Highlights
-
•
An extensive evaluation of the Abbott SARS-CoV-2 IgG assay.
-
•
The assay shows excellent sensitivity/specificity with little cross-reactivity.
-
•
Healthy volunteers and pre-pandemic samples had similar antibody cut-off indexes.
-
•
A lower, optimised limit for reactivity improves sensitivity in early disease.
Keywords: SARS-CoV-2, Antibodies, Assay evaluation, Reactivity index
Abbreviations: SARS-CoV-2, Novel severe acute respiratory syndrome coronavirus 2; COVID-19, Coronavirus disease 2019; CDC, Centers for Disease Control and Prevention; RCPA, Royal College of Pathologists of Australasia; RF, Rheumatoid factor; CAP, College of American Pathologists; FDA, Food and Drug Administration; ANA, anti-nuclear antibody; ds-DNA, double-stranded DNA antibody; HCW, Healthcare worker
Abstract
Introduction
We describe our evaluation of the Abbott SARS-CoV-2 IgG assay on the Architect immunoassay analyser.
Methods
We assessed assay precision, sensitivity, specificity, positive/negative predictive values (PPV/NPV), cross-reactivity (influenza/dengue/hepatitis B and C/rheumatoid factor/anti-nuclear/double-stranded DNA/syphilis) and sample throughput in samples from real-time polymerase chain reaction (RT-PCR) positive patients/healthcare workers (HCWs)/pre-pandemic samples. We compared the cut-off indexes (COIs) between all control samples (HCWs and pre-pandemic) to generate an optimised COI limit for reactivity.
Results
The assay specificity was 99.8% (n = 980) and sensitivity was 45.9–96.7% (n = 279). When tested ≥ 14 days post-positive RT-PCR (POS), the PPV/NPV was 96.4%/99.8%. The difference between the COIs of HCWs/pre-pandemic samples was small (0.01, p < 0.0001). There was minimal cross-reactivity with other antibodies. A lower COI limit for reactivity (≥0.55, using the 99th percentile COI of our controls and ROC analysis) improved diagnostic sensitivity, especially at 0–6 days POS (45.9–55.8%), with a small decrease in specificity (98.9%). The assay throughput was 100 samples in 70 min.
Conclusion
The Abbott SARS-CoV-2 IgG assay shows excellent performance in patients ≥ 14 days POS. The difference between the COIs of HCWs and pre-pandemic samples was numerically small. A lower COI limit improves assay sensitivity with a slight decrease in specificity.
1. Introduction
The novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can lead to mild or moderate coronavirus disease 2019 (COVID-19) with cough, fever, malaise, myalgias, gastrointestinal symptoms and anosmia [1], or a severe disease with acute respiratory distress syndrome [2]. The need for the serological diagnosis of SARS-CoV-2 has become acute. The Centers for Disease Control and Prevention (CDC) [3] and the Royal College of Pathologists of Australasia (RCPA) [4] stipulate that only viral testing (nucleic acid or antigen) is the standard test to diagnose acute infection. However, some studies [5], [6] show that the SARS-CoV-2 antibodies can increase as soon as the first week after infection. Antibody tests can act as an indirect marker for infection and help identify patients with prior infection/exposure [7].
Numerous serologic tests, especially rapid lateral flow immunoassays, were available in March 2020. The performance of these early tests was disappointing and a systematic review of point-of-care serologic studies up to end April 2020 does not support their use [8]. A Cochrane systematic review on SARS-CoV-2 antibody tests released on 25 June 2020 laments the low sensitivity (<30%) in the first week after symptom onset for it to be useful in COVID19 diagnosis [9]. However, the Cochrane report concedes that antibody testing may be useful if used after 15 days of symptom onset. All new tests must be rigorously evaluated and validated before use [10] as an unreliable test is worse than no test. Newer automated chemiluminescent immunoassays were released in June 2020. We report on our evaluation of the Abbott Architect chemiluminescent immunoassay for SARS-CoV-2-IgG including deriving an optimised cut-off index that may be useful in testing early Covid-19 samples.
2. Materials and methods
2.1. Participants
262 healthcare workers (HCWs) (laboratory staff, doctors, nurses, and housekeeping staff) from our hospital volunteered to provide serum for specificity testing. All volunteers provided informed consent, had no reported respiratory symptoms/fever at time of sampling, and agreed to a repeat (paired) SARS-CoV-2 IgG test 14 days after the first test. Stored samples (N = 718) from our staff health screening (HS) program in 2018 were also included as an additional control group.
Anonymised residual leftover sera (from other routine testing, e.g., renal panels, complete blood count) from subjects who had positive reverse transcription real-time polymerase chain reaction (RT-PCR) results for COVID-19 from April-June 2020 were recruited for analysis (n = 353). Our laboratory stores all residual serum samples at 4 °C for 10 days after testing as a standard operating procedure. The date of the 1st positive PCR test (POS) served as a surrogate for onset of disease. Results of COVID-19 serology was stratified according to days POS. There were 74 subjects who were not initially suspected of COVID-19. They were excluded from analysis as we would have to record disease onset as negative POS. In aggregate, we examined 279 samples from 160 individual SARS-CoV-2 RT-PCR positive patients for sensitivity; for specificity 262 HCW samples and 718 HS samples were evaluated (see Table 1 ).
Table 1.
Population demographics.
| Group | N | Mean age ± SD (range) | Males (%) | Females (%) |
|---|---|---|---|---|
| Sensitivity group | 279 | 50.3 ± 17.6 (23 to 98) | 234 (83.9%) | 45 (16.1%) |
| HS | 718 | 44.2 ± 13.4 (20 to 85) | 365 (50.8%) | 353 (49.2%) |
| HCWs | 262 | 39.0 ± 13.2 (20 to 67) | 29 (11.1%) | 233 (88.9%) |
| HCWs + HS | 980 | 42.8 ± 13.5 (20 to 85) | 394 (40.2%) | 586 (59.8%) |
HS: Health screening samples, HCWs: Healthcare worker samples.
We also evaluated the assay for any cross-reactivity with other antibodies. Of the 262 HCW volunteers, 229 had received the latest influenza vaccination (Southern hemisphere) within four weeks of their first SARS-CoV-2 IgG test. Samples that tested positive for dengue fever (N = 46) and 51 other antibody-positive subjects [Anti-HCV – 4, Hepatitis B – 29, anti-nuclear antibody (ANA) – 11, double-stranded DNA antibody (ds-DNA) – 1, rheumatoid factor (RF) – 5, syphilis – 1] were also included for cross-reactivity analysis.
2.2. Instrumentation and materials
For RT-PCR testing, our hospital molecular laboratory employs a duplex real-time RT-PCR that targets the N and E genes using a Qiagen EZ1 extraction system and Rotor Gene Q amplification system. The Abbott SARS-CoV-2 IgG assay is a qualitative chemiluminescent microparticle immunoassay used for the detection of IgG antibodies to SARS-CoV-2 (undisclosed epitope on the viral nucleocapsid) in human serum/plasma on the Architect i2000 System. The sample, SARS-CoV-2 antigen coated paramagnetic microparticles, and assay diluent are combined and incubated. Thereafter, anti-human IgG acridinium-labelled conjugate is added to create a reaction mixture. Following the addition of pre-trigger and trigger solutions, the resulting chemiluminescent reaction is directly proportional to the amount of IgG antibodies. When compared to the mean chemiluminescent signal of a calibrator, an IgG index is derived with a stated cut-off index (COI) of 1.4.
The assay claims an inter-assay CV of 5.9% and 1.2% for negative and positive controls at a mean COI of 0.04 and 3.53 respectively. The reported assay specificity is 99.6% and sensitivity of 100% for samples ≥ 14 days post symptom onset. To prevent potential interferences, the manufacturer advises instrument maintenance to be performed just before and following analysing a batch of 50 SARS-CoV-2 IgG samples. All samples were collected in Vacutainer Gel separator plain serum tubes (Becton Dickinson SST tubes with silica clot activator, polymer gel, silicone-coated interior).
2.3. Statistical analysis
For precision analysis, 5 pools (including negative and positive Abbott controls, and 3 positive serum samples over a range of COI values) were run 5 times daily over 5 days as per the CLSI EP15-A3 protocol [11]. As the Abbott assay is a qualitative test, the diagnostic specificity of the test is represented by the negative percentage agreement (NPA) between antibody negativity against all control subjects; diagnostic sensitivity is represented by the positive percentage agreement (PPA) between antibody positivity against all PCR positive patients tested. The positive predictive value (PPV) and negative predictive value (NPV) of the assay was also assessed. 95% CIs for sensitivity and specificity were calculated according to Clopper and Pearson (“exact” method) with standard logit confidence intervals for predictive values.
We compared the COIs of both populations of possible control subjects (262 HCW samples and 718 HS samples). We used the Mann-Whitney U test for any significant differences between medians, as the COI results of both the HS and HCW populations were not normally distributed despite log transformation. A p < 0.05 was considered to be statistically significant. We explored deriving an optimized COI for reactivity from the COIs of our entire COVID-19-naive population, using the 99th percentile of our control population and ROC analysis. We subsequently examined the influence of this optimized COI limit for reactivity on the diagnostic sensitivity/specificity/PPV/NPV of the assay. No cases with indeterminate or missing results were used in our study.
Statistical analyses were performed using MedCalc software v19.3.1 (MedCalc, Ostend, Belgium). For the 95% confidence interval in groups with 100% NPV, we used Stata 14 software (StataCorp. 2015. Stata Statistical Software: Release 14). As this work was part of evaluating new diagnostic assays and seroprevalence surveillance, it was deemed exempt by our institutional review board. Compliance with STARD guidelines is enclosed (see Supplemental Table 1).
3. Results
3.1. Performance analysis
The Abbott assay showed excellent precision, with a CV of 3.4% (negative control, COI = 0.06) and 1.6% (serum sample, COI = 8.6) (See Supplementary Table 2). The samples assessed covered a range of COIs from 0.05 to 8.84. The Architect was able to analyse 100 specimens for SARS-CoV-2 IgG in 1 h, 9 min and took 2 h, 9 min to analyse 250 specimens.
3.2. Comparing control populations
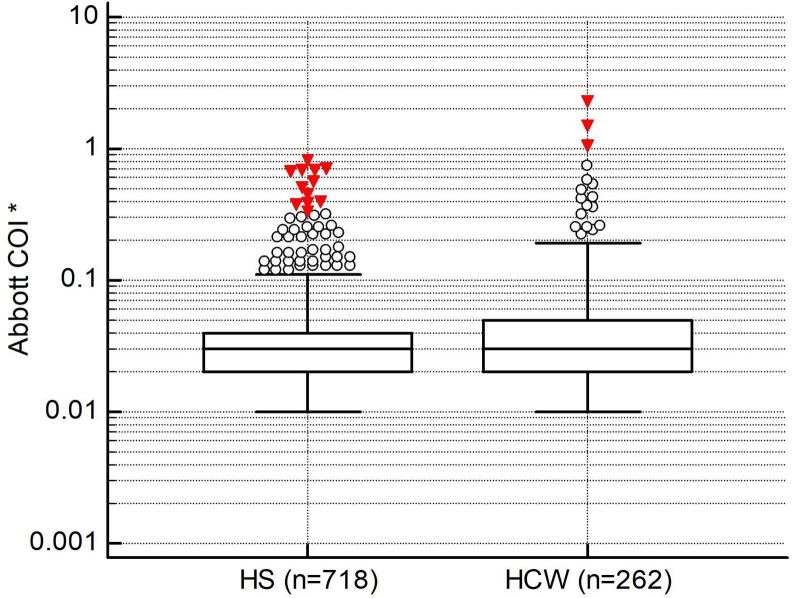
COIs between samples from a pre-pandemic health screening (2018) and currently healthy healthcare workers with two consecutive SARS-CoV-2 IgG tests were compared. None of the health screening samples were reactive. Only 2 samples from our current healthcare workers were reactive: one with COI 1.5 (a repeat test gave a COI of 1.45) and the other with COI 2.3 (a repeat test gave a COI of 2.36). When both reactive HCW samples were retested on the Roche Cobas e801 analyser using the Roche SARS-CoV-2 antibody assay, both were not reactive (COIs 0.08 and 0.09 respectively). These two samples were considered as false positive cases on the Abbott assay. The next lowest COIs in the HCW population were 1.05, 0.74 and 0.58. The difference between the consecutive HCW paired serum samples was minimal (Bland-Altman analysis: COI mean diff −0.001, 95% CI −0.004 to 0.002, p = 0.70). Indeed, 60% of the 2nd readings were identical to the 1st readings. The COIs of the HCW samples were also skewed rightwards and ranged from 0.01 to 2.30 (99th percentile of 1.01). On Mann-Whitney U testing, the median difference between the COIs of the 718 HS samples and the 262 HCW samples was 0.01 (95% CI 0.00–0.01, p < 0.0001. HS median 0.03, 95% CI 0.02–0.03, Inter-quartile range (IQR) 0.02–0.04; HCW median 0.03, 95% CI 0.03–0.04, IQR 0.02–0.05) (see Fig. 1 ). Although statistically significant, the median difference between both populations was numerically small. As such, both populations were combined to form one larger cohort for specificity analysis (n = 980).
Fig. 1.
Mann-Whitney U comparison between 2 populations of controls. (* COIs using logarithmic scale) (Abbreviations: HS: Health screen samples, HCW: Healthcare worker samples).
3.3. Optimized COI limit analysis
The 99th COI percentile from our control population was 0.57 (see Table 2 ). When we compared PCR positive cases (POS ≥ 14 days, from our sensitivity population) with the healthy population (specificity population) and performed ROC analysis, an associated COI criterion of COI 0.53 gave an AUC of 1.000 (95% CI 0.996 to 1.000), with sensitivity of 100.0% and specificity of 98.9%. Using the average of the 2 results, we determined that a COI of ≥0.55 could serve as an optimized COI for reactivity.
Table 2.
COIs of the specificity control group.
| Group | N | COI range | Median (CI) | IQR | 99th percentile |
|---|---|---|---|---|---|
| HS | 718 | 0.01–0.82 | 0.03 (0.02–0.03) | 0.02–0.04 | 0.47 |
| HCWs | 262 | 0.01–2.30 | 0.03 (0.03–0.04) | 0.02–0.05 | 1.01 |
| Total | 980 | 0.01–2.30 | 0.03 (0.03–0.03) | 0.02–0.04 | 0.57 |
HS: samples from health screening. HCWs: samples from healthcare workers.
3.4. Specificity analysis
Out of 980 control samples were reactive on the assay, with a resulting specificity of 99.80% (95% CI 99.27–99.98).
3.5. Cross-reactivity analysis
Two HCWs with a recent influenza vaccination tested positive for SARS-CoV-2 IgG. These two cases were the two false-positive cases from our HCW population elaborated earlier. All other cases with other antibodies (dengue, influenza, hepatitis C, hepatitis B, syphilis, ANA, ds-DNA and RF) tested negative for SARS-CoV-2 IgG on the Abbott assay (See Table 3 ).
Table 3.
Cross-reactivity analysis for the Abbott anti-SARS-CoV-2 assay.
| N | Negative | Positive | Median (IQR) | |
|---|---|---|---|---|
| Recent influenza vaccination | 229 | 227 | 2 | 0.03 (0.02–0.05) |
| Dengue fever | 46 | 46 | 0 | 0.02 (0.02–0.04) |
| Anti-HCV | 4 | 4 | 0 | 0.04 (0.03–0.06) |
| Hepatitis B* | 29 | 29 | 0 | 0.04 (0.03–0.06) |
| Syphilis antibodies | 1 | 1 | 0 | 0.16 (NA) |
| Anti-nuclear antibodies | 11 | 11 | 0 | 0.03 (0.02–0.04) |
| Anti-dsDNA | 1 | 1 | 0 | 0.03 (NA) |
| RF positive | 5 | 5 | 0 | 0.02 (0.02–0.04) |
| Total | 326 | 324 | 2 | 0.03 (0.02–0.05) |
Hepatitis B cases includes various combinations of HBsAg, anti-HBc IgM, HBeAg and anti-HBe.
3.6. Sensitivity analysis
Of the 353 RT-PCR samples, 74 were excluded as these inpatients were not initially suspected of having COVID-19 but tested positive for SARS-CoV-2 RT-PCR in their subsequent work-up. Interestingly, 35 of these 74 subjects (47.3%) were positive for SARS-Cov-2-IgG prior to having a positive RT-PCR test indicating an early antibody response. Of the remaining 279 samples (from 160 individual patients) the PPA increased stepwise from 45.9% in week 1, to 83.0% by week 2 and 96.7% after 14 days post positive RT-PCR (POS) (see Table 4 ). When we applied a COI of 0.55 as a limit for reactivity to our sensitivity analysis, the diagnostic sensitivity of the assay improved in all categories especially for subjects with early infection (cases between POS 0–6 days) (see Table 4). Chi-squared testing between the classification of cases between POS 0–6 days showed a significant difference between using the manufacturer recommended COI and our optimized COI (p < 0.0001).
Table 4.
Abbott assay sensitivity (positive percentage agreement) by days post positive RT-PCR.
| With a reactivity limit of COI ≥ 1.4 | |||||
|---|---|---|---|---|---|
| Days POS | N | IgG positive | IgG negative | PPA (%) | 95% CI |
| 0–6 | 172 | 79 | 93 | 45.93 | 38.32–53.68 |
| 7–13 | 47 | 39 | 8 | 82.98 | 69.19–92.35 |
| ≥7 | 107 | 97 | 10 | 90.65 | 83.48–95.43 |
| ≥14 | 60 | 58 | 2 | 96.67 | 88.47–99.59 |
| With a reactivity limit of COI ≥ 0.55 | |||||
| Days POS | N | IgG positive | IgG negative | PPA (%) | 95% CI |
| 0–6 | 172 | 96 | 76 | 55.81 | 48.06–63.37 |
| 7–13 | 47 | 40 | 7 | 85.11 | 71.69–93.80 |
| ≥7 | 107 | 100 | 7 | 93.46 | 86.99–97.33 |
| ≥14 | 60 | 60 | 0 | 100.00 | 94.04–100.00 |
COI: Cut-off index, POS: Post-positive RT-PCR, PPA: Positive percentage agreement.
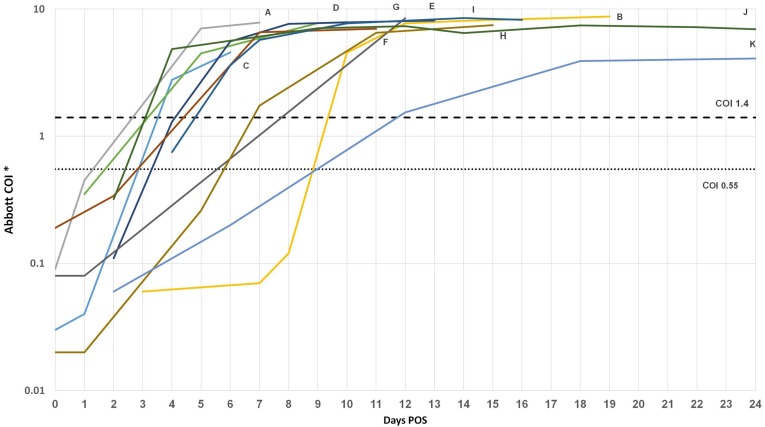
Eleven subjects were PCR positive subjects who initially had a negative SARS-CoV-2 IgG test, and subsequently became reactive later. Using a COI of 0.55 also allowed us to predict when such samples became reactive earlier than if a COI of 1.4 was employed (see Fig. 2 ). The interval between the last negative COI to first positive COI improved from day 2–11+ to 1–8+ when the optimized COI for reactivity was used – an improvement of 1–3 days.
Fig. 2.
Serial COIs of RT-PCR positive patients over days post-positive RT-PCR. Each curve labelled by a Latin letter represents an individual patient. (*COI values displayed on logarithmic scale.) (Abbreviations: COI: Cut-off index, POS: days post-positive RT-PCR.)
Using the optimized COI for reactivity, 9 more subjects out of 980 controls would be positive, and the specificity would be lower at 98.9% (95% CI 98.0–99.4).
3.7. Positive and negative predictive values
If the manufacturer’s recommended COI limit for reactivity (COI ≥ 1.4) is used (and assuming a disease prevalence of 5%), in subjects ≥ 14 days POS (specificity 99.80%, sensitivity 96.67%), the positive predictive value (PPV) of the Abbott assay would be 96.14% (95% CI 86.19–99.01) and the negative predictive value (NPV) would be 99.83% (95% CI 99.32–99.96). However, if examining subjects 0–6 days POS (sensitivity 45.93%), the PPV would be 92.22% (95% CI 74.61–97.95) and the NPV would be 97.23% (95% CI 96.83–97.58). If we use an optimized COI of ≥ 0.55 and assume a disease prevalence of 5%, in subjects 0–6 days POS (sensitivity 55.81%, specificity 98.9), the NPV is 97.70% (95% CI 97.29–98.05) but the PPV is 72.35% (95% CI 58.89–82.70). The changes in PPV and NPV with disease prevalence using the two different COI limits is displayed in Table 5 .
Table 5.
Changes to PPV/NPV with disease prevalence, using manufacturer and optimized COIs.
| Prevalence | Days POS |
COI ≥ 1.4 |
COI ≥ 0.55 |
||
|---|---|---|---|---|---|
| PPV (95% CI) | NPV (95% CI) | PPV (95% CI) | NPV (95% CI) | ||
| 1% | 0–6 | 69.45 (36.06–90.16) | 99.46 (99.38–99.53) | 33.43 (21.57–47.85) | 99.55 (99.47–99.62) |
| ≥7 | 81.78 (52.88–94.72) | 99.91 (99.83–99.95) | 45.68 (31.80–60.27) | 99.93 (99.86–99.97) | |
| ≥14 | 82.71 (54.49–95.03) | 99.97 (99.87–99.99) | 47.37 (33.33–61.83) | 100.00 (99.62–100.00) | |
| 5% | 0–6 | 92.22 (74.61–97.95) | 97.23 (96.83–97.58) | 72.35 (58.89–82.70) | 97.70 (97.29–98.05) |
| ≥7 | 95.90 (85.40–98.94) | 99.51 (99.12–99.73) | 81.42 (70.84–88.77) | 99.65 (99.29–99.83) | |
| ≥14 | 96.14 (86.19–99.01) | 99.83 (99.32–99.96) | 82.42 (72.26–89.41) | 100.00 (99.62–100.00) | |
| 10% | 0–6 | 96.16 (86.12–99.02) | 94.32 (93.54–95.02) | 84.67 (75.15–90.99) | 95.27 (94.45–95.97) |
| ≥7 | 98.01 (92.51–99.50) | 98.97 (98.16–99.43) | 90.25 (83.69–94.35) | 99.27 (98.52–99.64) | |
| ≥14 | 98.14 (92.94–99.53) | 99.63 (98.57–99.91) | 90.83 (84.62–94.69) | 100.00 (99.62–100.00) | |
4. Discussion
Overall, the Abbott SARS-CoV-2 IgG assay shows excellent performance, with a CV of 3.4% and 1.9% for the negative and positive calibrators. The specificity of the assay is 99.8%, with a diagnostic sensitivity of 96.7% ≥14 days POS. The PPV/NPV was 96.14%/99.83% in subjects ≥ 14 days POS (assuming a seroprevalence of 5%). This is in close agreement with the manufacturer’s specifications. The US CDC and the Food and Drug Administration (FDA) have stipulated a sensitivity of 90% and specificity of 95% for an acceptable SARS-CoV-2 serology test [12], [13]. The Abbott assay easily falls within these requirements. Furthermore, the Abbott assay has sufficient throughput, comparable to other SARS-CoV-2 immunoassays and may be of practical interest for centres handling many samples for COVID-19 screening. If desired, 2 Abbott Architect i2000s can be linked together in a single unit as the Architect i4000, effectively doubling the throughput. In the latest revision of the package insert (Abbott SARS-CoV-2 IgG package insert, H70891R03, May 2020), it is heartening to note that the requirement for batch analysis and instrument maintenance pre- and post-testing has been rectified.
Like the other reports [14], [15], we had access to pre-pandemic sera collected from healthy adults in 2018 as a control population for specificity testing. The COIs of the pre-pandemic subjects from 2018 and the baseline COIs of the healthy HCWs differed very slightly. The findings suggest that both populations can be used as control populations. However, to be considered COVID-19 naive, these HCWs had to meet 3 important criteria: no symptoms of upper respiratory tract infection/fever, two serial antibody testing 14 days apart that were both non-reactive, and negligible difference in COIs between the two results. This is helpful for centers that do not have access to stored pre-pandemic samples, as they can take active steps in the present to recruit samples using the above criteria. Samples used for cross-reactivity testing also had low COIs (≤0.4), well below the manufacturer stated COI of 1.4 or the optimised COI of 0.55.
When we used an optimized COI of 0.55 as a threshold for reactivity in sensitivity testing, it significantly improved the sensitivity in cases 0–6 days POS, up to 56% (Chi-squared test p < 0.0001). Expectedly, it had less impact on the already high assay sensitivity in subjects with POS ≥ 14 days (97–100% sensitivity). This is similar to the results of other studies that have advocated a lower threshold COI for reactivity on the Abbott assay; a lower COI of 0.8 and 1.0 has been suggested [16]. The validity of this lower COI for reactivity is reinforced, as we have used both ROC analysis and the 99th percentile of our healthy population to derive it. It is noteworthy that a recent document [17] that analysed ROC curves for the Abbott assay results ≥20 days and ≥30 days after the appearance of first symptoms found that an optimized COI of between 0.42 and 0.57 improved the assay sensitivity from 92.7% to 97.6% at ≥20 days and 93.5% to 98.4% at ≥30 days. Our optimized COI falls within the range of their suggested optimal COI.
The call for a lower COI threshold to improve the sensitivity/negative likelihood ratio of SARS-CoV-2 antibody assays is growing [16], [18], [19]. As shown in our study, a lower COI limit for reactivity can improve the sensitivity especially in early cases. This would be most useful when the antibody test is used in screening large populations with an unknown disease onset and/or low prevalence. While the sensitivity can be improved by an optimized threshold COI, it is typically done at the expense of the assay specificity. Further larger studies are required to examine how much of an impact a lower COI limit for reactivity will affect assay specificity, but in our hands, the specificity only decreased slightly from 99.8% to 98.9% when the optimized COI limit was used. Guidance is needed from learned societies with regards to how these optimized threshold COIs are obtained, whether they be ROC analysis, 99th percentiles of healthy populations, or maximum COI values from a healthy population.
Notwithstanding these considerations, the sensitivity and specificity of an assay should also be interpreted with pre-test probability and population prevalence in mind [20]. We have observed that seroprevalence has the greatest impact on the PPV of the assay, which is also reported in other studies [21]. In our study, when using a COI limit of ≥1.4, the PPV changed from 98.1% when seroprevalence was 10%, to 82.7% when the seroprevalence was 1%. This was even more pronounced when the optimized COI limit of ≥0.55 was used (90.8% at 10%, 47.4% at 1%). The composition of the test population also impacts the interpretation of the test’s diagnostic quality since different populations would have different rates of seroprevalence. The pre-test probability and population prevalence of SARS-CoV-2 is also unknown in many populations. This hampers the effective comparison between higher or lower COI thresholds.
We elected to use days POS rather than symptom onset for our study as subjects who are asymptomatic [22] or pre-symptomatic [23] would be otherwise excluded; using days POS mitigates against this pitfall. We found 47.3% (35/74) of the patient samples taken before a positive RT-PCR test were positive for SARS-CoV-2 IgG. This finding underscores the fact that SARS-CoV-2 antibody development may occur earlier than expected in some cases. Our study also supports the claim that SARS-CoV-2 antibodies tend to rise only after several days (assay sensitivity only > 90% after 14 days POS), which is in agreement with several other studies [14], [24], [25], [26], [27] that have evaluated the Abbott SARS-CoV-2 antibody assays (see Table 6 ). Our results further reinforce the fact that SARS-CoV-2 antibodies should not be used to diagnose acute infections but may complement RT-PCR results especially later in the disease evolution.
Table 6.
Studies evaluating the Abbott SARS-CoV-2 IgG assay.
| Study | N | Early sensitivity | Later sensitivity |
|---|---|---|---|
| Studies evaluating the assay by days post symptom onset | |||
| Theel ES, et al. [15] | 38 | 10.5% ≤7 days | 91.8% ≥15 days |
| 49.5% 8–14 days | |||
| Chew KL, et al. [16] | 177 | 8.6% ≤6 days | 84% 14–20 days |
| 43.6% 7–13 days | 84.4% ≥21 days | ||
| Public Health England [17] | 536 | Nil, all subjects were ≥20 days | 92.7% ≥20 days, 93.5% ≥30 days |
| Public Health England [24] | 96 | 92.9% when ≤10 days | 93.9% ≥14 days |
| 93.4% ≥21 days | |||
| Tang MS, et al. 25 | 103 | <47.83% when <14 days | 93.75% ≥14 days |
| Bryan A, et al. [26] | 125 | 53.1% at 7 days | 96.9% at 14 days |
| 82.4% at 10 days | 100% at 17 days | ||
| Nicol T, et al. [27] | 141 | 46.9% 0–7 days | 100% >14 days |
| 69.0% 8–14 days | |||
| Studies evaluating the assay by days post-positive RT-PCR | |||
| Theel ES, et al. [15] | 38 | 18.2% ≤7 days | 95.7% ≥20 days |
| Tang MS, et al. [25] | 103 | 47.62–69.57% when <14 days | 81.25% ≥14 days |
| Bryan A, et al. [26] | 125 | 88.7% at 7 days | 100% at 14 days |
| 97.2% at 10 days | |||
There have been concerns about assay cross-reactivity with other flu-like viruses, e.g. coronavirus. The minimal cross reactivity of SARS-CoV-2 antibodies in HCWs who had recent influenza vaccination is reassuring (only two cases). Dengue fever, endemic in many tropical climates including ours, can have a similar clinical presentation to COVID-19 (fever and myalgia). In fact, in this country, 2 cases with positive dengue serology results and later found to have SARS-CoV-2 infection have been reported [28]; the original dengue antibody results were confirmed to be false-positives on further investigation. It is noteworthy that positive dengue serology in 46 patients in our study did not cause any false positive SARS-CoV-2 IgG results. Other reports of the Abbott assay also demonstrate little cross-reactivity in patients with previous viral infections, haemodialysis [24], and rheumatologic conditions (n = 358) [21].
Novel findings in this study are, samples from healthy volunteers, with 2 sero-negative antibody tests at least 2 weeks apart, were tested with no significant difference between the 2 results, which could potentially serve as valid control subjects. An optimized COI limit for reactive samples (COI ≥ 0.55) (concordant values between 99th percentile of healthy controls and ROC analysis) improved the sensitivity of the assay in early infection (45.9–55.8% in subjects 0–6 days POS) but with a small decrease in specificity (99.8–98.9%). We found no evidence that the test's specificity might be markedly impaired in patients with previous dengue infection. We report the throughput of this assay, which would be of interest to centres that are considering analysis of large numbers of samples from COVID-19 screening. A strength of our study is that we have managed to gather a larger number of subjects for sensitivity/specificity testing. We have also provided a comprehensive comparison with prior studies of the Abbott assay for the benefit of readers (see Table 6).
A limitation of our study is that it is a single center study, and we do not have the seroprevalence of SARS-CoV-2 in our community. We have shown that the overall performance of the SARS-CoV-2 IgG assay is greatly affected by the prevalence of the disease in a population, and further studies are required to ascertain the true seroprevalence of SARS-CoV-2 in our country. The applied sensitivity and specificity cohorts might differ relevantly in terms of sex, age, and comorbidities. We do not have any data regarding symptom severity in our sensitivity cohort. We were also unable to perform cross-reactivity analysis with other commonly encountered coronaviruses (SARS/Mers-CoV), or patients with other active viral infections (e.g. active influenza infection). Nevertheless, the assay did not cross-react with sera from HCWs with a recent influenza vaccination. In addition, more extensive evaluations of other viruses [23] and the manufacturer show no cross-reactivity. We had few samples for cases before and after 0–6 days POS, and further studies on larger populations would be desirable.
Although recommended as the standard test to diagnose acute infection [3], [4], the RT-PCR is not perfect, and has variable performance [29] depending on the primer-probe sets and testing kits with variable sensitivities depending on the number of viral copies per reaction, with some studies report a 79% sensitivity in initial RT-PCR testing [30]. One study [31] found that in days 1 through 7 post-symptom onset, 40% of throat swabs were falsely negative on PCR testing. Another study [32] reported that only 67% of subjects were tested positive on RT-PCR testing, and a systemic review found that false negatives from RT-PCR testing can range from 2 to 29% [33]. When our optimized COI limit is used, within one week of a positive RT-PCR test, the antibody assay performance has a sensitivity of 55.8% and a specificity of 98.9%. When combined with RT-PCR testing, a highly specific, reliable and rapid serology test like the Abbott SARS-CoV-2 IgG assay may be a useful complementary tool to improve PPV and reduce false positive rates in low prevalence areas.
5. Conclusion
Our results show that the Abbott SARS-CoV-2 IgG assay has excellent performance and is highly comparable both with the manufacturer’s information and other published studies. We also show that the difference between the COIs of a healthy, disease-free pre-pandemic population and those of healthy subjects with paired sera (two weeks apart) is small. A lower COI limit for reactivity of ≥0.55, based on the 99th COI percentile of a COVID19-naive population and ROC analysis, improves assay sensitivity in subjects with early disease with a minimal decrease in specificity. We look forward to larger studies over longer periods of time to further evaluate the use of serology in the management of COVID-19.
CRediT authorship contribution statement
C.S. Lau: Conceptualization, Data curation, Final analysis, Writing - original draft , Writing - review & editing. H.M.L. Oh: Investigation, Data curation. S.P. Hoo: Investigation, Data curation. Y.L. Liang: Investigation, Data curation. S.K. Phua: Investigation, Data curation. T.C. Aw: Conceptualization, Final analysis, Supervision, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Temasek Holdings Pte Ltd and Abbott Diagnostics, Singapore, for sponsoring the test kits used in this study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cca.2020.09.005.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Gandhi R.T., Lynch J.B., Del Rio C. Mild or Moderate Covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMcp2009249. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 2.Berlin D.A., Gulick R.M., Martinez F.J. Severe Covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMcp2009575. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention, Evaluating and Testing Persons for Coronavirus Disease 2019 (COVID-19). Available at: https://www.cdc.gov/coronavirus/2019-ncov/hcp/clinical-criteria.html (accessed May 22, 2020).
- 4.The Royal College of Pathologists of Australia, Position Statement, COVID19 IgG/IgM Rapid POCT Tests. Available at: https://www.rcpa.edu.au/getattachment/bf9c7996-6467-44e6-81f2-e2e0cd71a4c7/COVID19-IgG-IgM-RAPID-POCT-TESTS.aspx (accessed May 22, 2020).
- 5.W. Liu, L. Liu, G. Kou, et al., Evaluation of Nucleocapsid and Spike Protein-based ELISAs for 2 detecting antibodies against SARS-CoV2, J. Clin. Microbiol. (March 30, 2020). [Epub ahead of print] Doi: 10.1128/JCM.00461-20. [DOI] [PMC free article] [PubMed]
- 6.Long Q.X., Liu B.Z., Deng H.J. Antibody Responses to SARS-CoV-2 in Patients with COVID-19. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 7.U.S. Food and Drug Administration, Important Information on the Use of Serological (Antibody) Tests for COVID-19 - Letter to Health Care Providers. Available at: https://www.fda.gov/medical-devices/letters-health-care-providers/important-information-use-serological-antibody-tests-covid-19-letter-health-care-providers (accessed May 22, 2020).
- 8.Bastos M.L., Tavaziva G., Abidi S.K. Diagnostic accuracy of serological tests for covid-19: systematic review and meta-analysis. BMJ. 2020;370 doi: 10.1136/bmj.m2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.J.J. Deeks, J. Dinnes, Y. Takwoingi, et al, Antibody tests for identification of current and past infection with SARS-CoV-2, Cochr. Database Systemat. Rev. 6 (2020) CD013652. 10.1002/14651858.CD013652. [DOI] [PMC free article] [PubMed]
- 10.Beeching N.J., Fletcher T.E., Beadsworth M.B.J. Covid-19: testing times. BMJ. 2020;369 doi: 10.1136/bmj.m1403. [DOI] [PubMed] [Google Scholar]
- 11.Clinical and Laboratory Standard Institute (CLSI), User Verification of Precision and Estimation of Bias; Approved Guideline-third ed., CLSI document EP15-A3, PA Wayne PA: CLSI, 2014.
- 12.Centers for Disease Control and Prevention, Interim Guidelines for COVID-19 Antibody Testing. Available at: https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antibody-tests-guidelines.html (accessed June 8, 2020).
- 13.US Food and Drug Administration, EUA Authorized Serology Test Performance. Available at: https://www.fda.gov/medical-devices/emergency-situations-medical-devices/eua-authorized-serology-test-performance (accessed May 29, 2020).
- 14.E.S. Theel, J. Harring, H. Hilgart, D. Granger, Performance Characteristics of Four High-Throughput Immunoassays for Detection of IgG Antibodies against SARS-CoV-2, J Clin Microbiol (June 8, 2020). [Epub ahead of print]. 10.1128/JCM.01243-20. [DOI] [PMC free article] [PubMed]
- 15.A. Stroemer, O. Grobe, R. Rose, H. Fickenscher, T. Lorentz, A. Krumbholz, Diagnostic accuracy of six commercial SARS-CoV-2 IgG/total antibody assays and identification of SARS-CoV-2 neutralizing antibodies in convalescent sera, medRxiv (June 17, 2020). [Preprint] 10.1101/2020.06.15.20131672. [DOI]
- 16.K.L. Chew, S.S. Tan, S. Saw, et al., Clinical evaluation of serological IgG antibody response on the Abbott Architect for established SARS-CoV-2 infection, Clin. Microbiol. Infect. (June 9, 2020). [Epub ahead of print]. 10.1016/j.cmi.2020.05.036. [DOI] [PMC free article] [PubMed]
- 17.Public Health England, Evaluation of sensitivity and specificity of four commercially available SARS-CoV-2 antibody immunoassays. Available at: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/898437/Evaluation__of_sensitivity_and_specificity_of_4_commercially_available_SARS-CoV-2_antibody_immunoassays.pdf (accessed July 16, 2020).
- 18.M. Plebani, A. Padoan, D. Negrini, B. Carpinteri, L. Sciacovelli, Diagnostic performances and thresholds: The key to harmonization in serological SARS-CoV-2 assays? Clin. Chim. Acta May 30, 2020. [Epub ahead of print] 10.1016/j.cca.2020.05.050. [DOI] [PMC free article] [PubMed]
- 19.J. Favresse, C. Eucher, M. Elsen, T.H. Marie, J.M. Dogne, J. Douxfils, Clinical performance of the Elecsys electrochemiluminescent immunoassay for the detection of SARS-CoV-2 total antibodies, Clin Chem June 2, 2020. [Epub ahead of print] 10.1093/clinchem/hvaa131. [DOI] [PMC free article] [PubMed]
- 20.J. Watson, P.F. Whiting, J.E. Brush, Interpreting a covid-19 test result, BMJ May 12, 2020. [Epub ahead of print] 10.1136/bmj.m1808. [DOI] [PubMed]
- 21.T. Perkmann, N. Perkmann-Nagele, M.K. Breyer, et al., Side by side comparison of three fully automated SARS-CoV-2 antibody assays with a focus on specificity, Clin. Chem. 10 August, 2020. [Published online ahed of print] 10.1093/clinchem/hvaa198. [DOI] [PMC free article] [PubMed]
- 22.K. Mizumoto, K. Kagaya, A. Zarebski, G. Chowell, Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020, Euro Surveill March 12, 2020. [Epub ahead of print]. 10.2807/1560-7917.ES.2020.25.10.2000180. [DOI] [PMC free article] [PubMed]
- 23.Wei W.E., Li Z., Chiew C.J., Yong S.E., Toh M.P., Lee V.J. Presymptomatic Transmission of SARS-CoV-2 — Singapore, January 23–March 16. MMWR Morb. Mortal. Wkly Rep. 2020;69(2020):411–415. doi: 10.15585/mmwr.mm6914e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Public Health England, Evaluation of the Abbott SARS-CoV-2 IgG for the detection of anti-SARSCoV-2 antibodies. Available at: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/887221/PHE_Evaluation_of_Abbott_SARS_CoV_2_IgG.pdf (accessed May 26, 2020).
- 25.M.S. Tang, K.G. Hock, N.M. Logsdon, et al., Clinical Performance of Two SARS-CoV-2 Serologic Assays, Clin. Chem. May 13, 2020. [Epub ahead of print] 10.1093/clinchem/hvaa120. [DOI] [PMC free article] [PubMed]
- 26.A. Bryan, G. Pepper, M.H. Wener, et al., Performance Characteristics of the Abbott Architect 1 SARS-CoV-2 IgG Assay and Seroprevalence in Boise, Idaho, J. Clin. Microbiol. May 7, 2020. [Epub ahead of print] 10.1128/JCM.00941-20. [DOI] [PMC free article] [PubMed]
- 27.T. Nicol, C. Lefeuvre, O. Serri, et al., Assessment of SARS-CoV-2 serological tests for the diagnosis of COVID-19 through the evaluation of three immunoassays: Two automated immunoassays (Euroimmun and Abbott) and one rapid lateral flow immunoassay (NG Biotech), J. Clin. Virol. June 15, 2020. [Epub ahead of print] 10.1016/j.jcv.2020.104511. [DOI] [PMC free article] [PubMed]
- 28.G. Yan, C.K. Lee, L. Lam, et al., Covert COVID-19 and false-positive dengue serology in Singapore, Lancet March 4 (2020). [Epub ahead of print] 10.1016/S1473-3099(20)30158-4. [DOI] [PMC free article] [PubMed]
- 29.A.K. Nalla, A.M. Casto, M.W. Huang, et al., Comparative Performance of SARS-CoV-2 Detection Assays Using Seven Different Primer-Probe Sets and One Assay Kit, J. Clin. Microbiol. May 26 (2020). [Epub ahead of print] 10.1128/JCM.00557-20. [DOI] [PMC free article] [PubMed]
- 30.J.L. He, L. Luo, Z.D. Luo, et al., Diagnostic performance between CT and initial real-time RT-PCR for clinically suspected 2019 coronavirus disease (COVID-19) patients outside Wuhan, China, Respir Med April 21, 2020. [Published online ahead of print] 10.1016/j.rmed.2020.105980. [DOI] [PMC free article] [PubMed]
- 31.Y. Yang, M. Yang, C. Shen, et al., Evaluating the accuracy of different respiratory specimens in the laboratory diagnosis and monitoring the viral shedding of 2019-nCoV infections, medRxiv 17 February, 2020. [Preprint] 10.1101/2020.02.11.20021493. [DOI]
- 32.J.J. Zhao, Q. Yuan, H.Y. Wang, et al., Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019, Clin. Infect Dis. 28 March (2020). [Published online ahead of print] 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed]
- 33.I. Arevalo-Rodriguez, D. Buitrago-Garcia, D. Simancas-Racines, et al., False-negative results of initial RT-PCR assays for covid-19: a systematic review, medRxiv 13 August (2020). [Preprint] 10.1101/2020.04.16.20066787. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.