Abstract
Protein glycosylation is essential to trafficking and immune functions of human neutrophils. During granulopoiesis in the bone marrow, distinct neutrophil granules are successively formed. Distinct receptors and effector proteins, many of which are glycosylated, are targeted to each type of granule according to their time of expression, a process called “targeting by timing.” Therefore, these granules are time capsules reflecting different times of maturation that can be used to understand the glycosylation process during granulopoiesis. Herein, neutrophil subcellular granules were fractionated by Percoll density gradient centrifugation, and N- and O-glycans present in each compartment were analyzed by LC–MS. We found abundant paucimannosidic N-glycans and lack of O-glycans in the early-formed azurophil granules, whereas the later-formed specific and gelatinase granules and secretory vesicles contained complex N- and O-glycans with remarkably elongated N-acetyllactosamine repeats with Lewis epitopes. Immunoblotting and histochemical analysis confirmed the expression of Lewis X and sialyl-Lewis X in the intracellular granules and on the cell surface, respectively. Many glycans identified are unique to neutrophils, and their complexity increased progressively from azurophil granules to specific granules and then to gelatinase granules, suggesting temporal changes in the glycosylation machinery indicative of “glycosylation by timing” during granulopoiesis. In summary, this comprehensive neutrophil granule glycome map, the first of its kind, highlights novel granule-specific glycosylation features and is a crucial first step toward a better understanding of the mechanisms regulating protein glycosylation during neutrophil granulopoiesis and a more detailed understanding of neutrophil biology and function.
Keywords: neutrophil, granule, targeting by timing, N-glycan, O-glycan, LacNAc, Lewis epitope, liquid chromatography, mass spectrometry (MS), neutrophil, glycosylation, N-linked glycosylation, plasma membrane
Neutrophils are central cells of innate immunity, primarily dedicated to the killing of invading microbes. They are nondividing, short-lived white blood cells that, in large numbers and with impressive specificity, can perform numerous functions. It is well-established that mature neutrophils in circulation contain different granule subsets that harbor distinct proteins and other biomolecules (1, 2). Sequential mobilization of granules allows for the cells to rapidly change their surface receptor repertoire and release matrix-destroying proteases to facilitate transmigration from the blood, extravasation into infected and inflamed tissues, and phagocytosis of microbes and cellular debris. Therefore, a distinct separation of proteins between the different granules and vesicles is important for neutrophils to appropriately respond to inflammation and infection.
The neutrophil granules are sequentially formed during granulopoiesis, the process of neutrophil maturation in the bone marrow (1). Early during the promyelocyte stage, the azurophil granules (AG) are formed, followed by the specific and gelatinase granules (SG and GG), which develop sequentially during the myelocyte/metamyelocyte and band cell stages, respectively. Finally, the so-called secretory vesicles (SV) are formed from the plasma membrane (PM) of mature neutrophils through endocytosis. Because no subsequent exchange of proteins between fully synthesized compartments are thought to occur, the neutrophil proteins that are synthesized during the different stages of granulopoiesis end up in the corresponding granule type, a process known as “targeting by timing.” This was elegantly exemplified in studies correlating transcriptomics and proteomic data of maturing neutrophils and their granules, SV, and PM (1, 3, 4).
The majority of granule and cell surface proteins are glycosylated (1). Glycosylation adds structural and functional heterogeneity to proteins by the attachment of oligosaccharides to specific amino acid residues. The N-glycans are attached to amino acid consensus sequences Asn-Xaa-Ser/Thr (where Xaa ≠ Pro), whereas O-glycans (mucin-type, O-GalNAc-type) are attached to Ser and Thr. Specific glycan structures and/or epitopes are involved in multiple aspects of the immune response, including neutrophil function. The glycosylation of neutrophil proteins is diverse and functionally important for the inflammatory response, e.g. by binding to immune regulating lectins such as selectins, siglecs, galectins, C-type lectins, and microbial adhesins (5–9).
Earlier MS studies documenting protein glycosylation in whole intact neutrophils showed the presence of Galβ1–4GlcNAc, i.e. N-acetyllactosamine chains (LacNAc) linked to lipids or protein (10, 11) and that some of the LacNAc moieties were modified with fucose and/or terminal sialic acid residues, generating epitopes such as Lewis X (Lex; Galβ1–4(Fucα1–3)GlcNAcβ-R) and sialyl-Lewis X (sLex; Neu5Acα2–3Galβ1–4(Fucα1–3)GlcNAcβ-R). These epitopes are the main ligands of the endothelial selectins involved in neutrophil capture and rolling on the endothelium, enabling transmigration to the peripheral tissue (12, 13). Deficiency in the sLex antigen presentation on the neutrophil cell surface causes leukocyte adhesion deficiency 2, associated with recurrent infection, persistent leukocytosis, and severe mental and growth retardation (14, 15). The interest in the roles played by Lex and sLex antigens in cell recruitment has resulted in more specific analyses of these neutrophil epitopes, on a subcellular level.
Detailed analyses of glycan structures of neutrophil granule glycoproteins have been carried out (9, 16–18). Although glycosylation is known to be essential to neutrophil function (12, 13, 19), a comprehensive mapping of the granule and cell surface glycome is still lacking even of healthy resting (nonactivated) neutrophils. Recently, we have identified paucimannosidic glycans (Man1–3GlcNAc2Fuc0–1) carried by glycoproteins in the sputum of cystic fibrosis patients (20). These glycoproteins were traced back to the AG of human neutrophils (21), for example, neutrophil elastase, where the unusually truncated glycans were shown to influence the binding of elastase to mannose-binding lectin and ɑ1-antitrypsin often present within inflamed tissues (22).
Herein, we provide a comprehensive characterization of the N- and O-glycans in neutrophil granules, isolated using subcellular fractionation and analyzed by porous graphitized carbon (PGC)–LC–MS/MS. We show vast differences in glycosylation between different granules. By building on the targeting-by-timing hypothesis, the granule-specific glycan differences therefore suggest that the glycosylation machinery undergoes alterations during granulopoiesis. The differences in glycosylation between granules could impact the immune modulating functions of neutrophils during inflammation.
Results
The neutrophil subcellular organelles are characterized by unique N-glycan signatures
Subcellular organelles of healthy resting neutrophils were first separated by two-layer Percoll gradients into three fractions; AG, SG + GG, and SV + PM (Fig. 1a). Organelle lysis and extraction and solubilization of organelle proteins were followed by enzymatic release of N-glycans, and MS analyses enabled the unambiguous identification of 69 unique N-glycans. The obtained profiles between the three fractions differed vastly from each other (Fig. 1b). Paucimannosidic and complex N-glycans were predominantly identified in AG and SG + GG, respectively, whereas SV + PM contained oligomannose and complex glycans, at similar levels (40–50%). The analysis identified 36 N-glycans in AG, 60 in SG + GG, and 61 in SV + PM. Of the total 69 N-glycans, 27 were present in all organelles, and SG + GG and SV + PM were mostly similar, sharing 50 N-glycans (Fig. 1c). These qualitative differences were corroborated also on the quantitative level (Fig. 1d). Hence, the different organelles all had unique sets of glycan structures that were in some parts overlapping. The detailed list of N-glycans and their relative abundances in each fraction is found in Table S1.
Figure 1.
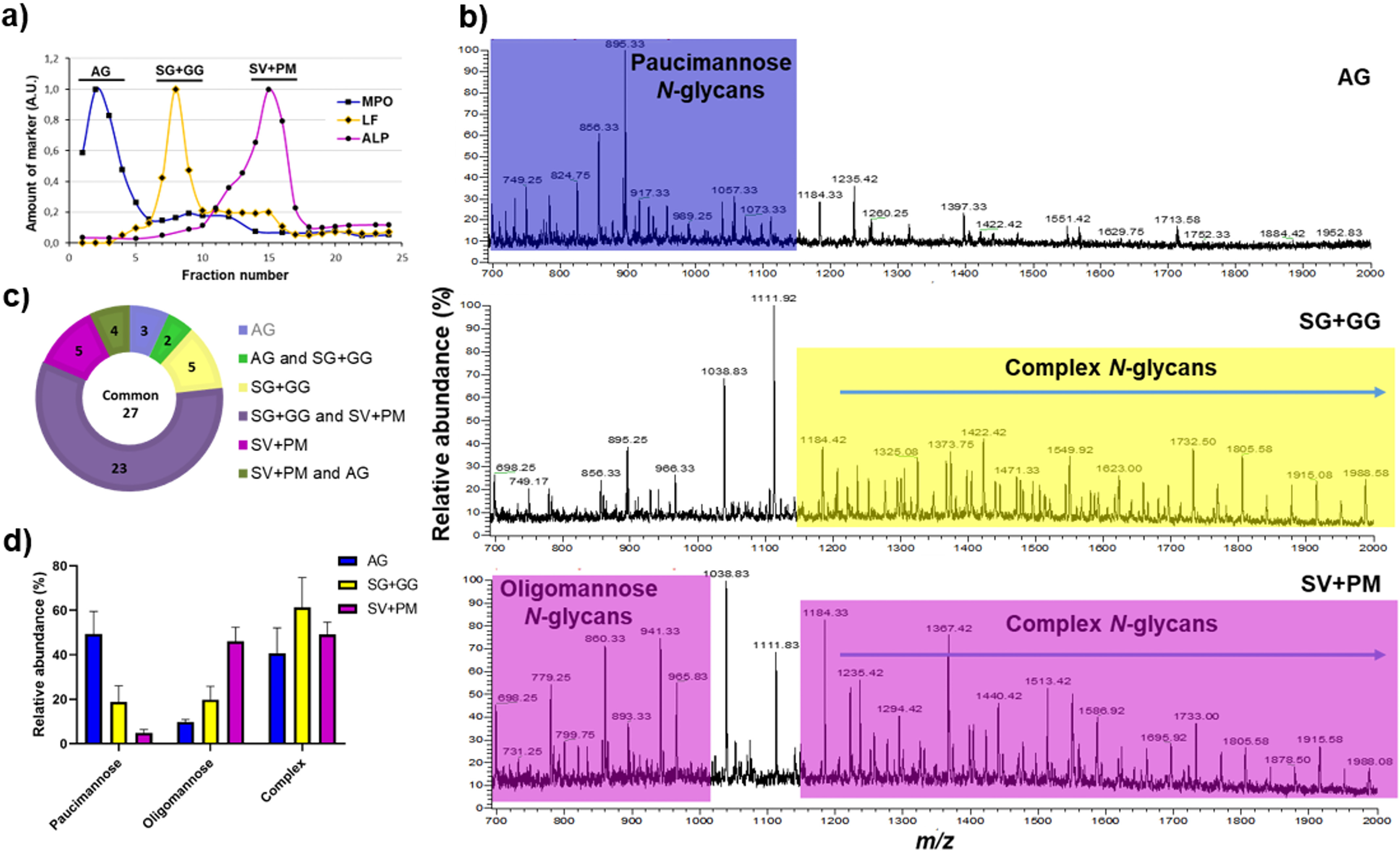
Validation of neutrophil subcellular fractionation and organelle-specific neutrophil N-glycan profiling. a, the granules of naive (resting) neutrophils were fractionated using a two-layer Percoll gradient. Fractions were assigned to distinct neutrophil organelles based on the level of known granule-specific protein markers as determined using immunoblot and activity assays (y axis, arbitrary units, A.U.): MPO activity (marker for AG, filled squares); LF (marker for SG, filled diamonds); and total ALP activity (measured in the presence of detergent; marker for SV + PM, filled circles). The data shown are representative from three separate, independent experiments. b, mass spectral profiles of N-glycans released from neutrophil granule protein extracts showing dramatic qualitative N-glycan profile differences between the AG, SG + GG, and SV + PM fractions. Characteristic N-glycan types for each granule fraction have been color-coded to highlight the most prominent differences. c, distribution of the identified N-glycan structures in the different neutrophil subcellular compartments. d, the N-glycans identified in the different subcellular fractions were grouped into the major N-glycan types, i.e. paucimannosidic, oligomannosidic, and complex N-glycans and their relative abundance were determined for each granule subset. The data points are plotted as the mean ± S.E. (n = 3).
Paucimannosidic and oligomannosidic-type N-glycans are abundant on proteins in the AG and SV + PM, respectively
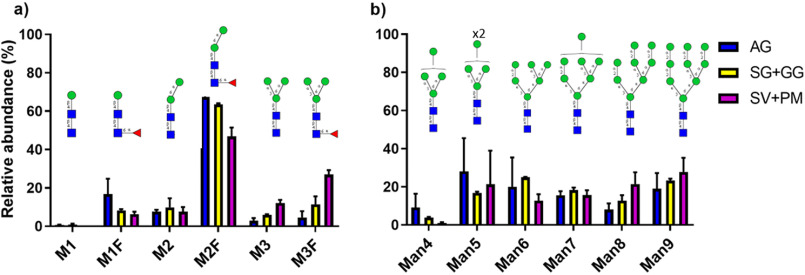
The hallmark of AG was the presence of paucimannosidic glycans, with a relative abundance of 49.4 ± 5.8% of all N-glycans. In contrast, only 18.7 ± 5.2% and 4.9 ± 0.9%, respectively, of all N-glycans were paucimannosidic in SG + GG and SV + PM (Fig. 1d). Six different paucimannosidic glycans were identified in AG, with the most abundant (65% of the paucimannose glycans) containing two mannoses and a core fucose (M2F; m/z 895.31−) (Fig. 2a and Fig. S1). In fact, within the class of paucimannosidic-type N-glycans, the M2F was clearly the most abundant paucimannosidic glycan species in all organelles (Fig. 2a), but the overall abundance of M2F in SV + PM and SG + GG was <5% of total N-glycans as compared with 40% in the AG. Only the α1,6-isomer of the M2F glycan was observed in agreement with our previous reports (21, 22). Another striking feature of AG is the high abundance of core fucosylation (80.4%) as compared with antenna fucosylation (19.6%).
Figure 2.
Distribution of mannose-terminating N-glycans in AG and SV + PM. a and b, relative abundance of the observed paucimannosidic (a) and oligomannose (b) N-glycans in AG, SG + GG, and SV + PM fractions. The relative distribution of N-glycans is plotted as the mean ± S.E. (n = 3). Blue squares, GlcNAc; green circles, mannose; red triangles, fucose.
Oligomannose and complex type N-glycans predominate in the SV + PM fraction, with a relative abundance of 46.1 ± 3.7% and 49.1 ± 3.2%, respectively (Fig. 1, b and d). Oligomannose N-glycans in the SV + PM fraction showed low abundance of Man4 glycan and a varied distribution of Man5 to Man9 glycans (Fig. 2b). Oligomannose glycans were present also in AG and SG + GG, however at much lower levels (9.7 ± 0.7% and 19.9 ± 4.2%, respectively). The distribution between Man4 to Man9 glycans was similar in these organelles as in SV + PM (Fig. 2b).
Complex N-glycans with LacNAc repeats are characteristic of the SG + GG and SV + PM
Complex N-glycans were observed in all fractions, with a higher abundance in the SG + GG (61.4 ± 9.5% of all N-glycans) as compared with the AG (40.7 ± 6.5%) and SV + PM (49.1 ± 3.2%) (Fig. 1d). Conventional glycans carrying single LacNAcs on both antennae were observed in AG, albeit in lower abundance (5.8 ± 1.8%) as compared with the other fractions. In contrast, one of the key glycosylation features in SG + GG and SV + PM was the presence of unusual, very-high-molecular-mass complex glycans extended with repeating LacNAc units (Fig. 3a). Because these glycans were characterized by broad peaks of extracted precursor ions caused by unresolved multiple isomers, their abundances were not considered during our initial analysis.
Figure 3.
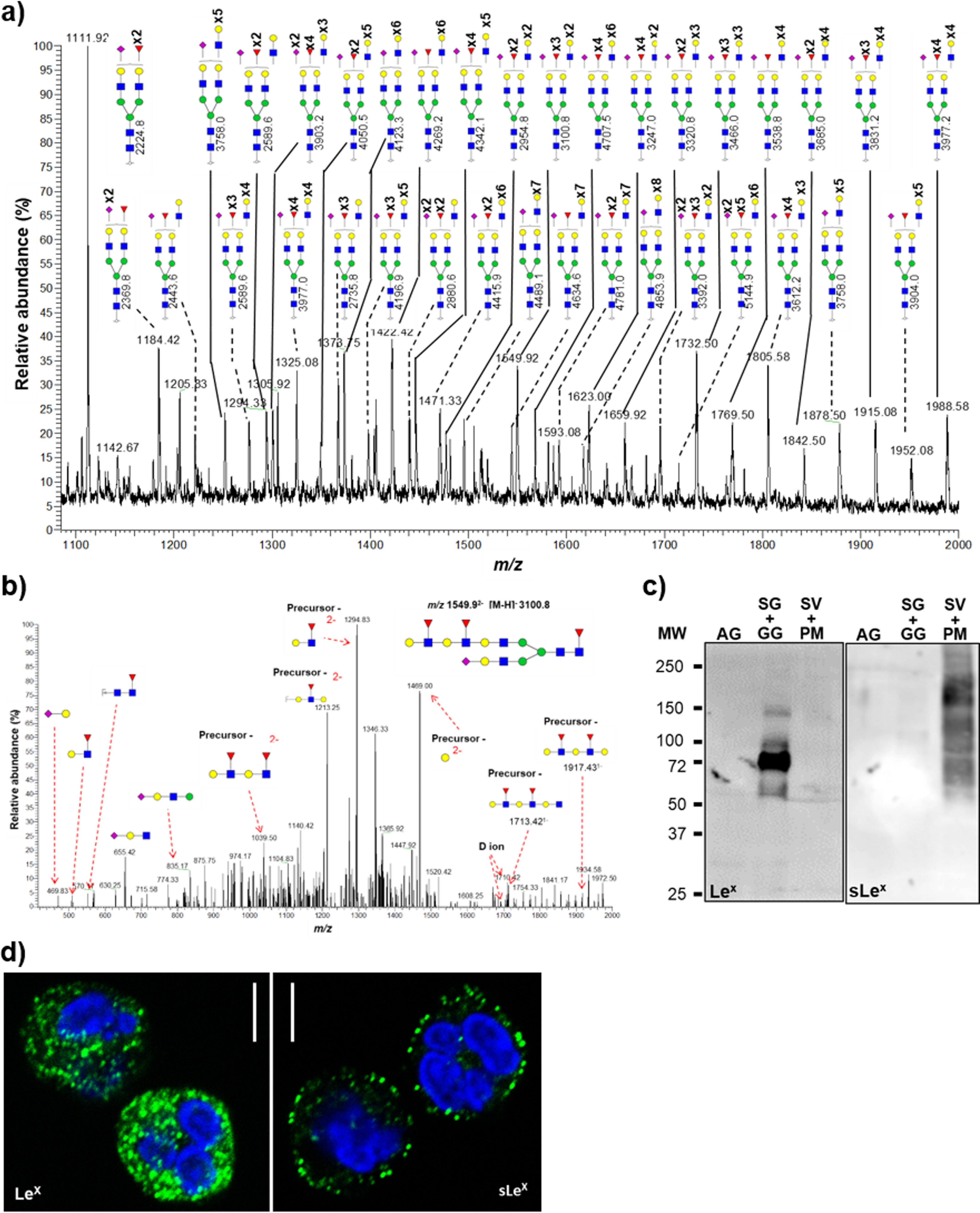
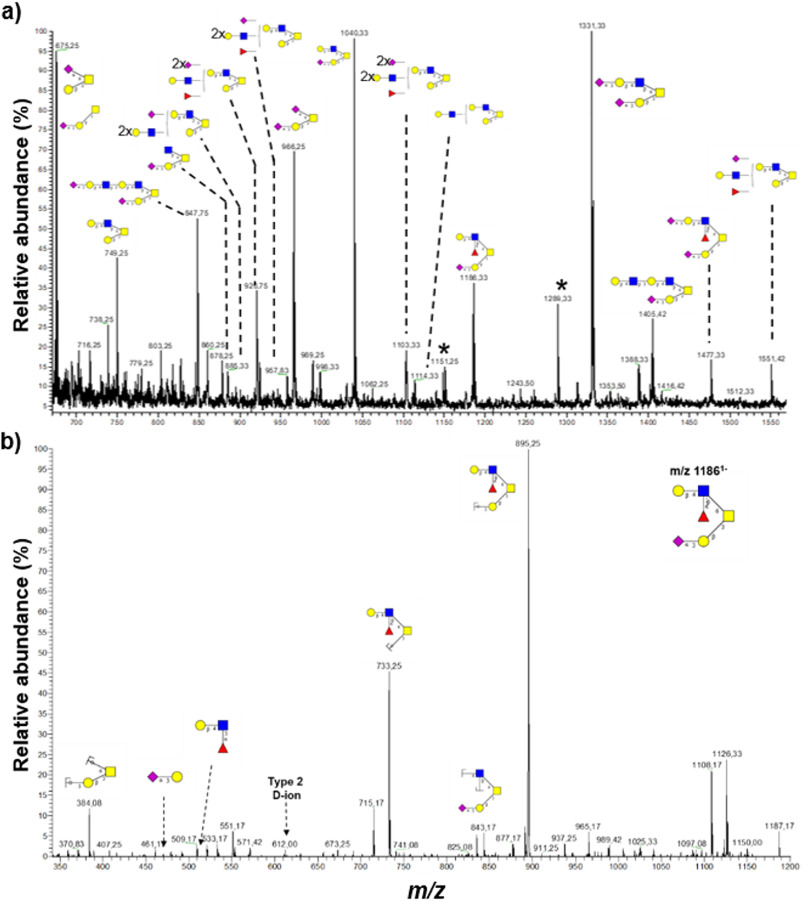
Unusual LacNAc-elongated complex N-glycans reside in the SG and GG fractions. a, representative mass spectral profile of SG + GG N-glycans illustrating the presence of unusual LacNAc-elongated N-glycans, up to 10 repeats, with and without fucose and/or sialic acid capping, forming Lewis epitopes. b, N-glycans were characterized using CID–MS/MS in negative polarity as exemplified by the annotated MS/MS fragmentation spectrum of the precursor ion m/z 15502− corresponding to an N-glycan containing four LacNAc units with fucosyl and sialyl decorations. Diagnostic ions confirmed the presence of LacNAc elongation as opposed to antenna branching. c, immunoblot analysis of Lex and sLex epitopes on proteins from different organelles. Equal protein amounts were loaded for each fraction as determined by BCA assay. d, representative images of immunofluorescence-stained neutrophils demonstrating the localization of Lex (left panel) and sLex (right panel) epitopes. Scale bar, 5 μm. Blue squares, GlcNAc; green circles, mannose; red triangles, fucose; yellow circles, galactose; purple diamonds, sialic acid (N-acetylneuraminic acid).
Apart from oligomannose structures, the glycan profile of SV + PM was similar to that of the SG + GG with elongated complex N-glycans (Fig. S2). In SG + GG, complex-type N-glycans were predominantly biantennary fucosylated glycans with or without terminal sialic acid. The most abundant glycan (m/z 1111.92−) identified in SG + GG was a biantennary monosialylated and difucosylated glycan with both core and a single antenna fucosylation (Fig. 3a). The identified sialylated glycans were found to be mainly α2,6 linked to galactose on the 3′ arm and α2,3 linked forming sLex on the 6′ arm. In conclusion, complex N-glycans with LacNAc repeats were abundant in SG + GG and SV + PM but absent from AG.
The LacNAc repeats are mainly found on biantennary N-glycans
The structures of the extended LacNAcs were confirmed based on their composition related to their molecular mass, along with supporting PGC–LC retention time data and MS/MS spectral data that concertedly were able to confirm the sequence and branch points of the extended structures. Spectra were manually annotated for B-/Y- and C-/Z- ion series as well as diagnostic ions for various glycoepitopes (23). A representative MS/MS fragmentation of a four-LacNAc-containing glycan at m/z 15502− is shown in Fig. 3b. The presence of various fragment ions, including m/z 1039.52−, 1213.22−, 1294.82−, 1469.02−, 1713.41−, and 1917.41− and D-ions of 1692.41− and 1710.41−, suggests exclusive elongation of 6′ arm rather than 3′ arm elongation (24, 25) and as opposed to branched tri- or tetra-antennary glycans. These fragment ions were observed in the MS/MS fragmentation spectra of other glycans as well (Fig. S3), which support and confirm our assessment. Furthermore, the monosaccharide composition of glycans with additional LacNAc repeats contains either one or two terminal sialic acids, supporting the presence of elongated biantennary glycans instead of higher branched glycans. Broad signal peaks of higher m/z values suggest that tri- and tetra-antennary glycans elongated with LacNAc units may also be present but at a lower abundance.
Information about the degree of LacNAc extensions could be obtained from the combined intensities of the broad LC peaks, which corresponds to structures potentially carrying three to ten LacNAc units (Fig. S4, top panel). It could be seen that structures containing six to eight LacNAc units were the most abundant in both SG + GG and SV + PM. Simple extrapolation of the data suggests that structures even longer than 10 LacNAc units could be present in both of these fractions, but they were due to structural ambiguity not included here.
The unusually long LacNAc repeats of SG + GG and SV + PM are decorated with Lewis epitopes
Fucosylation of the GlcNAc moiety in the LacNAc chains, building Lewis type epitopes, was observed in the SG + GG and SV + PM fractions. It should be noted that not all LacNAcs in an antenna were fucosylated. For the high-molecular-mass glycans, we calculated the number of Lewis epitopes per structure and their corresponding relative intensities (Fig. S4, bottom panel). Simple structures containing one to three Lewis epitopes were more abundant than structures containing higher numbers of fucose. Structures were found to be decorated with a maximum of seven fucose residues. The data also show that the amount of fucosylation per LacNAc was decreasing with the increasing number of Lewis epitopes.
When analyzing the fractions for Le epitopes by immunoblotting analyses, the Lex antigens were mainly found in the SG + GG fraction, whereas sLex epitopes were detected predominantly in the SV + PM fraction (Fig. 3c). Using confocal microscopy, the presence of Lex was detected in intracellular granules, whereas sLex was mainly localized to vesicles close to the cytoplasmic membrane, suggesting that sLex is carried by glycans that are associated with the SV + PM (Fig. 3d). Overall, these data show that neutrophils extensively express complex N-glycans with elongated LacNAc repeats that are decorated with fucose (Lex; in SG + GG) and capped with sialic acid (sLex; in SV + PM).
Gelatinase granule proteins carry N-glycans with higher numbers of LacNAc repeats than specific granules
Using crude granule separation, the above have demonstrated that the neutrophil granules, in addition to their difference in size, density, and protein composition, also differs in their glycan composition (Fig. 1, b and d) (3, 26) and indicated some peculiar N-glycosylation features in the SG + GG fraction. We wanted to investigate the granule-specific glycosylation further using a higher resolution granule separation technique. The SG and GG fractions were separated using a three-layer Percoll gradient (Fig. 4a) and analyzed for high molecular mass N-glycans.
Figure 4.
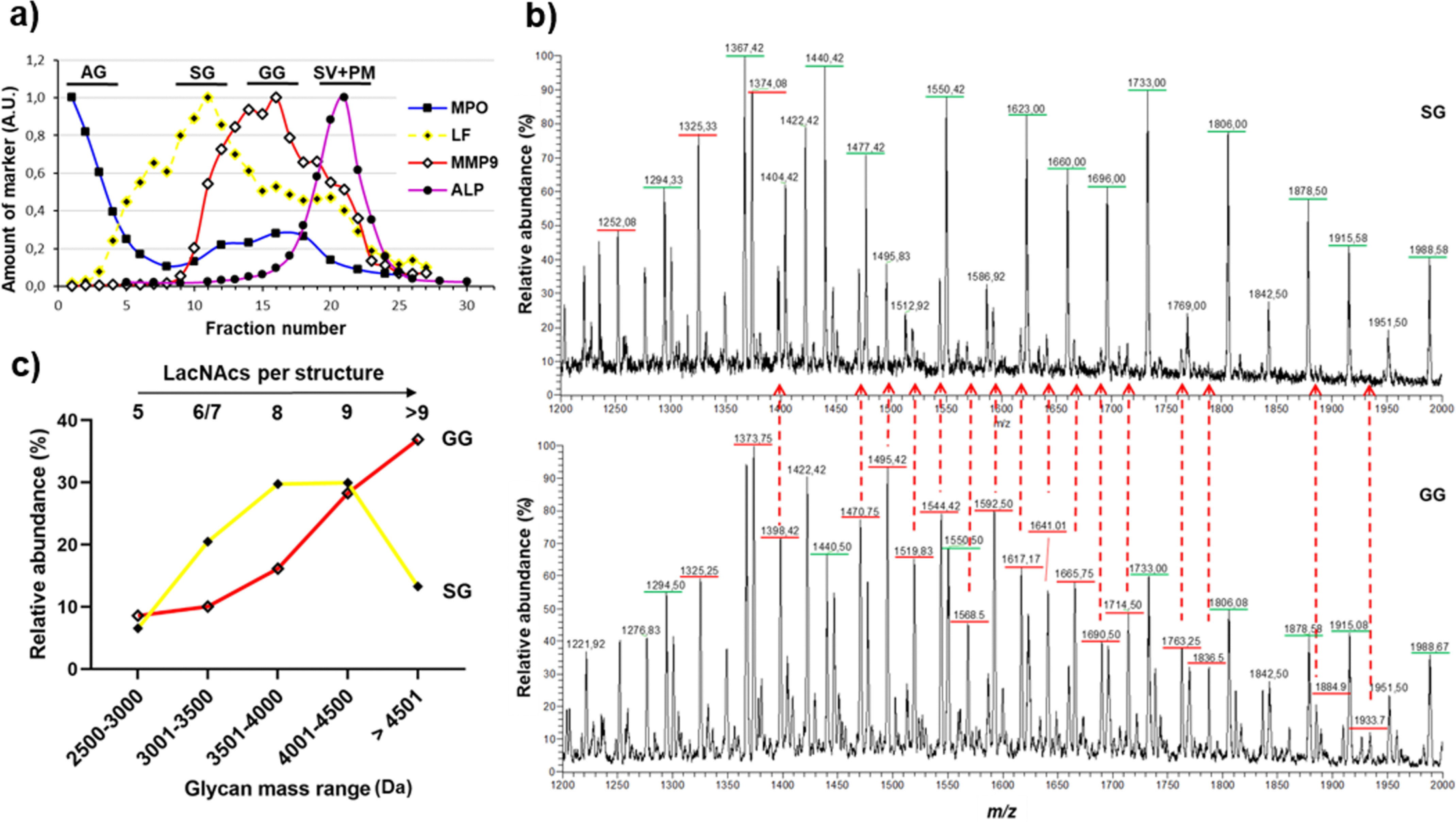
The elongation length of LacNAc-containing N-glycans differs in the SG and GG. a, the granules of naive neutrophils were fractionated using a three-layer Percoll gradient. Fractions were assigned to distinct neutrophil granule fractions based on the level of known granule-specific protein markers as determined using immunoblots and activity assays (y axis, arbitrary units, A.U.): MPO activity (marker for AG, filled squares); LF (marker for SG, filled diamonds); MMP9 (marker for GG, open diamonds); and total ALP activity (measured in the presence of detergent; marker for SV + PM, filled circles). The data are representative of three separate, independent experiments. The bars indicate which fraction numbers were pooled together for the glycan analysis. b, representative mass spectra focusing on the high m/z region encompassing the elongated LacNAc repeat–containing N-glycans released from SG (top panel) and GG (bottom panel) protein extracts, respectively. The m/z values underlined in green and red indicate doubly and triply charged ions, respectively. Many triply charged glycans found in the GG spectrum are either low in abundance or absent in the SG spectrum, indicated by the red dashed arrows. c, the relative intensities of glycan structures identified in SG and GG (based on predicted monosaccharide compositions) and divided into different mass ranges, showing that the GG have more glycans of higher mass than the SG. The number of potential LacNAcs in each mass range is shown.
Mass spectra of the N-glycans in the m/z 1200–2000 region revealed an enrichment of triply charged ions in the GG, whereas doubly charged ions were more abundant in the SG (Fig. 4b). It is important to note that the detection of more high-molecular-mass glycans in the isolated GG fraction (three-layer gradient) as compared with in the combined SG + GG fraction (two-layer gradient) is a consequence of absence of SG in the GG fraction. Consequently, the precursor ion (m/z 1933.73−) corresponding to eleven LacNAcs, four fucoses, and one sialic acid (, the longest glycan detected, was identified only in the GG fraction of the three-layer gradient.
The relative intensities of glycans, based on their predicted monosaccharide compositions, were grouped into different mass ranges. It is evident that the relative intensities of glycans containing five to eight LacNAcs per structure were more abundant in SG as compared with GG (Fig. 4c). In contrast, structures containing nine or more LacNAcs were more abundant in the GG fraction. Taken together, the data strongly suggest that the glycans in GG are further elongated and larger than those in the SG.
The glycan types are differently distributed between the membrane and luminal protein extracts
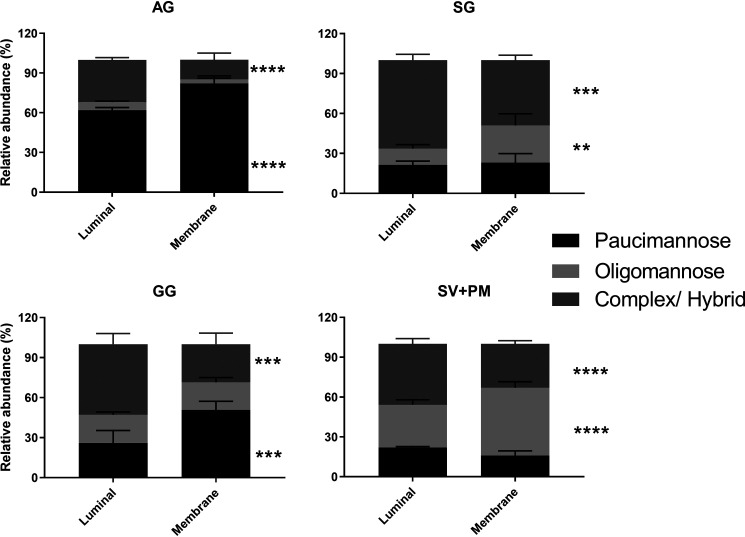
The glycans identified in the granule fractions may originate either from the granule membrane or luminal (i.e. soluble) proteins. For SV, the lumen contains plasma proteins taken up during invagination of the plasma membrane during neutrophil terminal differentiation (27). To address which of the identified glycans are displayed on membrane-associated proteins, the membrane protein extracts were separated from the soluble protein extracts for each granule population (AG, SG, and GG) and the SV + PM obtained from the three-layered Percoll isolation method. Paucimannosidic glycans were enriched in the AG membrane protein fraction, whereas oligomannose glycans were enriched in the membrane fraction of SG and SV + PM (Fig. 5). In contrast, complex glycans were observed in high abundance on the soluble protein fraction in all granules.
Figure 5.
The N-glycan signatures differ on soluble (luminal) and membrane proteins in the different neutrophil organelles. Quantitative glycomics was performed for N-glycans released from separated membrane and lumen (soluble) protein extracts from isolated neutrophil granules and SV + PM. The identified glycans have been grouped and quantified according to their N-glycan type, i.e. paucimannosidic, oligomannosidic, and complex N-glycans. Note that the glycans from the soluble proteins in the SV + PM fraction are not of neutrophil origin but come from plasma proteins. The data represent the means ± S.E., and unpaired two-way Student's t test was used for statistical analysis. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
O-Glycans are absent in AG, whereas extended LacNAc-rich core 2 O-glycans are consistently found in the other fractions
Apart from N-glycan analysis, proteins were also subjected to reductive β-elimination, after which the released O-glycans were analyzed. Notably, no O-glycans were detected in the AG, whereas SG + GG (Fig. 6a) and SV + PM (Fig. S5) demonstrated the presence of similar O-glycans, with a total of 17 O-glycans identified. The most abundant O-glycans were mono- and disialylated core 1 and core 2 glycans (Fig. 6a). Analogous to the LacNAc extension of the N-glycans, only one of the branches was elongated with LacNAcs on O-glycans. Sialic acid linked to galactose was found on the 3′ arm but not on the 6′ arm, whereas the fucose moiety was located on the 6′ arm together with LacNAc extensions as elucidated using molecular mass, PGC–LC retention time, and MS/MS fragmentation (Fig. 6b). Similar to N-glycans, O-glycans with Lex epitopes were identified, with fucosylation on 6′ arm and sialylation capping on the other arm (Fig. 6b). In conclusion, neutrophil granule proteins are modified with core 1 and core 2 O-glycans carrying LacNAc chains and Lex antigens, whereas sLex epitopes were not commonly observed.
Figure 6.
Dominant core 2 O-glycosylation in SG and GG fractions. a, core 2 O-glycans were commonly observed with LacNAc extensions on the 6′ arm and sialic acid capping on the 3′ arm galactose in SG + GG fractions. b, LC–MS/MS spectra of core 2 O-glycan (m/z 11861−) with sialic acid capping on the 3′ arm and Lewis epitope on the 6′ arm. Asterisks represent contaminating signals of unknown origin. Blue squares, GlcNAc; yellow squares, GalNAc; red triangles, fucose; yellow circle, galactose; purple diamonds, sialic acid (N-acetylneuraminic acid).
Granule glycosylation during granulopoiesis
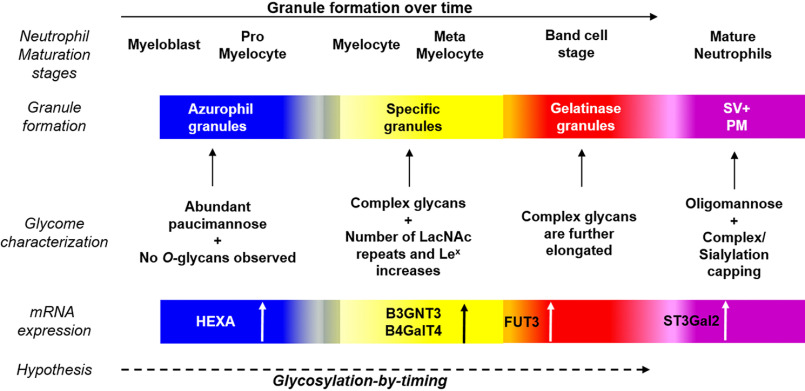
Different granules are sequentially formed at different stages during granulopoiesis: myeloblasts or promyelocytes (AG), myelocytes/metamyelocytes (SG), and band cell stage (GG). The quantitative comparison of the glycan composition revealed a shift from paucimannosidic N-glycans and a lack of O-glycans in the AG to LacNAc-containing complex glycans in the SG and GG (Fig. 7). This suggests a change in glycosylation machinery over time during granulopoiesis. We therefore compared the glycomic data with the corresponding mRNA expression profiles (from the publicly available Bloodspot database (28)) of enzymes responsible for glycan synthesis during granulopoiesis. We noticed a higher expression of HEXA encoding the N-acetyl-β-hexosaminidase subunit α, responsible for the formation of paucimannosidic glycans (21, 29), in promyelocytes as compared with cells from later stages of granulopoiesis (Fig. S6). In contrast, expression of B3GNT3 (GlcNAc transferase), B4GALT4 (galactosyl transferase), and FUT3 (fucosyl transferase), encoding glycosyltransferases responsible for the synthesis of LacNAc repeats and Lex, was expressed at higher levels in the later maturation stages as compared with the early maturation stage (28). Finally, ST3GAL4 encoding a sialyl transferase responsible for synthesizing sLex was found to be expressed predominantly during the final stages of maturation (28). In conclusion, the glycomic data together with database-mined mRNA expression data support our hypothesis that a change in the glycosylation machinery occurs over time during granulopoiesis.
Figure 7.
Overview of glycosylation-by-timing hypothesis. Granule formation during neutrophil maturation is a continuum, and granules carry a vast array of glycans. From the presence of shorter paucimannosidic glycans in AG, to ever-elongating complex glycans in SG, and subsequently in GG and the corresponding mRNA expression of different glycosidases and glycosyltransferases, suggests an evolution of granule glycosylation over time. Similar to the targeting-by-timing hypothesis describing the temporal protein synthesis of maturing neutrophils, we hypothesize a parallel glycosylation-by-timing granule glycan synthesis that takes place during neutrophil maturation.
Discussion
Defining molecular differences between neutrophil organelles, proteins, and finally their glycosylation, is a prerequisite for better understanding neutrophil biology and function. Mapping of the protein composition of neutrophil subcellular organelles (3, 26), as well as their gene and protein expression during granulopoiesis (4, 30–32), has provided a comprehensive qualitative and quantitative basis for the targeting-by-timing hypothesis, which appears to accurately describe the evolution of neutrophil protein composition over time during granulopoiesis. This work provides a platform on which maturation-associated post-translational modifications, not least glycosylation, can be studied. We have, as a first, characterized the N- and O-glycans in neutrophil granules, as well as in the SV/PM using a PGC–LC–MS/MS-based glycomics approach recognized for providing information of the glycan fine structures while also giving quantitative information at high sensitivity (33, 34).
The characteristic feature of the AG glycoproteins is the presence of paucimannosidic type N-glycans and the absence of O-glycans. Our developing knowledge of protein paucimannosylation in human biology including aspects covering its structure, function, and tissue expression has recently been extensively summarized (29). Sequential trimming of the glycoprotein intermediate, i.e. β1,2-GlcNAc–capped trimannosylchitobiose core with or without core fucose by Hex A and/or Hex B, and presumably a linkage-specific ɑ-mannosidase gives rise to paucimannosidic glycans of which one species (the ɑ1,6-isomer of M2F) represents >60% of the total amount of paucimannose. This was supported by a strong co-localization of Hex A and myeloperoxidase (MPO) in differentiated HL-60 cells using immunohistochemistry (21).
Upon inflammation in vivo, the AG proteins carrying paucimannosidic glycans are released or leak from activated neutrophils as a result of substantial cell activation or cell death. These encompass an ensemble of bioactive AG proteins, including MPO, neutrophil elastase, and other serine proteases (21, 22, 35, 36). The structure–function relationship of the paucimannosidic glycans, modulating the proinflammatory effects of these proteins, can at this point only be speculated upon, but both paucimannosidic and oligomannosidic-type N-glycans have been shown to bind to mannose recognizing C-type lectin receptors expressed by macrophages and dendritic cells (22, 37, 38). Also, the macrophage mannose-receptor has been shown to internalize neutrophil-derived MPO (39), playing a potential physiological role in regulating extracellular MPO levels. Although the preference of the mannose-receptor for specific glycan substrates has yet to be documented in neutrophils, the structures and localization in the various granule compartments identified here will be useful for future investigation.
Abundance of complex N- and O-glycans with extended LacNAc chains was a characteristic feature observed in the SG + GG, formed during the middle stages of granulopoiesis. The level of LacNAc chains identified are unique to neutrophils and have to our knowledge never been reported in other immune cells. In fact, the number of LacNAc repeats reported here is probably underestimated, because we incidentally identified structures with up to 16 LacNAcs in few of the spectra and the fact that the LC–MS/MS approach inherently favors the shorter structures. Given that the complex glycans in the SG, GG, and SV + PM fractions are mostly biantennary, these long LacNAc extensions placed close together on a cell surface could build a dense net for interactions with lectins, e.g. galactose-binding lectins such as galectins. In the context of neutrophil function and regulation, carbohydrate-dependent binding of galectin-3 can mediate cell adhesion, phagocytosis, and activation of the respiratory burst (40, 41). Interestingly, a prerequisite for the neutrophil response to galectin-3 is that the cells first are primed by an inflammatory stimulus and have mobilized the GG and SG, thereby having up-regulated granule-stored, LacNAc-bearing protein receptors to the cell surface for interactions with galectin-3 (42).
The LacNAc chain length may be biologically important in this context, as shown by a galectin-3 mutant carrying increased affinity for extended LacNAc structures, resulting in an enhanced respiratory burst response in primed neutrophils as compared with WT galectin-3 (43). Furthermore, the LacNAc chains in the SG, GG, and SV + PM were decorated with fucose and sialic acid residues, forming Lewis epitopes at the nonreducing end of N- and O-glycans. In addition to the well-characterized role of Lex and sLex in cell adhesion, cell priming, and phagocytosis (12, 19, 44, 45), the expression of Lea epitopes on neutrophils has recently been shown to enhance neutrophil transmigration and to play a role in regulating inflammation (46). Although we believe that the sialyl-Lewis epitopes identified in this study were predominantly sLex (Fig. 3, c and d), the presence of sLea especially on elongated structures needs to be explored.
Unlike AG and SG + GG, which are enriched in paucimannosidic and complex N-glycans, respectively, the SV + PM fraction contains both oligomannose and processed complex glycans. Although the soluble protein fraction of SV + PM carries proteins of non-neutrophil origin (i.e. plasma proteins invaginated into the SV (27)), the granule membrane protein glycosylation is of neutrophil origin. The SV + PM membrane fraction was predominantly composed of oligomannose glycans that are readily available recognition tags on the cell surface of resting or slightly primed cells (having mobilized the secretory vesicles) (38). Indeed, a historically well-known mannose-dependent interaction is that of the bacterial mannose-binding adhesin FimH, which recognizes several mannose structures on cell surfaces (47), mediating bacterial binding and phagocytosis (48).
Biosynthetically, protein synthesis and granule targeting are best described by the targeting-by-timing hypothesis, in which proteins synthesized at the same time during granulopoiesis end up in the same granules. How glycosylation fits into such a scheme is an interesting question with implications for cell regulation and function. Our data show distinct patterns of glycosylation for the different granules and organelles, and close analysis of these molecular features provides important insights; the granule-specific glycans and corresponding mRNA expression of enzymes responsible for glycan synthesis appear to co-emerge at different stages of granulopoiesis, which suggests a change in glycosylation machinery over time. The plasma membrane, which contains both unprocessed oligomannose and processed complex type glycans, is not formed within a defined time interval during neutrophil differentiation, as the granules are. This is in line with the fact that PM proteins are successively being modified with the glycans specific to each different developmental stage during granulopoiesis and are sorted to the cell surface. This results in the presence of oligomannose type glycans potentially produced during early stages of granulopoiesis, together with differently matured poly-LacNAc–containing glycans produced later on. The PM-targeted glycans will then be endocytosed and incorporated into the membranes of the SV during the final stages of cell maturation.
To this background, it is tempting to suggest that a protein synthesized at a certain time during granulopoiesis and targeted to a certain granule subtype, is specifically modified with a dedicated glycosylation pattern, synthesized at that same time by a glycosylation machinery that is concurrently expressed. Consequently, we have formulated a complementing hypothesis to targeting-by-timing, stating that protein glycosylation in neutrophils takes place through a “glycosylation-by-timing” process (Fig. 7).
Furthermore, we suggest that the variation of glycosylation during granulopoiesis may correlate to the evolution of the Golgi complex, for which the size and number of stacks vary greatly during granulopoiesis (49, 50), and/or the extensive glycan processing that may occur at different rates within the formed granules during neutrophil maturation, directions that we are currently pursuing. Also, the finding that the minute amounts of elastase found outside of AG and SG appear to be similarly glycosylated (22) may indicate that certain proteins (or glycans) do not follow glycosylation by timing. We are currently performing granular glycoproteomics to further investigate the specific protein carriers of the neutrophil glycans, which will give us better understanding of how protein glycosylation is evolving. In conclusion, we provide a map of subcellular glycans in healthy human neutrophils that displays peculiar granule-specific glycan signatures, and this first-time mapping of neutrophil granule glycans provides information on adaptations that may occur in the glycosylation machinery during granulopoiesis.
Materials and methods
Isolation of human neutrophils and subcellular fractionation
Human peripheral blood polymorphonuclear neutrophils were isolated from buffy coats (The Blood Center, Sahlgrenska University Hospital, Gothenburg, Sweden) obtained from healthy donors (51). The erythrocytes were removed by dextran (1%) sedimentation (1 × g). Monocytes and lymphocytes were removed by centrifugation on a Ficoll–Paque density gradient. The cells were then washed in Krebs–Ringer phosphate buffer containing glucose (10 mm) and Mg2+ (1.5 mm), and the remaining erythrocytes were removed by hypotonic lysis. These purified neutrophils (>95% pure) were fractionated according to Borregaard et al. (52). In short, 109 cells were treated with the serine protease inhibitor diisopropylfluorophosphate (final concentration, 8 mm), resuspended in 10 ml of homogenization buffer (100 mm KCl, 3 mm NaCl, 3.5 mm MgCl2, 10 mm PIPES, pH 7.4) with 1 mm ATP(Na)2, 0.5 mm phenylmethylsulfonyl fluoride and disrupted in a nitrogen bomb (Parr Instruments, Moline, IL, USA) at 400 psi (2500 kPa). Intact cells and nuclei were removed by successive centrifugation steps at 300 × g and 800 × g, respectively. The post nuclear supernatant was layered on top of a two-layer (1.05 and 1.12 g/liter) or a three-layer (1.05, 1.09 and 1.12 g/liter) Percoll gradient and centrifuged at 15,000 × g for 45 min in a fixed-angle JA-20 Beckman rotor. The material was typically collected in 25 fractions of 1.5 ml (two-layer gradients) or 32 fractions of 1.0 ml (three-layer gradients) from the bottom of the centrifuge tube.
Granule purity and fractionation
The distribution of four different, established markers (MPO, marker for the AG; lactoferrin [LF], marker for the SG; matrix metalloproteinase-9 [MMP9], marker for the GG; and alkaline phosphatase [ALP], marker for the SV and PM) was determined to establish the distribution of the different organelles in the gradient. Enzyme activity of MPO and ALP, respectively, was determined as previously described by Feuk-Lagerstedt et al. (53). The LF and MMP9 distribution was assessed by immunoblot using antibodies (anti-LF, Dako, rabbit polyclonal antibody, 1/2500; anti-MMP9, Calbiochem, rabbit polyclonal antibody 444236, 1/2000). When appropriate, fractions containing the different organelles were pooled, diluted in homogenization buffer, and ultracentrifuged (100,000 × g, 1 h) to concentrate the sample and to remove the Percoll. To separate the luminal (soluble) protein extracts from the granule membrane protein extracts, granule pellets were diluted in homogenization buffer, disrupted with a tip sonicator at one-third power (8 MHz) three times for 30 s on ice, and fractionated by ultracentrifugation (100,000 × g, 1 h, Beckman TLA 120.2 rotor). The luminal fraction was concentrated to ∼50 μl on a 3-kDa molecular mass cutoff Amicon Ultra 0.5-ml centrifugal filter, whereas membrane pellets were dissolved in 50 μl of TBS with 2% SDS. For subsequent glycome analyses, the protein concentration of total, soluble, and membrane protein fractions of the granules was determined by BCA assay (Thermo Scientific) using BSA as standard.
Detection of Lewis antigens by immunoblot and immunofluorescence
The primary antibodies used were directed against Lex (mouse monoclonal, clone HI98, Santa Cruz Biotechnology) and sLex (mouse monoclonal, clone CSLEX-1, BD Biosciences). For the detection of Lewis antigens by immunoblots, equal amounts of the protein extracts from the granules were blotted onto a polyvinylidene difluoride membrane, essentially as described (53). The membrane was blocked 1 h at room temperature in TBS, 0.1% BSA. Primary antibodies were diluted 1/500 in TBS, 0.01% Tween 20 and incubated at 4 °C overnight. Immunodetection was performed using secondary horseradish peroxidase-coupled rabbit anti-mouse (P0260, Dako, Denmark) with Clarity Western ECL substrate (170-5061, Bio-Rad). For the detection of Lewis antigens by immunofluorescence, isolated polymorphonuclear neutrophils were resuspended in cold PBS at 1.5 million cells/ml and attached to adhesion slides (Marienfeld, Germany) according to the manufacturer's instructions. The cells were then fixed in 3.5% paraformaldehyde in PBS and permeabilized with 1% Triton X-100 for 10 min. After extensive washing, blocking was performed 30 min, in 0.1% BSA in PBS. Primary antibodies were diluted 1/50 in 0.1% BSA in PBS and incubated for 1 h. Immunodetection was performed using secondary goat anti-mouse IgM Alexa 488 (A-11001, Thermo Fisher Scientific), diluted 1/300 in 0.1% BSA in PBS, and incubated 1 h. Imaging was performed with a confocal laser scanning LSM-700 microscope (Zeiss).
Release of N- and O-glycans from neutrophil granule proteins
Purified neutrophil granule (whole) proteins, and the soluble and membrane protein extracts (∼20 µg) were solubilized in homogenization buffer and reduced using 10 mm DTT in 100 mm ammonium bicarbonate (pH 8.4) for 45 min at 56 °C and alkylated using 25 mm iodoacetamide in 100 mm ammonium bicarbonate (pH 8.4) (both final concentration) for 30 min in the dark at 22 °C. N- and O-glycans from the solubilized proteins were released and processed as previously described (54). Proteins was then blotted on a primed 0.45-μm polyvinylidene difluoride membrane and stained with Direct Blue stain solution. The stained protein spots were excised, blocked with 1% (w/v) polyvinylpyrrolidone in 50% (v/v) methanol, and washed with MilliQ water. N-glycans were enzymatically released using 3.5 units of Flavobacterium meningosepticum N-glycosidase F (Promega) in 20 μl of water for 16 h at 37 °C. The unstable amino group (-NH2) of the reducing end GlcNAc residues of N-glycosidase F–released N-glycans were allowed to spontaneously convert to hydroxyl groups (-OH) in weak acid using 100 mm ammonium acetate, pH 5, for 1 h at 22 °C to facilitate subsequent quantitative reduction to glycan alditols. Reduction of the released N-glycans was carried out using 1 m sodium borohydride in 50 mm potassium hydroxide for 3 h at 50 °C. The O-glycans were then subsequently released by reductive β-elimination from the peptide:N-glycosidase F enzyme–treated proteins by incubating overnight in 0.5 m sodium borohydride in 50 mm potassium hydroxide. The reduction reaction was stopped by glacial acetic acid quenching. Dual desalting was then performed in micro-solid phase extraction formats using strong cation exchange/C18 (where N-glycans are not retained) and PGC (where N-glycans are retained) as stationary phases, respectively. The desalted N- and O-glycans were eluted from the PGC–solid phase extraction columns using 40% (v/v) acetonitrile (ACN) containing 0.1% (v/v) aqueous trifluoroacetic acid (TFA), dried, and dissolved in 10 μl of MilliQ water for N-glycan analysis. Bovine fetuin carrying sialo-N-glycans were included as a control glycoprotein to ensure efficient N-glycan release, clean-up, and LC–MS/MS performance.
PGC–LC–MS/MS–based N- and O-glycomics
Released N- and O-glycans were analyzed by liquid chromatography–tandem MS (LC–MS/MS) in two different laboratories. Glycans from the total granule protein extracts (nonseparated) were analyzed on PGC–LC using a 10-cm × 250-μm inner-diameter column (in-house), containing 5 μm PGC particle size (Thermo Scientific, Waltham, MA, USA) connected to an LTQ- ion trap mass spectrometer (Thermo Scientific). The N- and O-glycans were eluted using a linear gradient from 0 to 40% ACN in 10 mm ammonium bicarbonate over 40 min at a flow rate of 250 μl/min split down to 5–10 μl/min. The sample injection volume was 3 μl. Electrospray ionization–MS was performed in negative ion polarity with an electrospray voltage of 3.5 kV, capillary voltage of −33.0 V, and capillary temperature of 300 °C. The following scan events were used: MS full scan (m/z 380–2000) and data-dependent MS/MS scans after collision-induced dissociation (CID) on precursor ions at a normalized collisional energy of 35% with a minimum signal of 300 counts, isolated width of 2.0 m/z, and activation time of 30 ms. The data were viewed and manually analyzed using Xcalibur software (version 2.2, Thermo Scientific).
N-Glycans released from the soluble and membrane protein extracts from each granule were analyzed using capillary LC–MS/MS (Dionex Ultimate 3000) on an electrospray ionization linear ion trap mass spectrometer (LTQ Velos Pro, Thermo Scientific, Melbourne, Australia). The samples were injected onto a PGC capillary column (Hypercarb KAPPA, 5-μm particle size, 200 Å pore size, 180-μm inner diameter × 100-mm length; Thermo Scientific) kept at 50 °C. Separation of N-glycans was carried out over a linear gradient of 2–32% ACN, 10 mm ammonium bicarbonate for 75 min at a constant flow rate of 4 μl/min. The sample injection volume was 4 μl. The acquisition range was m/z 380–2000. The acquisition was performed in negative ionization polarity with an electrospray voltage of 4.0 kV, capillary voltage of −14.9 kV, and capillary temperature of 310 °C. Data-dependent acquisition was used where the top nine most abundant precursors in each full scan spectrum were selected for CID–MS/MS at a normalized collision energy of 35%. A minimum signal of 300 counts was required for MS/MS. The precursor isolation width was m/z 1.0, and the activation time was 10 ms. The mass spectrometer was calibrated using a tune mix (Thermo Scientific). Mass spectra were viewed and analyzed using Xcalibur V2.2 (Thermo Scientific).
Structural determination of N- and O-glycans
The monosaccharide compositions and each N- and O-glycan structural sequence and linkage were manually analyzed using monoisotopic mass, CID–MS/MS fragmentation and absolute and relative retention time on the PGC–LC–MS/MS data. Monoisotopic molecular masses of the identified glycans, which were generally within 0.5 Da of the theoretical masses, were matched against likely mammalian N- and O-glycan monosaccharide compositions using the GlycoMod web tool (https://web.expasy.org/glycomod/). GlycoWorkBench v2.1 was used to draw the proposed glycans and to generate in silico glycosidic and cross-ring fragments to assist in the structural interpretation based on the obtained CID–MS/MS data. (55).
Characterization of glycans was performed manually using PGC–LC retention time, monoisotopic precursor mass, and CID–MS/MS fragmentation patterns combined with the application of biosynthetic rules and several aiding software programs and approaches to enhance the confidence of each reported glycan structure (33, 56). The CID–MS/MS fragmentation data were compared with data available in UniCarb-DB (57) and UniCarb-DR (open-access database) and to in silico fragments generated using GlycoWork Bench software (58, 59). Further, the presence of diagnostic ions was used to identify structural motifs within the reported glycans, e.g. sLex, LacNAc repeats and core fucosylation (23, 60). The absolute and relative PGC retention behavior of each glycan (recorded in UniCarb-DB, UniCarb-DR, and Glycostore (https://glycostore.org)) was used to increase the confidence of the structures identified, especially for isomers. This includes the relative early elution of α2,6-sialylated versus α2,3-sialylated isomers (61) and core versus antennary fucosylation (60). For the larger glycan structures (>3200 Da) that did not produce high-quality fragment spectral data, we relied on biosynthetic rules of lactosamine extension and termination of lactosamines by sialylation or Lewis-type structures. For this reason, some structural ambiguity (indicated with brackets and open/unannotated glycosidic linkages) remained for the large glycan structures.
Statistical analysis
Statistical analysis was performed using GraphPad Prism version 8 software (La Jolla, CA, USA). The quantitative data are expressed as means ± S.E. and analyzed using two-tailed Student's t test, and p values < 0.05 were considered statistically significant.
Ethics statement
According to Swedish law on ethical conduct in human research, ethics approval for buffy coats was not needed because they were provided anonymously and could not be traced back to a specific individual.
Data availability
The raw MS/MS data files are submitted in GlycoPost (accession ID GPST000050).
Supplementary Material
This article contains supporting information.
Author contributions—V. V., R. D., J. B., M. T.-A., N. G. K., and A. K.-B. conceptualization; V. V., R. D., I. L., H. C. T., S. C., M. T.-A., N. G. K., and A. K.-B. data curation; V. V., I. L., H. C. T., S. C., and M. T.-A. formal analysis; V. V. and J. B. funding acquisition; V. V., R. D., J. B., M. T.-A., N. G. K., and A. K.-B. validation; V. V., R. D., J. B., M. T.-A., and A. K.-B. investigation; V. V., R. D., and N. G. K. methodology; V. V., R. D., I. L., H. C. T., S. C., M. T.-A., N. G. K., and A. K.-B. writing-original draft; V. V., R. D., I. L., HT, S. C., J. B., M. T.-A., N. G. K., and A. K.-B. writing-review and editing; R. D. and A. K.-B. project administration; J. B., M. T.-A., and A. K.-B. supervision.
Funding and additional information—This work was supported by Swedish Research Council Grants 2018-03077 and 2019-01123), the King Gustaf V 80-Year Memorial Foundation, the Swedish Heart–Lung Foundation, and the Swedish government under the agreement concerning research and education of physicians (ALF). The mass spectrometer (LTQ) was supported by the Knut and Alice Wallenberg Foundation.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- AG
- azurophil granule(s)
- SG
- specific granule(s)
- GG
- gelatinase granule(s)
- SV
- secretory vesicle(s)
- PM
- plasma membrane
- MPO
- myeloperoxidase
- LF
- lactoferrin
- MMP-9
- matrix metalloproteinase-9
- ALP
- alkaline phosphatase
- PGC
- porous graphitized carbon
- LacNAc
- N-acetyllactosamine
- Lex
- Lewis X
- sLex
- sialyl-Lewis X
- ACN
- acetonitrile
- CID
- collision-induced dissociation.
References
- 1. Borregaard N., and Cowland J. B. (1997) Granules of the human neutrophilic polymorphonuclear leukocyte. Blood 89, 3503–3521 10.1182/blood.V89.10.3503.3503_3503_3521 [DOI] [PubMed] [Google Scholar]
- 2. Faurschou M., and Borregaard N. (2003) Neutrophil granules and secretory vesicles in inflammation. Microbes Infect. 5, 1317–1327 10.1016/j.micinf.2003.09.008 [DOI] [PubMed] [Google Scholar]
- 3. Rørvig S., Østergaard O., Heegaard N. H., and Borregaard N. (2013) Proteome profiling of human neutrophil granule subsets, secretory vesicles, and cell membrane: correlation with transcriptome profiling of neutrophil precursors. J. Leukoc. Biol. 94, 711–721 10.1189/jlb.1212619 [DOI] [PubMed] [Google Scholar]
- 4. Le Cabec V., Cowland J. B., Calafat J., and Borregaard N. (1996) Targeting of proteins to granule subsets is determined by timing and not by sorting: the specific granule protein NGAL is localized to azurophil granules when expressed in HL-60 cells. Proc. Natl. Acad. Sci. U.S.A. 93, 6454–6457 10.1073/pnas.93.13.6454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Phillips M. L., Nudelman E., Gaeta F. C., Perez M., Singhal A. K., Hakomori S., and Paulson J. C. (1990) ELAM-1 mediates cell adhesion by recognition of a carbohydrate ligand, sialyl-Lex. Science 250, 1130–1132 10.1126/science.1701274 [DOI] [PubMed] [Google Scholar]
- 6. Walz G., Aruffo A., Kolanus W., Bevilacqua M., and Seed B. (1990) Recognition by ELAM-1 of the sialyl-Lex determinant on myeloid and tumor cells. Science 250, 1132–1135 10.1126/science.1701275 [DOI] [PubMed] [Google Scholar]
- 7. Yamaoka A., Kuwabara I., Frigeri L. G., and Liu F. T. (1995) A human lectin, galectin-3 (epsilon bp/Mac-2), stimulates superoxide production by neutrophils. J. Immunol. 154, 3479–3487 [PubMed] [Google Scholar]
- 8. Carlin A. F., Uchiyama S., Chang Y. C., Lewis A. L., Nizet V., and Varki A. (2009) Molecular mimicry of host sialylated glycans allows a bacterial pathogen to engage neutrophil Siglec-9 and dampen the innate immune response. Blood 113, 3333–3336 10.1182/blood-2008-11-187302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Graham S. A., Antonopoulos A., Hitchen P. G., Haslam S. M., Dell A., Drickamer K., and Taylor M. E. (2011) Identification of neutrophil granule glycoproteins as Lewisx-containing ligands cleared by the scavenger receptor C-type lectin. J. Biol. Chem. 286, 24336–24349 10.1074/jbc.M111.244772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Babu P., North S. J., Jang-Lee J., Chalabi S., Mackerness K., Stowell S. R., Cummings R. D., Rankin S., Dell A., and Haslam S. M. (2009) Structural characterisation of neutrophil glycans by ultra sensitive mass spectrometric glycomics methodology. Glycoconj. J. 26, 975–986 10.1007/s10719-008-9146-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Karlsson A., Miller-Podraza H., Johansson P., Karlsson K. A., Dahlgren C., and Teneberg S. (2001) Different glycosphingolipid composition in human neutrophil subcellular compartments. Glycoconj. J. 18, 231–243 10.1023/A:1013183124004 [DOI] [PubMed] [Google Scholar]
- 12. Foxall C., Watson S. R., Dowbenko D., Fennie C., Lasky L. A., Kiso M., Hasegawa A., Asa D., and Brandley B. K. (1992) The three members of the selectin receptor family recognize a common carbohydrate epitope, the sialyl Lewisx oligosaccharide. J. Cell Biol. 117, 895–902 10.1083/jcb.117.4.895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nimrichter L., Burdick M. M., Aoki K., Laroy W., Fierro M. A., Hudson S. A., Von Seggern C. E., Cotter R. J., Bochner B. S., Tiemeyer M., Konstantopoulos K., and Schnaar R. L. (2008) E-selectin receptors on human leukocytes. Blood 112, 3744–3752 10.1182/blood-2008-04-149641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Etzioni A., Frydman M., Pollack S., Avidor I., Phillips M. L., Paulson J. C., and Gershoni-Baruch R. (1992) Recurrent severe infections caused by a novel leukocyte adhesion deficiency. N. Engl. J. Med. 327, 1789–1792 10.1056/NEJM199212173272505 [DOI] [PubMed] [Google Scholar]
- 15. Etzioni A., Harlan J. M., Pollack S., Phillips L. M., Gershoni-Baruch R., and Paulson J. C. (1993) Leukocyte adhesion deficiency (LAD) II: a new adhesion defect due to absence of sialyl Lewis X, the ligand for selectins. Immunodeficiency 4, 307–308 [PubMed] [Google Scholar]
- 16. Lucka L., Fernando M., Grunow D., Kannicht C., Horst A. K., Nollau P., and Wagener C. (2005) Identification of Lewis x structures of the cell adhesion molecule CEACAM1 from human granulocytes. Glycobiology 15, 87–100 10.1093/glycob/cwh139 [DOI] [PubMed] [Google Scholar]
- 17. Poland D. C., García Vallejo J. J., Niessen H. W., Nijmeyer R., Calafat J., Hack C. E., Van Het Hof B., and Van Dijk W. (2005) Activated human PMN synthesize and release a strongly fucosylated glycoform of α1-acid glycoprotein, which is transiently deposited in human myocardial infarction. J. Leukoc. Biol. 78, 453–461 10.1189/jlb.1004566 [DOI] [PubMed] [Google Scholar]
- 18. Theilgaard-Mönch K., Jacobsen L. C., Rasmussen T., Niemann C. U., Udby L., Borup R., Gharib M., Arkwright P. D., Gombart A. F., Calafat J., Porse B. T., and Borregaard N. (2005) Highly glycosylated α1-acid glycoprotein is synthesized in myelocytes, stored in secondary granules, and released by activated neutrophils. J. Leukoc. Biol. 78, 462–470 10.1189/jlb.0105042 [DOI] [PubMed] [Google Scholar]
- 19. Brazil J. C., Sumagin R., Cummings R. D., Louis N. A., and Parkos C. A. (2016) Targeting of neutrophil Lewis X blocks transepithelial migration and increases phagocytosis and degranulation. Am. J. Pathol. 186, 297–311 10.1016/j.ajpath.2015.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Venkatakrishnan V., Thaysen-Andersen M., Chen S. C., Nevalainen H., and Packer N. H. (2015) Cystic fibrosis and bacterial colonization define the sputum N-glycosylation phenotype. Glycobiology 25, 88–100 10.1093/glycob/cwu092 [DOI] [PubMed] [Google Scholar]
- 21. Thaysen-Andersen M., Venkatakrishnan V., Loke I., Laurini C., Diestel S., Parker B. L., and Packer N. H. (2015) Human neutrophils secrete bioactive paucimannosidic proteins from azurophilic granules into pathogen-infected sputum. J. Biol. Chem. 290, 8789–8802 10.1074/jbc.M114.631622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Loke I., Østergaard O., Heegaard N. H. H., Packer N. H., and Thaysen-Andersen M. (2017) Paucimannose-rich N-glycosylation of spatiotemporally regulated human neutrophil elastase modulates its immune functions. Mol. Cell. Proteomics 16, 1507–1527 10.1074/mcp.M116.066746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Everest-Dass A. V., Abrahams J. L., Kolarich D., Packer N. H., and Campbell M. P. (2013) Structural feature ions for distinguishing N- and O-linked glycan isomers by LC–ESI–IT MS/MS. J. Am. Soc. Mass Spectrom. 24, 895–906 10.1007/s13361-013-0610-4 [DOI] [PubMed] [Google Scholar]
- 24. Harvey D. J. (2005) Fragmentation of negative ions from carbohydrates: part 3. Fragmentation of hybrid and complex N-linked glycans. J. Am. Soc. Mass Spectrom. 16, 647–659 10.1016/j.jasms.2005.01.006 [DOI] [PubMed] [Google Scholar]
- 25. Harvey D. J., Royle L., Radcliffe C. M., Rudd P. M., and Dwek R. A. (2008) Structural and quantitative analysis of N-linked glycans by matrix-assisted laser desorption ionization and negative ion nanospray mass spectrometry. Anal. Biochem. 376, 44–60 10.1016/j.ab.2008.01.025 [DOI] [PubMed] [Google Scholar]
- 26. Lominadze G., Ward R. A., Klein J. B., and McLeish K. R. (2006) Proteomic analysis of human neutrophils. Methods Mol. Biol. 332, 343–356 10.1385/1-59745-048-0:343 [DOI] [PubMed] [Google Scholar]
- 27. Borregaard N., Kjeldsen L., Rygaard K., Bastholm L., Nielsen M. H., Sengeløv H., Bjerrum O. W., and Johnsen A. H. (1992) Stimulus-dependent secretion of plasma proteins from human neutrophils. J. Clin. Invest. 90, 86–96 10.1172/JCI115860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bagger F. O., Kinalis S., and Rapin N. (2019) BloodSpot: a database of healthy and malignant haematopoiesis updated with purified and single cell mRNA sequencing profiles. Nucleic Acids Res. 47, D881–D885 10.1093/nar/gky1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tjondro H. C., Loke I., Chatterjee S., and Thaysen-Andersen M. (2019) Human protein paucimannosylation: cues from the eukaryotic kingdoms. Biol. Rev. Camb. Philos. Soc. 94, 2068–2100 10.1111/brv.12548 [DOI] [PubMed] [Google Scholar]
- 30. Mora-Jensen H., Jendholm J., Fossum A., Porse B., Borregaard N., Borregaad N., and Theilgaard-Mönch K. (2011) Technical advance: immunophenotypical characterization of human neutrophil differentiation. J. Leukoc. Biol. 90, 629–634 10.1189/jlb.0311123 [DOI] [PubMed] [Google Scholar]
- 31. Theilgaard-Mönch K., Jacobsen L. C., Borup R., Rasmussen T., Bjerregaard M. D., Nielsen F. C., Cowland J. B., and Borregaard N. (2005) The transcriptional program of terminal granulocytic differentiation. Blood 105, 1785–1796 10.1182/blood-2004-08-3346 [DOI] [PubMed] [Google Scholar]
- 32. Hoogendijk A. J., Pourfarzad F., Aarts C. E. M., Tool A. T. J., Hiemstra I. H., Grassi L., Frontini M., Meijer A. B., van den Biggelaar M., and Kuijpers T. W. (2019) Dynamic transcriptome–proteome correlation networks reveal human myeloid differentiation and neutrophil-specific programming. Cell Rep. 29, 2505–2519.e4 10.1016/j.celrep.2019.10.082 [DOI] [PubMed] [Google Scholar]
- 33. Ashwood C., Lin C. H., Thaysen-Andersen M., and Packer N. H. (2018) Discrimination of isomers of released N- and O-glycans using diagnostic product ions in negative ion PGC–LC–ESI–MS/MS. J. Am. Soc. Mass Spectrom. 29, 1194–1209 10.1007/s13361-018-1932-z [DOI] [PubMed] [Google Scholar]
- 34. Hinneburg H., Chatterjee S., Schirmeister F., Nguyen-Khuong T., Packer N. H., Rapp E., and Thaysen-Andersen M. (2019) Post-column make-up flow (PCMF) enhances the performance of capillary-flow PGC–LC–MS/MS–Based glycomics. Anal. Chem. 91, 4559–4567 10.1021/acs.analchem.8b05720 [DOI] [PubMed] [Google Scholar]
- 35. Loke I., Packer N. H., and Thaysen-Andersen M. (2015) Complementary LC-MS/MS-based N-glycan, N-glycopeptide, and intact N-glycoprotein profiling reveals unconventional Asn71-glycosylation of human neutrophil cathepsin G. Biomolecules 5, 1832–1854 10.3390/biom5031832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Reiding K. R., Franc V., Huitema M. G., Brouwer E., Heeringa P., and Heck A. J. R. (2019) Neutrophil myeloperoxidase harbors distinct site-specific peculiarities in its glycosylation. J. Biol. Chem. 294, 20233–20245 10.1074/jbc.RA119.011098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Feinberg H., Mitchell D. A., Drickamer K., and Weis W. I. (2001) Structural basis for selective recognition of oligosaccharides by DC-SIGN and DC-SIGNR. Science 294, 2163–2166 10.1126/science.1066371 [DOI] [PubMed] [Google Scholar]
- 38. Loke I., Kolarich D., Packer N. H., and Thaysen-Andersen M. (2016) Emerging roles of protein mannosylation in inflammation and infection. Mol. Aspects Med. 51, 31–55 10.1016/j.mam.2016.04.004 [DOI] [PubMed] [Google Scholar]
- 39. Shepherd V. L., and Hoidal J. R. (1990) Clearance of neutrophil-derived myeloperoxidase by the macrophage mannose receptor. Am. J. Respir. Cell Mol. Biol. 2, 335–340 10.1165/ajrcmb/2.4.335 [DOI] [PubMed] [Google Scholar]
- 40. Karlsson A., Christenson K., Matlak M., Björstad A., Brown K. L., Telemo E., Salomonsson E., Leffler H., and Bylund J. (2009) Galectin-3 functions as an opsonin and enhances the macrophage clearance of apoptotic neutrophils. Glycobiology 19, 16–20 10.1093/glycob/cwn104 [DOI] [PubMed] [Google Scholar]
- 41. Kuwabara I., and Liu F. T. (1996) Galectin-3 promotes adhesion of human neutrophils to laminin. J. Immunol. 156, 3939–3944 [PubMed] [Google Scholar]
- 42. Feuk-Lagerstedt E., Jordan E. T., Leffler H., Dahlgren C., and Karlsson A. (1999) Identification of CD66a and CD66b as the major galectin-3 receptor candidates in human neutrophils. J. Immunol. 163, 5592–5598 [PubMed] [Google Scholar]
- 43. Salomonsson E., Carlsson M. C., Osla V., Hendus-Altenburger R., Kahl-Knutson B., Oberg C. T., Sundin A., Nilsson R., Nordberg-Karlsson E., Nilsson U. J., Karlsson A., Rini J. M., and Leffler H. (2010) Mutational tuning of galectin-3 specificity and biological function. J. Biol. Chem. 285, 35079–35091 10.1074/jbc.M109.098160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Polley M. J., Phillips M. L., Wayner E., Nudelman E., Singhal A. K., Hakomori S., and Paulson J. C. (1991) CD62 and endothelial cell-leukocyte adhesion molecule 1 (ELAM-1) recognize the same carbohydrate ligand, sialyl-Lewis x. Proc. Natl. Acad. Sci. U.S.A. 88, 6224–6228 10.1073/pnas.88.14.6224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Beauharnois M. E., Lindquist K. C., Marathe D., Vanderslice P., Xia J., Matta K. L., and Neelamegham S. (2005) Affinity and kinetics of sialyl Lewis-X and core-2 based oligosaccharides binding to L- and P-selectin. Biochemistry 44, 9507–9519 10.1021/bi0507130 [DOI] [PubMed] [Google Scholar]
- 46. Brazil J. C., Sumagin R., Stowell S. R., Lee G., Louis N. A., Cummings R. D., and Parkos C. A. (2017) Expression of Lewis-a glycans on polymorphonuclear leukocytes augments function by increasing transmigration. J. Leukoc. Biol. 102, 753–762 10.1189/jlb.1MA0117-013R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sauer M. M., Jakob R. P., Luber T., Canonica F., Navarra G., Ernst B., Unverzagt C., Maier T., and Glockshuber R. (2019) Binding of the bacterial adhesin FimH to its natural, multivalent high-mannose type glycan targets. J. Am. Chem. Soc. 141, 936–944 10.1021/jacs.8b10736 [DOI] [PubMed] [Google Scholar]
- 48. Tewari R., MacGregor J. I., Ikeda T., Little J. R., Hultgren S. J., and Abraham S. N. (1993) Neutrophil activation by nascent FimH subunits of type 1 fimbriae purified from the periplasm of Escherichia coli. J. Biol. Chem. 268, 3009–3015 [PubMed] [Google Scholar]
- 49. Bainton D. F., and Farquhar M. G. (1966) Origin of granules in polymorphonuclear leukocytes: two types derived from opposite faces of the Golgi complex in developing granulocytes. J. Cell Biol. 28, 277–301 10.1083/jcb.28.2.277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bainton D. F., Ullyot J. L., and Farquhar M. G. (1971) The development of neutrophilic polymorphonuclear leukocytes in human bone marrow. J. Exp. Med. 134, 907–934 10.1084/jem.134.4.907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bøyum A., Løvhaug D., Tresland L., and Nordlie E. M. (1991) Separation of leucocytes: improved cell purity by fine adjustments of gradient medium density and osmolality. Scand. J. Immunol. 34, 697–712 10.1111/j.1365-3083.1991.tb01594.x [DOI] [PubMed] [Google Scholar]
- 52. Borregaard N., Heiple J. M., Simons E. R., and Clark R. A. (1983) Subcellular localization of the b-cytochrome component of the human neutrophil microbicidal oxidase: translocation during activation. J. Cell Biol. 97, 52–61 10.1083/jcb.97.1.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Feuk-Lagerstedt E., Movitz C., Pellmé S., Dahlgren C., and Karlsson A. (2007) Lipid raft proteome of the human neutrophil azurophil granule. Proteomics 7, 194–205 10.1002/pmic.200600482 [DOI] [PubMed] [Google Scholar]
- 54. Jensen P. H., Karlsson N. G., Kolarich D., and Packer N. H. (2012) Structural analysis of N- and O-glycans released from glycoproteins. Nat. Protoc. 7, 1299–1310 10.1038/nprot.2012.063 [DOI] [PubMed] [Google Scholar]
- 55. Ceroni A., Maass K., Geyer H., Geyer R., Dell A., and Haslam S. M. (2008) GlycoWorkbench: a tool for the computer-assisted annotation of mass spectra of glycans. J. Proteome Res. 7, 1650–1659 10.1021/pr7008252 [DOI] [PubMed] [Google Scholar]
- 56. Hinneburg H., Pedersen J. L., Bokil N. J., Pralow A., Schirmeister F., Kawahara R., Rapp E., Saunders B. M., and Thaysen-Andersen M. (2020) High-resolution longitudinal N- and O-glycoprofiling of human monocyte-to- macrophage transition. Glycobiology, in press 10.1093/glycob/cwaa020 [DOI] [PubMed] [Google Scholar]
- 57. Campbell M. P., Nguyen-Khuong T., Hayes C. A., Flowers S. A., Alagesan K., Kolarich D., Packer N. H., and Karlsson N. G. (2014) Validation of the curation pipeline of UniCarb-DB: building a global glycan reference MS/MS repository. Biochim. Biophys. Acta 1844, 108–116 10.1016/j.bbapap.2013.04.018 [DOI] [PubMed] [Google Scholar]
- 58. Damerell D., Ceroni A., Maass K., Ranzinger R., Dell A., and Haslam S. M. (2012) The GlycanBuilder and GlycoWorkbench glycoinformatics tools: updates and new developments. Biol. Chem. 393, 1357–1362 10.1515/hsz-2012-0135 [DOI] [PubMed] [Google Scholar]
- 59. Damerell D., Ceroni A., Maass K., Ranzinger R., Dell A., and Haslam S. M. (2015) Annotation of glycomics MS and MS/MS spectra using the GlycoWorkbench software tool. Methods Mol. Biol. 1273, 3–15 10.1007/978-1-4939-2343-4_1 [DOI] [PubMed] [Google Scholar]
- 60. Karlsson N. G., Schulz B. L., and Packer N. H. (2004) Structural determination of neutral O-linked oligosaccharide alditols by negative ion LC–electrospray–MSn. J. Am. Soc. Mass Spectrom. 15, 659–672 10.1016/j.jasms.2004.01.002 [DOI] [PubMed] [Google Scholar]
- 61. Pabst M., Bondili J. S., Stadlmann J., Mach L., and Altmann F. (2007) Mass + retention time = structure: a strategy for the analysis of N-glycans by carbon LC–ESI–MS and its application to fibrin N-glycans. Anal. Chem. 79, 5051–5057 10.1021/ac070363i [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw MS/MS data files are submitted in GlycoPost (accession ID GPST000050).