Abstract
Objective
Cholesterol plays a pivotal role in mitochondrial steroidogenesis, membrane structure, and respiration. Mitochondrial membranes are intrinsically low in cholesterol content and therefore must be replenished with cholesterol from other subcellular membranes. However, the molecular mechanisms underlying mitochondrial cholesterol transport remains poorly understood. The Aster-B gene encodes a cholesterol binding protein recently implicated in cholesterol trafficking from the plasma membrane to the endoplasmic reticulum (ER). In this study, we investigated the function and underlying mechanism of Aster-B in mediating mitochondrial cholesterol transport.
Methods
CRISPR/Cas9 gene editing was carried out to generate cell lines deficient in Aster-B expression. The effect of Aster-B deficiency on mitochondrial cholesterol transport was examined by both confocal imaging analysis and biochemical assays. Deletion mutational analysis was also carried out to identify the function of a putative mitochondrial targeting sequence (MTS) at the N-terminus of Aster-B for its role in targeting Aster-B to mitochondria and in mediating mitochondrial cholesterol trafficking.
Results
Ablation of Aster-B impaired cholesterol transport from the ER to mitochondria, leading to a significant decrease in mitochondrial cholesterol content. Aster-B is also required for mitochondrial transport of fatty acids derived from hydrolysis of cholesterol esters. A putative MTS at the N-terminus of Aster-B mediates the mitochondrial cholesterol uptake. Deletion of the MTS or ablation of Arf1 GTPase which is required for mitochondrial translocation of ER proteins prevented mitochondrial cholesterol transport, leading to mitochondrial dysfunction.
Conclusions
We identified Aster-B as a key regulator of cholesterol transport from the ER to mitochondria. Aster-B also coordinates mitochondrial cholesterol trafficking with uptake of fatty acids derived from cholesterol esters, implicating the Aster-B protein as a novel regulator of steroidogenesis.
Keywords: GRAMD1b, Cholesterol transport, Mitochondria, Arf1, Fatty acids
Abbreviations: Arf1, ADP Ribosylation Factor 1; GRAMD1B, GRAM Domain-Containing Protein 1B; COPI, Coat Protein Complex I; COPA, Coatomer Subunit Alpha; CPT-1, Carnitine Palmitoyltransferase I; CRISPR/Cas 9, Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-Associated Protein 9; DMEM, Dulbecco's Modified Eagle's Medium; ER, Endoplasmic Reticulum; GAPDH, Glyceraldehyde 3-Phosphate Dehydrogenase; KRPH, Kreb's Ringer Phosphate Buffer with HEPES; MAM, Mitochondria-Associated Membrane; MCD, Methyl-Beta Cyclodextrin; MTS, Mitochondrial Targeting Sequence; OCR, Oxygen Consumption Rate; PBS, Phosphate-Buffered saline; STARD1, Steroidogenic Acute Regulatory Protein; VASt domain, VAD1 analog of StAR-Related Lipid Transfer Domain.
Highlights
-
•
Ablation of Aster-B impairs cholesterol transport from the ER to mitochondria.
-
•
Aster-B couples mitochondrial cholesterol transport with uptake of fatty acids derived from hydrolysis of cholesterol ester.
-
•
An MTS at the N-terminus of Aster-B is required for the mitochondrial targeting and cholesterol transport.
-
•
Ablation or inhibition of Arf1 disrupts the mitochondrial cholesterol transport and causes mitochondrial dysfunction.
1. Introduction
Mitochondrial cholesterol transport plays an important role in maintaining mitochondrial membrane fluidity, permeability, and respiration. Mitochondrial membrane cholesterol content is intrinsically low when compared with other cellular membranes. This unique feature is required for the synthesis of steroids, oxysterols, and hepatic bile acids [1,2]. Cholesterol transport to mitochondria is believed to be primarily mediated through non-vesicular pathways, since mitochondria are completely disconnected from vesicular transport routes [3]. This transport has also been shown to be stimulated by the presence of fatty acids present on cholesterol, known as cholesterol esters [4]. Toward this end, mitochondria form multiple contact sites (MCSs) with other organelles, such as the endoplasmic reticulum (ER) and lysosomes, which serve as the major route for mitochondrial cholesterol import. For example, the ER forms multiple membrane contacts with mitochondria, also known as mitochondria-associated membranes (MAMs), which are implicated in the transport of phospholipids and cholesterol from the ER to mitochondria [5]. In support of this notion, cholesterol content in the MAM is much higher than in the rest of the ER [6,7]. Accordingly, depletion of mitofusin-2, a mitochondrial GTPase that tethers the ER with mitochondria, led to decreased progesterone synthesis [2,8]. Moreover, depletion of STARD1, a cholesterol transporter at the MAM, also prevented cholesterol transport from the ER to mitochondria [9]. However, the molecular mechanisms underlying the role of the MAM in cholesterol transport to mitochondria remains poorly understood.
Aster-B, also known as GRAMD1B (GRAM Domain-Containing 1B), is a member of a highly conserved family of proteins that contain both a GRAM domain and a steroidogenic acute regulatory protein-related lipid transfer (StART) domain [10]. The GRAM domain-containing proteins are known to regulate organelle contacts, whereas the StART domain is implicated in redistribution of phospholipids to different organelles in yeast [11]. These StART domains have also been shown to be activated upon fatty acid stimulation [4]. The GRAMD family of proteins is highly conserved from yeast to humans, suggesting an important functional role. Aster-B shares homology with a family of yeast lipid transfer at contact site (Ltc) proteins, which are also implicated in cholesterol trafficking [12,13]. Aster-B also contains an ASTER domain which shares homology with the VAD1 analog of the StAR-related lipid transfer (VASt) domain commonly found in six yeast Ltc proteins. The yeast Ltc-1 was recently reported to be an ER resident protein that forms tethers between both the ER-mitochondria and ER-vacuole contact sites via the mitochondrial import receptors Tom70/71 and the vacuolar protein Vac8, respectively [13]. Ltc1 was also shown to bind sterols and facilitate their membrane transfer in an in vitro assay [14]. In contrast, very little is known about the subcellular localization of Aster-B, nor its role in regulating mitochondrial function in mammalian cells, although targeted deletion of Aster-B was recently reported to facilitate cholesterol transport from the plasma membrane to the ER. Aster-B deficiency also caused defective steroidogenesis in mice, but the underlying causes remain elusive [12]. In researching the lipid transport activity of StART-domain proteins, we unexpectedly identified Aster-B as a key regulator of mitochondrial cholesterol trafficking and fatty acid transport from cholesterol esters to the mitochondria. Fatty acids have been shown to be critical regulators of mitochondrial cholesterol uptake, but their mechanism of delivery remains elusive [4,15]. Additionally, we demonstrated that ADP-ribosylation factor 1 (Arf1), a GTPase recently implicated in mitochondrial trafficking of ER proteins in yeast, is also required for mitochondrial cholesterol trafficking and fatty acid uptake into the mitochondria [16]. These findings provided key insights into the potential importance of Aster proteins in regulating the synthesis of steroids, oxysterols, and hepatic bile acids.
2. Materials and methods
2.1. Generation of Aster-B, Aster-C, and Arf1 KO cells using clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9 (CRISPR/Cas9) gene editing
Aster-B and Aster-C genes were knocked out in C2C12 cells, and Arf1 was knocked out in HeLa cells using CRISPR/Cas9 mouse plasmids from Santa Cruz (Aster-B: #sc-433439 and #sc-433439-HDR) (Aster-C: #sc-431521 and #sc-431521-HDR) (Arf1: #sc-401608 and #sc-401608-HDR) and then purified through green fluorescent protein (GFP) and red fluorescent protein (RFP) fluorescence by the UT Health San Antonio Flow Cytometry Core and cultured in complete media. Cells were then kept under selection with 10 μg/mL of puromycin (#sc-205821, Santa Cruz). Vector control cells were created by transfecting C2C12 cells with an empty pBABE-puro vector backbone (#1764, Addgene) and selected and kept in culture medium with 10 μg/mL of puromycin.
2.2. Cell culture
Cells were maintained at 37 °C with 5% CO2, cultured in high-glucose Dulbecco's Modified Eagle's Medium (DMEM, #D5796, Sigma) supplemented with 10% fetal bovine serum (FBS, #S11550H, Atlanta Biologicals) and 50 μg/mL of penicillin/streptomycin (#15140122, Invitrogen).
2.3. Cell transfection and confocal analysis
C2C12 transfections were performed using Viafect (E4981) from Promega. Cells were seeded overnight in 6-cm culture dishes and then transfected with 2.5 μg of plasmid DNA in DMEM. For confocal microscopy, cells were seeded after 24 h into 35-mm glass bottom confocal dishes (FD35) from Fluorodish and then imaged after another 24 h. Imaging of cells was performed using a Zeiss LSM 710 confocal microscope. All imaging was performed using live cells. Nuclei were stained with Hoechst Blue 33342 (#62249 Sigma), and mitochondria were stained with Mitotracker Green FM (#M7514 Fisher) or Mitotracker Red FM (#M22425 Fisher) or transfected with a mito-BFP plasmid (#49151 Addgene) or a mito-GFP plasmid (#44385 Addgene). ER was labeled by transfecting cells with an ER-localized gene expressing plasmid DsRed2-ER-5 (#55836 Addgene).
2.4. RNA extraction, reverse transcription, and real-time polymerase chain reaction (PCR)
Aster-C gene expression in C2C12-Vector and Aster-C KO cells was measured by quantitative real-time PCR. In brief, cells were seeded on 6-well plates for 24 h and then washed once with ice-cold phosphate-buffered saline (PBS). Total RNA was then extracted using TRIzol reagent (#15596018, Invitrogen). Two micrograms of total RNA was reverse transcribed to complementary DNA (cDNA) using SuperScript IV reverse transcriptase (#18090010, Invitrogen). cDNAs were then used as templates for quantification of Aster-C gene expression by real-time PCR analysis, which was performed using SYBR green PCR master mix (#4309155, Thermo Fisher Scientific) and the 7300 Real-Time PCR System (Applied Biosystems). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the internal control.
2.5. Cell treatment, cholesterol depletion, cholesterol measurement, nutrient starvation, and re-stimulation
For the cholesterol uptake assay, C2C12-Vector and C2C12-Aster-B KO cells were pre-treated with 0.5% methyl-beta cyclodextrin (MCD) from Sigma (C4555) in DMEM supplemented with 0.5% fatty acid free bovine serum albumin (BSA) from Gemini (700-107P) for 2 h. Cell nuclei were stained for the final 30 min with 400 nM of Hoechst Blue (62249) from Thermo Fisher. Cells were then incubated with 25-NBD-Cholesterol from Avanti Polar Lipids (DO-005189) which was pre-complexed with 0.1% MCD in DMEM supplemented with 0.5% fatty acid free BSA for 30 min. For NBD-Phosphatidic Acid and NBD-Phosphatidyl Choline uptake assays, Vector Control, and Knockout Cells were starved in a standard Krebs Ringer Phosphate HEPES medium (KRPH, 140 mM NaCl, 2 mM Na2HPO4, 4 mM KCl, 1 mM MgCl2, 1.5 mM CaCl2, 10 mM HEPES, pH7.4) without any added nutrients (such as serum, glucose, and amino acids) for 1 h. 50 μM of NBD-Phosphatidic Acid (810176) or 50 μM of NBD-Phosphatidyl Choline (810133P) from Avanti was then added, and cells were imaged.
For cholesteryl ester treatment, Vector Control and Aster-B Knockout Cells were stained with Mitotracker Green FM (#M7514 Fisher) and then starved for 30 min in KRPH buffer. Cells were then treated with 10 μM of CholEsteryl BODIPY™ 542/563 C11 (#C12680 Fisher) for 10 min. Permeabilized cells were starved for 30 min in KRPH and then pre-treated with 10 μM of Digitonin (D5628) from Sigma in KRPH buffer, washed twice in KRPH buffer and then treated with 10 μM CholEsteryl BODIPY™ 542/563 C11 or 10 μM 16:0 TopFluor® cholesterol (#810288P, Avanti Polar Lipids) for 10 min. For non-fluorescent cholesterol ester assays, 17:0 cholesteryl ester (700186) from Avanti Polar Lipids was used. Cholesterol (C3045) from Sigma was used in non-fluorescent free cholesterol stimulation assays.
2.6. Subcellular fractionation
Subcellular fractionation was performed according to the protocol described previously [17]. In brief, cells were transfected for 48 h using Xtreme Gene HP DNA Transfection reagent on three 15-cm plates per group using 2:1 μl of transfection reagent/μg protein using 20 μl/10 μg per plate. Cells were then scraped, and the organelles were isolated according to the above protocol.
2.7. Cholesterol measurement
Cells were pre-treated with 0.5% MCD from Sigma (C4555) in DMEM supplemented with 0.5% fatty acid free BSA from Gemini (700-107P) for 2 h. Cells were then treated for 2 h with vehicle or with 50 μM of free cholesterol (C3045) from Sigma, which was pre-complexed with 0.1% MCD in DMEM supplemented with 0.5% fatty acid free BSA for 30 min. Cells were collected and washed with ice-cold PBS twice and then subjected to subcellular fractionation. Cellular cholesterol was measured using the Cholesterol/Cholesterol Ester-Glo Assay Kit from Promega (J3190) according to the manufacturer's instructions.
2.8. Oxygen consumption rate analysis
Oxygen consumption rate (OCR) was measured using a Seahorse XF24 analyzer according to the manufacturer's instructions. To determine basal OCR, C2C12 Vector Control and Aster-B Knockout Cells were seeded in a 24-well microplate overnight. Cells were washed 3 times using KRPH supplemented with 10 mM of glucose and 1 mM of pyruvate. To determine OCR under cholesterol stimulation, cells were depleted of cholesterol for 2 h using 0.5% MCD as previously described and then treated with 25 μM of cholesterol ester. Measurements were taken at baseline and following injection of port A: 1.5 μg/mL of oligomycin, Port B: 1 μM of FCCP, Port C: 2 μM of rotenone plus 2 μM of antimycin A1.
2.9. Immunoblotting
For immunoblotting, C2C12 Vector Control, Aster-B Knockout cells, and cells transfected with a WT Aster-B expressing plasmid (RG219269) from Origene or Mutant Plasmid Δ2-31 were depleted and re-stimulated with cholesterol in the same manner as described above. Proteins were then isolated from cells and run on sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gels. Antibodies used include GRAMD1B 24905-1-AP (Proteintech), GFP ab13970 (Abcam) Tom20 sc-17764 (Santa Cruz), BiP C50B12 (Cell Signal), Calnexin ab22595 (Abcam), α-tubulin 11224 (Proteintech), and OPA1 D7C1A (Cell Signal).
2.10. Cell lysis and immunoprecipitation
HEK293A cells were transfected with plasmids as indicated in each experiment using X-tremeGENE™ HP DNA transfection reagent according to the manufacturer's instructions. After 48 h of transfection, cells were depleted of cholesterol with MCD and then re-stimulated with cholesterol as described above or treated with Exo-2 (10 μM). Cells were then rinsed twice with ice-cold PBS and lysed in ice-cold NP-40 lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.5% NP-40, 1 mM PMSF, and one tablet of EDTA-free protease inhibitors per 50 mL). The soluble fractions from cell lysates were isolated by centrifugation at 12,000 rpm for 10 min at 4 °C. Protein concentration was measured using a Pierce™ BCA protein assay kit (ThermoFisher, Cat#23225). For immunoprecipitation, primary anti-GFP antibody was added to an equal amount of total protein from each lysate and incubated with rotation for 2 h at 4 °C. Thirty microliters of a 50% slurry of protein A/G sepharose beads was added and continuously incubated with rotation at 4 °C overnight. The immunoprecipitates were then washed 5 times with lysis buffer, and 30 μL of SDS sample buffer was added to the precipitates and boiled for 5 min to denature the proteins. The eluates were then resolved by 10% SDS-PAGE and analyzed by western blot analysis.
2.11. Quantification and statistical analysis
Data were routinely represented as mean ± SD. Statistical significance was assessed by Student's t-test or one-way analysis of variance (ANOVA) using GraphPad Prism 6.0. Differences were considered statistically significant at p < 0.05, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
3. Results
3.1. Aster-B is required for plasma membrane uptake of cholesterol, but not other phospholipids
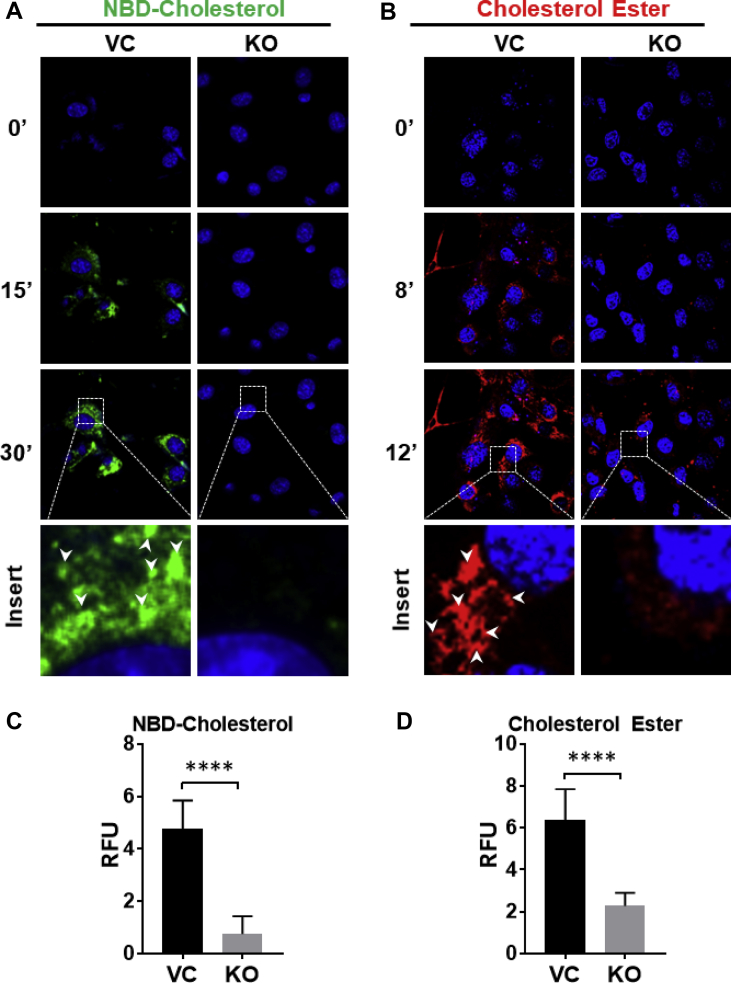
To determine the function of Aster-B in lipid transport, we first generated a C2C12 stable cell line that is deficient in Aster-B expression by CRISPR/Cas9-mediated gene editing and verified by western blot (Figure S1A). We next analyzed the effect of Aster-B deficiency on plasma membrane uptake of cholesterol and cholesterol esters. After depletion of the endogenous cholesterol content with 0.5% MCD [18], the knockout (KO) and vector control (VC) C2C12 cells were treated with 50 μM of 25-NBD-Cholesterol or 10 μM of CholEsteryl BODIPY™ 542/563 C11 and analyzed for cholesterol uptake by confocal microscopy. The results show that ablation of Aster-B significantly reduced the uptake of both cholesterol and cholesterol esters relative to the VC over a 30-minute period (Figure 1A and B, quantified in relative fluorescent units (RFU) in 1C and 1D, respectively). In contrast, deletion of Aster-C, which is an isoform of Aster-B, as verified by qPCR analysis (Figure S1B), did not significantly affect the 25-NBD-cholesterol uptake (Figure S1C), suggesting that Aster-B is specifically required for cholesterol uptake [14]. In support of this notion, Aster-B deficiency did not significantly affect the uptake of phospholipids, including phosphatidic acid (PA) and phosphatidylcholine (PC) (Figure S2).
Figure 1.
Ablation of Aster-B impairs uptake of cholesterol and cholesterol esters. (A) Time-lapse confocal imaging analysis depicting the cholesterol uptake in C2C12 vector control (VC) and Aster-B knockout (KO) cells. Cells were depleted of endogenous cholesterol using 0.5% methyl-β-cyclodextrin treatment for 2 hr and then treated with 50 μM of 25-NBD-Cholesterol for the times indicated. Nuclei are stained with Hoechst 33342. (B) Time-lapse confocal imaging analysis depicting the cholesterol ester uptake in C2C12 VC and Aster-B KO cells. Cells were starved in KRPH buffer for 30 min and treated with 10 μM of CholEsteryl BODIPY™ 542/563 C11 for the times indicated. Nuclei are stained with Hoechst 33342. (C) Fluorescence density analysis of green fluorescent cholesterol treated VC and KO cells at 30 minutes, n=10. (D) Fluorescence density analysis of CholEsteryl BODIPY™ 542/563 C11-treated VC and KO cells at 12 min n = 10. ∗∗∗∗p < 0.0001 by student's t-test.
3.2. Aster-B coordinates mitochondrial cholesterol transport with uptake of fatty acids hydrolyzed from cholesterol esters
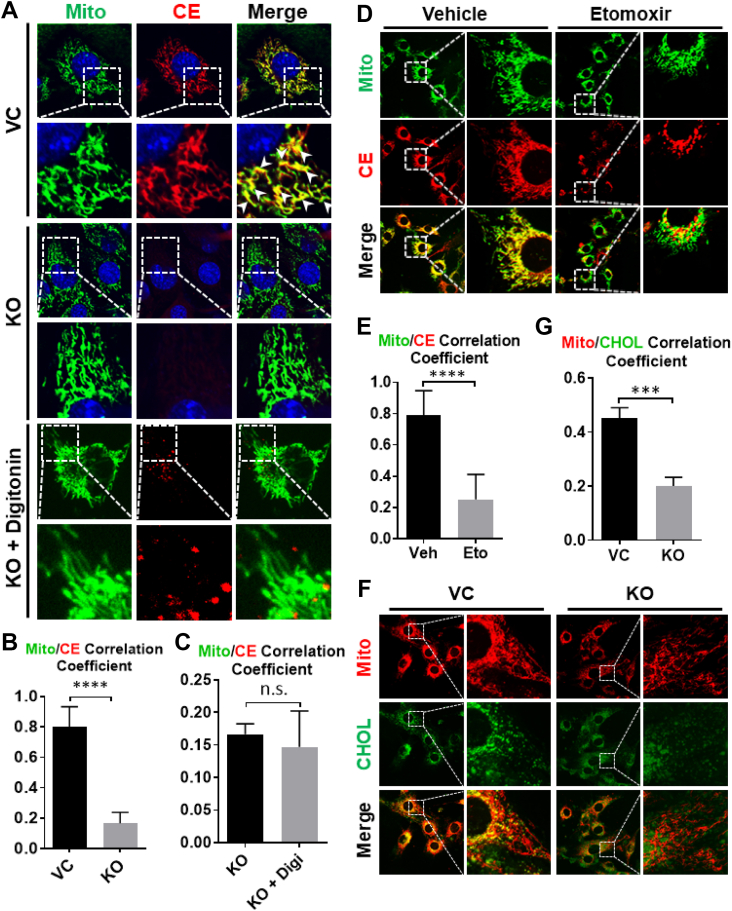
The yeast Ltc1 protein, an ortholog of Aster-B, tethers between the ER and mitochondria [14], suggesting a role for Aster-B in mitochondrial cholesterol transport. To test this hypothesis, we next investigated the effect of Aster-B deficiency on cholesterol trafficking to mitochondria in C2C12 cells by confocal imaging analysis. Aster-B KO and the VC cells were treated with 10 μM of CholEsteryl BODIPY™ 542/563 C11, stained with Mitotracker green, and analyzed for cholesterol uptake in mitochondria by confocal microscopy. The results show that the cholesterol esters were rapidly transported into the vector C2C12 cells, and the majority were co-localized with the mitochondria. In contrast, Aster-B deficiency prevented this phenomenon in Aster-B KO cells (Figure 2A, highlighted by arrowheads, Pearson's Correlation Coefficient quantified in 2B). To assess whether this was caused by defective cholesterol uptake from the plasma membrane, we next permeabilized the cells using digitonin, followed by treatment with 10 μM CholEsteryl BODIPY™ 542/563 C11. Digitonin selectively permeabilizes plasma membranes, allowing cholesterol and other lipids to freely enter the cell [19]. Again, Aster-B deficiency significantly impaired the mitochondrial transport of cholesterol, as evidenced by accumulation of punctas, which did not co-localize with mitochondria in Aster-B KO cells (Figure 2A, Pearson's Correlation Coefficient quantified in 2C). We next performed time-lapse confocal microscopy and demonstrated that Aster-B deficiency significantly delayed, but did not completely ablate mitochondrial cholesterol trafficking in Aster-B KO cells, likely due to the presence of other mitochondrial cholesterol transporters (Figure S3, the co-localization with mitochondria is highlighted by arrowheads). CholEsteryl BODIPY™ 542/563 C11 has the Bodipy fluorescent tag on the fatty acid portion of the cholesterol ester, and it is widely accepted that cholesterol esters are hydrolyzed to free cholesterol and free fatty acids before they are transported to the mitochondria for steroidogenesis [20]. To address this issue, C2C12 cells were pre-treated with vehicle or etomoxir, a CPT-1 inhibitor which blocks fatty acids transport to mitochondria for oxidation, and then incubated with 10 μM CholEsteryl BODIPY™ 542/563 C11. The results show that etomoxir significantly blocked co-localization of the Bodipy signal with mitochondria (Figure 2D, Pearson's Correlation Coefficient quantified in 2E), indicating that the Bodipy signal in mitochondria is likely from the Bodipy-fatty acid derived from hydrolysis of Bodipy-cholesterol esters, but not Bodipy-cholesterol ester itself. These results prompted us to investigate whether Aster-B also mediates fatty acids uptake or only fatty acids derived from hydrolysis of cholesterol esters. To answer this question, C2C12 vector control and Aster-B KO cells were incubated with 1 μM of BODIPY™ 558/568 C12 (Red C12), a saturated free fatty acid analog with a tail composed of 12 carbons and a BODIPY 558/568 fluorophore covalently bound at the hydrophobic end, using the pulse-chase method described previously [21]. The Red C12 fatty acid was readily transported to the mitochondria, but there was no significant difference between the VC and Aster-B KO cells (Figure S4A, Pearson's Correlation Coefficient quantified in S4B), suggesting that Aster-B is only required for mitochondrial transport of fatty acids released from hydrolysis of cholesterol ester, but not free fatty acids. We further validated this concept using 16:0 TopFluor® cholesterol ester, in which the Bodipy fluorescent is tagged on the cholesterol portion for the analysis of cholesterol transport in digitonin permeabilized vector control and Aster-B KO cells. Again, Aster-B deficiency prevented the co-localization of fluorescent Bodipy with mitochondria in Aster-B deficient cells when compared with vector controls (Figure 2F, Pearson's Correlation Coefficient quantified in 2G). This finding is highly reminiscent of what was found in the Aster-B KO cells incubated with CholEsteryl BODIPY™ 542/563 C11 after digitonin permeabilization (Figure 2A). Together, these findings suggest that Aster-B is required for mitochondrial cholesterol transport as well as mitochondrial uptake of free fatty acids released from cholesterol esters, which is likely used as an energy source for steroidogenesis.
Figure 2.
Aster-B deficiency impairs the transport of cholesterol and fatty acids derived from cholesterol esters into mitochondria. (A) Confocal imaging analysis depicting the mitochondrial transport of cholesterol ester in vector control (VC) and Aster-B knockout (KO) cells. Cells were starved in KRPH buffer for 30 min and then treated with 10 μM of CholEsteryl BODIPY™ 542/563 C11 for 10 min. Digitonin-treated cells were starved in KRPH buffer for 20 min and then pre-treated with 10 μM of digitonin for 10 min, washed twice in KRPH buffer, and then treated with 10 μM of CholEsteryl BODIPY™ 542/563 C11 for 10 min. Mitochondria and nuclei were stained by Mitotracker-Green and Hoechst 33342, respectively. (B) Pearson's Co-localization Coefficient of Mitotracker Green and CholEsteryl BODIPY™ 542/563 C11 in the VC and KO cells. n = 20. ∗∗∗∗p < 0.0001 by student's t-test. (C) Pearson's Co-localization Coefficient of Mitotracker Green and CholEsteryl BODIPY™ 542/563 C11 in vehicle and digitonin-treated Aster-B KO cells. n = 20. (D) Confocal images of C2C12 cells pre-treated with vehicle or 300 nM of etomoxir for 6 h. Cells were then starved for 30 min and treated with CholEsteryl BODIPY™ 542/563 C11 for 10 min. Mitochondria were stained with Mitotracker-Green. (E) Pearson's Co-localization Coefficient of Mitotracker-Green and CholEsteryl BODIPY™ 542/563 C11 in D. n = 20. ∗∗∗∗p < 0.0001 by student's t-test. (F) Confocal imaging analysis depicting the co-localization of cholesterol with mitochondria in VC and Aster-B KO cells. Cells were treated with MCD, permeabilized by digitonin, and then incubated with 16:0 TopFluor® cholesterol. Mitochondria were stained with Mitotracker-Red. (G) Pearson's Co-localization Coefficient of Mitotracker-Red and 16:0 TopFluor® cholesterol in F. n = 20. ∗∗∗p < 0.001 by student's t-test.
3.3. Aster-B deficiency reduces mitochondrial cholesterol content
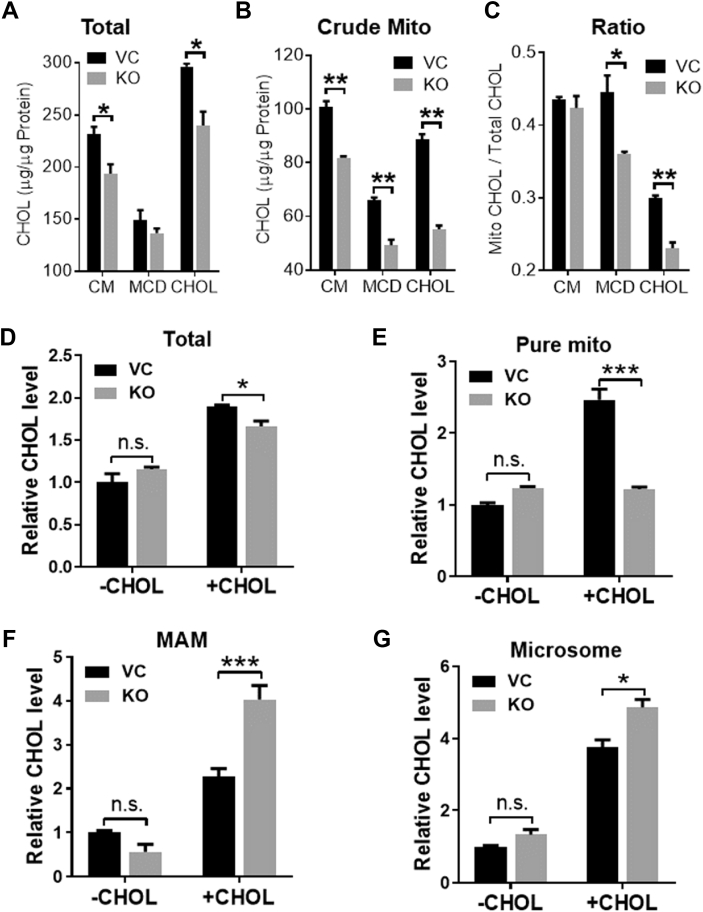
To further confirm that Aster-B is required for mitochondrial cholesterol transport, we next analyzed the effect of Aster-B on both total cholesterol and mitochondrial cholesterol content. Aster-B deficiency significantly decreased the total (Figure 3A) and mitochondrial cholesterol (Figure 3B) content as well as the ratio of mitochondrial cholesterol to total cholesterol (Figure 3C) in Aster-B deficient cells relative to vector control under normal culture conditions as well as in response to cholesterol stimulation. We next analyzed the effect of Aster-B deficiency on the subcellular distribution of cholesterol in further subfractionated organelles in permeabilized cells. In contrast to a moderate decrease in total cellular cholesterol content (Figure 3D), Aster-B deficiency significantly decreased purified mitochondrial cholesterol content (Figure 3E) concurrently with a dramatic increase of cholesterol content both in the mitochondrial-associated membrane (MAM) and microsomes (Figure 3F,G). These findings lend further support to Aster-B being required for the cholesterol transport from the ER to mitochondria.
Figure 3.
Aster-B deficiency reduces mitochondrial cholesterol content. (A–B) Total cholesterol level (A) and mitochondrial cholesterol level (B) in vector control (VC) and Aster-B knockout (KO) cells. Cells in normal culture condition (CM), treated with MCD (MCD) or incubated with 50 μM of cholesterol after MCD treatment (CHOL). n = 3. ∗p < 0.05, ∗∗p < 0.01 by two-way ANOVA. (C) The ratio of mitochondrial cholesterol to the total cholesterol in VC and Aster-B KO cells. n = 3. ∗p < 0.05, ∗∗p < 0.01 by two-way ANOVA. (D–G) Total cholesterol levels (D) and cholesterol levels in pure mitochondria (E), mitochondrial-associated membrane (MAM) (F) and microsomes (G) in VC and Aster-B KO cells. Cells were treated with MCD (-CHOL) or pemeabilized by digitonin after MCD treatment and re-incubated with 50 μM of cholesterol (+CHOL). n = 3. ∗p < 0.05, ∗∗∗p < 0.001 by two-way ANOVA.
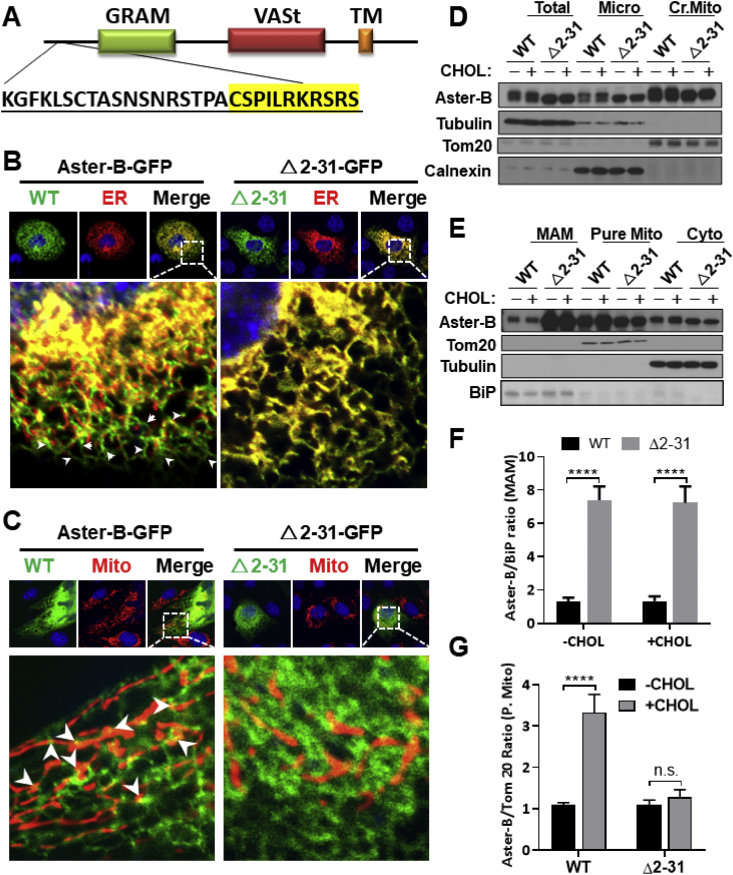
3.4. A mitochondrial targeting sequence (MTS) is required for the tethering of Aster-B at ER and mitochondrial contact sites
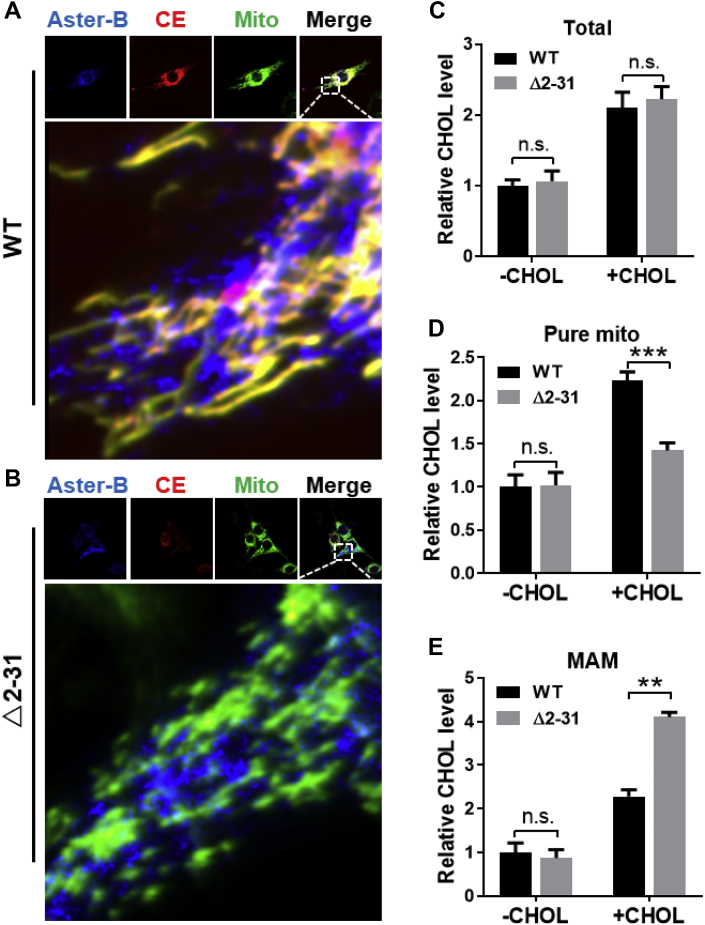
Using MitoFate, a software program that predicts mitochondrial pre-sequences, we identified a potential mitochondrial localization domain between amino acids 2–31 at the N-terminus of the Aster-B protein (Figure 4A, MTS highlighted in yellow). There are no MTS's predicted in either Aster-A or Aster-C proteins. We hypothesized that the MTS is responsible for the tethering of Aster-B with mitochondria. To test the hypothesis, we generated an Aster-B mutant plasmid which carries a deletion of this region (Δ2-31) and analyzed the effect on its subcellular localization by confocal imaging analysis upon non-fluorescent cholesterol ester stimulation. Consistent with the subcellular localization of yeast Ltc1, the Aster-B protein appeared to be co-localized at the contact sites between the ER and mitochondria (Figure 4B,C, highlighted by arrows). In support of a role of the MTS in mitochondrial tethering, deletion of the MTS rendered Aster-B completely co-localized within the ER membrane (Figure 4B). To confirm the results from the confocal analysis, we next analyzed the effect of ablation of the MTS on the subcellular localization of Aster-B in response to stimulation with non-fluorescent cholesterol esters in 293A cells transiently expressing the wild-type (WT) or the Δ2-31 deletion mutant of Aster-B protein. The total cell lysates were fractionated into different subcellular fractions, including cytosol, microsomes, and crude mitochondria, which were further fractionated into the MAM and pure mitochondria. The results show that the Aster-B protein was primarily distributed in the microsomes and crude mitochondrial fractions, with relatively low levels in the cytosol (Figure 4D,E). In contrast, deletion of the MTS led to significant retention of Aster-B in the MAM relative to the WT control (Figure 4E, quantified in Figure 4F). Additionally, treatment with cholesterol esters significantly increased Aster-B protein levels in the pure mitochondrial fraction, which was ablated by the deletion of the MTS (Figure 4E, quantified in Figure 4G). Our findings are corroborated by a recent report on the yeast Ltc1 protein that mutation of the mitochondrial tethering arms caused the Ltc1 protein to be completely localized in the ER [14]. To test whether this phenomenon was exclusive to the MTS, we created a plasmid with a mutation of the GRAM domain of Aster-B. Confocal imaging analysis showed that deletion of the GRAM domain did not significantly modify its subcellular localization in Aster-B KO cells transfected with the mutant plasmid and stimulated with cholesterol esters (Figure S5A). Subcellular fractionation and immunoblot analysis show that deletion of the GRAM domain decreased the Aster-B protein in MAM, but not in the pure mitochondria (Figure S5B), suggesting that the GRAM domain is not required for the mitochondrial targeting of Aster-B.
Figure 4.
MTS is required for the tethering of Aster-B at ER and mitochondrial contact sites. (A) Schematic representation of Aster-B protein showing deleted region (2–31) of plasmid with MTS highlighted in yellow. (B–C) Confocal images of C2C12 Aster-B knockout (KO)cells transfected with GFP labeled Aster-B or Δ2-31-Aster-B and an ER localized DsRed-ER protein (B) or a mitochondria resident protein, Mito-DsRed (C), respectively. Cells were starved for 30 min in KRPH buffer followed by 10 min of 10 μM of CholEsteryl BODIPY™ 542/563 C11 stimulation. (D–E) Subcellular fractionation and Western blot analysis of 293A cells transfected with either wild-type (WT) or Δ2-31 Aster-B plasmids. Cells were starved for 30 min in KRPH buffer and then treated with 10 μM of non-fluorescent cholesterol esters for 10 min. Micro, microsome; Cr. Mito, crude mitochondria, pure mito, pure mitochondria; cyto, cytosol. (F) Densitometric analysis showing the fold difference of WT Aster-B to Δ2-31 in the MAM, using Bip as an internal control, n = 3. ∗∗∗∗p < 0.0001 by two-way ANOVA. (G) Densitometric analysis showing the fold difference of WT Aster-B to Δ2-31 in the purified mitochondrial fraction using Tom20 as an internal control n = 3. ∗∗∗p < 0.001 by two-way ANOVA.
3.5. Deletion of MTS of Aster-B impairs mitochondrial cholesterol trafficking
The findings that the MTS is required for mitochondrial tethering with the ER prompted us to question whether the MTS is also required for mitochondrial cholesterol transport. We answered this question by determining whether the re-introduction of mutant Δ2-31 back to the Aster-B deficient cells would restore cholesterol transport when compared with WT Aster-B. The results show that transient expression of the WT Aster-B restored mitochondrial fatty acid transport upon stimulation with CholEsteryl BODIPY™ 542/563 C11 for 10 min in permeabilized Aster-B-deficient cells (Figure 5A, highlighted by arrows). In contrast, overexpression of mutant Δ2-31 in permeabilized Aster-B deficient cells failed to restore the fatty acid transport to the mitochondria (Figure 5B), further supporting a role for the MTS of Aster-B in mitochondrial cholesterol transport. We next determined whether reintroduction of the Δ2-31 deletion mutant to Aster-B deficient cells would restore mitochondrial cholesterol content relative to WT Aster-B. The results show that although the total cellular cholesterol levels were similar between WT and the Δ2-31 deletion mutant (Figure 5C), re-expression of the Δ2-31 deletion mutant failed to restore the cholesterol content in mitochondria (Figure 5D) relative to WT Aster-B control, leading to significant accumulation of cholesterol in the MAM (Figure 5E) of Aster-B KO cells, further suggesting that the MTS is required for mitochondrial cholesterol transport by Aster-B.
Figure 5.
Deletion of MTS of Aster-B impairs mitochondrial cholesterol trafficking. (A–B) Confocal imaging analysis depicting the mitochondrial cholesterol transport in C2C12 Aster-B knockout (KO) cells re-expressing GFP-Aster-B WT (A) or Δ2-31 mutant (B). Mitochondria were labeled by transfecting cells with mito-BFP. Cells were starved for 20 min in KRPH buffer, followed by 10 min of permeabilization with 10 μM of digitonin, and then treated with 10 μM of CholEsteryl BODIPY™ 542/563 C11 for 10 min. (C–E) Total cholesterol level (C) and cholesterol levels in pure mitochondria (D) and MAM (E) in Aster-B KO cells re-expressing GFP-Aster-B WT or Δ2-31 mutant. Cells were treated with MCD (-CHOL) or permeabilized by digitonin after MCD treatment and re-incubated with 50 μM of cholesterol (+CHOL). n = 3. ∗∗p < 0.01, ∗∗∗p < 0.001 by two -ANOVA.
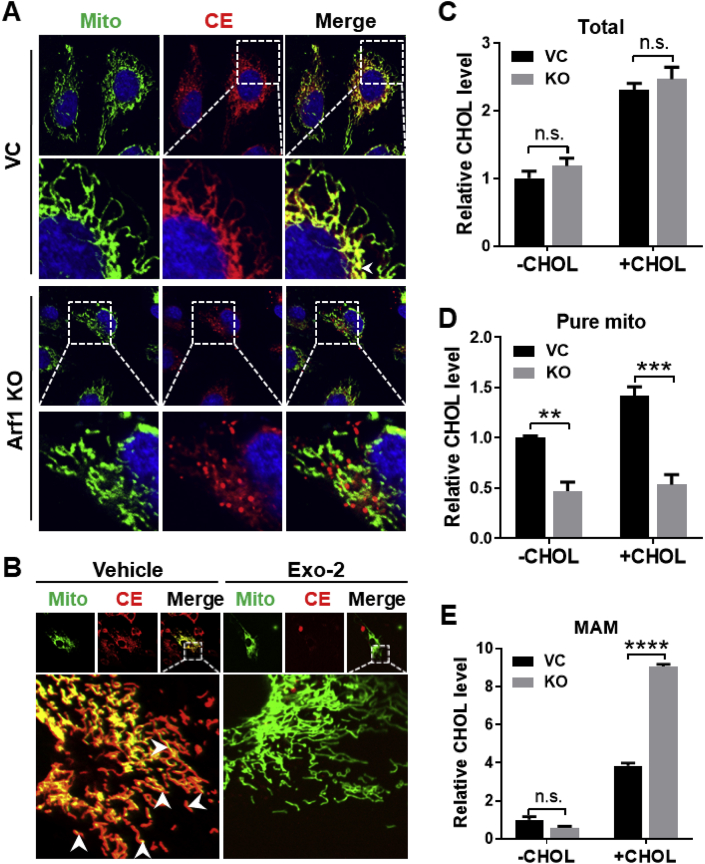
3.6. Arf1 is required for mitochondrial uptake of cholesterol and fatty acids hydrolyzed from cholesterol esters
Arf1, a GTPase required for COPI vesicle assembly, has recently been implicated in making connections to the ER with other organelles, including lipid droplets and mitochondria [16]. We have recently shown that Aster-C directly interacts with COPI [22]. Since the MTS is required for tethering Aster-B between the ER and mitochondria [23], we first analyzed the interaction between Aster-B and Arf1 by Co-IP analysis in 293A cells transiently expressing GFP tagged Aster-B. The results show that Aster-B was not associated with Arf1, but specifically interacted with COPA, a component of COPI vesicles (Figure S6A). However, the interaction between Aster-B and COPA was significantly weakened by Exo-2, a potent and specific inhibitor of Arf1 (Figure S6B). We next questioned whether Arf1 also plays a role in cholesterol transport from the ER to mitochondria. To test our hypothesis, we generated a stable HeLa cell line that is deficient in Arf1 expression using the CRISPR/Cas9 technique and determined the effect of Arf1 depletion on mitochondrial cholesterol uptake. Ablation of Arf1 was confirmed by western blot analysis (Figure S7). Consistent with the proposed role of Arf1 in ER-mitochondrial tethering [24], Arf1 deficiency prevented the mitochondrial fatty acid transport derived from CholEsteryl BODIPY™ 542/563 C11 relative to the vector control cells (Figure 6A, highlighted by arrows). In contrast to Aster-B, Arf1 was not required for uptake of cholesterol esters, however Arf1 deficiency did prevent the transport of fatty acids from the cholesterol esters to the mitochondria (Figure 6A). In support of this notion, we further show that pharmacological inhibition of Arf1 by Exo-2 also prevented the fatty acid uptake from cholesterol esters to the mitochondria relative to the vehicle-treated cells (Figure 6B). We further investigated this phenomenon by measuring the cholesterol levels in subcellular fractionated vector control and Arf1 KO cells permeabilized with digitonin. Although Arf1 deficiency did not significantly affect the total amount of cellular cholesterol (Figure 6C), Arf1 depletion led to a significant reduction of mitochondrial cholesterol content in the purified mitochondrial fraction (Figure 6D), which is accompanied by a significant increase of cholesterol content in the MAM fraction (Figure 6E).
Figure 6.
Arf1 is required for cholesterol transport into mitochondria. (A) Confocal imaging analysis depicting the mitochondrial cholesterol transport in HeLa vector control (VC) and Arf1 knockout (Arf1-KO) cells. Cells were starved in KRPH buffer for 30 min and then treated with 10 μM of CholEsteryl BODIPY™ 542/563 C11 for 10 min. (B) Confocal imaging analysis depicting the mitochondrial cholesterol transport in C2C12 cells treated with 10 μM of Exo-2 for 2 h followed by incubation with 10 μM of CholEsteryl BODIPY™ 542/563 C11 for 10 min. Nuclei were stained with Hoechst 33342, and mitochondria were stained with Mitotracker-Green. (C–E) Total cholesterol level (C) and cholesterol levels in pure mitochondria (D) and MAM (E) in HeLa VC and Arf1 KO cells. Cells were treated with MCD (-CHOL), or pemeabilized by digitonin after MCD treatment and re-incubated with 50 μM of cholesterol (+CHOL). n = 3. ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 by two-way ANOVA.
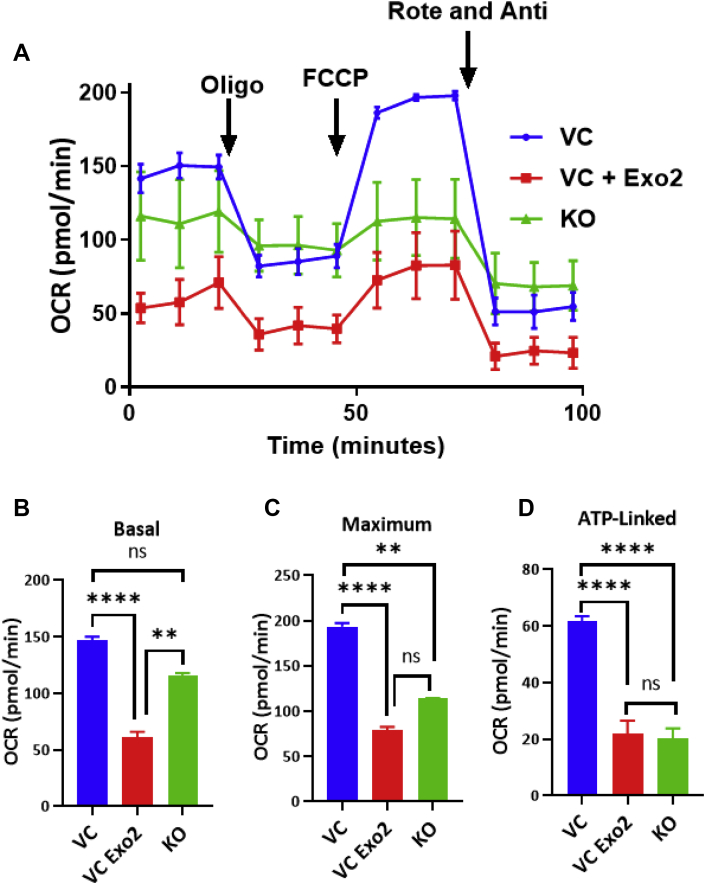
3.7. Ablation of Aster-B or inhibition of Arf1 causes mitochondrial dysfunction
As Aster-B is required for mitochondrial cholesterol transport, we hypothesized that this would also cause mitochondrial dysfunction as cholesterol is required for both mitochondrial biogenesis and membrane maintenance [25]. To test this hypothesis, we next examined the effects of Aster-B ablation on mitochondrial respiration in C2C12 cells using a Seahorse XF24 analyzer. The results showed that ablation of Aster-B significantly impaired mitochondrial respiration (Figure 7A, quantified in Figure 7B–D) in C2C12 cells which were first depleted of cholesterol by MCD, followed by stimulation using non-fluorescent cholesterol esters. Likewise, inhibition of Arf1 by Exo-2 also caused defective mitochondrial respiration to a similar degree to the Aster-B KO cells. Our findings are corroborated by a previous report which shows that Arf1 is required for both mitochondrial function and morphology [26].
Figure 7.
Ablation of Aster-B or inhibition of Arf1 causes mitochondrial dysfunction. (A) Seahorse XF analysis showing oxygen consumption rate (OCR) of C2C12 vector control with vehicle (VC) or 10 μM of Exo-2 (VC Exo-2) and Aster-B knockout (KO) cells treated with MCD for 2 hr and then treated with 25 μM non-fluorescent cholesterol ester for 1 h. Cells were treated with vehicle or 10 μM of Exo-2 throughout the assay. (B) Basal, (C) maximum, (D) ATP-linked respiration of the Seahorse XF analysis is shown. n = 5. ∗∗p < 0.01, ∗∗∗∗p < 0.0001 by student's t-test.
4. Discussion
Cholesterol transport to mitochondria is believed to be achieved primarily through the activity of lipid transfer proteins at membrane contact sites, and fatty acids have been shown to be crucial to this process possibly through StAR domains [4,15], since mitochondria are not connected with the vesicular trafficking networks. Mitochondria and the ER form multiple membrane contact sites known as the MAM, which plays an important role in phospholipid transport [27]. Although the ER is the major site of de novo cholesterol synthesis, only 0.5–1% of total cellular cholesterol is present in the ER [28]. Hormonal stimulation significantly increases the number of ER-mitochondrial contact sites in steroidogenic MA-10 Leydig cells [29]. Although the role of the MAM in cholesterol trafficking remains elusive, the ER and mitochondria move into close contact through tethers at the MAM during cholesterol flux [30]. Additionally, the MAM is highly enriched with cholesterol relative to the ER/mitochondria [29], which is also confirmed by the current study. Accordingly, several MAM proteins have been implicated in mitochondrial cholesterol transport and steroidogenesis [9]. In this study, we identified Aster-B as a key regulator of cholesterol trafficking to the mitochondria, which is supported by multiple lines of evidence. We show that Aster-B is localized at the MAM, where it facilitates cholesterol transport to mitochondria. Aster-B translocates to mitochondria in response to stimulation with cholesterol, which is mediated by an MTS at the N-terminus of Aster-B. Consequently, deletion of the MTS not only impairs mitochondrial cholesterol transport, but also leads to retention of Aster-B and a significant increase in cholesterol content in the MAM. The MTS is only found in Aster-B, but not in other isoforms of the Aster family, including Aster-A and Aster-C. Our findings are corroborated by a previous report which showed through an in vitro lipid transfer assay that Ltc1, a yeast ortholog of Aster-B, selectively transfers sterols but not phospholipids [14]. In addition to the MAM, the yeast Ltc1 is also found at other ER membrane contact sites, including ER-endosome and vacuole-mitochondria sites [10,12]. Yeast vacuoles are functionally similar to mammalian lysosomes. A previous study showed that deletion of the mitochondrial tethering sequences renders Ltc1 trapped in the ER, although the functional importance of Ltc1 at the contact sites remains elusive [14]. Therefore, the Aster family of proteins may also play a similar role in regulating lysosomal function.
Arf1 is a small GTPase that is best known for its critical roles in membrane traffic by initiating the recruitment of coat proteins [31]. However, cumulative evidence has also highlighted the importance of Arf1 in regulating other cellular functions, including cholesterol trafficking to lipid droplets, mitochondrial morphology, and sterol regulatory-element binding protein (SREBP) maturation [23,32,33]. Arf1 was also shown to directly interact with vacuolar v-GTPase, which is required for mTORC1 activation in yeast [34]. In further support for Aster-B in mitochondrial cholesterol trafficking, this study identified an unexpected role for Arf1 in regulating mitochondrial cholesterol transport. We show that genetic deletion or pharmacological inhibition of Arf1 not only prevented mitochondrial cholesterol transport, but also impaired mitochondrial uptake of fatty acids released from hydrolysis of cholesterol esters, leading to a significant increase in cholesterol content in the MAM. Our findings are corroborated by a recent report that the yeast homolog of Arf1, Arf-like protein I (Arl-I) is directly involved in the trafficking of proteins from the ER to the mitochondrial surface via an ER surface retrieval pathway termed ER-SURF [16]. Arl-I was identified by a genome-wide screening of regulators of the ER-SURF system, which allows for ER proteins to be targeted to the mitochondria. Furthermore, Arf1 was shown to contribute to the tethering of proteins at the ER mitochondrial contact site known as the ERMES complex in yeast, which plays a critical role in transporting lipids and Ca2+ between the ER and mitochondria [24].
Cholesterol transport to mitochondria is the rate-limiting step for steroidogenesis. Aster-B has also been shown to be heavily expressed in Leydig cells where testosterone is produced [35] as well as in the adrenal glands [12]. Mitochondrial cholesterol in the steroidogenic cells of the adrenal glands plays a critical role in the synthesis of hormones, including androgens, estrogens, mineralocorticoids, and glucocorticoids [36]. In final support of our findings, a recent report showed that mice lacking in Aster-B were deficient in both steroidogenesis and adrenal cholesterol [12]. Together, this study identifies Aster-B as a novel regulator of mitochondrial cholesterol and fatty acid transport, indicating an important role for Aster-B in steroidogenesis. Excess mitochondrial cholesterol has also been a consistent biomarker for several diseases, including Alzheimer's disease, steatohepatitis, and carcinogenesis [28]. As abnormal mitochondrial cholesterol is a common phenotype in these diseases, manipulation and further investigation of Aster-B could prove to be a powerful tool in understanding and developing therapeutics for these disorders.
Author contributions
Y.S. conceived the project and designed the research plan. J-P.A, J.Z., and H.S. performed the experiments. Y.S., J-P.A J.Z., H.S., and J.N. analyzed the data. Y.S. and J-P.A wrote the manuscript, and all authors edited it.
Acknowledgments
This work was partly sponsored by grants from the American Diabetes Association (1-18-IBS-329 (Y.S.), the National Institutes of Health (R01AG055747, Y.S.), Joe and Teresa Long Endowment (Y.S.), the T32 Aging Training Grant at UT Health San Antonio (J.R.A. and J.N.), and technical assistance from UT Health San Antonio Flow Cytometry Core for help with the CRISPR/Cas9 selection.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2020.101055.
Conflict of interest
The authors declare no competing interests.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Maxfield F.R., Wüstner D. Intracellular cholesterol transport. Journal of Clinical Investigation. 2002;110(7):891–898. doi: 10.1172/JCI16500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duarte A., Poderoso C., Cooke M., Soria G., Cornejo Maciel F., Gottifredi V. Mitochondrial fusion is essential for steroid biosynthesis. PloS One. 2012;7(9) doi: 10.1371/journal.pone.0045829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elustondo P., Martin L.A., Karten B. Mitochondrial cholesterol import. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2017;1862(1):90–101. doi: 10.1016/j.bbalip.2016.08.012. [DOI] [PubMed] [Google Scholar]
- 4.Petrescu A.D., Gallegos A.M., Okamura Y., Strauss J.F., 3rd, Schroeder F. Steroidogenic acute regulatory protein binds cholesterol and modulates mitochondrial membrane sterol domain dynamics. Journal of Biological Chemistry. 2001;276(40):36970–36982. doi: 10.1074/jbc.M101939200. [DOI] [PubMed] [Google Scholar]
- 5.Fujimoto M., Hayashi T. New insights into the role of mitochondria-associated endoplasmic reticulum membrane. International Review Cell and Molecular Biology. 2011;292:73–117. doi: 10.1016/B978-0-12-386033-0.00002-5. [DOI] [PubMed] [Google Scholar]
- 6.Area-Gomez E., Del Carmen Lara Castillo M., Tambini M.D., Guardia-Laguarta C., de Groof A.J.C., Madra M. Upregulated function of mitochondria-associated ER membranes in Alzheimer disease. The EMBO Journal. 2012;31(21):4106–4123. doi: 10.1038/emboj.2012.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujimoto M., Hayashi T., Su T.P. The role of cholesterol in the association of endoplasmic reticulum membranes with mitochondria. Biochemical and Biophysical Research Communications. 2012;417(1):635–639. doi: 10.1016/j.bbrc.2011.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marriott K.S., Prasad M., Thapliyal V., Bose H.S. sigma-1 receptor at the mitochondrial-associated endoplasmic reticulum membrane is responsible for mitochondrial metabolic regulation. Journal of Pharmacology and Experimental Therapeutics. 2012;343(3):578–586. doi: 10.1124/jpet.112.198168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prasad M., Kaur J., Pawlak K.J., Bose M., Whittal R.M., Bose H.S. Mitochondria-associated endoplasmic reticulum membrane (MAM) regulates steroidogenic activity via steroidogenic acute regulatory protein (StAR)-voltage-dependent anion channel 2 (VDAC2) interaction. Journal of Biological Chemistry. 2015;290(5):2604–2616. doi: 10.1074/jbc.M114.605808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Besprozvannaya M., Dickson E., Li H., Ginburg K.S., Bers D.M., Auwerx J. GRAM domain proteins specialize functionally distinct ER-PM contact sites in human cells. Elife. 2018;7 doi: 10.7554/eLife.31019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gatta A.T., Wong L.H., Sere Y.Y., Calderon-Norena D.M., Cockcroft S., Menon A.K. A new family of StART domain proteins at membrane contact sites has a role in ER-PM sterol transport. Elife. 2015;4 doi: 10.7554/eLife.07253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sandhu J., Li S., Fairall L., Pfisterer S.G., Gurnett J.E., Xiao X. Aster proteins facilitate nonvesicular plasma membrane to ER cholesterol transport in mammalian cells. Cell. 2018;175(2):514–529. doi: 10.1016/j.cell.2018.08.033. e520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Besprozvannaya M., Dickson E., Li H. vol. 7. 2018. GRAM domain proteins specialize functionally distinct ER-PM contact sites in human cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murley A., Sarsam R.D., Toulmay A., Yamada J., Prinz W.A., Nunnari J. Ltc1 is an ER-localized sterol transporter and a component of ER–mitochondria and ER–vacuole contacts. The Journal of Cell Biology. 2015;209(4):539–548. doi: 10.1083/jcb.201502033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jefcoate C. High-flux mitochondrial cholesterol trafficking, a specialized function of the adrenal cortex. Journal of Clinical Investigation. 2002;110(7):881–890. doi: 10.1172/JCI16771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen K.G., Aviram N., Laborenz J., Bibi C., Meyer M., Spang A. An ER surface retrieval pathway safeguards the import of mitochondrial membrane proteins in yeast. Science. 2018;361(6407):1118–1122. doi: 10.1126/science.aar8174. [DOI] [PubMed] [Google Scholar]
- 17.Wieckowski M.R., Giorgi C., Lebiedzinska M., Duszynski J., Pinton P. Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nature Protocols. 2009;4(11):1582–1590. doi: 10.1038/nprot.2009.151. [DOI] [PubMed] [Google Scholar]
- 18.Castellano B.M., Thelen A.M. Lysosomal cholesterol activates mTORC1 via an SLC38A9-Niemann-Pick C1 signaling complex. 2017;355(6331):1306–1311. doi: 10.1126/science.aag1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore M.S., Schwoebel E.D. Nuclear import in digitonin-permeabilized cells. Current Protocols Cell Biology Chapter. 2001;11:Unit 11.17 doi: 10.1002/0471143030.cb1107s05. [DOI] [PubMed] [Google Scholar]
- 20.Shen W.J., Azhar S., Kraemer F.B. Lipid droplets and steroidogenic cells. Experimental Cell Research. 2016;340(2):209–214. doi: 10.1016/j.yexcr.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rambold A.S., Cohen S., Lippincott-Schwartz J. Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Developmental Cell. 2015;32(6):678–692. doi: 10.1016/j.devcel.2015.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J., Andersen J.P., Sun H., Liu X., Sonenberg N., Nie J. Aster-C coordinates with COP I vesicles to regulate lysosomal trafficking and activation of mTORC1. EMBO Reports. 2020 doi: 10.15252/embr.201949898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilfling F., Thiam A.R., Olarte M.-J., Wang J., Beck R., Gould T.J. Arf1/COPI machinery acts directly on lipid droplets and enables their connection to the ER for protein targeting. Elife. 2014;3 doi: 10.7554/eLife.01607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davis D.A. Arf1 and ER-mitochondrial tethering – a new trick for an old dog. FEBS Journal. 2018;285(11):1985–1987. doi: 10.1111/febs.14490. [DOI] [PubMed] [Google Scholar]
- 25.Martin L.A., Kennedy B.E., Karten B. Mitochondrial cholesterol: mechanisms of import and effects on mitochondrial function. Journal of Bioenergetics and Biomembranes. 2016;48(2):137–151. doi: 10.1007/s10863-014-9592-6. [DOI] [PubMed] [Google Scholar]
- 26.Ackema K.B., Hench J., Böckler S., Wang S.C., Sauder U., Mergentaler H. The small GTPase Arf1 modulates mitochondrial morphology and function. The EMBO Journal. 2014;33(22):2659–2675. doi: 10.15252/embj.201489039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tatsuta T., Scharwey M., Langer T. Mitochondrial lipid trafficking. Trends in Cell Biology. 2014;24(1):44–52. doi: 10.1016/j.tcb.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 28.Garcia-Ruiz C., Mari M., Colell A., Morales A., Caballero F., Montero J. Mitochondrial cholesterol in health and disease. Histology & Histopathology. 2009;24(1):117–132. doi: 10.14670/HH-24.117. [DOI] [PubMed] [Google Scholar]
- 29.Fujimoto M., Hayashi T., Su T.-P. The role of cholesterol in the association of endoplasmic reticulum membranes with mitochondria. Biochemical and Biophysical Research Communications. 2012;417(1):635–639. doi: 10.1016/j.bbrc.2011.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Helle S.C., Kanfer G., Kolar K., Lang A., Michel A.H., Kornmann B. Organization and function of membrane contact sites. Biochimica et Biophysica Acta. 2013;1833(11):2526–2541. doi: 10.1016/j.bbamcr.2013.01.028. [DOI] [PubMed] [Google Scholar]
- 31.Beck R., Sun Z., Adolf F., Rutz C., Bassler J., Wild K. Membrane curvature induced by Arf1-GTP is essential for vesicle formation. Proceedings of the National Academy of Sciences of the U S A. 2008;105(33):11731–11736. doi: 10.1073/pnas.0805182105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ackema K.B., Hench J., Bockler S., Wang S.C., Sauder U., Mergentaler H. The small GTPase Arf1 modulates mitochondrial morphology and function. The EMBO Journal. 2014;33(22):2659–2675. doi: 10.15252/embj.201489039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smulan L.J., Ding W., Freinkman E., Gujja S., Edwards Y.J.K., Walker A.K. Cholesterol-independent SREBP-1 maturation is linked to ARF1 inactivation. Cell Reports. 2016;16(1):9–18. doi: 10.1016/j.celrep.2016.05.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dechant R., Saad S., Ibanez A.J., Peter M. Cytosolic pH regulates cell growth through distinct GTPases, Arf1 and Gtr1, to promote Ras/PKA and TORC1 activity. Molecular Cell. 2014;55(3):409–421. doi: 10.1016/j.molcel.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 35.McDowell E.N., Kisielewski A.E., Pike J.W., Franco H.L., Yao H.H.C., Johnson K.J. A transcriptome-wide screen for mRNAs enriched in fetal Leydig cells: CRHR1 agonism stimulates rat and mouse fetal testis steroidogenesis. PloS One. 2012;7(10):e47359. doi: 10.1371/journal.pone.0047359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu J., Zhang Z., Shen W.J., Azhar S. Cellular cholesterol delivery, intracellular processing and utilization for biosynthesis of steroid hormones. Nutrition and Metabolism. 2010;7:47. doi: 10.1186/1743-7075-7-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.