ABSTRACT
Mucus is an important host innate defense factor that lines most epithelial cell layers of the body and provides crucial physical and biological protection against pathogenic microorganisms. Mucins are the main glycoproteins of mucus that are responsible for interacting with microorganisms and are critical for the antimicrobial properties of mucus. The mechanisms by which microorganisms interact with mucins are poorly understood, especially in terms of fungi, and these interactions are continually evolving. Work in bacterial pathogens has shown that mucins inhibit bacterial virulence traits, including quorum sensing, toxin secretion and biofilm formation. Among the fungal clade, the common opportunistic human fungal pathogen and commensal Candida albicans engages in constant battle with the host innate immune system. This battle creates strong selective pressures for C. albicans to evolve in response to the host. Recent work in C. albicans found that mucins inhibit specific virulence traits, such as surface adherence, filamentation, biofilm formation and the production of secreted proteases. Here we review the current knowledge of microbial interactions with mucins, with a special emphasis on the interactions between C. albicans and mucins.
Keywords: mucus, mucins, mucin monomer, Candida albicans, innate immunity, biofilms, host–pathogen interactions, mucosal surface, epithelial cell layer, viscoelasticity
This review describes the current knowledge on the interactions between microorganisms and host mucins, with a focus on the opportunistic human fungal pathogen and commensal Candida albicans.
INTRODUCTION
The healthy human microbiota is composed of hundreds of trillions of diverse microorganisms, including bacteria, fungi and archaea, that share and compete for nutrients and environmental niches in the body (Ursell et al. 2013; Wang et al. 2017). Over the course of millions of years of evolution, these microorganisms have coevolved in a mutually symbiotic relationship with the host, where they contribute to host physiological processes; the host, in turn, provides a hospitable environment for these microorganisms to reside (Relman 2008; Chow et al. 2010; Pickard et al. 2017). In addition to providing physiological benefits to the host (e.g. through nutrient acquisition and synthesis), these microorganisms also protect the host from invading pathogens that may enter and colonize the host from the outside environment (Chow et al. 2010; Buffie and Pamer 2013; Sassone-Corsi and Raffatellu 2015; Chiu et al. 2017). Although the majority of members of the microbiota typically behave as mutualists or commensals, some of these microorganisms can have pathogenic potential under certain circumstances (Casadevall and Pirofski 2001; Casadevall 2017; Libertucci and Young 2019). Most research to date has focused on studying bacterial members of the microbiota, but fungal members have been found to play increasingly important roles in interacting with the host and in shaping the functions of the microbiota (Underhill and Iliev 2014; Lukeš et al. 2015; Kumamoto 2016). Both members of the microbiota and invading pathogens from the environment typically make contact with and often reside on mucosal membranes covering epithelial cell layers found in the body, such as the oral cavity, eyes, nose, ears, respiratory, gastrointestinal and reproductive tracts (Fig. 1; Linden et al. 2008; Hansson 2012; Frenkel and Ribbeck 2015). Regardless of where they originated from (the microbiota or the outside environment), those microorganisms that can breach the mucosal barrier have the potential to cause systemic infections in the host (Linden et al. 2008; Belkaid and Hand 2014). Consistently, defects in mucus production, mucus physiochemical properties, or in the expression of mucins can lead to several disease states in humans, such as cystic fibrosis, inflammatory bowel disease, ulcerative colitis, Sjogren's syndrome, cancer and preterm birth, which are all associated with microbial dysbiosis and often the presence of an infection (Henke et al. 2004, 2007; Kim and Ho 2010; Williams, Ranjendran and Ramage 2016; Wagner, Wheeler and Ribbeck 2018; Schroeder 2019). In this review, we focus on the interactions between mucins, the major components of mucus responsible for mediating microbial interactions with the host, and microorganisms with pathogenic potential, with an emphasis on Candida albicans, a common opportunistic fungal pathogen and commensal of humans.
Figure 1.
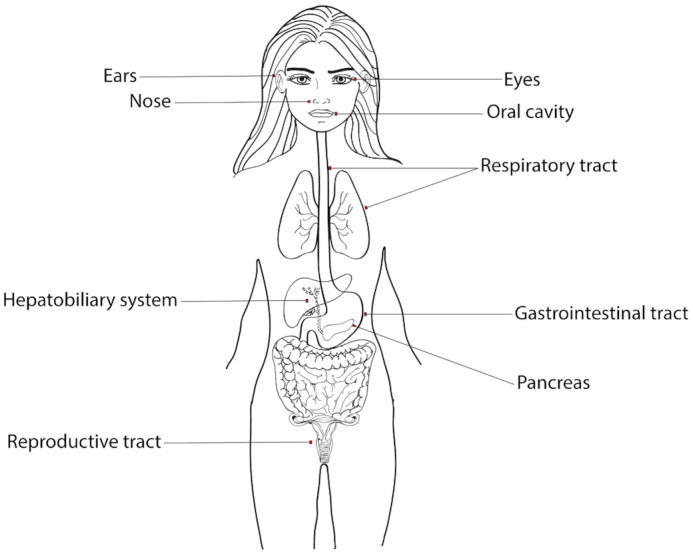
Mucosal membranes covering epithelial cell layers of the human body. Microorganisms are typically associated with mucosal membranes of the eyes, nose, oral cavity, respiratory, gastrointestinal and reproductive tracts. These microorganisms can include both commensals and pathogens. Figure adapted from (Frenkel and Ribbeck 2015)
It is estimated that fungal diseases cost the United States approximately $7.2 billion annually, where Candida infections account for approximately 20% of these costs (Benedict et al. 2019). Candida albicans, the most commonly isolated fungal pathogen from clinical settings, typically resides as a commensal fungus in the microbiota of the skin, vagina, gastrointestinal and urogenital tracts of humans (Kennedy and Volz 1985; Kumamoto 2002, 2011; Achkar and Fries 2010; Ganguly and Mitchell 2011). When alterations to the host microbiota occur, such as by changes in pH or residing microorganisms, C. albicans can overgrow, become invasive and cause a wide range of infections (Odds 1987; Kim and Sudbery 2011; Nobile and Johnson 2015). These infections, collectively referred to as candidiasis, can range from superficial skin infections to severe bloodstream infections, the latter of which typically occur in immunocompromised individuals and can be life-threating (Haynes 2001; Kullberg and Oude 2002; Kim and Sudbery 2011). C. albicans possesses numerous virulence traits that contribute to its pathogenicity, such as the production of host recognition molecules, the ability to undergo morphological transitions, and the release of secreted aspartyl proteases and phospholipases that can damage host cells (Calderone and Fonzi 2001). In addition, the ability to form biofilms, recalcitrant communities of cells encased in extracellular matrices, is another important virulence trait of C. albicans that enhances its survival in the host (Kumamoto 2002; Douglas 2003; Ganguly and Mitchell 2011; Nobile and Johnson 2015; Gulati and Nobile 2016; Lohse et al. 2018).
C. albicans biofilm formation in a host setting begins when C. albicans cells colonize a mucosal surface covering a layer of epithelial cells or an implanted medical device. The C. albicans biofilm life cycle consists of four basic stages (Chandra et al. 2001; Douglas 2003; Gulati and Nobile 2016). In the first stage, round yeast form cells adhere to a solid surface (e.g. the intestinal mucosa or an implanted central venous catheter; Kennedy et al. 1987; Hawser and Douglas 1994; Baillie and Douglas 1999; Nobile and Johnson 2015). This is followed by proliferation of the adhered cells and early stage filamentation (Baillie and Douglas 1999; Nobile and Johnson 2015). As the biofilm matures, extensive filamentation takes place along with the production of the extracellular matrix, which is comprised of proteins, polysaccharides, nucleic acids and lipids (Baillie and Douglas 1999; Zarnowski et al. 2014; Pierce et al. 2017). As a result, the mature biofilm architecture is such that it provides structural protection to the cells within the biofilm from both chemical and mechanical insults (Mitchell, Zarnowski and Andes 2016). In addition to this structural protection, cells within the biofilm upregulate drug efflux pumps, further enhancing the resistance and tolerance of biofilms to inhibitory compounds. In the final stage of the C. albicans biofilm life cycle, round yeast form cells disperse from the biofilm to colonize new sites (Uppuluri et al. 2010; Nobile and Johnson 2015). Taken together, C. albicans biofilms can not only be highly resistant and tolerant to chemical and mechanical perturbations but can also act as reservoirs to seed new sites of infection. Interestingly, recent work in C. albicans found that specific fungal virulence traits, such as surface adherence, filamentation and biofilm formation, are compromised in the presence of mucins (Kavanaugh et al. 2014).
In the following sections of this review, we begin by discussing the properties, functions and structures of mucins. We then review the types of mucins present in the human body and examine their production and biosynthesis. Lastly, we review known interactions between microorganisms and mucins, with a focus on the interactions between C. albicans and mucins.
Mucus and mucins—an overview
Mucus is a viscoelastic hydrogel that is comprised of 95% water, 3% mucin glycoproteins and 2% other small molecules, including immunoglobulin A (IgA), lipids and antimicrobial peptides (Celli et al. 2005). Mucus provides lubrication and hydration to epithelial linings and is a critical innate defense factor that protects the host against infection; this protection is largely attributed to large glycoproteins called mucins, which can be secreted or membrane-bound (Gendler 1995; Liévin-le Moal and Servin 2017; Petrou and Crouzier 2018). The unique physiochemical properties of mucus are such that mucus limits microbial penetration through the epithelial cell layer, while at the same time permits the passage of water and gases (Cone 2009; Bakshani et al. 2018). In addition to these properties, mucosal surfaces continuously regenerate, allowing for the efficient removal of contaminants, and preventing them from reaching the underlying epithelial cell layer (Cone 2009). Mucins within mucus are known to mediate physical interactions with microorganisms, serve as receptor binding sites for the adhesion of molecules, act as nutrient sources for microorganisms, serve as biochemical signals, and support gaseous exchange and nutrient absorption between host cells (Wagner, Wheeler and Ribbeck 2018). Some of these roles depend on where the mucins are produced and localized in the body. For example, a major role of lung mucins is to support gaseous exchange between host cells, while for gut mucins, it is to support nutrient absorption (Corfield 2015). Lastly, it is known that mucins can inhibit virulence traits, such as biofilm formation, motility and cellular morphology changes in opportunistic pathogens, thus maintaining them in a commensal state (Ogasawara et al. 2007; Celli et al. 2009; Caldara et al. 2012; Kavanaugh et al. 2014; Co et al. 2018).
Mucin monomer molecular weights range from 0.5–20 MDa (Bansil and Turner 2006; Balabushevich et al. 2018), where approximately 80% of the molecular weight of a mucin monomer comes from polysaccharides that are attached to the protein core, including N-acetylgalactosamine, N-acetylglucosamine, sialic acid, fucose and galactose (Bansil and Turner 2006; Brockhauser, Schachter and Stanley 2009). The protein core consisting of proline, threonine and serine (called the PTS domain) makes up the remaining 20% of the molecular weight of a mucin monomer, and is the main glycosylated region of the protein (Fig. 2; Bansil and Turner 2006; Brockhauser, Schachter and Stanley 2009).
Figure 2.
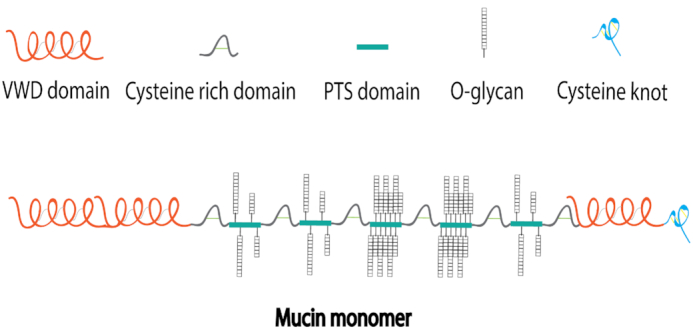
Schematic drawing of a mucin monomer. Each mucin monomer consists of a protein core called the PTS domain that is comprised primarily of proline, threonine and serine. O-glycosylation of polysaccharides occurs at PTS domains between cysteine rich domains. Typically, the C-terminus of the protein backbone contains a cysteine knot and the N-terminus contains several von Willebrand D (VWD) domains.
Human mucin glycoproteins belong to the MUC protein family, which is currently known to consist of 21 secreted and membrane-bound mucins (Corfield 2015, 2018). The five major secreted gel-forming mucins in the human body, which are important contributors to the viscoelasticity of mucus are MUC2, MUC5AC, MUC5B, MUC6 and MUC19 (Thornton, Rousseau and McGuckin 2008). MUC5AC and MUC5B are structurally similar proteins but are found in different niches of the host. For example, MUC5AC is found in mucus of the gastrointestinal and respiratory tracts, and MUC5B is found in salivary and cervical mucus. Currently, there are three known secreted but non-gel forming MUC proteins, MUC7, MUC8 and MUC9 (Corfield 2018). Unlike the gel-forming secreted mucins, MUC7 and MUC8 are not implicated in mediating viscoelasticity, however they have been shown to exhibit microbe-binding and anti-inflammatory properties (Xu et al. 2016; Cha and Song 2018). MUC7 specifically has been shown to possess fungicidal activity via a histatin-like domain found at its N-terminal region (Gururaja et al. 1999; Puri and Edgerton 2014). The remainder of the MUC protein family is made up of large membrane-bound mucins (also called tethered or cell surface-associated mucins) that form the glycocalyx mucus barrier and are known to mediate adherence to mucosal surfaces and to limit access of microorganisms to epithelial cell layers (Linden et al. 2008; Roy et al. 2014). These membrane-bound mucins include MUC1, MUC3A/B, MUC4, MUC12, MUC13, MUC15, MUC16, MUC17, MUC20, MUC21 and MUC22 (Linden et al. 2008; Pelaseyed and Hansson 2020).
Production of mucus and biosynthesis of mucins
Mucus is produced by mucus cells found in the surface epithelium, such as in goblet epithelial cells, in mucus glands, and in mixed glands containing mucus and serous cells (Lillehoj et al.2013; Pelaseyed et al. 2014). From there, mucus is secreted onto the epithelial cell layer forming the mucosal surface (Linden et al. 2008; Pelaseyed et al. 2014; Corfield 2015).
Polymerization and secretion of gel-forming secreted mucins into the mucosa is critical for creating the viscoelastic properties of mucus, while localization of membrane-bound mucins to the cell membrane is critical for forming the glycocalyx mucus barrier. For secreted mucins, oligomerization occurs by rapid dimerization of mucin monomers in the endoplasmic reticulum (ER) (Linden et al. 2008; Corfield 2015). This is followed by O-glycosylation in the Golgi apparatus (Linden et al. 2008; Corfield 2015). For membrane-bound mucins, which are monomeric, proper synthesis in the ER is dependent on cleavage of an SEA domain into two subunits by autoproteolysis concurrent with N-glycosylation (Macao et al. 2006). For membrane-bound mucins, similar to secreted mucins, O-glycosylation also takes place in the Golgi apparatus, but for membrane-bound mucins, the newly synthesized mucin monomer is then tethered to the cell membrane (Linden et al. 2008; Corfield 2015).
Microbial interactions with mucins
Mucins in mucus are critical in the host's defense against invading microorganisms. Consistent with this concept, host genes encoding mucins have been found to be upregulated in the presence of invading microorganisms (Wagner, Wheeler and Ribbeck 2018). In one example, exposure of ear epithelial cells to the otitis media causing bacteria Streptococcus pneumoniae, Haemophilus influenza and Moraxella catarrhalis led to an upregulation of host genes encoding MUC2, MUC5AC and MUC5B (Kerschner et al. 2014; Wagner, Wheeler and Ribbeck 2018). In another example, exposure of lung epithelial cells to Pseudomonas aeruginosa, Escherichia coli, Staphylococcus aureus, Staphylococcus epidermidis and Streptococcus pyogenes led to an upregulation of host genes encoding MUC5AC and MUC2 (Dohrman et al. 1998; Wagner, Wheeler and Ribbeck 2018). From the pathogen perspective, in response to host mucins, invading microorganisms can defend themselves in a number of different ways. They can, for example, secrete hydrolytic enzymes, such as proteases and glycosidases that degrade mucins and promote microbial invasion into the underlying host epithelial cell layer (Derrien et al. 2010). For example, in response to mucins, E. coli secretes the metalloprotease SslE, Vibrio cholerae secretes the metalloprotease TagA, and C. albicans secretes the aspartyl protease Sap2, which can all degrade mucins, potentially allowing for microbial penetration of the mucosal barrier (Colina et al. 1996; Szabady et al. 2011; Luo et al. 2014; Valeri et al. 2015).
Interestingly, other than invading microorganisms, some symbiotic members of the microbiota can also degrade mucins and are important in maintaining healthy host metabolic processes (Derrien et al. 2010; Tailford et al. 2015). For example, Akkermansia muciniphila, a bacterial colonizer of the mucosal layer of the intestinal tract that can degrade mucins is depleted in individuals with metabolic disorders, such as in diabetic and obese individuals (Collado et al. 2007; Cani and de Vos 2017; Shin et al. 2019; Xu et al. 2020). A. muciniphila is known to degrade and utilize mucins as a nutrient source by producing sialidases, fucosidases, N-acetyl-β-glucosaminidases and GlcNAc-sulfatases, which breakdown mucin monomers (Derrien et al. 2004; Ottman et al. 2016; Geerlings et al. 2018; Xu et al. 2020). Using mucins as a nutrient source is particularly useful in the colon, where carbon sources are extremely limited (Derrien et al. 2008). Interestingly, A. muciniphila colonizes the mucosal layer, but does not invade it, which is an intriguing distinction from most of the mucin degrading microorganisms that have pathogenic potential. In the case of A. muciniphila, the host may support its colonization of the mucosal layer in exchange for the benefits it provides to the host in preventing metabolic disorders.
In addition to these mucin degrading enzymes, microorganisms can also directly bind to mucins, an interaction that is continuously evolving between the host and pathogen. Many commensal and pathogenic bacterial species, such as Helicobacter pylori, Yersinia enterocolitica, P. aeruginosa and E. coli, to name a few, are known to physically bind to mucins (Sajjan and Forstner 1990; Wanke et al. 1990; Mantle and Husar 1994; Lindén et al. 2009; Dhar and McAuley 2019; Wheeler et al. 2019; Hoffman, Lalsiamthara and Aballay 2020). From the host perspective, microbial binding to mucins can be beneficial by allowing for the removal of pathogens through mucus flow and excretion, and even by ‘mucin shedding’ (Linden et al. 2008; Van Putten and Strijbis 2017). In an example of the latter concept, the human opportunistic bacterial pathogen, H. pylori, which typically colonizes the digestive tract, has been found to bind via its BabA and SabA adhesins to Lewisb, sialyl Lewisa and sialyl Lewisx extracellular domain antigens of the carbohydrate portion of the membrane-bound mucin MUC1 (Lindén et al. 2009; Dhar and McAuley 2019). This binding event induces shedding of the microbe-bound MUC1 from the gastric epithelial cell layer, thereby preventing H. pylori from adhering to host epithelial cells (Lindén et al. 2009; Dhar and McAuley 2019). This is followed by excretion of the microbe bound MUC1 into the stomach, where it is digested by stomach acids. Mucin shedding can significantly limit disease progression by pathogens, which in the case of H. pylori, is the development of chronic peptic ulcers in the host.
In another example of microbe-mucin binding, the opportunistic bacterial pathogen, P. aeruginosa, commonly found in the respiratory tract of humans is known to physically bind via multiple strain dependent surface adhesins, including flagellins, to the sialyl Lewisx extracellular domain antigens of the carbohydrate portion of airway mucins (Carnoy et al. 1994; Scharfman et al. 1999; Lillehoj, Kim and Kim 2002). From the host perspective, this binding interaction can lead to attenuation of P. aeruginosa virulence by inducing a downregulation of numerous bacterial virulence genes, including genes involved in quorum sensing (e.g. lasR), toxin secretion (e.g. pcrV) and siderophore biosynthesis (e.g. pvdA), as well as by inducing active biofilm dispersion (Caldara et al. 2012; Wheeler et al. 2019). From the pathogen perspective, on the other hand, P. aeruginosa can use mucin binding to its advantage to cause disease by enhancing its ability to adhere to and colonize the mucosal surface, and by aiding its penetration to the underlying epithelial cell layer (Derrien et al. 2010; Hoffman, Lalsiamthara and Aballay 2020). When this occurs, P. aeruginosa binds mucins primarily at the N-acetylgalactosamine and N-acetylglucosamine polysaccharide portions of mucin monomers, leading to their degradation, thus allowing P. aeruginosa to access the underlying epithelial cell layer (Hoffman, Lalsiamthara and Aballay 2020).
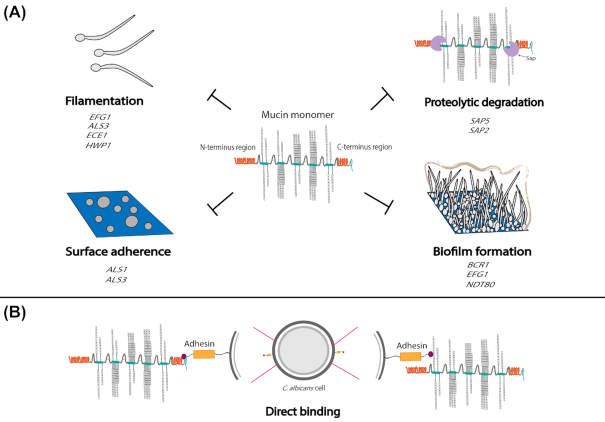
In terms of pathogenic fungi, there are many unanswered questions on how fungi interact with mucins, but the most mechanistic information is known for C. albicans (Fig. 3). One study determined that the gel-forming mucins MUC5AC, MUC5B and MUC2 prevent C. albicans from transitioning from the round yeast cell state to the elongated hyphal cell state that is critical for this fungus to invade the host epithelial cell layer and is an important structural feature of its biofilms (Kavanaugh et al. 2014; Basmaciyan et al. 2019). In addition, when hyphal C. albicans cells were exposed to mucins under hyphal inducing conditions, newly budded cells from these hyphae were in the round yeast form rather than the elongated hyphal form. In contrast, in the absence of mucins, newly budded cells under these conditions were always in the hyphal form. In addition, methylcellulose, a viscosity control used in this study to mimic the viscosity of mucins did not affect filamentation, suggesting that the biological properties of mucins, rather than their physical properties, are important in their ability to suppress C. albicans filamentation. Taken together, these results indicate that mucins suppress the development of hyphae from yeast cells and also suppress the development of new hyphae from existing hyphal cells. Not surprisingly in this study, several C. albicans genes involved in filamentation were downregulated in the presence of mucins, including EFG1, which encodes a major transcriptional regulator of filamentation, as well as ALS3, ECE1 and HWP1 (Kavanaugh et al. 2014). Interestingly, mucins appear to induce C. albicans cells to transition into a novel yeast morphology that phenotypically resembles the oval mating competent opaque cell type of this fungus, but that is functionally distinct since the morphology induced in the presence of mucins is not mating competent (Kavanaugh et al. 2014). This morphology also bears some resemblance to the C. albicans gastrointestinally induced transition (GUT) cell type identified as a commensal cell state that occurs when C. albicans cells are passaged through the gastrointestinal tract of a mouse (Pande, Chen and Noble 2013; Noble, Gianetti and Witchley 2017). It is possible that this novel mucin induced morphology may occur uniquely and specifically in response to mucins.
Figure 3.
Summary of known and hypothesized interactions between C. albicans and mucins. (A) Known interactions. These include the suppression of C. albicans adherence, filamentation, biofilm formation and secreted protease production in the presence of mucins. Several C. albicans genes encoding important virulence processes are known to be downregulated in the presence of mucins, including ALS1 and ALS3 (adherence); EFG1, ALS3, ECE1 and HWP1 (filamentation); BCR1, EFG1 and NDT80 (biofilm formation); and SAP5 and SAP2 (proteolytic degradation). (B) Hypothesized interactions. C. albicans adhesins may directly bind to a mucin monomer at the C-terminus of the PTS domain and/or at a glycan monosaccharide.
Another study found that C. albicans filamentation was inhibited by salivary mucins in a dose dependent manner (Ogasawara et al. 2007). In this study, C. albicans cells were grown for 24 hours under hyphal inducing conditions in the absence and presence of different concentrations of salivary mucins ranging from 125 to 1000 µg/mL (Ogasawara et al. 2007). In the absence of mucins, C. albicans cells formed long and numerous hyphae, while in the presence of mucins, there was a clear dose dependent reduction in hyphal formation as higher concentrations of mucins were used, and at 1000 µg/mL of mucins, no hyphae were observed whatsoever in the culture (Ogasawara et al. 2007). No differences in growth rates were observed in the absence versus the presence of mucins at concentrations up to 1000 µg/mL of mucins (Ogasawara et al. 2007). The expression of RAS1, which encodes the Ras1 GTPase that regulates the cAMP and MAP kinase pathways involved in the induction of hyphal formation, was also measured in this study (Feng et al. 1999; Leberer et al. 2001; Ogasawara et al. 2007). In the absence of mucins under hyphal inducing conditions, RAS1 expression levels were increased throughout the course of the experiment, while in the presence of mucins under the same conditions, RAS1 expression levels were significantly repressed (Ogasawara et al. 2007). This repression of RAS1 also correlated with a decrease in the expression of EFG1. Taken together, these results indicate that salivary mucins suppress the development of hyphae from yeast cells.
Other than filamentation, it has been shown that mucins, specifically MUC5AC, inhibit adherence of C. albicans cells to abiotic (polystyrene) and biotic (human epithelial cell) surfaces (Kavanaugh et al. 2014). The adherence inhibitory effects of mucins on these surfaces was observable after 30 minutes and increased significantly over the course of a 1-hour adhesion assay. Interestingly, the methylcellulose viscosity control also inhibited surface adhesion, suggesting that the physical properties of mucins contribute to their anti-adherence properties. Consistent with the finding that mucins inhibit adherence, a number of C. albicans genes involved in adherence were downregulated in the presence of mucins, such as the adhesins ALS1 and ALS3 (Kavanaugh et al. 2014). Therefore, by preventing C. albicans cells from adhering to surfaces, mucins impede the ability of C. albicans to achieve the first step necessary in the process of breaching the epithelial cell layer.
Since filamentation and adherence are important processes during C. albicans biofilm formation, it follows that biofilm formation would also be inhibited in the presence of mucins. Indeed, over the course of a 48-hour biofilm experiment in the presence of mucins, biofilm formation was severely constrained, where few hyphae were observed throughout the rudimentary (∼60 µm thick) biofilm formed (Kavanaugh et al. 2014). This is in contrast to the robust (∼500 µm thick) biofilm formed in the absence of mucins, containing long and extensive hyphae. The methylcellulose viscosity control in this experiment also inhibited biofilm formation, suggesting that the physical properties of mucins contribute to their antibiofilm properties. Consistent with the finding that mucins inhibit biofilm development, the genes BCR1, EFG1 and NDT80, encoding three of the six core C. albicans biofilm master regulators, were downregulated in the presence of mucins (Nobile et al. 2012; Kavanaugh et al. 2014).
Other than inhibiting filamentation, adherence and biofilm formation in C. albicans, mucins also suppress the expression of C. albicans secreted aspartyl protease encoding genes, such as SAP5 and SAP2 (Kavanaugh et al. 2014). By suppressing the expression of these hydrolytic enzymes, which are known C. albicans virulence factors that are similar to those produced by bacterial pathogens, mucins protect themselves from degradation and limit the ability of C. albicans to colonize the mucosal surface and invade the underlying epithelial cell layer (Colina et al. 1996; Naglik, Challacombe and Hube 2003; Nikou et al. 2019).
Although the molecular mechanisms for C. albicans direct binding to mucins are unknown, the adhesin Als1 is known to bind via its N-terminal region to fucose-containing glycans (Donohue et al. 2011). Based on these findings and the fact that mucins are heavily comprised of fucose glycans, it seems feasible that mucins could directly bind to Als1 as well as to the structurally similar protein Als3. Additionally, the hyphal specific cell surface protein Hwp1 is another candidate for mucin binding that is already known to interact with the host. Hwp1, which has a domain that resembles mammalian transglutaminase substrates, can bind to and form stable bonds of attachment to mammalian transglutaminases on the surfaces of host buccal epithelial cells (Staab et al. 1999). Hwp1 is also known to have complementary surface adhesion functions with Als1 and Als3 (Nobile et al. 2008). Taken together, it is plausible that Hwp1 could also directly bind to mucins. From the pathogen perspective, binding of C. albicans surface proteins and/or adhesins to mucins could increase adherence to the mucosal surface, allowing for C. albicans to penetrate to the epithelial cell layer. From the host perspective, binding of C. albicans surface adhesins to mucins could allow for mucin shedding to occur, where the C. albicans cells bound to mucins could be excreted in mucus flow, thereby reducing the number of C. albicans cells available to invade the host epithelial cell layer.
Another study assessing the abilities of several Candida species to bind to small intestinal mucins observed a hierarchy of mucin binding capabilities that appears to correlate with the abilities of the different species to cause disease in mammals, suggesting that direct mucin binding is an important virulence factor in the Candida clade (De Repentigny et al. 2000; Hirayama et al. 2020). The authors found that C. albicans, Candida dubliniensis and Candida tropicalis strongly adhered to mucins; Candida parapsilosis and Candida lusitaniae moderately adhered to mucins; and Candida krusei and Candida glabrata weakly adhered to mucins (De Repentigny et al. 2000). S. cerevisiae, which was used as a non-pathogenic outlier species, adhered to mucins the weakest relative to the Candida species tested. The binding of Candida species to mucins in this study appeared to be, in part, dependent on the C. albicans secreted aspartyl protease Sap2 (De Repentigny et al. 2000). The authors suggest that the C-terminal glycosylated region of small intestinal mucins may be involved in the direct binding of Candida adhesins to mucins, and that this region of the mucin monomer is a substrate specifically for Sap2 (De Repentigny et al. 2000). Taken together, although the mucosal surface acts as a barrier to Candida species from accessing the epithelial cell layer, mucins are likely substrates for several Candida secreted proteases that can degrade mucins, allowing Candida cells to access the underlying epithelial cell layer (De Repentigny et al. 2000).
Finally, it has been postulated that C. albicans cell type heterogeneity in the gastrointestinal tract, which is lined with mucus, can be modulated by the immune status of the host (Kumamoto and Pierce 2011; Pierce and Kumamoto 2012). This model predicts that C. albicans produces phenotypic variants with two distinct functions: one optimized for persistence as a commensal in the host and one optimized for pathogenic interactions with the host (Kumamoto and Pierce 2011; Koh 2013). When alterations in the host's immune status occur, the levels of these phenotypic variants are postulated to shift, changing the pathogenic potential of the population (Kumamoto and Pierce 2011). This model is supported by studies showing that variability in the levels of the transcriptional regulators Efg1 and Efh1 in mouse infection models can shift the C. albicans population between the commensal and pathogenic states (Pierce and Kumamoto 2012). Specifically, if a change in the host status selects for C. albicans cells with low Efg1 activity, then the C. albicans cell population shifts to become pathogenic, while if this change selects for cells with low Efh1 activity, then the population shifts to become commensal, and vice versa (White et al. 2007; Pierce and Kumamoto 2012). In terms of heterogeneity in cell morphology, another study showed that the C. albicans yeast form over other morphological forms is the commensal morphological form in the gastrointestinal tract in a monocolonized gnotobiotic mouse model (Böhm et al. 2017). This finding is logical given that the filamentous form is the morphological form that can breach the mucosal barrier (Basmaciyan et al. 2019). Interestingly, when the mice were treated with antibiotics, a morphologically heterogeneous population of cell types containing yeast and filamentous forms was formed, thereby increasing the pathogenic potential of the population (Böhm et al. 2017). This study also identified three transcriptional regulators, Zcf8, Zfu2 and Try4, that are required for maintaining this yeast form morphology and thus the commensal state of C. albicans in the mouse gastrointestinal tract (Böhm et al. 2017). Interestingly, these regulators also promote the adherence of C. albicans to mucin coated surfaces as well as to mucus producing intestinal epithelial cells (Böhm et al. 2017).
Much less is known about the interactions of the non-Candida fungal pathogens with mucins. Aspergillus fumigatus, an opportunistic human fungal pathogen that can colonize the respiratory tract and cause aspergillosis, is known to degrade mucins using hydrolytic enzymes, including proteases and glycosidases (St. Leger and Screen 2000; Oguma et al. 2011; Cowley et al. 2017). One study found that approximately 75% of the protein portions and 40% of the polysaccharide portions of mucins were degraded by A. fumigatus secreted proteases and glycosidases, respectively, that were produced under in vitro growth conditions in the presence of mucins (St. Leger and Screen 2000). Consistent with this finding, another study found that the A. fumigatus secreted serine protease Alp1 was highly upregulated at both the protein and transcript level in the presence of mucins (Farnell et al. 2012). From the pathogen perspective, degrading mucins using secreted proteases and glycosidases can allow A. fumigatus cells to access the underlying epithelial cell layer. In addition, studies have suggested that A. fumigatus likely uses mucins as a nutrient source (St. Leger and Screen 2000; Oguma et al. 2011; Cowley et al. 2017). From the host perspective, in response to A. fumigatus secreted proteases, the host compensates by upregulating the expression of MUC5AC in airway epithelial cells, which can be a double-edged sword (Cowley et al. 2017). The upregulation of MUC5AC could be protective against infection by inhibiting fungal colonization of the mucosal layer, but if MUC5AC becomes highly upregulated or upregulated for too long, this can lead to diseases related to mucus hypersecretion, such as allergic bronchopulmonary aspergillosis, which typically occurs in individuals with asthma or cystic fibrosis (Oguma et al. 2011; Gao et al. 2012).
CONCLUDING REMARKS
Mucus is an important host innate defense factor that lines most epithelial cell layers of the body and provides crucial physical and biological protection against pathogenic microorganisms. Mucins are the main glycoproteins of mucus that are responsible for interacting with microorganisms and are critical for the antimicrobial properties of mucus. The physiochemical properties of mucins can suppress key virulence traits in microorganisms, maintaining them in a commensal state. In the opportunistic human fungal pathogen C. albicans, adherence, filamentation, biofilm formation and the production of secreted proteases are suppressed by mucins, although the molecular mechanisms behind this suppression are unknown. In general, further work is needed to elucidate the molecular mechanisms involved in microbe–mucin interactions, which will be essential for our understanding of how a healthy mucosal barrier is maintained. In order to carry out such studies, there is a need for tractable mucus model systems that can be used to study microbe–mucin interactions. Mucosal model systems would also be helpful in the development of novel therapeutics to treat mucosal diseases, such as cystic fibrosis, inflammatory bowel disease and ulcerative colitis. Finally, given that mucins are such critical players against infection, mechanistically understanding their biological and physical properties will be useful in the development of novel therapeutic strategies against pathogenic microorganisms.
ACKNOWLEDGEMENTS
We thank members of the Nobile lab and Mohammad N. Qasim for insightful discussions on the topics covered in this manuscript. We also thank the two anonymous reviewers for their helpful suggestions that improved the manuscript.
Contributor Information
Ashley Valle Arevalo, Department of Molecular and Cell Biology, University of California – Merced, 5200 North Lake Rd., Merced, CA 95343, USA; Quantitative and Systems Biology Graduate Program, University of California – Merced, 5200 North Lake Rd., Merced, CA 95343, USA.
Clarissa J Nobile, Department of Molecular and Cell Biology, University of California – Merced, 5200 North Lake Rd., Merced, CA 95343, USA.
FUNDING
This work was supported by the National Institutes of Health (NIH) National Institute of Allergy and Infectious Diseases (NIAID) and National Institute of General Medical Sciences (NIGMS) awards R21AI125801 and R35GM124594, respectively, to C.J.N., and by a Pew Biomedical Scholar Award from the Pew Charitable Trusts to C.J.N. This work was also supported by the Kamangar family in the form of an endowed chair to C.J.N. A.V.A. was supported by a diversity supplement fellowship to parent grant R21AI125801. The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
Conflicts of Interest
Clarissa J. Nobile is a cofounder of BioSynesis, Inc., a company developing inhibitors and diagnostics of biofilm formation.
REFERENCES
- Achkar JM, Fries BC.. Candida infections of the genitourinary tract. Clin Microbiol Rev. 2010;23:253–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillie GS, Douglas LJ.. Role of dimorphism in the development of Candida albicans biofilms. J Med Microbiol. 1999;48:671–9. [DOI] [PubMed] [Google Scholar]
- Bakshani CR, Morales-Garcia AL, Althaus Met al. Evolutionary conservation of the antimicrobial function of mucus: a first defence against infection. npj Biofilms Microbiomes. 2018;4:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balabushevich NG, Sholina EA, Mikhalchik EVet al. Self-assembled mucin-containing microcarriers via hard templating on CaCO3 crystals. Micromachines. 2018;9:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansil R, Turner BS. Mucin structure, aggregation, physiological functions and biomedical applications. Curr Opin Colloid Interface Sci. 2006;11:164–70. [Google Scholar]
- Basmaciyan L, Bon F, Paradis Tet al. “Candida albicans interactions with the host: crossing the intestinal epithelial barrier.”. Tissue Barriers. 2019;7:1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y, Hand T. Role of microbiota in immunity and inflammation. Cell. 2014;157:121–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedict K, Jackson BR, Chiller Tet al. Estimation of direct healthcare costs of fungal diseases in the United States. Clin Infect Dis. 2019;68:1791–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockhauser I, Schachter H, Stanley P. Chapter 9: O-GalNAc Glycans. Essentials of Glycobiology. 2nd ed.Cold Spring Harbor: Cold Spring Harbor Laboratory Press, 2009. [PubMed] [Google Scholar]
- Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. 2013;13:790–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhm L, Torsin S, Tint SHet al. The yeast form of the fungus Candida albicans promotes persistence in the gut of gnotobiotic mice. PLoS Pathog. 2017;13:1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldara M, Friedlander RS, Kavanaugh NLet al. Mucin biopolymers prevent bacterial aggregation by retaining cells in the free-swimming state. Curr Biol. 2012;22:2325–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderone RA, Fonzi WA.. Virulence factors of Candida albicans. Trends Microbiol. 2001;9:327–35. [DOI] [PubMed] [Google Scholar]
- Cani PD, de Vos WM. Next-generation beneficial microbes: the case of Akkermansia muciniphila. Front Microbiol. 2017;8:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnoy C, Scharfman A, Van Brussel Eet al. Pseudomonas aeruginosa outer membrane adhesins for human respiratory mucus glycoproteins. Infect Immun. 1994;62:1896–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A, Pirofski L. Host‐pathogen interactions: the attributes of virulence. J Infect Dis. 2001;184:337–44. [DOI] [PubMed] [Google Scholar]
- Casadevall A. The pathogenic potential of a microbe. mSphere. 2017;2:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli J, Gregor B, Turner Bet al. Viscoelastic properties and dynamics of porcine gastric mucin. Biomacromolecules. 2005;6:1329–33. [DOI] [PubMed] [Google Scholar]
- Celli JP, Turner BS, Afdhal NHet al. Helicobacter pylori moves through mucus by reducing mucin viscoelasticity. Proc Natl Acad Sci U S A. 2009;106:14321–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha H-J, Song K.. Effect of MUC8 on airway inflammation: a friend or a foe? J Clin Med. 2018;7:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra J, Kuhn DM, Mukherjee PKet al. Biofilm formation by the fungal pathogen. Society. 2001;183:5385–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu L, Bazin T, Truchetet MEet al. Protective microbiota: From localized to long-reaching co-immunity. Front Immunol. 2017;8:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow J, Lee SM, Shen Yet al. Host–bacterial symbiosis in health and disease. Adv Immunol. 2010;107:243–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Co JY, Cárcamo-Oyarce G, Billings Net al. Mucins trigger dispersal of Pseudomonas aeruginosa biofilms. npj Biofilms Microbiomes. 2018;4:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colina AR, Aumont F, Deslauriers Net al. Evidence for degradation of gastrointestinal mucin by Candida albicans secretory aspartyl proteinase. Infect Immun. 1996;64:4514–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado MC, Derrien M, Isolauri Eet al. Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl Environ Microbiol. 2007;73:7767–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone RA. Barrier properties of mucus. Adv Drug Deliv Rev. 2009;61:75–85. [DOI] [PubMed] [Google Scholar]
- Corfield AP. Mucins: a biologically relevant glycan barrier in mucosal protection. Biochim Biophys Acta Gen Subj. 2015;1850:236–52. [DOI] [PubMed] [Google Scholar]
- Corfield AP. The interaction of the gut microbiota with the mucus barrier in health and disease in human. Microorganisms. 2018;6:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley AC, Thornton DJ, Denning DWet al. Aspergillosis and the role of mucins in cystic fibrosis. Pediatr Pulmonol. 2017;52:548–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Repentigny L, Aumont F, Bernard Ket al. Characterization of binding of Candida albicans to small intestinal mucin and its role in adherence to mucosal epithelial cells. Infect Immun. 2000;68:3172–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien M, Collado MC, Ben-Amor Ket al. The mucin degrader Akkermansia muciniphila is an abundant resident of the human intestinal tract. Appl Environ Microbiol. 2008;74:1646–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien M, van Passel MWJ, van de Bovenkamp JHBet al. Mucin-bacterial interactions in the human oral cavity and digestive tract. Gut Microbes. 2010;1:254–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien M, Vaughan EE, Plugge CMet al. Akkermansia municiphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol. 2004;54:1469–76. [DOI] [PubMed] [Google Scholar]
- Dhar P, McAuley J.. The role of the cell surface mucin MUC1 as a barrier to infection and regulator of inflammation. Front Cell Infect Microbiol. 2019;9:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohrman A, Miyata S, Gallup Met al. Mucin gene (MUC 2 and MUC 5AC) upregulation by Gram-positive and Gram-negative bacteria. Biochim Biophys Acta - Mol Basis Dis. 1998;1406:251–9. [DOI] [PubMed] [Google Scholar]
- Donohue DS, Ielasi FS, Goossens KVYet al. The N-terminal part of Als1 protein from Candida albicans specifically binds fucose-containing glycans. Mol Microbiol. 2011;80:1667–79. [DOI] [PubMed] [Google Scholar]
- Douglas LJ. Candida biofilms and their role in infection. Trends Microbiol. 2003;11:30–6. [DOI] [PubMed] [Google Scholar]
- Farnell E, Rousseau K, Thornton DJet al. Expression and secretion of Aspergillus fumigatus proteases are regulated in response to different protein substrates. Fungal Biol. 2012;116:1003–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Summers E, Guo Bet al. Ras signaling is required for serum-induced hyphal differentiation in Candida albicans. J Bacteriol. 1999;181:6339–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel ES, Ribbeck K.. Salivary mucins in host defense and disease prevention. J Oral Microbiol. 2015;7:29759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly S, Mitchell AP.. Mucosal biofilms of Candida albicans. Curr Opin Microbiol. 2011;14:380–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao FS, Gao YY, Liu MJet al. Chronic Aspergillus fumigatus exposure upregulates the expression of Mucin 5AC in the airways of asthmatic rats. Exp Lung Res. 2012;38:256–65. [DOI] [PubMed] [Google Scholar]
- Geerlings S, Kostopoulos I, de Vos Wet al. Akkermansia muciniphila in the human gastrointestinal tract: when, where, and how? Microorganisms. 2018;6:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendler S. Epithelial mucin genes. Annu Rev Physiol. 1995;57:607–34. [DOI] [PubMed] [Google Scholar]
- Gulati M, Nobile CJ.. Candida albicans biofilms: development, regulation, and molecular mechanisms. Microbes Infect. 2016;18:310–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gururaja TL, Levine JH, Tran DTet al. Candidacidal activity prompted by N-terminus histatin-like domain of human salivary mucin (MUC7). Biochim Biophys Acta Protein Struct Mol Enzymol. 1999;1431:107–19. [DOI] [PubMed] [Google Scholar]
- Hansson GC. Role of mucus layers in gut infection and inflammation. Curr Opin Microbiol. 2012;15:57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawser SP, Douglas LJ. Biofilm formation by Candida species on the surface of catheter materials in vitro. 1994;62:915–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes K. Virulence in Candida species. Trends Microbiol. 2001;9:591–6. [DOI] [PubMed] [Google Scholar]
- Henke MO, John G, Germann Met al. MUC5AC and MUC5B mucins increase in cystic fibrosis airway secretions during pulmonary exacerbation. Am J Respir Crit Care Med. 2007;175:816–21. [DOI] [PubMed] [Google Scholar]
- Henke MO, Renner A, Huber RMet al. MUC5AC and MUC5B mucins are decreased in cystic fibrosis airway secretions. Am J Respir Cell Mol Biol. 2004;31:86–91. [DOI] [PubMed] [Google Scholar]
- Hirayama T, Miyazaki T, Ito Yet al. Virulence assessment of six major pathogenic Candida species in the mouse model of invasive candidiasis caused by fungal translocation. Sci Rep. 2020;10:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman CL, Lalsiamthara J, Aballay A. Host mucin is exploited by Pseudomonas aeruginosa to provide monosaccharides required for a successful infection. MBio. 2020;11:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanaugh NL, Zhang AQ, Nobile CJet al. Mucins suppress virulence traits of Candida albicans. MBio. 2014;5:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy MJ, Volz PA, Edwards CAet al. Mechanisms of association of Candida albicans with intestinal mucosa. J Med Microbiol. 1987;24:333–41. [DOI] [PubMed] [Google Scholar]
- Kennedy MJ, Volz PA.. Effect of various antibiotics on gastrointestinal colonization and dissemination by Candida albicans. Med Mycol. 1985;23:265–73. [DOI] [PubMed] [Google Scholar]
- Kerschner JE, Hong W, Khampang Pet al. Differential response of gel-forming mucins to pathogenic middle ear bacteria. Int J Pediatr Otorhinolaryngol. 2014;78:1368–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Sudbery P.. Candida albicans, a major human fungal pathogen. J Microbiol. 2011;49:171–7. [DOI] [PubMed] [Google Scholar]
- Kim YS, Ho SB.. Intestinal goblet cells and mucins in health and disease: recent insights and progress. Curr Gastroenterol Rep. 2010;12:319–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh AY. Murine models of Candida gastrointestinal colonization and dissemination. Eukaryot Cell. 2013;12:1416–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullberg BJ, Oude LAML.. Epidemiology of opportunistic invasive mycoses. Eur J Med Res. 2002;7:183–91. [PubMed] [Google Scholar]
- Kumamoto CA, Pierce JV.. Immunosensing during colonization by Candida albicans: does it take a village to colonize the intestine? Trends Microbiol. 2011;19:263–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto CA. Candida biofilms. Curr Opin Microbiol. 2002;5:608–11. [DOI] [PubMed] [Google Scholar]
- Kumamoto CA. Inflammation and gastrointestinal Candida colonization. Curr Opin Microbiol. 2011;14:386–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto CA. The fungal mycobiota: small numbers, large impacts. Cell Host Microbe. 2016;19:750–1. [DOI] [PubMed] [Google Scholar]
- Leberer E, Harcus D, Dignard Det al. Ras links cellular morphogenesis to virulence by regulation of the MAP kinase and cAMP signalling pathways in the pathogenic fungus Candida albicans. Mol Microbiol. 2001;42:673–87. [DOI] [PubMed] [Google Scholar]
- Libertucci J, Young VB.. The role of the microbiota in infectious diseases. Nat Microbiol. 2019;4:35–45. [DOI] [PubMed] [Google Scholar]
- Lillehoj EP, Kato K, Wenju Luet al. Cellular and molecular biology of airway mucins. Int Rev Cell Mol Biol. 2013;303:139–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillehoj EP, Kim BT, Kim KC. Identification of Pseudomonas aeruginosa flagellin as an adhesin for Muc1 mucin. Am J Physiol - Lung Cell Mol Physiol. 2002;282:751–6. [DOI] [PubMed] [Google Scholar]
- Linden SK, Sutton P, Karlsson NGet al. Mucins in the mucosal barrier to infection. Mucosal Immunol. 2008;1:183–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindén SK, Sheng YH, Every ALet al. MUC1 limits Helicobacter pylori infection both by steric hindrance and by acting as a releasable decoy. PLoS Pathog. 2009;5, DOI: 10.1371/journal.ppat.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liévin-le Moal V, Servin AL. The front line of enteric host defense against unwelcome intrusion of harmful microorganisms : mucins, antimicrobial peptides, and microbiota. Clin Microbiol Rev. 2017;19:315–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse MB, Gulati M, Johnson ADet al. Development and regulation of single-and multi-species Candida albicans biofilms. Nat Rev Microbiol. 2018;16:19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukeš J, Stensvold CR, Jirků-Pomajbíková Ket al. Are human intestinal eukaryotes beneficial or commensals? PLoS Pathog. 2015;11:7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Q, Kumar P, Vickers TJet al. Enterotoxigenic Escherichia coli secretes a highly conserved mucin-degrading metalloprotease to effectively engage intestinal epithelial cells. Infect Immun. 2014;82:509–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macao B, Johansson DGA, Hansson GCet al. Autoproteolysis coupled to protein folding in the SEA domain of the membrane-bound MUC1 mucin. Nat Struct Mol Biol. 2006;13:71–6. [DOI] [PubMed] [Google Scholar]
- Mantle M, Husar SD.. Binding of Yersinia enterocolitica to purified, native small intestinal mucins from rabbits and humans involves interactions with the mucin carbohydrate moiety. Infect Immun. 1994;62:1219–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell KF, Zarnowski R, Andes DR. Fungal super glue: the biofilm matrix and its composition, assembly, and functions. PLoS Pathog. 2016;12:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naglik JR, Challacombe SJ, Hube B. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol Rev. 2003;67:400–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikou SA, Kichik N, Brown Ret al. Candida albicans interactions with mucosal surfaces during health and disease. Pathogens. 2019;8:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Fox EP, Nett JEet al. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell. 2012;148:126–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Johnson AD.. Candida albicans biofilms and human disease. Annu Rev Microbiol. 2015;69:71–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Schneider HA, Nett JEet al. Complementary adhesin function in C. albicans biofilm formation. Curr Biol. 2008;18:1017–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble SM, Gianetti BA, Witchley JN. Candida albicans cell-type switching and functional plasticity in the mammalian host. Nat Rev Microbiol. 2017;15:96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odds FC. Candida infections: an overview. Crit Rev Microbiol. 1987;15:1–5. [DOI] [PubMed] [Google Scholar]
- Ogasawara A, Komaki N, Akai Het al. Hyphal formation of Candida albicans is inhibited by salivary mucin. Biol Pharm Bull. 2007;30:284–6. [DOI] [PubMed] [Google Scholar]
- Oguma T, Asano K, Tomomatsu Ket al. Induction of mucin and MUC5AC expression by the protease activity of Aspergillus fumigatus in airway epithelial cells. J Immunol. 2011;187:999–1005. [DOI] [PubMed] [Google Scholar]
- Ottman N, Huuskonen L, Reunanen Jet al. Characterization of outer membrane proteome of Akkermansia muciniphila reveals sets of novel proteins exposed to the human intestine. Front Microbiol. 2016;7:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pande K, Chen C, Noble SM. Passage through the mammalian gut triggers a phenotypic switch that promotes Candida albicans commensalism. Nat Genet. 2013;45:1088–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaseyed T, Bergström JH, Gustafsson JKet al. The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol Rev. 2014;260:8–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaseyed T, Hansson GC.. Membrane mucins of the intestine at a glance. J Cell Sci. 2020;133, DOI: 10.1242/jcs.240929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrou G, Crouzier T.. Mucins as multifunctional building blocks of biomaterials. Biomater Sci. 2018;6:2282–97. [DOI] [PubMed] [Google Scholar]
- Pickard JM, Zeng MY, Caruso Ret al. Gut microbiota: role in pathogen colonization, immune responses and inflammatory disease. Immunol Rev. 2017;279:70–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce CG, Vila T, Romo JAet al. The Candida albicans biofilm matrix: composition, structure and function. J Fungi. 2017;3, DOI: 10.3390/jof3010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce JV, Kumamoto CA.. Variation in Candida albicans EFG1 expression enables host-dependent changes in colonizing fungal populations. MBio. 2012;3:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri S, Edgerton M. How does it kill?: understanding the candidacidal mechanism of salivary histatin 5. Eukaryot Cell. 2014;13:958–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relman DA. “Til death do us part”: coming to terms with symbiotic relationships. Nat Rev Microbiol. 2008;6:721–4. [DOI] [PubMed] [Google Scholar]
- Roy MG, Livraghi-butrico A, Fletcher AAet al. Muc5b is required for airway defence. Nature. 2014;505:412–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajjan SU, Forstner JF.. Characteristics of binding of Escherichia coli serotype O157:H7 strain CL-49 to purified intestinal mucin. Infect Immun. 1990;58:860–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassone-Corsi M, Raffatellu M. No vacancy: how beneficial microbes cooperate with immunity to provide colonization resistance to pathogens. J Immunol. 2015;194:4081–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharfman A, Degroote S, Beau Jet al. Pseudomonas aeruginosa binds to neoglycoconjugates bearing mucin carbohydrate determinants and predominantly to Sialyl-Lewis x conjugates. Glycobiology. 1999;9:757–64. [DOI] [PubMed] [Google Scholar]
- Schroeder BO. Fight them or feed them: how the intestinal mucus layer manages the gut microbiota. Gastroenterol Rep. 2019;7:3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Noh JR, Chang DHet al. Elucidation of Akkermansia muciniphila probiotic traits driven by mucin depletion. Front Microbiol. 2019;10:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. Leger RJ, Screen SE.. In vitro utilization of mucin, lung polymers, plant cell walls and insect cuticle by Aspergillus fumigatus, Metarhizium anisopliae and Haematonectria haematococca. Mycol Res. 2000;104:463–71. [Google Scholar]
- Staab JF, Bradway SD, Fidel PLet al. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science. 1999;283:1538. [DOI] [PubMed] [Google Scholar]
- Szabady RL, Yanta JH, Halladin DKet al. TagA is a secreted protease of Vibrio cholerae that specifically cleaves mucin glycoproteins. Microbiology. 2011;157:516–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tailford LE, Crost EH, Kavanaugh Det al. Mucin glycan foraging in the human gut microbiome. Front Genet. 2015;6, DOI: 10.3389/fgene.2015.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton DJ, Rousseau K, McGuckin MA. Structure and function of the polymeric mucins in airways mucus. Annu Rev Physiol. 2008;70:459–86. [DOI] [PubMed] [Google Scholar]
- Underhill DM, Iliev ID.. The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol. 2014;14:405–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppuluri P, Chaturvedi AK, Srinivasan Aet al. Dispersion as an important step in the Candida albicans biofilm developmental cycle. PLoS Pathog. 2010;6, DOI: 10.1371/journal.ppat.1000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursell LK, Metcalf JL, Wegener Parfrey Let al. Defining the human microbiome. Natr Rev. 2013;70:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valeri M, Paccani SR, Kasendra Met al. Pathogenic E. coli exploits SslE mucinase activity to translocate through the mucosal barrier and get access to host cells. PLoS One. 2015;10:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Putten JPM, Strijbis K. Transmembrane mucins: signaling receptors at the intersection of inflammation and cancer. J Innate Immun. 2017;9:281–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner CE, Wheeler KM, Ribbeck K. Mucins and their role in shaping the functions of mucus barriers. Annu Rev Cell Dev Biol. 2018;34:189–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Yao M, Lv Let al. The human microbiota in health and disease. Engineering. 2017;3:71–82. [Google Scholar]
- Wanke CA, Cronan S, Goss Cet al. Characterization of binding of Escherichia coli strains which are enteropathogens to small-bowel mucin. Infect Immun. 1990;58:794–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler KM, Cárcamo-Oyarce G, Turner BSet al. Mucin glycans attenuate the virulence of Pseudomonas aeruginosa in infection. Nat Microbiol. 2019;4:2146–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SJ, Rosenbach A, Lephart Pet al. Self-regulation of Candida albicans population size during GI colonization. PLoS Pathog. 2007;3:1866–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C, Ranjendran R, Ramage G. Pathogenesis of fungal infections in cystic fibrosis. Curr Fungal Infect Rep. 2016;10:163–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Pavlidis P, Thamadilok Set al. Recent evolution of the salivary mucin MUC7. Sci Rep. 2016;6:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wang N, Tan HYet al. Function of Akkermansia muciniphila in obesity: interactions with lipid metabolism, immune response and gut systems. Front Microbiol. 2020;11:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnowski R, Westler WM, Lacmbouh GAet al. Novel entries in a fungal biofilm matrix encyclopedia. MBio. 2014;5:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]