Abstract
Mounting literatures have revealed the crucial effects of long noncoding RNA (lncRNA) in various cancers, including glioma. HNF1A‐AS1, a novel lncRNA, is reported to modulate tumorigenesis and development of multiple cancers. However, the tumorigenic function of lncRNA HNF1A‐AS1 in glioma remains largely unknown. quantitative reverse transcription and polymerase chain reaction and western blot assays were applied to evaluate the expression of relevant mRNAs and proteins. 5‐Ethynyl‐2’‐ deoxyuridine, terminal deoxynucleotidyl transferase dUTP nick‐end labeling, flow cytometry, and transwell assays were conducted for examining the influence of HNF1A‐AS1 on glioma cell functions. The relationship among RNAs was investigated by mechanical experiments. The results demonstrated that HNF1A‐AS1 was predominantly highly expressed in glioma cell lines compared with nontumor glial epithelial cell, which was associated with the stimulation of transcription factor myelocytomatosis oncogene. Knockdown of HNF1A‐AS1 remarkably inhibited glioma cells proliferation, migration, and invasion, while accelerating cell apoptosis in vitro. Mechanically, HNF1A‐AS1 served as a miR‐32‐5p sponge. Moreover, SOX4 was discovered as a target of miR‐32‐5p. Inhibited miR‐32‐5p or upregulated SOX4 could markedly counteract the inhibitory effects of silencing HNF1A‐AS1 on glioma malignant biological behaviors. HNF1A‐AS1 exerted oncogenic property in glioma progression via upregulating miR‐32‐5p–mediated SOX4 expression, suggesting potential novel therapeutic target for future glioma treatment.
Keywords: glioma, HNF1A‐AS1, miR‐32‐5p, SOX4
Silencing HNF1A‐AS1 dampens glioma cell proliferation, migration, and invasion, while facilitating apoptosis. Myelocytomatosis oncogene induces overexpression of HNF1A‐AS1 via transcription activation. HNF1A‐AS1 directly binds to miR‐32‐5p and miR‐32‐5p inhibition could reverse the restraining effects of HNF1A‐AS1 knockdown on glioma.
1. INTRODUCTION
Glioma is the most common intracranial malignant tumor and is one of the leading causes of cancer deaths. 1 The occurrence of brain tumors is 21/100 000, which is much lower than other type of cancers, occupying only approximately 2% of all human cancers. However, the morbidity of glioma accounts for almost 60% among all kinds of brain tumors. 2
Accumulating evidence highlighted the importance of abundantly transcribed noncoding transcripts, among which long noncoding RNAs (lncRNAs) emerged as critical mediator in many processes of cell biology. Recently, HNF1A‐AS1, as a novel identified lncRNA, gained much attention. HNF1A‐AS1 is correlated with tumorigenic functions including cancer cell proliferation, migration, and invasion. For instance, HNF1A‐AS1 was revealed to facilitate colon cancer metastatic progression via regulating miR‐34a/SIRT1/p53 feedback loop. 3 Besides, overexpressed HNF1A‐AS1 activates the Wnt/β‐catenin signaling pathway to promote colorectal cancer carcinogenesis. 4
Substantial studies manifested that lncRNAs can function as competing endogenous RNAs (ceRNAs) in regulating the protein production of mRNA via binding to shared miRNAs post‐transcriptionally. 5 This discovery deepened the understanding of human genome and highlighted the important gene regulatory role of lncRNAs, which were previously thought as “transcription noise”. 6 Long noncoding RNAs could mediate various cellular biological functions including cell proliferation, apoptosis, migration, and invasion in pathology evolvement. 7 , 8 Increasing studies revealed the implication of lncRNAs in progression of brain tumors. 9 , 10 To date, the molecular relationship between lncRNA and glioma onset and progression has been unveiled. For instance, LINC00909 serves as a ceRNA to positively mediate the expression of MUC1‐C via binding to miR‐194, and consequently promotes glioma cell migration and invasion. 11 Long noncoding RNA UBE2R2‐AS1 serves as a ceRNA against miR‐877‐3p to upregulate TLR4, and inhibits the malignant phenotype of glioblastoma cells. 12 These studies indicated that lncRNAs are essential for glioma progression. Nevertheless, as a novel lncRNA, the biological role and molecular mechanisms of HNF1A‐AS1 in glioma has not been explored yet. This study aimed to reveal the underlying molecular mechanism of HNF1A‐AS1 in glioma progression.
2. MATERIALS AND METHODS
2.1. Tissue samples
In total, 35 glioma tissues and 10 normal tissues (excised from patients with other brain diseases) were obtained from Gaozhou People's Hospital with the approval from the Ethics Committee of Gaozhou People's Hospital. Each participant has signed informed consent. No patients have received chemo‐ or radiotherapy before surgery. After surgical resection, fresh tissues were instantly frozen in liquid nitrogen and stored at −80°C.
2.2. Cell lines
Human glioma cell lines (LN229, A172, SHG‐44, and U87) and normal brain glial cell line (HEB) were both procured from American Type Culture Collection (ATCC; Manassas, VA) and cultured under 95% air, 5% CO2 at 37°C. Dulbecco's Modified Eagle's medium (Invitrogen) adding 10% fetal bovine serum were applied for cell culture.
2.3. Quantitative reverse transcription and polymerase chain reaction (qRT‐PCR)
Total RNA from A172 and U87 cells were extracted by TRIzol reagent from Invitrogen in line with the guidebook. A Reverse Transcription Kit (Toyobo) was applied for synthesizing cDNA for conducting qRT‐PCR with SYBR Green Super Mix (Bio‐Rad). All RNA levels were measured via 2−ΔΔCT method, with U6 or glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) as the internal reference.
2.4. Cell transfection
Cells of A172 and U87 were seeded in the 24‐well plates for 48 hours of transfection with Lipofectamine 2000 (Invitrogen). The duplex sh‐RNAs of HNF1A‐AS1 (sh‐HNF1A‐AS1#1/2) and negative control (NC; termed short hairpin‐negative control [sh‐NC]), pcDNA3.1/myelocytomatosis oncogene (MYC), pcDNA3.1/SOX4, and NC (pcDNA3.1), as well as the miR‐32‐5p mimics and NC mimics, miR‐32‐5p inhibitor and NC inhibitor, all these were designed and synthesized by RiboBio.
2.5. 5‐Ethynyl‐2’‐ deoxyuridine (EdU) staining
A172 and U87 cells on sterile coverslips were transfected and plated in the 24‐well plates for treatment with the EdU incorporation assay kit (Ribobio). Cell nucleus was counterstained with EdU and DAPI (Beyotime), and visualized with laser confocal microscopy (Olympus).
2.6. Terminal deoxynucleotidyl transferase dUTP nick‐end labeling assay
Glioma cells after transfection were cultured with the terminal deoxynucleotidyl transferase dUTP nick‐end labeling (TUNEL) Apoptosis Assay Kit (Beyotime) in light of the user guide. After treating with DAPI, apoptotic cells were analyzed under confocal microscopy.
2.7. Flow cytometry of apoptosis
70% cold ethanol on ice was used to fix transfected glioma cells for 1 hour. Then, cells were treated with Annexin V‐APC and 7‐AAD staining in the dark. A flow cytometer (BD Biosciences) was used to assess cell apoptosis.
2.8. Transwell assay
Transfected cells were reaped and planted into the upper chamber of transwell inserts (Corning Incorporated) coating with or without Matrigel membrane (BD Biosciences) for invasion or migration analysis. After 48 hours, the invaded and migrated cells were stained in crystal violet. Then, a microscope (magnification, ×200) was used to count the cells.
2.9. Chromatin immunoprecipitation assay
Cells of A172 and U87 were fixed for 10 minutes to generate DNA‐protein cross‐links. After shearing into 200‐ to 1000‐bp chromatin fragments, cell lysates were immunoprecipitated with anti‐MYC or normal control anti‐immunoglobulin G (IgG) antibody (Millipore). The retrieved chromatin DNA by beads was subjected to qRT‐PCR analysis.
2.10. Dual‐luciferase reporter assays
HEK‐293T cells (ATCC) in the 24‐well plates were co‐transfected with the pGL3 vector containing HNF1A‐AS1 promoter, pRL‐TK‐Renilla plasmid (Promega Corporation), and pcDNA3.1/MYC or pcDNA3.1. Besides, the wild‐type (WT) or mutant (MUT) reporter constructs of HNF1A‐AS1 and SOX4 were generated by pmirGLO vectors and co‐transfected into HEK‐293T cells with miR‐32‐5p mimics and NC mimics. The Dual‐Luciferase Reporter Assay System (Promega) was employed to evaluate luciferase intensity after 48 hours of transfection.
2.11. Subcellular fractionation
A PARIS™ Kit (Invitrogen) was applied for isolating nuclear and cytoplasmic RNAs in 1 × 106 glioma cells, which were previously rinsed in precooled PBS. The isolated HNF1A‐AS1 was analyzed via qRT‐PCR. U6 and GAPDH, respectively, served as the nuclear and cytoplasmic controls.
2.12. Fluorescence in situ hybridization assay
After fixation, the glioma cells were collected for incubation with RNA Fluorescence in situ hybridization (FISH) probe for HNF1A‐AS1 (RiboBio) in the hybridization buffer. After slides were cultured with DAPI solution, cells were observed under a Olympus microscope.
2.13. RNA pull down
The protein extracts from glioma cells were obtained for mixing with biotin‐labeled HNF1A‐AS1 (Biotin HNF1A‐AS1 WT/MUT) and Biotin NC, as well as the beads for 1 hour. qRT‐PCR was followed for analyzing the mixture of pull‐down.
2.14. RNA immunoprecipitation assay
1 × 107 glioma cells in RNA immunoprecipitation lysis buffer were collected and immunoprecipitated with anti‐Ago2 or normal mouse control anti‐IgG antibody (Millipore). Finally, the purified RNA was analyzed by qRT‐PCR.
2.15. Western blot
The cellular protein samples from glioma cells were prepared for separation with SDS‐PAGE (10%) and electrotransferred to PVDF membranes. After culturing with 5% skimmed milk, membranes were incubated with primary antibodies (Abcam) against SOX4 and control GAPDH all night, then with horseradish peroxidase‐tagged secondary antibody for 2 hours. The band intensity was determined by enhanced chemiluminescence reagent (Santa Cruz Biotechnology).
2.16. Animal study
Six‐week‐old male nude mice was maintained in the SPF‐grade animal laboratory, acquired from the National Laboratory Animal Center (Beijing, China). All procedures were approved by the Animal Research Ethics Committee of Gaozhou People's Hospital. Animal study was achieved through subcutaneous inoculation of 5 × 106 A172 cells to mice for 4 weeks. Tumor volume was recorded every 4 days. After killing mice, tumors were excised and weighed for analysis.
2.17. Statistical analysis
All data were analyzed statistically via a t test or one‐way ANOVA by using PRISM 6 (GraphPad), with P < .05 as significant level. Results were given as the mean ± SD of three or more repetitive assays.
3. RESULTS
3.1. Silencing HNF1A‐AS1 dampens glioma cell proliferation, migration, and invasion, while facilitating apoptosis
To understand the expression status of HNF1A‐AS1 in glioma, we performed qRT‐PCR. It was revealed that HNF1A‐AS1 was significantly upregulated in glioma tissues than in normal tissues (Figure S1A). We also noticed that HNF1A‐AS1 was significantly overexpressed in glioma cells (LN229, A172, SHG‐44, and U87) in contrast to normal brain glial cell line (HEB) (Figure 1A). We also noticed that A172 and U87 cells contained the most significant upregulation of HNF1A‐AS1; thus they were used for following assays. For loss‐of‐function assays, we utilized two sh‐RNAs directly targeting HNF1A‐AS1 and verified the inhibitory efficiency through qRT‐PCR (Figure 1B). The EdU assay showed that silencing HNF1A‐AS1 significantly inhibited cell proliferation, as illustrated in Figure 1C. The TUNEL assay indicated that HNF1A‐AS1 knockdown could markedly promote glioma cell apoptosis (Figure 1D). Flow cytometry data showed notably elevated cell apoptosis after silencing HNF1A‐AS1, which further confirmed the pro‐apoptosis effects of HNF1A‐AS1 knockdown (Figure 1E). Besides, silencing HNF1A‐AS1 could significantly restrain cell migration and invasion, as shown in transwell assays (Figure 1F,G). Together, HNF1A‐AS1 played an oncogenic role in glioma progression.
FIGURE 1.
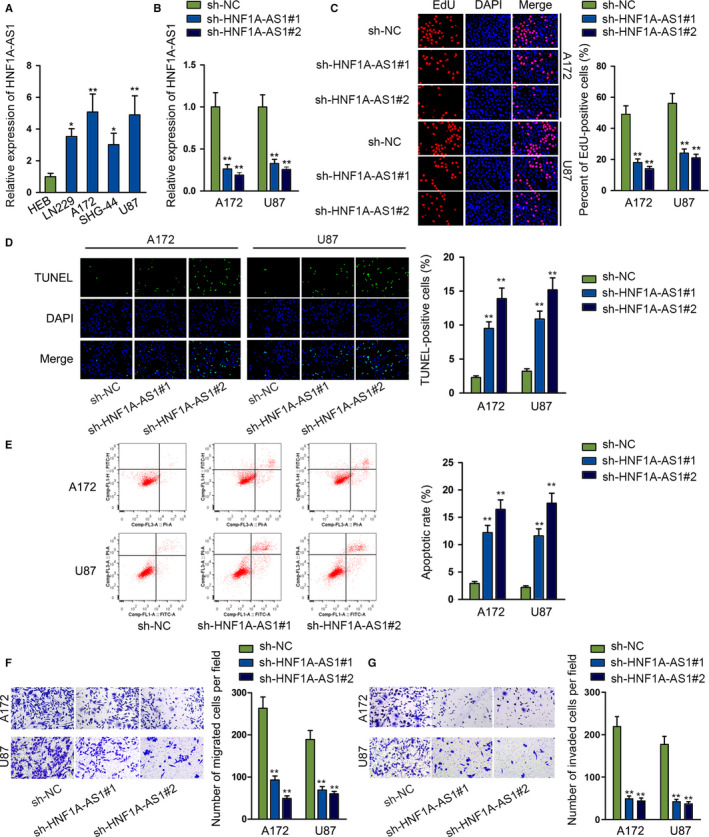
Silencing HNF1A‐AS1 dampens glioma cells proliferation, migration, and invasion, while facilitating apoptosis. A, qRT‐PCR analysis was used to determine the expression profile of HNF1A‐AS1 in glioma cell lines and normal brain glial cell. B, The transfection efficiency of sh‐HNF1A‐AS1#1 and sh‐HNF1A‐AS1#2 were detected by qRT‐PCR analysis. C, EdU assay was performed to assess glioma cells proliferation ability. D and E, TUNEL and flow cytometry assays were performed to determine cell apoptosis. F and G, Transwell migration and invasion assays were conducted to assess glioma cells migration and invasion ability. TUNEL, terminal deoxynucleotidyl transferase dUTP nick‐end labeling. *P < .05, **P < .01
3.2. MYC induces overexpression of HNF1A‐AS1 via transcription activation
We hypothesized that transcription factor might be associated with HNF1A‐AS1 overexpression in glioma cells. MYC might bind to HNF1A‐AS1 promoter region by prediction of UCSC (http://genome.ucsc.edu/) and Jaspar (http://jaspardev.genereg.net/) database jointly. The influence of MYC on HNF1A‐AS1 expression was subsequently detected. We observed that pcDNA3.1/MYC could force the overexpression of MYC efficiently (Figure 2A). We noticed that HNF1A‐AS1 expression was elevated after overexpressing MYC (Figure 2B). With the aid of Jaspar, we detected the putative MYC motif in human HNF1A‐AS1 promoter region, as shown on Figure 2C. Then, we found five concrete putative MYC binding sequences in HNF1A‐AS1 promoter by Jaspar. We divided the promoter region into five sectional area from −2500 bp according to putative binding sites predicted by Jaspar (Figure 2D). Then, the binding capacity of MYC to HNF1A‐AS1 promoter was investigated by chromatin immunoprecipitation (ChIP) assay. Its findings depicted that MYC bound to P5 section (Figure 2E). Moreover, we constructed pGL3 luciferase vector containing HNF1A‐AS1 full promoter region (P FL) and HNF1A‐AS1 promoter P5 deleted region (P D) (Figure 2F). Deletion of P5 abrogated the increased promoter enrichment in anti‐MYC, indicating that P5 fragment in HNF1A‐AS1 promoter region was indispensable for the interaction of HNF1A‐AS1 promoter with MYC (Figure 2G). This phenomenon was further elucidated by luciferase reporter assay on HEK‐293T cell. It indicated that pcDNA3.1/MYC elevated the promoter activity of pGL3‐FL, not that of pGL3‐D (Figure 2H). These data represented that MYC interacted with HNF1A‐AS1 promoter at approximately −2500 to −1900 bp upstream transcription start site (TSS). We found that there was only one specific binding site (−2237 to −2226) predicted by Jaspar in P5 fragment. Therefore, we intended to verify whether MYC bound to these sequences in P5 segment. We constructed P5‐WT and P5‐MUT (which mutated −2237 to −2226 sequence) accordingly (Figure 2I). Promoter activity of P5‐WT was strengthened after overexpressing MYC, while P5‐MUT, which lacked the sequence form −2237 to −2226, showed no alternation in luciferase activity after co‐transfection with pcDNA3.1/MYC (Figure 2J). Collectively, these data provided substantial proof for the activation of HNF1A‐AS1 transcription activity by MYC in glioma cells.
FIGURE 2.
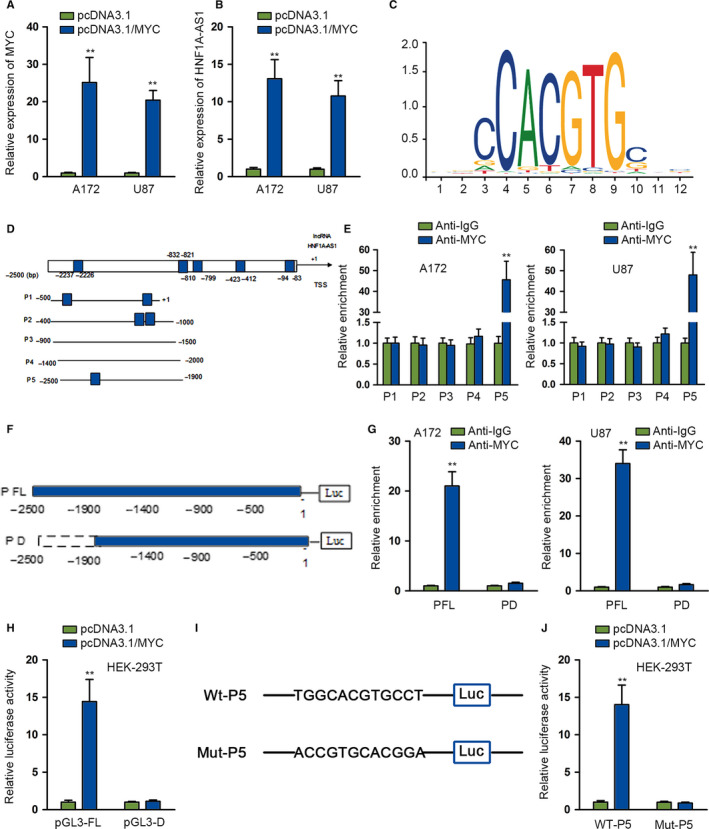
MYC induces overexpression of HNF1A‐AS1 via transcription activation. The expression of MYC was determined by quantitative RT‐PCR (qRT‐PCR) analysis after transfecting with pcDNA3.1 and pcDNA3.1/MYC plasmids. B, qRT‐PCR was performed to study the effects of overexpressing MYC on HNF1A‐AS1 expression. C, The putative MYC binding motif in human HNF1A‐AS1 promoter. D, HNF1A‐AS1 promoter was divided into four sections according to potential MYC binding sites. E, Chromatin immunoprecipitation (ChIP) assay using antibody targeting MYC and IgG was conducted to detect the affinity of MYC with HNF1A‐AS1 promoter. F, The full HNF1A‐AS1 promoter (P FL) and HNF1A‐AS1 P5 deleted (P D) were subcloned into pGL3 luciferase reporter vector. G, ChIP assay was used to investigate the relative enrichment of HNF1A‐AS1 promoter. H, Luciferase reporter assay was performed in HEK‐293T. I, pGL3‐WT‐P5, pGL3‐MUT‐P5 luciferase vectors were constructed accordingly. J, HEK‐293T was co‐transfected with pcDNA3.1 and pcDNA3.1/MYC and a luciferase reporter was performed between WT‐P5 and MUT‐P5. **P < .01
3.3. HNF1A‐AS1 directly binds to miR‐32‐5p and miR‐32‐5p inhibition could reverse the restraining effects of HNF1A‐AS1 knockdown on glioma
Many cytoplasmic lncRNAs have been found as ceRNAs in the carcinogenesis of cancers. HNF1A‐AS1 was found mainly situated in cytoplasm through subcellular fraction and FISH assays, as illustrated in Figure 3A,B. By browsing starBase tool, 13 eight miRNAs were found to interact with HNF1A‐AS1 (clip data: low stringency ≥ 1). Among them, miR‐32‐5p was upregulated significantly in A172 after silencing HNF1A‐AS1 compared with other miRNA candidates from qRT‐PCR results (Figure 3C). Subsequently, we detected an abnormal low expression status of miR‐32‐5p in glioma cells (Figure 3D). By browsing starBase, putative miR‐32‐5p binding site with HNF1A‐AS1 was detected. We mutated the putative binding site sequence correspondingly, as shown in Figure 3E. Both HNF1A‐AS1‐WT and HNF1A‐AS1‐MUT were subcloned into pmirGLO luciferase reporter vector. After co‐transfecting miR‐32‐5p mimics with pmirGLO vector containing HNF1A‐AS1‐WT or HNF1A‐AS1‐MUT, the luciferase activity of HNF1A‐AS1‐WT was markedly attenuated, instead of HNF1A‐AS1‐MUT in HEK‐293T, as illustrated in Figure 3F. MiR‐32‐5p was only significantly pulled down by Bio‐HNF1A‐AS1‐WT group, which verified the interaction between HNF1A‐AS1 and miR‐32‐5p (Figure 3G). HNF1A‐AS1 bound with miR‐32‐5p and negatively modulated the expression of miR‐32‐5p in glioma cells.
FIGURE 3.
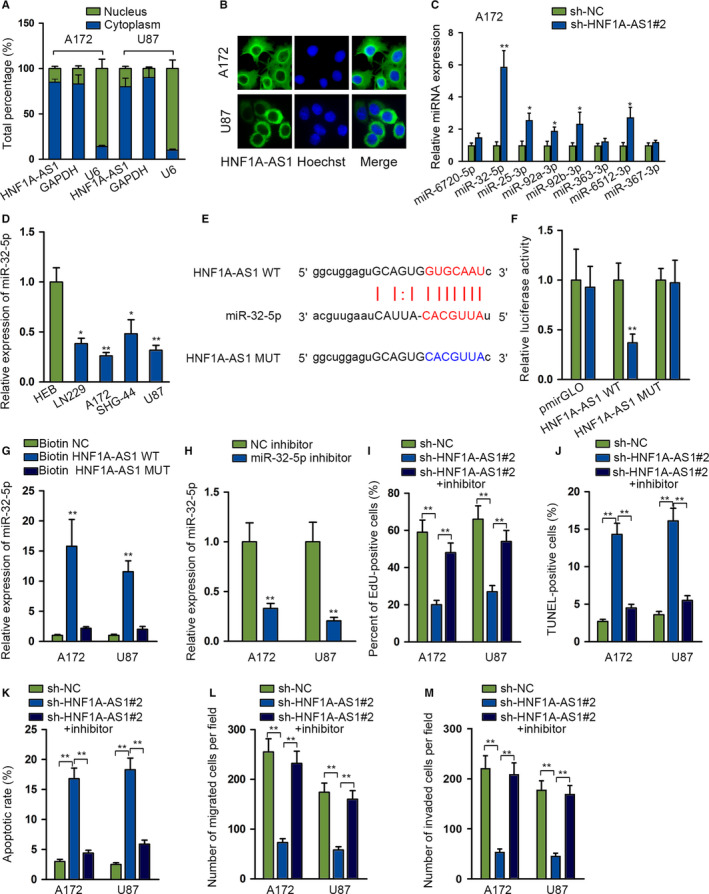
HNF1A‐AS1 directly binds to miR‐32‐5p and miR‐32‐5p inhibition could reverse the restraining effects of HNF1A‐AS1 knockdown on glioma. A and B, Subcellular fractionation and FISH assays were performed to determine the location of HNF1A‐AS1. C, Quantitative RT‐PCR (qRT‐PCR) analysis was performed to determine miRNAs expression after HNF1A‐AS1 depletion. D, The expression status of miR‐32‐5p was detected in glioma cell lines and normal brain glial cell lines. E, Putative wild and mutant miR‐32‐5p binding sites with HNF1A‐AS1. F, Luciferase reporters were performed in HEK‐293T. MiR‐32‐5p mimics could impair the luciferase activity of HNF1A‐AS1‐WT. G, RNA pull down showed enriched miR‐32‐5p by biotin‐labeled HNF1A‐AS1‐WT. H, qRT‐PCR was used to examine the transfection efficiency of miR‐32‐5p inhibitor. I‐M, Functional experiments were conducted in A172 and U87 cell lines to examine the rescue effects of silenced miR‐32‐5p on silenced HNF1A‐AS1 in cell proliferation, apoptosis, migration, and invasion. *P < .05, **P < .01
After that, we carried out rescue assays to determine whether HNF1A‐AS1 contributed to glioma progression via inhibiting miR‐32‐5p. MiR‐32‐5p expression was silenced via transfection of plasmids containing miR‐32‐5p inhibitor into A172 and U87 cell lines (Figure 3H). MiR‐32‐5p inhibition obviously restored the oncogenic effects of HNF1A‐AS1 in glioma cell proliferation (Figure 3I), apoptosis (Figure 3J,K; Figure S1B), migration (Figure 3L; Figure S1C), and invasion (Figure 3M; Figure S1D). These results elucidated that HNF1A‐AS1 mediated glioma development via downregulating miR‐32‐5p.
3.4. SOX4 is a direct target gene of miR‐32‐5p
We aimed to find mRNA targets of miR‐32‐5p to better understand its role in glioma. Aided by starBase, we screened three target genes (SOX4, ZDHHC5, ZFHX3) according to below circumstance (clip data: strict stringency ≥ 5; degradome data: high stringency ≥ 3; program number: five; predicted program: microT, miRanda, miRmap, PITA, PicTar). We found that SOX4 was significantly downregulated in the presence of miR‐32‐5p mimics compared with the other two mRNA candidates (Figure 4A). From qRT‐PCR results, we found a significant elevation of SOX4 in four glioma cells (Figure 4B). This expression status is contrary to that of miR‐32‐5p in glioma cells, thus we chose SOX4 to study. The putative miR‐32‐5p binding site in the 3′‐untranslated region (3ʹUTR) sequence of SOX4 (Figure 4C) was verified by the results of dual luciferase reporter assays, which showed significant decreased luciferase activity only in SOX4‐WT by miR‐32‐5p upregulation (Figure 4D). RNA immunoprecipitation assay data showed significant enriched miR‐32‐5p, HNF1A‐AS1, SOX4 in the anti‐Ago2 group instead of the IgG group (Figure 4E). The expression of SOX4 was examined after silencing HNF1A‐AS1 and then inhibiting miR‐32‐5p in A172 and U87. It was manifested that SOX4 was downregulated after silencing HNF1A‐AS1, yet increased again in the presence of miR‐32‐5p inhibition (Figure 4F). Together, these results confirmed the ceRNA role of HNF1A‐AS1 in glioma via regulating miR‐32‐5p/SOX4 axis.
FIGURE 4.
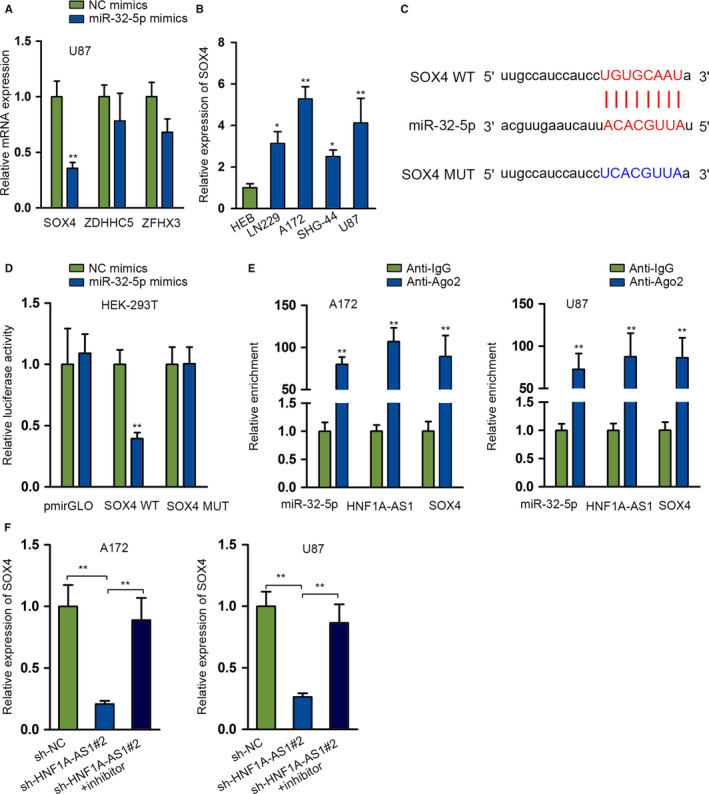
SOX4 is a direct target gene of miR‐32‐5p. A, The expression of potential target genes was detected by quantitative RT‐PCR (qRT‐PCR) after transfecting miR‐32‐5p mimics. B, The expression profile of SOX4 in cell lines was detected by qRT‐PCR. C, Putative wild and mutant miR‐32‐5p binding sites in 3ʹUTR sequence of SOX4 based on starBase data. D. Luciferase reporter assays were performed to investigate the association between miR‐32‐5p and SOX4. E, RIP was used performed to verify the mechanical relationship among miR‐32‐5p, HNF1A‐AS1 and SOX4. F. The effects of silenced miR‐32‐5p on silenced HNF1A‐AS1 in SOX4 expression were studied by qRT‐PCR. *P < .05, **P < .01
3.5. SOX4 enrichment restores the carcinogenesis role of HNF1A‐AS1
To further determine whether HNF1A‐AS1 facilitated glioma via acting as a ceRNA in regulating miR‐32‐5p/SOX4 network, we performed rescue experiments functionally. First, we validated the overexpression efficiency of SOX4 (Figure 5A). Next, we co‐transfected pcDNA3.1/SOX4 into HNF1A‐AS1–depleted A172 and U87 cell lines. The mRNA and protein levels of SOX4 were inhibited after silencing HNF1A‐AS1, while recovered again after co‐transfecting with pcDNA3.1/SOX4 form qRT‐PCR and western blot assays (Figure 5B,C). Based on the findings of functional rescue experiments, SOX4 upregulation could reverse HNF1A‐AS1 silencing produced biological effects on cell proliferation (Figure 5D), apoptosis (Figure 5E,F; Figure S1E), migration (Figure 5G; Figure S1F), and invasion (Figure 5H; Figure S1G). Furthermore, we constructed xenograft mice models by inoculating A172 cell lines transfected with sh‐NC, sh‐HNF1A‐AS1#2, and sh‐HNF1A‐AS1#2 + pcDNA3.1/SOX4. We found that silencing HNF1A‐AS1 could suppress tumor growth, but this effect was reversed by overexpressing SOX4 (Figure 5I; Figure S1H). Tumor volume and weight were reduced by silenced HNF1A‐AS1 while co‐transfection of SOX4 rescued the suppressive influence of silenced HNF1A‐AS1 on tumor volume and weight (Figure 5J,K). To sum up, HNF1A‐AS1 contributed to glioma progression by targeting miR‐32‐5p/SOX4 axis.
FIGURE 5.
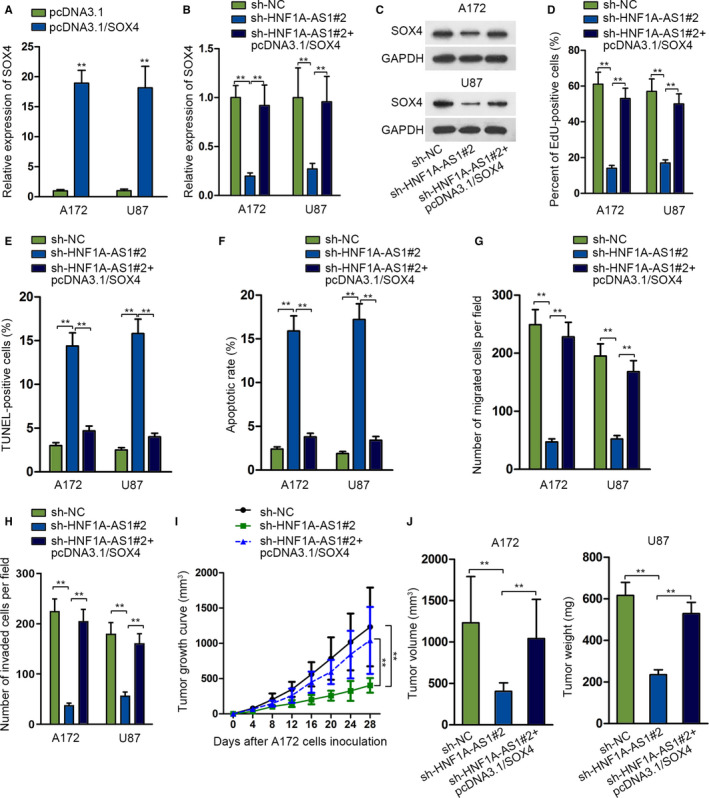
SOX4 enrichment restores the carcinogenesis role of HNF1A‐AS1. A, The expression of SOX4 after transfecting with pcDNA3.1/SOX4 was studied through quantitative RT‐PCR (qRT‐PCR) assay. B‐C, qRT‐PCR and western blot assays were used to detect the expression of SOX4 mRNA and protein expression level. D‐H, Rescue functional experiments were conducted among sh‐NC, sh‐HNF1A‐AS1#2, and sh‐HNF1A‐AS1#2 + pcDNA3.1/SOX4 groups in A172 and U87 cells. I‐K, Xenograft tumor growth curve, volume, and weight were compared among sh‐NC, sh‐HNF1A‐AS1#2, and sh‐HNF1A‐AS1#2 + pcDNA3.1/SOX4 groups. **P < .01
4. DISCUSSION
Glioma is one of the most common types of intracranial tumors which initiate from the central nervous system. The prognosis of patients with high‐grade glioma is much worse than those with low‐grade glioma. 14 Substantial studies supported the crucial regulator roles of lncRNAs in the initiation, development, and progression of various diseases, including tumors. 15 , 16 , 17 HNF1A‐AS1 has been reported to exhibit carcinogenesis property in gastric cancer 18 and non–small cell lung cancer. 19 In the present study, we found the aberrant overexpression of HNF1A‐AS1 in glioma cell lines, which was in accordance with HNF1A‐AS1 overexpression in urothelial carcinoma of the bladder. 20 We noticed that the overexpression of HNF1A‐AS1 was activated by transcription factor MYC. Previously, MYC amplification has been discovered to promote homologous recombination via targeting CDK18 in glioblastoma. 21 We found that HNF1A‐AS1 knockdown could suppress glioma cells proliferation, migration, and invasion abilities, while markedly enhance apoptosis capacity, which revealed the crucial oncogenic role of HNF1A‐AS1 in glioma progression. These findings accord with the function of HNF1A‐AS1 in oral squamous cell carcinoma. 22
Over the last decade, the ceRNA crosstalk is prevalent. Numerous reports have found that a large number of lncRNAs were involved in the ceRNA network. 23 LncRNAs could mediate the protein production of genes via acting as ceRNA according to substantial documents. Here, we detected the cytoplasmic localization of HNF1A‐AS1 through subcellular fractionation and FISH assays. We screened downstream combinable miRNAs with HNF1A‐AS1 and selected miR‐32‐5p through multiple channel. Inhibited miR‐32‐5p counteracted the curbing influence of inhibited HNF1A‐AS1 in glioma progression. We initially uncovered that HNF1A‐AS1 bound with miR‐32‐5p and decreased its expression in glioma cells. MiR‐32‐5p also presents low expression and involve in the ceRNA axis of SNHG14/miR‐32‐5p/SKIL in colorectal cancer. 24 MiR‐32‐5p targets HOXB8 to repress the cellular malignant behavior in cervical cancer cells. 25 LncRNA GAS5 modulates miR‐32‐5p/PTEN axis to suppress pancreatic cancer metastasis. 26
Subsequently, we screened SOX4 as the most potential target gene of miR‐32‐5p with putative binding site in its 3ʹUTR sequences. SOX4 has been reported to be an oncogene supported by multiple lines of evidence. SOX4 gene has been reported to be frequently amplified and upregulated in over 20 types of malignant tumors. 27 , 28 In the field of glioma, SOX4 has been found to be significantly elevated in glioblastoma multiforme. 29 Herein, we also detected the abnormal upregulation of SOX4 in glioma cells in comparison to normal control cell, which was the same expression profile previously revealed in medulloblastoma. 30 We clarified the oncogenic role of SXO2 in glioma via a collection of functional experiments. These data showed that SOX4 overexpression could dampen cells proliferation, migration, and invasion, while stimulating apoptosis in glioma cells. Last but not least, SOX4 overexpression could significantly reduce the biological effects induced by silencing HNF1A‐AS1 in glioma cells, which further confirmed the ceRNA role of SOX4.
All in all, we initially found that MYC induced HNF1A‐AS1 overexpression promoted glioma progression via modulating miR‐32‐5p/SOX4 axis, providing potent reference value for targeting HNF1A‐AS1 in future therapeutic strategies of glioma.
CONFLICTS OF INTERESTS
None.
AUTHORS' CONTRIBUTIONS
Jianheng Wu designed this study. Rong Li interpreted data. Linfan Li recorded data. Yimian Gu and Hui Zhan were responsible for preparation and investigaton. Jianheng Wu, Rong Li and Changbao Zhou devoted to data curation and methods. Chuanhong Zhong wrote the manuscript. All authors reviewed the manuscript.
ETHICAL APPROVAL
All procedures were approved by the Ethics Committee of Gaozhou People's Hospital.
Supporting information
Figure S1
ACKNOWLEDGMENT
We are very grateful to all individuals and groups involved in this study.
Wu J, Li R, Li L, et al. MYC-activated lncRNA HNF1A-AS1 overexpression facilitates glioma progression via cooperating with miR-32-5p/SOX4 axis. Cancer Med. 2020;9:6387–6398. 10.1002/cam4.3186
Funding information
Project Program of Neurosurgical Clinical Research Center of Sichuan Province; Science and Technology Foundation of Southwest Medical University (2017‐ZRQN‐180, 2017‐ZRQN‐110).
DATA AVAILABILITY STATEMENT
Research data and material are not shared.
REFERENCES
- 1. Fouladseresht H, Ziaee SM, Erfani N, Doroudchi M. Serum levels of APRIL increase in patients with glioma, meningioma and schwannoma. Asian Pac J Cancer Prev. 2019;20(3):751‐756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ahammed Muneer KV, Rajendran VR, Paul Joseph K. Glioma tumor grade identification using artificial intelligent techniques. J Med Syst. 2019;43(5):113. [DOI] [PubMed] [Google Scholar]
- 3. Fang C, Qiu S, Sun F, et al. Long non‐coding RNA HNF1A‐AS1 mediated repression of miR‐34a/SIRT1/p53 feedback loop promotes the metastatic progression of colon cancer by functioning as a competing endogenous RNA. Cancer Lett. 2017;410:50‐62. [DOI] [PubMed] [Google Scholar]
- 4. Zhang X, Xiong Y, Tang F, Bian Y, Chen Y, Zhang F. Long noncoding RNA HNF1A‐AS1 indicates a poor prognosis of colorectal cancer and promotes carcinogenesis via activation of the Wnt/beta‐catenin signaling pathway. Biomed Pharmacother. 2017;96:877‐883. [DOI] [PubMed] [Google Scholar]
- 5. Denzler R, Agarwal V, Stefano J, Bartel DP, Stoffel M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol Cell. 2014;54(5):766‐776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hu X, Hong Y, Shang C. Knockdown of long non‐coding RNA SNHG5 inhibits malignant cellular phenotypes of glioma via Wnt/CTNNB1 signaling pathway. J Cancer. 2019;10(5):1333‐1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang C, Qu Y, Xiao H, et al. LncRNA SNHG3 promotes clear cell renal cell carcinoma proliferation and migration by upregulating TOP2A. Exp Cell Res. 2019;384(1):111595. [DOI] [PubMed] [Google Scholar]
- 8. Zhang C, Liu J, Zhang Y, et al. LINC01210 accelerates proliferation, invasion and migration in ovarian cancer through epigenetically downregulating KLF4. Biomed Pharmacother. 2019;119:109431. [DOI] [PubMed] [Google Scholar]
- 9. Zhang Y, Yang G, Luo Y. Long non‐coding RNA PVT1 promotes glioma cell proliferation and invasion by targeting miR‐200a. Exp Ther Med. 2019;17(2):1337‐1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dong H, Cao W, Xue J. Long noncoding FOXD2‐AS1 is activated by CREB1 and promotes cell proliferation and metastasis in glioma by sponging miR‐185 through targeting AKT1. Biochem Biophys Res Comm. 2019;508(4):1074‐1081. [DOI] [PubMed] [Google Scholar]
- 11. Liu Z, Lu C, Hu H, et al. LINC00909 promotes tumor progression in human glioma through regulation of miR‐194/MUC1‐C axis. Biomed Pharmacother. 2019;116:108965. [DOI] [PubMed] [Google Scholar]
- 12. Xu W, Hu GQ, Da Costa C, et al. Long noncoding RNA UBE2R2‐AS1 promotes glioma cell apoptosis via targeting the miR‐877‐3p/TLR4 axis. Onco Targets Ther. 2019;12:3467‐3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA‐ceRNA, miRNA‐ncRNA and protein–RNA interaction networks from large‐scale CLIP‐Seq data. Nucleic Acids Res. 2014;42(D1):D92‐D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ordys BB, Launay S, Deighton RF, McCulloch J, Whittle IR. The role of mitochondria in glioma pathophysiology. Mol Neurobiol. 2010;42(1):64‐75. [DOI] [PubMed] [Google Scholar]
- 15. Liu Y, Yin L, Chen C, Zhang X, Wang S. Long non‐coding RNA GAS5 inhibits migration and invasion in gastric cancer via interacting with p53 protein. Dig Liver Dis. 2020;52(3):331‐338. [DOI] [PubMed] [Google Scholar]
- 16. Vafadar A, Shabaninejad Z, Movahedpour A, et al. Long non‐coding RNAs: epigenetic regulators in cancer. Curr Pharm Des. 2019;25(33):3563‐3577. [DOI] [PubMed] [Google Scholar]
- 17. Xu YH, Deng JL, Wang G, Zhu YS. Long non‐coding RNAs in prostate cancer: functional roles and clinical implications. Cancer Lett. 2019;464:37‐55. [DOI] [PubMed] [Google Scholar]
- 18. Liu HT, Liu S, Liu L, Ma RR, Gao P. EGR1‐mediated transcription of lncRNA‐HNF1A‐AS1 promotes cell‐cycle progression in gastric cancer. Can Res. 2018;78(20):5877‐5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu L, Chen Y, Li Q, Duan P. lncRNA HNF1A‐AS1 modulates non‐small cell lung cancer progression by targeting miR‐149‐5p/Cdk6. J Cell Biochem. 2019;120(11):18736‐18750. [DOI] [PubMed] [Google Scholar]
- 20. Wang YH, Liu YH, Ji YJ, Wei Q, Gao TB. Upregulation of long non‐coding RNA HNF1A‐AS1 is associated with poor prognosis in urothelial carcinoma of the bladder. Eur Rev Med Pharmacol Sci. 2018;22(8):2261‐2265. [DOI] [PubMed] [Google Scholar]
- 21. Ning J‐F, Stanciu M, Humphrey MR, et al. Myc targeted CDK18 promotes ATR and homologous recombination to mediate PARP inhibitor resistance in glioblastoma. Nat Commun. 2019;10(1):2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu Z, Li H, Fan S, Lin H, Lian W. STAT3‐induced upregulation of long noncoding RNA HNF1A‐AS1 promotes the progression of oral squamous cell carcinoma via activating Notch signaling pathway. Cancer Biol Ther. 2019;20(4):444‐453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gou L, Zou H, Li B. Long noncoding RNA MALAT1 knockdown inhibits progression of anaplastic thyroid carcinoma by regulating miR‐200a‐3p/FOXA1. Cancer Biol Ther. 2019;20(11):1355‐1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ye T, Zhang N, Wu W, et al. SNHG14 promotes the tumorigenesis and metastasis of colorectal cancer through miR‐32‐5p/SKIL axis. In Vitro Cell Dev Biol Anim. 2019;55(10):812‐820. [DOI] [PubMed] [Google Scholar]
- 25. Liu YJ, Zhou HG, Chen LH, et al. MiR‐32‐5p regulates the proliferation and metastasis of cervical cancer cells by targeting HOXB8. Eur Rev Med Pharmacol Sci. 2019;23(1):87‐95. [DOI] [PubMed] [Google Scholar]
- 26. Gao Z‐Q, Wang J‐F, Chen D‐H, et al. Long non‐coding RNA GAS5 suppresses pancreatic cancer metastasis through modulating miR‐32‐5p/PTEN axis. Cell Biosci. 2017;7:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moreno CS. SOX4: the unappreciated oncogene. Semin Cancer Biol. 2019;S1044‐579X(18):30145‐30147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ha Thi HT, Kim HY, Kim YM, Hong S. MicroRNA‐130a modulates a radiosensitivity of rectal cancer by targeting SOX4. Neoplasia (New York, NY). 2019;21(9):882‐892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lin B, Madan A, Yoon J‐G, et al. Massively parallel signature sequencing and bioinformatics analysis identifies upregulation of TGFBI and SOX4 in human glioblastoma. PLoS One. 2010;5(4):e10210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. de Bont, JM Kros JM, Passier MMCJ, et al. Differential expression and prognostic significance of SOX genes in pediatric medulloblastoma and ependymoma identified by microarray analysis. Neuro‐oncology. 2008;10(5):648‐660. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Data Availability Statement
Research data and material are not shared.


