Abstract
Fluorescent detection of transcripts using RNAscope has quickly become a standard in situ hybridization (ISH) approach in neuroscience, with over 400 publications since its introduction in 2012. RNAscope’s sensitivity and specificity allows to simultaneously detect up to three low abundance mRNAs in single cells (i.e., multiplexing) and, in contrast to other ISH techniques, it is performed in one day. BaseScope, a newer ultrasensitive platform, uses improved amplification chemistry of single oligonucleotide probe pairs (~50 bases). This technique allows discrimination of single nucleotide polymorphisms or splice variants that differ by short exons. A present limitation of BaseScope is that expression analysis is limited to a single gene (i.e. single-plexing). This unit outlines detailed protocols for both RNAscope and BaseScope in neuronal tissue. We discuss how to perform ISH experiments using either fresh-frozen or formalin-fixed paraffin-embedded sections, as well as dissociated cultured neurons. We also outline how to obtain quantitative data from hybridized tissue sections.
Keywords: RNA expression, RNAscope, BaseScope, transcriptome, alternative splicing, in situ hybridization
INTRODUCTION
Numerous in situ hybridization (ISH) approaches have been used in the field of neuroscience for the past decades, that were predominantly based on the use of complementary oligonucleotides or RNA probes directly labeled with radionucleotides for radioactive detection or coupled to enzymes for colorimetric detection (see (Young et al., 2018)). Those approaches had limitations, such as: high background, lack of sensitivity for low abundance transcripts, long turn-around times and analysis restricted to single gene products per section. A recent advancement to circumvent these limitations, has been the development of novel chemical reagents and signal amplification techniques that, due to their nature (see Figure 1), are highly specific, sensitive, easy to use and simultaneously detect expression of several genes in single cells. The technology for these products, promoted by ThermoFisher Scientific (ViewRNA) and Advance Cell Diagnostics (ACD; RNAscope), is based on the hybridization of approximately 20 “probe pair sets”. The sequential amplification via the tail region of the probe creates independent amplification branches or trees for the detection of several transcripts that are subsequently labeled with distinct fluorophores. A second related recent advancement in signal amplification, enhances sensitivity to the point that a single probe pair (~50 bases) can be used for detection. Using this novel ultrasensitive ISH approach, denoted as BaseScope (ACD), it is possible to investigate at a single-cell level in tissue, the expression of previously undetecable RNAs that differ by short nucleotide stretches or single bases, such as splice variants that vary by short exons (<50 bases), non-coding RNAs and single nucleotide polymorphisms (Baker et al., 2017; Erben et al., 2018). Because of the extreme heterogeneity of neural cells and transcriptome complexity of the brain, and the aforementioned simplicity and fast turn-around time of RNAscope/ViewRNA, these approaches are rapidly becoming the methods of choice to study the co-expression of genes, RNA isoforms and polymorphism in specific neural cell types.
Figure 1.
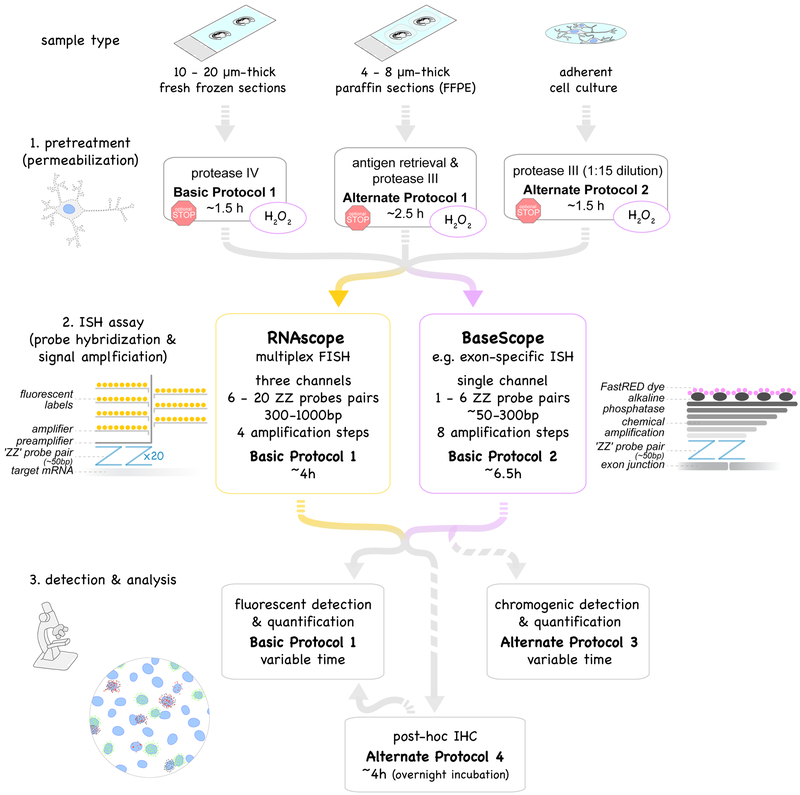
Overview of protocols described in this unit. Multiplex fluorescent in situ hybridization, RNAscope (yellow box, Basic Protocol 1) and exon-specific in situ hybridization, BaseScope (purple box, Basic Protocol 2) use probe pairs (called “ZZ” pairs) each targeting about 50 bases of target mRNA. While RNAscope uses 6 - 20 ZZ pairs (targeting about 300 - 1000 bases), improved signal amplification allows BaseScope assays to target short RNA sequences (50 - 300 bases) by using only 1 - 6 ZZ probe pairs. Signal amplification of both assays are schematically represented and involve 4 and 8 amplification steps, respectively. Fluorescent labels in RNAscope are detected using a fluorescent microscope (Basic Protocol 1). The FastRED dye used in BaseScope and converted by alkaline phosphatase is both visible under fluorescent and light microscope (Basic Protocol 2). Both RNAscope and BaseScope are compatible with different sample types: 10-20μm-thick fresh-frozen sections (Basic Protocol 1), 4–8μm-thick formalin-fixed paraffin-embedded (FFPE) sections (Alternate Protocol 1) and adherent cell cultures such as neuronal cultures (Alternate Protocol 2). Pretreatment and permeabilization to allow the probes to perfuse into the tissue to the target RNA need to be adjusted accordingly. BaseScope assays require an additional pretreatment step with H2O2 to saturate endogenous peroxidase activity (purple circles). Protocols can be interrupted at optional stopping points during pretreatments (red stop sign). Post-assay immunostaining (Basic Protocol 3) is optional.
In this unit, we describe protocols for both RNAscope (Basic Protocol 1) and BaseScope (Basic Protocol 2). These ISH assays comprise three sections: 1. pretreatment of sample depending on sample type, 2. the ISH assay itself (probe hybridization and signal amplification) and 3. the detection and analysis using high-resolution fluorescent images (20–63X). Additionally, we provide a protocol for the combination of ISH and immunohistochemistry (Alternate Protocol 4), and a detailed description of step-by-step analysis of ISH signal applicable for ISH assays beyond those described in this unit (see section Understanding Results & Statistical Analyses in Commentary). An overview of the protocols (Figure 1) and short print-out of the protocols covered in this unit (Figure 3) are provided as convenient guides to users.
Figure 3.
Short print-out protocol for RNAscope and BaseScope assays on fresh-frozen and paraffin sections as well as on adherent cell culture samples. For detailed protocols please refer to the text. Drops illustrate washing steps of varying lengths with indicated solutions, and the pen indicates the drawing of the hydrophobic barrier. The BaseScope assay requires an additional H2O2 pretreatment step (highlighted in light violet in top panels). Protocols can be paused (red stop signs).
NOTE: All protocols using animal or human tissue must first be reviewed and approved by an Institutional Animal Care and Use Committee (IACUC) and must follow officially approved procedures for the care and use of animals.
Strategic planning: Probe Design
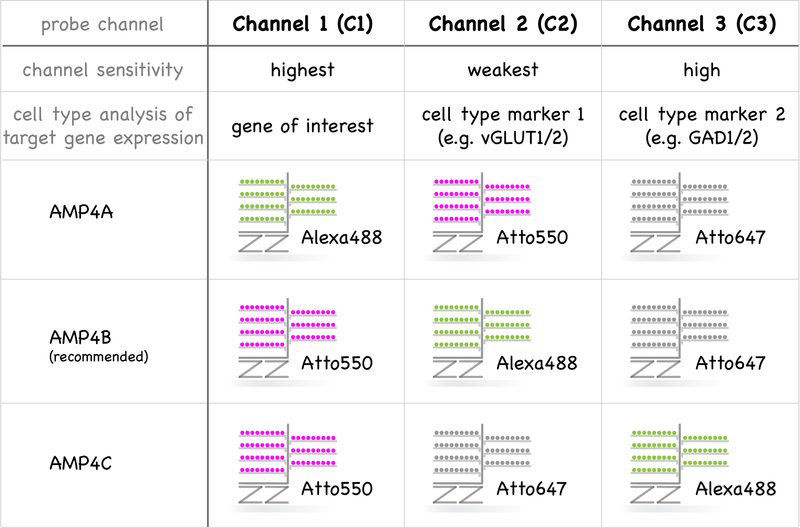
While probes are designed by the vendor, investigators need to consider a couple of important points, as discussed below, before requesting probes that are either synthesized by the vendor or purchased as an already existing probe from the catalog. First and foremost, both RNAscope and BaseScope probes need to be comprised of anti-sense sequences that perfectly match the RNA sequence of the species under investigation. No additional planning for channel consideration is required for single-plex BaseScope, but further steps are required to combine the different RNAscope probes effectively. The relative abundance of the targeted RNAs, the sensitivity of the different channels and background fluorescence need to be considered (see Figure 2). Probes designed for use in Channel 1 are most sensitive, closely followed by Channel 3. Hybridizations signals, which appear as dots, are also slightly larger for Channel 1 probes relative to probes on other channels. For these reasons, we regularly assign probes targeting the lower abundance transcripts – frequently our genes of interest – to Channel 1. Probes on Channel 2 show the lowest sensitivity, and we therefore assign probes targeting the most abundant transcripts to this channel (e.g. cell type-specific markers). Depending on which Amplification solution 4 is used, the assignment of three different fluorophores, Alexa488, Atto550 and Atto647, can be switched between the three Channels (see Figure 2). Based on our experience, we recommend using AMP4B for standard applications that results in detection of Atto550 (red fluorescence) on Channel 1, Alexa488 (green fluorescence) on Channel 2 and Atto6447 (far-red fluorescence) on Channel 3. However, it is important to remember that autofluorescence from accumulated lipofuscin granules or fixatives is most prominent in the green fluorescent range (Del Castillo et al., 1989; Dowson, 1982). The use of tissue from younger animals ameliorates autofluorescence artifacts associated with lipofuscin accumulation (Schnell et al., 1999).
Figure 2.
Multiplexing of the three channels in RNAscope. Detected fluorophores of the three channels (C1, C2, C3) can be adjusted by distinct amplification solution 4 (AMP4A, AMP4B, AMP4C). The sensitivity of the three channels is C1 > C2 >> C3. Therefore, we recommend examining the expression of a target gene (lowest expected expression) in different cell types using a channel 1 (C1) probe against this gene of interest and cell type-specific marker genes in Channel 2 & 3.
BASIC PROTOCOL 1: MULTIPLEX FLOURESCENT IN SITU HYBRIDIZATION (RNASCOPE) USING FRESH-FROZEN SECTIONS
The extremely high sensitivity and specificity of RNAscope is based on the probe design and the amplification of the signal (Wang et al., 2012). The probes for each RNA target are comprised of 6–20 oligonucleotide pairs that are denoted as “ZZ pairs” (each oligonucleotide is 18–25 bases long), and each ZZ pair is complementary to approximately 50 contiguous bases in the targeted RNA. The hybridized ZZ probe pairs are bound by the preamplifier via a 28-base tail region that will be used for signal amplification (see Figure 1). Importantly, hybridization conditions are such that stable binding of the preamplifier requires both Z probes of a pair to hybridize adjacent RNA sequences; off-target hybridization to non-specific RNA sequences does not result in signal amplification and therefore does not contribute to non-specific hybridization. This requirement, for the physical proximity of two specific probes to generate signal, differentiates RNAscope from other traditional ISH hybridization protocols that use either labeled single oligonucleotides or cRNAs. Following binding of the preamplifiers to each of the pairs (6–20 ZZ pairs per targeted RNA), signal amplification is achieved in the next steps: one preamplifier binds 20 amplifiers, and in turn, each amplifier has 20 binding sites for fluorescent labels. This sequential amplification scheme can theoretically yield an 8000-fold increase in signal per target, thus allowing detection of single transcripts. Multiplexing, ergo the simultaneous detection of several gene products, is possible because the reagents used for signal amplification in each channel are unique and are ultimately labeled with distinct fluorescent labels.
Basic Protocol 1 describes the use of the RNAscope assay on 10–20μm thick fresh-frozen sections; this is the preferred type of section because of its better preservation of RNA. Examples of RNAscope on fresh-frozen sections can be found in Figure 4 and 5.
Figure 4.
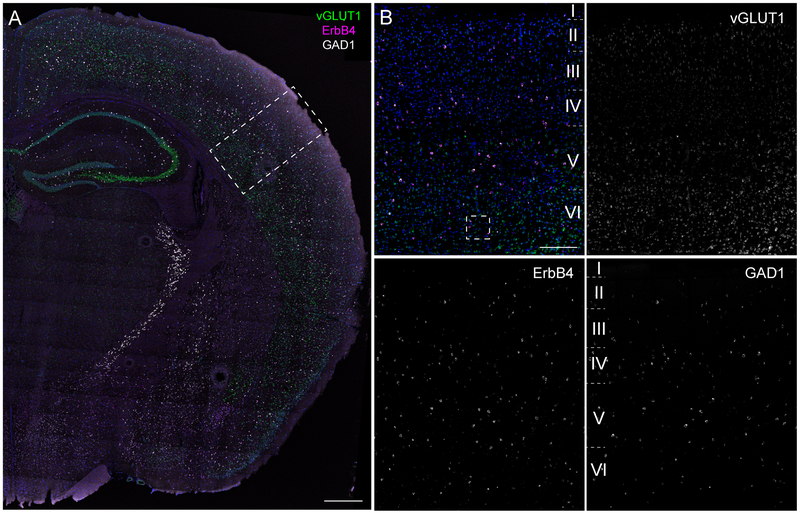
Example for RNAscope on fresh-frozen section (Basic Protocol 1). Expression of ErbB4 (magenta; Channel 1) was analyzed in excitatory neurons labeled by vesicular glutamate transporter 1 (vGLUT1; green; Channel 2) and in GABAergic neurons labeled by glutamate decarboxylase 1 (GAD1; white; Channel 3). (A) Overview of an adult coronal wildtype (WT; C57BL/6J) mouse brain section. (B) Magnification in the primary somatosensory cortex. Scale bars 500μm in A, 200μm in B. See figure 5 for analysis.
Figure 5.
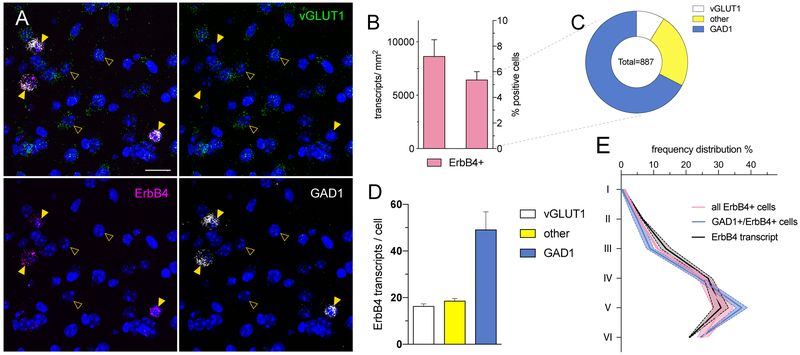
Quantitative analysis of an exemplary RNAscope experiment. (A) Inset for cellular resolution of ErbB4 (magenta; Channel 1) expression in the somatosensory cortex from Figure 4B. ErbB4 transcripts are confined to GABAergic interneurons (GAD1; white; Channel 3) in the cortex (arrowheads) and absent from glutamatergic neurons (vGLUT1; green; Channel 2; open arrowheads). (B-E) Quantification of ErbB4 expression in the somatosensory cortex using CellProfiler (see Understanding Results & Statistical Analyses; n=18429 cells; N=2 animals; bilateral). (B) ErbB4 transcript expression analyzed per area (right) and percentage of positive cells (left). (C) Fraction of ErbB4+ cells that overlap with glutamatergic (white), GABAergic (blue) signal or are none of both (yellow). (D) ErbB4 transcript levels per positive cell subdivided into the three cell populations. (E) Histogram analyses of ErbB4 expression in different cortical layers. Scale bar 20μm.
Note: All steps up to the probe hybridization should be performed under RNase-free conditions.
Materials
- RNAscope reagents (ACD):
- RNAscope® fluorescent multiplex kit (Cat No. 320851)
- Pretreatment kit (Cat No. 322380)
- 50x wash buffer (Cat No. 310091)
-
Target probes in three different channels for manual RNAscope assay (C1, C2, C3 species-specific, varying Cat No.)Note: Channel 1 probes are provided in dropper bottles (3mL) and serve as diluent for the other channels. Channel 2 and 3 probes are provided as a 50x stock in Eppendorf tubes (60μl); and are diluted 50-fold into the Channel 1 probe mix.
- Optional: Positive and negative probes (e.g. for mouse: 3-plex positive probe Cat No. 320881: Polr2a-C1, Ppib-C2, Ubc-C3; 3-plex negative probe bacterial DapB Cat No. 320871)
- Optional: Probe diluent (Cat No. 300041)
Sample: 10–20μm-thick fresh-frozen sections on Superfrost slides (for preparation see (Gerfen, 2003))
RNase AWAY (e.g. Molecular BioProducts, Fisher Scientific, Cat No. MBP#7000)
Paraformaldehyde (PFA; e.g. 16% PFA, Electron Microscopy Sciences, Cat No. 15710)
100% Ethanol (200 proof), 50% and 70% ethanol prepared with RNase-free water
RNase-free water (e.g. DEPC-treated water; KD Medical, Cat No. RGF-3050)
8 vertical glass Coplin jars (e.g. Ted Pella, Cat No. 432–1)
1x PBS (RNase-free)
Hydrophobic barrier pen (e.g. ImmEdge hydrophobic barrier pen; Vector Laboratories, Cat No. H-4000)
Absorbent paper (e.g. Whatman paper; GE Healthcare, Cat No. 10427804)
Humidifying chamber (e.g. ACD, Cat No. 310012)
Horizontal slide rack (e.g. ACD, Cat No. 310017)
Oven (40°C; e.g. HybEZ™ / HybEZ™ II Oven; ACD, Cat No. 321710/20 (110/220 V))
Water bath (40°C)
Distilled water
1.5 mL RNase-free Eppendorf tubes
Kimwipes (e.g. Kimtech Science Precision wipes; Kimberly-Clark Professional, Cat No. 05511)
Cover glass (24×50mm; e.g. Fisher Scientific, Cat No. 12–548-5M)
Aqueous mounting medium (e.g. Mowiol DABCO – see reagents and solutions)
Fluorescent microscope (e.g. Zeiss LSM710/780)
Preparations
-
1
Prepare fresh 4% paraformaldehyde (PFA) with RNase-free water and pre-chill in Coplin jar at 4°C the night before. Alternatively, 10% normal buffered formalin can be used.
-
2
The day of the assay, pre-heat and equilibrate the oven to 40°C. Assemble a humidifying chamber using absorbent paper and RNase-free water, and pre-warm it in the oven.
Pretreatment of fresh-frozen sections
-
3
Transfer slides quickly from their slide box stored at −80°C directly into pre-chilled 4% PFA. Sections should not be allowed to thaw and need to be completely submerged. Incubate for 15min at 4°C.
-
4
Wash slides in 1x PBS at room temperature (RT) by transferring slides to a Coplin Jar filled with 1x PBS. Move slides up and down five times and incubate 2min. Repeat wash once with fresh 1x PBS.
-
5
To dehydrate the sections, transfer the slides to a Coplin jar filled with 50% ethanol. Move slides up and down five times and incubate 5min at RT. Repeat one time in 70% ethanol, and twice with fresh 100% ethanol. (Note: slides can be stored for up to one week at −20°C in 100% ethanol. Prolonged storage may result in RNA degradation and suboptimal results.)
-
6
Carefully dry the bottom-side of the slide (i.e., side without section) with a Kimwipe and using a quick flick of the wrist remove excess liquid over the tissue, air-dry slides with sections facing upwards on an even surface for 5min at RT.
-
7
Draw a circle or rectangle around each section with a hydrophobic barrier pen, leaving about 5mm of space between the section and the barrier, and allow the barrier to completely dry before continuing (~1min).
-
8
Cover sections completely with protease IV pretreatment solution (provided with RNAscope pretreatment kit; e.g. 3 drops for coronal adult mouse brain section) and incubate 30min at RT. Cover sections with lid to avoid dust falling onto samples.
-
9
During incubation of the protease pretreatment, prepare probe mix: Preheat probes for 10min at 40°C in a water bath or oven. Swirl channel 1 probe and briefly spin down Channel 2 and Channel 3 probes. Wipe-down the tubes with RNase AWAY. In an RNase-free Eppendorf tube, prepare probe mix. Drip two drops of Channel 1 (~50μl) per section into the tube, and then mix-in Channel 2 and Channel 3 probes at 1:50 (i.e., 1μl per section). For weaker probes, instead use 1.5μl of Channel 2 or Channel 3 probes per section. The probe mix is prepared fresh for each experiment.
-
10
Remove the pretreatment solution by carefully blotting the side of the slide with an absorbent paper and giving the slide a quick flick of the wrist. Transfer slides into a Coplin Jar filled with 1x PBS at RT, and wash by gently moving slides up and down in the solution five times and incubating for 2min. Repeat the wash with fresh 1x PBS.
Probe Incubation & Amplification
-
11
Remove slides from Coplin jar. Blot and remove excess 1x PBS wash from the slides as described above. Transfer each slide to a horizontal slide rack in the pre-warmed humidifying chamber, pipette onto each section the ~50μl of probe mix (see step 9), being careful not to let the sections dry out.
-
12
Hybridize the sections for 2h at 40°C in the sealed humidified chamber in the oven.
-
13
During incubation of the probes prepare 1x wash buffer: Incubate 50x wash buffer for 10min at 40°C in a water bath or oven. Prepare 1L of 1x wash buffer using distilled water. Note, after the hybridization of the probes, it is no longer necessary to continue to work under RNase precaution. Unused 1x wash buffer can be stored at RT.
-
14
Remove the excess hybridization solution as described above and transfer each slide to a Coplin jar filled with 1x wash buffer at RT. As before, wash the sections by gently moving the slides up and down five times, and incubating for 2min. Repeat the wash once with fresh 1x wash buffer.
-
15
To begin the amplification process, sequentially remove from each slide the excess 1x wash buffer (as described in step 10) and transfer it to the humidified chamber. Completely cover each section with the Amplification solution 1 (AMP1; provided with RNAscope fluorescent multiplex kit; ~3 drops for an adult coronal mouse brain section). Incubate in the humidified chamber for 30min at 40°C.
-
16
Wash slides twice in 1x wash buffer at RT, as in step 14.
-
17
Remove excess wash, add Amplification solution 2 (AMP2; provided with RNAscope fluorescent multiplex kit) and incubate 15min at 40°C.
-
18
Wash slides twice in 1x wash buffer at RT, as in step 14.
-
19
Remove excess wash, add Amplification solution 3 (AMP3; provided with RNAscope fluorescent multiplex kit) and incubate 30min at 40°C.
-
20
Wash slides twice in 1x wash buffer at RT, as in step 14.
-
21
To label the amplified hybridizations with fluorophores, add Amplification solution 4 (AMP4A, AMP4B or AMP4C; provided with RNAscope fluorescent multiplex kit) and incubate 15min at 40°C.
Annotation: The different AMP4 solutions assign different fluorophores each channel (see strategic planning). We recommend using AMP4B for standard applications.
-
22
Wash slides twice in 1x wash buffer at RT, as in step 14.
-
23
One slide at the time, counterstain cell nuclei by adding ~2 drops of DAPI (provided with RNAscope fluorescent multiplex kit) and incubating for 30sec at RT.
-
24
Remove excess of DAPI as described above, add aqueous mounting medium like Mowiol DABCO (~10μl per coronal section – see reagents and solutions) and cover-slip each section carefully to avoid trapping air bubbles. Store slides horizontally in the dark overnight at 4°C to dry.
Detection & Analysis
-
25
Examine dried slides within a few days of preparation on a fluorescent (confocal) microscope (magnification 20–63X). The signals obtained from low abundance transcripts might be only visible at higher magnification (40–63X) and single dot resolution necessary for quantitative data analysis is only obtained at higher magnification (40–63X). Digital image analysis using a camera is highly recommended.
-
26
Signal quantification can be achieved by simple semi-quantitative scoring of the signal (ACD), using standard free image analysis software such as ImageJ (NIH) and CellProfiler (Carpenter et al. 2006), or commercial image analysis software such as Imaris (Bitplane) or specific ISH quantification software such as HALO (Indica Labs) and Aperio (Leica). For more details to data interpretation and signal quantification refer to Basic Protocol 4 and the commentary section Understanding Results & Statistical Analyses.
Alternate Protocol 1: Use of formalin-fixed paraffin-embedded sections
The protocol described here is for ISH analysis using 4–8um-thick formalin-fixed paraffin-embedded (FFPE) sections, which differs from Basic Protocol 1 used to analyze fresh-frozen sections. Although fresh frozen sections are generally thought to provide the highest sensitivity, from better RNA preservation, there are instances that either experimental conditions or access to cryostats may preclude the use of fresh-frozen sections. Moreover, FFPE sections allow for consistent serial sectioning and have frequently been used to archive human tissue because of their ease for long-term storage. Because the preparation of FFPE sections for RNAscope analysis requires different pretreatment steps than those described for fresh-frozen sections in Basic Protocol 1, here we provide an alternate protocol for FFPE sections. Probe hybridization and signal amplification, as well as detection are the same as in Basic Protocol 1. Fig. 6A depicts an example of RNAscope on FFPE sections of embryos.
Figure 6.
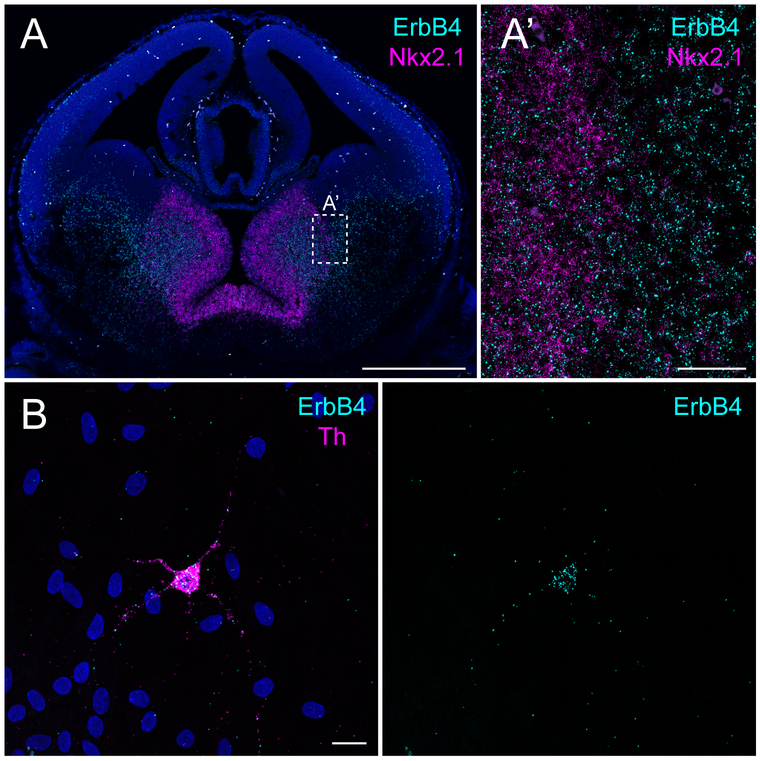
Example for RNAscope on FFPE sections (Alternate Protocol 1) and adherent cell culture (Alternate Protocol 2). (A) Expression of ErbB4 (C1, cyan) and Nkx2.1 (magenta, C3) on a coronal paraffin section of an E14.5 WT embryo. (A’) Magnification at the border between the medial and lateral ganglionic eminence. (B) Expression of ErbB4 (cyan, C1) in primary cultured dopaminergic neurons (prepared as in (Skirzewski et al., 2017)) identified by tyrosine hydroxylase (Th, C3, magenta) at DIV13. Scale bars 500μm in A and 50μm in A’, 20μm in B.
Materials
Materials list from Basic Protocol 1
Additional Materials
Sample: 4–8 μm-thick paraffin sections on Superfrost slides (for preparation see (Zeller, 2001))
Beaker
Heat plate
Optional: thermometer (100°C)
Xylene (e.g. Macron; Cat No. 8668–16)
Drying oven (60°C)
Preparations
-
1
The day of the assay, pre-heat and equilibrate the oven to 40°C. Assemble a humidifying chamber using absorbent paper and RNase-free water, and pre-warm it in the oven.
Pretreatment of formalin-fixed paraffin-embedded sections
-
2
Bake sections for 1h in an oven at 60°C. This can be done up to one week in advance and slides be stored at RT with desiccants.
-
3
During baking of the sections, prepare 1x antigen retrieval: Prepare 1x antigen retrieval from 10x stock (pretreatment II; provided with the RNAscope pretreatment kit) with RNase-free water. Unused 1x antigen retrieval can be stored at RT. On a heat plate, pre-heat Coplin jar filled with 1x antigen retrieval in a water-filled beaker brought to boil. Temperature can be controlled with thermometer and should be between 98°C and 102°C. Alternatively, antigen retrieval can also be performed in a pre-heated steamer.
-
4
In a fume hood, remove paraffin from sections in a Coplin jar filled with xylene. Move slides up and down five times and incubate for 5min at RT. Repeat once with fresh xylene.
-
5
Transfer slides to a Coplin jar filled with 100% ethanol. Move slides up and down five times and incubate 3min at RT. Repeat once with fresh 100% ethanol before drying.
-
6
After carefully drying with a Kimwipe the bottom-side of the slice (i.e., side without section), and giving the slide a quick flick of the wrist, air-dry the slides with sections facing upwards on an even surface for 5min at RT.
-
7
With forceps, slowly place slides into pre-heated 1x antigen retrieval (see step 3) and incubate for 15min.
-
8
Carefully remove slides with forceps and transfer into a Coplin jar filled with RNase-free water at RT. Wash sections by gently moving the slides up and down five times and incubate for 2min. Repeat the wash once with fresh RNase-free water.
-
9
Transfer slides into a Coplin jar filled with 100% ethanol, incubate ~15sec and air-dry for 5min as described above in step 6.
-
10
Draw a circle or rectangle around each section with a hydrophobic barrier pen, leaving about 5mm of space between the section and the barrier, and allow the barrier to completely dry before continuing (~1min). Optional: Dried slides can be stored overnight, and protocol continued the following day.
-
11
Place slides on slide rack in the pre-warmed humidifying chamber and cover completely with protease treatment III (provided with RNAscope pretreatment kit; e.g. ~3 drops per section for coronal adult mouse brain section). Incubate for 30min at 40°C in the sealed humidified chamber in the oven.
-
12
During incubation of the protease pretreatment, prepare probe mix: Preheat probes for 10min at 40°C in a water bath or oven. Swirl channel 1 probe and briefly spin down Channel 2 and Channel 3 probes. Wipe-down the tubes with RNase AWAY. In an RNase-free Eppendorf tube, prepare probe mix. Drip two drops of Channel 1 (~50μl) per section into the tube, and then mix-in Channel 2 and Channel 3 probes at 1:50 (i.e., 1μl per section). For weaker probes, instead use 1.5μl of Channel 2 or Channel 3 probes per section. The probe mix is prepared fresh for each experiment.
-
13
Remove the pretreatment solution by carefully blotting the side of the slide with absorbent paper and giving the slide a quick flick of the wrist. Transfer each slide into a Coplin jar filled with RNase-free water at RT. As before, wash the sections by gently moving the slides up and down five times, and incubating for 2min. Repeat the wash once with fresh RNase-free water.
Amplification, Detection & Analysis
-
14
Continue with steps 11–26 of Basic Protocol 1 for target probe incubation, signal amplification and detection.
Alternate Protocol 2: In situ hybridization in cultured adherent cells
Multiplex fluorescent ISH with RNAscope can also be performed on cultured adherent cells, such as established cell lines or primary neuronal cultures. Cells need to be plated on glass coverslips or alternatively chamber slides can be used. If mitotic active cultures are used, they should be 80–90% confluent at the time of fixation. As in the case of fresh-frozen and FFPE sections, cell culture samples need unique pretreatment conditions for performing RNAscope. The specific steps are described below; otherwise, the protocol follows closely Basic Protocol 1. Examples for RNAscope in primary cultured neurons are presented in Fig. 6B and Fig. 8D.
Figure 8.
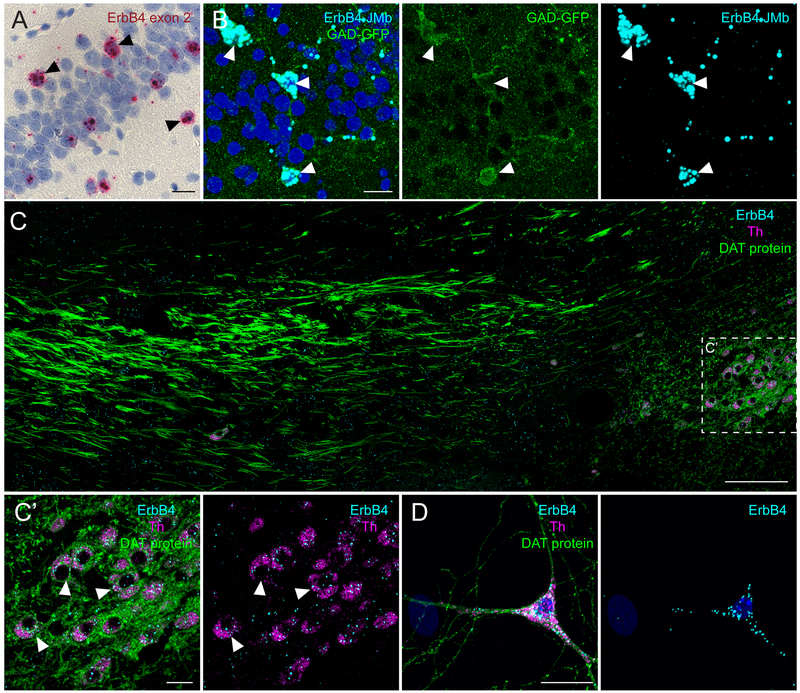
Examples for chromogenic BaseScope detection (Basic Protocol 2) and ISH-IHC combination (Basic Protocol 3). (A) Hippocampal section of an adult WT mice, labeled using BaseScope for ErbB4 with a single probe pair targeting the exon boundary exon 1/ exon 2. The section was counterstained with hematoxylin and signal detected with light microscopy. Arrowheads indicate ErbB4+ GABAergic interneurons. (B) Detection of one of the four ErbB4 splice variants (JMb; cyan) by BaseScope in GABAergic interneurons (GAD-GFP; green; arrowheads) in a section of an adult GAD-GFP mouse (kindly provided by Dr. Yuchio Yanagawa). GFP signal was amplified after the ISH assay with an anti-GFP antibody (NeuroMab; N86/8; RRID: AB_10671444). (C) ISH for ErbB4 (C1; cyan) and Th (C3; magenta) was combined with IHC with an antibody against DAT (green; Santa Cruz, sc-32258; RRID: AB_627400) in a sagittal FFPE section from an adult WT mouse (depicted substantia nigra compacta (SNc) on the left and dopaminergic medial forebrain bundle). (C’) Magnification of ErbB4-expressing dopaminergic neurons (arrowheads) in the SNc. (D) In primary mesencephalic cultures (DIV8), DAT (green) immunostaining was performed post-hoc to RNAscope for ErbB4 (C1; cyan) and Th (C3; magenta). Scale bars 100μm in B, 20μm in other panels.
NOTE: General handling procedure
The use of coverslips involves some handling challenges. With exception of the RNAscope reagents, all other steps (i.e. washes, fixation, dehydration) are performed by placing the coverslips inside a multi-well cell culture dish that is shaken on a horizontal shaker. Next, transfer the coverslips onto glass slides that were previously marked with circles of hydrophobic barrier, and completely cover with RNAscope reagents. Chamber slides can be used as an alternative to coverslips; drawing of the hydrophobic barrier is difficult. However, an advantage of using chamber slides, is that the procedure can be performed in Coplin jars as described earlier.
Materials
Materials list from Basic Protocol 1
Additional Materials
sample: adherent cell cultures (e.g. dissociated neurons) on glass coverslips
10% normal buffered formalin (NBF; e.g. Sigma-Aldrich; Cat No. HT5011)
Horizontal rocking plate
Glass slides (75×25mm; e.g. C&A Scientific, Premiere; Cat No. 8201)
Preparations
-
1
The day of the assay, pre-heat and equilibrate the oven to 40°C. Assemble a humidifying chamber using absorbent paper and RNase-free water, and pre-warm it in the oven.
-
2
Prepare glass slides with hydrophobic barrier pen circles with a diameter slightly exceeding the coverslip size used. Glass slides can be washed and dried between incubations and reused. Redraw hydrophobic barrier if necessary.
Pretreatment of adherent cell culture samples
-
3
Remove cell culture medium and rinse cultures carefully with 1x PBS.
-
4
Replace PBS with 10% NBF and fix cultures by incubating for 30min at RT while agitating on a horizontal shaker. All subsequent washes should be performed while agitating on shaker.
-
5
Remove NBF and replace with 1x PBS for 5min at RT. Repeat wash once with fresh 1x PBS.
-
6
To dehydrate cultures, remove PBS and substitute with 50% ethanol for 5min at RT, then with 70% ethanol once, and twice with fresh 100% ethanol. (Note: fixed cultures can be stored for up to six months at −20°C in 100% ethanol.)
-
7
To rehydrate culture samples, replace 100% ethanol with 70% ethanol and incubate for 2min at RT. Repeat these steps with 50% ethanol and then wash in 1x PBS at RT for 10min.
-
8
Freshly prepare a 1:15 dilution of protease III (provided with RNAscope pretreatment kit) in RNase-free 1x PBS (~100μl per 12mm coverslip)
-
9
Remove coverslips one-by-one from cell culture plate and place cultures-facing up onto the prepared glass slide that contain hydrophobic barrier circles (see preparation step 2). Samples should not dry out. Cover immediately with prepared 1:15 dilution of protease III. Incubate for 10min at RT. Cover sections with lid to avoid dust falling onto samples.
-
10
During incubation of the protease pretreatment, prepare probe mix: Preheat probes for 10min at 40°C in a water bath or oven. Swirl channel 1 probe and briefly spin down Channel 2 and Channel 3 probes. Wipe-down the tubes with RNase AWAY. In an RNase-free Eppendorf tube, prepare probe mix. Drip two drops of Channel 1 (~50μl) per coverslip into the tube, and then mix-in Channel 2 and Channel 3 probes at 1:50 (i.e., 1μl per section). For weaker probes, instead use 1.5μl of Channel 2 or Channel 3 probes per section. The probe mix is prepared fresh for each experiment.
-
11
Move coverslips back into culture well plate filled with 1x PBS and wash for 2min at RT. Repeat once with fresh 1x PBS.
Amplification, Detection & Analysis
-
12
Continue with steps 11–26 of Basic Protocol 1 for target probe incubation, signal amplification and detection. Perform washes in cell culture plate and probe incubation (~50μl per 12mm coverslip), as well as amplification steps (2–3 drops per 12mm coverslip) with coverslips placed on glass slides with barrier circles to safe solutions. Sample mounting has to be on glass slides with ~10μl aqueous mounting medium (e.g. Mowiol DABCO) per 12mm coverslip.
BASIC PROTOCOL 2: SINGLE-PAIR PROBE IN SITU HYBRIDIZATION (BASESCOPE)
Single-pair probe ISH, BaseScope, is a novel approach that allows the use of a single probe pair (“ZZ”). BaseScope relies on the same probe design as RNAscope. However, due to the improved amplification chemistry, it is possible to use only a single probe pair that hybridizes to about 50 bases of target RNA. Splice variants can be distinguished by targeting exon junctions ((Erben et al., 2018; Guo et al., 2018; Zhu et al., 2018); see Figure 1). Other BaseScope applications include the detection of long non-coding RNAs, microRNAs, short or very similar sequences that can distinguish up to a single nucleotide and the validation of short knockout sequences (Anderson et al., 2018; Baker et al., 2017).
BaseScope uses 1–6 “ZZ” probes and is currently only a single-plex assay; ergo only one target RNA can be detected at a time. The Fast-RED dye used can be visualized both by fluorescence and chromogenically when counterstained with DAPI and hematoxylin, respectively (see below). In practical terms, the amplification of the signal requires more amplification steps than RNAscope (4 vs 8 amplification steps) and an additional H2O2 treatment step (during the pretreatment) to block intrinsic enzyme activity and allow the signal detection by alkaline phosphatase. Pretreatment conditions are described here for fresh-frozen and FFPE sections, as well as for adherent cultures. A representative BaseScope experiment on FFPE sections showing the high sensitivity and specificity is shown in Figure 7, and an example detected with light microscopy is shown in Fig. 8A.
Figure 7.
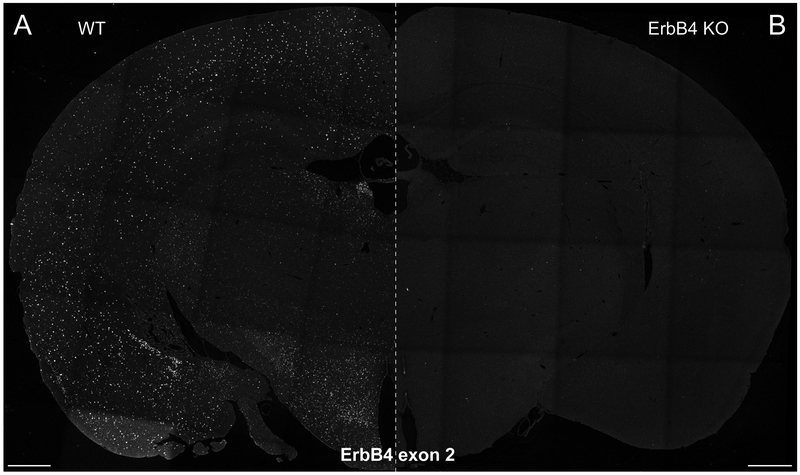
Exon-specific BaseScope experiment on FFPE sections (Basic Protocol 2). Exon 2 of the ErbB4 transcript was detected in coronal sections of adult C57BL/6J WT mice (A) but not in sections of ErbB4 mutant mice (ErbB4 KO; RRID:MGI:5318192) (B) that lack this exon (Tidcombe et al., 2003) (for details see (Erben et al., 2018)). The absence of signal in the single exon mutant confirms the high specificity of the assay. Scale bars 500μm.
NOTE: Even if reagents of RNAscope and BaseScope kits have the same name (e.g. AMP1) these reagents are not the same and not interchangeable! Only use reagents from one kit and do not mix.
Materials
- BaseScope reagents (ACD):
- 1–6 ZZ target probes (C1, species-specific; varying Cat No.)
- BaseScope™ reagent kit-RED (ACD; version 2 Cat No. 323910)
- Pretreatment kit (Cat No. 322380)
- 50x wash buffer (Cat No. 310091)
- Optional: positive and negative probes (e.g. for mouse: positive probes Polr2a Cat No. 701101 (1ZZ) or 701091 (3ZZ) and negative probes bacterial DapB Cat No. 701011 (1ZZ) or 701021 (3ZZ))
Sample: 10–20μm-thick fresh-frozen sections on Superfrost slides (for preparation see (Gerfen, 2003)) or 4–8 μm-thick paraffin sections on Superfrost slides (for preparation see (Zeller, 2001)) or adherent cultures on glass coverslips
RNase AWAY (e.g. Molecular BioProducts, Fisher Scientific, Cat No. MBP#7000)
Paraformaldehyde (PFA; e.g. 16% PFA, Electron Microscopy Sciences, Cat No. 15710)
100% Ethanol (200 proof), 50% and 70% ethanol prepared with RNase-free water
RNase-free water (e.g. DEPC-treated water; KD Medical, Cat No. RGF-3050)
12 vertical glass Coplin jars (e.g. Ted Pella, Cat No. 432–1)
1x PBS (RNase-free)
Hydrophobic barrier pen (e.g. ImmEdge hydrophobic barrier pen; Vector Laboratories, Cat No. H-4000)
Absorbent paper (e.g. Whatman paper; GE Healthcare, Cat No. 10427804)
Humidifying chamber (e.g. ACD, Cat No. 310012)
Horizontal slide rack (e.g. ACD, Cat No. 310017)
Oven (40°C; e.g. HybEZ™ / HybEZ™ II Oven; ACD, Cat No. 321710/20 (110/220 V))
Water bath (40°C)
Distilled water
1.5 mL RNase-free Eppendorf tubes
Kimwipes (e.g. Kimtech Science Precision wipes; Kimberly-Clark Professional, Cat No. 05511)
Cover glass (24×50mm; e.g. Fisher Scientific, Cat No. 12–548-5M)
DAPI (e.g. Invitrogen; Cat No. D3571)
Aqueous mounting medium (e.g. Mowiol DABCO – see reagents and solutions)
Fluorescent microscope (e.g. Zeiss LSM710/780)
Beaker
Heat plate
Optional: thermometer (100°C)
Xylene (e.g. Macron; Cat No. 8668–16)
Drying oven (60°C)
10% normal buffered formalin (NBF; e.g. Sigma-Aldrich; Cat No. HT5011)
Horizontal rocking plate
Glass slides (75×25mm; e.g. C&A Scientific, Premiere; Cat No. 8201)
Hematoxylin (e.g. Electron Microscopy Sciences; Cat No. 26030–10)
0.02% Ammonium water (see reagents and solutions) Ammonium Hydroxide (e.g. Sigma; Cat No. 221228)
Parafilm
Light microscope
Preparations
-
1
The day of the assay, pre-heat and equilibrate the oven to 40°C. Assemble a humidifying chamber using absorbent paper and RNase-free water, and pre-warm it in the oven.
Note: Depending on sample type (fresh-frozen sections (steps 2–12), formalin-fixed paraffin-embedded sections (steps 13–27) or adherent cell culture (steps 28–39) perform one of the following pretreatment protocols, skip steps of other protocols and continue with step 40 probe incubation.
1. Pretreatment of fresh-frozen sections:
Additional Preparations
-
2
Prepare fresh 4% paraformaldehyde (PFA) with RNase-free water and pre-chill in Coplin jar at 4°C the night before. Alternatively, 10% normal buffered formalin can be used.
Pretreatment
-
3
Transfer slides quickly from their slide box stored at −80°C directly into pre-chilled 4% PFA. Sections should not be allowed to thaw and need to be completely submerged. Incubate for 15min at 4°C.
-
4
Wash slides in 1x PBS at room temperature (RT) by transferring slides to a Coplin Jar filled with 1x PBS. Move slides up and down five times and incubate 2min. Repeat wash once with fresh 1x PBS.
-
5
To dehydrate the sections, transfer the slides to a Coplin jar filled with 50% ethanol. Move slides up and down five times and incubate 5min at RT. Repeat one time in 70% ethanol, and twice with fresh 100% ethanol. (Note: slides can be stored for up to one week at −20°C in 100% ethanol. Prolonged storage may result in RNA degradation and suboptimal results.)
-
6
Carefully dry the bottom-side of the slide (i.e., side without section) with a Kimwipe and using a quick flick of the wrist remove excess liquid over the tissue, air-dry slides with sections facing upwards on an even surface for 5min at RT.
-
7
Draw a circle or rectangle around each section with a hydrophobic barrier pen, leaving about 5mm of space between the section and the barrier, and allow the barrier to completely dry before continuing (~1min).
-
8
Cover section completely with 2–3 drops of H2O2 pretreatment (pretreatment 1 provided with pretreatment kit) and incubate for 10min at RT. Cover sections with lid to avoid dust falling onto sections.
-
9
Remove the pretreatment solution by carefully blotting the side of the slide with an absorbent paper and giving the slide a quick flick of the wrist. Transfer slides into a Coplin Jar filled with 1x PBS at RT, and wash by gently moving slides up and down in the solution five times and incubating for 2min. Repeat the wash with fresh 1x PBS.
-
10
Remove excess of PBS and cover sections completely with protease IV pretreatment solution (provided with pretreatment kit; e.g. 3 drops for coronal adult mouse brain section) and incubate 30min at RT. Cover sections with lid to avoid dust falling onto samples.
-
11
During incubation of the protease pretreatment, preheat BaseScope probes provided in ready-to-use dropper bottle for 10min at 40°C in a water bath or oven. Swirl probe to mix.
-
12
Remove the pretreatment solution and wash slides twice in 1x PBS at RT as described above.
2. Pretreatment of formalin-fixed paraffin-embedded sections:
-
13
Bake sections for 1h in an oven at 60°C. This can be done up to one week in advance and slides be stored at RT with desiccants.
-
14
During baking of the sections, prepare 1x antigen retrieval: Prepare 1x antigen retrieval from 10x stock (pretreatment II; provided with the pretreatment kit) with RNase-free water. Unused 1x antigen retrieval can be stored at RT. On a heat plate, pre-heat Coplin jar filled with 1x antigen retrieval in a water-filled beaker brought to boil. Temperature can be controlled with thermometer and should be between 98°C and 102°C. Alternatively, antigen retrieval can also be performed in a pre-heated steamer.
-
15
In a fume hood, remove paraffin from sections in a Coplin jar filled with xylene. Move slides up and down five times and incubate for 5min at RT. Repeat once with fresh xylene.
-
16
Transfer slides to a Coplin jar filled with 100% ethanol. Move slides up and down five times and incubate 3min at RT. Repeat once with fresh 100% ethanol before drying.
-
17
After carefully drying with a Kimwipe the bottom-side of the slice (i.e., side without section), and giving the slide a quick flick of the wrist, air-dry the slides with sections facing upwards on an even surface for 5min at RT.
-
18
Draw a circle or rectangle around each section with a hydrophobic barrier pen, leaving about 5mm of space between the section and the barrier, and allow the barrier to completely dry before continuing (~1min). Optional: Dried slides can be stored overnight, and protocol continued the following day.
-
19
Cover section completely with 2–3 drops of H2O2 pretreatment (pretreatment 1 provided with pretreatment kit) and incubate for 10min at RT. Cover sections with lid to avoid dust falling onto sections.
-
20
Remove the pretreatment solution by carefully blotting the side of the slide with an absorbent paper and giving the slide a quick flick of the wrist. Transfer slides into a Coplin Jar filled with RNase-free water at RT, and wash by gently moving slides up and down in the solution five times and incubating for 2min. Repeat the wash with fresh RNase-free water.
-
21
With forceps, slowly place slides into pre-heated 1x antigen retrieval (see step 3) and incubate for 15min.
-
22
Carefully remove slides with forceps and transfer into a Coplin jar filled with RNase-free water at RT. Wash sections by gently moving the slides up and down five times and incubate for 2min. Repeat the wash once with fresh RNase-free water.
-
23
Transfer slides into a Coplin jar filled with 100% ethanol, incubate ~15sec and air-dry for 5min as described above in step 6.
-
24
If necessary, reapply hydrophobic barrier.
-
25
Place slides on slide rack in the pre-warmed humidifying chamber and cover completely with protease treatment III (provided with pretreatment kit; e.g. ~3 drops per section for coronal adult mouse brain section). Incubate for 30min at 40°C in the sealed humidified chamber in the oven.
-
26
During incubation of the protease pretreatment, preheat BaseScope probes provided in ready-to-use dropper bottle for 10min at 40°C in a water bath or oven. Swirl probe to mix.
-
27
Remove the pretreatment solution and wash slides twice in RNase-free water at RT as described above.
3. Pretreatment of adherent cell culture samples:
Additional Preparations
-
28
Prepare glass slides with hydrophobic barrier pen circles with a diameter slightly exceeding the coverslip size used. Glass slides can be washed and dried between incubations and reused. Redraw hydrophobic barrier if necessary.
Pretreatment
-
29
Remove cell culture medium and rinse cultures carefully with 1x PBS.
-
30
Replace PBS with 10% NBF and fix cultures by incubating for 30min at RT while agitating on a horizontal shaker. All subsequent washes should be performed while agitating on shaker.
-
31
Remove NBF and replace with 1x PBS for 5min at RT. Repeat wash once with fresh 1x PBS.
-
32
To dehydrate cultures, remove PBS and substitute with 50% ethanol for 5min at RT, then with 70% ethanol once, and twice with fresh 100% ethanol. (Note: fixed cultures can be stored for up to six months at −20°C in 100% ethanol.)
-
33
To rehydrate culture samples, replace 100% ethanol with 70% ethanol and incubate for 2min at RT. Repeat these steps with 50% ethanol and then wash in 1x PBS at RT for 10min.
-
34
Remove coverslips one-by-one from cell culture plate and place cultures-facing up onto the prepared glass slide that contain hydrophobic barrier circles (see preparation step 2). Samples should not dry out. Cover immediately with 2–3 drops of H2O2 pretreatment (pretreatment 1; provided with pretreatment kit) and incubate for 10min at RT. Cover coverslips with lid to avoid dust falling onto sections.
-
35
Move coverslips back into culture well plate filled with 1x PBS and wash for 2min at RT. Repeat once with fresh 1x PBS.
-
36
Freshly prepare a 1:15 dilution of protease III (provided with pretreatment kit) in RNase-free 1x PBS (~100μl per 12mm coverslip)
-
37
Place coverslips cultures-facing up onto the prepared glass slide (as described above) and cover immediately with prepared 1:15 dilution of protease III. Incubate for 10min at RT. Cover sections with lid to avoid dust falling onto samples.
-
38
During incubation of the protease pretreatment, preheat BaseScope probes provided in ready-to-use dropper bottle for 10min at 40°C in a water bath or oven. Swirl probe to mix.
-
39
Move coverslips back into culture well plate filled with 1x PBS and wash for 2min at RT. Repeat once with fresh 1x PBS.
Probe Incubation & Amplification (for all sample types)
-
40
Individually transfer each slide (or coverslip) to a horizontal slide rack in the pre-warmed humidifying chamber, after blotting and removing the excess of wash solution from the slides as described above. Carefully cover sections with target probe (~2 drops), being careful not to let the sections dry out.
-
41
Hybridize the sections for 2h at 40°C in the sealed humidified chamber in the oven.
-
42
During incubation of the probes prepare 1x wash buffer: Incubate 50x wash buffer for 10min at 40°C in a water bath or oven. Prepare 2L of 1x wash buffer using distilled water. Note, after the hybridization of the probes, it is no longer necessary to continue to work under RNase precaution. Unused 1x wash buffer can be stored at RT.
-
43
Remove excess of hybridization solution as described above and transfer each slide into a Coplin jar filled with 1x wash buffer at RT. As before, wash the sections by gently moving the slides up and down five times, and incubating for 2min. Repeat the wash once with fresh 1x wash buffer.
-
44
To begin the amplification process (amplification solutions AMP1–8 provided in BaseScope reagent kit-RED), sequentially remove from each slide the excess 1x wash buffer and transfer it to the humidified chamber. Completely cover each section with the Amplification solution 1 (AMP1; ~3 drops for an adult coronal mouse brain section). Incubate in the humidified chamber for 30min at 40°C.
-
45
Wash slides twice in 1x wash buffer at RT, as in step 43.
-
46
Remove excess wash, add Amplification solution 2 (AMP2) and incubate 30min at 40°C.
-
47
Wash slides twice in 1x wash buffer at RT, as in step 43.
-
48
Remove excess wash, add Amplification solution 3 (AMP3) and incubate 15min at 40°C.
-
49
Wash slides twice in 1x wash buffer at RT, as in step 43.
-
50
Remove excess wash, add Amplification solution 4 (AMP4) and incubate 30min at 40°C.
-
51
Wash slides twice in 1x wash buffer at RT, as in step 43.
-
52
Remove excess wash, add Amplification solution 5 (AMP5) and incubate 30min at 40°C.
-
53
Wash slides twice in 1x wash buffer at RT, as in step 43.
-
54
Remove excess wash, add Amplification solution 6 (AMP6) and incubate 15min at 40°C.
-
55
Wash slides twice in 1x wash buffer at RT, as in step 43.
-
56
Remove excess wash, add Amplification solution 7 (AMP7) and incubate 30min at RT.
-
57
Wash slides twice in 1x wash buffer at RT, as in step 43.
-
58
Remove excess wash, add Amplification solution 8 (AMP8) and incubate 15min at RT.
-
59
Wash slides twice in 1x wash buffer at RT, as in step 43.
-
60
Prepare fresh FastRED solution provided with BaseScope reagent kit-RED (~60μl per coronal section). Dilute FastRED-B 1:60 in FastRED-A (e.g. 1μl FastRED-B in 60μl of FastRED-A).
-
61
Remove excess wash buffer and cover sections completely with FastRED solution. Incubate 10min at RT and cover sections with lid to avoid dust falling onto sections.
-
Remove excess FastRED solution and wash slides twice for 2min in tap water, as described in step 7.
Note: The FastRED dye used in the BaseScope assay is visible both in fluorescent and light microscopes. For fluorescent visualization use DAPI (outlined below, step 62), for chromogenic detection, sections need to be counterstained with hematoxylin (skip to step 63).
-
62
Counterstaininig with DAPI: One slide at the time, counterstain cell nuclei by carefully adding DAPI (1μg/mL in PBS; ~100μl) and incubating for 30sec at RT. Remove excess DAPI as described above. Skip to step 68 for mounting.
-
63
Counterstaining with Hematoxylin: Prepare 50% hematoxylin staining solution by mixing equal amounts of hematoxylin with distilled water in a Coplin jar.
-
64
Move slides into Coplin jar filled with 50% hematoxylin and incubate 2min at RT. Sections will turn purple.
-
65
Immediately, wash slides in tap water by moving slides up and down. Repeat washes with fresh tap water until the water becomes clear.
-
66
One-by-one incubate slides ~15sec in 0.02% Ammonia water. Move slides up and down five times. And then place into fresh tap water. Sections are turning blue.
-
67
Replace with fresh tap water and wash for 2min at RT. Remove excess of solution.
-
68
Add aqueous mounting medium (e.g. Mowiol DABCO; ~10μl per coronal section – see reagents and solutions) and cover-slip each section carefully to avoid trapping air bubbles. Store slides horizontally in the dark overnight at 4°C to dry.
Detection & Analysis
-
69
Examine dried slides within a few days of preparation on a fluorescent (confocal) microscope for DAPI-counterstained and on a light microscope for hematoxylin-counterstained sections (magnification 10–40X). Digital image analysis using a camera is highly recommended. For automated signal quantification, a magnification of at least 20X is recommended..
-
70
Signal quantification can be achieved by simple semi-quantitative scoring of the signal (ACD), using standard free image analysis software such as ImageJ (NIH) and CellProfiler (Carpenter et al. 2006), or commercial image analysis software such as Imaris (Bitplane) or specific ISH quantification software such as HALO (Indica Labs) and Aperio (Leica). In case of hematoxylin-counterstain, Spotstudio (ACD) software (see Table 1) can be used to analyze specifically bright-field single-plex data. For more details to data interpretation and signal quantification refer to Basic Protocol 4 and the section Understanding Results & Statistical Analyses.
Table 1.
Software to analyze ISH signal.
| software | source | website | chromogenic/ fluorescent analysis |
download | RRID |
|---|---|---|---|---|---|
| HALO | Indica Labs |
http://www.indicalab.com/products/multiplex-fish/ http://www.indicalab.com/products/sish-dual-cish-quantification/ |
both | ||
| Aperio | Leica | https://www.leicabiosystems.com/digital-pathology/analyze/ish-fish/ | both | ||
| Spotstudio | Advanced Cell Diagnostics | https://acdbio.com/rnascope®-spotstudio-software | chromogenic only | ||
| Imaris | Bitplane | http://www.bitplane.com/imaris | both | SCR_007370 | |
| ImageJ/Fiji | NIH | http://fiji.sc | both | http://fiji.sc/#download | SCR_002285 |
| CellProfiler | Broad Institute | http://cellprofiler.org | both | https://cellprofiler.org/releases/ | SCR_007358 |
BASIC PROTOCOL 3: POST-HOC IMMUNOSTAININGS
In situations when it is important to identify cell types expressing the RNA of interest, the multiplexing nature of RNAscope is advantageous over BaseScope. Nevertheless, BaseScope can be combined with immunohistochemistry (IHC) to identify different cell types expressing the RNA of interest; immunostaining is also compatible with RNAscope.
In our experience, the combination of BaseScope or RNAscope with immunohistochemistry only succeeds when the ISH is performed first, as the immunostaining interferes with the specificity and sensitivity of the ISH. However, this approach is challenging, as the success of the ISH depends on proper permeabilization to allow the probes to access the target RNAs and changing the conditions results frequently in off-target signal or reduced sensitivity. The permeabilization is mainly achieved using proteases, which in turn can destroy antigens of the antibodies used in the immunostaining. In our hands, only about 10% of the antibodies tested were compatible with performing ISH using protease digestion; largely independent of section type and corresponding pretreatment conditions. This success rate is consistent with reports from others (Dual ISH-IHC; ACD). In general, we had better success with antibodies against membrane-bound proteins than soluble proteins. If the combination with immunostaining is necessary, it may require a large number of different antibodies to identify one that is compatible with ISH. However, we established a post-hoc immunohistochemistry protocol using cell type-specific transgenic GFP mice that allows for easy identification of cell type-specific expression if transgenic GFP mice are available. GFP expression was destroyed and could not be observed directly after the ISH protocol, but performing a post-hoc immunostaining using antibodies against GFP restored the signal. Of note, not all GFP antibodies tested retrieved the GFP expression. Here, we describe a BaseScope/RNAscope-immunohistochemical protocol using a mouse monoclonal anti-GFP antibody from NeuroMab (clone N86/8; Davis CA). The protocol follows the ISH protocols for RNAscope (Basic Protocol 1) or BaseScope (Basic Protocol 2), but before counterstaining for DAPI continues with the post-hoc immunostaining. An example on sections of transgenic GFP mice is represented in Fig. 8B. Fig. 8C+D show examples of ISH-IHC combination with an antibody against the dopamine transporter DAT (clone 6–5G10; SantaCruz; sc-32258; 1:200; RRID: AB_627400) which epitope is not destroyed by the pretreatment.
Another combinatory approach useful in neuroscience is to combine retrograde-tracing with RNAscope and BaseScope. In this case, fluorescent ISH can be coupled with fluorescent Retrobeads or choleratoxin that are transported retrogradely because the fluorescent properties of these markers are not lost during sample preparation.
Materials
In situ hybridization prepared samples (from Basic Protocol 1 or Basic Protocol 2)
Normal donkey or goat serum (e.g. Sigma-Aldrich; D9663)
Triton X-100 (e.g. Thermo-Fisher; Cat No. 28314)
Mouse monoclonal anti-GFP antibody (clone N86/8; NeuroMab Cat No. 73–131, UC Davis CA; RRID: AB_10671444)
Anti-mouse fluorescent-conjugated secondary antibody (e.g. donkey-anti mouse Alexa488; Invitrogen, Cat No. A-21202; RRID: AB_141607)
In situ hybridization
-
1
Use samples of transgenic GFP mice on which ISH was performed according to Basic Protocol 1 or Basic Protocol 2 and corresponding pretreatment conditions for the sample type analyzed (fresh frozen or FFPE sections, adherent cultures; see Figure 1).
Note: Perform all steps until counterstain with DAPI (e.g. step 1–22 in Basic Protocol 1, RNAscope, and step 1–61 in Basic Protocol 2, BaseScope).
Post-hoc immunostaining
-
2
Immediately following the ISH, place sections in blocking solution (10% Normal Donkey/Goat Serum, 0.3% TritonX-100 in 1x PBS) for 1h at RT. This is performed on the slide rack in the humidifying chamber.
-
3
Slide by slide, remove excess of solution and replace with 1μg/mL mouse monoclonal anti-GFP antibody (clone N86/8, NeuroMab) in blocking solution. Place back in humidifying chamber and incubate overnight at 4°C.
-
4
Wash sections in 1x PBS + 0.25% Triton X-100 for 10min at RT. Repeat twice with fresh wash buffer. The washes can be performed in Coplin jars. Annotation: The washes with Triton X-100 might dissolve the hydrophobic barrier pen. In this case the pen needs to be carefully reapplied without letting sections dry out.
-
5
One-by-one remove excess of solution, place slide in humidifying chamber and cover with secondary antibody solution (e.g. 1μg/mL anti-mouse A488 in blocking solution). Incubate in the dark (closed humidifying chamber) for 2h at RT.
-
6
Wash sections in 1x PBS for 10min at RT. Repeat twice with fresh wash buffer.
-
7
One slide at the time, counterstain cell nuclei by covering sections with DAPI (either from RNAscope kit or 1μg/mL in 1x PBS (Invitrogen)) and incubating for 30sec at RT.
-
8
Remove excess of DAPI as described above, add aqueous mounting medium (e.g. Mowiol DABCO; ~10μl per coronal section – see reagents and solutions) and cover-slip each section carefully to avoid trapping air bubbles. Store slides horizontally in the dark overnight at 4°C to dry.
Detection & Analysis
-
9
Examine dried slides within a few days of preparation on a fluorescent (confocal) microscope (magnification 20–63X). The signals obtained from low abundance transcripts might be only visible at higher magnification (40–63X). Digital image analysis using a camera is highly recommended. Signal can be quantified and analyzed as described in Basic Protocol 4 and the section Understanding Results & Statistical Analyses.
BASIC PROTOCOL 4: AUTOMATED QUANTIFICATION OF FLUORESCENT ISH SIGNAL USING CELL-PROFILER
It is often desirable to support qualitative image data with quantitative analysis to describe reproducibility between samples and distribution within the data or area analyzed, and to compare different signals and samples statistically. Automated quantification of the signal is objective and accurate way to analyze the data. Different software, both commercial (HALO, Aperio, Spotstudio, Imaris) and freeware (ImageJ, CellProfiler), are available (see Table 1), and custom-made programs can be written for analysis (Haring et al., 2018). The general principles used by all software are similar (see Figure 9); that is: the signal is filtered by intensity above a background threshold, and this signal is assigned to cells that are designated as an area around the DAPI (or hematoxylin) nuclear staining to account for somatic localization of RNA transcripts. While commercial software is user-friendly, it is expensive and often limited to certain applications. Therefore, we have routinely analyzed RNAscope and BaseScope data using the module-based free open-source software CellProfiler (Carpenter et al., 2006) and, below, provide a step-by-step explanation of how to use our custom-made pipeline (see Figure 9).
Figure 9.
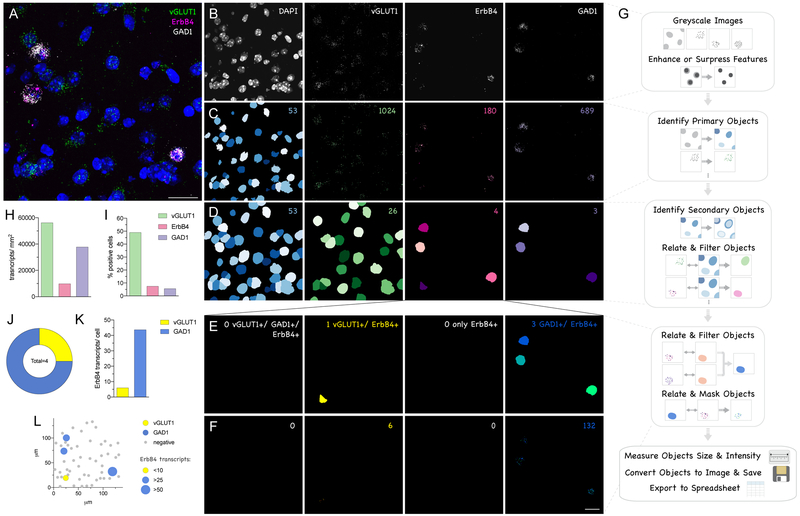
Quantification of multiplex fluorescent ISH signal (e.g. RNAscope) with CellProfiler. (A) Original RNAscope image analyzing ErbB4 expression (C1, magenta), in vGLUT1 (C2, green) and GAD1 (C3, white) positive neurons in the somatosensory cortex (see Figure 5). (B) For analysis with CellProfiler the image is split into individual fluorescent channels and converted to greyscale. (C) Objects, “nuclei” (blue) and “dots” in each channel (green, magenta, purple) are identified using the module “IdentifyPrimaryObjects” by intensity and size. (D) “Cells” (blue) are computed by increasing the size of the nuclei (“IdentifySecondaryObjects”). Cells positive for one of the signals are subsequently filtered based on the related number of dots. (E) Double- and triple-positive cells were analyzed for the subpopulation of ErbB4+ cells by filtering: triple-positive cells (NA), vGLUT1+ and ErbB4+ cells (yellow), only ErbB4+ (NA) and GAD1+ and ErbB4+ (blue). (F) ErbB4 expression levels of these subpopulations were obtained after masking the signal on these different cell types. (G) Schematic overview of CellProfiler pipeline used. (H) Transcript expression levels analyzed per area for vGLUT1 (green), ErbB4 (magenta) and GAD1 (purple). (I) Percentage of positive cells relative to all cells. (J) Subpopulations of ErbB4+ cells: double-positive for ErbB4 and vGLUT1 (yellow), and for ErbB4 and GAD1 (blue). (K) ErbB4 transcripts expressed in these subpopulations. (L) Representation of the original image with ErbB4+/vGLUT1+ in yellow, ErbB4+/GAD1+ in blue, and ErbB4- in grey. The size of the symbols represents ErbB4 transcript levels in each individual cell. Numbers in the right upper corner indicate the number of objects identified (B-F). Scale bars 20μm.
As an example, we are using RNAscope data from Figure 5 that corresponds to hybridization of ErbB4 transcripts expressed in vGLUT1+ pyramidal neurons and GAD1+ GABAergic interneurons in the adult mouse brain cortex. The example image shown in Figure 9A was taken at 63X magnification. Of note, for single-dot resolution and successful performance of the quantification with these pipelines, magnification of at least 40X for RNAscope and 20X for BaseScope is required. Pipelines (macros) for RNAscope and BaseScope analysis similar to that used for this example are freely available on the website of CellProfiler (http://cellprofiler.org/examples/published_pipelines; see (Erben et al., 2018)).
File input
-
1
Split different fluorescent channels (DAPI, green, red, far-red) of multi-channel image using image-manipulating software (e.g. ImageJ) and save individually as tiff files (recommended format). Note: CellProfiler recognizes the different channels of an image by the name of the file (e.g. contains C1 for DAPI, C2 for green, C3 for red, C4 for far-red).
-
2
Import image-set (all channels of one image) into CellProfiler by a simple drag-and-drop.
-
3
With the module “ColorToGrey”, convert image to greyscale (if not already pre-converted in ImageJ), see Fig. 9B.
-
4
Optional: Enhance the intensity features relative to the background with the module “EnhanceOrSuppressFeatures”, which can help to identify objects. We recommend enhancing the RNAscope signal with the feature “Speckle”. The size of the “Speckles” is in pixel units, as all subsequent objects, and therefore depends on the resolution of the image.
Object identification:
This is the key of the pipeline to identify both the RNAscope signal as well as DAPI-positive nuclei using the module “IdentifyPrimary Objects” (Fig. 9C). Objects are identified by intensity threshold that can be a manual value (not recommended), based on a previous measurement or an algorithm. The signal can be additionally restricted in size (in pixels). Finally, clumped objects can be distinguished by shape or intensity.
-
5
Measure background intensity of DAPI channel using the module “MeasureImageIntensity”.
-
6
Use the module “IdentifyPrimaryObjects” to identify DAPI-positive nuclei. We recommend identifying “nuclei” based on the previous measured mean intensity of background. The size filter of the objects in our example was 50–300 pixels and declumping for nuclei was done based on shape.
-
7
Identify RNAscope signal/ dots of one channel (e.g. green) using the same module. For dot identification in each channel both previous measurements or algorithm work generally well. For our example, we used the global Otsu algorithm, size exclusion of 2–20 pixels and distinguished clumped objects by intensity.
-
8
Repeat the identification of objects for each of the RNAscope channels (e.g. red and white = far-red) signal using the module “IdentifyPrimaryObjects” as described in step 7.
Identification of cells and filtering for single, double and triple positive cells
-
9
Enlarge the size of the nuclei using the module “IdentifySecondaryObjects”. This accounts for the somatic accumulation of RNAs. Use function “Distance – N” to increase the object size by a certain number of pixels until colliding with another object (Fig. 9D). We typically call these objects “cells”. In our example the nuclei were expanded by 25 pixels.
-
10
Relate the RNAscope signal (“children”) for one channel (e.g. green) to the cell (“parent”) with the module “RelateObjects”.
-
11
Repeat step 10 for the remaining RNAscope signals (e.g. red and white channel).
-
12
Filter positive cells for one channel (e.g. green) based on the number of dots assigned to this cell using the module “FilterObjects”. The threshold needs to be set based on expression levels of each probe to exclude false-positive cells. In our example, we were using a minimum of 10 dots for vGLUT1 (green), 5 dots for ErbB4 (red), and 25 dots for GAD1 (white).
-
13
Repeat step 12 for the remaining channels (e.g. red and white).
-
14
Relate the RNAscope signals (“RelateObjects”) to the newly identified cell population (e.g. red cells) and filter (“FilterObjects”) into double-positive (e.g. “red and green cells”, “red and white cells”) and triple positive (green, red and white) as in steps 10–13. In our example, we analyzed the population of ErbB4+ cells (red cells) for co-expression of one of the other markers (Fig. 9E).
Analysis of transcript expression levels
-
15
Re-relate signals back to the newly identified objects (single, double and triple positive cells) using the “RelateObjects” module as described in steps 10 and 11 to identify single-cell expression levels.
-
16
Mask the RNAscope signal with the positive cells to sort for transcripts expressed in the positive cells using the “MaskObjects” module (see Fig. 9F).
Measurements and data export
-
17
Optional: Use the modules “MeasureObjectSizeShape” and “MeasureObjectIntensity” to measure size and intensity of objects.
-
18
Optional: With the module “ConvertObjectsToImage”, convert objects to an image for subsequent export using the module “SaveImages”. This can serve as quality control, when the pipeline is run in the background mode.
-
19
Export your data in csv format with the module “ExportToSpreadsheet”. The summary analysis is stored in the file titled “_Image”, whereas details about each individual objcect are found in the respective files (see Fig. 9G).
Data representation:
Typical values obtained with this analysis are: expression levels per area (Fig. 9H; area measured e.g. in ImageJ), percentage of positive cells per all cells analyzed (Fig. 9I), subpopulations of positive cells (Fig. 9J), and the average transcript expression level in different cell populations (Fig. 9K). Another advantage of this analysis is that values per individual object are exported. This information allows one to determine: expression levels per cell, X/Y positions of each cell, size and intensity of each signal. This data can be used to plot the expression in one (e.g. cortical layer, see Fig. 5E) or two dimensions (Fig. 9L). In addition, the expression per cell data can be plotted either as histogram/frequency distribution or cumulative probability to determine the cut-off between background and signal (see (Erben et al., 2018)).
Note: The mentioned pipeline settings are optimized for this example only and will need to be adjusted for individual experiments depending on microscope magnification and settings, RNAscope probes and the sample itself (e.g. age, section type, brain area analyzed). When setting up a pipeline, we recommend adjusting the pipeline with a variety of small cropped areas (similar to the example image in Fig. 9A) across the entire region of interest and different samples in the test mode, before running the entire data-set in the background mode (“Hide All Windows on Run”).
REAGENTS AND SOLUTIONS
Mowiol DABCO mounting medium
Preparation of 25mL
In a 50mL Falcon tube, add slowly (over hours) 2.4g mowiol (Calbiochem, Cat No. 3475904) to 6g glycerol while mixing. Add 6mL of water and mix at RT overnight.
Add 12mL of 0.2M Tris-HCl (pH 6.8) and warm up in a water bath (beaker) on heat plate to 50°C while stirring.
After mowiol is dissolved, add 0.625g DABCO to 2.5% (Sigma, Cat No. D2522).
Clarify the solution by centrifugation at 5000g for 15min.
Aliquote supernatant and freeze at −20°C for up to 6 months.
Before using warm up shortly to 37°C. Store aliquot in use at 4°C.
0.02% (w/v) Ammonium water
Preparation of 50mL
In a fume hood, add 33μl of 30% Ammonium Hydroxide (e.g. Sigma; Cat No. 221228) to 50mL of distilled water in a Coplin jar.
Seal with parafilm and mix by inverting five times.
Prepare fresh for each experiment.
COMMENTARY
Background information
ISH is a widely used technique to analyze gene expression. The high specificity and reproducibility of ISH techniques that allow multiplexing, such as RNAscope and ViewRNA, can be used to determine developmental and cellular patterns of protein expression before resorting to the more complicated immunological approaches. Expression of non-coding RNAs can also be analyzed by ISH (Tan and Yang, 2018). In recent years numerous advancements in high through-put RNA sequencing from tissues and even from single cells has led to the identification of numerous neuronal subtypes, usually categorized by the neurotransmitter they synthesize, and a better understanding of the dynamic changes in transcriptomes during development and in disease (De Sa Nogueira et al., 2018; Guillaumet-Adkins et al., 2017; Johnson and Walsh, 2017). These methods are extremely powerful for rapidly identifying biomarkers and differentially expressed genes. However, in contrast to ISH approaches, these methods fail to provide the expression of genes in single cells in the context of intact tissue. This is one reason that ISH approaches, like RNAscope, are frequently used to validate and supplement RNAseq data (Ayata et al., 2018; Cembrowski et al., 2018; Haring et al., 2018; Shrestha et al., 2018). Next-generation ISH techniques, such as BaseScope, are also useful to complement transcriptome sequencing data that identifies short RNA sequences, splice variants and single nucleotide polymorphisms. Whereas quantitative real time reverse transcription (RT)-PCR is a faster and more quantitative method than BaseScope to analyze relative levels of distinct RNA splice variants, it also lacks cellular resolution in tissue. Importantly, we recently have shown that semi-quantitative analysis of splice variants using BaseScope provides similar results to quantitative real time RT-PCR, but with the added advantage of studying splice variants with cellular resolution in intact tissue (Erben et al., 2018; Zhu et al., 2018).
Critical Parameters & Troubleshooting:
Preparing sections:
A critical parameter for successful results is the overall quality of the sections, as regards the preservation of RNA, cell morphology, and tears/wrinkles; see general recommendations for high-quality tissue section preparation (Gerfen, 2003; Zeller, 2001). Avoid moisture and freeze-thawing cycles that are permissive for RNase activity and use RNase-free equipment and solutions where indicated. To reduce or avoid detachment of sections, which most commonly occurs during antigen retrieval, use charged Superfrost slides and increase the baking time. When using paraffin-embedded tissue, it is important to use fresh ethanol and xylene with agitation to fully remove paraffin; otherwise, residual wax can result in unspecific staining.
Reducing edge-artifacts and background:
To avoid edge-artifacts and background one should keep sections from drying-out, unless where it is specified in the protocol. To avoid these problems, it is important to ensure that the humidifying tissue and the hybridization chamber used throughout the procedure remain moist at all times; preferably, use RNase-free water. In addition, to reduce the risk sections drying-out we recommend working on a single slide at a time, maintain a manageable number of slides processed in a single experiment, submerge with reagent every section on a slide before proceeding to the next slide and, do not skimp on reagents, always use sufficient amounts to completely submerge each section (see also Troubleshooting Guide; ACD).
Optimization of conditions:
Follow the protocols with regards to incubation times and temperatures during probe hybridization and amplification, as these have been optimized for each reagent. On the other hand, pretreatment conditions may need to be optimized if samples are not properly prepared or fixed using other protocols (e.g. human tissue). In these cases, run preliminary assay(s) with negative and positive controls to evaluate morphology and staining. If the signals are weak, but the morphology of tissue is well preserved as indicated by strong nuclear staining, this could suggest that probe accessibility is impaired due to over-fixation or under-digestion. To address this point, pretreatment conditions can be adjusted as follows: Incubation time in target retrieval increased in increments of 5min and/or digestion time with protease treatment increased incrementally by 10min. On the other hand, if the preservation of morphology is poor (weak nuclear stain) and either high background staining or non-uniform signals are observed, it could suggest that tissue sections could have been over-digested or under-fixed. To address this point, decrease incubation times incrementally or dilute the protease (ACD; Troubleshooting Guide).
Control probes & slides:
Positive and negative control probes (see protocols) and slides (e.g. mouse 3T3 pellet, Cat # 310023, ACD) are available from the vendor and can help to evaluate signal-to-noise and trouble-shoot conditions. In cases where no signal is detected we direct the reader to follow the workflow suggested by the vendor (see recommended workflow in Troubleshooting Guide, ACD). Once RNAscope and BaseScope are successfully set-up, the hybridization of a well-known distinctively expressed gene can also be used as reference (e.g. GAD1/2, vGLUT1/2, TH). The signal should be positive in expected areas, but low/no signal should be observed where the gene is not expected to be expressed. The best negative control is a transgenic mouse that lacks the transcript. However, the transgene needs to lack the whole target sequence (in case of RNAscope ~1000 bases), otherwise residual transcript might result in signal.
Understanding Results & Statistical Analyses
There are important differences between RNAscope and BaseScope that affect the interpretation of results; therefore, we begin by separately discussing these differences below.
Under optimal conditions, RNAscope has single molecule resolution and one dot represents one transcript (Wang et al., 2012). The number of dots therefore equals the number of transcripts expressed. However, the size and intensity of dots, which can vary between channels and the number of fluorescent labels and probe pairs bound to a transcript, are not related to the number of transcripts. A single dot per cell in RNAscope, especially when observed in only a few cells (<10%), is considered background (ACD; see Scoring Guidelines in Troubleshooting Guide). Most housekeeping and cell marker genes are expressed at high levels (>10 dots per cell), to the extent that in some cases it is difficult to resolve single dots (clusters).
In contrast to RNAscope, which uses fluorescent labels, BaseScope uses an enzymatic dye reaction to amplify the signal and results in bigger dot sizes that sometimes fuse. In addition, the single-probe pair used in BaseScope cannot guarantee the same single transcript resolution, as compared to the 6–20 “ZZ” probes used in RNAscope. However, because the detection threshold in BaseScope is higher, in some cases single dots might represent actual signal and cannot be discarded as background in absence of additional evaluation. We have previously evaluated the background signal of BaseScope in single exon mutant mice and rarely observed cells that were positive for a single dot (Erben et al., 2018). Finally, due to short target sequences in BaseScope, variability in hybridization efficiency can be observed between different probes targeting distinct transcripts or regions within a transcript. These differences originate from variations in nucleotide sequences and accessibility of the transcript (e.g. protein binding target sequence); and are not observed in RNAscope that targets longer sequences (~1000 bases) (Baker et al., 2017; Erben et al., 2018).
Considering these general differences in the nature of the hybridization signal, the overall evaluation and quantification approaches are similar for RNAscope and BaseScope. The vendor suggests a simple semi-quantitative scoring to evaluate the ISH staining. A score from 0–4 is assigned depending on the average expression levels per cell (0: <1 dot per 10 cells; 1: 1–3 dots per cell; 2: 4–9 dots per cell; 3: 10–15 dots per cell; 4: >15 dots per cell) and can be compared to the expression levels of control genes (see Scoring Guidelines in Trouble Shooting Guide). While this method is helpful to quickly evaluate the overall hybridization success between samples and batches, it carries little quantitative information.
Automated quantification results in less subjective quantitative data. Software specialized for ISH analysis (HALO, Aperio, Spotstudio) is available, but similar analysis can be achieved using generic image anlaysis asoftware (e.g. Imaris) and open source software (e.g. ImageJ, CellProfiler) (see Table 1). Basic Protocol 4 outlines step-by-step how to apply a freely available custom-made CellProfiler pipeline to analyze multiplex fluorescent ISH data.
The described quantification approach can encounter a couple of issues that originate from the nature of the ISH technique itself. In brain areas where cells are very dense (e.g. hippocampal pyramidal cell layers), single cell quantification can be difficult. Distinguishing single nuclei is not always possible, and even if successful, transcripts from one cell can often overlap with the next cell area, resulting in false positive cells. Similarly, RNA transcripts that are transported into neuronal dendrites or axons (Holt and Schuman, 2013; Sahoo et al., 2018), are likely to be incorrectly assigned to the wrong cell. These shortcomings are common to all ISH techniques and practically cannot be avoided unless, at a single-cell level, an entire cell/neuron and its processes are counter labeled (e.g., filled with fluorophore). However, the transport of transcripts corresponds to a small fraction of the transcriptome, and in most brain areas single cell quantification works extremely well.
For statistical analyses, multiple samples hybridized under the same conditions and imaged with same parameters need to be compared. However, in histological analyses, such as ISH approaches, small sample numbers (n=2–4 animals; n=1–3 sections) are generally sufficient for statistical analyses (Erben et al., 2018; Haring et al., 2018; Li et al., 2018). Even with small sample numbers thousands of cells are analyzed in one region of interest, resulting in robust data. It is important to note that if different samples (e.g. animals) shall be compared, it is best to hybridize and analyze all samples at the same time to avoid inter-assay variability. If this is not possible because of a large number of samples, samples should be randomized (e.g. one control sample and one knock-out/treatment sample at the time) and imaging parameters kept identical.
Time Considerations
A major advantage of multiplex (RNAscope) and single-plex (BaseScope), relative to other ISH techniques (i.e., using radioactivity), is their extremely fast turn-around time. The approaches are standardized for optimal signal detection and do not have varying developing times, as ISH techniques using radioactive-labels and colorimetric substrates. In addition, the probes are custom-made, designed and generated by the vendor. Therefore, the hands-on time consists only of the ISH assay itself and the subsequent analysis. Depending on the sample-type, pretreatment varies between approximately 1.5 and 2.5 hours (see Figure 1). There are optional stopping points during the pretreatment to split the assay into two days. The ISH assay needs to be completed in one session and takes 4 hours for RNAscope and 6.5 hours for BaseScope. Slides are dried overnight and can be imaged with a microscope the next day or within a few days. The time for detection and analysis varies depending on the application, from brief examination to overview scanning and detailed quantification with statistical analysis. If a post-hoc immunostaining is desired (Basic Protocol 3) an additional hour for blocking the day of the ISH assay needs to be considered with an overnight incubation and 3 additional hours the following day.
The use of ISH assays described in this unit is relatively simple and do not pose major safety considerations (i.e., radiation exposure and contamination). Therefore, the intrinsic properties of these assays make them amenable, not only to trained scientists, but also to inexperienced trainees.
Significance Statement:
A major challenge in neuroscience has been to ascribe the expression of distinct RNAs and splice variants to specific neuronal and glial subtypes in tissue. A recent advancement to address this issue, has been the development of fluorescent in situ hybridization using novel “branched” amplification chemistry that provides the specificity, low background and sensitivity to simultaneously detect several gene transcripts in different fluorescent channels in a single cell. However, quantitative analysis of the signals is still challenging to many users. Here, we focus on the use of two methodologies, RNAscope and BaseScope, and demonstrate how data can be analyzed to derive quantitative results that rival RNA quantification from tissue extracts using quantitative real-time reverse transcription PCR.
ACKNOWLEDGMENT
This work was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD; ZIA-HD000711). We would like thank Dr. Vincent Schram from the NICHD Microscopy and Imaging Core; and Drs. Mario Penzo and Hugo Tejeda for critical review of the manuscript.
Footnotes
CONLFICTS OF INTEREST
LE and AB declare no competing financial interests.
INTERNET RESSOURCES
https://acdbio.com See the website of Advanced Cell Diagnostics (ACD), the vendor of both RNAscope and BaseScope in situ hybridization assays for product information, target probe catalog, user manuals, troubleshooting guide (https://acdbio.com/technical-support/solutions), insights on dual ISH-IHC (https://acdbio.com/science/applications/research-solutions/dual-ish-and-ihc) and data analysis with ImageJ (ACD; TS 46–003).
LITERATURE CITED
- Anderson CM, Laeremans A, Wang XM, Wu X, Zhang B, Doolittle E, Kim J, Li N, Pimentel HXY, Park E, et al. (2018). Visualizing Genetic Variants, Short Targets, and Point Mutations in the Morphological Tissue Context with an RNA In Situ Hybridization Assay. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayata P, Badimon A, Strasburger HJ, Duff MK, Montgomery SE, Loh YE, Ebert A, Pimenova AA, Ramirez BR, Chan AT, et al. (2018). Epigenetic regulation of brain region-specific microglia clearance activity. Nat Neurosci 21, 1049–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker AM, Huang W, Wang XM, Jansen M, Ma XJ, Kim J, Anderson CM, Wu X, Pan L, Su N, et al. (2017). Robust RNA-based in situ mutation detection delineates colorectal cancer subclonal evolution. Nat Commun 8, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. (2006). CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 7, R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cembrowski MS, Phillips MG, DiLisio SF, Shields BC, Winnubst J, Chandrashekar J, Bas E, and Spruston N (2018). Dissociable Structural and Functional Hippocampal Outputs via Distinct Subiculum Cell Classes. Cell 173, 1280–1292 e1218. [DOI] [PubMed] [Google Scholar]
- De Sa Nogueira D, Merienne K, and Befort K (2018). Neuroepigenetics and addictive behaviors: Where do we stand? Neurosci Biobehav Rev. [DOI] [PubMed] [Google Scholar]
- Del Castillo P, Llorente AR, and Stockert JC (1989). Influence of fixation, exciting light and section thickness on the primary fluorescence of samples for microfluorometric analysis. Basic Appl Histochem 33, 251–257. [PubMed] [Google Scholar]
- Dowson JH (1982). The evaluation of autofluorescence emission spectra derived from neuronal lipopigment. J Microsc 128, 261–270. [DOI] [PubMed] [Google Scholar]
- Erben L, He MX, Laeremans A, Park E, and Buonanno A (2018). A Novel Ultrasensitive In Situ Hybridization Approach to Detect Short Sequences and Splice Variants with Cellular Resolution. Mol Neurobiol 55, 6169–6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerfen CR (2003). Basic neuroanatomical methods. Curr Protoc Neurosci Chapter 1, Unit 1 1. [DOI] [PubMed] [Google Scholar]
- Guillaumet-Adkins A, Yanez Y, Peris-Diaz MD, Calabria I, Palanca-Ballester C, and Sandoval J (2017). Epigenetics and Oxidative Stress in Aging. Oxid Med Cell Longev 2017, 9175806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Zhao Y, Nguyen H, Liu T, Wang Z, and Lou H (2018). Quantitative Analysis of Alternative Pre-mRNA Splicing in Mouse Brain Sections Using RNA In Situ Hybridization Assay. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haring M, Zeisel A, Hochgerner H, Rinwa P, Jakobsson JET, Lonnerberg P, La Manno G, Sharma N, Borgius L, Kiehn O, et al. (2018). Neuronal atlas of the dorsal horn defines its architecture and links sensory input to transcriptional cell types. Nat Neurosci 21, 869–880. [DOI] [PubMed] [Google Scholar]
- Holt CE, and Schuman EM (2013). The central dogma decentralized: new perspectives on RNA function and local translation in neurons. Neuron 80, 648–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MB, and Walsh CA (2017). Cerebral cortical neuron diversity and development at single-cell resolution. Curr Opin Neurobiol 42, 9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Illenberger JM, McLaurin KA, Mactutus CF, and Booze RM (2018). Identification of Dopamine D1-Alpha Receptor Within Rodent Nucleus Accumbens by an Innovative RNA In Situ Detection Technology. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo PK, Smith DS, Perrone-Bizzozero N, and Twiss JL (2018). Axonal mRNA transport and translation at a glance. J Cell Sci 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell SA, Staines WA, and Wessendorf MW (1999). Reduction of lipofuscin-like autofluorescence in fluorescently labeled tissue. J Histochem Cytochem 47, 719–730. [DOI] [PubMed] [Google Scholar]
- Shrestha BR, Chia C, Wu L, Kujawa SG, Liberman MC, and Goodrich LV (2018). Sensory Neuron Diversity in the Inner Ear Is Shaped by Activity. Cell 174, 1229–1246 e1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirzewski M, Karavanova I, Shamir A, Erben L, Garcia-Olivares J, Shin JH, Vullhorst D, Alvarez VA, Amara SG, and Buonanno A (2017). ErbB4 signaling in dopaminergic axonal projections increases extracellular dopamine levels and regulates spatial/working memory behaviors. Mol Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, and Yang L (2018). Long noncoding RNA VPS9D1-AS1 overexpression predicts a poor prognosis in non-small cell lung cancer. Biomed Pharmacother 106, 1600–1606. [DOI] [PubMed] [Google Scholar]
- Tidcombe H, Jackson-Fisher A, Mathers K, Stern DF, Gassmann M, and Golding JP (2003). Neural and mammary gland defects in ErbB4 knockout mice genetically rescued from embryonic lethality. Proc Natl Acad Sci U S A 100, 8281–8286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, Wu X, Vo HT, Ma XJ, and Luo Y (2012). RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn 14, 22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young WS, Song J, and Mezey E (2018). Hybridization Histochemistry of Neural Transcripts. Curr Protoc Neurosci 82, 1 3 1–1 3 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller R (2001). Fixation, embedding, and sectioning of tissues, embryos, and single cells. Curr Protoc Pharmacol Appendix 3, 3D. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Sharp A, Anderson CM, Silberstein JL, Taylor M, Lu C, Zhao P, De Marzo AM, Antonarakis ES, Wang M, et al. (2018). Novel Junction-specific and Quantifiable In Situ Detection of AR-V7 and its Clinical Correlates in Metastatic Castration-resistant Prostate Cancer. Eur Urol 73, 727–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
KEY REFERENCES
- Wang et al. (2012). See above. This publication was first introducing the RNAscope multiplex fluorescent in situ hybridization assay and describes in detail the technology and principles of RNAscope.
- Baker et al. (2017), Erben et al. (2018), Zhu et al. (2018). See above. These publications were first to implement the BaseScope in situ hybridization assay to detect splice variants and single nucleotide polymorphisms; and describe its principles as well as validate its sensitivity and specificity.