ABSTRACT
Lactic acid bacteria (LAB) encompasses industrially relevant bacteria involved in food fermentations as well as health-promoting members of our autochthonous microbiota. In the last years, we have witnessed major progresses in the knowledge of the biology of their cell wall, the outermost macrostructure of a Gram-positive cell, which is crucial for survival. Sophisticated biochemical analyses combined with mutation strategies have been applied to unravel biosynthetic routes that sustain the inter- and intra-species cell wall diversity within LAB. Interplay with global cell metabolism has been deciphered that improved our fundamental understanding of the plasticity of the cell wall during growth. The cell wall is also decisive for the antimicrobial activity of many bacteriocins, for bacteriophage infection and for the interactions with the external environment. Therefore, genetic circuits involved in monitoring cell wall damage have been described in LAB, together with a plethora of defence mechanisms that help them to cope with external threats and adapt to harsh conditions. Since the cell wall plays a pivotal role in several technological and health-promoting traits of LAB, we anticipate that this knowledge will pave the way for the future development and extended applications of LAB.
Keywords: peptidoglycan, cell wall polysaccharides, bacteriocins, bacteriophages, stress response, resistance mechanisms
This review covers the most recent advances in the cell wall structure of lactic acid bacteria (LAB), its role as the target for bacteriocins, bacteriophages and host factors and how LAB sense and respond to cell wall damage.
INTRODUCTION
Lactic acid bacteria (LAB) are a diverse group of low G + C Gram-positive bacteria encompassing members of the families Aerococcaceae, Carnobacteriaceae, Enterococcaceae, Lactobacillaceae, Leuconostocaceae and Streptococcaceae (Vandamme, De Bruyne and Pot 2014). Phenotypically, LAB are non-sporulating, relatively aerotolerant, acidophilic anaerobes which are mainly unified by the production of, mainly, lactic acid from carbohydrate fermentation. LAB are widespread in nature, colonizing plants and the mammalian mucosae from which they reach raw food materials and promote their fermentation (George et al. 2018). The core genera Lactococcus, Lactobacillus, Leuconostoc, Pediococcus, Oenococcus and the species Streptococcus thermophilus constitute a group of industrially important bacteria which are widely used as starters and adjunct cultures for the production of fermented vegetables, dairy products, processed meats, alcoholic beverages, etc. Due to their long history of safe use, LAB are generally regarded as safe with notable exceptions in the case of the genera Streptococcus and Enterococcus, which include both commensal and pathogenic members. Specific species of Lactobacillus have also been linked to health-promoting (probiotic) properties (Puebla-Barragan and Reid 2019).
As Gram-positive bacteria, LAB cells are encased in a cell wall (CW) characterized by a thick peptidoglycan (PG) layer, which functions as a scaffold for the attachment of other CW components such as teichoic acids, polysaccharides and proteins (Chapot-Chartier and Kulakauskas 2014). Far from being a static and rigid structure, the CW is instead highly dynamic. It is implicated in several essential cell functions, including cell division and cell shape and is required to counteract turgor pressure. Moreover, as the outermost macrostructure of the bacterial cell, the CW is the main sensory interface between the cell and the external environment and it is crucial for survival. Accordingly, bacteria have evolved a plethora of mechanisms to monitor CW integrity and transmit the signal to the cytoplasm to mount a response, often through transcriptional activation. This response usually implies changes in the CW structure to neutralise CW damage and adapt to the new conditions.
The study of the CW has been fostered in the field of pathogenic bacteria, mostly because the CW is a prime target for antibiotic action (Schneider and Sahl 2010). Conversely, the CW is also a key element for many important technological and probiotic traits of LAB, comprising bacteriophage resistance, texturing, cheese ripening (flavour development), stress tolerance, adhesion and host cross-talk. Hence, CW structure and function has been the topic of two landmark reviews within the LAB field by Delcour et al. (1999) and Chapot-Chartier and Kulakauskas (2014). As a follow-up, this review will describe recent advances we have witnessed since then. We have specifically selected topics where research on the LAB CW has made fundamental contributions to the field and those which have an impact on the technological applications of LAB. A focus is made on the new insights into CW structure, the role of CW as the target for bacteriocins, bacteriophages and host factors (the threats), the genetic circuits involved in sensing and responding to CW damage, collectively known as the cell envelope stress response, and how LAB defeat these CW active antimicrobials (the defences). Alongside the study of the rather conserved PG biosynthetic machinery, shared by LAB and other Gram-positive bacteria, unveiling the biosynthetic routes of cell wall polysaccharides that explain their biochemical diversity and define phage-host recognition, has been a ground-breaking discovery of fundamental and applied importance. On the other hand, and for the first time, nucleotide intracellular pools have been shown to coordinate CW plasticity during cell growth. Moreover, pioneering studies on the second messenger c-di-AMP in Lactococcus lactis have been instrumental to understand the physiological response to osmotic stress in bacteria and a link to CW biosynthesis has been established. These are just some examples of the on-going activities in the LAB CW field which will be tackled in this review. Novel applications that emanate from the current knowledge and future directions will be also discussed. It should be noted that the role of CW in pathogenic LAB virulence will not be covered.
STRUCTURE AND ARCHITECTURE OF THE LAB CELL WALL
As Gram-positive bacteria, LAB have a thick CW comprising several glycopolymers, with PG providing a scaffold onto which teichoic acids and polysaccharides are attached (Fig. 1). The CW contains also proteins linked covalently to PG or through specific binding domains to CW glycopolymers and, in a few cases, bacteria may be surrounded by a proteinaceous S-layer. Despite the conserved general composition of the Gram-positive CW, the constituting glycopolymers exhibit structural diversity between bacterial species and even between strains in the case of teichoic acids and polysaccharides. This structural diversity often modulates susceptibility or resistance to CW antimicrobials. Moreover, the biosynthesis pathways of these CW glycopolymers exhibit conserved features or intermediates that can be the target of antimicrobials as described in this review.
Figure 1.
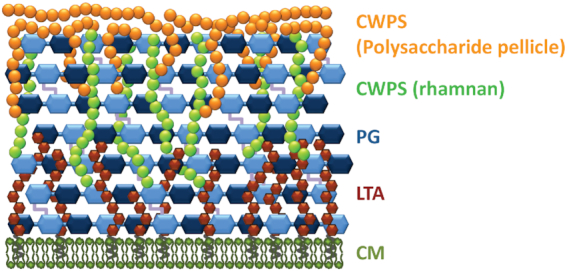
Schematic representation of the cell wall of Lactococcus lactis. Note that cell wall proteins are not shown. CWPS, cell wall polysaccharides; PG, peptidoglycan; LTA, lipoteichoic acids; CM, cytoplasmic membrane.
Nowadays PG structural analysis relies on powerful analytical tools including ultra-high performance liquid chromatography (UHPLC) and high resolution tandem mass spectrometry (MS/MS). Furthermore, nuclear magnetic resonance (NMR) spectroscopy allows studying the structure of polysaccharides and teichoic acids at the atomic level. Structural analysis combined with mutant construction specifically designed to target genes with vital functions (e.g. recombineering and CRISPS-Cas9-assisted recombineering) has been instrumental to decipher the biosynthetic pathways of the CW components and their modifications as described below. Moreover, the spatial localization of the constituting glycopolymers inside the CW also varies among LAB species, which are either ovococci (e.g. L. lactis and S. thermophilus) or bacilli (e.g. Lactobacillus sp.). Thus, recent insights into CW architecture provided by atomic force microscopy (AFM) and solid state NMR, both imaging techniques with high resolution power for visualizing CW polymers in whole (viable) bacterial cells, will be also summarized.
Peptidoglycan structure and biosynthesis
PG is a complex macromolecule made of linear glycan chains cross-linked by short peptide chains. It is produced by extracellular polymerization of disaccharide-pentapeptide subunits synthesized in the cytoplasm. The resulting glycan chains consist of alternating N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) that are linked via β-1,4-bonds. The peptidic chain is branched by its N-terminus on the lactyl group of MurNAc and is made of alternating D- and L-isomers, which vary among bacterial species at certain positions. A portion of the peptides of adjacent PG chains is cross-linked, interconnecting the glycan chains and thus, forming a meshwork surrounding the bacterial cell (Fig. 1). The variable structure of the peptide was used to define the PG chemotype that is characteristic of a bacterial species (Schleifer and Kandler 1972). The most common sequence for the stem peptide chain is L-Ala-γ-D-Glu-X-D-Ala-D-Ala, with X being a di-amino acid, most often L-Lys (such as in L. lactis and most lactobacilli), meso-diaminopimelic acid in Lactobacillus plantarum, or L-ornithine in Lactobacillus fermentum. Notably, in L. plantarum, Lactobacillus casei and other lactobacilli, the C-terminal D-Ala is replaced by D-lactate, which confers intrinsic vancomycin resistance (Delcour et al. 1999). Another variable feature is present at the level of the bridges linking peptide stems. Cross-linking is established by an amide bond most often between D-Ala in position 4 of one chain and the free amino group of the di-amino acid in position 3 of another chain. It can be either direct between the two peptide chains (e.g. in L. plantarum) (Bernard et al. 2011) or through short interpeptide bridges made of one D-amino acid, e.g. D-Asp or D-Asn in L. lactis and several lactobacilli (Courtin et al. 2006; Regulski et al. 2012) or a few L-amino acids, e.g. L-Ala2 or L-Ala3 in S. thermophilus (Layec et al. 2009). In addition to these differences occurring between bacterial species, structural variation in the PG molecule may also be encountered within a given LAB species (Chapot-Chartier and Kulakauskas 2014). These secondary modifications are found on glycan chains such as O-acetylation (found in all LAB studied) and N-deacetylation of MurNAc or/and GlcNAc (in L. lactis). Also modifications on peptide chains such as amidation of the carboxyl group of D-Glu and mDAP in stem peptides (seen in all LAB studied and in L. plantarum, respectively) or amidation of D-Asp in cross-bridges in L. lactis and L. casei. These modifications may concern only a portion of the sugar or amino acid residues, but they play essential roles in bacterial physiology and in particular, as covered in more detail below, in cell wall homeostasis and response to antimicrobials.
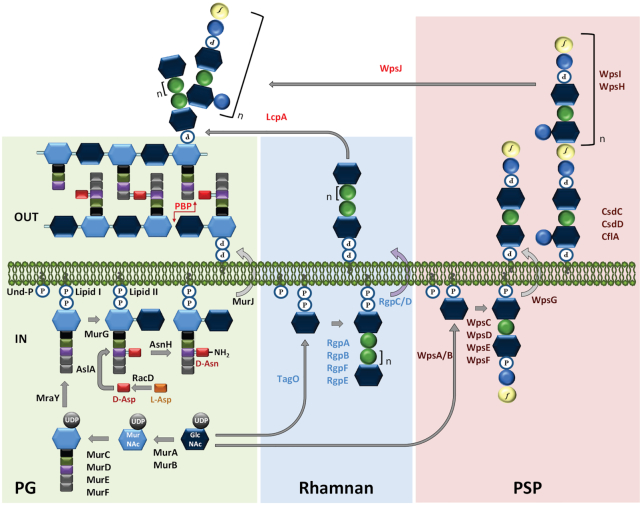
PG biosynthesis starts in the cytoplasm with the production of the soluble precursor UDP-MurNAc-pentapeptide (or pentadepsipeptide), also named Park nucleotide (Fig. 2). Several ligases (MurC, D, E, F) catalyze the sequential addition of amino acid residues on UDP-MurNAc, with the terminal two added by MurF as a D-Ala-D-Ala dipeptide or D-Ala-D-Lac depsipeptide, whose synthesis requires a D-D-ligase (Ddl) (Barreteau et al. 2008). The next step, catalyzed by the integral membrane protein MraY, results in the transfer of phospho-MurNAc-pentapeptide onto the lipid carrier undecaprenyl-phosphate (Und-P) to form lipid I (Bouhss, Trunkfield, Bugg et al. 2008). GlcNAc is then linked to MurNAc by MurG giving rise to lipid II consisting of the disaccharide-pentapeptide linked by a pyrophosphate bond to undecaprenol, which is a crucial intermediate of PG synthesis. Inside the cytoplasm, the soluble or the lipid-bound precursors may be the target of modifications including the addition of the peptide cross-bridge (e.g. D-Asp or L-Ala2–3) on the ε-amino group of the third amino acid or the several types of amino acid modifications described above.
Figure 2.
Schematic representation of the main proposed steps for the biosynthesis of peptidoglycan (PG, green background) and cell wall polysaccharides (CWPS), including rhamnan (blue background) and polysaccharide pellicle (PSP, pink background) in L. lactis. Hexagons represent MurNAc (light blue) and GlcNAc (dark blue). Circles represent sugars: rhamnose (green), glucose (blue), galactofuranose (yellow). Rectangles represent amino acids: L-Ala (black), D-Glu (green), L-Lys (purple), D-Ala (grey), D-Asp (red). P, phosphate. The biosynthesis of the three cell wall glycopolymers starts in the cytoplasm (IN). For PG, a soluble precursor, UDP-MurNAc-pentapeptide is first synthesized then transferred onto the lipid carrier undecaprenyl-phosphate (Und-P) by MraY (forming lipid I) and further assembled by MurG to form the lipid II precursor. The rhamnan chain and the PSP subunit are both assembled on the lipid carrier, Und-P. In our model, rhamnan synthesis is initiated by the transfer of GlcNAc-P onto Und-P by TagO, whereas PSP subunit synthesis is initiated by the transfer of GlcNAc onto Und-P. The three lipid-bound precursors are translocated to the outer side of the cytoplasmic membrane. MurJ is the flippase involved in the translocation of lipid II and WpsG is the presumed flippase involved in the export of lipid-linked PSP subunit. RgpC/D is an ABC-transporter involved in the transport of lipid-linked rhamnan chains. At the outer side of the membrane (OUT), PG subunits are polymerized by PBPs and possibly by SEDS (shape, elongation, division and sporulation) proteins (not shown). LcpA is proposed as the main transferase involved in anchoring rhamnan onto PG and WpsJ is a membrane glycosyltranferase with a GT-C fold proposed to be involved in attaching PSP onto rhamnan. The nature of the bond between PSP and rhamnan chains is unknown. The three proteins CsdC, CflA and CsdD are involved in the addition of side chain Glc onto PSP subunits, most probably at the outer face of the membrane. See text for further details. This figure is adapted from Sadovskaya et al. 2017 and Theodorou et al. 2019.
Next, lipid II is transferred to the extracellular face of the cytoplasmic membrane by a flippase protein that, after a long time of controversy and according to the most recent available data, is identified as MurJ, an integral membrane protein of the multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) superfamily (Sham et al. 2014). The last steps of PG synthesis involve monomer polymerization via transglycosylation and transpeptidation reactions taking place outside the cytoplasmic membrane (Fig. 2). The main proteins involved, named penicillin-binding proteins (PBPs) because they are the targets of penicillin and other β-lactam antibiotics, comprise different types of enzymes which may have both transglycosylase and transpeptidase activity (class A PBPs) or have only transpeptidase activity (class B PBP). The transglycosylation reaction incorporates the disaccharide unit into the pre-existing PG chain with the formation of a β-1,4 glycosidic bond and the lipid carrier is dephosphorylated and recycled. The transpeptidation reactions create covalent bonds between the carboxyl group of the fourth amino acid (D-Ala) of a stem peptide and the free amino group of the third amino acid of a neighbouring peptide chain or of the attached interpeptide bridge. These bonds determine the degree of PG reticulation. The proteins RodA and FtsW belonging to the SEDS (shape, elongation, division and sporulation) family, which were previously proposed to act as lipid II flippases, constitute a second class of PG polymerases with transglycosylase activity. Both were shown to work with a cognate class B PBP endowed with transpeptidase activity (Meeske et al. 2016). Whereas RodA is rather involved in the synthesis of the side wall PG in rod-shaped bacteria, FtsW contributes to septal PG synthesis during cell division.
Following synthesis, glycan chains may be modified by O-acetylation of MurNAc on the C6 OH group, as found at various levels in the different LAB studied. O-acetylation was also found on GlcNAc but only in L. plantarum up to now (Bernard et al. 2011). Another post-synthetic modification is the N-deacetylation of GlcNAc as found in L. lactis (Meyrand et al. 2007) but not in other LAB under physiological conditions so far. In addition, anchoring of glycopolymers including wall teichoic acids and polysaccharides (the structure of which is described below) can also be regarded as modifications of PG and they may compete with O-acetylation for the same site on MurNAc residues. Finally, the last modifications of PG that can be mentioned are the cleavages of PG by specific peptidoglycan hydrolases, that are involved in daughter cell separation after cell division but also in the insertion of new PG subunits, in turnover and remodelling processes (Chapot-Chartier 2010).
Teichoic acid structure and biosynthesis
Teichoic acids are anionic polymers made of alditol-phosphate repeating units and are classified into two groups: wall teichoic acids (WTAs) which are covalently linked to PG strands and lipoteichoic acids (LTAs) that are anchored to the cytoplasmic membrane through a lipid anchor. Whereas LTAs are present in all LAB studied, WTAs appear to be absent from certain lactobacilli such as L. casei and Lactobacillus rhamnosus, in agreement with the absence of biosynthesis tag or tar genes. Regarding L. lactis, teichoic acid fragments made of glycerol phosphate (Gro-P) subunits and partially substituted with Ala and Gal were purified after acid extraction (Vinogradov et al. 2018a). However, since the obtained structure is similar to that of the LTA chains, it was not possible to discern whether they were originated from LTA or WTA chains. Putative tag genes can be identified in L. lactis genomes except tagA, involved in the synthesis of the linkage unit, that is absent in most of them, thus suggesting that most L. lactis strains may not be able to synthesize WTAs.
Wall teichoic acids
WTAs exhibit variable structures between bacterial species (Brown, Santa Maria and Walker 2013). They usually contain alditols mainly glycerol (Gro) or ribitol (Rbo), and phosphate, the most studied WTAs being a poly(Gro-P) and poly(Rbo-P) backbone found in Bacillus subtilis and Staphylococcus aureus, respectively. Among LAB, they were best characterized in L. plantarum where both types, Gro-containing or Rbo-containing, are found. A given strain is able to synthesize only one type of WTA, although certain strains have the genetic potential to synthesize both types of WTAs (Tomita et al. 2010). Remarkably, L. plantarum has the capacity to switch its WTA alditol backbone, i.e. when the synthesis of poly(GroP) chains is abolished, poly(Rbo-P) type WTA can be synthesized (Bron et al. 2012).
The general biosynthesis pathway for WTAs is based on the B. subtilis model. WTA chains are synthesized inside the cytoplasm and synthesis is initiated by the enzyme TagO catalyzing the transfer of GlcNAc-P onto undecaprenyl phosphate, thus competing for the same lipid carrier as PG synthesis. Notably, TagO is homologous to MraY. A second sugar ManNAc is then added by TagA and subsequently, a Gro-P unit by TagB thus completing the linkage unit. TagF catalyzes WTA polymerization by adding Gro-P taken from CDP-Gro to the nascent WTA chain that can reach up to 60 residues. A similar pathway has been described for Rbo-P containing WTA with tar genes involved. After the intracellular synthesis is completed, the chain is translocated outside the cytoplasmic membrane by an ABC transporter TagGH. Finally, the chain is covalently linked to the C6-OH of MurNAc by enzymes belonging to the LytR-CpsA-Psr (LCP) family, as shown in B. subtilis (Kawai et al. 2011).
Lipoteichoic acids
LTA structures have been determined for several LAB including L. lactis (Kramer et al. 2008), L. plantarum (Grangette et al. 2005) and L. rhamnosus (Claes et al. 2012) and all of them were found to consist of poly(Gro-P) chains (type I LTAs). Whereas chemical structures of WTA and LTA chains can be identical, their biosynthesis pathways differ completely (Percy and Grundling 2014). The typical synthesis of type I LTA starts with the addition of two Glc residues from UDP-Glc onto diacylglycerol by YpfP, which will then constitute the glycolipid anchor. The resulting diglucosyl-diacylglycerol is then transferred from the inner to the outer side of the membrane by LtaA. The LTA chain is then polymerized by LtaS that adds in most species Gro-1-P units, from phopho-diacylglycerol donor substrate.
Modifications of teichoic acids
Two types of modifications may occur on teichoic acid (TA) chains. The first one is the well-known D-alanylation resulting from esterification of alditol groups of the TA chains with D-Ala, involving enzymes encoded by the dltABCD operon (Perego et al. 1995). In L. plantarum, an additional dltX gene encoding a small membrane protein is present in the dlt operon like in other bacilli. In Bacillus cereus, dltX was shown to be required for TA D-alanylation (Kamar et al. 2017). There is also another gene named pbpX2 located upstream of dltXABCD in L. plantarum, although its specific role in D-alanylation remains unknown (Matos et al. 2017). D-Ala substituents provide protonated amino groups that serve as counterions of negatively charged phosphate groups of the TA chains, thus lowering their global and local charge. The present model suggests that LTAs are substrate for Dlt proteins whereas WTA alanylation would occur through transfer of D-Ala from LTA to WTA chains. D-alanylation has a major impact on TA functionality, in particular in response to cationic antimicrobial peptides as detailed below.
The second type of modifications is represented by the addition of sugar substituents on the free hydroxyl groups of Gro or Rbo of the TA chains. Regarding LTAs, D-Ala was found as the only detectable substituent in L. plantarum and L. rhamnosus, whereas in L. lactis galactose (Gal) substituents were also found in addition to D-Ala (Kramer et al. 2008). Recently, the genes involved in Gal transfer onto TA chains were identified and shown to encode a three component glycosylation system, allowing extra-cytoplasmic modification of glycopolymers (Theodorou et al. 2020). In such a model, Gal would be first transferred from UDP-Gal onto undecaprenyl-phosphate, then flipped to the outside of the cytoplasmic membrane with the help of a four transmembrane segment flippase, and finally transferred onto TA chains by an integral membrane glycosyltranferase with a GT-C fold (Mann and Whitfield 2016).
Regarding WTAs, Glc residues present in all the determined structures play a key role in structural diversity within L. plantarum strains (Tomita, Tanaka and Okada 2017). One or two α-D-Glc substituents are present on the 1,5-linked poly(Rbo-P) chains. Moreover, in the case of Gro-P containing WTAs, Glc residue is part of the repeating unit in L. plantarum, made of chains of poly(1-α-D-glucosyl-GroP), and these chains can be substituted with other Glc residues. However, the genes involved in the synthesis have not been characterized yet. Notably, two types of WTA/LTA glycosylation mechanisms have been described in B. subtilis, either through the activity of an intracellular glycosyltranferase, such as TagE, for WTA or through a three-component glycosylation mechanism allowing extracellular glycosylation of LTA (Rismondo, Percy and Grundling 2018). Although glycosylation of TA is a widespread feature, the exact role in the bacterial physiology remains elusive whereas in Lactobacillus delbrueckii (Munsch-Alatossava and Alatossava 2013), as well as several pathogenic species, they play crucial roles in bacterial interactions with infecting phages or with their host.
Cell wall polysaccharides structure and biosynthesis
In most bacteria, polysaccharides (PS) are also found as components of the CW (Fig. 1), covalently bound to PG, which we will name here cell wall polysaccharides (CWPS). When they can be visualized by electron microscopy as a thick layer outside the cell such as in pathogenic streptococci and enterococci, they are usually named capsular polysaccharides (CPS). Bacteria, and particularly LAB, can also produce exopolysaccharides (EPS) that are loosely attached to the cell surface or released into the surrounding medium and that we will not describe here (for a recent review see Zeidan et al. 2017). Of note, there may have some ambiguity regarding the nomenclature used in the literature, since EPS can also mean extracellular polysaccharide, which may also reflect existing ambiguity at the experimental level. In our view, CWPS or CPS are PS covalently attached to the PG and require harsh acid treatment (such as TCA or HF treatment) to be extracted from the CW, whereas EPS can be purified from culture supernatant.
In the last five years, significant advances have revealed the structural diversity of L. lactis CWPS between strains and the complex biosynthesis pathway that can account for this chemical diversity have been unveiled. A singular component known as the polysaccharide pellicle (PSP) was first discovered in L. lactis MG1363 (Chapot-Chartier et al. 2010). This structure was characterized as a thin outer layer of the bacterial cell envelope composed of hexasaccharide repeating units linked by phosphodiester bonds. Similar, yet distinct in their composition, polymeric chains made of phosphate-oligosaccharide repeating units have been identified in two other L. lactis strains, 3107 (Ainsworth et al. 2014) and SMQ-388 (Farenc et al. 2014). Later on, it was shown that both L. lactis MG1363 and 3107 produce an additional neutral PS, made of linear polyrhamnose chains and known as the rhamnan, which together with the PSP forms part of the CWPS (Fig. 1). In addition, biochemical data strongly suggest that the two chains are covalently linked together (Sadovskaya et al. 2017). Regarding the spatial organization of CWPS inside the CW, PSP is exposed at the bacterial surface, whereas rhamnan appears to be trapped and embedded within the PG network.
A large genetic cluster (named cwps) comprising more than 20 genes encoding CWPS biosynthesis is present in the genomes of all L. lactis strains that have been sequenced to date, albeit with a large degree of sequence and gene content diversity, particularly at its 3’ end (Ainsworth et al. 2014; Sadovskaya et al. 2017). Based on sequence analysis, L. lactis strains have been classified into three distinct cwps genotypes, i.e. type A, B, and C (Mahony et al. 2013). According to this classification, the three strains mentioned above that synthesize the phospho-PS pellicle belong to the type-C group. When the chemical structures of CWPS from both an A-type strain, L. lactis UC509.9 and a B-type strain, L. lactis IL1403, were determined, they were shown to consist of a unique component made a rhamnose-rich or a rhamnan backbone chain, respectively, substituted with short oligosaccharide substituents (Vinogradov et al. 2018a; Vinogradov et al. 2018b).
The more conserved 5’-end of the cwps cluster encodes the proteins required for rhamnan biosynthesis (Sadovskaya et al. 2017) and the more variable 3’-end for PSP biosynthesis in C-type strains (Ainsworth et al. 2014) and, most likely, for side chain oligosaccharide in type A and B strains. Recently, a comprehensive biosynthesis scheme was proposed based on a mutational analysis of the genes with a CRISPR-Cas9 based method combined with structural analysis of the mutant CWPS by mass spectrometry and bioinformatic analysis of the proteins encoded in the gene cluster (Theodorou et al. 2019). This scheme was supported also by transmission electron microscopy and phage sensitivity assays. In this model scheme, the two CWPS components, rhamnan and PSP, are assembled independently from two distinct lipid-sugar precursors, undecaprenyl-pyrophosphate-GlcNAc synthesized by TagO and undecaprenyl-monophosphate-GlcNAc by WpsA/B, respectively (Fig. 2). Rhamnan synthesis follows an ABC-transporter dependent pathway, with an intracellular elongation of the polyrhamnose chains that is exported outside the membrane by an ABC-transporter and anchored onto PG by an LCP protein (Sadovskaya et al. 2017) (Fig. 2). The PSP subunit is synthesized as an oligosaccharide linked to undecaprenyl-phosphate, then flipped outside the cytoplasmic membrane and further polymerized by the GT-C fold membrane glycosyltransferase WpsI assisted by WpsH. Finally, PSP would be linked covalently to rhamnan at the extracellular face of the cytoplasmic membrane (Theodorou et al. 2019) by the membrane-embedded glycosyltranferase with a GT-C fold, WpsG (Fig. 2). The proposed scheme encompasses a mechanism involved in the extracytoplasmic modification of bacterial glycoconjugates (Mann and Whitfield 2016) and in this particular case, it would be dedicated to add complex substituents (polymeric PSP in C-type strains or shorter oligosaccharides in A- and B-type strains) onto the rhamnan chains. Interestingly, this model can account for the large structural diversity observed in L. lactis CWPS. Nevertheless, additional structural diversity of PSP subunits or rhamnan results from the activity of glycosyltranferases located outside the cwps gene cluster that are part of three component mechanisms, which catalyze the addition of single Glc as side chains of PSP subunits or rhamnan (Theodorou et al. 2020). In L. lactis, CWPS act as receptors for numerous bacteriophages and their structural diversity explains, at least partially, the narrow host range of a number of these phages (Mahony, Cambillau and van Sinderen 2017a).
S. thermophilus strains are often selected for their ability to produce EPS for which several structures are available. Evidence has been provided for the synthesis of a cell wall rhamnose-rich polysaccharide that could be similar to the rhamnose-glucose polysaccharide of pathogenic streptococci, which is a rhamnan chain with Glc substituents (Thevenard et al. 2014). The chemical structure of a complex branched rhamnose-glucose polysaccharide with a backbone tetrasaccharide repeating units made of rhamnose and glucose, carrying di-, tri- and tetrasaccharide side chains was recently established in S. thermophilus ST64987. Notably, this strain also synthesizes a second cell wall associated polysaccharide (named EPS) composed of pentasaccharide repeating units composed of galactose and glucose (McDonnell et al. 2020). In this strain, this second polysaccharide was found to be required for phage adsorption.
Lactobacilli also synthesize CWPS with structures varying between strains, although there is, in several cases, some ambiguity regarding the nomenclature of CWPS versus EPS. Regarding L. casei, several different structures are available of acid-extracted PS, which revealed to be highly rich in Rha such as in strain BL23 (Vinogradov et al. 2016). Their diversity was also revealed by a lectin microarray developed to compare the surface glycomes of a range of L. casei strains (Yasuda et al. 2011). In L. rhamnosus, PSs were identified in strain GG that were shown to be associated with the cell surface by AFM. These PS (named EPS for extracellular PS) comprised a long galactose-rich PS with known structure and a second Glc-rich PS making shorter chains (Francius et al. 2009). Other L. rhamnosus PS with an established structure were purified from culture supernatants thus rather corresponding to EPS. The diversity of CWPS structures between strains was also exemplified in three Lactobacillus helveticus strains (Vinogradov et al. 2013). In L. plantarum several putative gene clusters encoding PS biosynthesis—in this case named CPS—were identified in the genome of strain WCFS1 (Remus et al. 2012), but to our knowledge, the corresponding structures are not available. The lactobacilli CWPS with known structures are heteropolysaccharides that appear to be synthesized via a Wzy-dependent pathway, which is characterized by the synthesis of the repeating unit inside the cytoplasm on a lipid carrier. Then, the building block is flipped outside the cytoplasmic membrane by a Wzx flippase before polymerization by a Wzy polymerase. Synthesis is most likely regulated by the activity of a tyrosine (BY) kinase (Lebeer et al. 2009).
Cell wall proteins
Proteins are also basic components of the LAB CW. They can be either covalently linked to PG via sortase A (LPXTG proteins), associated to the CW through the interaction of CW binding domains (autolysins, S-layer) or linked to the cytoplasmic membrane via transmembrane domains or lipid anchors (Chapot-Chartier and Kulakauskas 2014). Their study has been approached by in silico and experimental procedures such as proteolytic digestion followed by liquid chromatography-MS/MS peptide analyses and labelling with fluorescent dyes and further fractionation (Mercier-Bonin and Chapot-Chartier 2017).
The CW binding domains of non-covalently bound CW proteins are diverse and bind to different CW polymers (Chapot-Chartier and Kulakauskas 2014). Among them, LysM is the most commonly found and known to bind to the GlcNAc-X-GlcNAc motif present in polysaccharides such as bacterial PG (Mesnage et al. 2014). Of note, CW binding domains can be exploited for surface display of recombinant proteins (antigens, antibodies and enzymes) for medical and industrial applications (Visweswaran, et al. 2014; Michon et al. 2016). Not surprising the screening of new CW binding domains from different sources and with distinct binding affinities is still on-going (Plavec, Strukelj and Berlec 2019).
Several membrane-located proteins, as yet mentioned, are involved in the biosynthesis and modification of CW polymers and together with PG hydrolases (autolysins), cytoskeletal elements and the cell divisome, ultimately drive cell morphogenesis and shape as reviewed elsewhere (Yang, Blair and Salama 2016; Egan et al. 2017). Although this topic is out of the scope of this review, it is worth mentioning that L. lactis is regarded as a good model to study morphogenesis of ovoid shape bacteria and some mechanistic insights have already been gained. Precisely, it has been shown that L. lactis can transition from ovoid to rod‐shaped when septum biosynthesis is disrupted (Pérez-Núñez et al. 2011) and that PBP2b participates in cell elongation and cell division (David et al. 2018). Similarly, the role in morphogenesis of the PG hydrolases LytA and LytB of L. plantarum have been recently demonstrated (Duchêne et al. 2019). Otherwise, the function of several LAB PG hydrolases in daughter cell separation and autolysis is well-established with AcmA and Acm2, being the main autolysins in L. lactis and L. plantarum, respectively (Buist et al. 1995; Rolain et al. 2012). Some autolysins seem to be functionally associated with the S-layer in L. acidophilus as AcmB (and orthologues thereof) which are exclusively found in S-layer-forming lactobacilli (Johnson and Klaenhammer 2016). Indeed, the S-layer lattice behaves as an important scaffolding structure for the display of numerous surface proteins that vary during growth (Klotz et al. 2017). Membrane proteins also served as receptors for certain bacteriocins and bacteriophages, defining their spectrum of inhibition and host range, respectively (Diep et al. 2007; Millen and Romero 2016).
On the other hand, CW proteins that are surface-exposed are often involved in adhesion to biotic and abiotic surfaces and responsible for multiple interactions of LAB within the external environment. Therefore, several CW proteins from specific LAB strains, i.e. particular PG hydrolases (p45 and p70), S-layers, mucus binding proteins and/or protein surface appendages (pili) are recognized as key probiotic effector molecules, mediating adhesion and persistence in the gut and immunoregulatory interactions with the host (Lebeer et al. 2018 and references therein).
Investigating the molecular architecture of the cell wall
Probing the spatial organization of the glycopolymer constituents inside the CW requires powerful biophysical techniques (Rohde 2019) and several of them have been successfully applied to LAB. AFM allows to probe cellular structures at nanometer resolution on living cells and was used to explore the microbial cell surface (Dufrêne 2014). One advantage of AFM is that bacteria may be immobilized in porous membranes without chemical fixation, thus allowing preservation of the native structure and organization of the macromolecules of the bacterial surface. AFM analysis relies on sensing small forces acting between a sharp tip and the sample surface. By scanning the whole surface, it is then possible to generate a three-dimensional image at (near) molecular resolution. In addition, AFM single-molecule force spectroscopy (SMFS) allows quantifying the forces between the tip and the sample. The tip can be further functionalized with a biomolecule (antibody or ligand) to localize or manipulate a specific molecule of the bacterial surface. Also, AFM can probe the nano-mechanical properties of the CW in situ in living bacterial cells (Tripathi et al. 2012). Furthermore, when CW mutants lacking outer CW components are studied by AFM, it is possible to reveal the organization of inner CW constituents. As an example, topographic imaging of L. lactis surface by AFM showed that wild-type cells had a smooth surface whereas mutants devoid of CWPS exhibited a rough surface, revealing the disappearance of an outer layer identified as PSP (Chapot-Chartier et al. 2010). In addition, 25 nm-wide concentric rings were detected running parallel to the short axis of the cells on the PSP mutant. With tips functionalized with LysM domain that specifically binds PG, it was possible to conclude that these periodic bands correspond to PG chains, thus revealing the nanoscale organization of PG (Andre et al. 2010). Besides, AFM was used to measure the rigidity of the CW of a L. lactis pyrB mutant which was found to be increased compared to that of wild-type cells (Solopova et al. 2016). AFM also helped to detect L. lactis pili (Meyrand et al. 2013) and to unravel the mechanical and adhesive properties of pili from L. lactis (Castelain et al. 2016) and L. rhamnosus at the nanoscale (Tripathi et al. 2013). In a very recent study, AFM single-cell force spectroscopy was used to quantify the force of the homotypic pili interactions between individual bacterial cells, using different L. lactis strains (Dramé et al. 2020). Moreover, in L. rhamnosus GG cells, two types of CWPS were identified, localized and stretched by SMFS with the use of lectin-functionalized tips (Francius et al. 2009). This allowed characterization of a mannose/glucose-rich PS with moderate extension and a galactose-rich PS with much longer extension. Regarding L. plantarum, AFM was combined with fluorescence microscopy and with lectins to study the spatial organization of WTAs. Topographic images of wild-type bacteria revealed a polarized surface morphology, with smooth poles and rougher side walls. With SMFS and a tip functionalized with lectins, WTAs were shown to be absent from the cell poles and localized on the side walls (Andre et al. 2011). These data are in agreement with the proposed model for CW structural organization in bacilli with both types of TAs such as B. subtilis, with WTAs protruding at the bacterial surface whereas LTAs would be rather located inside the CW, close to the cytoplasmic membrane (Percy and Grundling 2014).
Another powerful technique that was used to assess the localization of the CWPS in L. lactis is solid state high resolution NMR under magic angle sample spinning (HR-MAS NMR). By comparing wild-type L. lactis and a mutant devoid of PSP, HR-MAS NMR allowed to detect the surface exposed flexible PSP in the wild-type cells whereas rhamnan was not observed. In contrast, in the PSP-negative mutant, rhamnan was detected suggesting that it became exposed and/or more flexible in the mutant and thus telling that rhamnan is embedded inside the cell wall in the wild-type cells (Sadovskaya et al. 2017). As shown with other Gram positive bacteria, solid state NMR could also be a valuable tool for the detection and structural analysis of WTA (Kern et al. 2010) or for investigating PG architecture (Kim, Chang and Singh 2015).
The development of metabolic labelling with efficient small-molecule probes, consisting of fluorescent D-amino acids (FDAAs) or clickable D-amino acids, has allowed imaging of PG synthesis in live bacteria (Kuru et al. 2012). New PG incorporation can then be visualized with time-lapse microscopy or super-resolution microscopy (Radkov et al. 2018). As an example, incorporation of FDAA in L. lactis cells was clearly observed at the sites of cell division in actively dividing cells (Kuru et al. 2012). More generally, metabolite derivatives to label other glycopolymers of the cell envelope of LAB would be extremely valuable tools to track their biosynthesis (Siegrist et al. 2015).
THE LAB CELL WALL AS A TARGET FOR BACTERIOCINS, BACTERIOPHAGES AND HOST FACTORS
The inherent structural and physiological properties of the bacterial CW make it an excellent target for effective killing (Schneider and Sahl 2010). In the context of pathogenic bacteria, the CW and its biosynthetic pathway are indeed targeted by numerous antibiotics including fosfomycin, D-cycloserine, beta-lactams, glycopeptides, and other peptide antibiotics classes (lipopeptides, depsipeptides) (Grein, Schneider and Sahl 2019). For industrial LAB, bacteriocins and bacteriophages may be regarded as important CW antimicrobials in the context of food fermentations, whereas probiotic or commensal LAB are exposed to innate immunity factors such as lysozyme and host antimicrobial peptides (defensins, cathelicidins). All of them target particular structures in the CW as summarized in figure 3. Undoubtedly, both industrial LAB and probiotics are confronted with several abiotic and biotic stresses during starter production, food fermentation and in the host environment, which may have an impact on the CW. The reader is referred to the comprehensive review on the stress physiology of LAB for detail description on other stressors and defence mechanisms which will not be described here (Papadimitriou et al. 2016).
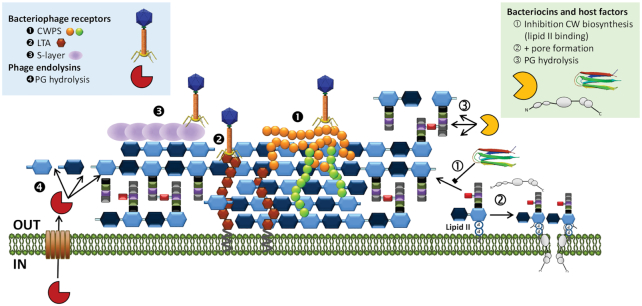
Figure 3.
Cell wall structures targeted by bacteriophages, bacteriocins and host factors (antimicrobial peptides and lysozyme). Molecules are depicted as in figures 1 and 2. CW, cell wall; PG, peptidoglycan; CWPS, cell wall polysaccharides; LTA, lipoteichoic acid.
Cell wall-targeting bacteriocins
Bacteriocins are a highly diverse group of ribosomally synthesized antimicrobial peptides, which are widely recognized by their role in intra- and interspecies competition in complex microbial communities (Kommineni et al. 2015; Chikindas et al. 2018). The use of nisin as a food biopreservative worldwide, the inclusion of bacteriocin-producers in protective cultures and the use of undefined fermentates as food shelf-life extensors contribute to the presence of bacteriocins in food, thereby acting as stressors not only for pathogens but also for beneficial LAB. Moreover, genomic and metagenomics studies of niches where LAB thrive underscore the common occurrence of bacteriocin clusters (Zheng et al. 2015; Tracanna et al. 2017; Garcia-Gutierrez et al. 2019).
Due to the large heterogeneity in terms of physicochemical properties, structure, spectrum of activity and mode of action, classification of bacteriocins is controversial but most of them fall into three major classes (Cotter, Hill and Ross 2005; Alvarez-Sieiro et al. 2016; Acedo et al. 2018). Class I (< 10 kDa) are ribosomally produced and post-translationally modified peptides (RiPPs). The class Ia lantibiotics (or lanthipeptides with antibiotic activity) are characterized by the presence of the non-proteogenic amino acids lanthionine and ß-methyl lanthionine which are introduced by the LanCB (type I) or LanM (type II) modification enzymes (for a detailed overview of all lanthipeptide synthesis routes, see Repka et al. 2017). Another modification frequently found is a head-to-tail peptide bond that links the N- and C-terminal residues, rendering a circular molecule (class Ib). Other RiPPs with antimicrobial activity are the sactibiotics or cysteine sulphur-to-α-carbon bridges containing peptides (class Ic), linear azol(in)e-containing peptides (LAPs) (class Id), glycocins decorated with carbohydrates (class Ie) and lasso peptides (class If) (Arnison et al. 2013; Alvarez-Sieiro et al. 2016). Examples of class Ia lantibiotics that target the CW are shown in Table 1.
Table 1.
LAB bacteriocins that target the cell wall.
| Bacteriocin | Producer | Class | Mode of action | References |
|---|---|---|---|---|
| Nisin | L. lactis | Ia Elongated lanthipeptide type I (LanBC modified) | Binding to lipid II + pore formation | Brötz et al. 1998 |
| Lacticin 481 | L. lactis | Ia Globular lanthipeptide type II (LanM modified) | Binding to lipid II + PG transglycosylation inhibition | Knerr et al. 2012 |
| Salivaricin A | S. salivarius | Ia Globular lanthipeptide type II (LanM modified) | Low affinity binding to lipid II | Geng et al. 2018 |
| Salivaricin B | S. salivarius | Ia Globular lanthipeptide type II (LanM modified) | Accumulation of PG soluble precursors | Barbour et al. 2016 |
| Plantaricin C | L. plantarum | Ia Globular lanthipeptide type II (LanM modified) | Binding to lipid II + pore formation (strain dependent) | Wiedemann et al. 2006a |
| Lacticin 3147 | L. lactis | Two-peptide lantibiotic type II (LanM modified) | Binding lipid II by LctA1 + pore formation LctA2 | Wiedemann et al. 2006b |
| Lcn972 | L. lactis | IId, non-modified | Lipid II binding, putative co-target | Martínez et al. 2008 |
| Enterolysin A | E. faecalis | III, bacteriolysin | Endopeptidase (stem and interpeptide bridge) | Khan et al. 2013 |
| Zoocin A | S. equi | III, bacteriolysin | D-alanyl-L-alanine Zn2+‐metallopeptidase | Gargis et al. 2009b |
| Millericin B | S. milleri | III, bacteriolysin | Endopeptidase (stem and interpeptide bridge) | Beukes et al. 2000 |
| Bac41 | E. faecalis | III, two-component bacteriolysin | D-isoglutamyl-L-lysine endopeptidase (BacL1) | Kurushima et al. 2013 |
Class II is composed of non-modified bacteriocins and are also subdivided in several subclasses. Class IIa or pediocin-like bacteriocins share a N-terminal conserved YGNGV motif and show very potent anti-Listeria activity; class IIb are bacteriocins that depend on the concerted action of two peptides; class IIc includes leaderless bacteriocins, and class IId comprises other small antimicrobial peptides with distinct properties. Non-modified bacteriocins do not require modification enzymes but structural genes are often located in clusters with genes encoding immunity factors, dedicated transporters and regulatory functions. Other large proteins (>10 kDa) or protein complexes with antimicrobial activity are included in class III. There are only a few members of this class that had been studied but some show a lytic mode of action (bacteriolysins) and will be described here because they act on the CW.
Generally, pore formation by insertion into the cytoplasmic membrane or through the interaction with receptors or docking molecules results in disruption of selective membrane permeability and rapid killing, this being the main mode of action of many LAB bacteriocins (Cotter et al. 2005). As for the CW active bacteriocins, two main groups can be defined: lipid II binders that inhibit CW biosynthesis, frequently in combination with pore formation, and lytic bacteriocins that degrade pre-existing PG (Table 1).
Lipid II binding bacteriocins
The CW precursor lipid II plays key roles in the physiology of the bacterial cell, not just as a PG building block, but also as a contributing element for organization of membrane domains and precise localization of the PG biosynthetic machinery (Scheffers and Tol 2015). Thus, it is not surprising that a plethora of natural compounds, including LAB bacteriocins, target lipid II (Grein et al. 2019).
Nisin is among the first lantibiotics for which lipid II binding was demonstrated (Brötz et al. 1998). Subsequent detailed analysis revealed the unique combination of CW biosynthesis inhibition and pore formation as its mode of action (Breukink et al. 1999). Structural studies identified the lipid II pyrophosphate as the primary site recognized by the N-terminal lanthionine rings A and B of nisin and related lantibiotics (Hsu et al. 2004). The complete MurNAc moiety, the pentapeptide unit and a membrane environment further strengthen this interaction (‘t Hart et al. 2016). Upon binding of lipid II, the flexible hinge region facilitates insertion of the nisin C-terminal domain into the bacterial membrane and lipid II becomes an integral part of the pore with a peptide:lipid II stoichiometry 2:1 (Hasper, de Kruijff and Breukink 2004) (Fig. 3). Alongside pore formation, nisin displaces lipid II from its functional locations, so that it becomes unavailable for CW biosynthesis and septum formation (Hasper et al. 2006). Furthermore, lipid II-nisin aggregates induce severe structural membrane deformations (Scherer et al. 2015), all in all resulting in potent antimicrobial activity in the nM range.
Other subgroup of lantibiotics, some of which have been shown to bind to lipid II, share the lipid II-binding motif of mersacidin, a lantibiotic that inhibits transglycosylation and prevents the incorporation of PG units into glycan strands without pore formation (Brötz et al. 1997) (Fig. 3). This group encompasses several natural analogues including lacticin 481, salivaricins and plantaricin C, among others (Table 1). Lacticin 481 inhibits PBP1b-catalyzed PG synthesis and forms a strong complex with lipid II in vitro (Böttiger et al. 2009; Knerr et al. 2012). Recent studies also suggest that salivaricin A2 from Streptococcus salivarius may also belong to this group of lipid II binders, although affinity for lipid II in vitro appears to be rather low (Geng et al. 2018). Similarly, salivaricin B appears to interfere with CW biosynthesis as judged by the accumulation of the soluble UDP-MurNAc-pentapeptide precursor and lack of interaction with bacterial membrane vesicles (Barbour et al. 2016). Plantaricin C binds strongly to lipid II in vitro and is also able to form pores, depending on the target strain (Wiedemann et al. 2006a).
The two-component lantibiotic lacticin 3147 also relies on lipid II binding for membrane poration. In this case, binding of the LctA1 peptide to lipid II induces conformational changes that facilitate recruitment of LctA2 and pore formation. Otherwise, the peptides alone are not, or just modestly, inhibitory (Wiedemann et al. 2006b).
Within the non-modified bacteriocins, Lcn972 is the only one described so far that inhibits CW biosynthesis upon binding to lipid II but it does not form pores (Martínez et al. 2008) (Fig. 3). Notably, despite binding to the rather conserved lipid II, Lcn972 is only active against other lactococci. Besides, inhibition of PG synthesis seems to occur only at the septum during cell division (Martínez, Rodríguez and Suárez 2000). Taken together, it has been argued that a putative co-target or docking molecule is required for Lcn972 killing (Martínez et al. 2008). Moreover, Lcn972 is a well-structured peptide in aqueous solutions with a ß-sandwich fold comprising two three-stranded antiparallel ß-sheets (Turner et al. 2013). This particular structure may represent a novel lipid II binding domain.
There is as yet another way by which some bacteriocins might interfere with the lipid II cycle and, consequently, hinder CW biosynthesis. The two component bacteriocin lactococcin G, and likely other close relatives, uses the membrane protein UppP as a receptor for targeted pore formation (Kjos et al. 2014). UppP (or BacA) is an undecaprenyl pyrophosphate phosphatase required for the synthesis and/or recycling of the lipid carrier, the molecule that ferries CW precursors through the membrane. Mutations in the uppP gene were found in L. lactis lactococcin G-resistant mutants and expression of this gene in Streptococcus pneumoniae rendered this strain, otherwise insensitive, susceptible to lactococcin G. It remains to be explored if UppP is in fact inactivated upon lactococcin G binding and what are the consequences, if any, on CW biosynthesis.
Bacteriolytic bacteriocins
Among the heat-labile class III bacteriocins, there is a group, also known as bacteriolysins, that hydrolyze the bacterial PG and provoke lysis of the target bacterial cells. In general, bacteriolysins are often organized in functional modules, bearing catalytic domains that cleave specific bonds of the PG and substrate recognition domains. Not many LAB bacteriolysins have been characterized so far but they may be more common than anticipated. Class III bacteriocins are frequently identified in (meta-) genomic datasets (Walsh et al. 2015; Collins et al. 2017), although their bacteriolytic mode of action remains to be demonstrated.
The best studied bacteriolysin is lysostaphin, a bacteriolysin produced by Staphylococcus simulans that hydrolyzes the pentaglycine cross-bridge of the S. aureus CW (Kumar 2008). Within LAB bacteriolysins (Table 1), enterolysin A is an endopeptidase that cleaves the rather conserved peptide bond between position 1 (L-Ala) and 2 (D-Glu) of the stem peptide and between L‐Lys and D‐Asp of the cross-bridge, both present in the PG of L. lactis, Pedicoccus pentosaceus and Lactobacillus delbrueckii subsp. bulgaricus (Khan, Flint and Yu 2013). Zoocin A and Millericin B, both synthesized by streptococcal species, display selective activity against streptococci and also target the stem peptide (Table 1). Zoocin A belongs to the M23 peptidase family and cleaves the bond between D-Ala of the stem peptide and L-Ala of the cross-bridge in streptococcal PG (Gargis et al. 2009b). The structure of a Cys74 to Ala74 mutant of its catalytic domain has been recently resolved by NMR (Xing, Simmonds and Timkovich 2017). The structure resembles that of lysosthaphin and LytM catalytic domains but with a wider substrate binding groove and no tyrosine residue in the active site. Millericin B seems to cleave at the glutamic acid of the stem peptide and at the N-terminus of the last residue of the cross-bridge (Beukes et al. 2000). For both bacteriolysins, immunity is provided by Fem-like proteins that modify the peptidoglycan structure of the producer. The zoocin A immunity protein (Zif) lengthens the streptococcal PG cross-bridge by incorporating an additional L-Ala (Gargis et al. 2009a). MilF is postulated to incorporate leucine into the cross-bridge, in conjunction with a putative leucyl-tRNA, for self-protection against millericin B (Beukes and Hastings 2001). Another bacteriolytic bacteriocin is the enterococcal Bac41 which consists of two extracytoplasmic proteins. BacL1, a D-isoglutamyl-L-lysine endopeptidase with a SH3 cell wall binding domain, lyses enterococcal cells only when the accessory protein BacA is present (Kurushima et al. 2013; Kurushima et al. 2015).
Bacteriophages, bacterial viruses that target the cell wall twice
Bacteriophages (or phages) are viruses that prey on bacteria. In the process of infection, phages entering a lytic cycle subvert the metabolic machinery of the host towards the production of viral particles which are ultimately released, in most cases, after lysis of the host. Temperate phages are able to integrate their genome into the bacterial chromosome as prophages (lysogenic cycle). Prophages can be activated, mainly by the SOS response, and enter into the lytic cycle. Phage infection poses a threat for biotechnological processes that rely on bacteria. Milk fermentations are particularly prone to phage infection owing to the prevailing non-sterile environment and the use of few starter strains that are cultivated in bulk quantities (Garneau and Moineau 2011). The economic impact of fermentation failure has fostered intensive research on phages infecting LAB, leading to ground-breaking concepts in phage biology as, for example, the role of CRISPR/Cas in adaptive phage resistance (Barrangou et al. 2007).
LAB phages are double‐stranded DNA tailed phages of the order Caudovirales. Most of them belong to the Siphoviridae family with long non-contractile tails and isometric or elongated icosahedral capsids but members of Podoviridae (short non-contractile tail) and Myoviridae (long contractile tail) have also been reported, the latter mostly represented by phages infecting Lactobacillus spp. (Mahony and van Sinderen 2014). Ten lactococcal phage species have been defined, with those belonging to the c2, 936 and P335 being the most prevalent in cheese factories (Deveau et al. 2006). By contrast, phages infecting S. thermophilus are more homogenous. There are two dominating types classified as pac- (headful packaging) and cos- (cohesive ends), according to their mode of DNA packaging, and the recently described 5093 and 987 phage types with distinct morphology (McDonnell et al. 2016). This grouping has been further confirmed by a large comparative pangenomic study encompassing 142 S. thermophilus phages (Szymczak et al. 2019). Nevertheless, as genomic studies progress, novel distinct genetic groups emerge within phages infecting S. thermophilus (Philippe et al. 2020) and other LAB species (Pujato et al. 2017; Kyrkou et al. 2019).
As pointed out by Chapot-Chartier (2014), the bacterial CW is key for a successful phage infection cycle as it forms a barrier that phages encounter twice. At the beginning of the infection, phages recognize their hosts through the presence of receptors on the bacterial surface (Fig. 3). Phage DNA must be then translocated into the cytoplasm and some phages make use of specific PG hydrolytic activities of their tail fibers. Later on, after propagation inside the host, newly formed viral particles have to be released and phages rely on the activity of endolysins to degrade the PG layer (Fig. 3).
Host recognition to initiate phage infection
The host range of LAB phages is, in general, highly specific and in some cases, phages infecting particular strains within a bacterial species are isolated. The molecular basis of this exquisite specificity has been recently unveiled by multidisciplinary approaches that combined phage/host genomics, structural biology and CW biochemistry studies (Mahony et al. 2017a). Host recognition takes place through phage tail components, organized as a straight tip or a baseplate, where the receptor binding protein (RBP) dictates the type of the interactions between the phage and the host (Veesler and Cambillau 2011). Indeed, RBP phylogenetic studies often correlate with host range, while pioneering phage domain shuffling experiments and adsorption/binding of recombinant RBPs to bacterial cells, further confirmed the role of RBPs in host recognition (Dupont et al. 2004; Murphy et al. 2016; Szymczak et al. 2019). Besides, reconstruction at the atomic level of the baseplate topology by electron microscopy and X-ray crystallography of several siphophages revealed the conserved RBP modularity, distinct assembling and activation strategies upon binding to the host, and provided meaningful insights into the function of other baseplate components, such as the contribution of additional binding domains in the distal and tail proteins and phages with dual RBPs (Veesler et al. 2012; Dieterle et al. 2017; Mahony et al. 2017b; Hayes et al. 2018).
The discovery of the CWPS as an integral component of the CW of several LAB species has represented a major step forward in the field of phage-host interactions, because it explains the narrow host range of LAB phages owing to the CWPS biochemical diversity (see above). In fact, swapping of glycosyltransferase-encoding genes from chemically distinct CWPS loci, also swapped the susceptibility to phages, i.e. hosts that are not infected by a given phage become susceptible to it when the CWPS glycosyltransferases of a susceptible host are synthesized (Ainsworth et al. 2014). Moreover, phage escape mutants, that are able to infect engineered lactococcal strains with altered CWPS, acquired mutations within the baseplate-encoding region, confirming their adaption to new host receptors (Theodorou et al. 2019). CWPSs have been unequivocally proven as receptors for several 936 and P335 phages, as well as for the less common 949 and 1358 phage groups (Farenc et al. 2014; Mahony et al. 2016; Mahony et al. 2017a and references therein). The rhamnose-rich CWPS is also the receptor of phage J-1 infecting L. casei/paracasei (Dieterle et al. 2017). Likewise, several streptococcal phages, most likely, make use of the streptococcal rhamnan PS to recognize their host (Szymczak et al. 2018), whereas others rather use an EPS component of the CW as receptor (McDonnell et al. 2020). Importantly, linking phage RBPs to CWPS genotypes provides an excellent tool for setting up a rational for programming starter rotation schemes (Mahony et al. 2013).
Other components of the LAB CW have also been identified as phage receptors. Lactococcal phages belonging to the group c2 bind to their hosts by a two-step process involving, first, reversible adsorption to an unidentified saccharide motif and later, irreversible binding to the membrane phage infection protein (Pip) or Pip-like proteins such as YjaE. Pip proteins are not essential for the host and mutations are easily selected under phage pressure (Millen and Romero 2016). L. delbrueckii subsp. lactis LTA is the receptor of phage LL-H, where the phage tail fibers are thought to reversibly adsorb to the surface exposed Glc substitutions of LTA, and irreversibly to the negatively charged poly(Gro-P) backbone of LTA (i.e. without or with few D-Ala substitutions). According to the model proposed by Munsch-Alatossava and Alatossava (2013), a stable calcium-LTA channel is formed, which is enlarged by the activity of a virion-associated PG hydrolase, to provide access for a membrane-interacting protein that guides the transfer of the phage DNA. The S-layer of L. helveticus seems to be involved in host recognition as several phage-resistant mutants carried point mutations or small deletions in the S-layer gene (Ventura, Callegari and Morelli 1999; Zago et al. 2017).
The need of PG hydrolytic activity for delivery phage DNA is another example of how the structure of the CW, in this case that of the PG, may determine phage infectivity. The tail-associated lysin (Tal) of the P335 phages TP901.1 and Tuc2009 undergo proteolytic processing and mature virions with either full-length or a C-terminally truncated Tal protein are produced. Stockdale et al. (2013) demonstrated that virions with the full length Tal are able to infect stationary phase cells with a highly cross-linked PG, while those with the truncated version infect exponentially growing cells better. Other virion-associated enzymatic activity might also contribute to infection by locally degrading other CW components. Specifically, a glycerol phosphodiesterase activity able to hydrolyse surface-associated carbohydrate polymers was located in the baseplate of L. delbrueckii Ld17 phage (Cornelissen et al. 2016).
Host lysis to end phage infection
At the end of the phage cycle, once virion particles have been accumulated in the infected bacteria, the PG hydrolytic activity of phage endolysins is required (Fig. 3). These proteins get access to their substrate through pores made by the phage holin in the cytoplasmic membrane (canonical lysis) or they are secreted through the aid of a non-cleavable N-terminal signal peptide (SAR endolysins) or by the general Sec-pathway (Sec-dependent endolysins). The two latter endolysins still rely on holin pore formation to be activated (for a recent review, see Fernandes and São-José 2018). Similar to bacterial PG hydrolases (autolysins), endolysins from phages infecting Gram positive bacteria are often modular enzymes with both catalytic and CW binding domains (Schmelcher, Donovan and Loessner 2012). Catalytic domains identified in LAB phage endolysins encompass distinct enzymatic specificities targeting different PG bonds and include N-acetyl-muramyl-L-Ala-amidases, γ-D-Glu-L-Lys-endopeptidases, N-acetyl-muramidases, and CHAP (cysteine-histidine dependent amido-hydrolase/peptidase domain) with both amidase and/or peptidase activity (Chapot-Chartier 2014). CW binding domains, when present, are also shared with host autolysins and govern substrate affinity. LysM, SH3_5, PG-binding_3 and Lc-LysBD domains have been identified in LAB phage endolysins (Chapot-Chartier 2014).
Several (covalent) modifications of the PG or LTA have been demonstrated to influence both the catalytic activity and substrate affinity of host autolysins. For example, the activity of the N-acetylglucosaminidase AcmA, the main L. lactis autolysin, is inhibited by N-acetylglucosamine deacetylation (Meyrand et al. 2007), by amidation of D-Asp in the PG cross-bridge (Veiga et al. 2009) or by galactose substitutions in LTA (Steen et al. 2008). Despite functional homology between bacterial autolysins and phage endolysins, research on the impact of CW modifications on endolysin activities, and consequently on phage propagation, is scarce. Interestingly, the degree of PG O-acetylation restrains the in vitro activity of LysTP712, the endolysin of the lactococcal prophage TP712 (Escobedo et al. 2019). Moreover, inactivation of FtsH, a stress-responsive membrane protease, impairs host lysis after activation of the lytic cycle of TP712 (Roces et al. 2013; Roces et al. 2016). L. lactis ΔftsH cells bound less fluorescent mCherry protein tagged with the CW binding domain of LysTP712, suggesting that lack of cell lysis after infection might be a consequence of the reduced binding of the phage endolysin to an altered cell surface. The interest on phage endolysins as potential clinical antimicrobials and beyond is currently increasing (Rodríguez-Rubio et al. 2016). It is envisaged that deeper mechanistic insights into LAB phage endolysins will foster their utilization in different fields. In fact, their use as antimicrobials to control LAB contaminants in fuel ethanol fermentations has already been explored (Roach et al. 2013). Another example is their value as reagents to improve purity assays of probiotic preparations. Here, endolysins targeting probiotic Lactobacillus might be used to lyse the probiotic and simplify detection of contaminating bacteria by plate counts (Dreher-Lesnick, Schreier and Stibitz 2015). Moreover, prophage endolysins are known to contribute to starter autolysis, a process involved in cheese ripening (Lortal and Chapot-Chartier 2005; Visweswaran et al. 2017). Likewise, the CW binding domains of LAB phage endolysins represent a rich source of highly diverse cell surface anchoring domains for protein surface display (Regulski et al. 2013; Chapot-Chartier 2014).
Host factors acting on the LAB cell wall
As any other member of the human microbiota, host-associated LAB are exposed to the innate immune effectors that comprise CW active compounds such as antimicrobial peptides and lysozyme.
Antimicrobial peptides targeting the cell wall
Production of cationic antimicrobial peptides (AMPs) or host defence peptides is highly conserved across the living kingdoms and constitutes the first-line defence barrier against infection. Moreover, compiling evidences also support the notion that species-specific defence peptides aid to determine the composition of a beneficial microbial community and its spatial organization within a given host (Mergaert 2018). As far as we know, susceptibility of LAB to host AMPs has not been systematically addressed, but it might be important when seeking for potential live therapeutics. In fact, differences in AMP susceptibility have been observed within the two subspecies of L. delbrueckii (Hugo et al. 2012).
By analogy with bacteriocins, most host defence peptides alter membrane integrity but some rely on specific CW components for antimicrobial activity, such as the human defensins hNP1 and hBD3 that bind to lipid II and inhibit CW biosynthesis (reviewed by Grein et al. 2019). In general, while interaction with PG does not seem to have a negative impact on the antimicrobial activity of AMPs, LTAs may reduce their activity by entrapping AMPs, likely through electrostatic interactions, before they reach the bacterial membrane (Malanovic and Lohner 2016).
Lysozyme
Lysozyme (EC 3.2.1.17) is a muramidase that hydrolyzes the β (1→4) glycosidic bonds between MurNAc and GlcNAc of the bacterial PG resulting in its degradation and subsequent cell lysis (Fig. 3). Also, antimicrobial peptide activity of lysozyme was demonstrated with the catalytically inactivated enzyme, with peptides resulting from its digestion and with synthetic lysozyme-derived peptides (Ibrahim, Imazato and Ono 2011).
In humans, lysozyme is also part of the innate immune response against invading microorganisms. It is found in most body fluids, such as tears, breast milk, and respiratory and saliva secretions and is present in neutrophils, monocytes, macrophages, and epithelial cells. Lysozyme also shapes the immune response to infection by releasing PG fragments that activate phagocytes and, paradoxically, also helps to resolve inflammation (Ragland and Criss 2017). Interestingly, lysozyme-mediated lysis is required to induce release of superoxide dismutase (SodA) from the potential biotherapeutic L. lactis CNCM I-1631 to reduce gut oxidative stress and ameliorate colitis (Ballal et al. 2015).
Besides commensal and/or probiotic LAB, industrial LAB are also exposed to this antimicrobial enzyme. Lysozyme is an authorized food preservative (E1105) in ripened cheese and cheese products, beer and malt beverages, wine and several alcoholic drinks under the Regulation (EC) No 1333/2008 in Europe. Lysozyme is also used to accelerate ripening and prevent late blowing by Clostridium tyrobutyricum in cheese and to control LAB growth in wine and beer (Silvetti et al. 2017). Thereby, not only commensal and/or probiotic LAB are exposed to lysozyme but industrial LAB too.
MODULATING THE LAB CELL WALL DURING GROWTH AND STRESS
The bacterial CW is a highly dynamic structure and changes occur, both during physiological growth and as response to external stimuli to maintain an optimal ratio of CW firmness and plasticity. The PG polymer, for example, must be strong and rigid enough to support the internal turgor pressure but must also be relaxed to allow the incorporation of new PG monomers for cell growth and cell division. These processes are carried out by the concerted activities of various enzymes involved in CW synthesis and hydrolysis (Chapot-Chartier and Kulakauskas 2014; Egan et al. 2017). On the other hand, monitoring the integrity of the bacterial CW is crucial for survival when exposed to CW damaging agents such as bacteriocins and host factors. For this purpose, bacteria are endowed with several signal-transducing regulatory systems, such as two component systems (TCS) and extracytoplasmic function (ECF) sigma factors, which are collectively known as the Cell Envelope Stress (CES) response, together with the gene networks that they govern (Jordan, Hutchings and Mascher 2008). Eukaryotic-like serine/threonine kinases (SktP or PknB homologues), together with their cognate phosphatases, are also involved in CW homeostasis but they are so far poorly characterized in industrial and commensal LAB. Nevertheless, a recent phosphoproteomic study in S. thermophilus has identified proteins of the divisome as main targets of PknB kinase (Henry et al. 2019).
Within Gram positive bacteria, the CES response elicited by CW active antibiotics has been deeply characterized in B. subtilis (Radeck, Fritz and Mascher 2017). Both the number and the type of signal-transducing devices as well as the functions encoded by the CES responsive genes vary extensively depending on the species but, in general, they are essential for counteracting the damage and restore or maintain the functionality of the CW. Whereas ECF-sigma factors are almost absent in LAB, TCSs are key players in the regulation of several physiological processes (Monedero, Revilla-Guarinos and Zúñiga 2017), being sensing CW damage and signal transduction to the cytoplasm among them. Specifically, the main mechanisms involved in PG remodelling during growth and the CES response have been studied in L. lactis as described below. Recent studies suggest that other regulatory mechanisms, either directly or indirectly, may also be important for CW homeostasis.
Equilibrium between PG rigidity and plasticity: a link between nucleotide pools and the cell wall in L. lactis
A mechanism that relies on the utilization of L-Asp for both PG and pyrimidine synthesis to maintain the equilibrium between hydrolysis and synthesis of PG during growth of the culture has been described for L. lactis (Solopova et al. 2016). Transcriptional and genetic studies demonstrated the key role of an aspartate transcarbamoylase, encoded by pyrB, in this mechanism. As depicted in Fig. 4, PyrB converts L-Asp to L-carbamoyl-L-aspartate which is further utilized for pyrimidine biosynthesis (Kilstrup et al. 2005). However, the PyrB substrate L-Asp is also engaged in PG biosynthesis (Fig. 2), wherein it is converted to D-Asp by the RacD racemase, attached to the stem peptide of PG by the AslA ligase (Veiga et al. 2006), and converted to D-Asn by AsnH (Veiga et al. 2009).
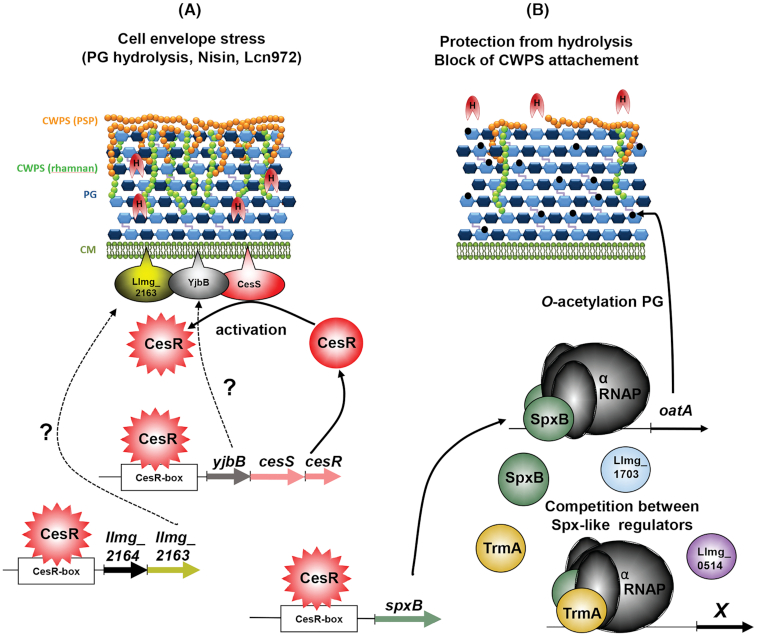
Figure 4.
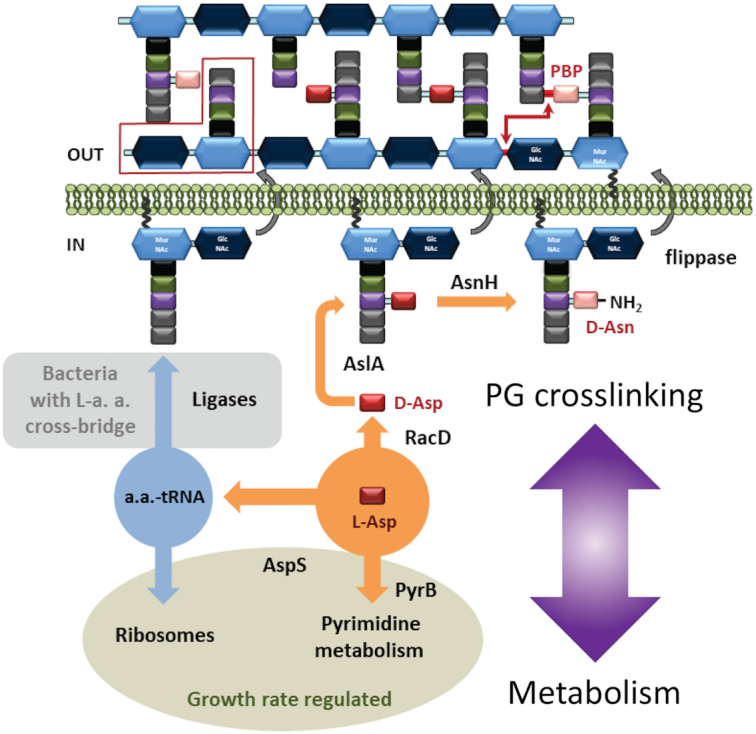
Proposed regulation of peptidoglycan (PG) plasticity in L. lactis and other LAB with D-Asp and other L- amino acids in the cross-bridge, respectively. Competition, based on the availability of precursors (double purple arrow), between PG crosslinking with pyrimidine metabolism in L. lactis (shadowed in orange) and with the protein synthesis machinery in other LAB (shadowed in blue) is shown. Both competing routes are growth rate regulated, leading to low PG crosslinking during the exponential phase and high PG crosslinking in the stationary phase. Representative D-Asp/Asp-less muropeptide is marked by a red box. Amino acids in PG stem peptides are presented as rectangles: L-Ala (black), D-Glu (green), L-Lys (purple), D-Ala (grey), D-Asp (red) and D-Asn (light red). N-acetylglucosamine is presented as dark blue hexagon, N-acetylmuramic acid as light blue hexagon. This figure has been adapted from the figure originally published in the Journal of Biological Chemistry. Solopova et al. 2016. © the American Society for Biochemistry and Molecular Biology.
It was proposed that conversion of L-Asp by PyrB reduces the amount of L-Asp available for PG synthesis (Fig. 4) and thus causes the appearance of Asp/Asn-less stem peptides in PG (Solopova et al. 2016). Presence of such stem peptides results in a decrease in PG crosslinking and, consequently, reduced PG thickness and rigidity. Expression of pyrimidine metabolism genes depends on pyrimidine availability in the medium, which is high during exponential growth and low in the stationary phase (Kilstrup et al. 2005). In this manner, a lower D-Asp/D-Asn content in PG ensures a flexible PG in exponential growth, while the lower expression of pyrB in the stationary phase results in a more rigid and thick PG. Therefore, simultaneous L-Asp utilization for both PG and pyrimidine biosynthesis determines the balance between CW flexibility and rigidity in exponential growth and the stationary phase.
This proposed regulatory mechanism allows L. lactis cells to avoid transcriptional control of the essential PG synthesis genes to ensure optimal CW plasticity during growth. This feature is in-line with the fact that, in general, essential genes are not regulated at the transcriptional level (Kobayashi et al. 2003). Secondly, autolysins, ensuring CW synthesis and potentially involved in CW rigidity regulation, are extracellularly located enzymes and their transcriptional regulation would be ineffective (Vollmer 2012). According to this reasoning, regulation of PG sensitivity to hydrolysis, i.e. autolysin activity, seems to occur through several mechanisms, including PG crosslinking in view of the proposed mechanism described above (Fig. 4).
Availability of L-Asp as a mechanism for regulating PG plasticity may also occur in other LAB which contain D-Asp/D-Asn in their PG, such as L. casei, L. delbrueckii, Lactobacillus brevis, Enterococcus faecium, among others. Furthermore, an analogous mechanism may apply to bacteria which contain L-amino acids in their PG cross-bridges, such as e.g. L-Ala-L-Ser or L-Ala-L-Ala. In this case, competition between the protein synthesis machinery and tRNA-dependent aminoacyl ligases for aminoacylated-tRNA would take the lead (Shepherd and Ibba 2013). Since ribosomal content is regulated in response to nutrient availability, it is higher in the exponential phase in comparison to the stationary phase (Wilson and Nierhaus 2007). This would enable to have more L-amino acids available for synthesis of the PG cross-bridges in stationary phase, and thus stronger PG. However, such mechanism is yet to be proved experimentally.
Counter-intuitively, pyrB was not down-regulated in cells treated with lysozyme (Solopova et al. 2016), indicating that this mechanism was not used by L. lactis MG1363 to respond to CW hydrolysis. However, a decrease of pyrB expression was observed after exposure to the bacteriocin Lcn972 (Martínez et al. 2007) and as a response to recombinant production of membrane proteins (Marreddy et al. 2011). This reflects the complexity of the CES response, in terms of signal perception (see below), combined with tight regulation of pyrB by other regulatory circuits related to nucleotide metabolism.
Cell envelope stress response: role of the CesSR regulon in L. lactis
The cascade of events was dissected in the L. lactis response to CW damage provoked by PG hydrolase activities, exemplified by lysozyme (Veiga et al. 2007), or by CW synthesis inhibition by the bacteriocin Lcn972 (Martínez et al. 2007), both leading to a stronger PG. The response is coordinated by the TCS CesSR, an orthologue of the well-characterized LiaRS of B. subtilis (Jordan et al. 2008). In this regulation scheme (Fig. 5), CW damage induces the CesSR genes and the membrane-anchored CesS sensor kinase activates the cognate response regulator CesR, most probably, by autophosphorylation and subsequent phosphate transfer.
Figure 5.
Overview of the lactococcal cell envelope stress response. Upon cell wall damage (A), the CesSR two component system (pink shapes) is activated. The response regulator CesR is phosphorylated by the histidine kinase CesS (black arrows) and triggers induction of the yjbBcesSR operon, llmg2164_2163 operon and spxB, among other genes. SpxB (green sphere) binds to RNAP and drives transcription of oatA. TmrA, Llmg_1703 and Llmg_0514 (yellow, blue and purple spheres, respectively) are SpxB paralogs which may fine tune SpxB binding to RNAP and transcription of other genes (X). Resistance to cell wall damage (B) is attained by SpxB-induced overproduction of O-acetyl-transferase OatA that acetylates peptidoglycan (i.e. protection from hydrolysis). Changes in the overall architecture of the cell wall, presumably produced by displacement of CWPSs from their attachment sites, may also contribute to resistance. The membrane proteins YjbB (grey) and Llmg2163 (golden yellow) could modulate the activity of CesS (dotted arrows) but their function remains to be elucidated (see text for further details). PG, peptidoglycan; CM, cytoplasmic membrane; H, PG hydrolase (lysozyme); α RNAP, RNA polymerase and α subunit; CWPS, cell wall polysaccharide; PSP, polysaccharide pellicle; black dots (•), sites of PG O-acetylation. Adapted from Veiga et al. 2007, published in the Journal of Biological Chemistry. © the American Society for Biochemistry and Molecular Biology.
The transcriptional activator CesR recognizes the CesR-box sequence in the promoter region of its own operon and that of a set of other genes. Based on the transcriptional analysis of L. lactis after Lcn972 treatment and comparison of the promoter regions of the highest up-regulated genes, a conserved inverted repeat TCAGHCTnnAGDCTGA (H = A/T/C; D = A/T/G) was defined as the CesR binding motif which is located, preferentially, at positions -73/-72 and -46 relative to the putative transcriptional start site (Martínez et al. 2007). L. lactis CesR is postulated to regulate, at least, up to 21 genes which likely help L. lactis to cope with CW damage. One of these genes (spxB) has been functionally characterized but, for most of the putative CesR gene targets, their role in L. lactis survival after CW damage remains to be elucidated.
Spx-like proteins were first discovered in L. lactis (Duwat, Ehrlich and Gruss 1999; Frees, Varmanen and Ingmer 2001) and characterized in B. subtilis, where Spx is a global regulatory factor that affects transcription of multiple genes in response to disulphide stress (Zuber 2004). It was shown that Spx exerts its positive or negative regulatory roles by binding to RpoA (the RNA polymerase α subunit), resulting in genome-wide changes in gene expression as part of the cellular response to toxic oxidants. The crystal structure of oxidized Spx reveals direct interactions between Spx and RpoA residues, which mediate interactions of the RNA polymerase (RNAP) with DNA, transcription activators, and σ factor subunits (Newberry et al. 2005). Spx is highly conserved among low G + C-content Gram-positive bacteria, including Enterococcus, Lactococcus, Streptococcus, and Lactobacillus. Another characteristic feature is that bacteria often possess multiple orthologues of Spx. For example, pathogens such as S. pneumoniae and Streptococcus agalactiae encode 4 paralogs, and Bacillus anthracis 3 paralogs. Among low G + C Gram-positive bacteria, the ‘champion’ is L. lactis, which contains 7 Spx paralogs, among them SpxB (Veiga et al. 2007) and TrmA (Duwat et al. 1999; Frees et al. 2001). The L. lactis TrmA was suggested to be regulated by the cell redox state (Frees et al. 2001), also indicating its possible involvement in response to oxidative stress.
As an outcome of the lactococcal CES response, SpxB binds to RpoA (RNAP, Fig. 5) and activates expression of oatA that encodes the PG O-acetyltransferase OatA, increasing PG resistance to hydrolysis. Furthermore, O-acetylation of PG could have a major impact into the overall CW architecture and provide further protection against the activity of CW antimicrobials. The rhamnan chains of CWPS are most probably covalently attached to PG at the same site that is acetylated by OatA (Sadovskaya et al. 2017) and, thus, PG O-acetylation could prevent CWPS attachment (Fig. 5). Bearing in mind that CWPS account for up to 50% of the L. lactis CW dry weight (Chapot-Chartier et al. 2010), major alteration on cell surface properties (surface net charge, permeability, etc.) are expected that may provide resistance to CW antimicrobials, e.g. by limiting access to their targets (Fig. 5).
The CesSR regulatory scheme may be fine-tuned by competition of SpxB with its paralog TrmA, and possibly other Spx-like paralogs of L. lactis (Veiga et al. 2007). Moreover, the RNA polymerase complex contains two RpoA subunits and may bind two different Spx-like molecules, each combination potentially directing the complex to different promoters (Fig. 5). The ability of both SpxB and TrmA to bind RpoA may be a sophisticated example of regulation, influencing multiple phenotypes, and among them PG resistance to hydrolysis or switch to dormant state. Such Spx-mediated response to CW damage may have evolved to assure coordinated responses with other regulatory networks and counteract multiple environmental stresses (Veiga et al. 2007). Moreover, it is possible that Spx-mediated interactions could trigger bistability, by which a genetically unique bacterial population can segregate into phenotypically distinct subpopulations (Dubnau and Losick 2006; Smits, Kuipers and Veening 2006).
Besides spx-mediated regulation, there might be other regulatory circuits in the CES response in L. lactis. For example, among the stress-responsive genes regulated by CesSR system is ftsH, encoding the conserved membrane protease who acts either as a chaperon or by degrading misassembled and damaged proteins, preferentially those located in the bacterial membrane (Dalbey, Wang and van Dijl 2012). In general, FtsH is required for a proper response to several environmental stresses (saline, temperature, acid) as evidenced in L. lactis, Oenococcus oeni, L. plantarum and L. rhamnosus (Nilsson, et al. 1994; Bourdineaud et al. 2003; Bove et al. 2012; Biswas, Keightley and Biswas 2019). Specifically, ftsH in L. plantarum is negatively regulated by CtsR, the transcriptional regulator of several molecular chaperone genes (Fiocco et al. 2009). FtsH also helps to cope with the stress imposed by recombinant membrane protein production in L. lactis (Pinto et al. 2011). Beyond FtsH contribution to the general stress response, it is unknown which specific role it may play in counteracting CW damage, but there are indirect observations pointing towards a putative function of FtsH in modulating the CW architecture of L. lactis. As already mentioned, the cell wall binding domain of a phage endolysin binds less tightly to L. lactis ΔftsH cells (Roces et al. 2016), and these ftsH cells are also resistant to lysozyme (B. Martínez, S. Kulakauskas, unpublished).
The precise stimulus that triggers CesSR and how it is perceived remains unclear. CesSR is induced by CW antimicrobials with distinct mode of action: lysozyme that hydrolyse PG and likely perturbs the cytoplasmic membrane through its antimicrobial peptide activity, and Lcn972 that binds to lipid II and inhibit CW biosynthesis (see above). Moreover, CesSR-regulon genes are induced by overexpression of membrane proteins (Marreddy et al. 2011; Pinto et al. 2011), in response to phage c2 infection in L. lactis IL1403, but not in L. lactis subsp. cremoris UC509.9 (Fallico et al. 2011; Ainsworth et al. 2013), and during bacterial emulsification (Tarazanova et al. 2019). While these puzzling observations may not help a priori to define the trigger, they underscore the relevance of CesSR in CW homeostasis of L. lactis.
Regarding stimulus perception and signal transduction, the induction mechanism of CesSR may be similar to that described for LiaRS and other TCSs, which are characterized by the presence of the so-called intramembrane-sensing kinases that require accessory membrane proteins to be activated (Mascher 2014). In B. subtilis, deletion of liaF leads to constitutive expression of the lia genes, suggesting a role as an inhibitor of the LiaS histidine kinase in the absence of stress (Jordan et al. 2006). In L. lactis, the first gene of the ces operon yjbB is homologous to liaF and could have the same function, since a lactococcal yjbB mutant is resistant to lysozyme, a phenotype that could be regarded as a sign of an activated CesSR (S. Kulakauskas, unpublished). Also, special attention should be paid to the lactococcal yth-operon (or llmg_2164–llmg_2163). Notably, this operon is among the highest up-regulated after treatment with Lcn972 and protects cells from its antimicrobial activity (Martínez et al. 2007; Roces et al. 2009). These genes are also necessary for resistance to acid stress in lactococci (Wu et al. 2018). However, mutations in this operon confer resistance to lysozyme and result in constitutive expression of the ces-operon and spxB in L. lactis MG1363 (Kulakauskas et al. 2017). Therefore, besides its role as countermeasures against damage of the cell envelope, the membrane located YthA (llmg_2163) might also modulate activation of CesSR through protein-protein interactions (Fig. 5) to prevent over-induction during the CES response. A similar function has been suggested for the B. subtilis LiaIH proteins, the sole effectors of the LiaRS response (Wolf et al. 2010) and functional counterparts of the Phage-Shock-Protein (Psp) response of E. coli (Manganelli and Gennaro 2017).
c-di-AMP and sRNAs, new players in cell wall homeostasis?
The so-called second messenger nucleotides, as cyclic adenosine monophosphate (c-AMP), cyclic guanosine monophosphate (c-GMP), cyclic di-guanosine monophosphate (c-di-GMP), and cyclic di-adenosine monophosphate (c-di-AMP) take part in control of cell growth, survival, and virulence. In particular, c-di-AMP participates in regulation of cell wall maintenance and potassium ion homeostasis in Gram-positive bacteria, (Commichau et al. 2018). c-di-AMP is synthesized by diadenylate cyclases (gene cdaA, or also called dacA) and degraded by phosphodiesterases (encoded by gdpP), these two enzymes thus participating in maintenance of proper balance of this second messenger. A model was proposed in which c-di-AMP decreases resistance to osmotic shock through inhibition of osmoprotectant transport (Pham et al. 2018). Remarkably, mutations in the phosphoglucosamine mutase gene glmM, encoded in the same operon as cdaA, were found to restore osmotolerance in L. lactis gdpP mutants. The mutations in glmM lowered the c-di-AMP level, and also resulted in reduction of the UDP-GlcNAc, which is a PG precursor (Zhu et al. 2016) (see Fig. 2). This result indicates c-di-AMP involvement in CW homeostasis not only through osmoregulation, but also through a direct effect on PG synthesis. Of note, UDP-GlcNAc is also a precursor of CWPS synthesis, suggesting that c-di-AMP levels could also impact CWPS synthesis.
sRNAs are widespread and ubiquitous key players in regulating gene expression in bacteria and are, very often, involved in stress responses. The identification and functional studies of sRNAs have just begun in LAB (Kok et al. 2017) and, so far, their participation in the CES response has yet to be demonstrated. These recent studies in L. lactis have shown that several sRNAs respond to different stresses, including osmotic shock (van der Meulen, de Jong and Kok 2017). Some of them might regulate the expression of CW genes, as judged by their location in intergenic regions or in the 3’ UTR of genes as pbp2A, involved in PG biosynthesis, and guaC, that takes part in nucleotide metabolism and thus may influence PG rigidity, as was shown for lysozyme resistant mutants guaA and pyrB (Solopova et al. 2016). Similarly, another sRNA is found upstream of the spxB operon and detected under pH and salt stress (van der Meulen et al. 2017). Although very preliminary and speculative, these examples support the notion that sRNAs might play a role in the CES response as described for enterobacteria and other microbes (Hobbs, Astarita and Storz 2010; Borgmann et al. 2018).
DEFEATING CELL WALL ACTIVE ANTIMICROBIALS: RESISTANCE MECHANISMS
LAB are endowed with an arsenal of defences to counteract the activity of CW antimicrobials. Resistance mechanisms have been discovered through the characterization of resistant mutants from several perspectives: identification of acquired mutations (genome sequence), altered gene expression (transcriptomics) and phenotypic and biochemical studies. Resistance can be intrinsic, owing to the inherent diversity of targets/receptors (e.g. CWPS and phage infection), or adaptive (e.g. stress-regulated CW component modifications and efflux pumps) which are the result of a transient or long-term exposure to the CW stressor. In this case, resistance mechanisms can be switched off, once the environmental conditions change, unless gain-of-function mutations are selected.
In this section, examples will be described pertaining to CW active antimicrobials and implying changes in the CW of LAB. Other defence mechanisms against pore-forming bacteriocins, host antimicrobial peptides and bacteriophages have been extensively reviewed. For pore-forming bacteriocins and host AMPs, changes in the composition of membrane phospholipids, membrane fluidity and downregulation, modification or loss of protein receptors are frequently involved in resistance (Kjos, Nes and Diep 2011; Bastos Mdo, Coelho and Santos 2015; Draper et al. 2015). For phages, several mechanisms have been described that interfere with the different steps of phage infection beyond host recognition (Samson and Moineau 2013; Dy et al. 2014). Interestingly, alongside ‘classical’ phage resistance mechanisms (e.g. restriction-modification, abortive infection, CRISPR/Cas), novel mechanisms are emerging by the study of non-model phage-host pairs and pangenome mining approaches (Ofir and Sorek 2018).
Modifications of the peptidoglycan structure
Polymeric PG is the most prominent constituent of the LAB CW and modifications in its chemical structure are essential for the cells to cope with CW antimicrobials and many other abiotic and biotic stresses. Changes in PG chemical structure can be achieved by altering the lipid-bound PG precursors or by post-synthetic modification of polymerized PG. Moreover, changes may occur in both the stem peptide and/or the sugar moieties, which can be transiently present (Chapot-Chartier and Kulakauskas 2014; Yadav, Espaillat and Cava 2018).
O-acetylation of MurNAc and GlcNAc
Addition of an acetyl moiety to the OH group of the C6 of MurNAc is a widespread PG modification that provokes lysozyme resistance. The presence of the bulky –CH3 group imposes a steric hindrance for binding of lysozyme and destabilizes the PG-lysozyme binding complex (Pushkaran et al. 2015). O-acetylation of MurNAc is catalyzed by OatA, a conserved integral membrane protein in Gram positive bacteria, which is regulated in L. lactis by the TCS CesSR in response to CW damage (Fig. 5). Nevertheless, under physiological conditions, a certain degree of MurNAc acetylation is observed in LAB, ranging from 3.2% in L. lactis MG1363 to 37% in L. plantarum NZ7100 (Chapot-Chartier and Kulakauskas 2014). As already mentioned, O-acetylation of GlcNAc is very infrequent, only described in L. plantarum and the enzyme involved is OatB (Bernard et al. 2011).
The degree of PG acetylation influences the activity of bacterial PG hydrolases (autolysins) and phage-encoded endolysins (Bernard et al. 2011; Escobedo et al. 2019). For the latter, inhibition was noted when the endolysin of the L. lactis phage TP712, with a lysozyme-like catalytic domain, was added externally to growing cells overexpressing spxB, while a L. lactis oatA mutant (without PG acetylation) was more susceptible than the wild-type strain. It is not clear if PG O-acetylation inhibits directly cationic bacteriocins and host AMPs or indirectly, as a result of changes in the overall CW architecture by preventing the attachment of negatively charged polymers (see Fig. 5).
Peptidoglycan N-deacetylation
By N-deacetylation, GlcNAc is converted to glucosamine by removal of the acetyl group at position C2 and this reaction is catalyzed by the PG-deacetylase PgdA. PgdA activity has been characterized in L. lactis and shown to inhibit autolysis by AcmA and lysozyme activity (Meyrand et al. 2007; Veiga et al. 2007). Remarkably, in the pathogenic LAB E. faecalis, PG N-deacetylation was detected only when bacteria were exposed to lysozyme, which triggered the expression of the PG GlcNAc-deacetylase (Benachour et al. 2012). Moreover, the content of N-deacetylated muropeptides is higher in biofilm (sessile) than in planktonic cells (Chang et al. 2018), underscoring the importance of the cell physiological state for PG modification.
Modifications in the stem peptide and cross-bridges
All the D-Glu residues present in the second position of the stem peptide in L. lactis, L. plantarum, L. casei and L. rhamnosus are amidated to D-iso-Gln. Amidation seems to occur intracellularly on the lipid-bound PG precursors by the activity of the S. aureus MurT/GadD homologs which have been identified in their genomes (Chapot-Chartier and Kulakauskas 2014). It is still unknown if there are changes in the levels of D-Glu amidation in LAB under particular stress conditions but, in S. aureus, reduced levels impair growth, increase sensitivity to beta-lactams and lysozyme, and also change affinity of lipid II binders such as oritavancin, vancomycin and the fungal defensin plectasin (Münch and Sahl 2015). On the other hand, D-Glu amidation results in a less negatively charged CW that necessarily reduces the initial electrostatic interactions with cationic antimicrobials including bacteriocins, host AMPs and lysozyme.
Amidation of D-Asp in the cross-bridge takes place in L. lactis through the activity of an asparagine synthetase encoded by asnH (Veiga et al. 2009). Lack of D-Asp amidation induces a higher autolytic rate and renders the cells sensitive to lysozyme and nisin, emphasizing the impact of the PG net charge in the bactericidal activity of these CW antimicrobials. Reduced adsorption of nisin was observed in the nisin producer L. lactis F44 overexpressing asnH (Hao et al. 2017) and the same occurs when the levels of PG O-acetylation or N-deacetylation are increased (Cao et al. 2018). On the other hand, it is also possible that modifications of the stem and/or the cross-bridge influences the activity of bacteriolysins, bearing in mind that self-immunity relies on such mechanisms (Beukes and Hastings 2001; Gargis et al. 2009a). However, resistance to bacteriolysins has not been comprehensively addressed so far.
Thickening peptidoglycan and modulating penicillin binding protein activity
As thickened PG is often observed in TEM micrographs of bacteriocin resistant mutants, it has been postulated that diffusion could be hindered by a more densely packed and thicker PG (Draper et al. 2015, and references therein). Several factors may account for PG thickening. Increased expression of genes encoding specific PBPs, pbp2A and pbpX, have been detected in the transcriptome of a nisin-resistant L. lactis IL1403 mutant and in L. lactis D1, a derivative of L. lactis MG1614 resistant to Lcn972, respectively (Kramer et al. 2006; Roces et al. 2012b). For the former, cells clearly showed a thicker septum, where lipid II is most abundant (Kramer et al. 2008). In the case of Lcn972 resistance, a reduced content of pentapeptide muropeptides was found that likely results in a denser PG mesh (Roces et al. 2012a). The importance of PG remodelling for Lcn972 resistance was emphasised by the higher sensitivity of L. lactis dacA, a mutant with a high content of pentapeptide muropeptides resulting from its inability to trim away the fifth D-Ala of the stem peptide (Roces et al. 2012a). Interestingly, infection of Lcn972 resistant mutants by the 936 phage sk1 is compromised. Fewer and consistently smaller lysis plaques were detected in standard overlay assays (Roces et al. 2012a), suggesting a sort of bacteriocin-bacteriophage cross-resistance mechanism. Whether interference occurs at the beginning (host recognition) or at the end (host lysis) is not known yet.
Lipoteichoic acid D-alanylation
Firstly described in S. aureus (Peschel et al. 1999), D-Ala esterification of LTA is a rather widespread defence mechanism against positively charged antimicrobial peptides, including CW active bacteriocins (nisin, Lct3147) and host-derived AMPs. LTA D-alanylation is responsible for the effective decrease of the net negative charge of the bacterial CW, contributing to resistance by repulsion forces. Proteins responsible for the incorporation of D-Ala residues into LTA are encoded by the dltABCD operon (see above) and are often upregulated in L. lactis and L. casei mutants resistant to nisin (Kramer et al. 2006; Revilla-Guarinos et al. 2013).
Not surprisingly, the level of LTA D-alanylation influences adsorption of the L. delbrueckii phage LL-H that uses LTA as receptor. In this case, the lower the D-Ala esterification, the higher the adsorption of the phage (Räisänen et al. 2007). LTA-D-alanylation is also important for colonization of the gastrointestinal tract by L. reuteri 100–23, as judged by the inability of a dlt mutant to compete in Lactobacillus-free mice (Walter et al. 2007). This mutant was more sensitive to nisin and could, theoretically, also be more sensitive to host AMPs, accounting for its poor survival in the gut.
Production of exopolysaccharides
Certain EPSs may provide a physical barrier for CW antimicrobials by blocking or shielding their target. The presence of EPS increased tolerance of L. lactis towards nisin and lysozyme, but not to other CW antimicrobials (penicillin G and vancomycin) (Looijesteijn et al. 2001). The same authors reported that a reduced phage titer and smaller lysis plaques were observed when comparing isogenic EPS producing vs non-EPS strains. Similarly, production of a plasmid-encoded hydrophilic EPS interfered with adsorption of phages 712 and c2 (Forde and Fitzgerald 2003). In other studies, the presence and composition of EPS did not correlate with phage sensitivity in L. lactis (Deveau, Van Calsteren and Moineau 2002), whereas a capsular PS is required for full phage adsorption in S. thermophilus (Rodríguez et al. 2008).
As a counter-measure, phages may carry EPS-hydrolyzing enzymes or depolymerases. In a prospective analysis of completely sequenced dsDNAphages, predicted depolymerase domains have been identified in phages infecting L. fermentum phiPYB5 (Glyco_hydro_66) and Leuconostoc phage phiLNTR3 (Pectate_lyase_3) (Pires et al. 2016). Whether these enzymes are required to reach the host receptor, to penetrate into biofilms or to facilitate virion diffusion after cell lysis remains to be elucidated. The temperate phage EV3 infecting Lactobacillus sanfranciscensis harbours a dextranase that enables EV3 lysogens to degrade dextran, thereby offering a competitive advantage in the sourdough environment (Picozzi et al. 2015).
Production of structural or storage polysaccharides appears to be also linked to self-protection and resistance to Lcn972. Presence of pBL1, the Lcn972-encoding plasmid, impairs cellobiose metabolism in L. lactis and leads to the accumulation of CW precursors and lower levels of UDP-activated sugars which are, supposedly, rerouted towards polysaccharide synthesis (Campelo et al. 2011).
Cell wall polysaccharide structure
Besides the role of the chemical diversity of CWPS as determinant of the host range in many LAB phages, i.e. defining phage resistance, it has been suggested that changes in CWPSs might reduce the antimicrobial activity of nisin (Xuanyuan et al. 2010). This claim was based on the observation that overexpression of rmlD, encoding a dTDP-4-dehydrorhamnose reductase involved in the synthesis of rhamnan in L. lactis, subtly enhances nisin resistance (2x MIC) in L. lactis MG1363. Otherwise, there is no further evidence that the chemical nature of the L. lactis CWPS or its abundance interferes with nisin. In S. thermophilus, however, modulation of the rhamnose-glucose polysaccharide synthesis contributes partly to bacitracin resistance. Exposure to bacitracin leads to induction of rmlC and rpgI, coding for a dTDP-4-dehydrorahamnose 3,5 epimerase and a putative glycosyltransferase for glucose branching, respectively. Moreover, rmlC mutants were roughly 5 times more susceptible to bacitracin than the wild-type (Thevenard et al. 2014).
Efflux pumps
ATP-binding cassette (ABC) transporters are often behind resistance to CW active antimicrobials in LAB. ABC-transporters use energy from ATP hydrolysis to pump a highly diverse (multidrug) or a specific set of compounds across biological membranes to import nutrients, export virulence factors and extrude toxic compounds such as antimicrobial peptides (Davidson and Maloney 2007). Phylogenetically, ABC-transporters of antimicrobial peptides in Firmicutes have been classified in 5 groups which are involved in synthesis (SunT and NisT), self-immunity (LanEFG) and resistance to antimicrobial peptides (BceAB and BcrAB) (Gebhard 2012).
BceAB-like transporters have been linked to nisin resistance in L. casei BL23 (ABC09 and ABC12) and in L. lactis (YsaCB) (Kramer et al. 2006; Revilla-Guarinos et al. 2013). BceAB-like transporters (named after the well-known bacitracin efflux BceAB transporter in B. subtilis) are characterized by a large permease (BceB) of approximately 650 aa with 10 transmembrane domains (TMD) and a large extracellular domain (≈ 200 aa) between TMD VII and VIII. Furthermore, they are often encoded in the neighborhood of a TCS to which they are functionally linked (Gebhard 2012). These transporters can function as: i) sensors that activate the cognate TCS which, in turn, triggers a response, ii) bona-fide transporters that export the drug and, thus, actively take part in resistance, and iii) both as sensors and resistance factors (Revilla-Guarinos et al. 2014). Such functional diversity is exemplified in L. casei BL23. In this strain, ABC09 is involved in sensing and detoxification, whereas ABC12 is required for activation of the TCS12 that governs the expression of the dlt operon, mrpF (lysinylation of membrane phospholipids), and of an orphan BceAB-like ABC transporter (Revilla-Guarinos et al. 2013). Less is known about L. lactis YsaCB and its putatively associated TCS (TCS-G). The genes are overexpressed in a nisin resistant mutant and confer resistance when cloned in L. lactis (Kramer et al. 2006). Interestingly, up-mutations in ysaCB are often selected in L. lactis after exposure to Lcn972 regardless of the strain background, suggesting a key role in resistance to this lipid II binding bacteriocin (López-González et al. 2018). In S. thermophilus LMD-9, a Bce-like detoxification module (TCS07-STER_1307–1308) is responsible for bacitracin resistance (Thevenard et al. 2014) but it is unknown if it confers resistance to other CW antimicrobials.
Miscellaneous resistance factors
Nisin resistance protein and associated transporter
The nisin resistance protein (NSR) was first identified in non-nisin producing lactococci and later shown to be a C-terminal processing peptidase that cleaves nisin at the peptide bond between MeLan28 and Ser29, removing the last six C-terminal amino acids (Sun et al. 2009). The truncated nisin 1–28 is 100-fold less active than full nisin, has a reduced affinity for membranes and a diminished pore-forming activity (Sun et al. 2009). As described for S. agalactiae ATCC 13 813 (Khosa, AlKhatib and Smits 2013), the gene nsr is often found in an operon structure with genes encoding a BceAB-like transporter (NsrFP) and a TCS (NsrRK), mirroring the lantibiotic immunity functions of NisI and NisEFG (Khosa, Lagedroste and Smits 2016). Recently, the transporter NsrFP was shown to function as a lantibiotic exporter and to confer nisin resistance when expressed in susceptible lactococci (Reiners et al. 2017). Other examples of what has been coined as ‘immune mimicry’ had already been described in E. faecium and Bacillus licheniformis that carry lacticin 3147 functional immunity homologues (Draper et al. 2009).
Nisinase
Nisinase is a dehydropeptide reductase that specifically reduced the C-terminal dehydroalanyl-lysine of nisin, and likely of other related lantibiotics, to alanyl-lysine. This nisin-degrading activity was first detected in the late 60 s in extracts of bacilli and later in several LAB such as L. brevis, L. plantarum, S. thermophilus and L. lactis as compiled by Draper et al. (2015). However, no further work has been pursued in the recent years.
llmg_2447, a putative L. lactis anti-sigma factor
The activity of many ECF sigma factors is negatively regulated by co-transcribed genes encoding anti-sigma factors, the majority of them being membrane proteins (Sineva, Savkina and Ades 2017). Anti-sigma factors keep ECF sigma factors bound in an inactive state until a signal triggers their release. Transcriptional analysis of L. lactis D1, a Lcn972 resistant mutant, revealed that llmg_2447 encoding a putative anti-sigma factor was highly induced. Cloning and expression of this gene in a sensitive strain increases resistance to Lcn972 by 16-fold without cross-resistance to other CW antimicrobials (Roces et al. 2012b). It was speculated that the ability to neutralize Lcn972 could have evolved from a primary function of this anti-sigma factor in sensing CW damage but it is not known yet how protection is accomplished.
CONCLUDING REMARKS AND FUTURE PERSPECTIVES
Revisiting the future directions compiled six years ago by Chapot-Chartier and Kulakauskas (2014), significant progress on the LAB CW has been made and, not surprisingly, new challenges have arisen (Fig. 6). Progress in our fundamental understanding of bacterial CW physiology is exemplified by the link between PG plasticity and nucleotide pools, that dictates the availability of L-amino acids for PG synthesis in L. lactis (Solopova et al. 2016). This brings us to a novel concept where the presence of amino acids in PG cross-bridges could be seen as an evolutionary adaptation that enables bacteria to keep optimal PG plasticity during growth and, likely, under stress. The question remains: how such a regulation could apply to other LAB having PG with different peptide cross-bridges or with a direct link between two stem peptides? On the other hand, in view of the structural diversity of the CWPSs and their specific interactions with phage RBPs, host-phage interactions are now better understood and this knowledge can be translated into practical solutions for the food industry (Mahony et al. 2017a). However, further decorations in CWPSs seem to occur and the responsible enzymes are yet to be identified. There have been also new insights into specific proteins involved in CW biogenesis and cell morphogenesis. We expect eagerly that a new scenario will emerge soon when molecular assembling, protein-protein interactions and multi-enzyme complexes replace single-protein function studies. These approaches will benefit from the progress of powerful fluorescent-based techniques and topographic imaging tools such as AFM (Turner, Hobbs and Foster 2016).
Figure 6.
Summary of the recent progress in the study of the LAB cell wall and future challenges.
Moreover, PG originating from probiotic or commensal LAB was anticipated to play an active role in the gut's immune balance (Chapot-Chartier and Kulakauskas 2014). In this context, major progress has been made to unveil the underlying mechanisms behind colonization resistance by E. faecium. This mechanism is mediated by the specific activity of SagA. This PG hydrolase induces the liberation of particular muropeptides that strongly boost the host innate immunity, resulting in host protection against enteric infections (Rangan et al. 2016; Kim et al. 2019). Also recently, TA D-alanylation in L. plantarum has been demonstrated to be fundamental for the commensal bacteria-host cross-talk in a Drosophila model (Matos et al. 2017). The interaction of CW components of commensal LAB and their host has not been covered in this review but intensive research is on-going to decipher this molecular dialogue. Progress in this field will eventually help to define new avenues to treat intestinal disorders or those related to undernutrition. Noteworthy, the CW state is very important for excretion of signal-peptide-less cytosolic proteins and the release of membrane vesicles which may carry effector molecules mediating some probiotic traits of the producing bacteria (Liu et al. 2018; Ebner and Götz 2019). Prophage endolysins seems to be important for the formation of membrane vesicles in B. subtilis, generating holes in the PG through which the membrane protrudes (Toyofuku et al. 2017). Will this mechanism apply to probiotic LAB?
Despite exhaustive research on the stress physiology of LAB, we are still investigating the mechanisms by which LAB monitors CW integrity and the genetic circuits that are activated. The CES response has been relatively well described in L. lactis (CesSR) and L. casei (Bce-like modules) so far. Still, the precise stimulus that triggers the cascade events in L. lactis CesSR, the function of many of the CesR gene targets and the putative (auto)regulatory functions and cross-talk to other stress-responsive mechanisms remain to be clarified. CesFSR (LiaFSR orthologues) are present in several Lactobacillus species and Leuconostoc but not in Weisella, Pediococcus and Oenococcus (Monedero et al. 2017). Therefore, we can expect species-specific regulatory networks responding to CW damage, which may differ from those of L. lactis.
Undeniably, one of the driving forces in LAB CW research is the importance of this cellular structure for the successful performance of LAB in already established applications as food fermentations, bulk starter production, interactions with phages and with the host, etc. However, new fields of application are emerging and the role of CW is under exploration. A recent example is the promising prospects of L. lactis as a cell factory for the production of chemicals or other commodities (van Tilburg et al. 2019). Knowledge on the LAB CW components and their biosynthetic routes helped to improve the efficiency of L. lactis for the production of coloured anthocyanins, which are valuable alternatives to replace synthetic colorants in the food industry. In this case, production yields were enhanced using a CWPS-less L. lactis mutant that performed better than the wild-type, most likely due to a higher substrate availability (Solopova et al. 2019). Likewise, PG modifications, namely O-acetylation and N-deacetylation, reduced nisin adsorption to producing cells and improved nisin productivity (Cao et al. 2018). Also, deep understanding on the mode of action of bacteriocins targeting the CW, their specific interactions with receptors and the mechanisms behind resistance allow the rational design of potent bacteriocin variants with wider spectra and able to circumvent resistance mechanisms (Li, Montalban-Lopez and Kuipers 2018; Cebrián et al. 2019; Field et al. 2019).
On the other hand, evolutionary engineering (or adaptive laboratory evolution/experimental evolution experiments) is gaining momentum in the LAB field with the aim to improve starter culture performance, select new phenotypes and study stress physiology as reviewed recently (Bachmann et al. 2017; Liu et al. 2019). This strategy may take advantage of the essentiality of the CW and its function as a first defence barrier against a wide range of stressors as exemplified by López-González et al. (2018). In this work, adaptive evolution under cell envelope stress was applied on industrial L. lactis using the CW-active bacteriocin Lcn972. Based on a preliminary characterization of the evolved cultures, diverse stable phenotypes depending on the strain background could be identified, underscoring the feasibility of this approach to introduce diversity within available industrial strains.
The properties of the LAB CW also determine the potential uses of LAB for decontamination purposes. Certain LAB strains have been shown to bind a wide range of chemicals (carcinogenic compounds, heavy metals, pesticides, mycotoxins, etc), mostly through physical binding to PG, although specific binding sites and affinities to PG and other CW components have not been clearly defined yet (Lili et al. 2018; Chiocchetti et al. 2019). The availability of CW mutants are instrumental for such mechanistic studies to guide future LAB-based bioremediation approaches (Alcántara et al. 2017).
The study of structure and function of the LAB CW has been lagging behind that of pathogenic bacteria but many recent achievements indicate that this scenario is changing. As demonstrated in numerous occasions, LAB offer unique opportunities to address both the fundamental understanding of CW biology and the translation of this knowledge into new applications.
ACKNOWLEDGEMENTS
We sincerely thank JE Suárez (University of Oviedo, Spain) and H.G. Sahl (University of Bonn, Germany) for critical reading of this manuscript and valuable suggestions. Finally, the authors are indebted to all past and present members of our research groups and all the scientists who have contributed to the field of the LAB CW. We acknowledge support of the publication fee by the CSIC Open Access Publication Support Initiative through its Unit of Information Resources for Research (URICI).
Contributor Information
Beatriz Martínez, DairySafe research group. Department of Technology and Biotechnology of Dairy Products. Instituto de Productos Lácteos de Asturias, IPLA-CSIC. Paseo Río Linares s/n. 33300 Villaviciosa, Spain.
Ana Rodríguez, DairySafe research group. Department of Technology and Biotechnology of Dairy Products. Instituto de Productos Lácteos de Asturias, IPLA-CSIC. Paseo Río Linares s/n. 33300 Villaviciosa, Spain.
Saulius Kulakauskas, Université Paris-Saclay, INRAE, AgroParisTech, Micalis Institute, 78350, Jouy-en-Josas, France.
Marie-Pierre Chapot-Chartier, Université Paris-Saclay, INRAE, AgroParisTech, Micalis Institute, 78350, Jouy-en-Josas, France.
FUNDING
The work of BM and AR was supported by Spanish State Research Agency and European Regional Development Funds [BIO2017–88147-R] and Asturias Innovation 2018–2020, Principado de Asturias, Spain, and European Regional Development Funds [IDI/2018/000119]. The work of MPCC and SK was supported by INRA, Région Ile de France and French National Research Agency (ANR-11-BSV8–004-01 and ANR-18-CE5–01-02).
Conflicts of Interests
None declared.
REFERENCES
- Acedo JZ, Chiorean S, Vederas JCet al. The expanding structural variety among bacteriocins from Gram-positive bacteria. FEMS Microbiol Rev. 2018;42:805–28. [DOI] [PubMed] [Google Scholar]
- Ainsworth S, Sadovskaya I, Vinogradov Eet al. Differences in lactococcal cell wall polysaccharide structure are major determining factors in bacteriophage sensitivity. mBio. 2014;5:e00880–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainsworth S, Zomer A, Mahony Jet al. Lytic infection of Lactococcus lactis by bacteriophages Tuc2009 and c2 triggers alternative transcriptional host responses. Appl Environ Microbiol. 2013;79:4786–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcántara C, Jadan-Piedra C, Vélez Det al. Characterization of the binding capacity of mercurial species in Lactobacillus strains. J Sci Food Agric. 2017;97:5107–13. [DOI] [PubMed] [Google Scholar]
- Alvarez-Sieiro P, Montalbán-López M, Mu Det al. Bacteriocins of lactic acid bacteria: extending the family. Appl Microbiol Biotechnol. 2016;100:2939–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre G, Deghorain M, Bron Pet al. Fluorescence and atomic force microscopy imaging of wall teichoic acids in Lactobacillus plantarum. ACS Chem Biol. 2011;6:366–76. [DOI] [PubMed] [Google Scholar]
- Andre G, Kulakauskas S, Chapot-Chartier MPet al. Imaging the nanoscale organization of peptidoglycan in living Lactococcus lactis cells. Nat Commun. 2010;1:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnison PG, Bibb MJ, Bierbaum Get al. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep. 2013;30:108–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann H, Molenaar D, Branco Dos Santos Fet al. Experimental evolution and the adjustment of metabolic strategies in lactic acid bacteria. FEMS Microbiol Rev. 2017;41:S201–S19. [DOI] [PubMed] [Google Scholar]
- Ballal SA, Veiga P, Fenn Ket al. Host lysozyme-mediated lysis of Lactococcus lactis facilitates delivery of colitis-attenuating superoxide dismutase to inflamed colons. Proc Natl Acad Sci USA. 2015;112:7803–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour A, Tagg J, Abou-Zied OKet al. New insights into the mode of action of the lantibiotic salivaricin B. Sci Rep. 2016;6:31749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau Het al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–12. [DOI] [PubMed] [Google Scholar]
- Barreteau H, Kovac A, Boniface Aet al. Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol Rev. 2008;32:168–207. [DOI] [PubMed] [Google Scholar]
- Bastos Mdo C, Coelho ML, Santos OC. Resistance to bacteriocins produced by Gram-positive bacteria. Microbiology. 2015;161:683–700. [DOI] [PubMed] [Google Scholar]
- Benachour A, Ladjouzi R, Le Jeune Aet al. The lysozyme-induced peptidoglycan N-acetylglucosamine deacetylase PgdA (EF1843) is required for Enterococcus faecalis virulence. J Bacteriol. 2012;194:6066–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard E, Rolain T, Courtin Pet al. Characterization of O-acetylation of N-acetylglucosamine: a novel structural variation of bacterial peptidoglycan. J Biol Chem. 2011;286:23950–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beukes M, Bierbaum G, Sahl H-Get al. Purification and partial characterization of a murein hydrolase, Millericin B, produced by Streptococcus milleri NMSCC 061. Appl Environ Microbiol. 2000;66:23–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beukes M, Hastings JW. Self-protection against cell wall hydrolysis in Streptococcus milleri NMSCC 061 and analysis of the millericin B operon. Appl Environ Microbiol. 2001;67:3888–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S, Keightley A, Biswas I. Characterization of a stress tolerance-defective mutant of Lactobacillus rhamnosus LRB. Mol Oral Microbiol. 2019;34:153–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgmann J, Schäkermann S, Bandow JEet al. A small regulatory RNA controls cell wall biosynthesis and antibiotic resistance. mBio. 2018;9:e02100–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouhss A, Trunkfield AE, Bugg TDet al. The biosynthesis of peptidoglycan lipid-linked intermediates. FEMS Microbiol Rev. 2008;32:208–33. [DOI] [PubMed] [Google Scholar]
- Bourdineaud JP, Nehme B, Tesse Set al. The ftsH gene of the wine bacterium Oenococcus oeni is involved in protection against environmental stress. Appl Environ Microbiol. 2003;69:2512–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bove P, Capozzi V, Garofalo Cet al. Inactivation of the ftsH gene of Lactobacillus plantarum WCFS1: effects on growth, stress tolerance, cell surface properties and biofilm formation. Microbiol Res. 2012;167:187–93. [DOI] [PubMed] [Google Scholar]
- Breukink E, Wiedemann I, van Kraaij Cet al. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science. 1999;286:2361–4. [DOI] [PubMed] [Google Scholar]
- Bron PA, Tomita S, van S IIet al. Lactobacillus plantarum possesses the capability for wall teichoic acid backbone alditol switching. Microb Cell Fact. 2012;11:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S, Santa Maria JP Jr., Walker S. Wall teichoic acids of gram-positive bacteria. Annu Rev Microbiol. 2013;67:313–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brötz H, Bierbaum G, Reynolds PEet al. The lantibiotic mersacidin inhibits peptidoglycan biosynthesis at the level of transglycosylation. Eur J Biochem. 1997;246:193–9. [DOI] [PubMed] [Google Scholar]
- Brötz H, Josten M, Wiedemann Iet al. Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin and other lantibiotics. Mol Microbiol. 1998;30:317–27. [DOI] [PubMed] [Google Scholar]
- Buist G, Kok J, Leenhouts KJet al. Molecular cloning and nucleotide sequence of the gene encoding the major peptidoglycan hydrolase of Lactococcus lactis, a muramidase needed for cell separation. J Bacteriol. 1995;177:1554‐63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttiger T, Schneider T, Martínez Bet al. Influence of Ca(2+) ions on the activity of lantibiotics containing a mersacidin-like lipid II binding motif. Appl Environ Microbiol. 2009;75:4427–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campelo AB, Gaspar P, Roces Cet al. The Lcn972 bacteriocin-encoding plasmid pBL1 impairs cellobiose metabolism in Lactococcus lactis. Appl Environ Microbiol. 2011;77:7576–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Liang D, Hao Pet al. The increase of O-acetylation and N-deacetylation in cell wall promotes acid resistance and nisin production through improving cell wall integrity in Lactococcus lactis. J Ind Microbiol Biotechnol. 2018;45:813–25. [DOI] [PubMed] [Google Scholar]
- Castelain M, Duviau MP, Canette Aet al. The nanomechanical properties of Lactococcus lactis pili are conditioned by the polymerized backbone pilin. PLoS One. 2016;11:e0152053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebrián R, Macia-Valero A, Jati APet al. Design and expression of specific hybrid lantibiotics active against pathogenic Clostridium spp. Front Microbiol. 2019;10:2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JD, Wallace AG, Foster EEet al. Peptidoglycan compositional analysis of Enterococcus faecalis biofilm by stable isotope labeling by amino acids in a bacterial culture. Biochemistry. 2018;57:1274–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapot-Chartier M-P. Bacterial autolysins. In: König H, Claus H, Varna A (eds). Prokaryotic cell wall compounds. Berlin, Heidelberg, Germany: Springer Verlag; 2010, 383–406. [Google Scholar]
- Chapot-Chartier MP, Kulakauskas S. Cell wall structure and function in lactic acid bacteria. Microb Cell Fact. 2014;13:Suppl 1: S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapot-Chartier MP, Vinogradov E, Sadovskaya Iet al. Cell surface of Lactococcus lactis is covered by a protective polysaccharide pellicle. J Biol Chem. 2010;285:10464–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapot-Chartier MP Interactions of the cell-wall glycopolymers of lactic acid bacteria with their bacteriophages. Front Microbiol. 2014;5:236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikindas ML, Weeks R, Drider Det al. Functions and emerging applications of bacteriocins. Curr Opin Biotechnol. 2018;49:23–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiocchetti GM, Jadán-Piedra C, Monedero Vet al. Use of lactic acid bacteria and yeasts to reduce exposure to chemical food contaminants and toxicity. Crit Rev Food Sci Nutr. 2019;59:1534–45. [DOI] [PubMed] [Google Scholar]
- Claes IJ, Segers ME, Verhoeven TLet al. Lipoteichoic acid is an important microbe-associated molecular pattern of Lactobacillus rhamnosus GG. Microb Cell Fact. 2012;11:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FWJ, O'Connor PM, O'Sullivan Oet al. Bacteriocin Gene-Trait matching across the complete Lactobacillus Pan-genome. Sci Rep. 2017;7:3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commichau FM, Gibhardt J, Halbedel Set al. A delicate connection: c-di-AMP affects cell integrity by controlling osmolyte transport. Trends Microbiol. 2018;26:175–85. [DOI] [PubMed] [Google Scholar]
- Cornelissen A, Sadovskaya I, Vinogradov Eet al. The Baseplate of Lactobacillus delbrueckii Bacteriophage Ld17 Harbors a Glycerophosphodiesterase. J Biol Chem. 2016;291:16816–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter PD, Hill C, Ross RP. Bacteriocins: developing innate immunity for food. Nat Rev Microbiol. 2005;3:777–88. [DOI] [PubMed] [Google Scholar]
- Courtin P, Miranda G, Guillot Aet al. Peptidoglycan structure analysis of Lactococcus lactis reveals the presence of an L,D-carboxypeptidase involved in peptidoglycan maturation. J Bacteriol. 2006;188:5293–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalbey RE, Wang P, van Dijl JM. Membrane proteases in the bacterial protein secretion and quality control pathway. Microbiol Mol Biol Rev. 2012;76:311–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David B, Duchêne MC, Haustenne GLet al. PBP2b plays a key role in both peripheral growth and septum positioning in Lactococcus lactis. PLoS One. 2018;13:e0198014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson AL, Maloney PC. ABC transporters: how small machines do a big job. Trends Microbiol. 2007;15:448–55. [DOI] [PubMed] [Google Scholar]
- Delcour J, Ferain T, Deghorain Met al. The biosynthesis and functionality of the cell-wall of lactic acid bacteria. Antonie Van Leeuwenhoek. 1999;76:159–84. [PubMed] [Google Scholar]
- Deveau H, Labrie SJ, Chopin MCet al. Biodiversity and classification of lactococcal phages. Appl Environ Microbiol. 2006;72:4338–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H, Van Calsteren MR, Moineau S. Effect of exopolysaccharides on phage-host interactions in Lactococcus lactis. Appl Environ Microbiol. 2002;68:4364–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diep DB, Skaugen M, Salehian Zet al. Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proc Natl Acad Sci U S A. 2007;104:2384–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterle ME, Spinelli S, Sadovskaya Iet al. Evolved distal tail carbohydrate binding modules of Lactobacillus phage J-1: a novel type of anti-receptor widespread among lactic acid bacteria phages. Mol Microbiol. 2017;104:608–20. [DOI] [PubMed] [Google Scholar]
- Dramé I, Formosa-Dague C, Lafforgue Cet al. Analysis of homotypic interactions of Lactococcus lactis pili using single-cell force spectroscopy. ACS Appl Mater Interfaces. 2020;12:21411–23. [DOI] [PubMed] [Google Scholar]
- Draper LA, Cotter PD, Hill Cet al. Lantibiotic resistance. Microbiol Mol Biol Rev. 2015;79:171–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper LA, Grainger K, Deegan LHet al. Cross-immunity and immune mimicry as mechanisms of resistance to the lantibiotic lacticin 3147. Mol Microbiol. 2009;71:1043–54. [DOI] [PubMed] [Google Scholar]
- Dreher-Lesnick SM, Schreier JE, Stibitz S. Development of phage lysin LysA2 for use in improved purity assays for live biotherapeutic products. Viruses. 2015;7:6675–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D, Losick R. Bistability in bacteria. Mol Microbiol. 2006;61:564–72. [DOI] [PubMed] [Google Scholar]
- Duchêne MC, Rolain T, Knoops Aet al. Distinct and specific role of NlpC/P60 endopeptidases LytA and LytB in cell elongation and division of Lactobacillus plantarum. Front Microbiol. 2019;10:713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufrêne YF Atomic force microscopy in microbiology: new structural and functional insights into the microbial cell surface. mBio. 2014;5:e01363–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont K, Vogensen FK, Neve Het al. Identification of the receptor-binding protein in 936-species lactococcal bacteriophages. Appl Environ Microbiol. 2004;70:5818–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duwat P, Ehrlich SD, Gruss A. Effects of metabolic flux on stress response pathways in Lactococcus lactis. Mol Microbiol. 1999;31:845–58. [DOI] [PubMed] [Google Scholar]
- Dy RL, Richter C, Salmond GPet al. Remarkable Mechanisms in Microbes to Resist Phage Infections. Annu Rev Virol. 2014;1:307–31. [DOI] [PubMed] [Google Scholar]
- Ebner P, Götz F. Bacterial excretion of cytoplasmic proteins (ECP): occurrence, mechanism, and function. Trends Microbiol. 2019;27:176–87. [DOI] [PubMed] [Google Scholar]
- Egan AJ, Cleverley RM, Peters Ket al. Regulation of bacterial cell wall growth. FEBS J. 2017;284:851–67. [DOI] [PubMed] [Google Scholar]
- Escobedo S, Campelo AB, Wegmann Uet al. Insight into the lytic functions of the lactococcal prophage TP712. Viruses. 2019;11:881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallico V, Ross RP, Fitzgerald GFet al. Genetic response to bacteriophage infection in Lactococcus lactis reveals a four-strand approach involving induction of membrane stress proteins, D-alanylation of the cell wall, maintenance of proton motive force, and energy conservation. J Virol. 2011;85:12032–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farenc C, Spinelli S, Vinogradov Eet al. Molecular insights on the recognition of a Lactococcus lactis cell wall pellicle by the phage 1358 receptor binding protein. J Virol. 2014;88:7005–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes S, São-José C. Enzymes and mechanisms employed by tailed bacteriophages to breach the bacterial cell barriers. Viruses. 2018;10:396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field D, Blake T, Mathur Het al. Bioengineering nisin to overcome the nisin resistance protein. Mol Microbiol. 2019;111:717–31. [DOI] [PubMed] [Google Scholar]
- Fiocco D, Collins M, Muscariello Let al. The Lactobacillus plantarum ftsH gene is a novel member of the CtsR stress response regulon. J Bacteriol. 2009;191:1688–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde A, Fitzgerald GF. Molecular organization of exopolysaccharide (EPS) encoding genes on the lactococcal bacteriophage adsorption blocking plasmid, pCI658. Plasmid. 2003;49:130–42. [DOI] [PubMed] [Google Scholar]
- Francius G, Alsteens D, Dupres Vet al. Stretching polysaccharides on live cells using single molecule force spectroscopy. Nat Protoc. 2009;4:939–46. [DOI] [PubMed] [Google Scholar]
- Frees D, Varmanen P, Ingmer H. Inactivation of a gene that is highly conserved in Gram-positive bacteria stimulates degradation of non-native proteins and concomitantly increases stress tolerance in Lactococcus lactis. Mol Microbiol. 2001;41:93–103. [DOI] [PubMed] [Google Scholar]
- Garcia-Gutierrez E, Mayer MJ, Cotter PDet al. Gut microbiota as a source of novel antimicrobials. Gut Microbes. 2019;10:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargis SR, Gargis AS, Heath HEet al. Zif, the zoocin A immunity factor, is a FemABX-like immunity protein with a novel mode of action. Appl Environ Microbiol. 2009a;75:6205–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargis SR, Heath HE, Heath LSet al. Use of 4-sulfophenyl isothiocyanate labeling and mass spectrometry to determine the site of action of the streptococcolytic peptidoglycan hydrolase zoocin A. Appl Environ Microbiol. 2009b;75:72–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau JE, Moineau S. Bacteriophages of lactic acid bacteria and their impact on milk fermentations. Microb Cell Fact. 2011;10:Suppl 1:S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhard S ABC transporters of antimicrobial peptides in Firmicutes bacteria - phylogeny, function and regulation. Mol Microbiol. 2012;86:1295–317. [DOI] [PubMed] [Google Scholar]
- Geng M, Austin F, Shin Ret al. Covalent structure and bioactivity of the type AII lantibiotic Salivaricin A2. Appl Environ Microbiol. 2018;84:e02528–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George F, Daniel C, Thomas Met al. Occurrence and Dynamism of Lactic Acid Bacteria in Distinct Ecological Niches: A Multifaceted Functional Health Perspective. Front Microbiol. 2018;9:2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grangette C, Nutten S, Palumbo Eet al. Enhanced antiinflammatory capacity of a Lactobacillus plantarum mutant synthesizing modified teichoic acids. Proc Natl Acad Sci USA. 2005;102:10321–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grein F, Schneider T, Sahl HG. Docking on Lipid II-A widespread mechanism for potent bactericidal activities of antibiotic peptides. J Mol Biol. 2019;431:3520–30. [DOI] [PubMed] [Google Scholar]
- Hao P, Liang D, Cao Let al. Promoting acid resistance and nisin yield of Lactococcus lactis F44 by genetically increasing D-Asp amidation level inside cell wall. Appl Microbiol Biotechnol. 2017;101:6137–53. [DOI] [PubMed] [Google Scholar]
- Hasper HE, de Kruijff B, Breukink E. Assembly and stability of nisin-lipid II pores. Biochemistry. 2004;43:11567–75. [DOI] [PubMed] [Google Scholar]
- Hasper HE, Kramer NE, Smith JLet al. An alternative bactericidal mechanism of action for lantibiotic peptides that target lipid II. Science. 2006;313:1636–7. [DOI] [PubMed] [Google Scholar]
- Hayes S, Duhoo Y, Neve Het al. Identification of dual receptor binding protein systems in lactococcal 936 group phages. Viruses. 2018;10:668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry C, Haller L, Blein-Nicolas Met al. Identification of Hanks-Type kinase PknB-specific targets in the Streptococcus thermophilus Phosphoproteome. Front Microbiol. 2019;10:1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs EC, Astarita JL, Storz G. Small RNAs and small proteins involved in resistance to cell envelope stress and acid shock in Escherichia coli: analysis of a bar-coded mutant collection. J Bacteriol. 2010;192:59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu ST, Breukink E, Tischenko Eet al. The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics. Nat Struct Mol Biol. 2004;11:963–7. [DOI] [PubMed] [Google Scholar]
- Hugo AA, Tymczyszyn EE, Gómez-Zavaglia Aet al. Effect of human defensins on lactobacilli and liposomes. J Appl Microbiol. 2012;113:1491–7. [DOI] [PubMed] [Google Scholar]
- Ibrahim HR, Imazato K, Ono H. Human lysozyme possesses novel antimicrobial peptides within its N-terminal domain that target bacterial respiration. J Agric Food Chem. 2011;59:10336–45. [DOI] [PubMed] [Google Scholar]
- Johnson BR, Klaenhammer TR. AcmB is an S-layer-associated beta-N-acetylglucosaminidase and functional autolysin in Lactobacillus acidophilus NCFM. Appl Environ Microbiol. 2016;82:5687–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan S, Hutchings MI, Mascher T. Cell envelope stress response in Gram-positive bacteria. FEMS Microbiol Rev. 2008;32:107–46. [DOI] [PubMed] [Google Scholar]
- Jordan S, Junker A, Helmann JDet al. Regulation of LiaRS-dependent gene expression in Bacillus subtilis: identification of inhibitor proteins, regulator binding sites, and target genes of a conserved cell envelope stress-sensing two-component system. J Bacteriol. 2006;188:5153–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamar R, Réjasse A, Jehanno Iet al. DltX of Bacillus thuringiensis is essential for D-Alanylation of teichoic acids and resistance to antimicrobial response in insects. Front Microbiol. 2017;8:1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai Y, Marles-Wright J, Cleverley RMet al. A widespread family of bacterial cell wall assembly proteins. EMBO J. 2011;30:4931–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern T, Giffard M, Hediger Set al. Dynamics characterization of fully hydrated bacterial cell walls by solid-state NMR: evidence for cooperative binding of metal ions. J Am Chem Soc. 2010;132:10911–9. [DOI] [PubMed] [Google Scholar]
- Khan H, Flint SH, Yu PL. Determination of the mode of action of enterolysin A, produced by Enterococcus faecalis B9510. J Appl Microbiol. 2013;115:484–94. [DOI] [PubMed] [Google Scholar]
- Khosa S, AlKhatib Z, Smits SH. NSR from Streptococcus agalactiae confers resistance against nisin and is encoded by a conserved nsr operon. Biol Chem. 2013;394:1543–9. [DOI] [PubMed] [Google Scholar]
- Khosa S, Lagedroste M, Smits SH. Protein defense systems against the lantibiotic nisin: function of the immunity protein NisI and the resistance protein NSR. Front Microbiol. 2016;7:504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilstrup M, Hammer K, Ruhdal Jensen Pet al. Nucleotide metabolism and its control in lactic acid bacteria. FEMS Microbiol Rev. 2005;29:555–90. [DOI] [PubMed] [Google Scholar]
- Kim B, Wang YC, Hespen CWet al. Enterococcus faecium secreted antigen A generates muropeptides to enhance host immunity and limit bacterial pathogenesis. Elife. 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Chang J, Singh M. Peptidoglycan architecture of Gram-positive bacteria by solid-state NMR. Biochim Biophys Acta. 2015;1848:350–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjos M, Nes IF, Diep DB. Resistance mechanisms against bacteriocins targeting the mannose phosphotransferase system. Appl Environ Microbiol. 2011;77:3335–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjos M, Oppegård C, Diep DBet al. Sensitivity to the two-peptide bacteriocin lactococcin G is dependent on UppP, an enzyme involved in cell-wall synthesis. Mol Microbiol. 2014;92:1177–87. [DOI] [PubMed] [Google Scholar]
- Klotz C, O'Flaherty S, Goh YJet al. Investigating the effect of growth phase on the surface-layer associated proteome of Lactobacillus acidophilus using quantitative proteomics. Front Microbiol. 2017;8:2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knerr PJ, Oman TJ, Garcia De Gonzalo CVet al. Non-proteinogenic amino acids in lacticin 481 analogues result in more potent inhibition of peptidoglycan transglycosylation. ACS Chem Biol. 2012;7:1791–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Ehrlich SD, Albertini Aet al. Essential Bacillus subtilis genes. Proc Natl Acad Sci USA. 2003;100:4678–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok J, van Gijtenbeek LA, de Jong Aet al. The evolution of gene regulation research in Lactococcus lactis. FEMS Microbiol Rev. 2017;41:S220–s43. [DOI] [PubMed] [Google Scholar]
- Kommineni S, Bretl DJ, Lam Vet al. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature. 2015;526:719–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer NE, Hasper HE, van den Bogaard PTet al. Increased D-alanylation of lipoteichoic acid and a thickened septum are main determinants in the nisin resistance mechanism of Lactococcus lactis. Microbiology. 2008;154:1755–62. [DOI] [PubMed] [Google Scholar]
- Kramer NE, van Hijum SA, Knol Jet al. Transcriptome analysis reveals mechanisms by which Lactococcus lactis acquires nisin resistance. Antimicrob Agents Chemother. 2006;50:1753–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulakauskas S, Buist G, Bulbarela-Sampieri Cet al. Selection for lysozyme resistant lactococci leads to antibiotic resistance and correlates with high SpxB expression. 12th International Symposium on Lactic Acid Bacteria, Egmond aan Zee, the Netherlands, 2017. Abstract B019, 41. Royal Netherlands Society for Microbiology and the Federation of European Microbiological Societies. [Google Scholar]
- Kumar JK. Lysostaphin: an antistaphylococcal agent. Appl Microbiol Biotechnol. 2008;80:555–61. [DOI] [PubMed] [Google Scholar]
- Kuru E, Hughes HV, Brown PJet al. In Situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angew Chem Int Ed Engl. 2012;51:12519–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurushima J, Hayashi I, Sugai Met al. Bacteriocin protein BacL1 of Enterococcus faecalis is a peptidoglycan D-isoglutamyl-L-lysine endopeptidase. J Biol Chem. 2013;288:36915–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurushima J, Nakane D, Nishizaka Tet al. Bacteriocin protein BacL1 of Enterococcus faecalis targets cell division loci and specifically recognizes L-Ala2-cross-bridged peptidoglycan. J Bacteriol. 2015;197:286–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrkou I, Carstens AB, Ellegaard-Jensen Let al. Expanding the diversity of Myoviridae phages infecting Lactobacillus plantarum-A novel lineage of Lactobacillus phages comprising five new members. Viruses. 2019;11:611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layec S, Gerard J, Legue Vet al. The CHAP domain of Cse functions as an endopeptidase that acts at mature septa to promote Streptococcus thermophilus cell separation. Mol Microbiol. 2009;71:1205–17. [DOI] [PubMed] [Google Scholar]
- Lebeer S, Bron PA, Marco MLet al. Identification of probiotic effector molecules: present state and future perspectives. Curr Opin Biotechnol. 2018;49:217–23. [DOI] [PubMed] [Google Scholar]
- Lebeer S, Verhoeven TL, Francius Get al. Identification of a gene cluster for the biosynthesis of a long, galactose-rich exopolysaccharide in Lactobacillus rhamnosus GG and functional analysis of the priming glycosyltransferase. Appl Environ Microbiol. 2009;75:3554–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lili Z, Junyan W, Hongfei Zet al. Detoxification of cancerogenic compounds by lactic acid bacteria strains. Crit Rev Food Sci Nutr. 2018;58:2727–42. [DOI] [PubMed] [Google Scholar]
- Li Q, Montalban-Lopez M, Kuipers OP. Increasing the antimicrobial activity of nisin-based lantibiotics against Gram-negative pathogens. Appl Environ Microbiol. 2018;84:e00052–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Chan SHJ, Chen Jet al. Systems Biology - A guide for understanding and developing improved strains of lactic acid bacteria. Front Microbiol. 2019;10:876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Defourny KAY, Smid EJet al. Gram-positive bacterial extracellular vesicles and their impact on health and disease. Front Microbiol. 2018;9:1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looijesteijn PJ, Trapet L, de Vries Eet al. Physiological function of exopolysaccharides produced by Lactococcus lactis. Int J Food Microbiol. 2001;64:71–80. [DOI] [PubMed] [Google Scholar]
- Lortal S, Chapot-Chartier MP. Role, mechanisms and control of lactic acid bacteria lysis in cheese. Int Dairy J. 2005;15:857–71. [Google Scholar]
- López-González MJ, Escobedo S, Rodríguez Aet al. Adaptive evolution of industrial Lactococcus lactis under cell envelope stress provides phenotypic diversity. Front Microbiol. 2018;9:2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony J, Cambillau C, van Sinderen D. Host recognition by lactic acid bacterial phages. FEMS Microbiol Rev. 2017a;41:S16–s26. [DOI] [PubMed] [Google Scholar]
- Mahony J, Kot W, Murphy Jet al. Investigation of the relationship between lactococcal host cell wall polysaccharide genotype and 936 phage receptor binding protein phylogeny. Appl Environ Microbiol. 2013;79:4385–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony J, McDonnell B, Casey Eet al. Phage-Host interactions of cheese-making lactic acid bacteria. Annu Rev Food Sci Technol. 2016;7:267–85. [DOI] [PubMed] [Google Scholar]
- Mahony J, Oliveira J, Collins Bet al. Genetic and functional characterisation of the lactococcal P335 phage-host interactions. BMC Genomics. 2017b;18:146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony J, van Sinderen D. Current taxonomy of phages infecting lactic acid bacteria. Front Microbiol. 2014;5:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malanovic N, Lohner K. Gram-positive bacterial cell envelopes: the impact on the activity of antimicrobial peptides. BBA-Biomembranes. 2016;1858:936–46. [DOI] [PubMed] [Google Scholar]
- Manganelli R, Gennaro ML. Protecting from envelope stress: variations on the Phage-Shock-Protein theme. Trends Microbiol. 2017;25:205–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann E, Whitfield C. A widespread three-component mechanism for the periplasmic modification of bacterial glycoconjugates. Can J Chem. 2016;94:883–93. [Google Scholar]
- Marreddy RK, Pinto JP, Wolters JCet al. The response of Lactococcus lactis to membrane protein production. PLoS One. 2011;6:e24060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez B, Böttiger T, Schneider Tet al. Specific interaction of the unmodified bacteriocin Lactococcin 972 with the cell wall precursor lipid II. Appl Environ Microbiol. 2008;74:4666–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez B, Rodríguez A, Suárez JE. Lactococcin 972, a bacteriocin that inhibits septum formation in lactococci. Microbiology. 2000;146:949–55. [DOI] [PubMed] [Google Scholar]
- Martínez B, Zomer AL, Rodríguez Aet al. Cell envelope stress induced by the bacteriocin Lcn972 is sensed by the lactococcal two-component system CesSR. Mol Microbiol. 2007;64:473–86. [DOI] [PubMed] [Google Scholar]
- Mascher T. Bacterial (intramembrane-sensing) histidine kinases: signal transfer rather than stimulus perception. Trends Microbiol. 2014;22:559–65. [DOI] [PubMed] [Google Scholar]
- Matos RC, Schwarzer M, Gervais Het al. D-Alanylation of teichoic acids contributes to Lactobacillus plantarum-mediated Drosophila growth during chronic undernutrition. Nat Microbiol. 2017;2:1635–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell B, Hanemaaijer L, Bottacini Fet al. A cell wall-associated polysaccharide is required for bacteriophage adsorption to the Streptococcus thermophilus cell surface. Mol Microbiol. 2020, DOI 10.1111/mmi.14494. [DOI] [PubMed] [Google Scholar]
- McDonnell B, Mahony J, Neve Het al. Identification and analysis of a novel group of bacteriophages infecting the lactic acid bacterium Streptococcus thermophilus. Appl Environ Microbiol. 2016;82:5153–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeske AJ, Riley EP, Robins WPet al. SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature. 2016;537:634–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier-Bonin M, Chapot-Chartier MP. Surface Proteins of Lactococcus lactis: Bacterial Resources for muco-adhesion in the gastrointestinal tract. Front Microbiol. 2017;8:2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergaert P Role of antimicrobial peptides in controlling symbiotic bacterial populations. Nat Prod Rep. 2018;35:336–56. [DOI] [PubMed] [Google Scholar]
- Mesnage S, Dellarole M, Baxter NJet al. Molecular basis for bacterial peptidoglycan recognition by LysM domains. Nat Commun. 2014;5:4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyrand M, Boughammoura A, Courtin Pet al. Peptidoglycan N-acetylglucosamine deacetylation decreases autolysis in Lactococcus lactis. Microbiology. 2007;153:3275–85. [DOI] [PubMed] [Google Scholar]
- Meyrand M, Guillot A, Goin Met al. Surface proteome analysis of a natural isolate of Lactococcus lactis reveals the presence of pili able to bind human intestinal epithelial cells. Mol Cell Proteomics. 2013;12:3935–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michon C, Langella P, Eijsink VGet al. Display of recombinant proteins at the surface of lactic acid bacteria: strategies and applications. Microb Cell Fact. 2016;15:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millen AM, Romero DA. Genetic determinants of lactococcal C2viruses for host infection and their role in phage evolution. J Gen Virol. 2016;97:1998–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monedero V, Revilla-Guarinos A, Zúñiga M. Physiological role of two-component signal transduction systems in food-associated lactic acid bacteria. Adv Appl Microbiol. 2017;99:1–51. [DOI] [PubMed] [Google Scholar]
- Munsch-Alatossava P, Alatossava T. The extracellular phage-host interactions involved in the bacteriophage LL-H infection of Lactobacillus delbrueckii ssp. lactis ATCC 15808. Front Microbiol. 2013;4:408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy J, Bottacini F, Mahony Jet al. Comparative genomics and functional analysis of the 936 group of lactococcal Siphoviridae phages. Sci Rep. 2016;6:21345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münch D, Sahl H-G. Structural variations of the cell wall precursor lipid II in Gram-positive bacteria — Impact on binding and efficacy of antimicrobial peptides. BBA-Biomembranes. 2015;1848:3062–71. [DOI] [PubMed] [Google Scholar]
- Newberry KJ, Nakano S, Zuber Pet al. Crystal structure of the Bacillus subtilis anti-alpha, global transcriptional regulator, Spx, in complex with the alpha C-terminal domain of RNA polymerase. Proc Natl Acad Sci U S A. 2005;102:15839–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson D, Lauridsen AA, Tomoyasu Tet al. A Lactococcus lactis gene encodes a membrane protein with putative ATPase activity that is homologous to the essential Escherichia coli ftsH gene product. Microbiology. 1994;140:2601–10. [DOI] [PubMed] [Google Scholar]
- Ofir G, Sorek R. Contemporary phage biology: from classic models to new insights. Cell. 2018;172:1260–70. [DOI] [PubMed] [Google Scholar]
- Papadimitriou K, Alegria A, Bron PAet al. Stress physiology of lactic acid bacteria. Microbiol Mol Biol Rev. 2016;80:837–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percy MG, Grundling A. Lipoteichoic acid synthesis and function in gram-positive bacteria. Annu Rev Microbiol. 2014;68:81–100. [DOI] [PubMed] [Google Scholar]
- Perego M, Glaser P, Minutello Aet al. Incorporation of D-alanine into lipoteichoic acid and wall teichoic acid in Bacillus subtilis. Identification of genes and regulation. J Biol Chem. 1995;270:15598–606. [DOI] [PubMed] [Google Scholar]
- Peschel A, Otto M, Jack RWet al. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J Biol Chem. 1999;274:8405–10. [DOI] [PubMed] [Google Scholar]
- Pham HT, Nhiep NTH, Vu TNMet al. Enhanced uptake of potassium or glycine betaine or export of cyclic-di-AMP restores osmoresistance in a high cyclic-di-AMP Lactococcus lactis mutant. PLos Genet. 2018;14:e1007574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe C, Levesque S, Dion MBet al. Genomic and morphological characterization of a novel genus of phages infecting Streptococcus thermophilus. Appl Environ Microbiol. 2020, DOI 10.1128/AEM.00227-20: AEM.00227-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picozzi C, Meissner D, Chierici Met al. Phage-mediated transfer of a dextranase gene in Lactobacillus sanfranciscensis and characterization of the enzyme. Int J Food Microbiol. 2015;202:48–53. [DOI] [PubMed] [Google Scholar]
- Pinto JP, Kuipers OP, Marreddy RKet al. Efficient overproduction of membrane proteins in Lactococcus lactis requires the cell envelope stress sensor/regulator couple CesSR. PLoS One. 2011;6:e21873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires DP, Oliveira H, Melo LDet al. Bacteriophage-encoded depolymerases: their diversity and biotechnological applications. Appl Microbiol Biotechnol. 2016;100:2141–51. [DOI] [PubMed] [Google Scholar]
- Plavec TV, Strukelj B, Berlec A. Screening for new surface anchoring domains for Lactococcus lactis. Front Microbiol. 2019;10:1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puebla-Barragan S, Reid G. Forty-five-year evolution of probiotic therapy. Microbial Cell. 2019;6:184–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujato SA, Guglielmotti DM, Martinez-Garcia Met al. Leuconostoc mesenteroides and Leuconostoc pseudomesenteroides bacteriophages: Genomics and cross-species host ranges. Int J Food Microbiol. 2017;257:128–37. [DOI] [PubMed] [Google Scholar]
- Pushkaran AC, Nataraj N, Nair Net al. Understanding the structure-function relationship of lysozyme resistance in Staphylococcus aureus by peptidoglycan O-acetylation using molecular docking, dynamics, and lysis assay. J Chem Inf Model. 2015;55:760–70. [DOI] [PubMed] [Google Scholar]
- Pérez-Núñez D, Briandet R, David Bet al. A new morphogenesis pathway in bacteria: unbalanced activity of cell wall synthesis machineries leads to coccus-to-rod transition and filamentation in ovococci. Mol Microbiol. 2011;79:759–71. [DOI] [PubMed] [Google Scholar]
- Radeck J, Fritz G, Mascher T. The cell envelope stress response of Bacillus subtilis: from static signaling devices to dynamic regulatory network. Curr Genet. 2017;63:79–90. [DOI] [PubMed] [Google Scholar]
- Radkov AD, Hsu YP, Booher Get al. Imaging bacterial cell wall biosynthesis. Annu Rev Biochem. 2018;87:991–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragland SA, Criss AK. From bacterial killing to immune modulation: Recent insights into the functions of lysozyme. PLoS Pathog. 2017;13:e1006512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan KJ, Pedicord VA, Wang YCet al. A secreted bacterial peptidoglycan hydrolase enhances tolerance to enteric pathogens. Science. 2016;353:1434–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regulski K, Courtin P, Kulakauskas Set al. A novel type of peptidoglycan-binding domain highly specific for amidated D-Asp cross-bridge, identified in Lactobacillus casei bacteriophage endolysins. J Biol Chem. 2013;288:20416‐26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regulski K, Courtin P, Meyrand Met al. Analysis of the peptidoglycan hydrolase complement of Lactobacillus casei and characterization of the major gamma-D-glutamyl-L-lysyl-endopeptidase. PLoS One. 2012;7:e32301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiners J, Lagedroste M, Ehlen Ket al. The N-terminal region of nisin is important for the BceAB-type ABC transporter NsrFP from Streptococcus agalactiae COH1. Front Microbiol. 2017;8:1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus DM, van Kranenburg R, van S IIet al. Impact of 4 Lactobacillus plantarum capsular polysaccharide clusters on surface glycan composition and host cell signaling. Microb Cell Fact. 2012;11:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repka LM, Chekan JR, Nair SKet al. Mechanistic understanding of lanthipeptide biosynthetic enzymes. Chem Rev. 2017;117:5457–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revilla-Guarinos A, Gebhard S, Alcántara Cet al. Characterization of a regulatory network of peptide antibiotic detoxification modules in Lactobacillus casei BL23. Appl Environ Microbiol. 2013;79:3160–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revilla-Guarinos A, Gebhard S, Mascher Tet al. Defence against antimicrobial peptides: different strategies in Firmicutes. Environ Microbiol. 2014;16:1225–37. [DOI] [PubMed] [Google Scholar]
- Rismondo J, Percy MG, Grundling A. Discovery of genes required for lipoteichoic acid glycosylation predicts two distinct mechanisms for wall teichoic acid glycosylation. J Biol Chem. 2018;293:3293–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach DR, Khatibi PA, Bischoff KMet al. Bacteriophage-encoded lytic enzymes control growth of contaminating Lactobacillus found in fuel ethanol fermentations. Biotechnol Biofuels. 2013;6:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roces C, Campelo AB, Escobedo Set al. Reduced binding of the endolysin LysTP712 to Lactococcus lactis DftsH contributes to phage resistance. Front Microbiol. 2016;7:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roces C, Campelo AB, Veiga Pet al. Contribution of the CesR-regulated genes llmg0169 and llmg2164-2163 to Lactococcus lactis fitness. Int J Food Microbiol. 2009;133:279–85. [DOI] [PubMed] [Google Scholar]
- Roces C, Courtin P, Kulakauskas Set al. Isolation of Lactococcus lactis mutants simultaneously resistant to the cell wall-active bacteriocin Lcn972, lysozyme, nisin and bacteriophage c2. Appl Environ Microbiol. 2012a;78:4157–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roces C, Pérez V, Campelo ABet al. The putative lactococcal extracytoplasmic function anti-sigma factor llmg2447 determines resistance to the cell wall-active bacteriocin lcn972. Antimicrob Agents Chemother. 2012b;56:5520–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roces C, Wegmann U, Campelo ABet al. Lack of the host membrane protease FtsH hinders release of the Lactococcus lactis bacteriophage TP712. J Gen Virol. 2013;94:2814–8. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Rubio L, Gutíerrez D, Donovan DMet al. Phage lytic proteins: biotechnological applications beyond clinical antimicrobials. Crit Rev Biotechnol. 2016;36:542–52. [DOI] [PubMed] [Google Scholar]
- Rodríguez C, Van der Meulen R, Vaningelgem Fet al. Sensitivity of capsular-producing Streptococcus thermophilus strains to bacteriophage adsorption. Lett Appl Microbiol. 2008;46:462–8. [DOI] [PubMed] [Google Scholar]
- Rohde M The Gram-positive bacterial cell wall. Microbiol Spectr. 2019;7: 10.1128/microbiolspec.GPP3-0044-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolain T, Bernard E, Courtin Pet al. Identification of key peptidoglycan hydrolases for morphogenesis, autolysis, and peptidoglycan composition of Lactobacillus plantarum WCFS1. Microb Cell Fact. 2012;11:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Räisänen L, Draing C, Pfitzenmaier Met al. Molecular interaction between lipoteichoic acids and Lactobacillus delbrueckii phages depends on D-alanyl and alpha-glucose substitution of poly(glycerophosphate) backbones. J Bacteriol. 2007;189:4135–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadovskaya I, Vinogradov E, Courtin Pet al. Another brick in the wall: a rhamnan polysaccharide trapped inside peptidoglycan of Lactococcus lactis. mBio. 2017;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson JE, Moineau S. Bacteriophages in food fermentations: new frontiers in a continuous arms race. Annu Rev Food Sci Technol. 2013;4:347–68. [DOI] [PubMed] [Google Scholar]
- Scheffers D-J, Tol MB. LipidII: just another brick in the wall?. PLoS Pathog. 2015;11:e1005213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer KM, Spille JH, Sahl HGet al. The lantibiotic nisin induces Lipid II aggregation, causing membrane instability and vesicle budding. Biophys J. 2015;108:1114–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleifer KH, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972;36:407–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelcher M, Donovan DM, Loessner MJ. Bacteriophage endolysins as novel antimicrobials. Future Microbiol. 2012;7:1147–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider T, Sahl HG. An oldie but a goodie - cell wall biosynthesis as antibiotic target pathway. Int J Med Microbiol. 2010;300:161–9. [DOI] [PubMed] [Google Scholar]
- Sham LT, Butler EK, Lebar MDet al. Bacterial cell wall. MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science. 2014;345:220–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd J, Ibba M. Direction of aminoacylated transfer RNAs into antibiotic synthesis and peptidoglycan-mediated antibiotic resistance. FEBS Lett. 2013;587:2895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist MS, Swarts BM, Fox DMet al. Illumination of growth, division and secretion by metabolic labeling of the bacterial cell surface. FEMS Microbiol Rev. 2015;39:184–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvetti T, Morandi S, Hintersteiner Met al. Use of hen egg white lysozyme in the food industry. In: Hester PY (ed). Egg innovations and strategies for improvements. San Diego: Academic Press, 2017, 233–42. [Google Scholar]
- Sineva E, Savkina M, Ades SE. Themes and variations in gene regulation by extracytoplasmic function (ECF) sigma factors. Curr Opin Microbiol. 2017;36:128–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits WK, Kuipers OP, Veening JW. Phenotypic variation in bacteria: the role of feedback regulation. Nat Rev Microbiol. 2006;4:259–71. [DOI] [PubMed] [Google Scholar]
- Solopova A, Formosa-Dague C, Courtin Pet al. Regulation of cell wall plasticity by nucleotide metabolism in Lactococcus lactis. J Biol Chem. 2016;291:11323–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solopova A, van Tilburg AY, Foito Aet al. Engineering Lactococcus lactis for the production of unusual anthocyanins using tea as substrate. Metab Eng. 2019;54:160–9. [DOI] [PubMed] [Google Scholar]
- Steen A, Buist G, Kramer NEet al. Reduced lysis upon growth of Lactococcus lactis on galactose is a consequence of decreased binding of the autolysin AcmA. Appl Environ Microbiol. 2008;74:4671‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockdale SR, Mahony J, Courtin Pet al. The lactococcal phages Tuc2009 and TP901-1 incorporate two alternate forms of their tail fiber into their virions for infection specialization. J Biol Chem. 2013;288:5581–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Zhong J, Liang Xet al. Novel mechanism for nisin resistance via proteolytic degradation of nisin by the nisin resistance protein NSR. Antimicrob Agents Chemother. 2009;53:1964–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymczak P, Filipe SR, Covas Get al. Cell wall glycans mediate recognition of the dairy bacterium Streptococcus thermophilus by bacteriophages. Appl Environ Microbiol. 2018;84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymczak P, Rau MH, Monteiro JMet al. A comparative genomics approach for identifying host-range determinants in Streptococcus thermophilus bacteriophages. Sci Rep. 2019;9:7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarazanova M, Huppertz T, Starrenburg Met al. Transcriptional response of Lactococcus lactis during bacterial emulsification. PLoS One. 2019;14:e0220048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- t Hart P, Oppedijk SF, Breukink Eet al. New insights into nisin's antibacterial mechanism revealed by binding studies with synthetic Lipid II analogues. Biochemistry. 2016;55:232–7. [DOI] [PubMed] [Google Scholar]
- Theodorou I, Courtin P, Palussière Set al. A dual-chain assembly pathway generates the high structural diversity of cell-wall polysaccharides in Lactococcus lactis. J Biol Chem. 2019;294:17612–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodorou I, Courtin P, Sadovskaya Iet al. Three distinct glycosylation pathways are involved in the decoration of Lactococcus lactis cell wall glycopolymers. J Biol Chem. 2020;295:5519–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevenard B, Besset C, Choinard Set al. Response of S. thermophilus LMD-9 to bacitracin: involvement of a BceRS/AB-like module and of the rhamnose-glucose polysaccharide synthesis pathway. Int J Food Microbiol. 2014;177:89–97. [DOI] [PubMed] [Google Scholar]
- Tomita S, Irisawa T, Tanaka Net al. Comparison of components and synthesis genes of cell wall teichoic acid among Lactobacillus plantarum strains. Biosci Biotechnol Biochem. 2010;74:928–33. [DOI] [PubMed] [Google Scholar]
- Tomita S, Tanaka N, Okada S. A rapid NMR-based method for discrimination of strain-specific cell wall teichoic acid structures reveals a third backbone type in Lactobacillus plantarum. FEMS Microbiol Lett. 2017;364. [DOI] [PubMed] [Google Scholar]
- Toyofuku M, Carcamo-Oyarce G, Yamamoto Tet al. Prophage-triggered membrane vesicle formation through peptidoglycan damage in Bacillus subtilis. Nat Commun. 2017;8:481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracanna V, de Jong A, Medema MHet al. Mining prokaryotes for antimicrobial compounds: from diversity to function. FEMS Microbiol Rev. 2017;41:417–29. [DOI] [PubMed] [Google Scholar]
- Tripathi P, Beaussart A, Alsteens Det al. Adhesion and nanomechanics of pili from the probiotic Lactobacillus rhamnosus GG. ACS Nano. 2013;7:3685–97. [DOI] [PubMed] [Google Scholar]
- Tripathi P, Beaussart A, Andre Get al. Towards a nanoscale view of lactic acid bacteria. Micron. 2012;43:1323–30. [DOI] [PubMed] [Google Scholar]
- Turner DL, Lamosa P, Rodríguez Aet al. Structure and properties of the metastable bacteriocin Lcn972 from Lactococcus lactis. J Mol Struct. 2013;1031:207–10. [Google Scholar]
- Turner RD, Hobbs JK, Foster SJ. Atomic Force Microscopy analysis of bacterial cell wall peptidoglycan architecture. Methods Mol Biol. 2016;1440:3–9. [DOI] [PubMed] [Google Scholar]
- Vandamme P, De Bruyne K, Pot B. Phylogenetics and systematics. In: Holzapfel WH, Wood BJ (eds). Lactic Acid Bacteria. John Wiley & Sons, Ltd, Chichester, United Kingdom., 2014, 31–44. [Google Scholar]
- van der Meulen SB, de Jong A, Kok J. Early transcriptome response of Lactococcus lactis to environmental stresses reveals differentially expressed small regulatory RNAs and tRNAs. Front Microbiol. 2017;8:1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tilburg AY, Cao H, van der Meulen SBet al. Metabolic engineering and synthetic biology employing Lactococcus lactis and Bacillus subtilis cell factories. Curr Opin Biotechnol. 2019;59:1–7. [DOI] [PubMed] [Google Scholar]
- Veesler D, Cambillau C. A common evolutionary origin for tailed-bacteriophage functional modules and bacterial machineries. Microbiol Mol Biol Rev. 2011;75:423–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veesler D, Spinelli S, Mahony Jet al. Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism. Proc Natl Acad Sci U S A. 2012;109:8954–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veiga P, Bulbarela-Sampieri C, Furlan Set al. SpxB regulates O-acetylation-dependent resistance of Lactococcus lactis peptidoglycan to hydrolysis. J Biol Chem. 2007;282:19342–54. [DOI] [PubMed] [Google Scholar]
- Veiga P, Erkelenz M, Bernard Eet al. Identification of the asparagine synthase responsible for D-Asp amidation in the Lactococcus lactis peptidoglycan interpeptide crossbridge. J Bacteriol. 2009;191:3752–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veiga P, Piquet S, Maisons Aet al. Identification of an essential gene responsible for D-Asp incorporation in the Lactococcus lactis peptidoglycan crossbridge. Mol Microbiol. 2006;62:1713–24. [DOI] [PubMed] [Google Scholar]
- Ventura M, Callegari ML, Morelli L. Surface layer variations affecting phage adsorption on seven Lactobacillus helveticus strains. Ann Microbiol. 1999;49:45–53. [Google Scholar]
- Vinogradov E, Sadovskaya I, Courtin Pet al. Determination of the cell wall polysaccharide and teichoic acid structures from Lactococcus lactis IL1403. Carbohydr Res. 2018a;462:39–44. [DOI] [PubMed] [Google Scholar]
- Vinogradov E, Sadovskaya I, Grard Tet al. Structural studies of the cell wall polysaccharide from Lactococcus lactis UC509.9. Carbohydr Res. 2018b;461:25–31. [DOI] [PubMed] [Google Scholar]
- Vinogradov E, Sadovskaya I, Grard Tet al. Structural studies of the rhamnose-rich cell wall polysaccharide of Lactobacillus casei BL23. Carbohydr Res. 2016;435:156–61. [DOI] [PubMed] [Google Scholar]
- Vinogradov E, Valence F, Maes Eet al. Structural studies of the cell wall polysaccharides from three strains of Lactobacillus helveticus with different autolytic properties: DPC4571, BROI, and LH1. Carbohydr Res. 2013;379:7–12. [DOI] [PubMed] [Google Scholar]
- Visweswaran GR, Kurek D, Szeliga Met al. Expression of prophage-encoded endolysins contributes to autolysis of Lactococcus lactis. Appl Microbiol Biotechnol. 2017;101:1099–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visweswaran GR, Leenhouts K, van Roosmalen Met al. Exploiting the peptidoglycan-binding motif, LysM, for medical and industrial applications. Appl Microbiol Biotechnol. 2014;98:4331–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer W. Bacterial growth does require peptidoglycan hydrolases. Mol Microbiol. 2012;86:1031–5. [DOI] [PubMed] [Google Scholar]
- Walsh CJ, Guinane CM, Hill Cet al. In silico identification of bacteriocin gene clusters in the gastrointestinal tract, based on the Human Microbiome Project's reference genome database. BMC Microbiol. 2015;15:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Loach DM, Alqumber Met al. D-alanyl ester depletion of teichoic acids in Lactobacillus reuteri 100–23 results in impaired colonization of the mouse gastrointestinal tract. Environ Microbiol. 2007;9:1750–60. [DOI] [PubMed] [Google Scholar]
- Wiedemann I, Bottiger T, Bonelli RRet al. Lipid II-based antimicrobial activity of the lantibiotic plantaricin C. Appl Environ Microbiol. 2006a;72:2809–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann I, Bottiger T, Bonelli RRet al. The mode of action of the lantibiotic lacticin 3147–a complex mechanism involving specific interaction of two peptides and the cell wall precursor lipid II. Mol Microbiol. 2006b;61:285–96. [DOI] [PubMed] [Google Scholar]
- Wilson DN, Nierhaus KH. The weird and wonderful world of bacterial ribosome regulation. Crit Rev Biochem Mol Biol. 2007;42:187–219. [DOI] [PubMed] [Google Scholar]
- Wolf D, Kalamorz F, Wecke Tet al. In-depth profiling of the LiaR response of Bacillus subtilis. J Bacteriol. 2010;192:4680–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Liu J, Miao Set al. Contribution of YthA, a PspC family transcriptional regulator of Lactococcus lactis F44 acid tolerance and nisin yield: a transcriptomic approach. Appl Environ Microbiol. 2018;84:e02483–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing M, Simmonds RS, Timkovich R. Solution structure of the Cys74 to Ala74 mutant of the recombinant catalytic domain of Zoocin A. Proteins. 2017;85:177–81. [DOI] [PubMed] [Google Scholar]
- Xuanyuan Z, Wu Z, Li Ret al. Loss of IrpT function in Lactococcus lactis subsp. lactis N8 results in increased nisin resistance. Curr Microbiol. 2010;61:329–34. [DOI] [PubMed] [Google Scholar]
- Yadav AK, Espaillat A, Cava F. Bacterial strategies to preserve cell wall integrity against environmental threats. Front Microbiol. 2018;9:2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DC, Blair KM, Salama NR. Staying in shape: the impact of cell shape on bacterial survival in diverse environments. Microbiol Mol Biol Rev. 2016;80:187–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda E, Tateno H, Hirabayashi Jet al. Lectin microarray reveals binding profiles of Lactobacillus casei strains in a comprehensive analysis of bacterial cell wall polysaccharides. Appl Environ Microbiol. 2011;77:4539–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zago M, Orrù L, Rossetti Let al. Survey on the phage resistance mechanisms displayed by a dairy Lactobacillus helveticus strain. Food Microbiol. 2017;66:110–6. [DOI] [PubMed] [Google Scholar]
- Zeidan AA, Poulsen VK, Janzen Tet al. Polysaccharide production by lactic acid bacteria: from genes to industrial applications. FEMS Microbiol Rev. 2017;41:S168–s200. [DOI] [PubMed] [Google Scholar]
- Zheng J, Ganzle MG, Lin XBet al. Diversity and dynamics of bacteriocins from human microbiome. Environ Microbiol. 2015;17:2133–43. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Pham TH, Nhiep THet al. Cyclic-di-AMP synthesis by the diadenylate cyclase CdaA is modulated by the peptidoglycan biosynthesis enzyme GlmM in Lactococcus lactis. Mol Microbiol. 2016;99:1015–27. [DOI] [PubMed] [Google Scholar]
- Zuber P. Spx-RNA polymerase interaction and global transcriptional control during oxidative stress. J Bacteriol. 2004;186:1911–8. [DOI] [PMC free article] [PubMed] [Google Scholar]