of oxysterols on CD64 and CD16a. Unlike CD206, which recognizes mannose residues on the surface of proteins, CD64 and CD16a are two members of the Fc receptor family which function in binding and phagocytosing antibody-coated pathogens (25). Even though these two proteins were not initially identified in our proteomic screen, we conducted Western blot analysis on a-SecoA–treated BAL macrophages to determine whether CD64 and CD16a also form SecoA-protein adducts. Fig. 6A shows that SecoA forms adducts with CD64, whereas CD16a was not found to be adducted by SecoA (Fig. 6B). Cell-free binding assays showed that CD64 binding to its ligand, human IgG, was impaired with SecoA and SecoB treatment in a dose-dependent manner (Fig. 6C). In contrast, SecoA or SecoB treatment did not affect the ability of CD16a to bind human IgG (Fig. 6D), suggesting that not all surface receptors are equally affected by oxysterol adduction.
Figure 6.
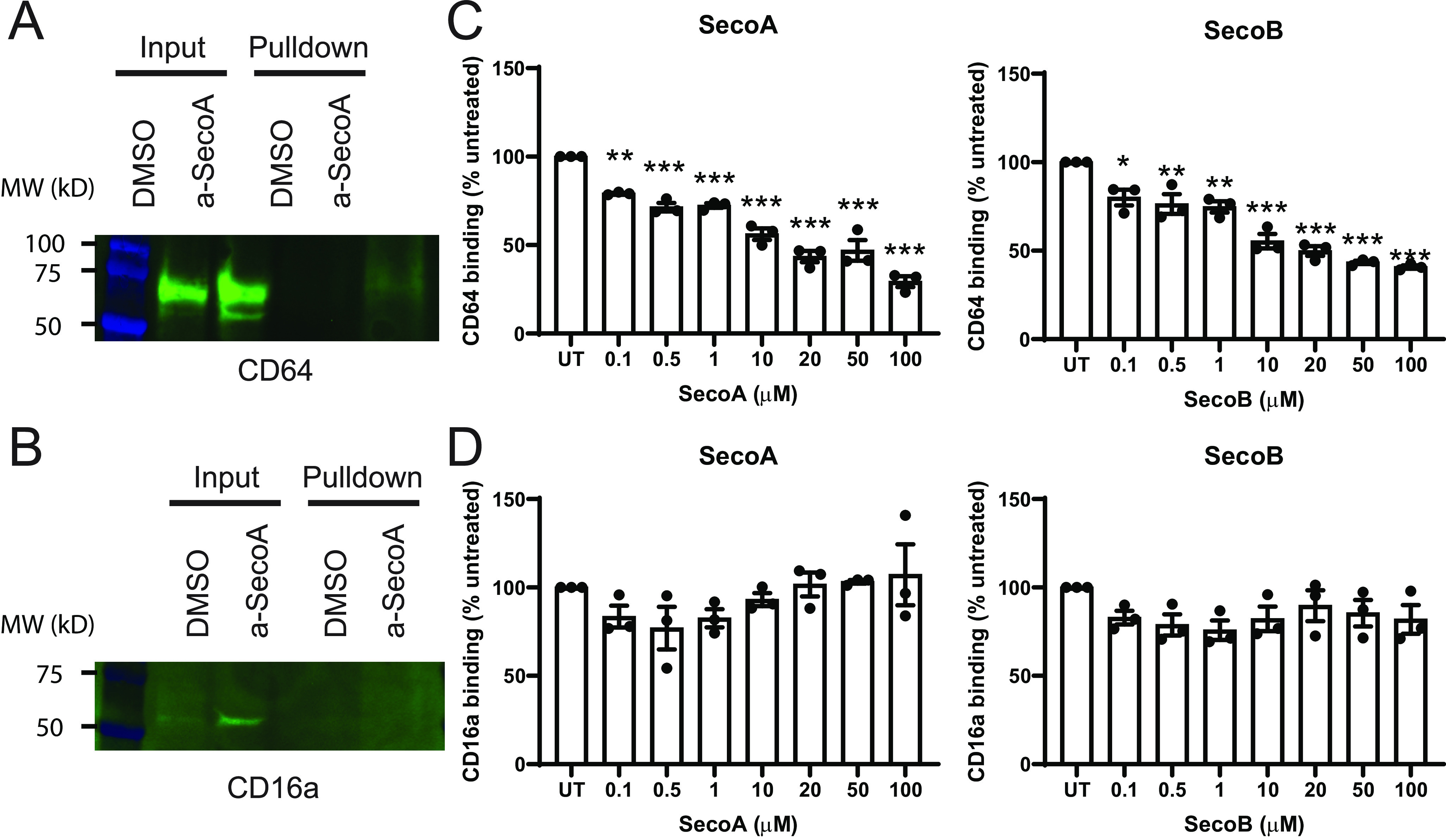
Oxysterols affect some, but not all, phagocytic receptors. BAL macrophages treated with vehicle or a-SecoA and adducts were isolated according to “Materials and methods.” A and B, isolated adducts were analyzed by Western blotting for (A) CD64 and (B) CD16a. C and D, the effect of oxysterol treatment on the binding of recombinant (C) CD64 and (D) CD16a to human IgG was tested by in house ELISA. Bars represent the mean (±) S.E. of three independent experiments with quadruplicate wells per treatment. *, p < 0.05; **, p < 0.01; ***, p < 0.001 by one-way ANOVA with Tukey post hoc test.
To determine the functional effect of oxysterol adduction, we investigated the uptake of both zymosan and antibody-coated bacterial bioparticles in primary human BAL macrophages. Similar to the THP-1 model, BAL macrophages that were treated with oxysterols showed decreased uptake of zymosan bioparticles (Fig. 7A). Incubation of BAL macrophages with SecoA or SecoB led to significant decreases in the uptake of antibody-coated bacterial particles (Fig. 7B), suggesting that oxysterols affect uptake antibody–coated particles even though binding of not all Fc receptors is affected.
Figure 7.
Oxysterols impair phagocytosis in human BAL macrophages. A and B, BAL macrophages were treated with the indicated concertation of oxysterol for 2 h before addition of pH-sensitive (A) zymosan or (B) IgG opsonized S. aureus bioparticles. Cells were incubated with particles for 4 h and then phagocytosis assessed by fluorescence. Each point represents the mean of values of triplicate wells from a single donor. Bars represent mean ± S.E. of five donors. Samples from each donor were considered matched values. *, p < 0.05 by one-way ANOVA with Dunnett's multiple comparison test.
Discussion
Ground-level ozone is one of the most common inhaled pollutants to which humans are exposed. Exposure to ozone has been linked to increases in inflammatory markers as well as increases in susceptibility to infection (6, 7). This is of particular concern to susceptible populations such as those with underlying lung diseases, like asthma, as decrease in respiratory host defense functions and increased susceptibility to infection can cause exacerbation of disease symptoms and lead to acute lung function decline (4, 26). Despite a wealth of human exposure data, the mechanisms linking ozone exposure to increased infectivity are not well known. Building on our previous studies demonstrating that ozone-derived oxysterols form protein adducts in lung epithelial cells and that the formation of these adducts significantly affects the function of these proteins (9), we set out to conduct an unbiased proteomic analysis to assess the adductome in BAL macrophages. Our data show that ozone-derived oxysterols form protein adducts with the surface receptor CD206 and that the adduction impaired CD206 function, which in turn was associated with decreased phagocytic function in these cells. Together, these data suggest that modification of surface receptors, such as CD206, by ozone-derived oxysterols could mediate the reduced phagocytic function of macrophages seen in the lung after exposure to ozone (12–14).
Much research has focused on the interaction of ozone with elements of the airway-lining fluid made up of phospholipids, proteins, and cholesterol as it is the first point of contact this reactive gas will encounter. Ozone exposure can lead to oxidation of phospholipids and surfactant proteins (27–30), which in turn can lead to detrimental effects on cell viability and host defense mechanisms. For example, neutrophils treated with the oxidized phospholipid 4HNE have impaired oxidative burst and impaired phagocytosis (31). Surfactant protein A exposed to ozone had impaired ability to stimulate inflammatory cytokines (32) and also impaired ability to stimulate the uptake of bacteria (33). Similarly, cholesterol can be oxidized by ozone to form many oxysterol species, including SecoA and SecoB (34–36). These oxysterol species are electrophilic and can attack lysine residues on proteins to form covalent oxysterol-protein adducts (9, 10).
We have previously shown that ozone-derived oxysterols form protein adducts in lung epithelial cells and that the formation of adducts on the liver X receptor leads to decreased liver X receptor function and increased inflammation (9). Although these previous studies were conducted to identify the role of oxysterol adduction of a specific protein in epithelial cells, it is unclear what the overall “oxysterol adductome” is and to what extent it affects the function of other lung cell types. Using alkynyl-tagged SecoA, click chemistry isolation technique, followed by MS proteomics (Fig. 1) we identified over 140 proteins that formed adducts with a-SecoA (Table S2). Adducted proteins were involved in a number of various cellular functions (Fig. 3), suggesting that the impact of oxysterol adduction likely has multiple functional effects on macrophages beyond the phagocytic functions analyzed in more detail here. However, based on our data, ozone-derived oxysterol adduction is likely to have a significant impact on macrophage surface receptor function.
Our proteomic analysis identified individual adducted lysine residues by the loss of a 512 m/z fragment diagnostic of a-SecoA/B adduction with b and y ions identifying the specific lysine site of adduction. Sequence motif analysis of adducted peptides found that acidic residues, like glutamate and aspartate, were overrepresented in positions −2 to +2 from adducted lysine residues as well as sites five and six residues away on either side of the adducted lysine (Fig. 3C). The first step in protein adduction of SecoA is imine formation on a lysine side-chain, a process that is generally subject to acid catalysis (37) (Fig. 2B). This suggests that acidic residues shown in the sequence motif play an essential catalytic role in the adduction process. Nearest neighbor side-chain lysine aspartate or lysine glutamate interactions are not unexpected, providing a structure framework for catalysis, but catalytic activity for acidic groups at the more remote sites five and six residues away from the reactive lysine is more difficult to rationalize. It seems reasonable to suggest that protein secondary structure plays a role in catalysis by remote acid side-chains, with helix structures bringing acid side-chains on remote residues into proximity with the reactive lysine. Unwinding the normal helix 3.6 residue repeat unit to accommodate sterol binding in a hydrophobic helix core and also bring remote acidic groups into the catalytic sphere would account for the preference for aspartate and glutamate residues at the remote sites. In this regard, it is noteworthy that proline is significantly underrepresented throughout the motif, suggesting that secondary structure (helix or sheet) surrounding the reactive lysine does favor adduct formation.
We were interested to find CD206 as one of the proteins identified in our proteomic screen of the oxysterol adductome as it is known to bind pathogens such as Candida albicans (16), Pneumocystis carinii (15), and Mycobacterium tuberculosis (38, 39). CD206 contains eight CTLD, however, intact receptor binding can be mimicked with only CTLD4–8 (21, 40). We identified adduction on lysine 502, which falls between CTLD2 and 3. Although these domains are not associated with strong carbohydrate binding, adduction could lead to secondary structure changes that would result in impairment with ligand interaction. It is also possible that CD206 is adducted at additional sites that were not identified in the proteomic screen but are capable of impacting ligand binding. After confirming the CD206-oxysterol adduction by Western blotting, we determined the functional consequences of this protein modification. Our results demonstrated that oxysterol treatment impairs the binding of recombinant CD206 to a known ligand, mannan (Fig. 4), and showed decreased uptake of zymosan particles in oxysterol-treated macrophages (Figs. 5 and 7). Interestingly, SecoA was more potent in reducing CD206 binding to mannan than SecoB, suggesting that oxysterols have varying impacts on protein function. Indeed, we have previously found that a-SecoA and a-SecoB form adducts more readily than other alkynyl-tagged ozone-derived oxysterols (10). Taken together, our data show that SecoA adduction of CD206 leads to impaired receptor function and consequently reduced macrophage phagocytosis.
It is unknown if CD206 adduction occurs in vivo and the levels of ozone-derived oxysterol levels in the lung fluid following ozone exposure is unclear. We have previously shown that oxysterol levels are increased in vitro and humans in vivo after ozone exposure (9); however, the measured values are underestimates as the process of obtaining lung lavage results in significant dilution of the lung-lining fluid components. Quantification of free oxysterols is hampered by the reactive nature of free SecoA/B and its high propensity to form protein adducts. Technical limitations prevent isolating and identifying neutral lipid-adducted proteins (with no alkynyl tag) by mass spectrometry so the extent of oxysterol adduction in vivo is not known. This limits the ability of our findings to be directly compared with human ozone exposure. To address the relevance of our findings, we performed click chemistry isolation on macrophages loaded with a-Chol followed by ozone exposure in vitro, which determines whether endogenously formed oxysterols can form protein adducts. Our data indicate that CD206 was adducted, albeit to a much lower extent, in response to ozone exposure (Fig. 4D). This mirrors our previous data from similar experiments performed in lung epithelial cells (9). The reduced abundance of CD206 adducts detected is likely because of the limited incorporation (14.5% of all cholesterol) of a-Chol into the macrophage samples leading to limited formation of endogenous alkynyl-tagged oxysterols and, thus, fewer detectable protein adducts. The captured pool of adducted proteins from this experiment is an underrepresentation of the total adducted proteins because those proteins adducted by ozone-derived oxysterols generated from untagged cholesterol would not be detected. Despite these limitations, CD206 adducts were still detectable, supporting the hypothesis that oxysterol-CD206 adducts are formed in vivo following ozone exposure.
Although the effect of SecoA/B on CD206 binding was striking, macrophages contain many receptors to aid in recognizing and phagocytosing pathogen-specific motifs such as terminal sugar moieties, lipoproteins, and opsonized particles. We expanded our investigation and investigated the role of oxysterol adduction and modification of the Fc receptor family members, CD64 and CD16a, which recognize the constant region of antibodies and aid in macrophage recognition of antibody-coated particles. Both CD64 and CD16a are expressed on macrophages and have high and low affinity for binding IgG, respectively (41). Similar to CD206, we found that CD64 was adducted by a-SecoA and that binding of CD64 was impaired with oxysterols (Fig. 6, A and C). Interestingly, CD16a was not identified to be adducted by oxysterols (Fig. 6B) and the binding of recombinant human CD16a to human IgG was unaffected by oxysterols (Fig. 6D), indicating that not all endocytic receptors have the same susceptibility to forming adducts with oxysterols.
As indicated above, our data show that SecoA/B adduction depends on the proximity of acidic residues surrounding the target lysine residue (Fig. 3C). Theoretical analysis of tryptic peptides generated from CD64 show that a few peptides match the proximate acid residue criteria, which is consistent with Western blot observations and impaired binding function. Interestingly, no CD64 peptides were found in the MS analysis. This may be because of the relatively low expression of CD64 compared with other macrophage proteins, modification of only a subset of CD64 molecules, or that SecoA/B-modified peptides were not suitable for detection by MS (too long or too hydrophobic). In contrast, CD16 does not contain theoretical tryptic peptides that meet the proximate acid residue sequence motif characterization that we identified, potentially explaining the lack of SecoA adducts found and the lack of functional defects in receptor binding (Fig. 6, B and D). Overall, the uptake of antibody-coated particles was impaired with oxysterol treatment (Fig. 7B) which shows that even though CD16a is unaffected by oxysterols, impairment of CD64 (and potentially other Fc receptors) is sufficient to impair the phagocytic function of macrophages.
It is important to state that although oxysterols did reduce macrophage phagocytosis, the suppression was not a complete inhibition. The adduction of SecoA is reversible in nature and adduct levels are likely to decline after removal of excess free oxysterol (42). Given this, it is unclear how much of the total available proteins are actually adducted following ozone exposure, but it is likely that only a fraction of susceptible phagocytic receptors form oxysterol adducts in vivo. In addition, cells can activate responses, such as the unfolded protein response and autophagy, which reduce and remove adduct-damaged proteins. The stability and longevity of SecoA-adducted proteins remains an active area of research. Lastly, the redundancy in ligand recognition between multiple endocytic receptors may explain why the endocytosis of mannan and opsonized Staphylococcus aureus particles was not completely eliminated with oxysterol treatment. This notion is supported by the fact that although CD64 was adducted by oxysterols and had impaired binding to IgG, CD16a, which also binds IgG, was unaffected.
Although our study focuses on the importance of cholesterol-derived ozone products, ozone also reacts with other components of the lung-lining fluid. Previous reports have shown that oxidized phospholipids, like 4HNE, can form protein adducts and have shown to disrupt oxidative burst involved in neutrophil phagocytosis (31, 43). However, unlike the other phospholipid-derived species which can attack arginine, lysine, and cysteine residues, SecoA and SecoB lack a Michael acceptor. As a result, adducts from SecoA and SecoB are formed exclusively on lysine residues. Thus, the impact of ozone exposure in the lung is a combination of ozone-induced oxidation of cholesterol, proteins, and other phospholipid species present in the lung-lining fluid. Further understanding of the combined role of oxidized proteins, oxidized lipid adducts, and oxysterol adducts on driving biological effects in macrophages will enhance the understanding of ozone-induced immune suppression in macrophages.
Our study is the first to definitively show that ozone-derived oxysterols form adducts with phagocytic receptors in human lung macrophages and that these effects correlate with functional defects in macrophage phagocytosis. This novel mechanism could be broadly applicable to the uptake of other cargo or the function of other surface receptors. Macrophages can recognize numerous cargo, including bacterial-derived lipoproteins, oxidized cholesterol, other sugar moieties, and damage associated signals from dead and dying cells. Adduction of surface receptors involved in the uptake of these ligands may be similarly affected by oxysterol adduction. This mechanism can help explain how the inhalation of ozone and subsequent formation of oxidized lipids leads to reduced phagocytosis in macrophages and the increased susceptibility to infection in the lung. What determines susceptibility to electrophilic attack of ozone-derived oxysterols on lysine residue remains unclear but our analysis suggests that acidic amino acid residues surrounding a lysine residue may enhance susceptibility to adduction. Other factors, like number of lysine residues and accessibility of the lysine residue to the exposed protein surface where it can interact with exogenous oxysterol may also be important. Further investigation combining identification of susceptible lysine residues with protein structural and functional information could lead to a predictive model identifying proteins and cellular functions affected by ozone-derived oxysterols beyond the targets shown here and in our previous studies (9).
Materials and methods
Cells and culture
THP-1 monocytes were kindly provided by Dr. Tim Moran (UNC Chapel Hill, NC) and were cultured in RPMI media (Gibco) supplemented with 10% fetal bovine serum, 1% l-glutamine, 1% penicillin-streptomycin (Life Technologies), 1% HEPES, and 0.1% β-mercaptoethanol (Gibco). THP-1 cells were differentiated to an M2-like phenotype essentially as described (22). Cells were incubated with 150 ng/ml PMA (Sigma) for 24 h and then allowed to recover in full-growth media for an additional 24 h before treatment with 20 ng/ml of both recombinant human IL-4 and IL-13 (R&D Systems) for 72 h. Primary human airway macrophages were obtained from the lung BAL of healthy volunteers in collaboration with the United States Environmental Protection Agency (EPA) using a protocol approved by the University of North Carolina at Chapel Hill Institutional Review Board (Chapel Hill, NC), as described previously (44) and that abides by the Declaration of Helsinki principles. BAL cells were cultured in RPMI media with 10% fetal bovine serum, 1% l-glutamine, 1% penicillin-streptomycin and allowed to adhere to tissue culture dishes for 1–2 h prior to alkyne tag treatment.
Cell labeling with a-SecoA and click chemistry isolation of adducts
a-SecoA was synthesized following the procedures published elsewhere (10, 45). Adducted proteins were identified as described previously (9). Essentially, primary human BAL macrophages from three individual donors were treated with vehicle or 20 μm a-SecoA in RPMI 1640 supplemented with 2% FBS for 4 h. Our previous study of oxysterol adducts in epithelial cells (9) showed that this time point and dose of a-SecoA allowed for sufficient adduction for reliable identification of adducted proteins without affecting cell viability. Cells were then lysed in 50 mm HEPES, 150 mm NaCl, 0.1% Triton X-100, 0.1% SDS in the presence of protease inhibitors. Lysates from the three donors were pooled and adducts were stabilized with the addition of NaBH4 (5 mm final concentration) on ice for 1 h to stabilize the Schiff base adducts before being reacted with azido-biotin. Adduction was confirmed by Western blotting and probing with IRDye 800CW streptavidin (925-32230, Li-Cor Biosciences, Lincoln, NE) and β-actin (Abcam, ab8299). Adducted proteins were isolated with streptavidin beads (GE Healthcare) and released from the beads with long wave UV light (365 nm). Isolated protein was loaded on a 10% NuPAGE Novex BisTris precast mini gel (Invitrogen) and run for 5 min at 200 V in MES buffer, stained with Simply Blue (Bio-Rad). Bands were excised for in-gel trypsin digestion for MS analysis. Briefly, the excised gels were reduced and alkylated with DTT and iodoacetamide, respectively. They were then sequentially dehydrated with 50% acetonitrile and 80 μl of 25 mm ammonium bicarbonate containing 1 μg trypsin was added (Promega V5111, Madison, WI) before proteins were identified by MS.
MS analysis
Trypsinized peptides were loaded onto a self-packed biphasic C18/SCX MudPIT column using a helium-pressurized cell (pressure bomb). The MudPIT column consisted of 360 μm outer diameter × 150 μm inner diameter fused silica, which was fitted with a filter-end fitting (IDEX Health & Science) and packed with 6 cm of Phenomenex Luna® SCX material (04A-4398, 5 μm, 100 Å) followed by 4 cm of Phenomenex Jupiter® C18 material (04A-4053, 5 μm, 300 Å). Once the sample was loaded, the MudPIT column was connected using an Inline MicroFilter union (M-520, IDEX Health & Science) to an analytical column (360 μm, outer diameter × 100 μm, inner diameter), equipped with a laser-pulled emitter tip and packed with 20 cm of C18 reverse phase material (Phenomenex Jupiter®, 3 μm, 300 Å, Phenomenex). Using a Dionex Ultimate 3000 RSLCNano and Autosampler, MudPIT analysis was performed with an 8-step salt pulse gradient (25, 50, 100, 200, 300, 500, 750 mm, and 1M ammonium acetate). Following each salt pulse, peptides were gradient-eluted from the reverse analytical column at a flow rate of 350 nl/min, with the mobile phase solvents consisting of 0.1% formic acid, 99.9% water (solvent A) and 0.1% formic acid, 99.9% acetonitrile (solvent B). For the peptides from the first eight SCX fractions, the reverse phase gradient consisted of 0–1 min 2% B → 5% B for 5 min → 60% B for 64 min → 99% B for 10 min → 99% B for 5 min followed by an equilibration at 2% B. Peptides were introduced via nano-electrospray into a Q ExactiveTM Plus hybrid Quadrupole-Orbitrap mass spectrometer (Thermo Scientific). The Q ExactiveTM was operated in the data-dependent mode acquiring HCD MS/MS scans (r = 17,500) after each MS1 scan (r = 70,000) on the 15 most abundant ions using an MS1 ion target of 3 × 106 ions and an MS2 target of 5 × 104 ions. The maximum ion time for MS/MS scans was set to 40 ms, the HCD-normalized collision energy was set to 27, dynamic exclusion was set to 30 s, and peptide match and isotope exclusion were enabled.
Protein and peptide identification
Thermo.RAW datafiles from data-dependent mode were converted to an mzML format using ProteoWizard (version 3.0.5211) (46, 47). The mzML files were searched using MS-GF+beta (version 9517) (48) against humanRefSeq_Version54 database (34,589 proteins). MS-GF+beta was configured allowing semi-tryptic search with maximum three missed cleavages. A fixed carbamidomethyl modification on Cys (+57.0215 Da) and three dynamic modifications on Met oxidation (+15.9949 Da), N-terminal Gln deamidation (−17.0265 Da), and Lys-aSA adduct (+541.3880 Da, aSA-triazole-hexanoic acid modification; C32H51N3O4) were allowed per peptide with a precursor tolerance 10 ppm and a fragment ion tolerance 20 ppm. Protein identifications required a minimum of two distinct identified peptides, two minimum spectra, and one additional peptide per protein. Maximum protein level false discovery rate was 1% calculated by diving the number of reverse sequence proteins identified by the total number of protein identified, multiplying by two and converting to a percentage. For protein assembly and peptide visualization, IDPicker (3.1.9288 64-bit) (49) was used. Detailed analysis view and all aSA (+541.3880) modified peptides and their protein ID and site of modification are included in Table S3, A–C. For the ambiguous assignment for the aSA modified peptides, the identified spectra (Table S3C) are manually inspected following the criteria: 1) the modified peptide precursor error within 10 ppm and 2) the modified peptides containing diagnostic fragment m/z 512. The final refined adducted peptides and their proteins are listed in Tables S1 and S2, respectively. The ambiguously identified proteins were used for functional and component analyses using Webgestalt (WEB-based GEne SeT AnaLysis Toolkit) (50, 51). The modified peptide sequence motif image was generated with pLogo (52).
All human bronchoalveolar lavage macrophage proteomics data are available via ProteomeXchange consortium via the PRIDE partner repository (53) with the dataset identifier PXD019317 and 10.6019/PXD019317. The list of peptides with known sites of modification and their proteins are provided in Tables S1, S2, and S3 (A–C).
In vitro ozone exposure
200,000 primary human BAL macrophages were plated in 12-well culture inserts (Corning) and allowed to adhere for 3 h before treatment with a-Chol (20 μm)–supplemented media for 48 h, at which time three inserts were harvested to quantify a-Chol levels in the total cholesterol pool as described (9). BAL macrophages were brought to the air–liquid interface immediately prior to ozone exposure and exposed to air or 0.4 ppm ozone for 4 h, as described previously (54, 55). Cells were harvested 1 h after air/ozone exposure and samples pooled for catch and release to confirm adducts as described above.
Phagocytosis assay
Analysis of macrophage phagocytosis was done as described (23). Briefly, M2-differentiated THP-1 cells (50,000 cells/well) or primary BAL macrophages (30,000 cells/well) were allowed to adhere on black-walled 96-well tissue culture plates (Corning) as described above. Cells were treated with 200 μl of the indicated concentrations of oxysterols in RPMI media supplemented with 2% FBS for 2 h. pH rhodo bioparticles from S. aureus, or pH rhodo zymosan particles (Thermo Fisher Scientific) were prepared according to the manufacturer's instructions. S. aureus particles were opsonized using bioparticle opsonizing reagent (Thermo Fisher Scientific) according to the manufacturer's protocol. After oxysterol treatment, 50 μl of bioparticles were added to each well and incubated for 4 h. Phagocytosis of bioparticles was quantified using a CLARIOstar fluorescent microplate reader (BMG Labtech, Offenburg, Germany) by assessing fluorescence emission at 585 nm.
Cell-free binding assays
96-well plates were coated with 10 μg/ml of mannan (for CD206, Sigma), or human IgG (1 μg/ml for CD64 or 2 μg/ml for CD16a) overnight in ELISA coating buffer (BioLegend). Plates were blocked with 5% BSA in Tris-buffered saline. Human recombinant CD206 (0.5 μg/ml), CD64 (125 ng/ml), or CD16a (1.5 μg/ml) with His6 tag (all from R&D Systems) were prepared in Tris-buffered saline and treated with the indicated concentrations on oxysterol at 37°C for 1 h before addition to the plate. Coated plates were washed three times with TBST and recombinant protein samples were allowed to bind for 2 h. Unbound protein was removed by washing three times with TBST and bound protein detected with mouse anti–His6-HRP secondary antibody (BioLegend) for 30 min and development with TMB solution (BD Biosciences). For the CD206 binding assay, all buffers were supplemented with 10 mm CaCl2•2H2O.
Western blotting
THP-1 cells were lysed in RIPA buffer (50 mm Tris HCl, 150 mm NaCl, 2 mm EDTA, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS) and 10 μg separated on 4–20% acrylamide gels (Bio-Rad) and transferred onto nitrocellulose membranes. Blots were probed with rabbit-anti CD206 (Cell Signaling Technology, 91992) and mouse-anti actin (Santa Cruz Biotechnology, sc47778). For Western blot confirmation of click chemistry adducts, isolated proteins following the streptavidin pulldown were separated on an acrylamide gel and transferred to a nitrocellulose membrane. The membranes were probed with rabbit-anti CD206 (Cell Signaling Technology, 91992), anti CD64 (Abcam, ab134073) or CD16a (Cell Signaling Technology, 19731). Bands were detected with an HRP conjugated anti-rabbit antibody (Santa Cruz Biotechnology, sc2357) or anti-mouse binding protein (Santa Cruz Biotechnology, sc516102), and detected with chemiluminescence. When necessary, the brightness and contrast of the Western blot image was adjusted uniformly across the image to highlight the difference between treatment groups.
Statistical analysis
Unless otherwise indicated, in vitro experiments were performed in triplicate wells and were repeated in at least three separate experiments. Samples from individual donors were considered matched values. Significant differences were determined by one-way ANOVA with Dunnett's multiple comparison of oxysterol-treated groups to vehicle-treated controls with a p < 0.05 threshold. See figure legends for additional information on statistical analyses.
Data Availability
Mass Spectrometry data are available through the ProteomeXchange consortium via the PRIDE partner repository with identifier PXD019317. All other data are contained within the paper.
Supplementary Material
Acknowledgments
We thank Joleen Soukup and Dr. Andy Ghio for help in obtaining the primary human BAL macrophages.
This article contains supporting information.
Author contributions—P. F. D., H.-Y. H. K., N. A. P., and I. J, conceptualization; P. F. D. and H.-Y. H. K. data curation; P. F. D., H.-Y. H. K., N. A. P., and I. J. formal analysis; P. F. D. and H.-Y. H. K. investigation; P. F. D., H.-Y. H. K., and N. A. P. methodology; P. F. D. writing-original draft; P. F. D., H.-Y. H. K., N. A. P., and I. J. writing-review and editing; H.-Y. H. K. validation; N. A. P. and I. J. funding acquisition; I. J. supervision.
Funding and additional information—This work was supported in part by the United States Environmental Protection Agency through cooperative agreement CR83346301 with the Center for Environmental Medicine, Asthma and Lung Biology at the University of North Carolina at Chapel Hill (to I. J.) and National Institutes of Health Grants T32ES007126 (to P. F. D.) and R01ES028269 (to H.-Y. H. K., N. A. P., and I. J.). The views expressed in this article are those of the authors and do not necessarily reflect the position or policy of the Environmental Protection Agency. The EPA does not endorse any products or commercial services mentioned in this publication. This article has not been formally reviewed by the EPA. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- SecoA
- secosterol A
- SecoB
- secosterol B
- BAL
- bronchoalveolar lavage
- a-SecoA
- alkynyl-tagged SecoA
- a-SecoB
- alkynyl-tagged SecoB
- a-Chol
- alkynyl-tagged cholesterol
- CTLD
- c-type lectin-binding domains
- PMA
- phorbol 12-myristate 13-acetate
- HCD
- higher energy collisional dissociation
- TBST
- Tris-buffered saline with Tween 20
- ANOVA
- analysis of variance.
References
- 1. Koren H. S., Devlin R. B., Graham D. E., Mann R., McGee M. P., Horstman D. H., Kozumbo W. J., Becker S., House D. E., and McDonnell W. F. (1989) Ozone-induced inflammation in the lower airways of human subjects. Am. Rev. Respir. Dis. 139, 407–415 10.1164/ajrccm/139.2.407 [DOI] [PubMed] [Google Scholar]
- 2. Graham D. E., and Koren H. S. (1990) Biomarkers of inflammation in ozone-exposed humans. Comparison of the nasal and bronchoalveolar lavage. Am. Rev. Respir. Dis. 142, 152–156 10.1164/ajrccm/142.1.152 [DOI] [PubMed] [Google Scholar]
- 3. Aris R. M., Christian D., Hearne P. Q., Kerr K., Finkbeiner W. E., and Balmes J. R. (1993) Ozone-induced airway inflammation in human subjects as determined by airway lavage and biopsy. Am. Rev. Respir. Dis. 148, 1363–1372 10.1164/ajrccm/148.5.1363 [DOI] [PubMed] [Google Scholar]
- 4. White M. C., Etzel R. A., Wilcox W. D., and Lloyd C. (1994) Exacerbations of childhood asthma and ozone pollution in Atlanta. Environ. Res. 65, 56–68 10.1006/enrs.1994.1021 [DOI] [PubMed] [Google Scholar]
- 5. Farhat S. C. L., Almeida M. B., Silva-Filho L., Farhat J., Rodrigues J. C., and Braga A. L. F. (2013) Ozone is associated with an increased risk of respiratory exacerbations in patients with cystic fibrosis. Chest 144, 1186–1192 10.1378/chest.12-2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kesic M. J., Meyer M., Bauer R., and Jaspers I. (2012) Exposure to ozone modulates human airway protease/antiprotease balance contributing to increased influenza A infection. PLoS One 7, e35108 10.1371/journal.pone.0035108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Purvis M. R., Miller S., and Ehrlich R. (1961) Effect of atmospheric pollutants on susceptibility to respiratory infection. I. Effect of ozone. J. Infect. Dis. 109, 238–242 10.1093/infdis/109.3.238 [DOI] [PubMed] [Google Scholar]
- 8. Uppu R. M., Cueto R., Squadrito G. L., and Pryor W. A. (1995) What does ozone react with at the air/lung interface? Model studies using human red blood cell membranes. Arch. Biochem. Biophys. 319, 257–266 10.1006/abbi.1995.1290 [DOI] [PubMed] [Google Scholar]
- 9. Speen A. M., Kim H. H., Bauer R. N., Meyer M., Gowdy K. M., Fessler M. B., Duncan K. E., Liu W., Porter N. A., and Jaspers I. (2016) Ozone-derived oxysterols affect liver X receptor (LXR) signaling: A potential role for lipid-protein adducts. J. Biol. Chem. 291, 25192–25206 10.1074/jbc.M116.732362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Windsor K., Genaro-Mattos T. C., Miyamoto S., Stec D. F., Kim H. Y., Tallman K. A., and Porter N. A. (2014) Assay of protein and peptide adducts of cholesterol ozonolysis products by hydrophobic and click enrichment methods. Chem. Res. Toxicol. 27, 1757–1768 10.1021/tx500229h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Devlin R. B., McDonnell W. F., Mann R., Becker S., House D. E., Schreinemachers D., and Koren H. S. (1991) Exposure of humans to ambient levels of ozone for 6.6 hours causes cellular and biochemical changes in the lung. Am. J. Respir. Cell Mol. Biol. 4, 72–81 10.1165/ajrcmb/4.1.72 [DOI] [PubMed] [Google Scholar]
- 12. Canning B. J., Hmieleski R. R., Spannhake E. W., and Jakab G. J. (1991) Ozone reduces murine alveolar and peritoneal macrophage phagocytosis: The role of prostanoids. Am. J. Physiol. 261, L277–L282 10.1152/ajplung.1991.261.4.L277 [DOI] [PubMed] [Google Scholar]
- 13. Driscoll K. E., Vollmuth T. A., and Schlesinger R. B. (1987) Acute and subchronic ozone inhalation in the rabbit: Response of alveolar macrophages. J. Toxicol. Environ. Health 21, 27–43 10.1080/15287398709531000 [DOI] [PubMed] [Google Scholar]
- 14. Jakab G. J., and Hemenway D. R. (1994) Concomitant exposure to carbon black particulates enhances ozone-induced lung inflammation and suppression of alveolar macrophage phagocytosis. J. Toxicol. Environ. Health 41, 221–231 10.1080/15287399409531838 [DOI] [PubMed] [Google Scholar]
- 15. O'Riordan D. M., Standing J. E., and Limper A. H. (1995) Pneumocystis carinii glycoprotein A binds macrophage mannose receptors. Infect. Immun. 63, 779–784 10.1128/IAI.63.3.779-784.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marodi L., Korchak H. M., and Johnston R. B. Jr. (1991) Mechanisms of host defense against Candida species. I. Phagocytosis by monocytes and monocyte-derived macrophages. J. Immunol. 146, 2783–2789 [PubMed] [Google Scholar]
- 17. Astarie-Dequeker C., N'Diaye E.-N., Le Cabec V., Rittig M. G., Prandi J., and Maridonneau-Parini I. (1999) The mannose receptor mediates uptake of pathogenic and nonpathogenic mycobacteria and bypasses bactericidal responses in human macrophages. Infect. Immun. 67, 469–477 10.1128/IAI.67.2.469-477.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Martinez-Pomares L. (2012) The mannose receptor. J. Leukoc. Biol. 92, 1177–1186 10.1189/jlb.0512231 [DOI] [PubMed] [Google Scholar]
- 19. Becker S., Madden M. C., Newman S. L., Devlin R. B., and Koren H. S. (1991) Modulation of human alveolar macrophage properties by ozone exposure in vitro. Toxicol. Appl. Pharmacol. 110, 403–415 10.1016/0041-008X(91)90042-D [DOI] [PubMed] [Google Scholar]
- 20. Gazi U., and Martinez-Pomares L. (2009) Influence of the mannose receptor in host immune responses. Immunobiology 214, 554–561 10.1016/j.imbio.2008.11.004 [DOI] [PubMed] [Google Scholar]
- 21. Taylor M. E., Bezouska K., and Drickamer K. (1992) Contribution to ligand binding by multiple carbohydrate-recognition domains in the macrophage mannose receptor. J. Biol. Chem. 267, 1719–1726 [PubMed] [Google Scholar]
- 22. Genin M., Clement F., Fattaccioli A., Raes M., and Michiels C. (2015) M1 and M2 macrophages derived from THP-1 cells differentially modulate the response of cancer cells to etoposide. BMC Cancer 15, 577 10.1186/s12885-015-1546-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Clapp P. W., Pawlak E. A., Lackey J. T., Keating J. E., Reeber S. L., Glish G. L., and Jaspers I. (2017) Flavored e-cigarette liquids and cinnamaldehyde impair respiratory innate immune cell function. Am. J. Physiol. Lung Cell. Mol. Physiol. 313, L278–L292 10.1152/ajplung.00452.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gordon S. (2016) Phagocytosis: An immunobiologic process. Immunity 44, 463–475 10.1016/j.immuni.2016.02.026 [DOI] [PubMed] [Google Scholar]
- 25. Ravetch J. V., and Kinet J. P. (1991) Fc receptors. Annu. Rev. Immunol. 9, 457–492 10.1146/annurev.iy.09.040191.002325 [DOI] [PubMed] [Google Scholar]
- 26. Kreit J. W., Gross K. B., Moore T. B., Lorenzen T. J., D'Arcy J., and Eschenbacher W. L. (1989) Ozone-induced changes in pulmonary function and bronchial responsiveness in asthmatics. J. Appl. Physiol. 66, 217–222 10.1152/jappl.1989.66.1.217 [DOI] [PubMed] [Google Scholar]
- 27. Santrock J., Gorski R. A., and O'Gara J. F. (1992) Products and mechanism of the reaction of ozone with phospholipids in unilamellar phospholipid vesicles. Chem. Res. Toxicol. 5, 134–141 10.1021/tx00025a023 [DOI] [PubMed] [Google Scholar]
- 28. Uhlson C., Harrison K., Allen C. B., Ahmad S., White C. W., and Murphy R. C. (2002) Oxidized phospholipids derived from ozone-treated lung surfactant extract reduce macrophage and epithelial cell viability. Chem. Res. Toxicol. 15, 896–906 10.1021/tx010183i [DOI] [PubMed] [Google Scholar]
- 29. Oosting R. S., van Greevenbroek M. M., Verhoef J., van Golde L. M., and Haagsman H. P. (1991) Structural and functional changes of surfactant protein A induced by ozone. Am. J. Physiol. 261, L77–L83 10.1152/ajplung.1991.261.2.L77 [DOI] [PubMed] [Google Scholar]
- 30. Wang G., Bates-Kenney S. R., Tao J. Q., Phelps D. S., and Floros J. (2004) Differences in biochemical properties and in biological function between human SP-A1 and SP-A2 variants, and the impact of ozone-induced oxidation. Biochemistry 43, 4227–4239 10.1021/bi036023i [DOI] [PubMed] [Google Scholar]
- 31. Chacko B. K., Wall S. B., Kramer P. A., Ravi S., Mitchell T., Johnson M. S., Wilson L., Barnes S., Landar A., and Darley-Usmar V. M. (2016) Pleiotropic effects of 4-hydroxynonenal on oxidative burst and phagocytosis in neutrophils. Redox Biol. 9, 57–66 10.1016/j.redox.2016.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang G., Umstead T. M., Phelps D. S., Al-Mondhiry H., and Floros J. (2002) The effect of ozone exposure on the ability of human surfactant protein a variants to stimulate cytokine production. Environ. Health Perspect. 110, 79–84 10.1289/ehp.0211079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mikerov A. N., Umstead T. M., Gan X., Huang W., Guo X., Wang G., Phelps D. S., and Floros J. (2008) Impact of ozone exposure on the phagocytic activity of human surfactant protein A (SP-A) and SP-A variants. Am. J. Physiol. Lung Cell. Mol. Physiol. 294, L121–L130 10.1152/ajplung.00288.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Murphy R. C., and Johnson K. M. (2008) Cholesterol, reactive oxygen species, and the formation of biologically active mediators. J. Biol. Chem. 283, 15521–15525 10.1074/jbc.R700049200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pulfer M. K., Taube C., Gelfand E., and Murphy R. C. (2005) Ozone exposure in vivo and formation of biologically active oxysterols in the lung. J. Pharmacol. Exp. Ther. 312, 256–264 10.1124/jpet.104.073437 [DOI] [PubMed] [Google Scholar]
- 36. Pulfer M. K., and Murphy R. C. (2004) Formation of biologically active oxysterols during ozonolysis of cholesterol present in lung surfactant. J. Biol. Chem. 279, 26331–26338 10.1074/jbc.M403581200 [DOI] [PubMed] [Google Scholar]
- 37. Lowry T. H., and Richardson K. S. (1987) Mechanism and Theory in Organic Chemistry, 3rd ed., Harper & Row, New York [Google Scholar]
- 38. Gaynor C. D., McCormack F. X., Voelker D. R., McGowan S. E., and Schlesinger L. S. (1995) Pulmonary surfactant protein A mediates enhanced phagocytosis of Mycobacterium tuberculosis by a direct interaction with human macrophages. J. Immunol. 155, 5343–5351 10.4049/jimmunol.169.7.3565 [DOI] [PubMed] [Google Scholar]
- 39. Schlesinger L. S. (1993) Macrophage phagocytosis of virulent but not attenuated strains of Mycobacterium tuberculosis is mediated by mannose receptors in addition to complement receptors. J. Immunol. 150, 2920–2930 [PubMed] [Google Scholar]
- 40. Taylor M. E., and Drickamer K. (1993) Structural requirements for high affinity binding of complex ligands by the macrophage mannose receptor. J. Biol. Chem. 268, 399–404 [PubMed] [Google Scholar]
- 41. Daeron M. (1997) Fc receptor biology. Annu. Rev. Immunol. 15, 203–234 10.1146/annurev.immunol.15.1.203 [DOI] [PubMed] [Google Scholar]
- 42. Finot P. A., Bujard E., Mottu F., and Mauron J. (1977) Availability of the true Schiff's bases of lysine. Chemical evaluation of the Schiff's base between lysine and lactose in milk. Adv. Exp. Med. Biol. 86b, 343–365 [DOI] [PubMed] [Google Scholar]
- 43. Cui Y., Li X., Lin J., Hao Q., and Li X. D. (2017) Histone ketoamide adduction by 4-oxo-2-nonenal is a reversible posttranslational modification regulated by Sirt2. ACS Chem. Biol. 12, 47–51 10.1021/acschembio.6b00713 [DOI] [PubMed] [Google Scholar]
- 44. Devlin R. B., McDonnell W. F., Becker S., Madden M. C., McGee M. P., Perez R., Hatch G., House D. E., and Koren H. S. (1996) Time-dependent changes of inflammatory mediators in the lungs of humans exposed to 0.4 ppm ozone for 2 hr: A comparison of mediators found in bronchoalveolar lavage fluid 1 and 18 hr after exposure. Toxicol. Appl. Pharmacol. 138, 176–185 10.1006/taap.1996.0111 [DOI] [PubMed] [Google Scholar]
- 45. Windsor K., Genaro-Mattos T. C., Kim H. Y., Liu W., Tallman K. A., Miyamoto S., Korade Z., and Porter N. A. (2013) Probing lipid-protein adduction with alkynyl surrogates: Application to Smith-Lemli-Opitz syndrome. J. Lipid Res. 54, 2842–2850 10.1194/jlr.M041061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chambers M. C., Maclean B., Burke R., Amodei D., Ruderman D. L., Neumann S., Gatto L., Fischer B., Pratt B., Egertson J., Hoff K., Kessner D., Tasman N., Shulman N., Frewen B., Baker T. A., Brusniak M. Y., Paulse C., Creasy D., Flashner L., Kani K., Moulding C., Seymour S. L., Nuwaysir L. M., Lefebvre B., Kuhlmann F., Roark J., Rainer P., Detlev S., Hemenway T., Huhmer A., Langridge J., Connolly B., Chadick T., Holly K., Eckels J., Deutsch E. W., Moritz R. L., Katz J. E., Agus D. B., MacCoss M., Tabb D. L., and Mallick P. (2012) A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol. 30, 918–920 10.1038/nbt.2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kessner D., Chambers M., Burke R., Agus D., and Mallick P. (2008) ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 24, 2534–2536 10.1093/bioinformatics/btn323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim S., and Pevzner P. A. (2014) MS-GF+ makes progress towards a universal database search tool for proteomics. Nat Commun. 5, 5277 10.1038/ncomms6277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ma Z. Q., Dasari S., Chambers M. C., Litton M. D., Sobecki S. M., Zimmerman L. J., Halvey P. J., Schilling B., Drake P. M., Gibson B. W., and Tabb D. L. (2009) IDPicker 2.0: Improved protein assembly with high discrimination peptide identification filtering. J Proteome Res. 8, 3872–3881 10.1021/pr900360j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang B., Kirov S., and Snoddy J. (2005) WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 33, W741–748 10.1093/nar/gki475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang J., Duncan D., Shi Z., and Zhang B. (2013) WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 41, W77–W83 10.1093/nar/gkt439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. O'Shea J. P., Chou M. F., Quader S. A., Ryan J. K., Church G. M., and Schwartz D. (2013) pLogo: a probabilistic approach to visualizing sequence motifs. Nat Methods. 10, 1211–1212 10.1038/nmeth.2646 [DOI] [PubMed] [Google Scholar]
- 53. Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D. J., Inuganti A., Griss J., Mayer G., Eisenacher M., Perez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yilmaz S., et al. (2019) The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 47, D442–D450 10.1093/nar/gky1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bauer R. N., Müller L., Brighton L. E., Duncan K. E., and Jaspers I. (2015) Interaction with epithelial cells modifies airway macrophage response to ozone. Am. J. Respir. Cell. Mol. Biol. 52, 285–294 10.1165/rcmb.2014-0035OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hatch G. E., Duncan K. E., Diaz-Sanchez D., Schmitt M. T., Ghio A. J., Carraway M. S., McKee J., Dailey L. A., Berntsen J., and Devlin R. B. (2014) Progress in assessing air pollutant risks from in vitro exposures: matching ozone dose and effect in human airway cells. Toxicol. Sci. 141, 198–205 10.1093/toxsci/kfu115 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mass Spectrometry data are available through the ProteomeXchange consortium via the PRIDE partner repository with identifier PXD019317. All other data are contained within the paper.