Abstract
Objectives
Diet-driven obesity is increasingly widespread. Its consequences pose major challenges to human health and health care systems. There are MAP kinase-interacting kinases (MNKs) in mice, MNK1 and MNK2. Studies have demonstrated that mice lacking either MNK1 or MNK2 were partially protected against high-fat diet (HFD)-induced weight gain and insulin resistance. The aims of this study were to evaluate the phenotype of mice lacking both MNKs when given an HFD, to assess whether pharmacological inhibition of MNK function also protects against diet-induced obesity (DIO) and its consequences and to probe the mechanisms underlying such protection.
Methods
Male wild-type (WT) C57Bl6 mice or mice lacking both MNK1 and MNK2 (double knockout, DKO) were fed an HFD or control diet (CD) for up to 16 weeks.
In a separate study, WT mice were also given an HFD for 6 weeks, after which half were treated with the recently-developed MNK inhibitor ETC-206 daily for 10 more weeks while continuing an HFD. Metabolites and other parameters were measured, and the expression of selected mRNAs and proteins was assessed.
Results
MNK-DKO mice were almost completely protected from HFD-induced obesity. Higher energy expenditure (EE) in MNK-DKO mice was observed, which probably reflects the changes in a number of genes or proteins linked to lipolysis, mitochondrial function/biogenesis, oxidative metabolism, and/or ATP consumption. The MNK inhibitor ETC-206 also prevented HFD-induced weight gain, confirming that the activity of the MNKs facilitates weight gain due to excessive caloric consumption.
Conclusions
Disabling MNKs in mice, either genetically or pharmacologically, strongly prevents weight gain on a calorie-rich diet. This finding likely results from increased energy utilisation, involving greater ATP consumption, mitochondrial oxidative metabolism, and other processes.
Keywords: Adipose tissue, Diet-induced obesity, MNK, Lipid metabolism, Mitochondria, Energy expenditure
Highlights
-
•
Knockout of MNK1/MNK2 protects mice against diet-induced obesity.
-
•
MNK1/2 DKO mice have higher energy expenditure.
-
•
MNK1/2 DKO increases the expression of genes of lipid and mitochondrial metabolism.
-
•
Pharmacological inhibition of MNKs has similar effects.
Abbreviations
- 4E-BP
eIF4E-binding protein
- AGPAT9
1-acylglycerol-3-phosphate O-acyltransferase 9
- AT
adipose tissue
- ATGL
adipose triglyceride lipase
- BMAL1
brain and muscle ARNT (Aryl hydrocarbon receptor nuclear translocator)-like protein 1
- CD
control diet
- CIDEA
cell death-inducing DFFA (DNA fragmentation factor subunit alpha)-like effector A
- DEC1
basic helix-loop-helix family member e40
- DIO
diet-induced obesity
- DKO
double knockout
- EE
energy expenditure
- eIF
eukaryotic initiation factor
- HFD
high-fat diet
- HLX
H2.0 like homeobox
- HOXA5
homeobox a5
- HSL
hormone-sensitive lipase
- MNK
MAP kinase-interacting kinase
- mTORC1
mechanistic target of rapamycin complex 1
- NAFLD
non-alcoholic fatty liver disease
- NEFA
non-esterified fatty acid
- NRF2
nuclear factor erythroid 2-related factor 2
- ORO
oil red O
- PER
period circadian protein
- PGC1α
peroxisome proliferator-activated receptor gamma coactivator 1-alpha
- PPARγ
peroxisome proliferator-activated receptor gamma
- RQ
respiratory quotient
- SERCA
sarco/endoplasmic reticulum Ca2+-ATPase
- SFRP5
secreted frizzled-related protein 5
- SLC29A1
solute carrier family 29 member 1
- TG
triglyceride
- UCP
uncoupling protein
- WNT
wingless/Int-1
- WT
wild-type
- YY1
yin yang 1 transcription factor
1. Introduction
Weight gain, obesity, and their consequences are increasingly prevalent in many societies, both in high- and middle-income countries [1]. Obesity is associated with non-alcoholic fatty liver disease (NAFLD), type-2 diabetes, and other related comorbidities [2]. The chronic diseases associated with obesity present a substantial challenge for human health, health care systems, society, and the economy. Understanding the physiological and molecular mechanisms involved in weight gain and its adverse consequences is necessary to provide insights into tackling the ‘epidemic’ of type 2 diabetes and associated NAFLD. We studied the roles of the MAP kinase-interacting kinases (MNKs), which are phosphorylated and activated by MAP kinases [3,4], as putative signalling molecules involved in the development of obesity and liver fat accumulation.
Mice (and humans) have 2 genes encoding MNKs, Mknk1 and Mknk2, that encode the proteins MNK1 and MNK2, which differ in their regulation and other properties (reviewed [5]). MNK1 activity is tightly controlled by ERK and p38 MAP kinase signalling [6,7], and MNK2 displays high basal activity [8]. The only validated in vivo MNK substrate is eukaryotic translation initiation factor (eIF4E), which it phosphorylates on Ser209 [9,10]; MNKs are the only kinases that phosphorylate eIF4E in vivo [9]. eIF4E plays a crucial role in protein synthesis and its control and is strongly implicated in cancers [11]. Indeed, most reports have concerned the role of eIF4E and its phosphorylation in cancer, although MNKs or phosphorylation of eIF4E is implicated in other physiological processes, for example, neurological disorders [[12], [13], [14]]. Our most recent study was the first to uncover MNKs' role in diet-induced obesity (DIO) [15].
The high-fat-fed male rodent is a widely used, accepted pre-clinical model of DIO and associated metabolic disorders such as liver fat accumulation [16]. We previously showed that male mice lacking either MNK1 or MNK2 are partially protected against the adverse effects of consuming a high-fat diet (HFD), such as weight gain and insulin resistance [15]. Notably, the phenotypes of HFD-fed MNK1-KO or MNK2-KO mice differed in several respects.
That study's results raised several important questions concerning the MNKs' roles in metabolic disease. For example, does knocking out both MNK isoforms (MNK double knockout; MNK-DKO) confer a stronger phenotype than a knockout of either isoform? Second, does pharmacological inhibition of MNKs also prevent DIO? In particular, can MNK inhibition protect against obesity and liver fat accumulation developed as a consequence of consuming the HFD? Third, through what molecular mechanisms does disabling MNK signalling protect against DIO? We attempted to answer these questions in this study.
Our data showed that knockout of both MNK isoforms prevents DIO and liver fat accumulation; increases energy expenditure (EE); and increases the expression in adipose tissue (AT) of key genes involved in lipolysis, mitochondrial function, and ATP-consuming futile cycling. Furthermore, inhibition of MNK activity with a specific pharmacological inhibitor in HFD-fed wild-type (WT) mice also prevents DIO (weight gain) and fatty liver and leads to changes in gene expression similar to those observed in MNK-DKO animals. These novel findings help explain the protection against DIO conferred by disabling MNKs.
2. Materials and methods
2.1. Animal studies
All animal procedures were conducted in the Bioresources Unit at the South Australian Health and Medical Research Institute (SAHMRI) in accordance with local regulations and with the approval of the SAHMRI Animal Ethics Committee (SAM-181, -324 and -416.19).
Male C57Bl6 mice were maintained under a 12 h light/dark cycle (lights on at 07:00 h) at 19 °C–23 °C with free access to food and water. Male mice have been widely used in studies such as these [16] to avoid complications caused by the oestrus cycle in female animals. In addition, this allowed a direct comparison with our earlier studies, which also used only males [15]. As we used mice in which there was a homozygous knockout of 2 genes on different chromosomes, it was unfeasible to use littermates (because the numbers of WT or Mknk1−/− Mknk2−/− animals produced in this manner would be prohibitively low). We recognised that this was not the best method, but in this case, it was unavoidable; we further elaborate on this topic in the discussion section.
At 4 weeks of age, mice were randomly allocated to groups and subsequently fed either an HFD or control diet (Table S1) for a further 8 or 16 weeks. The energy content of the diets was 13 kJ/g for the control and 19.3 kJ/g for the HFD. Notably, the HFD also had significant sucrose content, which provided 1.7 kJ/g energy in the HFD. Nevertheless, because it contains a markedly higher fat content, for simplicity, we termed it an HFD. Diets were γ-irradiated (25 kGy) at Steritech (Victoria, Australia) before delivery. Animals were monitored daily and weighed weekly.
Two study formats were adopted (detailed in the results). The first format assessed WT and MNK-DKO mice. At the end of the feeding period, mice were fasted overnight before blood and tissue collection at post-mortem. Whole blood was collected from mice under isoflurane anaesthesia, via cardiac puncture into K3 EDTA vacutainers. Blood was then centrifuged at 5000 rpm for 10 min at 4 °C for plasma collection. At post-mortem, tissues and organs were carefully dissected, weighed, immediately snap-frozen, and stored at −80 °C.
The second format used only WT mice, which were allocated randomly to either the HFD or CD. Six weeks after commencing the diet, the MNK inhibitor ETC-206 was administered daily to half of the mice in each diet group at 100 mg/kg (see the results for pilot data to establish this dose). The inhibitor was provided by the Experimental Therapeutics Centre (now Experimental Drug Development Centre), Biomedical Sciences Institutes, Singapore. Control mice were gavaged with a volume-matched amount of vehicle (0.5% (w/v) sodium carboxymethylcellulose and 0.5% Tween 80) by using fresh needles for each treatment or diet group to avoid possible transfer of material between animals. Mice were housed according to treatment (and diet). After 16 weeks on the diet, mice were fasted for 1 h, given ETC-206, and then fasted for a further 4–6 h before being anaesthetised using isoflurane. Blood and tissues were collected as described above.
2.2. Plasma and faecal lipids
Plasma lipids and cholesterol were measured by a Beckman Coulter AU480 chemistry analyser at the CSIRO research laboratories.
After 8 weeks on a diet, mice were housed individually and faeces was collected over 2 consecutive 24-h periods. Faecal samples were freeze-dried and then analysed for fat content by gas chromatography–mass spectrometry at the CSIRO research laboratories [17].
2.3. Indirect calorimetry and physical activity measurement
Indirect calorimetry and physical activity monitoring were performed by using the Promethion metabolic cage system (Sable Systems, NV, USA). Mice were housed individually with ad libitum access to food and water. Monitoring was performed over 48 h following a 24-h acclimatisation period.
2.4. Gene expression analysis
Total RNA was isolated from AT samples by using the TRIzol Plus RNA Purification Kit (Life Technologies). RNA was reverse-transcribed into cDNA by using the QuantiNova Reverse Transcription kit (Qiagen). Real-time quantitative polymerase chain reactions were performed using Taqman Fast Advanced Master Mix and Taqman Gene Expression Assays or PowerUp SYBR Green Master Mix (Life Technologies). Data were normalised to the endogenous control gene, B2m (β2-microglobulin), which was established using NormFinder (which yielded a value of 0.195). Changes in mRNA expression were determined by the ΔΔCt method.
2.5. Gel electrophoresis and immunoblotting
Tissues (100 mg) were homogenised in approximately 450 μL of RIPA lysis buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% sodium dodecyl sulphate, 50 mM β-glycerophosphate, 1 mM ethylenediaminetetra-acetic acid [EDTA], 0.5 mM NaVO3, 0.1% 2-mercaptoethanol and protease inhibitors [Roche]) by using a TissueRuptor (Qiagen). After lysis, insoluble material was removed by centrifugation at 13,200×g for 10 min at 4 °C.
Protein concentrations were measured by the Bradford assay (Bio-Rad). Denaturing gel electrophoresis and immunoblotting were performed as described previously [18]. Blots were visualized using an LI-COR Odyssey® Quantitative Imaging System, which was also employed for quantification of signals. Details of the primary antibodies used are provided in Table S2. Fluorescently-labelled secondary antibodies were from Thermo Fisher Scientific.
2.6. Liver Oil Red O staining
Liver samples were embedded in Tissue-Tek® optimum cutting temperature compound and cryosections were cut at 12 μm. Oil Red O (ORO) staining was performed as previously described [19].
2.7. Lipid uptake assays in vivo
Lipid absorption in vivo was determined using the 13C-labelled mixed triglyceride (13C-MTG) breath test [20,21]. Briefly, mice were fasted for 5 h in cages with a raised wire mesh to prevent coprophagia and housed in it for the duration of the experiment. 13C-MTG was dissolved in olive oil and administered at 15 mg/kg body weight by oral gavage. Breath samples were collected by placing the mouse in a gas-tight glass container for 120 s. Ten ml of breath were then syringed out of the chamber and injected into an evacuated 10 ml Exetainer tube (Labco). Mice were immediately returned to their cage between sample collections. Breath samples were collected at baseline and every 5 min until 60 min after 13C-MTG administration, followed by every 15 min until 240 min. The 13C content of breath samples was determined by isotope ratio mass spectrometry.
2.8. Statistical analyses
Statistical tests were performed by using GraphPad Prism (ver. 6). Data were analysed by ANOVA, and data are presented based on this test; in some cases, we also conducted comparisons using Student's t test, pairwise according to genotype and diet, and we commented on this in specific instances. Data throughout this manuscript are presented as mean ± SEM. ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001.
3. Results
3.1. MNK-DKO mice were strongly protected from diet-induced weight gain and liver fat accumulation
After 16 weeks on the HFD, WT mice showed substantial weight gain; by contrast, DKO mice showed no significant increase in weight compared to CD-fed DKO mice (Figure 1A–D; 1A,B show the weights and weight gains measured weekly for all groups). MNK-DKO mice also showed smaller or no increases in the weight of all fat depots analysed (Figure S1A–D). Some fat depots (gonadal, subcutaneous) also tended to be smaller in MNK-DKO mice than in WT mice on the CD (significant differences were observed by using Student's t test but not ANOVA). Furthermore, MNK-DKO mice did not show the increases in plasma triglycerides (TGs) or non-esterified fatty acids (NEFAs) observed in HFD-WT mice (Figure 1E,F). The increases in plasma cholesterol, LDL-C, and HDL-C observed in HFD-WT were substantially blunted in the HFD-DKO animals (Figure S1E–G). Staining for lipids in the liver using ORO revealed lower levels of hepatic fat in DKO mice after 8 weeks on either diet, and no significant increase on HFD in the DKOs (Figure 1G,H). These data demonstrate the better metabolic profile of DKO-HFD animals relative to WT-HFD controls.
Figure 1.
MNK-DKO mice were protected against HFD-induced obesity and other metabolic consequences. (A,B) Weekly body weight and net weight gains (compared to weight at start of the diet feeding period). (C) Body weights at 18 weeks of age. (D) Net weight gains at week 18 compared to the initial weight at week 4. (E,F) Plasma lipid levels. (G) Percentage area of ORO positive stain in liver at 12 weeks of age. (H) Representative images of ORO-stained sections of liver (scale bar, 1 mm). (I) Energy expenditure measured in metabolic cages. (n = 8) Data are represented as mean ± SEM. Solid bars: CD. Empty bars: HFD. See also Figures S1 and S2.
The lack of weight gain of MNK-DKO mice might simply reflect the impaired uptake of lipids (the main source of energy in the HFD) from the gut. Lipid absorption in vivo was therefore assessed by using the 13C-MTG breath test [20,21]. No differences were observed between WT and MNK-DKO mice on either the CD or HFD (data not shown). We collected faeces and measured fat content; as expected, fat content was higher in faeces from mice fed the HFD, but similar increases were observed in samples from WT and MNK-DKO mice on the HFD (Figure S1H), indicating the MNK-DKO animals do not excrete more fat. Thus, the absence of weight gain in DKO-HFD mice cannot be accounted for by defective lipid uptake.
3.2. MNK-DKO mice showed higher EE
Next, we assessed whether metabolic changes might have accounted for the striking protection against DIO observed in MNK-DKO mice, by housing the mice in ‘metabolic cages’. Compared to WT-HFD mice, DKO-HFD mice showed significantly elevated EE; (Figure 1I); EE also trended higher in MNK-DKO animals on the CD (significant by t test for WT vs DKO on the CD), suggesting that a higher metabolic rate is an intrinsic feature of MNK-DKO mice, rather than only being induced under conditions of excess caloric intake. The rates of carbon dioxide emission (VCO2) and oxygen consumption (VO2) were also higher in MNK-DKO mice (Figure S2A,B). Food intake and movement (activity) did not differ significantly between any of the groups (data not shown). The data for respiratory quotient (RQ; i.e. the ratio of CO2 produced to O2 consumed; Figure S2C) showed a reduction in this value for MNK-DKO mice on the HFD compared to the same mice on the CD, indicating that the former had higher levels of lipid utilisation. The higher EE of MNK-DKO mice probably contributes to their protection from DIO and to the smaller size of some fat depots on the CD.
3.3. HFD-fed MNK-DKO mice showed increased expression of genes of lipid metabolism in adipose tissue
To gain insights into the mechanisms underlying the resistance of MNK-DKO mice to DIO, we analysed the expression of genes in adipose tissue (AT). In gonadal fat, levels of mRNAs for several enzymes of lipid metabolism were increased in MNK-DKO compared to WT mice. mRNAs for the lipolytic enzymes including adipose triglyceride lipase (ATGL; Pnpla2) and hormone-sensitive lipase (HSL; Lipe) were higher or trended higher in DKO-HFD than in WT-HFD mice (Figure 2A,B), as was the level of Agpat9 (encoding the rate-limiting enzyme of TG storage, AGPAT9, 1-acylglycerol-3-phosphate O-acyltransferase 9 [22]; Figure 2C). Dgat1 (also involved in lipid storage; Figure 2D) also trended higher. Levels of all these mRNAs decreased or trended lower on the HFD in WT mice but were maintained at CD levels in DKO animals. Immunoblot analysis showed that the levels of ATGL and AGPAT9 proteins also trended higher in DKO-HFD mice than in WT-HFD controls (Figure 2E; quantified in Figure 2F,G). The Western blot data also confirmed that MNKs were the only kinases acting on eIF4E in AT (i.e. P-eIF4E was completely absent in AT from MNK-DKO mice, in line with the literature [9]). However, we observed no consistent change in P-eIF4E levels in AT in the HFD versus CD-fed animals (Figure 2E). Pnpla2 and Lipe mRNAs were also higher in subcutaneous AT from MNK-DKO mice than from WT animals (Figure S3A,B), in this case, on both diets. Thus, MNK-DKO mice may have greater capacity for TG breakdown in AT than WT animals.
Figure 2.
MNK-DKO mice showed increased expression of mRNAs and proteins involved in lipolysis and lipid storage in gonadal AT. (A–D) Expression of the indicated mRNAs. (n = 8) (E) Immunoblots of indicated proteins performed by using lysates of gonadal AT. (F,G) Quantification of ATGL and AGPAT9 protein levels. Signals were normalised to tubulin, and expression is shown as fold change relative to WT-CD (n = 3). Data are represented as mean ± SEM. Solid bars: CD. Empty bars: HFD. See also Figure S3.
We also analysed the expression of genes in the liver, but observed no consistent changes, indicating that the primary metabolic effect of knocking out the MNKs probably operates in AT.
3.4. Expression of genes involved in oxidative metabolism was elevated in AT of MNK-DKO mice
The observation that MNK-DKO mice showed higher EE than WT mice prompted us to test the expression of genes involved in mitochondrial oxidative metabolism in AT. We observed significantly higher levels of the mRNAs for several proteins involved in mitochondrial oxidative phosphorylation (Atp6, Nd1, Nd5, and Cox1; all encoded by mitochondrial DNA) in MNK-DKO mice than in WT mice on the HFD, both in gonadal (Figure 3A–D) and subcutaneous (Nd5, Cox1; Figure S4A–D) AT. Levels also trended higher (significant by t test for WT vs DKO on the CD) in gonadal AT of MNK-DKO mice than in WT mice on the CD and were significantly higher in subcutaneous AT from DKO than in WT mice on the CD (Figure S4A–D). The mRNA for PGC1α (Ppargc1a), a transcription factor that regulates mitochondrial biogenesis [23,24], was also elevated in MNK-DKO mice on the HFD (Figure 3E; Figure S4E). Nrf2 and Pparγ, encoding coactivators of PGC1α, were also higher in gonadal or subcutaneous AT of DKO-HFD mice versus WT-HFD controls (Figure 3F,G; Figure S4F,G); notably, in subcutaneous AT, they were also higher in DKO mice fed the CD. Yy1 (encoding yin yang 1 transcription factor), another positive regulator of mitochondria-related gene expression [25], was elevated in MNK-DKO mice on both diets, significantly so on the HFD (Figure 3H; Figure S4H). Immunoblot analyses revealed a trend towards higher levels of PGC1α protein in gonadal AT from DKO animals on either diet (Figure 3I, data quantified in 3J).
Figure 3.
MNK-DKO mice showed increased levels of mRNAs encoded by mitochondrial genes and regulators of mitochondrial biogenesis in gonadal AT. (A–D) Expression of mRNAs encoding proteins involved in mitochondrial oxidative phosphorylation. (E–H) mRNA expression of regulators of mitochondrial biogenesis. (n = 5–7) (I) Immunoblots of indicated proteins performed by using lysates of gonadal AT. (J) Quantification of PGC1α protein levels. Expression is shown as fold change relative to WT-CD (n = 3). Data are represented as mean ± SEM. Solid bars: CD. Empty bars: HFD. See also Figure S4.
Thus, MNK-DKO mice showed an overall increase in the expression of mRNAs encoding mitochondrial mRNAs and transcriptional regulators of mitochondrial biogenesis in gonadal and subcutaneous AT; in the latter, these mRNAs were elevated in MNK-DKO mice on either diet, again indicating that their regulation by MNKs is an intrinsic feature of MNK-DKO animals, not merely a response to caloric overload.
3.5. MNK-DKO mice showed increased expression of markers and regulators of AT browning and thermogenesis
Mitochondria play a key role in the energy-consuming process of thermogenesis, a phenomenon associated with white AT browning, which is the emergence of beige adipocytes within white AT [26,27]. Beige adipocytes and brown adipocytes are from different lineages; however, both cell types share several phenotypic similarities. Uncoupled mitochondrial respiration (due, e.g., to higher levels of uncoupling protein 1, encoded by Ucp1) contributes to energy wasting (e.g. as a key element of thermogenic mechanisms [28,29]). Despite our efforts, we did not detect consistent changes in the levels of UCP1 or its mRNA in gonadal AT (data not shown). In subcutaneous AT, however, its levels (Figure S5A), and those of the mRNA for the related protein UCP2 (Figure S5B) were elevated in MNK-DKO mice on the HFD, although the difference was not significant. We also assessed other markers of browning, such as the widely used marker Cidea (encoding cell death-inducing DFFA-like effector A) [30]; its expression trended higher in gonadal or subcutaneous AT from DKO animals on either diet (Figure 4A; Figure S5C). Slc29a1 (Solute carrier family 29 member 1) and Hoxa5 (Homeobox a5) were recently identified in cell type-specific profiling studies as being enriched in brown AT [31]; both were elevated in gonadal or subcutaneous AT of MNK-DKO mice on either diet, and more strongly in the latter (Figure 4B,C; Figure S5D,E), again indicating significant browning of AT in MNK-DKO mice. Hlx (H2.0-like homeobox) promotes the browning of white AT [32]. In gonadal AT, its expression trended higher in gonadal AT of MNK-DKO mice on the CD and was significantly elevated in subcutaneous AT of MNK-DKO mice on either diet (Figure 4D; Figure S5F). These data indicate that the ‘browning programme’ is activated in AT of MNK-DKO mice, probably leading to increased oxidative metabolism and ATP production.
Figure 4.
MNK-DKO mice showed increased expression of browning markers, specific transcriptional regulator, Atp2a2, and Sfrp5 in AT. (A–F) Expression of the indicated mRNAs in gonadal AT was assessed by RT-qPCR. (n = 5–6). Data are represented as mean ± SEM. Solid bars: CD. Empty bars: HFD. See also Figure S5.
Processes other than UCP1-mediated mitochondrial uncoupling can operate to consume ATP and enhance EE; one such mechanism involves the sarco/endoplasmic reticulum Ca2+-ATPase (SERCA) family of calcium pumps (SERCA1 and SERCA2, encoded by Atp2a1 and Atp2a2), which are normally expressed in muscle, where they play an important role in thermogenesis [33]; notably, they are also expressed in beige AT [34,35]. Atp2a1 expression was not significantly altered in subcutaneous AT from MNK-DKO mice (and was undetectable in gonadal AT; data not shown). By contrast, the levels of Atp2a2 were higher in gonadal and subcutaneous AT from MNK-DKO mice, more markedly on the HFD, than in the WT controls (Figure 4E; Figure S5G). Pollard et al. [35] demonstrated that upregulation of SERCA1 in white AT protected against DIO, and Ikeda et al. [34] reported that SERCA2 mediated UCP1-independent thermogenesis in beige adipose by acting as a ‘sink’ for metabolic energy.
3.6. Sfrp5 was strongly downregulated in AT from MNK-DKO mice
Which signalling pathways might be involved in the increased levels of proteins of mitochondrial oxidative metabolism and PGC1α and its coactivators? In adipocytes, stimulation of the WNT (Wingless/Int-1) pathway promotes the expression of several genes involved in mitochondrial biogenesis and function [36]; one well-established inhibitor of WNT signalling is secreted frizzled-related protein (SFRP5). Notably, Sfrp5 mRNA was far lower in both gonadal and subcutaneous AT of MNK-DKO mice than in HFD-WT animals (Figure 4F; Figure S5H). The lower levels of expression of Sfrp5 probably help explain or at least contribute to the observed increased expression of genes involved in mitochondrial metabolism in AT of DKO-HFD mice. Because inhibiting SFRP5 in obese mice improved glucose tolerance [37], these changes also probably contribute to the better metabolic ‘health’ of mice where MNK function has been disabled [15].
3.7. Possible role of changes in expression of factors linked to circadian rhythms
Notably, phosphorylation of eIF4E positively regulates the translation of mRNAs that encode certain proteins involved in circadian rhythms, notably, the Per1 and Per2 mRNAs (encoding period circadian protein homolog 1/2) [14], and these are thus linked to the control of energy metabolism (reviewed [38]). The PER proteins negatively regulate their own transcription by repressing the transcription factor Bmal1, brain and muscle ARNT (Aryl hydrocarbon receptor nuclear translocator)-like protein 1, which is normally a driver of the transcription of Per genes [39]. Because phosphorylation of eIF4E promotes the translation of Per1 and Per2 [14], PER1/2 protein levels and their inhibitory role should be reduced in MNK-DKO mice; thus, the transcription of the Per1, Per2, and Bmal1 mRNAs is expected to be higher in MNK-DKO mice. Consistent with this, we observed elevated Per1, Per2, and Bmal1 mRNA expression in gonadal AT of MNK-DKO mice (Figure S6A–C). The transcription factor Dec1 (encoding basic helix-loop-helix family member e40) is a downstream target for Bmal1 [38], and accordingly, its mRNA levels were also higher in MNK-DKO mice (Figure S6D). This result, however, is inconsistent with some reports indicating that Dec1 deficiency protects against DIO [40,41]. Thus, the altered Dec1 expression in MNK-DKO mice probably does not explain the protection of MNK-DKO mice against DIO.
3.8. Establishing the appropriate dose of MNK inhibitor for in vivo studies
Because of the robust protection against DIO observed in MNK-KO mice (in this study and [15]), we assessed whether pharmacological inhibition of the MNKs could confer similar benefits; to do so, we used a specific, potent MNK inhibitor: ETC-206 [42].
To determine the dose of ETC-206 required to achieve significant MNK inhibition in relevant tissues, 4-week-old C57Bl6 male mice were placed on an HFD for 4 weeks. Next, half the mice were given ETC-206 daily at 75 or 100 mg/kg by oral gavage, or vehicle as control while on the HFD for 2 more weeks. Mice were culled 4–6 h after the final dose, and AT was harvested for analysis. Plasma samples from blood collected by cardiac puncture at sacrifice were analysed for ETC-206; a dose of 100 mg/kg yielded somewhat higher plasma drug levels than 75 mg/kg (Figure S7A) did.
To assess the efficacy of ETC-206 in inhibiting MNK activity in vivo, AT samples were analysed by immunoblot for phosphorylated and total eIF4E. Because eIF4E is only phosphorylated by the MNKs (at Ser209) [9] (Figure 2E), its phosphorylation [10] provides a specific read-out of MNK activity. Although 75 mg/kg inhibited eIF4E phosphorylation, a slightly greater inhibition was observed at 100 mg/kg (Figure S7B); thus, we used the latter concentration in subsequent work; the study design is depicted in Figure S7C. We observed no adverse effects in drug-treated mice, including over a longer period of treatment (16 weeks).
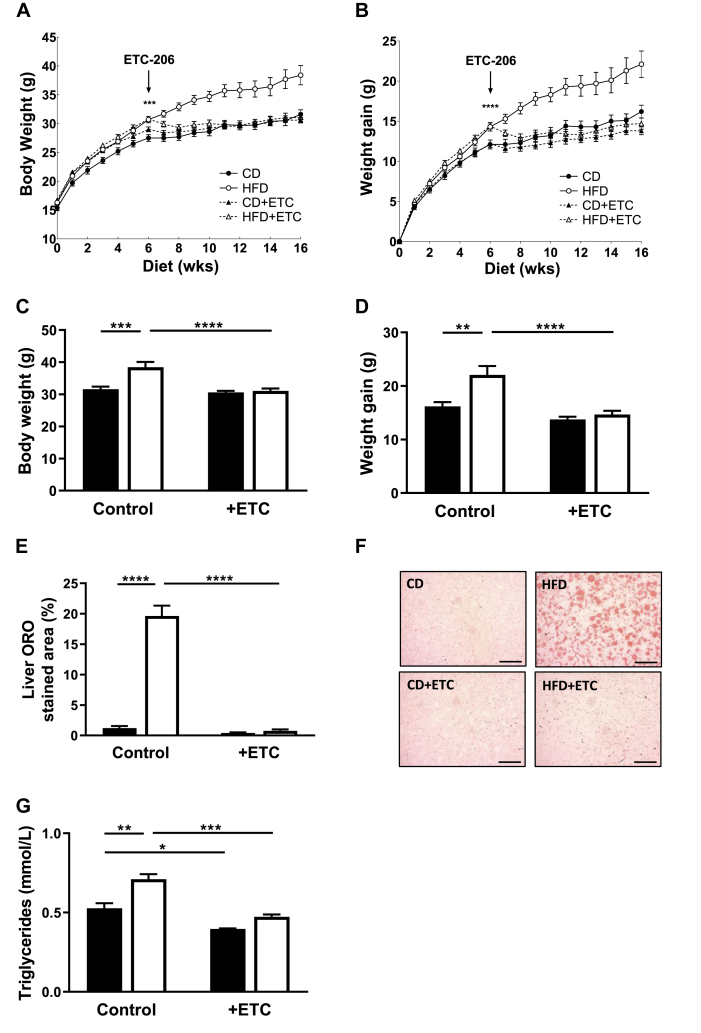
3.9. MNK inhibition substantially reduced weight gain on the HFD
As expected, vehicle-treated HFD-fed mice showed greater weight gain than corresponding CD-fed animals (Figure 5A–D). By contrast, HFD animals that received ETC-206 showed no greater weight gain than CD-fed controls, indicating that ETC-206 prevented further weight gain and was observed to ‘reverse’ the initial weight gain of HFD-fed animals during weeks 4–10 (i.e., the significant gain occurred before they started to receive the drug).
Figure 5.
Inhibition of MNKs reduced weight gain on an HFD. (A,B) Weekly body weights and net weight gains of ETC-206-treated mice. Mice were given the inhibitor or vehicle control daily after 6 weeks on diet. ∗ denotes significance, by t test, between of all CD versus all HFD-fed animals at 6 weeks on the CD/HFD. (C) Body weights at 20 weeks of age (after 16 weeks of HFD). (D) Weight gains at week 20 compared to the initial weights at week 4. (n = 10–12) (E) Percentage area of ORO positive stain in liver at 20 weeks of age. (n = 3–4) (F) Representative images of ORO-stained sections of liver (scale bar, 1 mm). (G) Plasma triglyceride levels. (n = 3–4) Data are represented as mean ± SEM. Solid bars: CD. Empty bars: HFD.
Regrettably, in this study, we did not measure metabolic parameters because housing mice in the ‘metabolic cages’ was incompatible with continued daily gavage (because the animals would have had to be removed from the cages to be gavaged with the drug, invalidating the respirometry data).
After 16 weeks on the HFD, WT mice showed a very marked increase in hepatic fat accumulation (assessed by ORO staining; Figure 5E,F), this effect was completely abolished in ETC-treated animals. HFD-fed mice also showed markedly increased plasma TGs (Figure 5G); these increases were not observed in those treated with ETC-206 (HFD + ETC), similar to the data for MNK-DKO mice (Figure 1E). Therefore, pharmacological inhibition of MNKs with ETC-206 protects HFD-fed mice from several key features of DIO including accumulation of fat in the liver.
3.10. MNK inhibition affected the expression of genes involved in lipid metabolism or oxidative metabolism in AT
Levels of Pnpla2, Lipe, Agpat9, and Dgat1 mRNAs were significantly higher in AT of HFD + ETC mice than in HFD controls (Figure 6A–D), which is broadly similar to our observations for MNK-DKO mice. Agpat9 and Dgat1 were also higher in ETC-treated mice on the CD. Levels of ATGL and AGPAT9 proteins tended to follow similar patterns to those of the corresponding mRNAs (Figure 6E; quantified in Figure 6F,G) and thus also mirrored the situation in MNK-DKO mice (Figure 2E–G).
Figure 6.
Inhibition of MNKs increased expression of genes and proteins involved in lipolysis and lipogenesis in gonadal AT. (A–D) Expression of the indicated mRNAs. (n = 3–4) (E) Immunoblots of indicated proteins performed by using lysates of gonadal AT. (F,G) Quantification of ATGL and AGPAT9 protein levels. Signals were normalised to tubulin, and expression is shown as fold change relative to CD (n = 3). Data are represented as mean ± SEM. Solid bars: CD. Empty bars: HFD.
Similar to the situation for MNK-DKO mice, levels of the Atp6, Nd1, Nd5, and Cox1 mRNAs were also elevated in gonadal AT of HFD + ETC mice compared to HFD controls, and in the cases of Nd1 and Cox1, and in drug-treated mice on the CD (Figure 7A–D). mRNAs for factors that promote mitochondrial biogenesis, Pgc1α, Nrf2, and Pparγ, also trended higher in ETC-treated HFD mice than in vehicle controls (Figure 7E–G). In line with data for MNK-DKO mice, ETC-206 treatment caused a marked decrease in the expression of Sfrp5 on the HFD (Figure 7H). PGC1α protein levels tended to be slightly elevated in samples from drug-treated animals (Figure 7I,J).
Figure 7.
Inhibition of MNKs increased the expression of specific genes in gonadal AT. (A–H) Expression of the indicated mRNAs in gonadal AT was assessed by RT-qPCR. (n = 3–4) (I) Immunoblots of indicated proteins performed by using lysates of gonadal AT. (J) Quantification of PGC1α protein levels. Signals were normalised to tubulin, and expression is shown as fold change relative to CD (n = 3). Data are represented as mean ± SEM. Solid bars: CD. Empty bars: HFD. See also Figure S4.
The data show that MNK inhibition and MNK knockout have similar effects on the response of mice consuming an HFD, implying that their kinase activity mediates these effects and that the phenotype does not depend on changes during development (when MNK activity was intact in the drug study).
4. Discussion
We have previously shown [15] that mice in which either the Mknk1 or the Mknk2 gene was deleted show partial protection against adverse effects of an HFD. In this study, we extended that initial work in key respects. Our data in this study showed for the first time that either genetic or pharmacological inhibition of both MNKs protected mice against DIO and liver fat accumulation. Our new findings provide key insights into the mechanisms that probably underlie protection, substantially add to an understanding of the physiological roles of the MNKs, and indicate a novel potential therapeutic option for treating weight gain and associated NAFLD.
First, we tested the effects of knocking out both Mknk genes on the response to consuming an HFD and showed that MNK-DKO mice were almost entirely protected against DIO and liver fat accumulation. MNK-DKO mice also had significantly decreased plasma triglycerides and cholesterol when fed either an HFD or chow diet, strongly suggesting that MNK-DKO mice had an improved metabolic profile. However, for practical reasons (breeding from MNK1+/− MNK2+/− parents generates far too few homozygous progeny for studies of this type), we did not use the ideal approach [43], namely, using WT and KO littermates, in these studies, and observed similar protection from DIO in MNK-KO mice in 5 cohorts of mice, in 2 different vivariums (in the United Kingdom and Australia, including following additional back-crossing between studies in the second location). Essentially, identical protection was also afforded by an MNK inhibitor. Thus, disabling the MNKs consistently prevents DIO.
Second, we showed that treating HFD-fed WT mice with a specific MNK inhibitor, ETC-206 [44], also prevented weight gain and liver fat accumulation, even after these mice commenced the HFD. These data demonstrate that the effect of disabling MNK function is due to the loss of its activity, not simply of the proteins performing another type of function (which has been observed for certain other MAP kinase-activated protein kinases, e.g., [45]), and importantly, that it does not reflect a change that occurs earlier during development.
Third, metabolic analysis of the mice revealed significantly higher EE in HFD-fed MNK-DKO mice than in WT mice; this difference may, at least in part, account for the smaller fat depots in the DKO animals. We then probed potential mechanisms that could account for higher EE and to explain why MNK-DKO animals are resistant to DIO.
Significantly, the expression of relevant genes and proteins involved in the mobilisation of energy stores was altered in the AT of DKO mice. The elevated levels of enzymes of lipid storage and breakdown in DKO-HFD mice may indicate a more rapid turnover of TG stores in these animals. In particular, faster lipolysis would increase the supply of free fatty acids and thus of substrates for oxidation in mitochondria. Reports have demonstrated that fatty acids may also increase energy metabolism through the activation of PPARs, which sustain mitochondrial function in the heart [46] and skeletal muscle [47].
The acetyl units generated by lipid breakdown are oxidised through the TCA cycle, and this is linked to the mitochondrial electron transport chain and production of ATP. Our data showed elevated expression of mRNAs for several mitochondria-encoded proteins involved in oxidative metabolism in HFD-fed DKO mice versus WT controls (Atp6, Nd1, Nd5, Cox1), suggesting higher levels of mitochondrial activity. The translation of mRNAs for several mitochondrial proteins is controlled by eIF4E [48] via its control by the eIF4E-binding proteins (4E-BPs), which inhibit eIF4E activity. Notably, knockout of the eIF4E-binding proteins (which normally inhibits eIF4E's function in translation) resulted in mice being more sensitive to DIO [49]. Thus, different regulatory inputs into eIF4E (phosphorylation, 4E-BPs) exert distinct effects on sensitivity to caloric excess. Thus, further research should examine whether disabling the MNKs counters the increased sensitivity of 4E-BP knockout mice to DIO.
Nrf2 mRNA levels were elevated in AT from HFD-fed MNK-DKO mice. This transcription factor was reported to protect against the adverse effects of excess caloric load (e.g. [50]), but this is controversial [51]. Nrf2 promotes mitochondrial oxidative metabolism and mechanisms that manage any resulting reactive oxygen species [52,53].
Signalling through mTORC1 (mechanistic target of rapamycin complex 1, a protein kinase) plays an important role in lipid metabolism [54] and has been reported to be modulated by MNKs [55,56]. However, we observed no differences in the phosphorylation of 4E-BP1, a specific substrate for mTORC1 between AT of WT and MNK-DKO mice, on either diet, making it unlikely that differences in mTORC1 signalling explain the resistance of MNK-DKO mice to DIO.
Browning of AT involves increased expression of genes for factors which drive mitochondrial function/oxidative metabolism, including PGC1α, PPARγ, and Yy1 [23,57]. Expression of all 3 genes was elevated in AT of DKO-HFD mice. In gonadal and subcutaneous AT, expression of several beige/brown fat markers [31,58] including Slc29a1 and Hoxa5 [59], was increased in DKO mice, even on the CD, consistent with the other changes in gene expression that we observed.
Furthermore, we observed a marked decrease in the expression of Sfrp5 in subcutaneous and gonadal AT of DKO or ETC-206-treated mice on the HFD. This is a potentially important contributor to the protection against DIO afforded by knocking out MNKs, because knockout of Sfrp5 has also protected mice against DIO [36,60] (although conflicting data have been published [61]). The high levels of Sfrp5 that usually occur in obese mice repress oxidative metabolism, perhaps by inhibiting WNT3a signalling [36]. Thus, the markedly decreased levels of Sfrp5 observed in MNK-DKO mice probably contributed to the increased expression of genes involved in oxidative metabolism, such as mitochondrial proteins [36]. Indeed, elevated expression of a number of mitochondrial genes was observed in MNK-DKO mice on the HFD, as was the increase in EE. The levels of several of these mRNAs were higher in DKO mice even on the CD, suggesting this is an intrinsic feature of disabling the MNKs, rather than a function of the MNKs triggered by ‘overnutrition’. Indeed, the observation that levels of P-eIF4E are not elevated in HFD conditions indicate that the HFD does not enhance MNK activity. This finding is consistent with the fact that MNK2, which has high basal activity, is the major MNK isoform in AT [15].
The higher EE and oxidative metabolism in MNK-DKO or drug-treated mice leads to the next question: What is this energy being used for? Our data revealed higher levels of the SERCA2 Ca2+-ion transporter, which, in common with SERCA1, forms part of an ‘energy-wasting mechanism’, a futile cycle that consumes ATP [62]. SERCA1 is upregulated in another mouse model, which is protected against DIO [35]. Thus, the higher expression of the ‘ATP sink’, together with the previous observations pointing to elevated mitochondrial biogenesis and function, probably contributes to the higher EE and decreased weight gain of DKO mice fed the HFD. Thus, to summarise our findings, the higher EE of MNK-DKO mice probably reflects increased ATP demand (by SERCA2) and associated higher mitochondrial oxidative phosphorylation, to fulfil this demand, with the substrates being provided, for example, by higher rates of lipolysis. This results in ‘burning’ of the excess calories provided by the HFD and thus protection against DIO and accumulation of fat in the liver; the effects in the liver are probably secondary to the changes in AT.
A major remaining question relates to how the loss of MNK function leads to these changes, in particular on an HFD. The key regulatory translation initiation factor eIF4E remains the only validated in vivo substrate for the MNKs [5,63]; nonetheless, the finding that levels of a range of transcripts are altered suggests that the loss of eIF4E phosphorylation affects the transcription or stability of these mRNAs; however, there is no known link between phosphorylation of eIF4E and any relevant transcription factor. Some additional proteins can be phosphorylated by the MNKs, at least in vitro [5], but these do not appear to be linked to the gene expression changes we observed. If eIF4E is the relevant substrate, the effects of knocking out or inhibiting MNKs on metabolism are probably due to the altered translation of certain mRNAs. Only one unbiased screen (in mouse embryonic fibroblasts) has thus far provided information on which mRNAs' translation is affected by eIF4E phosphorylation [64]. This did not reveal any candidates that seemed relevant to explaining the phenotype observed in this study. One approach to assessing whether eIF4E is the relevant MNK substrate with respect to protection from DIO would be to examine the response to a HFD of the mouse line that those authors used. In that line, the phosphorylation site in eIF4E, Ser209, was altered to a non-phosphorylatable alanine residue [64]. However, some data suggest that alanine at this position is an inaccurate mimic of non-phosphorylated serine [65]. To gain further insights into the physiological roles of the MNKs, it will be invaluable to screen for additional substrates, once a suitable method is developed. Similarly, it would be important to identify the tissue(s) in which MNK function mediates the responses to caloric overload, a goal that requires the development of conditional knockout mouse lines for Mknk1 and Mknk2, which are not available.
Further research could also determine whether knockout or inhibition of the MNKs also prevents or reduces weight gain in genetic models of obesity such as ob/ob mice [66].
Last, this study demonstrated that inhibition of the MNKs can prevent, and to some extent, reverse effects of caloric ‘overload’ in mice. MNKs are druggable targets [42,67] and, as non-essential enzymes [9], inhibiting their function will probably be low risk. Moreover, the MNK inhibitor was well tolerated by the mice; drug-treated mice showed no adverse effects. MNK inhibitors used in our study and others, have entered clinical trials (eFT-508; Tomivosertib; https://clinicaltrials.gov/). Thus, based on our novel data, MNK inhibitors may be valuable in the worldwide struggle against the development of metabolic disorders such as obesity and NAFLD that result from the weight gain associated with excessive caloric intake and other changes in lifestyle.
Author contributions
L.Y.S., W.X.K., X.W., K.B.J., D.W., and B.T. conducted the experiments and analysed the data. L.G., J.Z., and C.D.B. provided supervision. C.D.B. and A.J.P. provided advice. C.G.P. designed the overall project and provided supervision. L.Y.S., W.X.K., X.W., B.T., C.D.B, A.J.P., and C.G.P. contributed to writing the manuscript. C.G.P. acquired the funding.
Acknowledgements
We thank Cathryn Pape and Michael Adams for measuring the plasma and faecal lipids. We also thank Dr Roger Yazbek for analysing the breath test samples for lipid absorption experiments. These studies were supported by the South Australian Health & Medical Research Institute, Australia and the Experimental Therapeutics Centre (ETC), A-STAR, Singapore, who provided ETC-206. ETC played no role in the study design or collection/interpretation of the data; AUM Biosciences, to whom this compound is now licensed, approved the submission of this article. B.T. was supported in part by funding from the South Australian Government for a joint research laboratory with Shengli Hospital. C.D.B. was funded in part by the Southampton NIHR Biomedical Research Centre, UK. We thank Dr Beverley Mühlhäusler (CSIRO, Adelaide) for helpful advice.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2020.101054.
Conflict of interest
The authors declare no competing interests.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Collaboration, NCDRF Worldwide trends in diabetes since 1980: a pooled analysis of 751 population-based studies with 4.4 million participants. Lancet. 2016;387:1513–1530. doi: 10.1016/S0140-6736(16)00618-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Byrne C.D., Targher G. NAFLD: a multisystem disease. Journal of Hepatology. 2015;62:S47–S64. doi: 10.1016/j.jhep.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 3.Waskiewicz A.J., Flynn A., Proud C.G., Cooper J.A. Mitogen-activated kinases activate the serine/threonine kinases Mnk1 and Mnk2. The EMBO Journal. 1997;16:1909–1920. doi: 10.1093/emboj/16.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fukunaga R., Hunter T. Mnk1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. The EMBO Journal. 1997;16:1921–1933. doi: 10.1093/emboj/16.8.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buxade M., Parra-Palau J.L., Proud C.G. The Mnks: MAP kinase-interacting kinases (MAP kinase signal-integrating kinases) Frontiers in Bioscience. 2008;13:5359–5373. doi: 10.2741/3086. [DOI] [PubMed] [Google Scholar]
- 6.Wang X., Flynn A., Waskiewicz A.J., Webb B.L., Vries R.G., Baines I.A. The phosphorylation of eukaryotic initiation factor eIF4E in response to phorbol esters, cell stresses, and cytokines is mediated by distinct MAP kinase pathways. Journal of Biological Chemistry. 1998;273:9373–9377. doi: 10.1074/jbc.273.16.9373. [DOI] [PubMed] [Google Scholar]
- 7.Waskiewicz A.J., Johnson J.C., Penn B., Mahalingam M., Kimball S.R., Cooper J.A. Phosphorylation of the cap-binding protein eukaryotic translation initiation factor 4E by protein kinase Mnk1 in vivo. Molecular and Cellular Biology. 1999;19:1871–1880. doi: 10.1128/mcb.19.3.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scheper G.C., Morrice N.A., Kleijn M., Proud C.G. The mitogen-activated protein kinase signal-integrating kinase Mnk2 is a eukaryotic initiation factor 4E kinase with high levels of basal activity in mammalian cells. Molecular and Cellular Biology. 2001;21:743–754. doi: 10.1128/MCB.21.3.743-754.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ueda T., Watanabe-Fukunaga R., Fukuyama H., Nagata S., Fukunaga R. Mnk2 and Mnk1 are essential for constitutive and inducible phosphorylation of eukaryotic initiation factor 4E but not for cell growth or development. Molecular and Cellular Biology. 2004;24:6539–6549. doi: 10.1128/MCB.24.15.6539-6549.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flynn A., Proud C.G. Serine 209, not serine 53, is the major site of phosphorylation in initiation factor eIF-4E in serum-treated Chinese hamster ovary cells. Journal of Biological Chemistry. 1995;270:21684–21688. doi: 10.1074/jbc.270.37.21684. [DOI] [PubMed] [Google Scholar]
- 11.Proud C.G. Mnks, eIF4E phosphorylation and cancer. Biochimica et Biophysica Acta. 2015;1849:766–773. doi: 10.1016/j.bbagrm.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Amorim I.S., Kedia S., Kouloulia S., Simbriger K., Gantois I., Jafarnejad S.M. Loss of eIF4E phosphorylation engenders depression-like behaviors via selective mRNA translation. Journal of Neuroscience. 2018;38:2118–2133. doi: 10.1523/JNEUROSCI.2673-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moy J.K., Khoutorsky A., Asiedu M.N., Black B.J., Kuhn J.L., Barragan-Iglesias P. The MNK-eIF4E signaling Axis contributes to injury-induced nociceptive plasticity and the development of chronic pain. Journal of Neuroscience. 2017;37:7481–7499. doi: 10.1523/JNEUROSCI.0220-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao R., Gkogkas C.G., de Z.N., Blum I.D., Yanagiya A., Tsukumo Y. Light-regulated translational control of circadian behavior by eIF4E phosphorylation. Nature Neuroscience. 2015 doi: 10.1038/nn.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore C.E., Pickford J., Cagampang F.R., Stead R.L., Tian S., Zhao X. MNK1 and MNK2 mediate adverse effects of high-fat feeding in distinct ways. Scientific Reports. 2016;6:23476. doi: 10.1038/srep23476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Panchal S.K., Brown L. Rodent models for metabolic syndrome research. Journal of Biomedicine and Biotechnology. 2011;2011:351982. doi: 10.1155/2011/351982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lepage G., Roy C.C. Direct transesterification of all classes of lipids in a one-step reaction. The Journal of Lipid Research. 1986;27:114–120. [PubMed] [Google Scholar]
- 18.Liu R., Iadevaia V., Averous J., Taylor P.M., Zhang Z., Proud C.G. Impairing the production of ribosomal RNA activates mammalian target of rapamycin complex 1 signalling and downstream translation factors. Nucleic Acids Research. 2014;42:5083–5096. doi: 10.1093/nar/gku130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehlem A., Hagberg C.E., Muhl L., Eriksson U., Falkevall A. Imaging of neutral lipids by oil red O for analyzing the metabolic status in health and disease. Nature Protocols. 2013;8:1149–1154. doi: 10.1038/nprot.2013.055. [DOI] [PubMed] [Google Scholar]
- 20.Kalivianakis M., Elstrodt J., Havinga R., Kuipers F., Stellaard F., Sauer P.J. Validation in an animal model of the carbon 13-labeled mixed triglyceride breath test for the detection of intestinal fat malabsorption. The Journal of Pediatrics. 1999;135:444–450. doi: 10.1016/s0022-3476(99)70166-6. [DOI] [PubMed] [Google Scholar]
- 21.Roberts A.L., Howarth G.S., Liaw W.C., Moretta S., Kritas S., Lymn K.A. Gastrointestinal pathology in a mouse model of mucopolysaccharidosis type IIIA. Journal of Cellular Physiology. 2009;219:259–264. doi: 10.1002/jcp.21682. [DOI] [PubMed] [Google Scholar]
- 22.Takeuchi K., Reue K. Biochemistry, physiology, and genetics of GPAT, AGPAT, and lipin enzymes in triglyceride synthesis. American Journal of Physiology. Endocrinology and Metabolism. 2009;296:E1195–E1209. doi: 10.1152/ajpendo.90958.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uldry M., Yang W., St-Pierre J., Lin J., Seale P., Spiegelman B.M. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metabolism. 2006;3:333–341. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 24.Harms M., Seale P. Brown and beige fat: development, function and therapeutic potential. Nature Medicine. 2013;19:1252–1263. doi: 10.1038/nm.3361. [DOI] [PubMed] [Google Scholar]
- 25.Cunningham J.T., Rodgers J.T., Arlow D.H., Vazquez F., Mootha V.K., Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- 26.Cohen P., Spiegelman B.M. Cell biology of fat storage. Molecular Biology of the Cell. 2016;27:2523–2527. doi: 10.1091/mbc.E15-10-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Srivastava S., Veech R.L. Brown and brite: the fat soldiers in the anti-obesity fight. Frontiers in Physiology. 2019;10:38. doi: 10.3389/fphys.2019.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozak L.P., Anunciado-Koza R. UCP1: its involvement and utility in obesity. International Journal of Obesity (2005) 2008;32(Suppl 7):S32–S38. doi: 10.1038/ijo.2008.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nedergaard J., Golozoubova V., Matthias A., Asadi A., Jacobsson A., Cannon B. UCP1: the only protein able to mediate adaptive non-shivering thermogenesis and metabolic inefficiency. Biochimica et Biophysica Acta. 2001;1504:82–106. doi: 10.1016/s0005-2728(00)00247-4. [DOI] [PubMed] [Google Scholar]
- 30.Fischer A.W., Shabalina I.G., Mattsson C.L., Abreu-Vieira G., Cannon B., Nedergaard J. UCP1 inhibition in Cidea-overexpressing mice is physiologically counteracted by brown adipose tissue hyperrecruitment. American Journal of Physiology. Endocrinology and Metabolism. 2017;312:E72–E87. doi: 10.1152/ajpendo.00284.2016. [DOI] [PubMed] [Google Scholar]
- 31.Roh H.C., Tsai L.T.Y., Shao M., Tenen D., Shen Y., Kumari M. Warming induces significant reprogramming of beige, but not brown, adipocyte cellular identity. Cell Metabolism. 2018;27:1121–1137 e1125. doi: 10.1016/j.cmet.2018.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang L., Pan D., Chen Q., Zhu L.J., Ou J., Wabitsch M. Transcription factor Hlx controls a systematic switch from white to brown fat through Prdm16-mediated co-activation. Nature Communications. 2017;8:68. doi: 10.1038/s41467-017-00098-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gamu D., Juracic E.S., Hall K.J., Tupling A.R. The sarcoplasmic reticulum and SERCA: a nexus for muscular adaptive thermogenesis. Applied Physiology, Nutrition, and Metabolism. Physiologie Appliquée, Nutrition et Métabolisme. 2019;45:1–10. doi: 10.1139/apnm-2019-0067. [DOI] [PubMed] [Google Scholar]
- 34.Ikeda K., Kang Q., Yoneshiro T., Camporez J.P., Maki H., Homma M. UCP1-independent signaling involving SERCA2b-mediated calcium cycling regulates beige fat thermogenesis and systemic glucose homeostasis. Nature Medicine. 2017;23:1454–1465. doi: 10.1038/nm.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pollard A.E., Martins L., Muckett P.J., Khadayate S., Bornot A., Clausen M. AMPK activation protects against diet induced obesity through Ucp1-independent thermogenesis in subcutaneous white adipose tissue. Nature Metabolism. 2019;1:340–349. doi: 10.1038/s42255-019-0036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mori H., Prestwich T.C., Reid M.A., Longo K.A., Gerin I., Cawthorn W.P. Secreted frizzled-related protein 5 suppresses adipocyte mitochondrial metabolism through WNT inhibition. Journal of Clinical Investigation. 2012;122:2405–2416. doi: 10.1172/JCI63604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rulifson I.C., Majeti J.Z., Xiong Y., Hamburger A., Lee K.J., Miao L. Inhibition of secreted frizzled-related protein 5 improves glucose metabolism. American Journal of Physiology. Endocrinology and Metabolism. 2014;307:E1144–E1152. doi: 10.1152/ajpendo.00283.2014. [DOI] [PubMed] [Google Scholar]
- 38.Sato F., Kohsaka A., Bhawal U.K., Muragaki Y. Potential roles of Dec and Bmal1 genes in interconnecting circadian clock and energy metabolism. International Journal of Molecular Sciences. 2018;19:781. doi: 10.3390/ijms19030781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chaix A., Zarrinpar A., Panda S. The circadian coordination of cell biology. Journal of Cell Biology. 2016;215:15–25. doi: 10.1083/jcb.201603076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noshiro M., Kawamoto T., Nakashima A., Ozaki N., Ueno T., Saeki M. Deficiency of the basic helix-loop-helix transcription factor DEC1 prevents obesity induced by a high-fat diet in mice. Genes to Cells. 2018 doi: 10.1111/gtc.12607. [DOI] [PubMed] [Google Scholar]
- 41.Noshiro M., Kawamoto T., Nakashima A., Ozaki N., Saeki M., Honda K. DEC1 regulates the rhythmic expression of PPARgamma target genes involved in lipid metabolism in white adipose tissue. Genes to Cells. 2020 doi: 10.1111/gtc.12752. [DOI] [PubMed] [Google Scholar]
- 42.Yang H., Chennamaneni L.R., Ho M.W.T., Ang S.H., Tan E.S.W., Jeyaraj D.A. Optimization of selective mitogen-activated protein kinase interacting kinases 1 and 2 inhibitors for the treatment of blast crisis leukemia. Journal of Medicinal Chemistry. 2018;61:4348–4369. doi: 10.1021/acs.jmedchem.7b01714. [DOI] [PubMed] [Google Scholar]
- 43.Lancaster G.I., Kammoun H.L., Kraakman M.J., Kowalski G.M., Bruce C.R., Febbraio M.A. PKR is not obligatory for high-fat diet-induced obesity and its associated metabolic and inflammatory complications. Nature Communications. 2016;7:10626. doi: 10.1038/ncomms10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cherian J., Nacro K., Poh Z.Y., Guo S., Jeyaraj D.A., Wong Y.X. Structure-activity relationship studies of mitogen activated protein kinase interacting kinase (MNK) 1 and 2 and BCR-ABL1 inhibitors targeting chronic myeloid leukemic cells. Journal of Medicinal Chemistry. 2016;59:3063–3078. doi: 10.1021/acs.jmedchem.5b01712. [DOI] [PubMed] [Google Scholar]
- 45.Kotlyarov A., Yannoni Y., Fritz S., Laass K., Telliez J.B., Pitman D. Distinct cellular functions of MK2. Molecular and Cellular Biology. 2002;22:4827–4835. doi: 10.1128/MCB.22.13.4827-4835.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haemmerle G., Moustafa T., Woelkart G., Buttner S., Schmidt A., van de Weijer T. ATGL-mediated fat catabolism regulates cardiac mitochondrial function via PPAR-alpha and PGC-1. Nature Medicine. 2011;17:1076–1085. doi: 10.1038/nm.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meex R.C., Hoy A.J., Mason R.M., Martin S.D., McGee S.L., Bruce C.R. ATGL-mediated triglyceride turnover and the regulation of mitochondrial capacity in skeletal muscle. American Journal of Physiology. Endocrinology and Metabolism. 2015;308:E960–E970. doi: 10.1152/ajpendo.00598.2014. [DOI] [PubMed] [Google Scholar]
- 48.Morita M., Gravel S.P., Chenard V., Sikstrom K., Zheng L., Alain T. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metabolism. 2013;18:698–711. doi: 10.1016/j.cmet.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 49.Le Bacquer O., Petroulakis E., Paglialunga S., Poulin F., Richard D., Cianflone K. Elevated sensitivity to diet-induced obesity and insulin resistance in mice without 4E-BP1 and 4E-BP2. Journal of Clinical Investigation. 2007;117:387–396. doi: 10.1172/JCI29528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xue P., Hou Y., Chen Y., Yang B., Fu J., Zheng H. Adipose deficiency of Nrf2 in ob/ob mice results in severe metabolic syndrome. Diabetes. 2013;62:845–854. doi: 10.2337/db12-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Z., Zhou S., Jiang X., Wang Y.H., Li F., Wang Y.G. The role of the Nrf2/Keap1 pathway in obesity and metabolic syndrome. Reviews in Endocrine & Metabolic Disorders. 2015;16:35–45. doi: 10.1007/s11154-014-9305-9. [DOI] [PubMed] [Google Scholar]
- 52.Tonelli C., Chio I.I.C., Tuveson D.A. Transcriptional regulation by Nrf2. Antioxidants & Redox Signaling. 2018;29:1727–1745. doi: 10.1089/ars.2017.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holmstrom K.M., Baird L., Zhang Y., Hargreaves I., Chalasani A., Land J.M. Nrf2 impacts cellular bioenergetics by controlling substrate availability for mitochondrial respiration. Biology Open. 2013;2:761–770. doi: 10.1242/bio.20134853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lamming D.W., Sabatini D.M. A central role for mTOR in lipid homeostasis. Cell Metabolism. 2013;18:465–469. doi: 10.1016/j.cmet.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brown M.C., Gromeier M. MNK inversely regulates TELO2 vs. DEPTOR to control mTORC1 signaling. Molecular Cellular Oncology. 2017;4 doi: 10.1080/23723556.2017.1306010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brown M.C., Gromeier M. MNK controls mTORC1:substrate association through regulation of TELO2 binding with mTORC1. Cell Reports. 2017;18:1444–1457. doi: 10.1016/j.celrep.2017.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cheng C.F., Ku H.C., Lin H. PGC-1alpha as a pivotal factor in lipid and metabolic regulation. International Journal of Molecular Sciences. 2018;19:3447. doi: 10.3390/ijms19113447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ikeda K., Maretich P., Kajimura S. The common and distinct features of brown and beige adipocytes. Trends in Endocrinology and Metabolism. 2018;29:191–200. doi: 10.1016/j.tem.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cao W., Zhang T., Feng R., Xia T., Huang H., Liu C. Hoxa5 alleviates obesity-induced chronic inflammation by reducing ER stress and promoting M2 macrophage polarization in mouse adipose tissue. Journal of Cellular and Molecular Medicine. 2019;23:7029–7042. doi: 10.1111/jcmm.14600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rauch A., Mandrup S. Lighting the fat furnace without SFRP5. Journal of Clinical Investigation. 2012;122:2349–2352. doi: 10.1172/JCI64196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ouchi N., Higuchi A., Ohashi K., Oshima Y., Gokce N., Shibata R. Sfrp5 is an anti-inflammatory adipokine that modulates metabolic dysfunction in obesity. Science. 2010;329:454–457. doi: 10.1126/science.1188280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mottillo E.P., Ramseyer V.D., Granneman J.G. SERCA2b cycles its way to UCP1-independent thermogenesis in beige fat. Cell Metabolism. 2018;27:7–9. doi: 10.1016/j.cmet.2017.12.015. [DOI] [PubMed] [Google Scholar]
- 63.Xie J., Merrett J.E., Jensen K.B., Proud C.G. The MAP kinase-interacting kinases (MNKs) as targets in oncology. Expert Opinion on Therapeutic Targets. 2019;23:187–199. doi: 10.1080/14728222.2019.1571043. [DOI] [PubMed] [Google Scholar]
- 64.Furic L., Rong L., Larsson O., Koumakpayi I.H., Yoshida K., Brueschke A. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14134–14139. doi: 10.1073/pnas.1005320107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Beggs J.E., Tian S., Jones G.G., Xie J., Iadevaia V., Jenei V. The MAP kinase-interacting kinases regulate cell migration, vimentin expression and eIF4E/CYFIP1 binding. Biochemical Journal. 2015;467:63–76. doi: 10.1042/BJ20141066. [DOI] [PubMed] [Google Scholar]
- 66.Tschop M., Heiman M.L. Rodent obesity models: an overview. Experimental and Clinical Endocrinology & Diabetes. 2001;109:307–319. doi: 10.1055/s-2001-17297. [DOI] [PubMed] [Google Scholar]
- 67.Reich S.H., Sprengeler P.A., Chiang G.G., Appleman J.R., Chen J., Clarine J. Structure-based design of pyridone-aminal eFT508 targeting dysregulated translation by selective mitogen-activated protein kinase interacting kinases 1 and 2 (MNK1/2) inhibition. Journal of Medicinal Chemistry. 2018;61:3516–3540. doi: 10.1021/acs.jmedchem.7b01795. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.