Summary
SIRT7 is a member of the mammalian sirtuins and functions as an NAD+-dependent deacylase. Here we show that SIRT7 deficiency leads to a lowered histone acetyltransferase 1 (HAT1) activity and therefore decreased histone H4K5 and H4K12 acetylation. This in turn causes CENP-A dislocation at the centromere, which further affects chromatin assembly. SIRT7 ablation results in aneuploidy and aging phenotypes, including senescence and nucleolar expansion. Moreover, SIRT7 knockout mice are susceptible to DSS-induced colitis and alcohol-derived epithelial disturbance, revealing a disrupted intestinal epithelial homeostasis. Notably, absence of SIRT7 aggravates the susceptibility of colorectal cancer incidence in APCMin/+ mouse model and elicits further the Wnt signaling. Our findings indicate a tumor suppressive role of SIRT7 in the case of colorectal cancer. Together with the activities in maintaining genome integrity and intestinal homeostasis, activating SIRT7 may serve as a strategy to treat bowel diseases and colorectal cancer.
Subject Areas: Molecular Biology, Molecular Genetics, Cancer
Graphical Abstract
Highlights
-
•
SIRT7 deacetylates HAT1 further regulates CENP-A nucleosome assembly
-
•
SIRT7 preserves genome integrity and intestinal homeostasis
-
•
SIRT7-ablation leads to intestinal tumorigenesis
Molecular Biology; Molecular Genetics; Cancer
Introduction
Mammalian sirtuins SIRT1-7 were demonstrated to regulate several cellular and physiological functions such as cell survival, apoptosis, genomic stability, and metabolic activities via numerous pathways (Chalkiadaki and Guarente, 2015; Chang and Guarente, 2014; Imai and Guarente, 2014). Sirtuins are NAD+-dependent deacylases or ADP ribosyl-transferases that respond to NAD+ level and cellular energy status and further modulate biological processes in different cellular compartments through targeting histones, transcription factors, metabolic enzymes, and many protein substrates (Bonkowski and Sinclair, 2016; Finkel et al., 2009). SIRT7 is the only sirtuin that predominantly localizes in the nucleolus, and perhaps the least understood sirtuin member for its physiological roles. SIRT7 was shown to activate RNA polymerase I transcription of ribosomal DNA (Ford et al., 2006); this activity appears to favor ribosome biogenesis and cell growth. SIRT7 also regulates RNA polymerase II and III activities (Blank and Grummt, 2017; Tsai et al., 2014), collectively pointed out a decisive role of SIRT7 in sustaining transcription efficiencies in vitro. SIRT7 maintains metabolic homeostasis, and it was shown that SIRT7 overexpression could revert fatty liver disease, whereas SIRT7 deficiency led to hepatic steatosis (Yoshizawa et al., 2014). A related finding showed that SIRT7 deacetylates GABP-β1 in the liver, further proposed a mitochondrial link of explaining the liver pathology found in SIRT7 knockout mice (Ryu et al., 2014). SIRT7 was shown to associate with cancer incidence; however, the conclusion remains controversial. SIRT7 was demonstrated to deacetylate histone H3 at the K18 residue as the mechanism to support oncogenic transformation in a xenograft approach (Barber et al., 2012). Furthermore, SIRT7 was found to associate with human hepatocellular carcinoma via a mechanism related to MiR-125a-5p (Kim et al., 2013). On the other hand, SIRT7 has been found to repress the tumorigenic transcription factor, HIF-1 α (Hubbi et al., 2013). Consistent with the potential tumor suppressor function, SIRT7 was revealed to be down-regulated in head and neck squamous cell carcinoma in a case study (Lai et al., 2013). More recently, SIRT7 was validated to deacetylate and therefore facilitate the degradation of SMAD4; it further suppresses breast cancer cell metastasis (Tang et al., 2017).
Given this conflicting published landscape, we wished to obtain more evidence regarding cancer incidence at the loss of SIRT7, at the same time study SIRT7 activities in regulating genome stability, cell proliferation, and epithelial homeostasis in vivo. Here we focused on the intestinal epithelium and found the SIRT7 knockout mice were more susceptible to acute and chronic intestinal damages and displayed disrupted proliferation and regeneration abilities. We also obtained evidence that the loss of SIRT7 led to higher colorectal cancer incidence, proposing a tumor suppressive role of SIRT7 in the intestinal epithelium.
Results
SIRT7 Deficiency Leads to Aneuploidy and a Compromised CENP-A Nucleosome Assembly
SIRT7 deficiency has been demonstrated to cause impaired DNA damage response (Li et al., 2016; Tang et al., 2019; Vazquez et al., 2016) and showed compromised double-strand break repair upon irradiation treatment (Li et al., 2016; Vazquez et al., 2016). We found even in normal culturing condition without irradiation, SIRT7-ablated mouse embryonic fibroblast (MEF) and HeLa cell were prone to harbor irregular chromosome content, in both cases led to approximately 2-fold increase of aneuploidy cells (Figure 1A). We also observed elevated γH2AX level (Figure S1A), distorted nuclear structure (Figure S1B), and expanded nucleolar area (Figures S1C and S1D) in the SIRT7-ablated cells, confirming that SIRT7 plays crucial role to safeguard the genome integrity (Song et al., 2017) and suppresses aging phenotypes such as nucleolar expansion (Buchwalter and Hetzer, 2017; Tiku and Antebi, 2018). To reason the phenotypes of chromosomal segregation error and aneuploidy upon SIRT7 loss, we next explored the SIRT7-interacting proteins via immunoprecipitation. We chose to express C-terminally FLAG-tagged SIRT7 (SIRT7-FLAG) in HEK-293T cells, to avoid a steric hindrance in substrate-binding possibly resulting from the N-terminal tagging (Blank et al., 2017; Halasa et al., 2019; Priyanka et al., 2016). The purified SIRT7-interacting complexes were then analyzed by mass spectrometry.
Figure 1.
SIRT7 Facilitates CENP-A Assembly at the Centromere
(A) Wild-type and SIRT7 knockout MEF or HeLa cells were applied for metaphase spreads and chromosome counting post DAPI stain. The bar graph summary presents three independent experiments with ≥50 cells counted per experiment and the representative images were shown.
(B) Immunoprecipitation of SIRT7-FLAG-associated proteins from HEK293T cell extracts. An equal amount of starting materials was shown on the left. The purified protein complexes were resolved on SDS-PAGE and further visualized by silver staining, prior to mass spectrometry analysis.
(C) Immunoblot demonstration of nucleosome assembly proteins associated with SIRT7.
(D and E) (D) Immunofluorescence analysis of CENP-A (green) and (E) CENP-E (red) distribution in wild-type or SIRT7 KO HeLa cells. Cells with dislocated CENP-A foci from CENP-E were counted as abnormal. Bar graphs summarized three independent experiments with ≥50 cells scored in each experiment.
(F) Immunofluorescence analysis of CENP-A (green) and HP1β (red) distribution in wild-type or SIRT7 KO HeLa cells. Cells with dislocated CENP-A foci from HP1β were counted as abnormal. Bar graph summarized three independent experiments with ≥50 cells scored in each experiment.
Scale bars: (D–F) 5 μm, enlarged image in (E) 0.5 μm. Unpaired Student's t test was used, and data are shown as mean ± SD. See also Figure S1 and Table S1.
Gene ontology (GO) analysis of the immunoprecipitation results indicated that SIRT7 is involved in the nucleosome assembly process (Table S1). Among the identified SIRT7-interacting proteins, HAT1, RBBP4/7, NPM1, and histone H4 are components in the pre-nucleosomal complex for CENP-A loading (Figures 1B and 1C) (Foltz et al., 2009). CENP-A is a histone H3 variant and a key epigenetic determinant for centromere specification, formation, and maintenance. CENP-A is essential for kinetochore constituent recruitment and further facilitates mitosis (Allshire and Karpen, 2008; Black and Cleveland, 2011; Fukagawa and Earnshaw, 2014). To examine whether SIRT7 contributes to CENP-A nucleosome assembly, we performed the immunofluorescence analysis of detecting CENP-A in colchicine-arrested HeLa cells. We noticed a dispersed, abnormal CENP-A distribution in SIRT7-ablated cells (Figure 1D), indicating that CENP-A was improperly incorporated into the centromeric loci (Foltz et al., 2006). Consistent with the notion, a significant amount of CENP-A was found distally away from the centromeric motor protein CENP-E (Figure 1E), and similarly the case of dislocated CENP-A from the pericentromeric protein HP1β (Figure 1F). The results demonstrate that SIRT7 assists CENP-A incorporation into the centromere, which also propose a novel mechanism to explain the compromised chromosomal stability that occurred in SIRT7-deficient cells.
SIRT7 Deacetylates and Regulates HAT1 Activity at the CENP-A Nucleosome
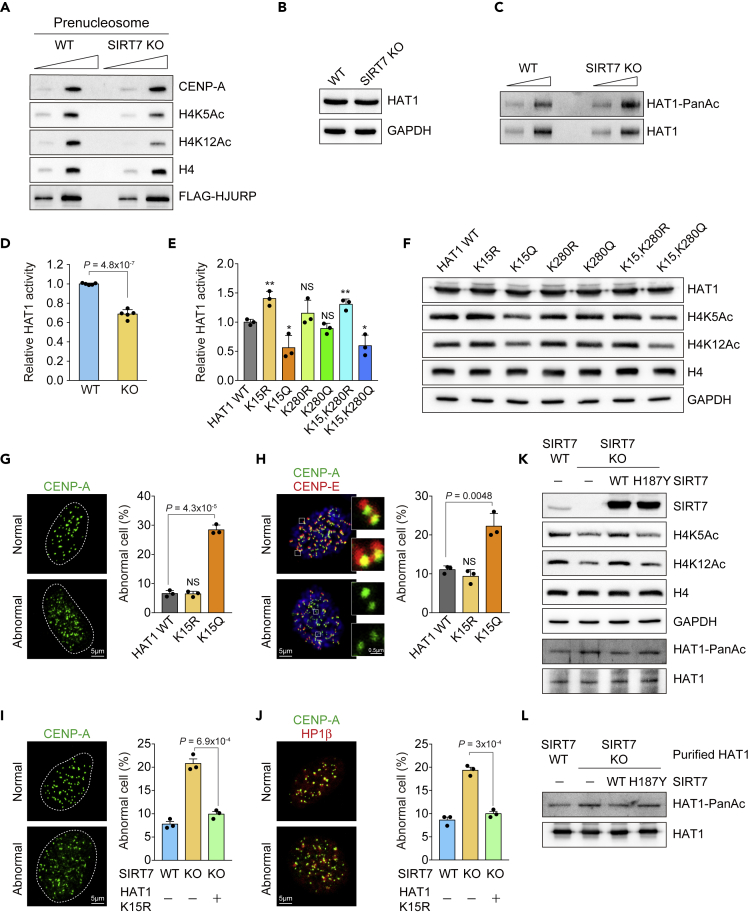
The correct deposition of CENP-A at the centromere is mediated by the specialized histone chaperone HJURP. HJURP recognizes newly synthesized CENP-A via the CENP-A Targeting Domain (CATD) and forms a pre-nucleosomal complex with the CENP-A:H4 tetramer (Dunleavy et al., 2009; Foltz et al., 2009). Histone H4 was shown to present additional epigenetic feature, i.e., H4K5Ac and H4K12Ac primarily in the pre-nucleosomal complex to promote centromere specification (Shang et al., 2016). As SIRT7 functions as a histone deacylase (Li et al., 2016) and was shown to deacetylate histone H3K18 (Barber et al., 2012), we first tested whether SIRT7 could as well deacetylate histone H4K5 and K12, hence regulate CENP-A nucleosome deposition into the centromere. Surprisingly, we instead detected decreased levels of acetylated histone H4K5 and H4K12 in the FLAG-HJURP purified pre-nucleosomal fraction from SIRT7 KO HeLa cells (Figure 2A).
Figure 2.
SIRT7 Deacetylates HAT1 Further Regulates Histone H4K5 and K12 Acetylation
(A) Reduced H4K5Ac and H4K12Ac in the purified pre-nucleosomal CENP-A-H4 complex from SIRT7 KO HeLa cells in comparison with WT cells. H4 and HJURP-FLAG were used as loading controls.
(B and C) (B) Unaltered HAT1 expression and (C) elevated acetylation level of HAT1 in SIRT7 KO HeLa cells revealed by immunoblot analysis.
(D) Comparison of HAT1 activity in WT versus SIRT7 KO HeLa cells. Wild-type activity was set as 1.
(E) Comparison of HAT1 activities in HeLa cells expressing HAT1 point mutations to mimic acetylated (K15Q, K280Q, and K15Q/K280Q) or unacetylated (K15R, K280R, and K15R/K280R) conditions.
(F) Immunoblot analysis of histone H4K5Ac and H4K12Ac levels from cells expressing HAT1 mutants.
(G) Immunofluorescence analysis of CENP-A (green) and (H) CENP-E (red) distribution in HAT1-WT, HAT1-K15R, or HAT1-K15Q-expressing HeLa cells. Cells with dislocated CENP-A foci from CENP-E were counted as abnormal. Bar graph summarized three independent experiments with ≥50 cells scored in each experiment.
(I) Immunofluorescence analysis of CENP-A (green) and (J) HP1β (red) distribution in WT, SIRT7 KO, or SIRT7 KO HeLa cells with HAT1-K15R expression as indicated. Cells with dislocated CENP-A foci from HP1β were counted as abnormal. Bar graph summarized three independent experiments with ≥50 cells scored in each experiment.
(K) Immunoblot analysis of histone H4K5, H4K12, and HAT1 acetylation levels in WT or SIRT7 KO HeLa cells that were re-expressing SIRT7-WT or SIRT7-H187Y mutant.
(L) In vitro deacetylation assay of using purified HAT1 from WT or SIRT7 KO HeLa cells, respectively. Purified hyperacetylated HAT1 was treated with SIRT7 or SIRT7-H187Y mutant as indicated, and detected with pan anti-acetyllysine immunoblot.
Scale bars: (G–J) 5 μm; enlarged image in (H) 0.5 μm. Unpaired Student's t test was used (NS, not significant, ∗p < 0.05, ∗∗p < 0.01), and data are shown as mean ± SD. See also Figure S1.
Histone acetyltransferase 1 (HAT1) was demonstrated to preferentially acetylate H4K5 and H4K12 in the pre-nucleosomal CENP-A:H4 complex (Shang et al., 2016); we further tested whether SIRT7 could deacetylate and modulate HAT1 activity. The expression level of HAT1 did not change (Figure 2B), but the level of acetylated HAT1 was evidently increased in SIRT7 knockout HeLa cells (Figure 2C), accompanied with lowered HAT1 activity (Figure 2D). To study how the acetylation status impacts on HAT1 function, we searched for frequently occurring acetylation sites in HAT1 (Mertins et al., 2013; Weinert et al., 2013) and made a series of corresponding point mutations to mimic acetylated (K15Q, K280Q, and K15Q/K280Q) or unacetylated (K15R, K280R, and K15R/K280R) states. The constitutively acetylated HAT1 mutants K15Q and K15Q/K280Q, but not K280Q, exhibited decreased acetyltransferase activities compared with wild-type HAT1 and other unacetylated mutants (Figures 2E and 2F), indicating that the K15 residue is a critical acetylation site for modulating HAT1 activity. Consistently, the expression of HAT1-K15Q in HeLa cells led to significant CENP-A deposition errors and CENP-A/CENP-E mis-localization (Figures 2G and 2H), in a manner similar to the loss of SIRT7 (Figures 1D–1F). Exogenous expression of HAT1-K15R lowered the CENP-A error in SIRT7 KO HeLa cells (Figures 2I and 2J). The reduced H4K5 and H4K12 acetylation in SIRT7 KO cells can be rescued by re-expression of wild-type SIRT7 but not the catalytic inactive mutant H187Y (Figure 2K). Consistently, purified hyperacetylated HAT1 can be deacetylated by wild-type SIRT7 (Figure 2L). To test whether other nuclear or cytosolic SIRTs play a role in regulation of HAT1 activity, we next overexpressed SIRT1/2/6 in SIRT7 KO cell lines, then examined histone H4K5 and H4K12 acetylations and the respective HAT1 activity changes. We found the alterations were SIRT7 specific (Figure S1E). Taken together, these data suggest that SIRT7 positively regulates HAT1 activity and subsequently augments the acetylation of histone H4K5 and H4K12.
Ablation of SIRT7 Causes Dysregulated Intestinal Proliferation and Elevated Senescence Associated with Aging
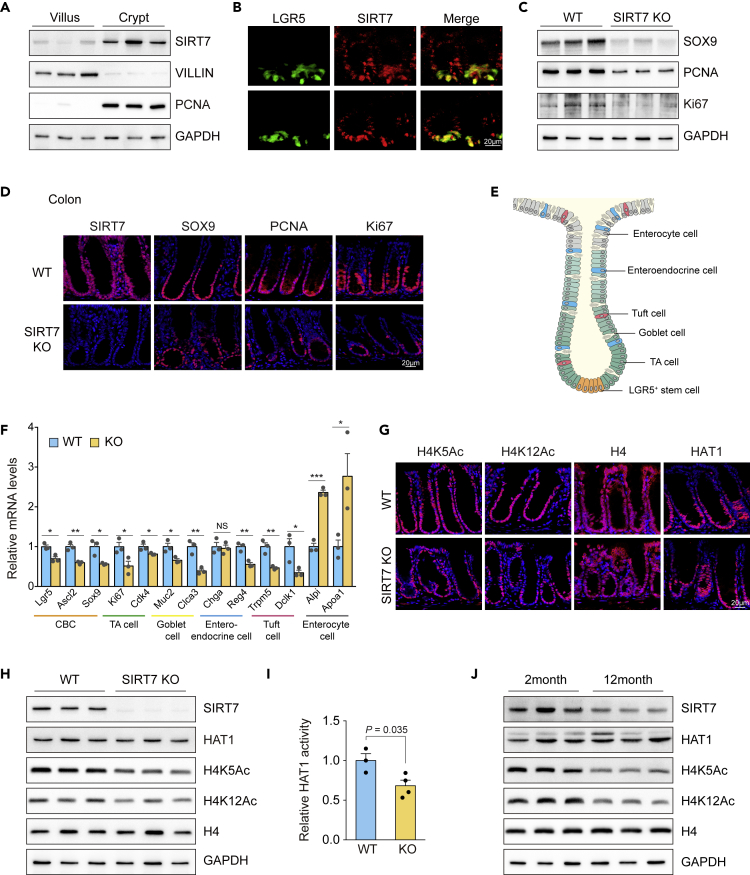
The adult intestinal epithelium undergoes rapid turnover and is replenished every 4–5 days (van der Flier and Clevers, 2009). Millions of epithelial cells are generated daily in the intestine, and the regeneration relies on small populations of adult stem cells that are enriched in the crypt base (Barker, 2014). The rapid proliferation and differentiation properties specify the intestine to be a suitable adult organ for investigating the consequence of SIRT7 deficiency with accumulated chromosomal instability. In the mouse small intestine, SIRT7 expresses more abundantly in the crypt (Figure 3A). A fraction of the crypt SIRT7 is expressed in the Lgr5+ (leucine-rich repeat-containing G protein-coupled receptor 5) intestinal adult stem cells (Schepers et al., 2012) (Figure 3B), suggesting that SIRT7 may play a role in supporting Lgr5+ cell function and intestinal epithelial proliferation. We analyzed the crypt lysates and found stem cell marker SOX9 and proliferation markers PCNA and Ki67 were markedly decreased in the SIRT7 knockout samples (Figure 3C) and associated with a shorter villus phenotype (Figure S2A). Similarly, SIRT7 is highly expressed in the colon crypt (Figure 3D). Absence of SIRT7 led to reduced expressions of SOX9, PCNA, and Ki67 (Figures 3D and S2B) and showed proliferative deficit as detected by the BrdU-labeled cells in the colon crypt (Figure 2C). The analysis of colon epithelial cell type markers for crypt base columnar (CBC) stem cell, transit-amplifying (TA) cell, goblet cell, tuft cell, enteroendocrine cell, and enterocyte cell (Haber et al., 2017) by RT-PCR revealed that SIRT7 KO mice were compromised for the specification of most cell types in the colon and shifted toward enterocytes (Figures 3E and 3F). The observation was also supported by the staining of alkaline phosphatase (ALP) and periodic acid-Schiff (PAS) for enterocyte and goblet cell, respectively (Figure S2C). Of note, acetylation of histone H4K5 and H4K12 was down-regulated in the SIRT7 KO crypt, corresponding to lower HAT1 activity, but not the HAT1 protein level (Figures 3G–3I), as found in vitro (Figure 2K). Immunoprecipitation assay revealed that, in both intestine and colon, HAT1 associated with SIRT7 (Figure S2D), similar to the results from SIRT7 KO HeLa cells (Figure 2C). SIRT7 was demonstrated to mediate histone H3K18 (Barber et al., 2012) and H3K36 deacetylation (Tanabe et al., 2018); however, we did not detect obvious hyperacetylation in the SIRT7 KO colon (Figures S2E and S2F), suggesting that the modulation of histone H4K5 and H4K12 acetylation via HAT1 could be a more important feature related to SIRT7 activity in the gut.
Figure 3.
Decreased H4K5Ac and H4K12Ac in SIRT7 Knockout Intestinal Crypt
(A) Immunoblot analysis of SIRT7 level from intestinal villus and crypt cells. VILLIN and PCNA were applied as markers for villus and crypt, respectively (n = 3).
(B) Immunohistostaining of LGR5 (green) and SIRT7 (red) in crypts. Representative images were shown.
(C) Immunoblot analysis of SOX9, PCNA, and Ki67 of intestinal crypt cells from wild-type and SIRT7 KO mice (n = 3).
(D) Immunohistostaining of crypt stem cell marker SOX9, and TA cell markers PCNA and Ki67 in colon samples from wild-type and SIRT7 KO mice.
(E) Graphic image of intestinal epithelium structure.
(F) Quantitative RT-PCR analysis of cell type specifying gene expressions in the colon samples from wild-type and SIRT7 KO mice (n = 3).
(G) Representative images of H4K5Ac and H4K12Ac levels in colon samples from wild-type and SIRT7 KO mice.
(H) Immunoblot analysis of the indicated proteins from wild-type and SIRT7 KO intestinal crypt lysates (n = 3).
(I) HAT1 activities in intestinal crypt lysates from wild-type and SIRT7 KO mice (n ≥ 3). (J) Immunoblot analysis of the indicated proteins from 2- and 12-month old mice (n = 3).
Scale bars: (B, D, G) 20 μm. Unpaired Student's t test was used (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001), and data are shown as mean ± SEM.
See also Figures S2 and S3.
SIRT7 ablation led to a higher senescence-associated (SA) β-galactosidase activity and elevated expressions of cellular senescence markers such as p16, p21, and p53, both in MEF and HeLa cells in our tests (Figures S3A–S3D), similar to the earlier reports in vitro (Serrano, 2016; Vazquez et al., 2016). Interestingly, all these senescence features were also preserved in SIRT7 KO intestinal tissues (Figures S3E and S3F), indicating a premature-aging phenotype in the intestine. Consistently, SIRT7 was decreased in the colon samples from 12-month-old mice in comparison with young adults, with the associated, reduced histone H4K5 and H4K12 acetylation (Figure 3J). Together, the results indicate that SIRT7 plays an important role in upholding intestinal epithelial homeostasis, and the decay maybe relevant to age-associated intestinal dysfunction.
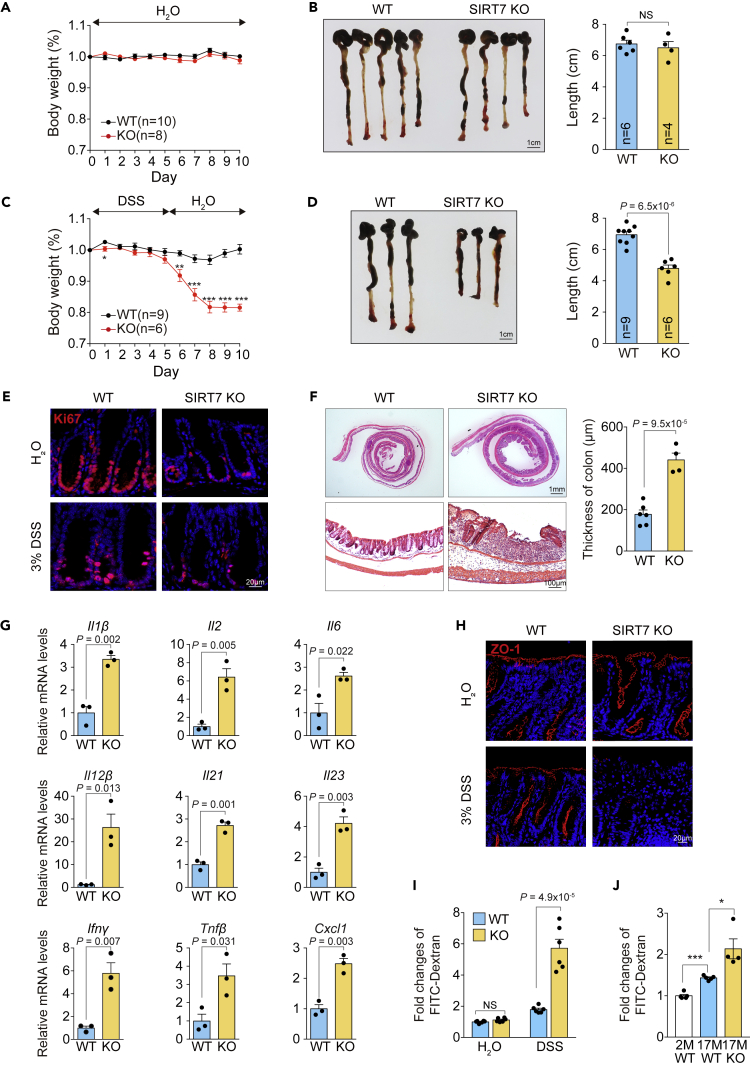
SIRT7-Deficient Mice Are Susceptible to DSS-Induced Colitis and Alcohol-Induced Epithelial Barrier Disruption
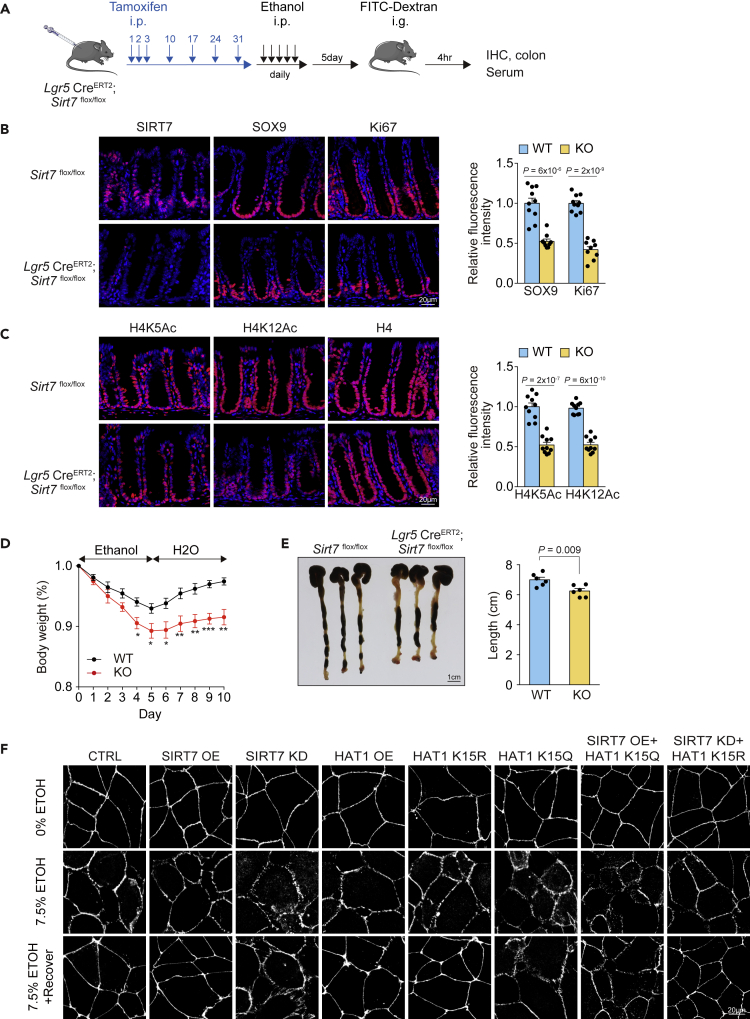
The abnormal morphology observed in the SIRT7 KO colon sections (Figures 3D, 3G, and S2E) raised the possibility that the SIRT7 KO mice may be susceptible to intestinal challenges such as dextran sulfate sodium (DSS)-induced colitis, a commonly used model to mimic inflammatory bowel disease (IBD). In our control test of providing normal drinking water, both wild-type and SIRT7 KO mice did not show body weight change or colon length difference (Figures 4A and 4B). When wild-type and SIRT7 KO mice were treated with 3% DSS in the drinking water ad libitum for 5 days, we found a marked weight loss in the SIRT7 KO cohort and an inability to recover in the subsequent 5 days (Figure 4C). The DSS-induced colon shortening was pronounced in the SIRT7 KO mice (Figure 4D), associated with a defect in proliferation (Figure 4E). In addition, the SIRT7 KO mice displayed extensive crypt loss and epithelial thickening in the distal-middle portion of the colon when compared with their wild-type littermates (Figure 4F), showing that the animals experienced severe colitis. The expression levels of pro-inflammatory cytokines, including Il1β, Il6, and Il12β, were significantly induced in SIRT7 KO colon samples upon DSS treatment (Figure 4G), whereas the basal levels of the same cytokines in the untreated cohorts were similar, except for Il12β (Figure S4). The phenotypes of inflammatory response and susceptible colitis suggested the loss of SIRT7 could result in a disrupted intestinal barrier function. We thus inspected the intestinal barrier function with tight junction associated protein Zonula Occludens-1 (ZO-1) (Stevenson et al., 1986) staining in control and SIRT7 KO mice. The distribution of ZO-1 appeared to be normal in both wild-type and SIRT7 KO groups in the drinking water test. Upon DSS challenge, SIRT7 KO mice failed to re-build the intestinal barrier as seen by the diminished ZO-1 (Figure 4H). In accordance with the severe ulceration, the mucosal permeability of SIRT7 KO mice was increased by 3-fold in a serum-based FITC-dextran measurement (Figure 4I). Of note, mucosal permeability increased with aging, in 17-month-old wild-type mice, and was further augmented in the SIRT7 KO (Figure 4J). These data demonstrated that SIRT7 is required for mucosal barrier function, in the case of normal aging, and when responding to acute colitis.
Figure 4.
SIRT7 KO Mice Are Susceptible to DSS-Induced Colitis
(A and B) (A) Wild-type and SIRT7 KO mice were fed with water for 10 days, and the body weight records and (B) colon length comparison on day 10 were shown.
(C and D) (C) Wild-type and SIRT7 KO mice were fed with 3% DSS then switched to regular water as indicated. The body weight records and (D) colon length comparison on day 10 were shown.
(E) Immunohistostaining of Ki67 in colon samples from wild-type and SIRT7 KO mice with water or 3% DSS as indicated.
(F) Colon sections were collected at day 10 for H&E staining, and the quantitative histopathology were shown.
(G) Quantitative RT-PCR analysis of inflammatory genes from colon homogenates of DSS-treated wild-type and SIRT7 KO mice (3% DSS for 3 days; n = 3 per group).
(H) ZO-1 staining of colon sections from mice that were treated with water or DSS.
(I) Intestinal permeability measured by the FITC-dextran leakage in the blood serum from wild-type and SIRT7 KO mice with water or 3% DSS as indicated (n = 6 per group).
(J) Intestinal permeability measured by the FITC-dextran leakage in the blood serum from 2- or 17-month-old mice as indicated (n = 4 per group).
Scale bars: (B and D) 1 cm; (E) 20 μm; (F) (left) 1 mm, (right) 100 μm; (H) 20 μm. Unpaired Student's t test was used (NS, not significant, ∗p < 0.05, ∗∗∗p < 0.001), and data are shown as mean ± SEM. See also Figure S4.
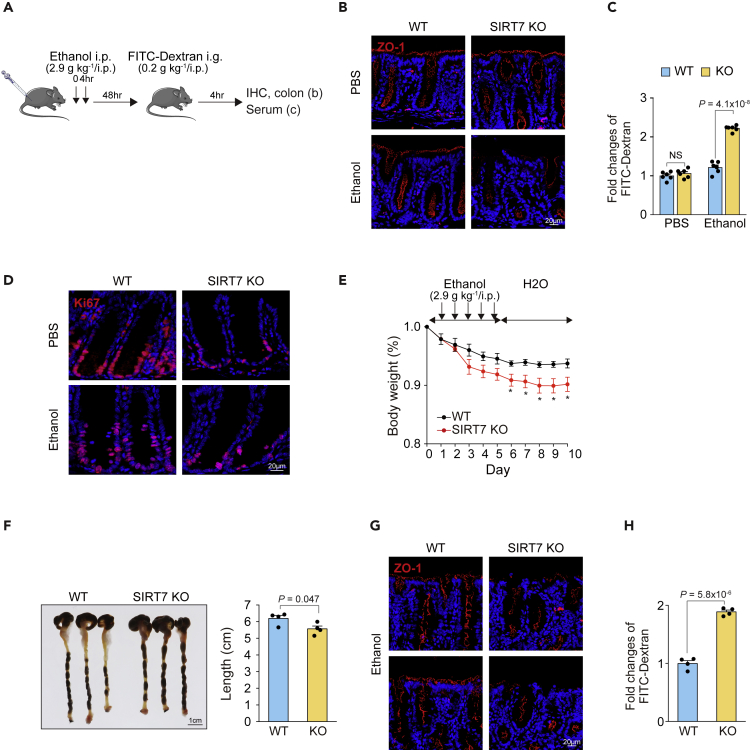
To validate further the activity of SIRT7 in upholding mucosal barrier function, we applied alcohol dosing as another epithelial challenge model. Alcohol consumption contributes to double-stranded DNA breaks and chromosome rearrangements (Garaycoechea et al., 2018; Roswall and Weiderpass, 2015), and the ingestion at long-term excess can lead to epithelial barrier damage (Chen et al., 2015). To determine whether the loss of SIRT7 could worsen alcohol-induced mucosal barrier dysfunction, we applied an acute ethanol exposure condition by intraperitoneal administration of ethanol (2.9 g kg−1) twice in 4 h (Figure 5A) (Garaycoechea et al., 2018). The treatment did not cause visible disturbance in the wild-type mice (Figure 5B), but in the SIRT7 KO mice, the same condition triggered a marked tight junction damage in the colon, as revealed by ZO-1 staining, and FITC-dextran measurement from the serum samples (Figures 5B and 5C), at the same time showed proliferation defect (Figure 5D). A chronic challenge of dosing ethanol daily for 5 days caused 10% weight loss and colon shortening in the SIRT7 KO cohort, whereas the wild-type mice were mildly affected under the same condition (Figures 5E and 5F). The ZO-1 distribution and FITC-dextran assays demonstrated again that the SIRT7 KO mice are susceptible to ethanol-induced epithelial disruption (Figures 5G and 5H), possibly due to the failure of epithelial regeneration.
Figure 5.
SIRT7 KO Mice Are Susceptible to Alcohol-Induced Epithelial Barrier Disruption
(A) Ethanol treatment scheme for intestine epithelium barrier assessment.
(B) ZO-1 staining of colon sections from mice that were post PBS or ethanol treatment for 48 h (n = 3 per group).
(C) Intestinal permeability measured by the concentration of FITC-dextran in the blood serum (n = 6 per group).
(D) Immunohistostaining of Ki67 in colon samples from wild-type and SIRT7 KO mice with PBS or ethanol treatment.
(E) Wild-type and SIRT7 KO mice were injected intraperitoneally with ethanol for 5 days, and body weights were scored daily (n = 4 per group).
(F) Colon length comparison of wild-type and SIRT7 KO mice on day 10.
(G) ZO-1 staining of colon sections from mice that were treated with ethanol for 5 days (n = 3 per group).
(H) Intestinal permeability measured by the FITC-dextran leakage in the blood serum (n = 4 per group).
Scale bars: (B, D, and F) 20 μm; (F) 1 cm. Unpaired Student's t test was used (NS, not significant, ∗p < 0.05), and data are shown as mean ± SEM.
SIRT7 Supports Epithelial Regeneration in the Lgr5+ Cells and Maintains Epithelial Homeostasis with HAT1
Lgr5 expresses selectively at the intestinal crypt base (Barker et al., 2007) and serves as a specific marker for proliferative stem cell and neoplastic cell origin (Barker et al., 2009; Schepers et al., 2012). To investigate whether the phenotypes are caused by intestinal epithelium, we generated a Sirt7 flox/flox strain (Figure S5A) and crossed that with Lgr5EGFP-ires-CreERT2 mice (Barker et al., 2007). The ablation of SIRT7 in Lgr5+ intestinal stem cells (SIRT7-iKO) was carried out by tamoxifen dosing for 7 times in 31 days (Mihaylova et al., 2018), before daily ethanol challenge in the subsequent 5 days (Figure 6A). The administration scheme allowed nearly complete ablation of SIRT7 in the crypt and led to the perturbed SOX9 and Ki67 expressions in the colon (Figure 6B). SIRT7-iKO mice had lower acetylation levels of histone H4K5 and H4K12 (Figure 6C), in a manner very similar to the SIRT7 whole-body knockouts (Figure 3G). Similar stem cell-depleting results were also found in the small intestine samples from SIRT7-iKO mice. In addition, lower PCNA levels suggested further that the loss of SIRT7 in Lgr5+ cells may lead to a limited proliferative capability (Figure S5B). The SIRT7-iKO mice responded to ethanol administration with similar symptoms as in the whole-body knockouts including body weight loss (Figure 6D), colon shortening (Figure 6E), and epithelial barrier dysfunction shown by ZO-1 expression and FITC-dextran assay (Figures S5C and S5D). In line with the results, in a culture model of examining tight junction disturbance at ethanol exposure (Ma et al., 1999), we found that the Sirt7 knockdown colorectal Caco-2 cells failed to re-establish tight junction from the ethanol treatment in a 2-h recovery period, as marked by ZO-1 (Figure 6F). Caco-2 cells with SIRT7 or HAT1 overexpression showed less disrupted intercellular borders upon ethanol exposure and re-assembled the tight junction in 2 h post ethanol removal. HAT1 mutant K15Q in Caco-2 cells showed compromised tight junction in a manner similar to Sirt7 knockdown, whereas HAT-K15R rescued the Sirt7 knockdown phenotype (Figure 6F), indicating that SIRT7 is important for intestinal epithelial homeostasis and barrier function via a synergistic action with HAT1. These results also specify SIRT7 function in Lgr5+ stem cells for epithelial regeneration, and at the same time suggest a plausible role of SIRT7 in maintaining intestinal stem cells.
Figure 6.
SIRT7 Is Critical for Intestinal Epithelium in Preventing Alcohol-Induced Colon Damage
(A) Experimental scheme of tamoxifen-induced Sirt7 conditional knock out in LGR5+ cells (SIRT7-iKO) and the subsequent ethanol treatment.
(B) Immunohistostaining of crypt stem cell marker SOX9, and TA cell marker Ki67 in colons from wild-type and SIRT7-iKO mice (n = 3, >3 sections were scored for each mouse).
(C) Immunohistostaining of H4K5Ac and H4K12Ac in colons from wild-type and SIRT7-iKO mice (n = 3).
(D and E) (D) Body weight record of wild-type and SIRT7-iKOmice (n = 6 per group), and (E) the colon length comparison on day 10.
(F) Tight junction disruption by ethanol treatment and the recovery were assayed via ZO-1 staining in human colorectal cancer Caco2 cells. Expressions of SIRT7 and HAT1 mutants were indicated.
Scale bars: (B, C, and F) 20 μm; (E) 1 cm. Unpaired Student's t test was used (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001), and data are shown as mean ± SEM. See also Figure S5.
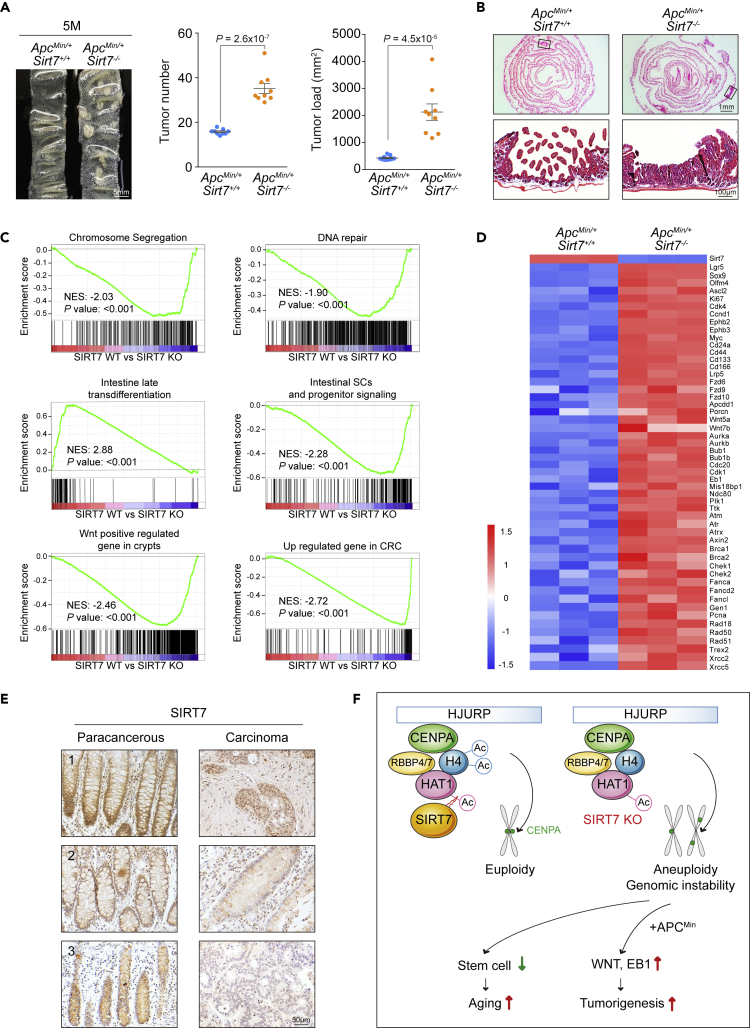
SIRT7 Deficiency Exacerbates Colorectal Tumor Malignance
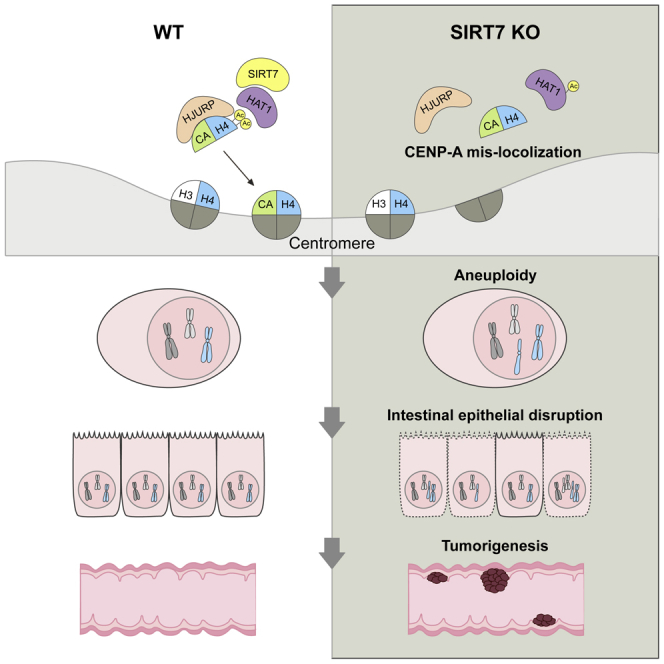
SIRT7 was reported to play an oncogenic role (Barber et al., 2012). On the other hand, the loss of SIRT7 leads to genetic instability (Li et al., 2016; Vazquez et al., 2016) and aneuploidy (Figures 1A and S1A), the hallmark that highly associates with multiple cancers (Hanahan and Weinberg, 2011; Tang and Amon, 2013). To address the conundrum, we investigate the consequence of SIRT7 deficiency in a commonly studied intestinal cancer model, Adenomatous polyposis coli (ApcMin/+) (Moser et al., 1990), to determine the outcome of cancer incidence in the intestine. In both wild-type and SIRT7 KO cohorts at 3 months of age, we did not find noticeable tumor in the small intestine (Figures S6A and S6B). However, when ApcMin/+ mutation was introduced in the Sirt7-null background, the compound mice exhibited profound increase in tumor number and tumor loads than the ApcMin/+ cohort at 3 months of age, indicating that SIRT7 deficiency triggered an accelerated tumor initiation (Figures S6C and S6D). At 5 months of age, more apparent differences were found in the compound ApcMin/+; Sirt7−/− mice with 2-fold increase in tumor number, near 5-fold increase in tumor loads, and significantly more polyps identified in the intestine samples (Figures 7A and 7B), with lower levels of histone H4K5 and H4K12 acetylation (Figure S6E). At 7 months of age, similar increases were also identified in the colon samples in the ApcMin/+; Sirt7−/− mice (Figure S6F). To examine gene expression changes associated to SIRT7 loss in colorectal tumorigenesis, we performed RNA sequencing analysis using intestinal tissues from 5-month-old ApcMin/+ and ApcMin/+; Sirt7−/− mice. Gene set enrichment analysis (GSEA) revealed that the loss of SIRT7 significantly enriched gene signatures associated to dysregulated chromosome segregation, DNA-repair response, and intestinal cell fate, at the same time with further raised Wnt signaling and colorectal cancer genes expressed (Figure 7C). This is also clear from the heatmap that the representative Wnt-responsive genes such as Lrp5, Fzds, Apcdd1, and Wnts, as well as DNA repair genes Atm, Atr, Brcas, Cheks, Fancs, Pcna, Rads, and Xrccs, were upregulated in the SIRT7 knockout intestine samples (Figure 7D). It is known that the human colorectal cancer-associated APC mutations and truncations often lead to APC inactivation and thus elicit constitutively active Wnt signal and prompt tumor initiation (Fodde et al., 2001). The gene expression differences between ApcMin/+ and ApcMin/+; Sirt7−/− samples indicate that SIRT7 deficiency stimulates the already elevated Wnt signaling associated to Apc mutation and at the same time worsens the progression of colorectal cancer led by chromosomal instability. The conclusion is coherent to clinical relevance, as a higher level of SIRT7 was scored in normal colon specimens, whereas considerably lower SIRT7 (Figure 7E and Table S2) and histone H4K5Ac and H4K12Ac levels were perceived in the human colorectal cancer biopsy in our immunohistochemistry analyses (Figure S7A). Our result of hypoacetylated histone H4K5/K12 correlated well with the clinical colorectal cancer dataset GSE28814 (Loboda et al., 2011) that patients with lower HAT1 levels were associated with decreased survival (Figure S7B). Taken together, our results demonstrate that SIRT7 deficiency leads to a compromised CENP-A centromeric nucleosome assembly and causes chromosomal instability. SIRT7 loss also triggers a disrupted epithelial homeostasis in the intestine and a dysregulated Wnt signaling that aggravates colorectal cancer incidence (Figure 7F).
Figure 7.
SIRT7 Ablation Aggravates Tumor Malignance in the APCMin/+ Mouse Model and Down-Regulation of SIRT7 in Human Colorectal Cancer
(A) Macroscopic image, tumor number, and tumor load from the small intestines of 5-month-old mice as indicated (n = 9 per group).
(B) Representative H&E images of the small intestines associated to (A).
(C) GSEA enrichment plots of differentially expressed genes from ApcMin/+; Sirt7−/− intestine samples.
(D) Heatmap summary of the RNA-seq results related to Wnt target and DNA repair genes in the ApcMin/+; Sirt7−/−intestine samples (n = 3).
(E) Representative SIRT7 immunohistostaining results of human CRC biopsy specimens.
(F) A schematic model of SIRT7 function in facilitating nucleosome assembly and genome stability.
Scale bars: (A) 5 mm; (B) (upper) 1 mm, (lower) 100 μm; (E) 50 μm. Unpaired Student's t test was used, and data are shown as mean ± SEM. See also Figures S6, S7, and Table S2.
Discussion
In the current study, we uncovered a novel activity of SIRT7 in regulating CENP-A nucleosome assembly. SIRT7 functions in concert with HAT1 to modulate the acetylation status of histone H4K5 and H4K12, further ensuring the proper docking of CENP-A on the pre-nucleosomal CENP-A:H4 complex. Cells that were devoid of SIRT7 exhibited dysregulated CENP-A nucleosome assembly, which is consistent with the higher aneuploidy level as a consequence (Yoda and Tomonaga, 2004) and agrees with the recent demonstration that CENP-A error led to centromeric chromatin decompaction in aged oocytes thus augmenting aneuploidy (Zielinska et al., 2019). Other nuclear sirtuins have been demonstrated to safeguard genomic stability as well. SIRT1 was shown to facilitate the loading of histone H1 and condensin I complex during mitosis (Fatoba and Okorokov, 2011). SIRT6 was reported to deacetylate histone H3K18 and further preserved pericentric heterochromatin silencing in maintaining mitotic fidelity (Fatoba and Okorokov, 2011). The cytosolic SIRT2 was demonstrated to shuttle from cytoplasm to nucleus during interphase (North and Verdin, 2007) and specifically deacetylate histone H4K16 and hence facilitate mitosis (Serrano et al., 2013; Vaquero et al., 2006). Our finding proposes a related, yet additional, mode of sirtuin activity in upholding genomic stability, via assisting the CENP-A nucleosome assembly. These findings, including the current observation, comprise target-specific and likely collaborative actions of sirtuins in ensuring cell cycle progression and genome integrity. Furthermore, our model also agrees with earlier finding that MEF cells derived from HAT1 knockout mice showed deficiency in DNA damage repair, and with increased aneuploidy (Nagarajan et al., 2013).
Genome instability and aneuploidy have been long recognized as hallmark of cancer (Hanahan and Weinberg, 2011; Tang and Amon, 2013) and a likely advantageous context for the adaptation of tumor cells during the selections of exposing to different types of cellular stresses (Chunduri and Storchova, 2019). Colorectal cancer is a suitable example representing the situation. The highly proliferative nature of intestinal epithelium enables sporadic chromosome segregation errors to accumulate, in an environment that commonly encounters oxidative and inflammatory damages. We found that SIRT7 executes critical activities in the intestine, including safeguarding the genome stability, maintaining the intestinal structure and the epithelial integrity, therefore protecting the intestine from environmental challenges such as ethanol and DSS. Moreover, higher colorectal cancer susceptibility at the loss of SIRT7 in the ApcMin/+ background points toward a tumor suppressive role of SIRT7. The loss of SIRT7 also exaggerates Wnt signaling (Figure 7D), a key mechanism that promotes intestinal tumorigenesis by turning normal intestinal stem cell self-renewal and differentiation to aberrant lineage decision, and incorrect expansion of progenitor cells (Gregorieff and Clevers, 2005). Whether the deficiency of SIRT7 leads to a general uncontrolled Wnt signaling, or is limited to certain tissues, such as in the intestine, remains to be clarified before the applications in other cancer types.
Our results raised other intriguing questions in intestinal homeostasis and aging. For instance, we noticed that the reduced acetylation levels of histone H4K5 and K12 were both detected in the SIRT7 KO intestines and in the Lgr5-specific SIRT7 KO samples, suggesting that histone H4K5 and K12 acetylation occurred during cell proliferation then preserved in the differentiated intestinal epithelium. Whether the acetylation of H4K5 and K12 involves in the cell lineage decision, the upholding of epithelial integrity, or even serves as markers for cancer predisposition, are important questions to peruse. Moreover, the lowered acetylation of histone H4K5 and H4K12 found in the aging intestine tissues, together with the decline in SIRT7 level, points out a stimulating application of targeting SIRT7 or HAT1 to treat bowel diseases. Most tellingly, our findings uncovered an unidentified role of SIRT7 in precluding intestinal tumorigenesis and therefore proposed a strategy of using SIRT7-activating compounds in treating human colorectal cancer.
Limitations of the Study
Our results indicate that SIRT7 deficiency is associated with aneuploidy consequence. From in vitro and biochemical approaches, we show that the phenotype is connected to the less active, hyperacetylated HAT1 state resulting from the absence of SIRT7. The change of HAT1 activity leads to lower histone H4K5/K12 acetylation and thus affects centromeric chromatin assembly. SIRT7 knockout mice also reveal similar, reduced histone H4K5/K12 acetylation in the intestine, together with dysregulated epithelial regeneration and higher cancer incidence in the ApcMin/+ background. Nonetheless, the conclusion would be strengthened if the aneuploidy level could be quantified in each cell type in the intestine. The better visualization of aberrant chromosome content ex vivo or in vivo, along with the studies on the proposed acetylation sites, e.g., histone H4K5, H4K12, and HAT1-K15, should be continued, for demonstrating the role of aneuploidy in epithelial pathology and cancer progression.
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to the lead contact Yun-Chi Tang (yctang@sibs.ac.cn).
Materials Availability
All material used in the current study are listed in Transparent Methods section and Key Resources Table in Supplemental Information, and materials are available upon request.
Data and Code Availability
The RNA-seq data have been deposited in the Gene Expression Omnibus database under accession numbers GSE138289, and NODE database with accession numbers OEP000258.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We thank Hua Feng of the Omics Core of Bio-Med Big Data Center, CAS-MPG Partner Institute for Computational Biology, SIBS, CAS for the assistance in RNA-seq and Ping Wu and Dr. Chao Peng of the Mass Spectrometry System at the National Facility for Protein Science in Shanghai (NFPS), Zhangjiang Lab, China for the mass spectrometry data collection and analysis.
This work was supported by grants from the National Key Research and Development Program of China (2017YFA0503600 to Y.-C.T.), General Program of National Natural Science Foundation of China (31471342 and 31671408 to Y.-C.T., 31671221 to H.-C.C).
Author Contributions
H.-C.C. and Y.-C.T. supervised the project and experiments. X.L., C.L., and Q.L. performed the experiments. X.L., H.-C.C., and Y.-C.T. analyzed and interpreted the data. H.-C.C. and Y.-C.T. wrote the manuscript. All authors reviewed and approved the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: September 25, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101461.
Contributor Information
Hung-Chun Chang, Email: hcchang@ion.ac.cn.
Yun-Chi Tang, Email: yctang@sibs.ac.cn.
Supplemental Information
References
- Allshire R.C., Karpen G.H. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat. Rev. Genet. 2008;9:923–937. doi: 10.1038/nrg2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber M.F., Michishita-Kioi E., Xi Y., Tasselli L., Kioi M., Moqtaderi Z., Tennen R.I., Paredes S., Young N.L., Chen K. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature. 2012;487:114–118. doi: 10.1038/nature11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol. 2014;15:19–33. doi: 10.1038/nrm3721. [DOI] [PubMed] [Google Scholar]
- Barker N., Ridgway R.A., van Es J.H., van de Wetering M., Begthel H., van den Born M., Danenberg E., Clarke A.R., Sansom O.J., Clevers H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Black B.E., Cleveland D.W. Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell. 2011;144:471–479. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank M.F., Chen S., Poetz F., Schnolzer M., Voit R., Grummt I. SIRT7-dependent deacetylation of CDK9 activates RNA polymerase II transcription. Nucleic Acids Res. 2017;45:2675–2686. doi: 10.1093/nar/gkx053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank M.F., Grummt I. The seven faces of SIRT7. Transcription. 2017;8:67–74. doi: 10.1080/21541264.2016.1276658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonkowski M.S., Sinclair D.A. Slowing ageing by design: the rise of NAD(+) and sirtuin-activating compounds. Nat. Rev. Mol. Cell Biol. 2016;17:679–690. doi: 10.1038/nrm.2016.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwalter A., Hetzer M.W. Nucleolar expansion and elevated protein translation in premature aging. Nat. Commun. 2017;8:328. doi: 10.1038/s41467-017-00322-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalkiadaki A., Guarente L. The multifaceted functions of sirtuins in cancer. Nat. Rev. Cancer. 2015;15:608–624. doi: 10.1038/nrc3985. [DOI] [PubMed] [Google Scholar]
- Chang H.C., Guarente L. SIRT1 and other sirtuins in metabolism. Trends Endocrinol. Metab. 2014;25:138–145. doi: 10.1016/j.tem.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Torralba M., Tan J., Embree M., Zengler K., Starkel P., van Pijkeren J.P., DePew J., Loomba R., Ho S.B. Supplementation of saturated long-chain fatty acids maintains intestinal eubiosis and reduces ethanol-induced liver injury in mice. Gastroenterology. 2015;148:203–214 e16. doi: 10.1053/j.gastro.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chunduri N.K., Storchova Z. The diverse consequences of aneuploidy. Nat. Cell Biol. 2019;21:54–62. doi: 10.1038/s41556-018-0243-8. [DOI] [PubMed] [Google Scholar]
- Dunleavy E.M., Roche D., Tagami H., Lacoste N., Ray-Gallet D., Nakamura Y., Daigo Y., Nakatani Y., Almouzni-Pettinotti G. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- Fatoba S.T., Okorokov A.L. Human SIRT1 associates with mitotic chromatin and contributes to chromosomal condensation. Cell Cycle. 2011;10:2317–2322. doi: 10.4161/cc.10.14.15913. [DOI] [PubMed] [Google Scholar]
- Finkel T., Deng C.X., Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodde R., Kuipers J., Rosenberg C., Smits R., Kielman M., Gaspar C., van Es J.H., Breukel C., Wiegant J., Giles R.H. Mutations in the APC tumour suppressor gene cause chromosomal instability. Nat. Cell Biol. 2001;3:433–438. doi: 10.1038/35070129. [DOI] [PubMed] [Google Scholar]
- Foltz D.R., Jansen L.E., Bailey A.O., Yates J.R., 3rd, Bassett E.A., Wood S., Black B.E., Cleveland D.W. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz D.R., Jansen L.E., Black B.E., Bailey A.O., Yates J.R., 3rd, Cleveland D.W. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- Ford E., Voit R., Liszt G., Magin C., Grummt I., Guarente L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 2006;20:1075–1080. doi: 10.1101/gad.1399706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T., Earnshaw W.C. The centromere: chromatin foundation for the kinetochore machinery. Dev. Cell. 2014;30:496–508. doi: 10.1016/j.devcel.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaycoechea J.I., Crossan G.P., Langevin F., Mulderrig L., Louzada S., Yang F., Guilbaud G., Park N., Roerink S., Nik-Zainal S. Alcohol and endogenous aldehydes damage chromosomes and mutate stem cells. Nature. 2018;553:171–177. doi: 10.1038/nature25154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorieff A., Clevers H. Wnt signaling in the intestinal epithelium: from endoderm to cancer. Genes Dev. 2005;19:877–890. doi: 10.1101/gad.1295405. [DOI] [PubMed] [Google Scholar]
- Haber A.L., Biton M., Rogel N., Herbst R.H., Shekhar K., Smillie C., Burgin G., Delorey T.M., Howitt M.R., Katz Y. A single-cell survey of the small intestinal epithelium. Nature. 2017;551:333–339. doi: 10.1038/nature24489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halasa M., Bartuzi D., Cieslak D., Kaczor A.A., Miziak P., Stepulak A., Matosiuk D. Role of N-terminus in function and dynamics of sirtuin 7: an in silico study. J. Biomol. Struct. Dyn. 2019;38:1–9. doi: 10.1080/07391102.2019.1600585. [DOI] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hubbi M.E., Hu H., Kshitiz, Gilkes D.M., Semenza G.L. Sirtuin-7 inhibits the activity of hypoxia-inducible factors. J. Biol. Chem. 2013;288:20768–20775. doi: 10.1074/jbc.M113.476903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S., Guarente L. NAD+ and sirtuins in aging and disease. Trends Cell Biol. 2014;24:464–471. doi: 10.1016/j.tcb.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.K., Noh J.H., Jung K.H., Eun J.W., Bae H.J., Kim M.G., Chang Y.G., Shen Q., Park W.S., Lee J.Y. Sirtuin7 oncogenic potential in human hepatocellular carcinoma and its regulation by the tumor suppressors MiR-125a-5p and MiR-125b. Hepatology. 2013;57:1055–1067. doi: 10.1002/hep.26101. [DOI] [PubMed] [Google Scholar]
- Lai C.C., Lin P.M., Lin S.F., Hsu C.H., Lin H.C., Hu M.L., Hsu C.M., Yang M.Y. Altered expression of SIRT gene family in head and neck squamous cell carcinoma. Tumour Biol. 2013;34:1847–1854. doi: 10.1007/s13277-013-0726-y. [DOI] [PubMed] [Google Scholar]
- Li L., Shi L., Yang S., Yan R., Zhang D., Yang J., He L., Li W., Yi X., Sun L. SIRT7 is a histone desuccinylase that functionally links to chromatin compaction and genome stability. Nat. Commun. 2016;7:12235. doi: 10.1038/ncomms12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loboda A., Nebozhyn M.V., Watters J.W., Buser C.A., Shaw P.M., Huang P.S., Van't Veer L., Tollenaar R.A., Jackson D.B., Agrawal D. EMT is the dominant program in human colon cancer. BMC Med. Genomics. 2011;4:9. doi: 10.1186/1755-8794-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T.Y., Nguyen D., Bui V., Nguyen H., Hoa N. Ethanol modulation of intestinal epithelial tight junction barrier. Am. J. Physiol. 1999;276:G965–G974. doi: 10.1152/ajpgi.1999.276.4.G965. [DOI] [PubMed] [Google Scholar]
- Mertins P., Qiao J.W., Patel J., Udeshi N.D., Clauser K.R., Mani D.R., Burgess M.W., Gillette M.A., Jaffe J.D., Carr S.A. Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat. Methods. 2013;10:634–637. doi: 10.1038/nmeth.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylova M.M., Cheng C.W., Cao A.Q., Tripathi S., Mana M.D., Bauer-Rowe K.E., Abu-Remaileh M., Clavain L., Erdemir A., Lewis C.A. Fasting activates fatty acid oxidation to enhance intestinal stem cell function during homeostasis and aging. Cell Stem Cell. 2018;22:769–778 e764. doi: 10.1016/j.stem.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser A.R., Pitot H.C., Dove W.F. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 1990;247:322–324. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- Nagarajan P., Ge Z., Sirbu B., Doughty C., Agudelo Garcia P.A., Schlederer M., Annunziato A.T., Cortez D., Kenner L., Parthun M.R. Histone acetyl transferase 1 is essential for mammalian development, genome stability, and the processing of newly synthesized histones H3 and H4. Plos Genet. 2013;9:e1003518. doi: 10.1371/journal.pgen.1003518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North B.J., Verdin E. Interphase nucleo-cytoplasmic shuttling and localization of SIRT2 during mitosis. PLoS One. 2007;2:e784. doi: 10.1371/journal.pone.0000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyanka A., Solanki V., Parkesh R., Thakur K.G. Crystal structure of the N-terminal domain of human SIRT7 reveals a three-helical domain architecture. Proteins. 2016;84:1558–1563. doi: 10.1002/prot.25085. [DOI] [PubMed] [Google Scholar]
- Roswall N., Weiderpass E. Alcohol as a risk factor for cancer: existing evidence in a global perspective. J. Prev. Med. Public Health. 2015;48:1–9. doi: 10.3961/jpmph.14.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu D., Jo Y.S., Lo Sasso G., Stein S., Zhang H., Perino A., Lee J.U., Zeviani M., Romand R., Hottiger M.O. A SIRT7-dependent acetylation switch of GABPbeta1 controls mitochondrial function. Cell Metab. 2014;20:856–869. doi: 10.1016/j.cmet.2014.08.001. [DOI] [PubMed] [Google Scholar]
- Schepers A.G., Snippert H.J., Stange D.E., van den Born M., van Es J.H., van de Wetering M., Clevers H. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 2012;337:730–735. doi: 10.1126/science.1224676. [DOI] [PubMed] [Google Scholar]
- Serrano L., Martinez-Redondo P., Marazuela-Duque A., Vazquez B.N., Dooley S.J., Voigt P., Beck D.B., Kane-Goldsmith N., Tong Q., Rabanal R.M. The tumor suppressor SirT2 regulates cell cycle progression and genome stability by modulating the mitotic deposition of H4K20 methylation. Genes Dev. 2013;27:639–653. doi: 10.1101/gad.211342.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M. Unraveling the links between cancer and aging. Carcinogenesis. 2016;37:107. doi: 10.1093/carcin/bgv100. [DOI] [PubMed] [Google Scholar]
- Shang W.H., Hori T., Westhorpe F.G., Godek K.M., Toyoda A., Misu S., Monma N., Ikeo K., Carroll C.W., Takami Y. Acetylation of histone H4 lysine 5 and 12 is required for CENP-A deposition into centromeres. Nat. Commun. 2016;7:13465. doi: 10.1038/ncomms13465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C., Hotz-Wagenblatt A., Voit R., Grummt I. SIRT7 and the DEAD-box helicase DDX21 cooperate to resolve genomic R loops and safeguard genome stability. Genes Dev. 2017;31:1370–1381. doi: 10.1101/gad.300624.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson B.R., Siliciano J.D., Mooseker M.S., Goodenough D.A. Identification of ZO-1: a high molecular weight polypeptide associated with the tight junction (zonula occludens) in a variety of epithelia. J. Cell Biol. 1986;103:755–766. doi: 10.1083/jcb.103.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe K., Liu J., Kato D., Kurumizaka H., Yamatsugu K., Kanai M., Kawashima S.A. LC-MS/MS-based quantitative study of the acyl group- and site-selectivity of human sirtuins to acylated nucleosomes. Sci. Rep. 2018;8:2656. doi: 10.1038/s41598-018-21060-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M., Li Z., Zhang C., Lu X., Tu B., Cao Z., Li Y., Chen Y., Jiang L., Wang H. SIRT7-mediated ATM deacetylation is essential for its deactivation and DNA damage repair. Sci. Adv. 2019;5:eaav1118. doi: 10.1126/sciadv.aav1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Shi L., Xie N., Liu Z., Qian M., Meng F., Xu Q., Zhou M., Cao X., Zhu W.G. SIRT7 antagonizes TGF-beta signaling and inhibits breast cancer metastasis. Nat. Commun. 2017;8:318. doi: 10.1038/s41467-017-00396-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y.C., Amon A. Gene copy-number alterations: a cost-benefit analysis. Cell. 2013;152:394–405. doi: 10.1016/j.cell.2012.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiku V., Antebi A. Nucleolar function in lifespan regulation. Trends Cell Biol. 2018;28:662–672. doi: 10.1016/j.tcb.2018.03.007. [DOI] [PubMed] [Google Scholar]
- Tsai Y.C., Greco T.M., Cristea I.M. Sirtuin 7 plays a role in ribosome biogenesis and protein synthesis. Mol. Cell Proteomics. 2014;13:73–83. doi: 10.1074/mcp.M113.031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Flier L.G., Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu. Rev. Physiol. 2009;71:241–260. doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- Vaquero A., Scher M.B., Lee D.H., Sutton A., Cheng H.L., Alt F.W., Serrano L., Sternglanz R., Reinberg D. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 2006;20:1256–1261. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez B.N., Thackray J.K., Simonet N.G., Kane-Goldsmith N., Martinez-Redondo P., Nguyen T., Bunting S., Vaquero A., Tischfield J.A., Serrano L. SIRT7 promotes genome integrity and modulates non-homologous end joining DNA repair. EMBO J. 2016;35:1488–1503. doi: 10.15252/embj.201593499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert B.T., Scholz C., Wagner S.A., Iesmantavicius V., Su D., Daniel J.A., Choudhary C. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 2013;4:842–851. doi: 10.1016/j.celrep.2013.07.024. [DOI] [PubMed] [Google Scholar]
- Yoda K., Tomonaga T. Centromere identity originates in the structure of CENP-A/H4 tetramer itself: a mechanism for aneuploidy. Lancet. 2004;364:1022–1024. doi: 10.1016/S0140-6736(04)17077-3. [DOI] [PubMed] [Google Scholar]
- Yoshizawa T., Karim M.F., Sato Y., Senokuchi T., Miyata K., Fukuda T., Go C., Tasaki M., Uchimura K., Kadomatsu T. SIRT7 controls hepatic lipid metabolism by regulating the ubiquitin-proteasome pathway. Cell Metab. 2014;19:712–721. doi: 10.1016/j.cmet.2014.03.006. [DOI] [PubMed] [Google Scholar]
- Zielinska A.P., Bellou E., Sharma N., Frombach A.S., Seres K.B., Gruhn J.R., Blayney M., Eckel H., Moltrecht R., Elder K. Meiotic kinetochores fragment into multiple lobes upon cohesin loss in aging eggs. Curr. Biol. 2019;29:3749–3765.e7. doi: 10.1016/j.cub.2019.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data have been deposited in the Gene Expression Omnibus database under accession numbers GSE138289, and NODE database with accession numbers OEP000258.