Abstract
Global health and food security constantly face the challenge of emerging human and plant diseases caused by bacteria, viruses, fungi, and other pathogens. Disease outbreaks such as SARS, MERS, Swine Flu, Ebola, and COVID-19 (on-going) have caused suffering, death, and economic losses worldwide. To prevent the spread of disease and protect human populations, rapid point-of-care (POC) molecular diagnosis of human and plant diseases play an increasingly crucial role. Nucleic acid-based molecular diagnosis reveals valuable information at the genomic level about the identity of the disease-causing pathogens and their pathogenesis, which help researchers, healthcare professionals, and patients to detect the presence of pathogens, track the spread of disease, and guide treatment more efficiently. A typical nucleic acid-based diagnostic test consists of three major steps: nucleic acid extraction, amplification, and amplicon detection. Among these steps, nucleic acid extraction is the first step of sample preparation, which remains one of the main challenges when converting laboratory molecular assays into POC tests. Sample preparation from human and plant specimens is a time-consuming and multi-step process, which requires well-equipped laboratories and skilled lab personnel. To perform rapid molecular diagnosis in resource-limited settings, simpler and instrument-free nucleic acid extraction techniques are required to improve the speed of field detection with minimal human intervention. This review summarizes the recent advances in POC nucleic acid extraction technologies. In particular, this review focuses on novel devices or methods that have demonstrated applicability and robustness for the isolation of high-quality nucleic acid from complex raw samples, such as human blood, saliva, sputum, nasal swabs, urine, and plant tissues. The integration of these rapid nucleic acid preparation methods with miniaturized assay and sensor technologies would pave the road for the “sample-in-result-out” diagnosis of human and plant diseases, especially in remote or resource-limited settings.
Keywords: Point-of-care diagnostics, DNA/RNA extraction, Nucleic acid amplification, Raw samples, Infectious diseases, Plant diseases
Graphical abstract
Highlights
-
•
This review mainly focuses on rapid, field-portable nucleic acid extraction technologies from raw human and plant samples.
-
•
In the article, miniaturized nucleic acid extraction methods are discussed for easily accessible sample types, such as human blood, oral/nasal samples, urine, and plant tissues.
-
•
The working principles, pros, and cons of various point-of-care sample preparation platforms are highlighted in the review.
1. Introduction
Emerging human and plant diseases are one of the major threats to global health and human civilization. Outbreaks such as the current COVID-19 pandemic upend daily life. According to Johns Hopkins University, above 25 million people are infected by the novel coronavirus, and more than 800,000 people have lost their lives worldwide when this article is written. The global economy and many other aspects of human activities have been severely impacted. Early, rapid, and accurate detection of diseases is crucial to maximizing crisis management efficiency, treatment outcomes, and economic stability. However, current practices of human and plant disease detection are mainly restricted to the centralized laboratories. Usually, patients or samples are taken to hospitals or diagnostic clinics for testing, and test results are returned within several days. Often, disease detection is delayed in developing countries or regions due to the shortage of skilled personnel and medical infrastructure. Moreover, even the healthcare systems of developed countries are facing an unprecedented challenge for laboratory-based disease detection during the ongoing COVID-19 outbreak. Therefore, the demand for portable, easy-to-use, and point-of-care (POC) diagnostic tests is increasing rapidly.
POC testing of infectious human and plant diseases frees crucial time for planning, preparing, and responding to stop or limit the spread of disease in a community or an agricultural field. In POC diagnosis, patients’ samples are immediately analyzed for disease screening at the sampling point. This type of testing requires a very small sample size for biomarker detection, which can be collected by patients themselves without assistance from medical personnel. After the addition of samples to the testing device, the results are displayed within a few minutes. In POC testing, various detection techniques such as nucleic acid testing (Batule et al., 2020; Leiske et al., 2015), lateral flow assays (LFA) (Fang et al., 2014; R. H. Tang et al., 2017), nanomaterial-based sensors (Li et al., 2020; Liu et al., 2014; Ngo et al., 2018; Padmavathy et al., 2012), colorimetric immunosensors (Ren et al., 2017), volatile organic compound sensors (Z. Li et al., 2019a, Li et al., 2019b), bio-optical sensors (Jin et al., 2018; Yoo and Lee, 2016), and electrochemical sensors (Dutta et al., 2018; W. Liu et al., 2018) have been applied for the rapid detection of a broad range of human and plant diseases. Among these techniques, molecular assays based on nucleic acid amplification (NAA) are widely preferred. NAA-based assays examine the genomic information of pathogens or cells and thus can accurately identify microorganisms as well as their pathogenic strains, which cannot be easily achieved with other techniques. Moreover, NAA-based assays are sensitive, specific, and often can be multiplexed for the simultaneous identification of multiple pathogens (Basha et al., 2017; Stumpf et al., 2016).
NAA-based human and plant disease detection involves three major steps: nucleic acid extraction/purification, amplification, and detection. For nucleic acid extraction, the first step is cell lysis, which releases nucleic acids and other intracellular molecules of interest. Several on-chip cell lysis techniques such as chemical lysis (Ma et al., 2019; Yoon et al., 2018), mechanical lysis (J. Choi et al., 2015; Mahalanabis et al., 2009), electrical lysis (Hügle et al., 2018; Kim et al., 2009; Nan et al., 2014), ultrasonic lysis (Branch et al., 2017), thermal lysis (Leiske et al., 2015; Wang et al., 2011; Zhang et al., 2016), and enzymatic lysis (Lounsbury et al., 2013; Petralia et al., 2013) have been demonstrated for rapid lysis of human and pathogen cells.
After cell lysis, the nucleic acids are separated from the lysate, which may contain proteins, cell debris, cell lysis chemicals, and other impurities. This step is typically referred to as nucleic acid purification or isolation. In the conventional liquid-liquid extraction protocols, cell lysate is mixed with an equal volume of phenol-chloroform mixture to remove proteins and cell debris in the organic phase (Ayoib et al., 2017). Then, the aqueous phase containing nucleic acid is transferred to a new tube to precipitate DNA or RNA by adding salt and alcohol. This conventional method is tedious and time-consuming. Moreover, this method requires toxic chemicals (e.g., organic solvents), which limits its applicability outside the laboratories. For POC applications, solid-phase extraction (SPE) is more widely used for nucleic acid isolation and purification (Kim et al., 2010; Price et al., 2009; Reinholt and Baeumner, 2014). In SPE, cell lysate is mixed with or passed through solid-phase materials, such as filter papers (R. Tang et al., 2017a), silica membranes or beads (Branch et al., 2017), polymer resins (Byrnes et al., 2013), organic ligands (Jin et al., 2017), nanomaterials (H. Liu et al., 2018), or magnetic particles (Fu et al., 2018; Kang et al., 2017; Neto et al., 2017) to selectively bind nucleic acids at a pH lower than 7.5 (Yoon et al., 2018), remove impurities, and elute DNA or RNA molecules from the solid phase at a higher pH (~pH 8).
By selecting proper cell lysis techniques (e.g., chemical-free) and sample matrices (e.g., inhibitor-free), direct amplification of nucleic acids from raw samples without extraction and purification steps has been demonstrated (Curtis et al., 2008; Estrela et al., 2019; Walker and Hsieh, 2019). However, such a strategy has several major drawbacks. For example, many raw samples like whole blood and mucus are dense biofluids, which makes it difficult to perform molecular assays without proper dilution (Yaren et al., 2017). Furthermore, not all nucleic acid amplification methods can amplify targets from raw samples (Bender et al., 2018; McFall et al., 2015). Without pre-concentrating the DNA or RNA via extraction steps, direct detection may suffer from a higher limit of detection (LOD) of the assays (Czilwik et al., 2015; Hassan et al., 2018; Hoos et al., 2017). Therefore, nucleic acid extraction and purification are crucial steps for sensitive and accurate detection of human and plant diseases (Van Heirstraeten et al., 2014).
After nucleic acid extraction and purification, many amplification and detection strategies have been developed in the past decades. Polymerase chain reaction (PCR) and its variants such as reverse transcription-polymerase chain reaction (RT-PCR) are still considered the gold-standard method for nucleic acid detection (Petralia and Conoci, 2017). PCR and RT-PCR assays are highly sensitive and specific. For POC applications, the PCR reagents can be lyophilized without sacrificing assay performance (Chen et al., 2010; Czilwik et al., 2015). Moreover, in multiplexed PCR (Cai et al., 2014; Czilwik et al., 2015) or RT-PCR assays (Chan et al., 2016a; Yaren et al., 2017; Yin et al., 2020), simultaneous amplification of multiple pathogens’ DNA or RNA is possible for high-throughput screening. Over the past few years, researchers have developed many modified versions of PCR for rapid NAA in resource-limited settings, such as continuous-flow PCR (Fu et al., 2018), digital PCR (Hindson et al., 2011; Yin et al., 2019a), droplet PCR (Cai et al., 2014; Chiou et al., 2013; Markey et al., 2010), insulated isothermal PCR (Tsai et al., 2019), and ultrafast photonic PCR (Son et al., 2016, 2015). Nevertheless, precise temperature control on a miniaturized thermal cycler is still a major challenge (Liu et al., 2017; Park et al., 2011). Due to this limitation, isothermal amplification methods are better suited for in-field disease detection. Representative methods include loop-mediated isothermal amplification (LAMP) (Lee et al., 2019; Ma et al., 2019; Park et al., 2017), nucleic acid sequence-based amplification (NASBA) (Tsaloglou et al., 2011), strand displacement amplification (SDA) (Fang et al., 2014; Lafleur et al., 2016), recombinase polymerase amplification (RPA) (Bender et al., 2018; Magro et al., 2017b; Rohrman and Richards-Kortum, 2012), and helicase dependent amplification (HDA) (Linnes et al., 2014; Magro et al., 2017a; Rosenbohm et al., 2020). Among these isothermal techniques, LAMP has been most widely researched for POC applications (Choi et al., 2016; Ma et al., 2019; Ye et al., 2018; Y. Zhang et al., 2014). Like RT-PCR, RT-LAMP combines reverse transcription and LAMP assays in the same pot for specific RNA amplification (Estrela et al., 2019; Rodriguez et al., 2015). In general, LAMP is more robust and inhibitor-tolerant than PCR (Damhorst et al., 2015; Kaneko et al., 2007). In addition, its isothermal reaction condition (65 °C) allows the use of a much simpler and lower-cost heating instrument to run the LAMP assay (Lu et al., 2016; Wang et al., 2020; Y. Zhang et al., 2014). Moreover, LAMP assays can directly amplify nucleic acids from raw samples such as whole blood (Lee et al., 2019; X. Liu et al., 2018) and swabs due to their robustness (Hoos et al., 2017). LAMP or RT-LAMP reagents can also be lyophilized to store at room temperature up to several months (Hayashida et al., 2015; Seok et al., 2017). Recently, several modified versions of the LAMP assay such as Tte UvrD Helicase-LAMP (New England Biolabs, USA) and UDG-LAMP (Hsieh et al., 2014) have been demonstrated to further improve the specificity and other drawbacks of the assay.
The final step for disease identification is the detection and quantification of amplicons. In the laboratory, gel electrophoresis is usually performed to confirm the amplicons based on their molecular sizes. For POC visualization of amplicon products, lateral flow strips can be used instead (Fu et al., 2018; Huang et al., 2013; Lee et al., 2019; Rodriguez et al., 2015). Lateral flow-based amplicon detection is sequence-specific, and a single strip can detect multiple amplicons simultaneously (Park et al., 2017). Another technique commonly used for laboratory-based assay quantification is real-time detection (e.g., real-time PCR or quantitative PCR (qPCR)). For real-time detection, DNA probes such as molecular beacons, TaqMan probes, or DNA intercalating dyes are included in the amplification mixture (Borysiak et al., 2015; Loo et al., 2017; Wu et al., 2013). However, conventional qPCR requires bulky, expensive, and sophisticated instruments. Alternatively, amplicon detection can also been achieved on cost-effective smartphone-based reader devices (Borysiak et al., 2015; Choi et al., 2016; Kaur et al., 2019; Ma et al., 2019; Wang et al., 2020; Zhu et al., 2020). Smartphone-based nucleic acid detection platforms have been used for rapid screening of both human and plant diseases in resource-limited settings (Hernández-Neuta et al., 2019; Kong et al., 2017). Finally, the LAMP assay can also be detected and quantified by turbidity or color change of the assay solution (Curtis et al., 2008; Estrela et al., 2019; M. Zhang et al., 2020a, Zhang et al., 2020b). For colorimetric detection of the LAMP assay, pH-sensitive dyes (Kaarj et al., 2018), metal ion indicators (Port et al., 2014; Seok et al., 2017), or functionalized gold nanoparticles (Choi et al., 2016) have been reported as color indicators in the literature.
For POC disease diagnostics, an ideal system should integrate all steps from raw sample processing to amplicon detection, and run the steps automatically with minimal human intervention. After the first demonstration of a miniaturized total chemical analysis system (μ-TAS) by Manz et al. (1990), researchers in the last 30 years have developed numerous molecular detection systems utilizing microfluidic (Kolluri et al., 2017; Koo et al., 2017; Liu et al., 2011; Q. Liu et al., 2018a, Liu et al., 2018c; Petralia et al., 2013; Wu et al., 2014; Yan et al., 2017; Yin et al., 2019b; Zhang et al., 2016) or paper-based devices (Choi et al., 2016; Deng et al., 2017; Liu et al., 2017; Loo et al., 2017; Rodriguez et al., 2015). Furthermore, several commercial platforms such as GeneXpert Systems (Cepheid, USA), ARIES Systems (Luminex, USA), BioFire FilmArray Torch (BioFire Diagnostics, USA), and Integrated Cycler (Focus Diagnostics, USA) have been developed for rapid molecular diagnosis of human diseases.
However, most of these detection systems do not incorporate a sample preparation step to isolate biomarkers of interest from raw sample matrices such as blood, saliva, urine, and plant tissue (Berry et al., 2014; Chan et al., 2016a; Stumpf et al., 2016). Because of the complex nature of raw samples, many systems still depend on off-chip sample preparation (Kaur et al., 2019; Liu et al., 2017; Magro et al., 2017b; Rodriguez et al., 2015; Wang et al., 2020) or sample pretreatment steps such as plasma (Kaarj et al., 2018; Yin et al., 2020) or serum separation (Estrela et al., 2019; Tsai et al., 2019; Zhang et al., 2019) before the actual assay reactions. As a result, so far only a few number of truly integrated “sample-in-answer-out” systems have been demonstrated for practical use in real-world settings. For POC disease detection, automatic and hand-free sample preparation is a prerequisite because sample purity and contaminants directly affect the detection performance (e.g., sensitivity, accuracy, etc.) (Van Heirstraeten et al., 2014). In this review, we have summarized emerging POC sample preparation techniques, which demonstrate great potential for easy integration with miniaturized nucleic acid amplification and detection platforms for on-site and rapid detection of human and plant diseases from raw samples (e.g., human blood, saliva, urine, and plant tissue). This review specifically focuses on rapid extraction methods for the isolation of high-quality nucleic acid targets due to their preferred analytical performance in disease detection. The extraction techniques are discussed and grouped based on the sample matrix types, including most commonly accessible samples such as human blood, oral/nasal samples, urine, and plant tissues. For each extraction technique, we discuss their principles, advantages, disadvantages, and applications for real patient samples.
2. Nucleic acid extraction from human blood
Blood is one of the most widely used body fluids for the molecular diagnosis of human diseases. Human whole blood consists of plasma (~55% of total blood volume), buffy coat (including white blood cells plus platelets, ~1% of total blood volume), and red blood cells (~45% of total blood volume) (Alberts et al., 2002). Undiluted plasma contains a high concentration of interfering proteins whose total concentration is typically 60–80 mg/mL, equivalent to a solution of 6–8% (w/w) BSA (Walker et al., 1990). More than 50% of the composition of serum proteins are albumins, followed by immunoproteins (e.g., IgG, IgA, IgM, IgD), transferrin, fibrinogen, clotting factors, etc. (Walker et al., 1990).
Nucleic acid extraction from whole blood is an essential step for DNA/RNA-based diagnosis. However, isolation of nucleic acids from whole blood is a multistep process, which is usually performed in well-equipped laboratories by skilled technicians. Standard laboratory extraction procedure involves three major steps: lysis of cell nucleus membranes with surfactants (e.g., SDS, CTAB, or Triton X-100), denaturation of proteins by proteases (e.g., proteinase K), and purification of nucleic acids (Basha et al., 2017; Kim et al., 2009). However, the actual extraction protocols vary significantly depending on the purposes of sample preparation. For example, for genomic DNA isolation, white blood cells need to be separated from the rest of the blood components (J. Choi et al., 2015). In contrast, for the detection of pathogenic nucleic acids or cell-free DNAs, either serum or pathogen-infected blood cells are separated before extraction (Zhang et al., 2019). Removing red blood cells also helps to reduce the inference of hemoglobin, which is one of the major sources of inhibitors for downstream NAA reactions (Magro et al., 2017a).
Several miniatured sample preparation techniques for whole blood, plasma, or serum have been reported for POC pathogen detection (Batule et al., 2020; Ganguli et al., 2017; L. Zhang et al., 2020a, Zhang et al., 2020b), short tandem repeat analysis (Gan et al., 2014; Lounsbury et al., 2013), single nucleotide polymorphism detection (Lu et al., 2016), cancer diagnosis (Zhang et al., 2010), forensic analysis (Duarte et al., 2010), and hereditary genetic testing (Zhuang et al., 2015). However, rapid sample preparation platforms for forensic analysis are beyond the scope of this review. In this section, miniaturized nucleic acid extraction systems for disease detection from human blood samples will be discussed. A summary of rapid nucleic acid extraction methods for human blood is presented in Table 1 .
Table 1.
Rapid nucleic acid isolation methods for pathogen detection in blood.
| Sample Type | Target Pathogen | Cell Lysis Method | Nucleic Acid Extraction Technique | Nucleic acid Amplification Method | LOD/Extraction Efficiency | Total Sample-To-Answer/Sample Preparation Time | Reference | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Human serum | Hepatitis B virus and E. coli | Laser lysis | Polystyrene coated magnetic beads absorbed proteins and cell debris after laser irradiation | Real-time PCR | 20 copies/μL | 32 min (sample-to-answer) | Lee et al. (2006) | |||
| Whole blood | Gram-positive and Gram-negative bacteria | Hybrid chemical and mechanical lysis | Silica bead/polymer composite SPE column | Benchtop RT-PCR | 102 CFU/mL for E. coli 103–104 CFU/mL for Bacillus subtilis and Enterococcus faecalis | ~1 h (sample preparation) | Mahalanabis et al. (2009) | |||
| Whole blood | E. coli | Surface-modified micropillar arrays captured bacterial cells | After washing PCR inhibitors, captured cells were directly used for PCR | Real-time PCR | 40% (cell capture efficiency) | 1 h (sample-to-answer) | Hwang et al. (2011) | |||
| Whole blood | Pseudomonas aeruginosa, Staphylococcus aureus and E. coli | Thermal lysis | No purification | Multiplex array PCR | ~103 CFU/mL | 3 h (sample-to-answer) | Cai et al. (2014) | |||
| Whole blood | HIV-1 | On-chip chemical lysis | No Purification | RT-LAMP | 670 copies/μL | ~45 min (sample-to-answer) | Damhorst et al. (2015) | |||
| Whole blood | Malaria parasite | On-chip chemical lysis | Dimethyl adipimidate (DMA)/Thin film Sample processing technique | Mach-Zehnder Interferometer-Isothermal solid-phase DNA Amplification (MZI-IDA) | Less than 1 parasite/μL | 60 min (sample-to-answer) | Liu et al. (2016) | |||
| Blood serum | E. coli | Off-chip chemical lysis | Surface modified microfluidic chip | Benchtop real-time PCR | 90% (extraction efficiency) | 30 min (sample preparation) | Choi et al. (2020) | |||
| Blood serum | Hepatitis B virus (HBV) and HIV | On-chip chemical cell lysis | Ultrasonic assisted magnetic beads based SPE | Benchtop real-time PCR | 103 copies HBV/mL 5 × 103 copies HIV/mL | Less than 1 min (sample preparation) | Zhang et al. (2019) | |||
| Whole blood | HIV | Off-chip chemical cell lysis | Polymerized acrylate based SPE | Off-chip real-time PCR | 1000 copies/mL | 35 min (sample preparation) | Byrnes et al. (2013) | |||
| Whole blood | E. coli | Small FTA disk in micropipette tip to lyse and trap DNA from blood | LAMP | 8 CFU per reaction | 1 h (sample-to-answer) | Lu et al. (2016) | ||||
| Whole blood | E. coliand HBV | Target separation and laser irradiated magnetic bead system (TS-LIMBS) | Benchtop real-time PCR | ~90% (capture efficiency for E. coli) | 12 min (sample preparation) | Cho et al. (2007) | ||||
| Serum | Staphylococcus warneri, Streptococcus agalactiae, E. coli, and Haemophilus influenzae | Chemical lysis | SPE using silica coated magnetic particles | Real-time PCR using freeze-dried reagents | 15 CFU/mL S. warneri, 1000 CFU/mL S. agalactiae, 25 CFU/mL E. coli and 10 CFU/mL H. influenzae | 35 min (sample preparation) 3 h and 45 min (sample-to-answer) | Czilwik et al. (2015) | |||
| Whole blood | Acinetobacter baumannii (Ab) | Chemical | SPE using silica membrane | RT-LAMP using preloaded reagents | 102 CFU/mL | 2 h (sample-to-answer) | Loo et al. (2017) | |||
| Whole blood | HBV | Chemical lysis | SPE using Magnetic beads | RT-PCR using lyophilized reagents | 102 copies/mL | 15 min (sample preparation) 48 min (sample-to-answer) | Zhang et al., 2020a, Zhang et al., 2020b | |||
| Whole blood | HIV-1 | Chemical lysis using Triton-X | Fusion 5 membrane trapped blood cells. After washing, the membrane was used as template for PCR amplification. | On-chip real-time PCR | 50 copies/μL | Less than 2 min (sample preparation time) | Jangam et al. (2013) | |||
| Plasma | E. coli | Whatman FTA paper | LAMP | 500 cells/mL | 1 h (Sample-to-answer) | Connelly et al. (2015) | ||||
| Whole blood, Water spinach | E. coli, S. pneumonia | Fast Technology Analysis (FTA) card was used for DNA extraction | Paper-based LAMP | 100 CFU/mL for E. coli | 1 h (sample-to-answer) | Choi et al. (2016) | ||||
| Whole blood Serum, Saliva | HBV | Chemical lysis | Fusion 5 membrane was used for DNA extraction | Benchtop PCR | 104 copies/mL | 2 min (Sample preparation time) | (R. Tang et al., 2017a) | |||
| Whole blood | S. aureus | Chemical lysis | Glass filter membrane (GF/F grade, Whatman, UK) captured DNA | 3 min (Sample preparation) | Seok et al. (2019) | |||||
| Plasma | Dengue virus | Chemical lysis | Chitosan modified Fusion 5 filter paper captured viral RNA | RT-PCR | 100 copies/mL | 90 min (sample-to-answer) | Yin et al. (2020) | |||
| Blood serum | Zika and dengue virus | Chemical lysis using Triton X | Sequence specific capture of nucleic acids on glass fiber membrane | Paper based RT-LAMP using dry reagents | 0.5 copies/μL | 5 min (sample preparation) 1 h (sample-to-answer) | Batule et al. (2020) | |||
2.1. Microfluidic devices for nucleic acid extraction from serum samples
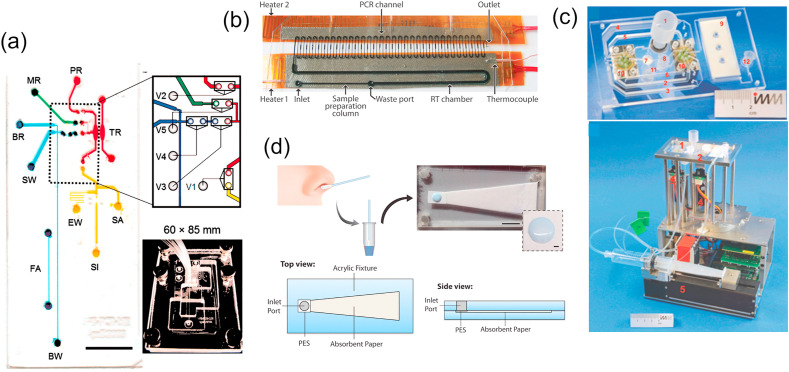
Blood serum is blood plasma without the clotting factors (in presence of anticoagulants) and is often preferred over whole blood as a better testing medium. For rapid pathogenic DNA isolation from serum, several microfluidic chips have been reported in the past. Lee et al. (2006) developed a Laser-Irradiated Magnetic Bead System (LIMBS) for pathogen DNA extraction from human serum by combining laser irradiation and carboxyl terminated magnetic beads. During the laser irradiation, the photothermal effect of the magnetic beads lysed hepatitis B viruses (HBV), E. coli, and Gram-positive bacteria within 40 s. Zhang et al. (2019) developed a microfluidic chip with pre-stored reagents to isolate nucleic acids from HBV and human immunodeficiency viruses (HIV) in less than a minute (Fig. 1 a). The microfluidic chip contained pressure-driven elastic membrane valves (PDEMV) to divide a serpentine microfluidic channel into several chambers for reagent pre-storage. In the reaction chamber, ultrasonic cell lysis and silica membrane-based SPE were integrated for rapid sample preparation from serum samples. Choi et al. (2020) reported a surface-modified polyethylene terephthalate (PET) microchip for nucleic acid extraction from various raw samples such as human serum, milk, and juice (Fig. 1b). To introduce positive charges on the inner surfaces of the microchip, poly (2-dimethylaminomethyl styrene) films were deposited on PET via an initiated chemical vapor deposition process. The microchip required ~30 min of incubation to capture 90% DNA from cell lysate.
Fig. 1.
Schematic illustrations of various microfluidic chips utilized for blood sample nucleic acid extraction: (a) PMMA microchip with two distinct regions for reagent storage and nucleic acid extraction (reproduced with permission from Ref (Zhang et al., 2019). © American Institute of Physics (AIP) 2019), (b) Surface-modified polyethylene terephthalate (PET) microchip for extraction of E. coli DNA from serum samples in 30 min (reproduced with permission from Ref (Choi et al., 2020)., © The Polymer Society of Korea and Springer (2019), (c) Cell lysis microchip for mixing whole blood and lysis buffer to lyse HIV-1 virus. (reproduced with permission from Ref. (Damhorst et al., 2015), © Engineering Sciences Press 2015), (d) Surface-modified micropillars-packed microchip for capturing E. coli cells from 50% whole blood (reproduced with permission from Ref (Hwang et al., 2011)., © Elsevier 2011), and (e) Dielectrophoresis chip for pathogen separation from diluted blood samples and PCR amplification (reproduced with permission from Ref. (Cai et al., 2014), © The Royal Society of Chemistry (2014).
2.2. Microfluidic devices for nucleic acid extraction from whole blood
Integrated microfluidic platforms have been developed to extract nucleic acids directly from whole blood, thereby skipping the steps of serum separation. For example, Mahalanabis et al. (2009) developed a disposable microfluidic chip for detecting Gram-positive and Gram-negative bacteria. In this microfluidic device, silica-impregnated porous polymer monoliths were fabricated to isolate pathogenic DNA via SPE. In addition to DNA binding, the silica-polymer composite column also generated mechanical shear to assist the chemical lysis of bacterial cells. For malaria detection, Liu et al. (2016) reported a dimethyl adipimidate/thin-film sample processing system for sample preparation in a microfluidic chip. To bind DNA from cell lysates, amine groups were introduced on the top and bottom surfaces of the chip to bring in positive charges. Damhorst et al. (2015) reported a sample-in-answer-out platform for HIV-1 detection from whole blood. In this microfluidic chip, blood and lysis buffer were passed through serpentine microfluidic channels for mixing and cell lysis (Fig. 1c). The lysed sample and RT-LAMP master mix were then injected onto microwells for LAMP amplification. The integrated system detected as low as 670 copies of HIV-1 per microliter of whole blood.
The detection of trace amounts of pathogens in clinical samples still remains a major challenge for NAA-based diagnostics. For early detection, processing of whole blood or plasma in a microfluidic device without pathogen enrichment may not yield an amplifiable signal. As a result, enrichment of target pathogen is often required for early disease detection. Hwang et al. (2008) developed surface-modified silicon pillars to capture bacterial cells such as Escherichia coli, Staphylococcus epidermidis, and Streptococcus mutans. The surface-modified pillars were arranged in arrays inside a microfluidic chamber for cell capturing (Fig. 1d). After cell capture, the captured cells were lysed directly on the pillars’ surface to extract pathogenic DNA. Later, the same group (Hwang et al., 2011) integrated on-chip PCR amplification with this rapid sample preparation technique to develop a complete “sample-in-answer-out” pathogen detection platform from whole blood. Cai et al. (2014) presented a dielectrophoretic technique in a microfluidic platform for pathogen separation from diluted whole blood, water, and other contaminated environmental samples. As shown in Fig. 1e, after dielectrophoretic separation, the pathogens captured in grooves were mixed with the droplets of preloaded PCR master mix. Then, the droplets were slipped away from the grooves to their original positions to run multiplex array PCR amplification for pathogen detection. The integrated device simultaneously detected Pseudomonas aeruginosa, Staphylococcus aureus, and Escherichia coli from blood within 3 h. For each pathogen, the platform had a LOD of ~103 CFU/mL. Jin et al. (2018) utilized positively charged homobifunctional imidoesters (HI), including dimethyl pimelimidate, dimethyl adipimidate, and dimethyl suberimidate, to capture pathogens from various raw samples, such as blood plasma, swab, saliva, and urine. A premixed solution of HIs and sample was added onto the surface-modified microfluidic chip for selective binding of HI-pathogen complexes. One potential drawback is that the HI reagents may bound non-specifically to any negatively charged molecules present in the sample.
2.3. Centrifugal microfluidic and other pump-free miniaturized devices
Conventional microfluidic devices depend on expensive syringe pumps for precise fluid manipulation. To eliminate the need for syringe pumps, several pump-free microfluidic devices such as hand-operated microfluidics (Byrnes et al., 2013; Li et al., 2012; Park and Park, 2018), vacuum-driven microfluidics (Yeh et al., 2017), digital microfluidics (Hung et al., 2017), centrifugal microfluidics or CDfuge devices (Kinahan et al., 2016), paperfuge (Bhamla et al., 2017) and capillary tube-based microfluidic devices (L. Zhang et al., 2014) have emerged in recent years.
Among these, centrifugal microfluidic (or CDfuge) devices with pre-stored extraction and assay reagents have become one of the most widely studied systems. In a centrifugal microfluidic device (also called LabDisk), the entire nucleic acid extraction process (including cell lysis, DNA binding to magnetic particles, washing steps to remove impurities, and DNA elution) can be carried out sequentially by varying the rotational speed of the disk. In this manner, these devices can replicate the standard steps of conventional laboratory nucleic acid extraction protocol. Moreover, in the centrifugal microfluidic platform, nucleic acid extraction and amplification can be easily integrated on the same disk chip for sample-to-answer detection of human diseases. For example, Cho et al. (2007) reported a centrifugal microfluidic device combining plasma separation and photothermal lysis for rapid DNA extraction from HBV and E. coli within 12 min. In this device, after plasma separation from whole blood, antibody-coated magnetic beads captured specific pathogens. Then, the captured pathogens were lysed by laser irradiation (Fig. 2 a). For total DNA extraction from human whole blood, Strohmeier et al. (2015) developed an integrated LabDisk by combining chemical cell lysis and magnetic bead-based DNA extraction. In this device, magnetic beads were pre-stored as a dry pellet. Then, blood and other DNA extraction reagents were loaded to the device before operation. Czilwik et al. (2015) presented a sample-in-answer-out platform by integrating chemical bacterial lysis, solid phase-based DNA extraction, and nested PCR detection on a centrifugal LabDisk for on-site and highly sensitive detection of both Gram-positive and Gram-negative bacteria from human serum. In this device, pathogen-specific primers and other PCR reagents were freeze-dried. Human serum, silica-coated magnetic particles, and other extraction reagents were manually pipetted to initiate the assay. All DNA extraction steps were completed automatically in less than 30 min. Extracted DNA was pre-amplified using consensus primer pairs and then split into 13 real-time PCR chambers for target amplification. Loo et al. (2017) performed hybrid mechanical-chemical bacterial lysis, silica membrane-based DNA extraction, and real-time RT-LAMP amplification on an integrated LabDisk. After sample loading, this integrated platform could detect minimally 100 CFU/mL Acinetobacter baumannii in whole blood within 2 h from 10 μL of sample. The integration of the isothermal LAMP method significantly lowered total assay time and simplified the heating system required to run the device. Yang et al., 2018a, Yang et al., 2018b reported a fully automated double rotation axes LabDisk for rapid POC diagnosis of HBV from 500 μL of whole blood (Fig. 2b). While a conventional LabDisk with one rotation axis only drives fluid radially outward, double rotation axes device, on the other side, enables fluid to be impelled in any arbitrary direction. Therefore, this unconstrained fluid movement capability increased the spatial utilization of the LabDisk. The device used pre-stored extraction and lyophilized PCR reagents and completed the whole process of plasma separation, cell lysis, DNA extraction, and real-time PCR amplification for the detection of HBV within 50 min. A LOD of as low as 100 copies/mL was reported. Despite their performance potential, centrifugal microfluidic devices have major drawbacks when scaling up. One of major drawbacks is high energy consumption, which limits its use in the field where the power supply is extremely limited. Furthermore, the fabrication of LabDisk devices is a complicated process, which presents a disadvantage for mass production and cost-effectiveness.
Fig. 2.
Various pump-free platforms for pathogen extraction and detection in blood. (a) The internal components of a portable centrifugal device developed for the extraction of HBV and E. coli DNA in 12 min (left), and a schematic diagram of target DNA extraction via laser irradiation (right) (reproduced with permission from Ref.(Cho et al., 2007), © The Royal Society of Chemistry © 2007). (b) Schematic of a centrifugal microfluid device showing the internal layout of various chambers used for sample preparation and nucleic acid amplification to detect HBV in 50 min (reproduced with permission from Ref. (L. Yang et al., 2018a, Yang et al., 2018b), © American Chemical Society 2019). (c) Schematic of a micropipette tip-based sample-to-answer E. coli detection system (reproduced with permission from Ref. (Lu et al., 2016), ©Elsevier 2016). (d) Schematic of a tube-based platform used for genomic DNA extraction from whole blood in 5 min (reproduced with permission from Ref. (Yin et al., 2019a), © The Royal Society of Chemistry (2019).
Other pump-free microfluidic extraction devices have also been demonstrated. Byrnes et al. (2013) developed a micropipette tip based-device for solid-phase DNA extraction. In this device, cell lysate, washing, and elution buffer were driven through a column of porous polymerized acrylate by hand-generated air pressure using a syringe. This hand-operated device extracted HIV-1 RNA from whole blood within 35 min. Lu et al. (2016) presented another micropipette tip-based sample-to-answer nucleic acid detection system from raw samples such as whole blood, bacteria, and plants (Fig. 2c). For sample preparation, a 600-μm diameter FTA disk was inserted into the micropipette tip. Yin et al. (2019a) reported a tube-based microfluidic device with pre-stored extraction reagents (Fig. 2d). In a PTFF tube with a 1-mm inner diameter, lysis buffer, washing buffer, and elution buffer were stored as droplets separated by mineral oil. An external magnetic field drove magnetic beads through these droplets for sequential nucleic acid binding, purification, and releasing. This capillary tube-based system extracted genomic DNA from whole blood in 5 min and can be easily applied to isolate pathogenic DNA.
2.4. Paper-based devices for nucleic acid extraction from human blood
In addition to microfluidic devices, paper-based devices are widely used for POC diagnostics (Chen et al., 2019; Magro et al., 2017a; Z. Yang et al., 2018), pathogen detection (J. R. Choi et al., 2015), food safety analysis (Liu et al., 2019; Trinh et al., 2019), and environmental monitoring (Sarwar et al., 2019; Seok et al., 2019; R. Tang et al., 2017b). Paper-based devices are easy to fabricate and inexpensive. They do not require an external pump for fluid manipulation. In paper devices, liquids flow spontaneously due to capillary action. Thus, nucleic acid extraction, amplification, and detection can be easily integrated on a single paper device without manual sample transfer steps.
For rapid sample preparation from raw samples (e.g., whole blood and plant leaf), Whatman FTA (GE Healthcare) cards are the most frequently used quick extraction technology and can pre-store dried proprietary lytic reagents (R. H. Tang et al., 2017). FTA cards lyse cells on contact and bind nucleic acids from cell lysate. Moreover, FTA cards contain chemical denaturants to denature proteins and prevent DNA degradation. For rapid nucleic acid isolation, Whatman FTA cards have been integrated into several microfluidic and paper-based devices. Connelly et al. (2015) fabricated a sliding-strip device by integrating FTA card-based DNA extraction and LAMP amplification for the rapid detection of pathogens from blood plasma (Fig. 3 a). Choi et al. (2016) developed a four-layer paper device for sample-to-answer detection of E. coli from various raw samples such as water, milk, blood, and spinach leaves (Fig. 3b). The device combines an FTA card for sample preparation, a glass membrane for LAMP amplification, and a lateral flow strip for amplicon detection. Different layers were initially separated by polyvinyl chloride (PVC) films. During operation, all sensor layers were combined by removing the intermediate PVC films to sequentially complete DNA extraction, amplification, and detection steps (Choi et al., 2017, 2016). A customized handheld heating device was also fabricated to run the amplification assay in resource-limited settings.
Fig. 3.
Various paper-based devices used for nucleic acid extraction from human blood samples. (a) Cross section view and operating procedure of a sliding-strip device for E. coli detection in blood plasma (reproduced with permission from Ref. (Connelly et al., 2015), © American Chemical Society 2015). (b) Four-layered paper-based biosensor used to detect E. coli in blood, water and milk samples (left), and disposable tape used for sealing the paper device for LAMP amplification (right) (reproduced with permission from Ref. (Choi et al., 2016), © The Royal Society of Chemistry (2016). (c) Schematic of the interaction of charge-switchable chitosan and nucleic acid in a pH-dependent manner (reproduced with permission from Ref. (Byrnes et al., 2015), © The Royal Society of Chemistry (2015). (d) Operating process of a handheld, lateral flow device for extraction of S. aureus DNA from blood in 3 min (reproduced with permission from Ref. (Seok et al., 2019), © IOP Publishing 2019). (e) Schematic illustration of the working principle of a paper-strip device used for viral RNA extraction from serum sample (reproduced with permission from Ref. (Batule et al., 2020), © Elsevier 2020).
Like the FTA card, the Fusion 5 membrane has also been reported for nucleic acid extraction from whole blood. For isolating HIV-1 proviral DNA from whole blood, Jangam et al. (2009) developed a Filtration Isolation of Nucleic Acids (FINA) method by using the Fusion 5 membrane. In the FINA method, whole blood was pipetted onto the Fusion 5 membrane to trap blood cells. NaOH was added to quickly lyse entrapped cells and wash away hemoglobin, cell debris, and other inhibitors. The FINA method has been modified to increase the limit of detection of HIV-1 from whole blood (Jangam et al., 2013; McFall et al., 2016, 2015). In the modified FINA method, whole blood lysed with Triton X was added onto the Fusion 5 membrane to capture nucleic acids. In a field trial for 61 patient samples, McFall et al. (2015) observed 100% detection sensitivity and specificity by using the modified FINA method. Tang et al. (2017a) compared the performance of the FTA card and Fusion 5 membrane for HBV detection from clinical blood samples and found that the detection limits were 103 copies/mL and 104 copies/mL for the FTA card and Fusion 5 membrane, respectively. To increase the nucleic acid extraction efficiency of the Fusion 5 membrane, Yin et al. (2020) proposed a method to modify the membrane surface with chitosan. Chitosan is a linear polysaccharide with a pKa ranging from 6.3 to 6.5 (Byrnes et al., 2015). As a result, chitosan exhibits pH-dependent interactions with negatively charged nucleic acids (Fig. 3c). Utilizing the chitosan-modified Fusion 5 membrane, the authors separated dengue virus (DENV) RNA from crude cell lysate. After RNA extraction, the modified Fusion 5 membrane was directly used for PCR amplification without inhibition.
Besides FTA card and Fusion 5 membrane disks, nitrocellulose and glass fiber membranes have also been reported for DNA extraction (Byrnes et al., 2015; Seok et al., 2019). Seok et al. (2019) utilized a glass filter membrane (GF/F grade, Whatman, UK) to develop a handheld lateral flow-based sample preparation device (Fig. 3d). In this device, the glass pad successfully captured Staphylococcus aureus DNA from lysates of various raw samples, such as blood, saliva, urine, water, and milk. Moreover, the purified DNA could be stored in the device up to 2 months before elution. To investigate the effect of paper pore size for chitosan modification, Byrnes et al. (2015) studied the interaction of low molecular weight (~5000 MW) chitosan with a nitrocellulose membrane with 5–10 μm pore diameter (FF80HP, GE Healthcare) and a glass fiber membrane with 10–100 μm pore diameter (Standard 17, GE Healthcare). The DNA capture capacity was lower in the modified nitrocellulose membrane than in the modified glass fiber membrane. The authors hypothesized that the integration of chitosan into smaller nitrocellulose pores reduced the amount of available chitosan for DNA binding in the nitrocellulose membrane. In addition, chitosan obstructed the convective transport of DNA in the smaller pores of the nitrocellulose membrane. As such, the authors recommended using a glass fiber membrane or other papers having large pores for chitosan modification. Batule et al. (2020) immobilized single-stranded DNA probes on a glass fiber membrane (or binding pad) for viral RNA enrichment and extraction in a paper strip device (Fig. 3e). In this device, three different probes specific to Zika, dengue, and chikungunya viruses captured viral RNAs from serum cell lysate for the early detection of mosquito-borne diseases. After hybridization, viral RNAs were eluted and manually transferred to another paper-chip device containing dry RT-LAMP reagents for amplification.
3. Nucleic acid extraction from oral and nasal samples
Oral and nasal specimens (e.g., saliva, sputum, and nasal swabs) have also been extensively used for human disease diagnosis. Oral and nasal samples are non-invasive and can be easily collected at resource-limited settings. In this section, miniaturized nucleic acid extraction systems for human disease detection from saliva, sputum, throat swab and nasal swab samples are discussed.
3.1. Nucleic acid extraction from saliva
Among oral specimens, saliva is an extracellular fluid containing >99% water. As a result, nucleic acid extraction from saliva is relatively simple and less tedious compared to other body fluids. Several miniaturized systems using saliva as a diagnostic fluid have been developed (Chen et al., 2010; Wand et al., 2018; H. Yang et al., 2018; Zhu et al., 2020). These integrated systems utilized SPE (Chen et al., 2010, 2013; H. Yang et al., 2018), paper-based extraction (Jiang et al., 2018; Seok et al., 2019; R. Tang et al., 2017a), or pH-responsive polymer-based extraction (Zhu et al., 2020) for rapid sample preparation from saliva (also see Table 2 ).
Table 2.
Rapid nucleic acid extraction for pathogen detection in oral samples.
| Sample Type | Target Pathogen | Cell Lysis Method | Nucleic Acid Extraction Technique | Nucleic Acid Amplification Method | LOD/Extraction Efficiency | Total Sample-To-Answer/Sample Preparation time | Reference |
|---|---|---|---|---|---|---|---|
| Saliva | B.cereus, HIV-1 virus | On-chip chemical lysis | SPE using silica membrane | On-chip PCR using dry reagents | 104 copies/mL | 1 h (sample-to-answer) | (Chen et al., 2010; Qiu et al., 2011) |
| Saliva | Methicillin-susceptible S. aureus and methicillin-resistant S. aureus | Thermal lysis | Aluminum oxide membrane-based DNA extraction | Real-time RT-PCR | ~100 copies of bacterial DNA per sample | Less than 2.5 h (sample-to-answer) | Oblath et al. (2013) |
| Saliva | Mycobacterium tuberculosis (M.tb) | Chemical lysis | SPE using magnetic bead | Digital RPA using preloaded liquid reagents | 91.3% (extraction efficiency of M.tb) | ~45 min (sample-to-answer) | (H. Yang et al., 2018) |
| Saliva | Zika virus | Chemical lysis | Cellulose paper-based RNA extraction | RT-LAMP combined with colorimetric detection | 3.5 plaque-forming units (PFU)/mL | 50 min (sample-to-answer) | Jiang et al. (2018) |
| Saliva | Zika virus | Chemical lysis | SPE using magnetic bead | Real-time RT-RPA using preloaded liquid reagents | 5 PFU/mL | Less than 15 min (sample preparation) | Chan et al. (2018) |
| Saliva | Zika virus | Chemical lysis | Chitosan-modified silicon dioxide capillaries | In-situ RT-PCR | 50 transducing units (TU)/mL | 25 min (sample preparation) 90 min (sample-to-answer) | Zhu et al. (2020) |
| Sputum (artificial) | E. coli | Paper-based microfluidic origami device | Benchtop PCR | 33 CFU/mL | 1.5 h (sample preparation) | Govindarajan et al. (2012) | |
| Sputum | M. tb | On-chip chemical cell lysis | SPE using photoactivated polycarbonate micropillars | Continuous flow PCR | 50 cells/mL | 30 min (sample-to-answer) | Wang et al. (2012) |
| Sputum | M. tb | Chemical lysis and magnetic bead-based nucleic acid extraction into a tube | Real-time LAMP using prestored reagents | 1000 cells/mL | 15 min (sample preparation) | Creecy et al. (2015) | |
| Sputum | M. tb | Liquefied sputum with 4% NaOH was directly added to LAMP mixture | LAMP | 2 copies/μL | 60 min (sample-to-answer) | Bentaleb et al. (2016) | |
| Sputum | M. tb | Chemical cell lysis and SPE using silica membrane on a LabDisk | Real-time LAMP using prestored reagents | 103 CFU/mL | 2 h (sample-to-answer) | Loo et al. (2017) | |
| Oropharyngeal swabs | S. pneumoniae and M. pneumoniae | On-chip chemical lysis | SPE using magnetic particles | LAMP amplification using prestored primers | 20 fg DNA per reaction | ~15 min (sample preparation) | Wang et al. (2019) |
| Throat swab | H1N1 Influenza Virus from | Antibody-coated magnetic beads captured viral ribonucleoprotein | RT-PCR | 10 TCID50 | ~3.5 h (sample-to-answer) | Ferguson et al. (2011) | |
3.1.1. Microfluidic and on-chip extraction platforms
For viral and bacterial pathogen detection from human saliva, Chen et al. (2010) integrated silica membrane-based DNA extraction, PCR amplification, and lateral flow-based amplicon detection in a microfluidic cassette (Fig. 4 a). All DNA extraction and PCR reagents were prestored in the microfluid cassette. After sample loading, the cassette was inserted into a custom-built analyzer to begin the operation (Qiu et al., 2011). This portable system was able to detect B. cereus and HIV-1 virus in saliva samples with a LOD of ~104 cells/mL. For multiplex detection of bacterial species in saliva, Oblath et al. (2013) combined aluminum oxide membrane-based DNA extraction and real-time PCR amplification on a microchip (Fig. 4b). For DNA extraction, the aluminum oxide membrane was sandwiched between reaction wells and a PDMS layer. Unlike the silica membrane, the aluminum oxide membrane-based DNA extraction method did not need the washing steps. For the simultaneous detection of antibodies and nucleic acids in saliva samples, Chen et al. (2013) developed a dual-path microfluidic chip. In this device, the loaded saliva sample was split into two different compartments for ELISA-based antibody detection and silica membrane-based nucleic acid extraction, respectively. Following nucleic acid purification, on-chip RT-PCR was performed using the dry illustra™ RT-PCR Beads (GE Healthcare). In a proof-of-concept application, the authors detected anti-HIV antibodies and HIV RNA simultaneously in the saliva sample. For Mycobacterium tuberculosis (M. tb) detection from saliva samples, Yang et al., 2018a, Yang et al., 2018b presented a low-cost and portable system by integrating magnetic bead-based solid-phase DNA extraction and a digital RPA assay. The integrated system had two components: a cartridge and a control instrument. In the cartridge, lysis buffer, magnetic beads, binding buffer, and washing buffers were prestored in different centrifuge tubes (Fig. 4c). During DNA extraction, these liquid reagents were automatically transferred from one tube to another to mimic a benchtop DNA extraction protocol. This integrated device demonstrated approximately 90% recovery of M. tb from spiked saliva samples. Chan et al. (2018) converted a 3D printer into a sample preparation device for saliva (Fig. 4d). The modified 3D printer automatically performed magnetic particle-based nucleic acid extraction from up to 12 samples simultaneously within 15 min. The authors also utilized the heated bed of the printer to run an RT-RPA assay for the rapid detection of Zika virus in saliva.
Fig. 4.
Various portable devices used for nucleic acid extraction from human saliva samples. (a) Schematic of an integrated microfluidic cassette for detecting HIV-1 in saliva (reproduced with permission from Ref. (Chen et al., 2010), © Springer 2010). (b) Hybrid PDMS/aluminum oxide membrane/glass microchip used for S. aureus detection (top), and a cross section view of the chip (bottom) (reproduced with permission from Ref. (Oblath et al., 2013), © The Royal Society of Chemistry (2013). (c) Schematic illustration of an integrated molecular diagnostic system for tuberculosis detection from saliva in 45 min (reproduced with permission from Ref. (H. Yang et al., 2018), © Springer 2018). (d) 3D printer-based Zika virus detection platform (reproduced with permission from Ref. (Chan et al., 2018), © Elsevier 2018). (e) Paper-based Zika virus RNA extraction device (top), and operating mechanism of the ball valve used in the device (bottom) (reproduced with permission from Ref. (Jiang et al., 2018), © Willey Online Library 2018). (f) Schematic illustration of a Zika virus detection chip embedded with chitosan-modified capillaries to capture viral RNA from saliva (reproduced with permission from Ref. (Zhu et al., 2020), © MDPI 2020).
3.1.2. Paper-based platforms for nucleic acid extraction from saliva
Paper-based devices have also been frequently used for nucleic acid extraction from saliva. Jiang et al. (2018) utilized cellulose papers to capture Zika viral RNA from saliva (Fig. 4e). The sample preparation device had two parts: a buffer unit (top) and an integrated mixing and detection unit (bottom). The buffer unit consisted of four reservoirs to store liquid reagents. The authors introduced bearing ball-based valves for reagent storage in these reservoirs. When the bottom part was slid under these reservoirs, a pin pushed the ball valves upward to open them and allow the prestored buffers to flow downward through the cellulose paper (Fig. 4e). Zhu et al. (2020) developed a microfluidic device embedded with chitosan-modified capillaries for the rapid extraction of Zika RNA from saliva (Fig. 4f). When the acidic lysate (pH = 5.5) was passed through the surface-modified capillary, positively charged chitosan molecules bond nucleic acids. Later, the PCR master mix (pH = 8.5) was introduced into the capillary to release the nucleic acids. As demonstrated by this device, charge-switchable ploymers could be a promising strategy to selectively capture and release nucleic acids from biofluids for rapid sample preparation at the detection site.
3.2. Nucleic acid extraction from sputum
3.2.1. Microfluidic and on-chip extraction platforms for sputum
Sputum is another oral fluid that is often used for the detection of a variety of human diseases such as tuberculosis (TB), influenza, and pneumonia (Table 2). Unlike saliva, sputum is a highly viscous fluid (Kaur et al., 2019). As a result, nucleic acid extraction from sputum samples involves additional pretreatment steps, including sample disinfection, liquefaction, and homogenization (Reed et al., 2016). In the laboratory, sample pretreatment steps are performed by skilled personnel inside a negative pressure biosafety cabinet to prevent infection. Thus, minimizing the risk of infection for on-site sputum sample preparation is another challenge. To ensure the safe processing of sputum samples in resource-limited settings, Park et al. (2018) developed a fully closed device for sputum sample pretreatment (Fig. 5 a). This portable device used simultaneous mechanical and chemical methods for sputum homogenization and cell lysis. However, an additional device was required for DNA extraction from homogenized sputum lysate. For rapid DNA extraction from sputum samples, Ferguson et al. (2016) developed a disposable bead blender, in which sample disinfection, liquefaction, mechanical cell lysis, and solid phase-based DNA extraction were performed semi-automatically in less than 20 min. In this miniaturized device, trisodium phosphate and povidone iodine were used as liquefaction reagent and disinfectant, respectively. However, during liquefaction and disinfection steps, a significant reduction (~104 fold) in viral load was observed for sputum samples spiked with M. tb . For the detection of antibiotic resistant M. tb. strains, Wang et al. (2012) developed a low-cost thermoplastic fluidic cartridge to extract pathogen DNA (Fig. 5b). In this sample cartridge, photoactivated polycarbonate micropillars capture nucleic acids from sputum lysate. Following DNA extraction, continuous-flow PCR amplification, ligase detection reaction, and a universal array assay were performed in the cartridge to detect single-base variations among M. tb strains. The system successfully detected as low as 50 M. tb cells in 1 mL of sputum samples, which is a 100-fold improvement from conventional detection methods.
Fig. 5.
Various sputum and oral swab sample preparation systems. (a) A handheld sputum collection device (reproduced with permission from Ref. (Park et al., 2018), © Springer Nature 2018). (b) 3D drawing of an integrated microfluidic cartridge used in tuberculosis detection, and a close-up image of the micropillar arrays utilized in SPE (inset) (reproduced with permission from Ref. (Wang et al., 2012), © Wiley Online Library 2012). (c) Tube-based automatic nucleic acid extraction and amplification system for M. tb detection (reproduced with permission from Ref. (Creecy et al., 2015), © PLoS One 2015). (d) Schematic layout of a centrifugal microfluidic device showing various chambers for DNA extraction and amplification for tuberculosis detection in 2 h (reproduced with permission from Ref. (Loo et al., 2017), © Elsevier 2017). (e) Paper microfluidic origami device for sputum sample preparation (reproduced with permission from Ref. (Govindarajan et al., 2012), © The Royal Society of Chemistry (2012). (f) Schematic diagram of an integrated microchip used in pneumoniae detection (left), and a photograph of the microchip (right) (reproduced with permission from Ref (Wang et al., 2019)., © Elsevier 2019).
Isothermal amplification methods such as LAMP have been widely used in the detection of pathogens in sputum samples (Bentaleb et al., 2016; Etchebarne et al., 2017; Wang et al., 2016). For tuberculosis detection, Bentaleb et al. (2016) demonstrated that the LAMP assay successfully amplified target pathogen from homogenized sputum samples with 4% NaOH solution. For Zika virus detection by RT-LAMP, Yang et al., 2018a, Yang et al., 2018b studied three different one-step sputum sample preparation methods: thermal lysis, NaOH lysis, and proteinase K lysis. Among these methods, the highest sensitivity was observed for the proteinase K lysis, which was comparable to the sensitivity of samples prepared by QIAamp viral RNA mini kit (Qiagen, USA). For highly sensitive detection of methicillin-resistant Staphylococcus aureus, Wang et al. (2011) utilized specific probe-conjugated magnetic beads to capture target nucleic acids from thermally lysed sputum samples. For rapid sample-to-answer detection of M. tb, Creecy et al. (2015) integrated magnetic bead-based DNA extraction and LAMP amplification in a fluorinated ethylene propylene (FEP) tube (Fig. 5c). Inside the FEP tube, DNA extraction reagents and LAMP master mix were stored sequentially separated by air columns. For sample preparation, an external magnet was used to move silica-coated magnetic beads through these buffer solutions. Loo et al. (2017) developed an integrated LabDisk for on-site detection of M. tb in sputum samples (Fig. 5d). After loading the homogenized sputum sample, LabDisk automatically performed cell lysis, silica membrane-based DNA extraction, and real-time LAMP amplification for target detection within 2 h. This device achieved a detection limit of 103 CFU/mL M. tb in sputum.
3.2.2. Paper-based platforms for nucleic acid extraction from sputum
Paper-based sample preparation methods for sputum have also been reported (Govindarajan et al., 2012; Guio et al., 2006; R. Tang et al., 2017a). To detect tuberculosis, Guio et al. (2006) utilized an FTA card for in-field nucleic acid extraction and storage. On the FTA card, the sputum sample can be lysed within 1 h at room temperature. After lysis, extensive washings were performed with FTA purification reagents and TE buffers to remove PCR inhibitors from the dried FTA card. The overall FTA card-based sample preparation time was about 2.5 h. For instrument-free nucleic acid extraction, Govindarajan et al. (2012) developed a paper-based origami device (Fig. 5e). In this device, a Fusion 5 membrane was used to capture nucleic acids from sputum lysate. For on-site applications, lysis buffer was dried on paper. This device was demonstrated to isolate E. coli DNA from spiked pig mucin (simulated sputum) with a low bacterial concentration of 33 CFU/mL. The total sample preparation time was 1.5 h in field settings. Obviously, one of the major drawbacks of paper-based methods for viscous sputum sample preparation is the relatively long sample preparation time due to the slow diffusion rate of molecules in the paper matrix. However, given the cost-effectiveness and simplicity of paper devices, their use has promising potential for POC sample preparation.
3.3. Nucleic acid extraction from oral swab samples
Instead of directly collecting oral fluids, swabs are often used to collect oral samples for human disease detection (Ferguson et al., 2011; Wang et al., 2019). To detect H1N1 influenza virus from a throat swab, Ferguson et al. (2011) developed a sample-to-answer, portable platform by integrating antibody-coated magnetic bead-based sample preparation, RT-PCR amplification, ssDNA generation, and sequence specific electrochemical detection in a monolithic device. The authors demonstrated that the LOD of this integrated system was 10 TCID50 (50% tissue culture infective dose), which is approximately 4 times lower than the clinical titer of the H1N1 influenza virus. To detect Streptococcus pneumoniae and Mycoplasma pneumoniae from oropharyngeal swabs, Wang et al. (2019) integrated magnetic particle-based nucleic acid extraction and LAMP amplification on a microfluidic chip. In this device, DNA binding, washing, and elution steps were performed manually by loading liquid reagents into the microchip using a micropipette (Fig. 5f). After DNA extraction, the eluted DNA was split to several microchambers with pre-stored primers for LAMP amplification.
3.4. Nucleic acid extraction from nasal samples
Besides oral samples, nasal specimens (e.g., nasal swab, nasopharyngeal swab, nasal wash, and nasopharyngeal aspirate) have been widely used for the molecular detection of human diseases in both laboratory and field settings (Lafleur et al., 2016; Wang et al., 2018). Table 3 summarizes the rapid nucleic acid extraction methods for biomarker detection in nasal specimens. For rapid on-site detection of respiratory pathogens, several groups demonstrated direct nucleic acid amplification methods in raw and unpurified nasal samples (Hoos et al., 2017; Létant et al., 2007; Sun et al., 2020). Nie et al. (2012) developed a boiling-based lysis method to detect human enterovirus 71 (EV71) in nasopharyngeal swabs. In this protocol, raw samples were heated to 95 °C for 30 s for viral lysis before LAMP amplification. The detection sensitivity of the LAMP assay using boiled samples was approximately 90%. Dou et al. (2019) directly added cell lysate to the LAMP mixture to detect Bordetella pertussis from nasopharyngeal swab samples. For cell lysis, an equal volume of sample and bacterial lysis buffer (50 mM Tris buffer, 4 M urea, and 0.1% Triton, pH 7.5) were mixed and incubated at room temperature for 10 min. Although the detection sensitivity of the direct amplification method (without nucleic acid extraction) may be lower than that of standard protocols with extraction, and may not be suitable for early detection of diseases without purifying and concentrating the targets, the simplicity of the procedure still makes it attractive for various POC applications.
Table 3.
Rapid nucleic acid extraction platforms for pathogen detection in nasal samples.
| SampleType | Target Pathogen | Cell Lysis Method | Nucleic Acid Extraction Technique | Nucleic Acid Amplification Method | LOD/Extraction Efficiency | Total Sample-To-Answer/Sample Preparation Time | Reference |
|---|---|---|---|---|---|---|---|
| Nasal aspirate | Bordetella pertussis | Chemical lysis | On-chip SPE using silica beads | On-chip PCR | – | Less than 30 min (sample-to-answer) | Easley et al. (2006) |
| Nasopharyngeal swab | Influenza A virus | Chemical lysis | SPE using silica/polymer composite | On-chip RT-PCR | 103 copies/mL | ~3 h (sample-to-answer) | Cao et al. (2012) |
| Nasopharyngeal swab | Human enterovirus 71 (EV71) | Thermal lysis | Benchtop real-time RT-LAMP | 1.6 TCID50 per reaction | ~1 min (sample preparation) | Nie et al. (2012) | |
| Nasopharyngeal swab | Influenza A virus, gram-positive and gram-negative bacteria | On-chip chemical lysis | SPE using porous membrane | Benchtop real-time RT-PCR | Higher or similar nucleic acid yields compared to commercial kits | ~10 min (sample preparation) | Van Heirstraeten et al. (2014) |
| Nasopharyngeal swab | Influenza A (H1N1) | Polyethersulfone (PES) membrane captures RNA-Glycoblue precipitate | Paper-based LAMP | 106 copies/mL | 45 min (sample-to-answer) | Rodriguez et al. (2015) | |
| Nasal swab | Methicillin-resistant S. aureus | Enzymatic lysis | Isothermal strand displacement amplification (iSDA) | ~5 × 103 copies per swab | 60 min (sample-to-answer) | Lafleur et al. (2016) | |
| Nasopharyngeal swabs | Respiratory syncytial virus | No nucleic acid extraction | Benchtop LAMP | ~2500 RNA copies per reaction | 30 min (sample-to-answer) | Hoos et al. (2017) | |
| Nasopharyngeal swabs and aspirates | Bordetella pertussis | After chemical lysis, crude lysate was used as template for DNA amplification | On-chip LAMP amplification | 5 CFU/reaction | 45 min (sample-to-answer) | Dou et al. (2019) | |
3.4.1. Microfluidic platforms for nucleic acid extraction from nasal samples
Nasal samples are frequently processed by microfluidic devices. For example, Easley et al. (2006) integrated solid phase-based DNA extraction, PCR amplification, and electrophoretic separation on a hybrid glass/PDMS microchip for Bordetella pertussis detection from the nasal aspirate (Fig. 6 a). In the microfluidic channel, silica beads of sizes varying from 5 μm to 30 μm were packed against the etched weir by applying a vacuum for nucleic acid extraction. The total sample preparation time for this microchip was less than 10 min. However, a syringe pump was required to deliver samples and reagents to the microchip, which limits its potential POC applications. Furthermore, the packed bed of silica beads could potentially be clotted by impure samples, which limits the total sample processing capacity of the microchip (e.g., ~1 μL of nasal aspirate). For influenza A virus detection in nasopharyngeal aspirate and swab specimens, Cao et al. (2012) integrated silica/polymer composite-based DNA extraction and continuous flow RT-PCR on a single microfluidic chip (Fig. 6b). For solid-phase nucleic acid extraction, porous polymer monoliths were formed in a microfluidic channel. Then, the formed monoliths were impregnated with silica beads (Bhattacharyya and Klapperich, 2006; Mahalanabis et al., 2009). This porous silica/polymer composite provided significantly low resistance to flow compared to the packed bed of silica and thus could process much larger sample volumes (e.g., 100 μL nasal samples) for better detection sensitivities. Yang et al., 2018a, Yang et al., 2018b developed a microfluidic cassette for instrument-free, on-site sample preparation from nasopharyngeal swab samples. For solid-phase DNA extraction, a silica membrane (80-μm thickness) was integrated into the microfluidic cassette (Fig. 6c). All reagents required for nucleic acid extraction were pre-stored in the cassette. During operation, a portable setup controlled microfluidic valves and flow of reagents through the silica membrane (Fig. 6c). The nucleic acid extraction yield for this microfluidic cassette was similar to that of commercial nucleic acid isolation kits (Quickgene DNA tissue kit S or RNA tissue kit SII, Fujifilm, Germany), thus demonstrating great promise for field applications.
Fig. 6.
Various microfluidic devices utilized to isolate nucleic acids from nasal specimens. (a) Schematic and photograph of an integrated microchip used in whooping cough detection. Yellow, red, green, and blue dyes indicate the domains for nucleic acid extraction, amplification, injection, and amplicon separation, respectively (reproduced with permission from Ref. (Easley et al., 2006), © PNAS 2006). (b) Microfluidic chip attached with two thin-film heaters for continuous flow PCR amplification to detect Influenza A virus (reproduced with permission from Ref. (Cao et al., 2012), © PLoS One 2012). (c) Images of a microfluidic cassette used for the extraction of viral and bacterial nucleic acid from nasal swab (top) and a portable controlling unit (bottom) (reproduced with permission from Ref. (Van Heirstraeten et al., 2014), © The Royal Society of Chemistry (2014). (d) A PES membrane-based paper device filtering RNA-Glycoblue precipitate from cell lysate for H1N1 detection (top), and schematic illustrations of the paper device (bottom) (reproduced with permission from Ref. (Rodriguez et al., 2015), © American Chemical Society 2015). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4.2. Paper-based devices for nucleic acid extractions from nasal samples
For paper-based processing of nasal samples, Rodriguez et al. (2015) utilized a polyethersulfone (PES) membrane to capture Influenza A RNA-Glycoblue precipitate from nasal sample lysate (Fig. 6d). After washing the captured precipitate, the LAMP reaction mixture was directly added onto the PES membrane for paper-based LAMP amplification. This paper device detected a viral load as low as 106 copies/mL, which is ten-fold lower than the LOD of conventional ELISA assays. Later, the authors further modified the design to minimize the number of manual interventions necessary for pathogen detection (Rodriguez et al., 2016). In the modified paper device, a plastic cover film was used to seal the PES membrane for LAMP amplification, which minimized cross-contamination and false-positive rates.
4. Nucleic acid extraction from urine samples
Urine is another common diagnostic fluid for human disease detection, which consists of ~95% water, 2% urea, various ions (chloride, sodium, potassium, etc.), and many other small molecules, which are the most valuable part for diagnosis. The analysis of urine biomarkers (e.g., proteins, glucose, electrolytes, and pathogens, etc.) can reveal the underlying health problems of a person at the early stages before symptom develops. The collection procedure of urine samples is relatively easy compared to that of other body fluids. Furthermore, urine samples are not limited by volume, and a large volume of urine samples can be enriched for highly sensitive detection of early disease markers. Table 4 summarizes the rapid nucleic acid extraction methods for biomarker detection in urine.
Table 4.
Rapid nucleic acid isolation systems for pathogen detection in urine.
| Sample Type | Target Pathogen | Cell Lysis Method | Nucleic Acid Extraction Technique | Nucleic Acid Amplification Method | LOD/Extraction Efficiency | Total Sample-To-Answer/Sample Preparation Time | Reference | |
|---|---|---|---|---|---|---|---|---|
| Urine | E. coli | On-chip hybrid chemical/mechanical cell lysis | SPE using silica impregnated polymer monolith | Benchtop real-time PCR | 10 CFU/mL | 40 min (sample preparation) | Kulinski et al. (2009) | |
| Urine | Chlamydia trachomatis | Chemical lysis | Paper-based SPE into a micropipette tip | HDA | 1000 cells/sample | ~50 min (sample-to-answer) | Linnes et al. (2014) | |
| Urine | Zika virus | Chemical lysis | Magnetic particles based SPE in a modified 3D printer | RT-RPA using prestored liquid reagents | 5 PFU/mL | 15 min (sample preparation) 25 min (sample to answer) | Chan et al. (2016b) | |
| Urine | Zika virus | Wax printed paper microfluidic chip was used to filter target pathogens | Paper based RT-LAMP | 1 copy/μL | 15 min (sample to answer) | Kaarj et al. (2018) | ||
| Urine | E. coli, Klebsiella pneumoniae and S. aureus | Chemical lysis | Chitosan-modified glass filter paper embedded in capillaries captured nucleic acids | LAMP amplification using prestored LAMP primers | 200 CFU per capillary | 85 min (sample-to-answer) | Hui et al. (2018) | |
| Urine | Zika Virus | Chemical lysis | Cellulose paper-based RNA extraction | RT-LAMP | 3.5 PFU/mL | 50 min (sample to answer) | Jiang et al. (2018) | |
| Urine | Brucella ovis | Pathogen enrichment, chemical cell lysis and functionalized Teflon filter-based DNA extraction | Benchtop real-time PCR | 1 CFU/mL | Less than 20 min (sample preparation) | Zhao et al. (2019) | ||
| Urine | M. tb | Chemical cell lysis | Magnetic bead-based DNA extraction into a transfer pipette | Benchtop real-time PCR | 90% (extraction efficiency) | ~20 min (sample preparation) | Pearlman et al. (2020) | |
| Urine | Trichomonas vaginalis (TV) | Chemical lysis | Chitosan-modified Fusion 5 membrane captured DNA | HDA | 7 genomic equivalents of TV DNA per mL | 2 min (sample preparation) | Rosenbohm et al. (2020) | |
4.1. Microfluidic and other miniaturized systems for nucleic acid extraction from urine
Kulinski et al. (2009) developed a simple microfluidic chip for the rapid preparation of urine samples. In this microchip, the microfluidic channels were filled with silica-impregnated porous polymer monolith for solid-phase nucleic acid extraction. However, samples flowed through the porous monolith at very high pressure (~150 psi). For low-pressure microfluidic sample preparation from urine, Shin et al. (2014) presented dimethyl adipimidate (DMA)-based solid-phase extraction in an amine-modified silicon microchip (Fig. 7 a). After cell lysis, DMA, a bifunctional imidoester, was mixed with cell lysate to electrostatically bind DNA to form a DMA-DNA complex. Then, the mixture passed through amine-modified microfluidic channels to capture DMA-DNA complexes from the lysate solution. The interaction between DMA and the amine-modified surface is pH-dependent. Therefore, after the washing steps, a high pH elution buffer was used to release the DMA-DNA complexes from the surfaces. Later, the authors developed a self-powdered nucleic acid extraction device to conduct on-site DMA-based nucleic acid extraction (Han et al., 2016). In this device, the negative pressure generated by a syringe-based vacuum actuator was used to pull urine and buffer solutions through amine-modified microchannels for sample preparation. Next, DMA and other homobifunctional imidoesters (HI) were used to enrich target pathogens from urine samples (Jin et al., 2018; Zhao et al., 2019). Zhao et al. (2019) utilized HIs and amine-functionalized diatomaceous earth (ADE) to concentrate Brucella ovis from a large volume (<50 mL) of urine in a syringe-based sample preparation system. In this system, a Teflon syringe filter was used to trap the ADE–HI–pathogen complexes for target enrichment, cell lysis, washing, and DNA elution. The detection limit of this combined pathogen enrichment and nucleic acid extraction system was 1 CFU/mL, which is a 100-fold improvement over the commercial kit-based methods. For Chlamydia trachomatis detection in urine samples, Chan et al. (2016a) presented a medium-throughput 3D printer-based molecular diagnostic system. The authors replaced the extruder of the printer with a tip-comb attachment to conduct magnetic particle-based DNA extraction. The tip-comb attachment consisted of either 8 or 12 tips depending on the number of samples required to be simultaneously processed, and small permanent magnets were placed inside these tips for magnetic particle manipulation. This modified 3D printer automatically extracted nucleic acids from 12 samples simultaneously within 15 min. Following automatic nucleic acid extraction, the authors also demonstrated a water bath-based two-step PCR amplification, where the 3D printer automatically transferred PCR tubes in between two water baths with different temperatures for thermal cycling. Later, the authors integrated an isothermal RPA assay with this 3D printer-based nucleic acid extraction system for Zika virus detection from urine samples (Chan et al., 2016b). During the RPA assay, the heated printer bed was used to directly heat the RPA tubes, which simplified the overall complexity of the system by eliminating two different temperature water baths and sample transportation between these baths. For on-site nucleic acid purification from urine samples, Pearlman et al. (2020) presented a high-gradient magnetic separation (HGMS) technique to capture DNA binding magnetic particles in the steel wool matrix (Fig. 7b). The steel wool matrix was placed into a transfer pipette, and when the mixture of sputum cell lysate and magnetic beads was pulled up through the pipette, the magnetic beads were captured on the magnetized steel wool matrix due to the dominance magnetic force exerted on beads over viscous drag. The authors demonstrated that the DNA capture efficiency of the system was ~90% from DNA-spiked urine samples, which is similar to that of Qiagen nucleic acid extraction kits. Moreover, the HGMS-based system processed a larger volume of samples compared to commercial kits.
Fig. 7.
Various portable urine sample preparation systems. (a) DMA-based nucleic acid extraction in an amine-modified silicon microchip (reproduced with permission from Ref (Shin et al., 2014)., © The Royal Society of Chemistry (2014). (b) Schematic of a high-gradient magnetic separation (HGMS) enabled nucleic acid extraction method (reproduced with permission from Ref. (Pearlman et al., 2020), © American Chemical Society 2020). (c) Schematic illustration of a chromatography paper-based nucleic acid extraction system for Chlamydia trachomatis detection (reproduced with permission from Ref (Linnes et al., 2014)., © The Royal Society of Chemistry (2014). (d) Schematic of the microfluidic filtration of small Zika RNA in a wax-printed cellulose paper (reproduced with permission from Ref (Kaarj et al., 2018)., © Springer Nature 2018). (e) A pipette-actuated capillary comb system for sample-to-answer bacterial pathogen detection in 85 min. Cross section view of the system (top left), actual photographs (top right), schematic of liquid handling through the system (bottom left), and a photograph of the assembled capillary comb and 1 mL pipette tip (bottom right) (reproduced with permission from Ref (Hui et al., 2018)., © The Royal Society of Chemistry (2018). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4.2. Paper-based systems for nucleic acid extraction from urine
Paper-based devices have also been extensively utilized to isolate pathogenic nucleic acids from urine samples (J. R. Choi et al., 2015; Seok et al., 2019). For Chlamydia trachomatis detection, Linnes et al. (2014) developed a simple molecular diagnostics device by inserting chromatography paper into a micropipette tip (Fig. 7c). In this device, the bacterial cells were trapped on the paper, and then the captured cells were directly used for in-situ HDA amplification in the tip. Similarly, Etchebarne et al. (2017) demonstrated a direct LAMP assay without DNA extraction from concentrated bacterial cells in urine samples. However, low detection sensitivity was observed for the direct amplification assays without nucleic acid extraction (Wand et al., 2018). Kaarj et al. (2018) presented a wax-printed paper microfluidic chip to filter Zika virus RNA from various raw samples such as urine, water, and diluted plasma (Fig. 7d). When the urine sample was loaded into the microchip, only small viral RNAs were able to flow through the microchannel at the same speed as the bulk liquid, and finally reached the opposite circular end for detection, while large molecules were stuck in the sample loading area. After microfluidic filtration, the detection area of the microchip was cut, and the RT-LAMP master mix was added onto it for a paper-based RT-LAMP amplification. By combining microfluidic filtration and paper-based DNA amplification, the authors successfully detected Zika virus at a titer as low as 1 CFU/mL in less than 15 min. Jiang et al. (2018) utilized cellulose chromatography paper to capture viral RNA from cell lysate. After washing the captured RNA, cellulose paper was directly used as the template for RT-LAMP amplification to detect Zika virus. The authors found no inhibitory effect of the cellulose paper on the RT-LAMP assay. To increase nucleic acid capture efficiency from the urine samples, chitosan-based surface modification of Fusion 5 membrane and glass filter paper has been reported (Hui et al., 2018; Rosenbohm et al., 2020). For Trichomonas vaginalis detection, Rosenbohm et al. (2020) utilized a chitosan-modified Fusion 5 membrane to capture DNA from a large sample volume (~50 mL). To extract DNA, cell lysate was loaded into a syringe and pumped through the chitosan-modified filter using a syringe pump. After this extraction, the chitosan functionalized filter was directly used as the template for an isothermal HDA assay to detect Trichomonas vaginalis. In the HDA assay, the authors found no inhibitory effect of chitosan. For the multiplex detection of bacterial pathogens in urine, Hui et al. (2018) presented a pipette-actuated capillary array comb (PAAC) system, where a chitosan-modified glass filter paper was used for DNA extraction (Fig. 7e). The PAAC system consisted of six capillaries. Each capillary was embedded with a functionalized glass filter paper to capture DNA from the cell lysate. Moreover, target-specific LAMP primers were preloaded onto these chitosan-modified filter papers for multiplexed amplification. The PAAC system demonstrated a very high λ DNA capture efficiency (~97%) from the MES (2-(N-morpholino) ethanesulfonic acid) buffer (pH = 5.0) for all samples tested in the concentration range of 0.2 ng/μL to 4 ng/μL. Furthermore, a portable smartphone device was integrated with this PAAC system for heating and endpoint fluorescence measurement of the LAMP assays. Thus, this integrated system has great potential for the molecular detection of pathogens in resource-limited settings.
5. Nucleic acid extraction from plants
Every year, plant diseases cause approximately 220 billion dollars of crop losses worldwide (Sarkozi, 2019). According to the Food and Agriculture Organization (FAO) of the United Nations, more than 30% of global crop production is lost annually due to plant pathogens (e.g., fungi, bacteria, and viruses) and pests (Sarkozi, 2019). For plant disease detection, molecular methods via nucleic acid amplification play a vital role in modern agriculture for protecting crops and increasing agricultural yield to fulfill the ever-growing demand for food supply (Lau and Botella, 2017; Martinelli et al., 2015). However, most nucleic acid-based methods for plant pathogen identification and genotyping are laboratory-based. At present, field samples need to be transported to a laboratory for nucleic acid extraction, amplification, and detection (Lau and Botella, 2017; Ristaino et al., 2020). Sample preparation from plant tissues is a complicated multistep process due to the rigid polysaccharide cell walls of plant cells. In the laboratory, the CTAB-based extraction method is widely used to extract nucleic acid from plant samples (Demeke and Jenkins, 2010). A typical CTAB extraction protocol involves mechanical grinding, chemical cell lysis, temperature-assisted incubation, and organic phase-based nucleic acid extraction (Mahuku, 2004; Murray and Thompson, 1980). Following DNA extraction, PCR or LAMP amplification is performed for pathogen detection. As a result, conventional methods are time-consuming, and they require expensive laboratory instruments and skilled technicians (Khiyami et al., 2014). On the other hand, plant diseases spread rapidly, and some diseases such as late blight can destroy entire field crops in a few days if not treated (Gugino et al., 2013). Therefore, rapid and accurate on-site molecular detection platforms are essential for plant disease management (Donoso and Valenzuela, 2018; Nezhad, 2014).
In recent years, several portable PCR systems (Julich et al., 2011; Koo et al., 2013; Radhakrishnan et al., 2019) and isothermal methods such as LAMP (Hodgetts et al., 2011; Ristaino et al., 2020), RPA (Cha et al., 2020; Lau et al., 2016; Mekuria et al., 2014) and HDA (Lau and Botella, 2017; Schwenkbier et al., 2015) have been reported for facilitating field diagnosis of plant diseases. However, a lack of simple, instrument-free nucleic acid extraction methods remains one of the major challenges when converting laboratory-based molecular diagnostics to sample-to-answer field tests for plant diseases (Ali et al., 2017; Nezhad, 2014).
5.1. Laboratory-based rapid nucleic acid extraction protocols for plant samples
In recent years, several rapid nucleic acid extraction methods have been reported for plant sample preparation (see Table 5 ). To simplify nucleic acid extraction from complex plant samples, Edwards et al. (1991) developed a sodium dodecyl sulfate (SDS)-based method to isolate PCR amplifiable genomic DNA in 15 min without using any hazardous chemicals such as chloroform or phenol. However, the protocol involved high-speed centrifugation for sample preparation. Thus, like the CTAB method, SDS-based DNA extraction is also confined to a laboratory setting. Nevertheless, an instrument-free, NaOH-based rapid DNA extraction method has been developed (Wang et al., 1993). In this NaOH-based nucleic acid extraction method, a small piece of sample (e.g., leaf, plantlet, callus) was homogenized with 0.5 N NaOH solution. Then, 5 μL of the homogenized solution was quickly transferred to a new tube containing 495 μL Tris buffer (pH = 8) for neutralization. While this NaOH-based method enables rapid sample preparation from plant samples, the neutralization step significantly dilutes the sample (e.g., 100x dilution), which reduces DNA concentration and thus detection sensitivity. Moreover, this NaOH method works best for young tissues, not aged or dry plant samples. Chomczynski and Rymaszewski (2006) developed an alkaline polyethylene glycol (PEG)-based one-step nucleic acid extraction method. Unlike the NaOH method, this PEG-based method did not involve a neutralization step. Cha et al. (2020) utilized both NaOH and PEG and developed a DNA extraction buffer to extract nucleic acids from pinewood chips. Tomlinson et al. (2010) combined chemical cell lysis and a lateral flow strip-based nucleic acid purification for rapid sample preparation. For chemical-free sample preparation, Hieno et al. (2019) tested a water-based extraction method to detect Phytophthora nicotianae from infected tomato, eggplant, and cucumber. In this water-based extraction method, fine slices of infected vegetables were vortexed with DI water. Then, the supernatant was used as a template for LAMP amplification. However, the non-specific amplification rate of the water-based extraction solution was higher than the conventional extraction buffer (Hieno et al., 2019).
Table 5.
Rapid nucleic acid extraction methods for plant pathogen detection.
| Sample Type | Target Pathogen | Cell Lysis Method | Nucleic Acid Extraction Technique | Nucleic Acid Amplification Method | LOD | Total Sample-To-Answer/Sample Preparation Time | Reference | ||
|---|---|---|---|---|---|---|---|---|---|
| Rhododendron plant Leaves | Phytophthora Species | Agdia sample extraction bag | RPA using lyophilized RPA reagents | 33 pg DNA/reaction | 30 min (sample-to-answer) | McCoy et al. (2020) | |||
| Rice leaf | Magnaporthe oryzae | Chemical lysis with SDS based buffer | Cellulose-based dipstick captured DNA from lysate | RPA | 3 min (sample preparation) | (Y. Zhang et al., 2020a, Zhang et al., 2020b) | |||
| Pinewood chips | Bursaphelenchus xylophilus | Quick DAP extraction buffer | RPA using lyophilized TwistAmp RPA kit (TwistDX, UK) | 6 pg DNA/reaction | 30 min (sample-to-answer) | Cha et al. (2020) | |||
| Plant leaves and woods | Xylella fastidiosa, Ceratocystis platani and Phytophthora ramorum | OptiGene Plant Material Lysis Kit | Real-time LAMP | 0.02 pg/μL DNA | 20 min (sample-to-answer) | Aglietti et al. (2019) | |||
| Tomato, eggplant and cucumber (fruit) | Phytophthora nicotianae | Water extraction method | Real-time LAMP | 10 fg DNA/reaction | 20 min (sample-to-answer) | Hieno et al. (2019) | |||
| Tomato leaves | Phytophthora infestans | Microneedle patch-based DNA extraction method | Benchtop real-time PCR | 1.2 pg DNA/reaction | Less than 1 min (sample preparation time) | Paul et al. (2019) | |||
| Leaf | Grapevine red blotch virus | Pin-prick method using micropipette tip | RT-LAMP | 5 min (sample preparation time) | Romero et al. (2019) | ||||
| Leaf and root | Candidatus Phytoplasma mali | Chemical lysis in nylon mesh bags (BIOREBA, Switzerland) | Real-time RPA using lyophilized TwistAmp RPA kit (TwistDX, UK) | 10 gene copies/reaction | 30 min (sample-to-answer) | Valasevich and Schneider (2017) | |||
| Potato tuber | Clavibacter michiganensis | FTA card-based DNA extraction inside of a micropipette tip | LAMP | 2 h (sample-to-answer) | Lu et al. (2016) | ||||
5.2. Commercial products for on-site nucleic acid extraction from plant samples
For field-based plant sample preparation, several commercial products such as the plastic vial-based plant material DNA extraction kit (OptiGene Ltd., UK) (De Jonghe et al., 2017; Tangkanchanapas et al., 2018), Spotcheck lateral flow extraction buffer (Neogen, USA) (Tangkanchanapas et al., 2018), plastic sample extraction bag (Agdia Inc., USA) (McCoy et al., 2020; Mekuria et al., 2014), and nylon mesh extraction bag (BIOREBA, Switzerland) (Valasevich and Schneider, 2017) have been developed. For DNA extraction using the OptiGene plant kit, a small piece of sample is placed inside of a plastic vial containing lysis buffer and bearing balls. After grinding the sample by vigorous shaking, 10 μL of lysate is transferred to another vial containing 2 mL dilution buffer. This DNA extraction kit has been demonstrated for on-site sample preparation to detect plant pathogens (e.g., Dickeya dianthicola, Xylella fastidiosa, Ceratocystis platani, and Phytophthora ramorum, etc.) via LAMP amplification (Aglietti et al., 2019; Ocenar et al., 2019). However, like the NaOH method, the lysate dilution step may lower the DNA yield and therefore the LOD. The Neogen lateral flow extraction buffer has been developed mainly for immunoassay-based pathogen detection. A 10x diluted ground sample with this buffer has also been demonstrated for LAMP amplification to detect Pepper chat fruit viroid from infected tomato and pepper plants (Tangkanchanapas et al., 2018). The sample extraction bag (Agdia) can also perform rapid nucleic acid extraction in the field (Fig. 8 a) (McCoy et al., 2020). For in-field applications, the plant sample is placed inside of a plastic mesh bag containing the Agdia general extraction buffer 2. Then, the outside of the bag is rubbed with a blunt object such as a pen (Mekuria et al., 2014). After sample maceration, the crude extract can be directly used for RPA amplification to detect Phytophthora species, which infect various food crops such as potato, tomato, and soybean, etc.
Fig. 8.
Rapid nucleic acid extraction methods from plant samples. (a) Photograph of an Agdia sample preparation bag with ground leaves inside (reproduced with permission from Ref (McCoy et al., 2020)., © MDPI 2020). (b) FTA card-based sample preparation. Pressing of infected leaves onto FTA card (left) and punching a small FTA disk for subsequent molecular analysis (right). (reproduced with permission from Ref. (Ndunguru et al., 2005), © BioMed Central 2005). (c) Photograph of a cellulose dipstick (left), and schematic illustration of the dipstick based nucleic acid purification from ground leaf sample (right) (reproduced with permission from Ref (Zou et al., 2017)., ©PLoS Biology 2017). (d) Schematic illustration of microneedle patch-based nucleic acid extraction method (reproduced with permission from Ref (Paul et al., 2019)., © American Chemical Society 2019).
5.3. Paper-based methods for plant DNA extraction
Besides commercial sample extraction bags and nucleic acid extraction kits, paper devices made of FTA card and cellulose paper are extensively used for on-site rapid plant sample preparation, including cell lysis, nucleic acid extraction, and storage (Grund et al., 2010; Ndunguru et al., 2005). For FTA card-based sample preparation, infected plant samples are pressed onto the FTA card (Fig. 8b). Alternatively, the homogenized sample with Tris-EDTA buffer can be added to the FTA card (Grund et al., 2010). After sample addition, the dried FTA card is usually shipped to a laboratory for nucleic acid purification and amplification. For on-site plant disease detection, Lu et al. (2016) integrated FTA card-based DNA extraction and LAMP amplification within a micropipette tip. A pre-inserted FTA disk captured Clavibacter michiganensis DNA from a ground potato sample. For E. coli detection from spinach leaves, Choi et al. (2016) combined FTA card-based DNA extraction, LAMP amplification, and lateral flow-based detection into a paper-based system. This paper-based system detected E. coli from a spinach leaf sample within an hour without utilizing heavy instruments.
In addition to FTA cards, cellulose-based papers such as Whatman No.1 paper (GE Healthcare, USA) have also been reported for DNA purification from crude cell lysate (Y. Zhang et al., 2020a, Zhang et al., 2020b; Zou et al., 2017). Zou et al. (2017) presented a cellulose dipstick-based nucleic acid purification system for several plants, including rice, tomato, wheat, soybean, barley, and sugarcane (Fig. 8c). The cellulose dipstick captured DNA from the crude lysate in less than 10 s. The total DNA purification time was approximately 30 s, and thus promising for field applications.
5.4. Microneedle-based rapid nucleic acid extraction from plant tissues
Every commercial product or existing extraction method mentioned above requires mechanical grinding of infected plant samples or homogenization. Often, mechanical grinding is manually performed by various tools such as disposable plastic pestles, beads, extraction bags, or FTA cards. However, sample homogenization via mechanical grinding completely disrupts the plant tissues and therefore generates an extra amount of impurities (e.g., cell and tissue debris, proteins, etc.) that must be separated from nucleic acids later for further analysis. To overcome this drawback, we recently (Paul et al., 2020, 2019) developed a new plant DNA/RNA extraction method without the need for cell or tissue lysis. This minimally invasive method utilized a polymeric microneedle (MN) patch to penetrate soft plant tissues (e.g., fresh leaves) and extract assay-ready nucleic acid samples in less than a minute (Fig. 8d). Microneedle patches were made of biocompatible and swellable polymers such as polyvinyl alcohol (PVA). Each patch contained 225 conical microneedles arranged in a 15 × 15 array. During the DNA extraction, these conical microneedles break plant cell walls and release nucleic acid and other intracellular molecules. Because PVA is a highly water-swellable polymer, it rapidly absorbs intracellular water molecules for depositing nucleic acid on the needle surfaces. The total DNA extraction time, including the washing step, was less than 1 min. The extracted DNA can directly be used as the template for PCR amplification without further purification. These microneedle patches successfully isolated genomic DNA from several plant species, including tomato, pepper, and potato (Paul et al., 2020). Moreover, to demonstrate the capability of low abundance pathogenic DNA isolation, the microneedle patches were applied to extract Phytophthora infestans DNA from both laboratory-inoculated and field-collected tomato leaves. For laboratory-inoculated samples, the microneedle patches isolated pathogen DNA after 1 day post-inoculation. For field-collected samples, a 100% detection sensitivity was achieved for the late blight disease detection. The integration of this rapid nucleic acid extraction method with a field-based amplification method (e.g., LAMP) would have great potential for direct molecular diagnosis of plant pathogens in the field. A different approach, presented by Romeo et al. (2019), is a micropipette tip-based rapid nucleic acid extraction method for on-site detection of grapevine red blotch virus. For sample preparation, an infected leaf was stabbed three to five times with a 10 μL micropipette tip. Then, the top of the tip was immersed in 10 μL of DI water for 5 min. Finally, 0.5 μL of samples was used as the template for running the LAMP assay.
6. Nucleic acid extraction from other raw samples
In addition to human blood, oral/nasal, urine, and plant tissue samples, POC nucleic acid extraction devices or methods have also been developed for various other raw sample types. Given the broadness of sample types, this section will only briefly summarize representative sample matrices such as stool samples and vaginal/cervical swabs to highlight the recent progress on POC nucleic acid extraction.
6.1. POC nucleic acid extraction from stool samples
Stool samples are non-invasive and can be used to simultaneously test for multiple enteropathogenic bacteria and diseases. Although the composition of feces is variable, human feces has a median moisture content of 75%, with bacterial biomass comprising 25–54% of the organic solids (Rose et al., 2015). The complex composition of feces poses a major challenge for direct testing. Freifeld et al. (2012) developed a single-use microreactor for stool sample preparation. In the microheater, a mixture of sample, lysis buffer, and resin beads was heated at 92 °C for 5 min for cell lysis. A polystyrene ssDNA-capture strip was also inserted into the microreactor to capture target ssDNA from cell lysate. Mosley et al. (2016) developed a microfluidic chip to extract Helicobacter pylori DNA from crude stool samples within 7 min. This device utilized a design of immiscible filtration assisted by surface tension (IFAST), in which silica-coated superparamagnetic particles were transferred between microwell chambers separated by alternating aqueous and mineral oil barriers. Kang et al. (2017) developed a fully integrated microfluidic cartridge to isolate viral and bacterial nucleic acid from fecal samples. In the device, magnetic vibration-based stool sample homogenization, chemical lysis and magnetic bead-based DNA extraction and purification were performed automatically in five different chambers for rapid sample preparation. The extraction efficiency of the device was comparable to that of laboratory-based methods. To demonstrate paper-based stool sample preparation, Ye et al. (2018) utilized a glass fiber paper disc to capture nucleic acids from chemically lysed stool samples. After washing the captured DNA, the filter disc was cut and transferred to the LAMP mix for rapid rotavirus A detection. The total sample-to-answer detection time of the system was 30 min, and the LOD was 1 × 103 copies of rotavirus A/mL.
6.2. Rapid nucleic acid extraction from vaginal and cervical swab samples
Vaginal and cervical swabs are typically used to diagnose sexually transmitted infections. Vaginal swabs obtain samples of vaginal discharge, which is composed of cervical mucus, shed cells, and bacteria. Gulliksen et al. (2012) designed an automated lab-on-a-chip platform for human papillomavirus (HPV) detection from cervical specimens, consisting of two separate chips for sample preparation and nucleic acid amplification. The sample preparation chip was used to perform target preconcentration, chemical lysis and silica filter-based nucleic acid extraction. Lee et al. (2016) demonstrated the isolation of HPV DNA from cervical specimens using polyethyleneimine-conjugated magnetic nanowires. After mechanical cell lysis, magnetic nanowires were added in the cell lysate to bind the released nucleic acids with the cationic polymer. Zhu et al. (2019) designed an integrated microfluidic chip for HPV detection from cervical specimens within 1 h. The microfluidic device combined thermal lysis method of nucleic acid extraction and solid-phase PCR for rapid target detection.
Furthermore, paper-based devices have been developed for nucleic acid extraction from vaginal and cervical swab samples. Rodriguez et al. (2016) developed a foldable paperfluidic chip for HPV detection from cervical specimens within 1 h. For DNA extraction, the sample was mixed with cell lysis and DNA precipitation buffers and loaded into a circular disc of PES membrane to capture the precipitated DNA from cell lysate. Later, Horst et al. (2018) utilized a similar PES membrane-based sample preparation technique to develop a paperfluidic device to detect sexually transmitted bacterium Neisseria gonorrhoeae from vaginal or urethral swab samples within 80 min.
7. Summary and future perspectives
Over the past few years, significant progress has been made to simplify and speed up the nucleic acid isolation process from various raw human samples (e.g., blood, saliva, urine, sputum, stool, etc.) and difficult-to-lyse plant tissues. This review highlights a few representative systems such as microfluidic chips, paper-based devices, microneedle patches, and extraction bags to showcase the current frontiers of various POC nucleic acid extraction platforms. The microfluidic platform is a very cost-effective solution for sample preparation because of the reduced sample and reagents size. However, most of the sample preparation microchips presented in the literature do not have on-chip reagent storage capability and depend on external syringe pumps for fluid manipulation, which limits their applicability to resource-limited settings. To overcome this limitation, several pump-free microfluidic methods such as centrifugal microfluidics, digital microfluidics, hand-operated microfluidic, and capillary-based microfluidic devices have emerged. Beside these, paper-based devices have become popular for rapid sample preparation. Paper devices are inexpensive and easy-to-fabricate. Sample preparation reagents can be easily dried and stored on the device for long-term use or transportation. Moreover, paper-based devices facilitate nucleic acid extraction by providing additional purification functions. For example, paper membranes can filter blood cells from whole blood and thus eliminate the need for centrifugation. Furthermore, paper disks with entrapped nucleic acids can be directly added to the amplification reactions without the need for nucleic acid elution, due to the inert chemical properties of the cellulose matrix. To further increase the nucleic acid extraction efficiency of the paper device, chitosan-based charge switching extraction methods can be integrated to selectively bind target DNA/RNA from complex biofluids for rapid sample preparation at the detection site.
There are also several challenges remaining to be solved for the development of future POC diagnostic devices, including 1) further speeding nucleic acid extraction and purification within minutes, 2) better POC methods for the extraction of fragile RNA molecules, and 3) robust extraction strategies that can be potentially applied to several different sample matrices (e.g., blood, saliva, and nasal swab). The last point is especially relevant for accurate diagnosis of newly emerging infectious diseases such as COVID-19, when the optimal diagnostic media has not been identified at the beginning of outbreaks or one specific sample matrix is subject to significant variation of false positive or false negative rates.
Some of the sample preparation methodologies reviewed here can in principle be applied to isolate protein or other biomarkers from body fluids or plants, in addition to nucleic acids. Many of these extraction platforms or methods have already been integrated with portable nucleic acid amplification and detection systems, such as smartphone devices, to generate fully integrated sample-to-answer detection platforms that have great potential for rapid screening of human diseases in resource-limited settings. In addition to NAA, many rapid nucleic acid extraction methodologies may also pave the road for POC DNA sequencing analysis in the future, which is expected to generate an even more profound impact on POC diagnostics and precision medicine.
Compared to human disease detection, in-field molecular diagnosis of plant pathogens is still lagging slightly behind. However, several promising recent progressions, such as rapid DNA isolation from infected plant leaves via a MN patch (Paul et al., 2019) and the Mobile And Real-time PLant disEase (MARPLE) diagnosis platform (Radhakrishnan et al., 2019), have been reported to speed field-based plant disease detection. The United Nations have also recently declared 2020 as the International Year of Plant Health (IYPH). Given the increased awareness of the importance of plant health and global food security, it is anticipated that the field of Agbio sensors and diagnostics will experience rapid and fruitful growth.
CRediT authorship contribution statement
Rajesh Paul: Writing - review & editing, developed the outline of the review, lead the human blood, oral/nasal, urine, and plant tissue sections. Emily Ostermann: Writing - review & editing, lead the other sample section. Qingshan Wei: Writing - review & editing, developed the outline of the review.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors gratefully acknowledge the funding support from the NCSU Chancellor's Faculty Excellence Program, the Kenan Institute for Engineering Technology & Science (KIETS), NCSU Game-Changing Research Incentive Program for the Plant Science Initiative (GRIP4PSI), USDA (Award No. 2019-67030-29311), NSF Career Award (Award No. 1944167), and BASF.
References
- Aglietti C., Luchi N., Pepori A.L., Bartolini P., Pecori F., Raio A., Capretti P., Santini A. Amb. Express. 2019;9:1–14. doi: 10.1186/s13568-019-0774-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts B., Johnson A., Lewis J., Raff M., Roberts K., Walter P. fourth ed. Garland Science; New York: 2002. Molecular Biology of the Cell. [Google Scholar]
- Ali N., Rampazzo R.D.C.P., Costa A., Di T., Krieger M.A. BioMed Res. Int. 2017:1–13. doi: 10.1155/2017/9306564. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ARIES Systems. https://www.luminexcorp.com/aries/(Accessed 3 June 2020).
- Ayoib A., Hashim U., Gopinath S.C.B., Md Arshad M.K. Appl. Microbiol. Biotechnol. 2017;101:8077–8088. doi: 10.1007/s00253-017-8493-0. [DOI] [PubMed] [Google Scholar]
- Basha I.H.K., Ho E.T.W., Yousuff C.M., Hamid N.H. Bin. Micromachines. 2017;8:266. doi: 10.3390/mi8090266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batule B.S., Seok Y., Kim M.-G. Biosens. Bioelectron. 2020;151 doi: 10.1016/j.bios.2019.111998. [DOI] [PubMed] [Google Scholar]
- Bender A.T., Borysiak M.D., Levenson A.M., Lillis L., Boyle D.S., Posner J.D. Anal. Chem. 2018;90:7221–7229. doi: 10.1021/acs.analchem.8b00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentaleb E.M., Abid M., El Messaoudi M.D., Lakssir B., Ressami E.M., Amzazi S., Sefrioui H., Ait Benhassou H. BMC Infect. Dis. 2016;16:1–11. doi: 10.1186/s12879-016-1864-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry S.M., Lavanway A.J., Pezzi H.M., Guckenberger D.J., Anderson M.A., Loeb J.M., Beebe D.J. J. Mol. Diagnostics. 2014;16:297–304. doi: 10.1016/j.jmoldx.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhamla M.S., Benson B., Chai C., Katsikis G., Johri A., Prakash M. Nat. Biomed. Eng. 2017;1:9. [Google Scholar]
- Bhattacharyya A., Klapperich C.M. Anal. Chem. 2006;78:788–792. doi: 10.1021/ac051449j. [DOI] [PubMed] [Google Scholar]
- BioFire FilmArray Torch. https://www.biofiredx.com/products/filmarray/(Accessed 3 June 2020).
- Borysiak M.D., Kimura K.W., Posner J.D. Lab Chip. 2015;15:1697–1707. doi: 10.1039/c4lc01479k. [DOI] [PubMed] [Google Scholar]
- Branch D.W., Vreeland E.C., McClain J.L., Murton J.K., James C.D., Achyuthan K.E. Micromachines. 2017;8:1–17. doi: 10.3390/mi8070228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes S., Fan A., Trueb J., Jareczek F., Mazzochette M., Sharon A., Sauer-Budge A.F., Klapperich C.M. Anal. Methods. 2013;5:3177. doi: 10.1039/C3AY40162F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes S.A., Bishop J.D., Lafleur L., Buser J.R., Lutz B., Yager P. Lab Chip. 2015;15:2647–2659. doi: 10.1039/c5lc00317b. [DOI] [PubMed] [Google Scholar]
- Cai D., Xiao M., Xu P., Xu Y.C., Du W. Lab a chip - miniaturisation. Chem. Biol. 2014;14:3917–3924. doi: 10.1039/c4lc00669k. [DOI] [PubMed] [Google Scholar]
- Cao Q., Mahalanabis M., Chang J., Carey B., Hsieh C., Stanley A., Odell C.A., Mitchell P., Feldman J., Pollock N.R., Klapperich C.M. PLoS One. 2012;7:1–12. doi: 10.1371/journal.pone.0033176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha D., Kim D., Choi W., Park S., Han H. PLoS One. 2020;15 doi: 10.1371/journal.pone.0227476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K., Coen M., Hardick J., Gaydos C.A., Wong K.Y., Smith C., Wilson S.A., Vayugundla S.P., Wong S. PLoS One. 2016;11:1–19. doi: 10.1371/journal.pone.0158502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K., Weaver S.C., Wong P.Y., Lie S., Wang E., Guerbois M., Vayugundla S.P., Wong S. Sci. Rep. 2016;6:1–12. doi: 10.1038/srep38223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K., Wong P.Y., Parikh C., Wong S. Anal. Biochem. 2018;545:4–12. doi: 10.1016/j.ab.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Mauk M., Qiu X., Liu C., Kim J., Ramprasad S., Ongagna S., Abrams W.R., Malamud D., Corstjens P.L.A.M., Bau H.H. Biomed. Microdevices. 2010;12:705–719. doi: 10.1007/s10544-010-9423-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Chen C., Liu Y., Du W., Feng X., Liu B.F. Sensor. Actuator. B Chem. 2019;283:472–477. [Google Scholar]
- Chen Z., Abrams W.R., Geva E., de Dood C.J., González J.M., Tanke H.J., Niedbala R.S., Zhou P., Malamud D., Corstjens P.L.A.M. BioMed Res. Int. 2013:1–12. doi: 10.1155/2013/543294. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou C.H., Jin Shin D., Zhang Y., Wang T.H. Biosens. Bioelectron. 2013;50:91–99. doi: 10.1016/j.bios.2013.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y.K., Lee J.-G., Park J.-M., Lee B.-S., Lee Y., Ko C. Lab Chip. 2007;7:565–573. doi: 10.1039/b616115d. [DOI] [PubMed] [Google Scholar]
- Choi J., Hyun J.C., Yang S. Sci. Rep. 2015;5:1–12. doi: 10.1038/srep15167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J.R., Hu J., Tang R., Gong Y., Feng S., Ren H., Wen T., Li X.J., Wan Abas W.A.B., Pingguan-Murphy B., Xu F. Lab Chip. 2016;16:611–621. doi: 10.1039/c5lc01388g. [DOI] [PubMed] [Google Scholar]
- Choi J.R., Tang R., Wang S.Q., Wan Abas W.A.B., Pingguan-Murphy B., Xu F. Biosens. Bioelectron. 2015;74:427–439. doi: 10.1016/j.bios.2015.06.065. [DOI] [PubMed] [Google Scholar]
- Choi J.R., Yong K.W., Tang R., Gong Y., Wen T., Yang H., Li A., Chia Y.C., Pingguan-Murphy B., Xu F. Adv. Healthc. Mater. 2017;6:1–9. doi: 10.1002/adhm.201600920. [DOI] [PubMed] [Google Scholar]
- Choi Y., Kim Y.T., Lee S.J., Lee E., Lee K.G., Im S.G. Macromol. Res. 2020;28:249–256. [Google Scholar]
- Chomczynski P., Rymaszewski M. Biotechniques. 2006;40:454–458. doi: 10.2144/000112149. [DOI] [PubMed] [Google Scholar]
- Connelly J.T., Rolland J.P., Whitesides G.M. Anal. Chem. 2015;87:7595–7601. doi: 10.1021/acs.analchem.5b00411. [DOI] [PubMed] [Google Scholar]
- Creecy A., Russ P.K., Solinas F., Wright D.W., Haselton F.R. PLoS One. 2015;10:1–15. doi: 10.1371/journal.pone.0130260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis K.A., Rudolph D.L., Owen S.M. J. Virol. Methods. 2008;151:264–270. doi: 10.1016/j.jviromet.2008.04.011. [DOI] [PubMed] [Google Scholar]
- Czilwik G., Messinger T., Strohmeier O., Wadle S., von Stetten F., Paust N., Roth G., Zengerle R., Saarinen P., Niittymäki J., McAllister K., Sheils O., O'Leary J., Mark D. Lab Chip. 2015;15:3749–3759. doi: 10.1039/c5lc00591d. [DOI] [PubMed] [Google Scholar]
- Damhorst G.L., Duarte-Guevara C., Chen W., Ghonge T., Cunningham B.T., Bashir R. Engineering. 2015;1:324–335. doi: 10.15302/J-ENG-2015072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jonghe K., De Roo I., Maes M. Eur. J. Plant Pathol. 2017;147:749–759. [Google Scholar]
- Demeke T., Jenkins G.R. Anal. Bioanal. Chem. 2010;396:1977–1990. doi: 10.1007/s00216-009-3150-9. [DOI] [PubMed] [Google Scholar]
- Deng H., Zhou X., Liu Q., Li B., Liu H., Huang R., Xing D. ACS Appl. Mater. Interfaces. 2017;9:41151–41158. doi: 10.1021/acsami.7b12637. [DOI] [PubMed] [Google Scholar]
- Donoso A., Valenzuela S. Plant Pathol. 2018;67:1451–1461. [Google Scholar]
- Dou M., Macias N., Shen F., Dien Bard J., Domínguez D.C., Li X.J. EClinicalMedicine. 2019;8:72–77. doi: 10.1016/j.eclinm.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte G.R.M., Price C.W., Littlewood J.L., Haverstick D.M., Ferrance J.P., Carrilho E., Landers J.P. Analyst. 2010;135:531–537. doi: 10.1039/b918996c. [DOI] [PubMed] [Google Scholar]
- Dutta G., Rainbow J., Zupancic U., Papamatthaiou S., Estrela P., Moschou D. Chemosensors. 2018;6:43. [Google Scholar]
- Easley C.J., Karlinsey J.M., Bienvenue J.M., Legendre L.A., Roper M.G., Feldman S.H., Hughes M.A., Hewlett E.L., Merkel T.J., Ferrance J.P., Landers J.P. Proc. Natl. Acad. Sci. 2006;103:19272–19277. doi: 10.1073/pnas.0604663103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K., Johnstone C., Thompson C. Nucleic Acids Res. 1991;19:1349. doi: 10.1093/nar/19.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrela P.F.N., Mendes G. de M., de Oliveira K.G., Bailão A.M., Soares C.M. de A., Assunção N.A., Duarte G.R.M. J. Virol. Methods. 2019;271 doi: 10.1016/j.jviromet.2019.113675. [DOI] [PubMed] [Google Scholar]
- Etchebarne B.E., Li Z., Stedtfeld R.D., Nicholas M.C., Williams M.R., Johnson T.A., Stedtfeld T.M., Kostic T., Khalife W.T., Tiedje J.M., Hashsham S.A., Hughes M.J. Front. Microbiol. 2017;8:2211. doi: 10.3389/fmicb.2017.02211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z., Wu W., Lu X., Zeng L. Biosens. Bioelectron. 2014;56:192–197. doi: 10.1016/j.bios.2014.01.015. [DOI] [PubMed] [Google Scholar]
- Ferguson B.S., Buchsbaum S.F., Wu T.T., Hsieh K., Xiao Y., Sun R., Soh H.T. J. Am. Chem. Soc. 2011;133:9129–9135. doi: 10.1021/ja203981w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson T.M., Weigel K.M., Lakey Becker A., Ontengco D., Narita M., Tolstorukov I., Doebler R., Cangelosi G.A., Niemz A. Sci. Rep. 2016;6:1–9. doi: 10.1038/srep19541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freifeld A.G., Simonsen K.A., Booth C.S., Zhao X., Whitney S.E., Karre T., Iwen P.C., Viljoen H.J. J. Mol. Diagnostics. 2012;14:274–279. doi: 10.1016/j.jmoldx.2012.01.003. [DOI] [PubMed] [Google Scholar]
- Fu Y., Zhou X., Xing D. Sensor. Actuator. B Chem. 2018;261:288–296. [Google Scholar]
- Gan W., Zhuang B., Zhang P., Han J., Li C.-X., Liu P. Lab Chip. 2014;14:3719–3728. doi: 10.1039/c4lc00686k. [DOI] [PubMed] [Google Scholar]
- Ganguli A., Ornob A., Yu H., Damhorst G.L., Chen W., Sun F., Bhuiya A., Cunningham B.T., Bashir R. Biomed. Microdevices. 2017;19:1–13. doi: 10.1007/s10544-017-0209-9. [DOI] [PubMed] [Google Scholar]
- GeneXpert Systems. https://www.cepheid.com/en_US/systems/GeneXpert-Family-of-Systems/GeneXpert-System (Accessed 3 June 2020).
- Govindarajan A.V., Ramachandran S., Vigil G.D., Yager P., Böhringer K.F. Lab Chip. 2012;12:174–181. doi: 10.1039/c1lc20622b. [DOI] [PubMed] [Google Scholar]
- Grund E., Darissa O., Adam G. J. Phytopathol. 2010;158:750–757. [Google Scholar]
- Gugino B.K., Johnson S.B., Judelson H., Ristaino J., Roberts P., Secor G., Seebold K., Snover-Clift K., Wyenandt A., Grünwald N.J., Smart C.D., Fry W.E., McGrath M.T., Seaman A., Zitter T.A., McLeod A., Danies G., Small I.M., Myers K., Everts K., Gevens A.J., Gugino B.K., Johnson S.B., Judelson H., Ristaino J., Roberts P., Secor G., Seebold K., Snover-Clift K., Wyenandt A., Grünwald N.J., Smart C.D. Plant Dis. 2013;97:296–306. doi: 10.1094/PDIS-08-12-0791-FE. [DOI] [PubMed] [Google Scholar]
- Guio H., Okayama H., Ashino Y., Saitoh H., Xiao P., Miki M., Yoshihara N., Nakanowatari S., Hattori T. Int. J. Tubercul. Lung Dis. 2006;10:906–910. [PubMed] [Google Scholar]
- Gulliksen A., Keegan H., Martin C., O'Leary J., Solli L.A., Falang I.M., Grønn P., Karlgård A., Mielnik M.M., Johansen I.-R., Tofteberg T.R., Baier T., Gransee R., Drese K., Hansen-Hagge T., Riegger L., Koltay P., Zengerle R., Karlsen F., Ausen D., Furuberg L. J. Oncol. 2012:1–12. doi: 10.1155/2012/905024. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K., Yoon Y.J., Shin Y., Park M.K. Lab Chip. 2016;16:132–141. doi: 10.1039/c5lc00891c. [DOI] [PubMed] [Google Scholar]
- Hassan M.M., Ranzoni A., Cooper M.A. Biosens. Bioelectron. 2018;99:150–155. doi: 10.1016/j.bios.2017.07.057. [DOI] [PubMed] [Google Scholar]
- Hayashida K., Kajino K., Hachaambwa L., Namangala B., Sugimoto C. PLoS Neglected Trop. Dis. 2015;9 doi: 10.1371/journal.pntd.0003578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Neuta I., Neumann F., Brightmeyer J., Ba Tis T., Madaboosi N., Wei Q., Ozcan A., Nilsson M. J. Intern. Med. 2019;285:19–39. doi: 10.1111/joim.12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieno A., Li M., Afandi A., Otsubo K., Suga H., Kageyama K. J. Phytopathol. 2019;167:174–184. [Google Scholar]
- Hindson B.J., Ness K.D., Masquelier D.A., Belgrader P., Heredia N.J., Makarewicz A.J., Bright I.J., Lucero M.Y., Hiddessen A.L., Legler T.C., Kitano T.K., Hodel M.R., Petersen J.F., Wyatt P.W., Steenblock E.R., Shah P.H., Bousse L.J., Troup C.B., Mellen J.C., Wittmann D.K., Erndt N.G., Cauley T.H., Koehler R.T., So A.P., Dube S., Rose K.A., Montesclaros L., Wang S., Stumbo D.P., Hodges S.P., Romine S., Milanovich F.P., White H.E., Regan J.F., Karlin-Neumann G.A., Hindson C.M., Saxonov S., Colston B.W. Anal. Chem. 2011;83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgetts J., Tomlinson J., Boonham N., González-Martín I., Nikolić P., Swarbrick P., Yankey E.N., Dickinson M. Bull. Insectol. 2011;64:41–42. [Google Scholar]
- Hoos J., Peters R.M., Tabatabai J., Grulich-Henn J., Schnitzler P., Pfeil J. J. Virol. Methods. 2017;242:53–57. doi: 10.1016/j.jviromet.2017.01.006. [DOI] [PubMed] [Google Scholar]
- Horst A.L., Rosenbohm J.M., Kolluri N., Hardick J., Gaydos C.A., Cabodi M., Klapperich C.M., Linnes J.C. Biomed. Microdevices. 2018;20:1–7. doi: 10.1007/s10544-018-0280-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh K., Mage P.L., Csordas A.T., Eisenstein M.S., Tom Soh H. Chem. Commun. 2014;50:3747–3749. doi: 10.1039/c4cc00540f. [DOI] [PubMed] [Google Scholar]
- Huang H., Jin L., Yang X., Song Q., Zou B., Jiang S., Sun L., Zhou G. Biosens. Bioelectron. 2013;42:261–266. doi: 10.1016/j.bios.2012.10.078. [DOI] [PubMed] [Google Scholar]
- Hügle M., Dame G., Behrmann O., Rietzel R., Karthe D., Hufert F.T., Urban G.A. RSC Adv. 2018;8:20124–20130. doi: 10.1039/c8ra02177e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui J., Gu Y., Zhu Y., Chen Y., Guo S.J., Tao S.C., Zhang Y., Liu P. Lab Chip. 2018;18:2854–2864. doi: 10.1039/c8lc00543e. [DOI] [PubMed] [Google Scholar]
- Hung P.-Y., Jiang P.-S., Lee E.-F., Fan S.-K., Lu Y.-W. Microsyst. Technol. 2017;23:313–320. [Google Scholar]
- Hwang K.Y., Jeong S.Y., Kim Y.R., Namkoong K., Lim H.K., Chung W.S., Kim J.H., Huh N. Sensor. Actuator. B Chem. 2011;154:46–51. [Google Scholar]
- Hwang K.Y., Lim H.K., Jung S.Y., Namkoong K., Kim J.H., Huh N., Ko C., Park J.C. Anal. Chem. 2008;80:7786–7791. doi: 10.1021/ac8012048. [DOI] [PubMed] [Google Scholar]
- Integrated Cycler. https://www.focusdx.com/product-catalog/integrated-cycler (Accessed 23 June 2020).
- Jangam S.R., Agarwal A.K., Sur K., Kelso D.M. Biosens. Bioelectron. 2013;42:69–75. doi: 10.1016/j.bios.2012.10.024. [DOI] [PubMed] [Google Scholar]
- Jangam S.R., Yamada D.H., McFall S.M., Kelso D.M. J. Clin. Microbiol. 2009;47:2363–2368. doi: 10.1128/JCM.r00092-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Loeb J.C., Manzanas C., Lednicky J.A., Fan Z.H. Angew. Chem. Int. Ed. 2018;57:17211–17214. doi: 10.1002/anie.201809993. [DOI] [PubMed] [Google Scholar]
- Jin C.E., Koo B., Lee E.Y., Kim J.Y., Kim S.H., Shin Y. Biosens. Bioelectron. 2018;111:66–73. doi: 10.1016/j.bios.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C.E., Lee T.Y., Koo B., Choi K.C., Chang S., Park S.Y., Kim J.Y., Kim S.H., Shin Y. Anal. Chem. 2017;89:7502–7510. doi: 10.1021/acs.analchem.7b01193. [DOI] [PubMed] [Google Scholar]
- Julich S., Riedel M., Kielpinski M., Urban M., Kretschmer R., Wagner S., Fritzsche W., Henkel T., Möller R., Werres S. Biosens. Bioelectron. 2011;26:4070–4075. doi: 10.1016/j.bios.2011.03.035. [DOI] [PubMed] [Google Scholar]
- Kaarj K., Akarapipad P., Yoon J.-Y. Sci. Rep. 2018;8:12438. doi: 10.1038/s41598-018-30797-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko H., Kawana T., Fukushima E., Suzutani T. J. Biochem. Biophys. Methods. 2007;70:499–501. doi: 10.1016/j.jbbm.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Kang J.S., Park C., Lee J., Namkung J., Hwang S.Y., Kim Y.S. Biochip J. 2017;11:76–84. [Google Scholar]
- Kaur N., Michael J.S., Toley B.J. Sci. Rep. 2019;9:1–13. doi: 10.1038/s41598-019-51873-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khiyami M.A., Almoammar H., Awad Y.M., Alghuthaymi M.A., Abd-Elsalam K.A. Biotechnol. Biotechnol. Equip. 2014;28:775–785. doi: 10.1080/13102818.2014.960739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Johnson M., Hill P., Gale B.K. Intregr. Biol. 2009;1:574. doi: 10.1039/b905844c. [DOI] [PubMed] [Google Scholar]
- Kim Jitae, Mauk M., Chen D., Qiu X., Kim Jungkyu, Gale B., Bau H.H. Analyst. 2010;135:2408–2414. doi: 10.1039/c0an00288g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinahan D.J., Kearney S.M., Kilcawley N.A., Early P.L., Glynn M.T., Ducrée J. PLoS One. 2016;11 doi: 10.1371/journal.pone.0155545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolluri N., Klapperich C.M., Cabodi M. Lab Chip. 2017;18:75–94. doi: 10.1039/c7lc00758b. [DOI] [PubMed] [Google Scholar]
- Kong J.E., Wei Q., Tseng D., Zhang J., Pan E., Lewinski M., Garner O.B., Ozcan A., Di Carlo D. ACS Nano. 2017;11:2934–2943. doi: 10.1021/acsnano.6b08274. [DOI] [PubMed] [Google Scholar]
- Koo C., Malapi-Wight M., Kim H.S., Cifci O.S., Vaughn-Diaz V.L., Ma B., Kim S., Abdel-Raziq H., Ong K., Jo Y.-K., Gross D.C., Shim W.-B., Han A. PLoS One. 2013;8 doi: 10.1371/journal.pone.0082704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo K.M., Wee E.J.H.H., Wang Y., Trau M. Lab Chip. 2017;17:3200–3220. doi: 10.1039/c7lc00587c. [DOI] [PubMed] [Google Scholar]
- Kulinski M.D., Mahalanabis M., Gillers S., Zhang J.Y., Singh S., Klapperich C.M. Biomed. Microdevices. 2009;11:671–678. doi: 10.1007/s10544-008-9277-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafleur L.K., Bishop J.D., Heiniger E.K., Gallagher R.P., Wheeler M.D., Kauffman P., Zhang X., Kline E.C., Buser J.R., Kumar S., Byrnes S.A., Vermeulen N.M.J., Scarr N.K., Belousov Y., Mahoney W., Toley B.J., Ladd P.D., Lutz B.R., Yager P. Lab Chip. 2016;16:3777–3787. doi: 10.1039/c6lc00677a. [DOI] [PubMed] [Google Scholar]
- Lau H.Y., Botella J.R. Front. Plant Sci. 2017;8:1–14. doi: 10.3389/fpls.2017.02016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau H.Y., Wang Y., Wee E.J.H., Botella J.R., Trau M. Anal. Chem. 2016;88:8074–8081. doi: 10.1021/acs.analchem.6b01551. [DOI] [PubMed] [Google Scholar]
- Lee H.J., Hwang N.R., Hwang S.H., Cho Y. Biosens. Bioelectron. 2016;86:864–870. doi: 10.1016/j.bios.2016.07.066. [DOI] [PubMed] [Google Scholar]
- Lee J.-G., Cheong K.H., Huh N., Kim S., Choi J.-W., Ko C. Lab Chip. 2006;6:886–895. doi: 10.1039/b515876a. [DOI] [PubMed] [Google Scholar]
- Lee J.W., Nguyen V.D., Seo T.S. Biochip J. 2019;13:243–250. [Google Scholar]
- Leiske M.N., Hartlieb M., Paulenz C., Pretzel D., Hentschel M., Englert C., Gottschaldt M., Schubert U.S. Adv. Funct. Mater. 2015;25:2458–2466. [Google Scholar]
- Létant S.E., Ortiz J.I., Tammero L.F.B., Birch J.M., Derlet R.W., Cohen S., Manning D., McBride M.T. J. Clin. Microbiol. 2007;45:3498–3505. doi: 10.1128/JCM.01712-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Miao B., Li Z., Sun Z., Peng N. ACS Sens. 2019;4:2738–2745. doi: 10.1021/acssensors.9b01270. [DOI] [PubMed] [Google Scholar]
- Li W., Chen T., Chen Z., Fei P., Yu Z., Pang Y., Huang Y. Lab Chip. 2012;12:1587–1590. doi: 10.1039/c2lc40125h. [DOI] [PubMed] [Google Scholar]
- Li Z., Paul R., Ba Tis T., Saville A.C.A.C.A.C., Hansel J.C., Yu T., Ristaino J.B., Wei Q. Nat. Plants. 2019;5:856–866. doi: 10.1038/s41477-019-0476-y. [DOI] [PubMed] [Google Scholar]
- Li Z., Yu T., Paul R., Fan J., Yang Y., Wei Q. Nanoscale Adv. 2020;2:3083–3094. doi: 10.1039/c9na00724e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnes J.C., Fan A., Rodriguez N.M., Lemieux B., Kong H., Klapperich C.M. RSC Adv. 2014;4:42245–42251. doi: 10.1039/C4RA07911F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Zhan F., Liu F., Zhu M., Zhou X., Xing D. Biosens. Bioelectron. 2014;62:38–46. doi: 10.1016/j.bios.2014.06.020. [DOI] [PubMed] [Google Scholar]
- Liu H., Zhao F., Jin C.E., Koo B., Lee E.Y., Zhong L., Yun K., Shin Y. Anal. Chem. 2018;90:5108–5115. doi: 10.1021/acs.analchem.7b05136. [DOI] [PubMed] [Google Scholar]
- Liu P., Li X., Greenspoon S.A., Scherer J.R., Mathies R.A. Lab Chip. 2011;11:1041–1048. doi: 10.1039/c0lc00533a. [DOI] [PubMed] [Google Scholar]
- Liu Q., Nam J., Kim S., Lim C.T., Park M.K., Shin Y. Biosens. Bioelectron. 2016;82:1–8. doi: 10.1016/j.bios.2016.03.050. [DOI] [PubMed] [Google Scholar]
- Liu Q., Zhang X., Chen L., Yao Y., Ke S., Zhao W., Yang Z., Sui G. Sensor. Actuator. B Chem. 2018;270:371–381. [Google Scholar]
- Liu S., Wei M., Liu R., Kuang S., Shi C., Ma C. Anal. Chim. Acta. 2019;1080:162–169. doi: 10.1016/j.aca.2019.07.011. [DOI] [PubMed] [Google Scholar]
- Liu W., Das J., Mepham A.H., Nemr C.R., Sargent E.H., Kelley S.O. Lab Chip. 2018;18:1928–1935. doi: 10.1039/c8lc00371h. [DOI] [PubMed] [Google Scholar]
- Liu W., Zhang M., Liu X., Sharma A., Ding X. Biosens. Bioelectron. 2017;96:213–219. doi: 10.1016/j.bios.2017.04.047. [DOI] [PubMed] [Google Scholar]
- Liu X., Zhang C., Zhao M., Liu K., Li H., Li N., Gao L., Yang X., Ma T., Zhu J., Hui W., Hua K., Cui Y. Biosens. Bioelectron. 2018;115:70–76. doi: 10.1016/j.bios.2018.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo J.F.C., Kwok H.C., Leung C.C.H., Wu S.Y., Law I.L.G., Cheung Y.K.Y.K.Y.K., Cheung Y.K.Y.K.Y.K., Chin M.L., Kwan P., Hui M., Kong S.K., Ho H.P. Biosens. Bioelectron. 2017;93:212–219. doi: 10.1016/j.bios.2016.09.001. [DOI] [PubMed] [Google Scholar]
- Lounsbury J.A., Karlsson A., Miranian D.C., Cronk S.M., Nelson D.A., Li J., Haverstick D.M., Kinnon P., Saul D.J., Landers J.P. Lab Chip. 2013;13:1384–1393. doi: 10.1039/c3lc41326h. [DOI] [PubMed] [Google Scholar]
- Lu W., Wang J., Wu Q., Sun J., Chen Y., Zhang L., Zheng C., Gao W., Liu Y., Jiang X. Biosens. Bioelectron. 2016;75:28–33. doi: 10.1016/j.bios.2015.08.016. [DOI] [PubMed] [Google Scholar]
- Ma Y.D., Li K.H., Chen Y.H., Lee Y.M., Chou S.T., Lai Y.Y., Huang P.C., Ma H.P., Lee G. Bin. Lab Chip. 2019;19:3804–3814. doi: 10.1039/c9lc00797k. [DOI] [PubMed] [Google Scholar]
- Magro L., Escadafal C., Garneret P., Jacquelin B., Kwasiborski A., Manuguerra J.C., Monti F., Sakuntabhai A., Vanhomwegen J., Lafaye P., Tabeling P. Lab Chip. 2017;17:2347–2371. doi: 10.1039/c7lc00013h. [DOI] [PubMed] [Google Scholar]
- Magro L., Jacquelin B., Escadafal C., Garneret P., Kwasiborski A., Manuguerra J.-C., Monti F., Sakuntabhai A., Vanhomwegen J., Lafaye P., Tabeling P. Sci. Rep. 2017;7:1347. doi: 10.1038/s41598-017-00758-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahalanabis M., Al-Muayad H., Kulinski M.D., Altman D., Klapperich C.M. Lab Chip. 2009;9:2811–2817. doi: 10.1039/b905065p. [DOI] [PubMed] [Google Scholar]
- Mahuku G.S. Plant Mol. Biol. Rep. 2004;22:71–81. [Google Scholar]
- Manz A., Widmers H.M., Graber N. Sensor. Actuator. B Chem. 1990;1:244–248. [Google Scholar]
- Markey A.L., Mohr S., Day P.J.R. Methods. 2010;50:277–281. doi: 10.1016/j.ymeth.2010.01.030. [DOI] [PubMed] [Google Scholar]
- Martinelli Federico, Scalenghe Riccardo, Davino Salvatore, Panno S., Scuderi Giuseppe, Ruisi Paolo, Villa Paolo, Stroppiana D., Boschetti Mirco, Goulart L.R., Davis Cristina E., Dandekar Abhaya M., Martinelli F., Scalenghe R., Davino S., Ruisi P., Scuderi G., Villa P., Stroppiana D., Boschetti M., Davis C.E., Dandekar A.M. Agron. Sustain. Dev. 2015;35:1–25. [Google Scholar]
- McCoy A.G., Miles T.D., Bilodeau G.J., Woods P., Blomquist C., Martin F.N., Chilvers M.I. Plants. 2020;9:466. doi: 10.3390/plants9040466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall S.M., Neto M.F., Reed J.L., Wagner R.L. J. Vis. Exp. 2016;2016 doi: 10.3791/54289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall S.M., Wagner R.L., Jangam S.R., Yamada D.H., Hardie D., Kelso D.M. J. Virol. Methods. 2015;214:37–42. doi: 10.1016/j.jviromet.2015.01.005. [DOI] [PubMed] [Google Scholar]
- Mekuria T.A., Zhang S., Eastwell K.C. J. Virol. Methods. 2014;205:24–30. doi: 10.1016/j.jviromet.2014.04.015. [DOI] [PubMed] [Google Scholar]
- Mosley O., Melling L., Tarn M.D., Kemp C., Esfahani M.M.N.N., Pamme N., Shaw K.J. Lab Chip. 2016;16:2108–2115. doi: 10.1039/c6lc00228e. [DOI] [PubMed] [Google Scholar]
- Murray M.G., Thompson W.F. Nucleic Acids Res. 1980;8:4321–4326. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan L., Jiang Z., Wei X. Lab Chip. 2014;14:1060. doi: 10.1039/c3lc51133b. [DOI] [PubMed] [Google Scholar]
- Ndunguru J., Taylor N.J., Yadav J., Aly H., Legg J.P., Aveling T., Thompson G., Fauquet C.M. Virol. J. 2005;2:45. doi: 10.1186/1743-422X-2-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neto M.F., Butzler M.A., Reed J.L., Rui X., Fisher M.J., Kelso D.M., McFall S.M. J. Virol. Methods. 2017;248:107–115. doi: 10.1016/j.jviromet.2017.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezhad A.S. Lab Chip. 2014;14:2887–2904. doi: 10.1039/c4lc00487f. [DOI] [PubMed] [Google Scholar]
- Ngo H.T., Freedman E., Odion R.A., Strobbia P., De Silva Indrasekara A.S., Vohra P., Taylor S.M., Vo-Dinh T. Sci. Rep. 2018;8:4075. doi: 10.1038/s41598-018-21615-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie K., Qi S., Zhang Yong, Luo L., Xie Y., Yang M., Zhang Yi, Li J., Shen H., Li Q., Ma X. PLoS One. 2012;7 doi: 10.1371/journal.pone.0052486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oblath E.A., Henley W.H., Alarie J.P., Ramsey J.M. Lab Chip. 2013;13:1325–1332. doi: 10.1039/c3lc40961a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocenar J., Arizala D., Boluk G., Dhakal U., Gunarathne S., Paudel S., Dobhal S., Arif M. PLoS One. 2019;14 doi: 10.1371/journal.pone.0218868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmavathy B., Vinoth Kumar R., Jaffar Ali B.M. J. Nanobiotechnol. 2012;10:8. doi: 10.1186/1477-3155-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B.H., Oh S.J., Jung J.H., Choi G., Seo J.H., Kim D.H., Lee E.Y., Seo T.S. Biosens. Bioelectron. 2017;91:334–340. doi: 10.1016/j.bios.2016.11.063. [DOI] [PubMed] [Google Scholar]
- Park H.J., Woo A., Cha J.M., Lee K.S., Lee M.Y. Sci. Rep. 2018;8:1–8. doi: 10.1038/s41598-018-34781-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Park J.-K. Lab Chip. 2018;18:1215–1222. doi: 10.1039/c7lc01128h. [DOI] [PubMed] [Google Scholar]
- Park S., Zhang Y., Lin S., Wang T.-H., Yang S. Biotechnol. Adv. 2011;29:830–839. doi: 10.1016/j.biotechadv.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul R., Ostermann E., Gu Z., Ristaino J.B., Wei Q. Curr. Protoc. Plant Biol. 2020;5 doi: 10.1002/cppb.20104. [DOI] [PubMed] [Google Scholar]
- Paul R., Saville A.C.A.C., Hansel J.C.J.C., Ye Y., Ball C., Williams A., Chang X., Chen G., Gu Z., Ristaino J.B.J.B., Wei Q. ACS Nano. 2019;13:6540–6549. doi: 10.1021/acsnano.9b00193. [DOI] [PubMed] [Google Scholar]
- Pearlman S.I., Leelawong M., Richardson K.A., Adams N.M., Russ P.K., Pask M.E., Wolfe A.E., Wessely C., Haselton F.R. ACS Appl. Mater. Interfaces. 2020;12:12457–12467. doi: 10.1021/acsami.9b21564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petralia S., Conoci S. ACS Sens. 2017;2:876–891. doi: 10.1021/acssensors.7b00299. [DOI] [PubMed] [Google Scholar]
- Petralia S., Verardo R., Klaric E., Cavallaro S., Alessi E., Schneider C. Sensor. Actuator. B. 2013;187:99–105. [Google Scholar]
- Port J.R., Nguetse C., Adukpo S., Velavan T.P. Malar. J. 2014;13:454. doi: 10.1186/1475-2875-13-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price C.W., Leslie D.C., Landers J.P. Lab Chip. 2009;9:2484–2494. doi: 10.1039/b907652m. [DOI] [PubMed] [Google Scholar]
- Qiu X., Chen D., Liu C., Mauk M.G., Kientz T., Bau H.H. Biomed. Microdevices. 2011;13:809–817. doi: 10.1007/s10544-011-9551-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan G.V., Cook N.M., Bueno-Sancho V., Lewis C.M., Persoons A., Mitiku A.D., Heaton M., Davey P.E., Abeyo B., Alemayehu Y., Badebo A., Barnett M., Bryant R., Chatelain J., Chen X., Dong S., Henriksson T., Holdgate S., Justesen A.F., Kalous J., Kang Z., Laczny S., Legoff J.-P., Lesch D., Richards T., Randhawa H.S., Thach T., Wang M., Hovmøller M.S., Hodson D.P., Saunders D.G.O. BMC Biol. 2019;17:65. doi: 10.1186/s12915-019-0684-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J.L., Walker Z.J., Basu D., Allen V., Nicol M.P., Kelso D.M., McFall S.M. Tuberculosis. 2016;101:114–124. doi: 10.1016/j.tube.2016.09.002. [DOI] [PubMed] [Google Scholar]
- Reinholt S.J., Baeumner A.J. Angew. Chem. Int. Ed. 2014;53:13988–14001. doi: 10.1002/anie.201309580. [DOI] [PubMed] [Google Scholar]
- Ren X., Yan J., Wu D., Wei Q., Wan Y. ACS Sens. 2017;2:1267–1271. doi: 10.1021/acssensors.7b00495. [DOI] [PubMed] [Google Scholar]
- Ristaino J.B., Saville A.C., Paul R., Cooper D.C., Wei Q. Plant Dis. 2020;104:708–716. doi: 10.1094/PDIS-06-19-1186-RE. [DOI] [PubMed] [Google Scholar]
- Rodriguez N.M., Linnes J.C., Fan A., Ellenson C.K., Pollock N.R., Klapperich C.M. Anal. Chem. 2015;87:7872–7879. doi: 10.1021/acs.analchem.5b01594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez N.M., Wong W.S., Liu L., Dewar R., Klapperich C.M. Lab Chip. 2016;16:753–763. doi: 10.1039/c5lc01392e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrman B.A., Richards-Kortum R.R. Lab Chip. 2012;12:3082–3088. doi: 10.1039/c2lc40423k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero Romero J.L., Carver G.D., Arce Johnson P., Perry K.L., Thompson J.R., Lucina J., Romero R., Gavriela, Carver D., Johnson P.A., Keith, Perry L., Thompson J.R. Arch. Virol. 2019;164:1453–1457. doi: 10.1007/s00705-019-04207-y. [DOI] [PubMed] [Google Scholar]
- Rose C., Parker A., Jefferson B., Cartmell E. Crit. Rev. Environ. Sci. Technol. 2015;45:1827–1879. doi: 10.1080/10643389.2014.1000761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbohm J.M., Robson J.M., Singh R., Lee R., Zhang J.Y., Klapperich C.M., Pollock N.R., Cabodi M. Anal. Methods. 2020;12:1085–1093. doi: 10.1039/c9ay02478f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkozi A. New Standards to Curb the Global Spread of Plant Pests and Diseases. 2019. http://www.fao.org/news/story/en/item/1187738/icode/
- Sarwar M., Leichner J., Naja G.M., Li C.Z. Microsys. NanoEng. 2019;5:1–10. doi: 10.1038/s41378-019-0096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenkbier L., Pollok S., König S., Urban M., Werres S., Cialla-May D., Weber K., Popp J. Anal. Methods. 2015;7:211–217. [Google Scholar]
- Seok Y., Jang H., Oh J., Joung H.-A., Kim M.-G. Biomed. Phys. Eng. Express. 2019;5 [Google Scholar]
- Seok Y., Joung H.-A., Byun J.-Y., Jeon H.-S., Shin S.J., Kim S., Shin Y.-B., Han H.S., Kim M.-G. Theranostics. 2017;7:2220–2230. doi: 10.7150/thno.18675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y., Perera A.P., Wong C.C., Park M.K. Lab Chip. 2014;14:359–368. doi: 10.1039/c3lc51035b. [DOI] [PubMed] [Google Scholar]
- Son J.H., Cho B., Hong S., Lee S.H., Hoxha O., Haack A.J., Lee L.P. Light Sci. Appl. 2015;4 e280–e280. [Google Scholar]
- Son J.H., Hong S., Haack A.J., Gustafson L., Song M., Hoxha O., Lee L.P. Adv. Healthc. Mater. 2016;5:167–174. doi: 10.1002/adhm.201500708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohmeier O., Keil S., Kanat B., Patel P., Niedrig M., Weidmann M., Hufert F., Drexler J., Zengerle R., von Stetten F. RSC Adv. 2015;5:32144–32150. [Google Scholar]
- Stumpf F., Schwemmer F., Hutzenlaub T., Baumann D., Strohmeier O., Dingemanns G., Simons G., Sager C., Plobner L., von Stetten F., Zengerle R., Mark D. Lab Chip. 2016;16:199–207. doi: 10.1039/c5lc00871a. [DOI] [PubMed] [Google Scholar]
- Sun F., Ganguli A., Nguyen J., Brisbin R., Shanmugam K., Hirschberg D.L., Wheeler M.B., Bashir R., Nash D.M., Cunningham B.T. Lab Chip. 2020;20:1621–1627. doi: 10.1039/d0lc00304b. [DOI] [PubMed] [Google Scholar]
- Tang R., Yang H., Choi J.R., Gong Y., Hu J., Wen T., Li X.J., Xu B., Mei Q., Xu F. Microchim. Acta. 2017;184:2141–2150. [Google Scholar]
- Tang R., Yang H., Gong Y., You M.L., Liu Z., Choi J.R., Wen T., Qu Z., Mei Q., Xu F. Lab Chip. 2017;17:1270–1279. doi: 10.1039/c6lc01586g. [DOI] [PubMed] [Google Scholar]
- Tang R.H., Yang H., Choi J.R., Gong Y., Feng S.S., Pingguan-Murphy B., Huang Q.S., Shi J.L., Mei Q.B., Xu F. Crit. Rev. Biotechnol. 2017;37:411–428. doi: 10.3109/07388551.2016.1164664. [DOI] [PubMed] [Google Scholar]
- Tangkanchanapas P., Höfte M., De Jonghe K. J. Virol. Methods. 2018;259:81–91. doi: 10.1016/j.jviromet.2018.06.003. [DOI] [PubMed] [Google Scholar]
- Tomlinson J.A., Boonham N., Dickinson M. Plant Pathol. 2010;59:465–471. [Google Scholar]
- Trinh K.T.L., Trinh T.N.D., Lee N.Y. Biosens. Bioelectron. 2019;135:120–128. doi: 10.1016/j.bios.2019.04.011. [DOI] [PubMed] [Google Scholar]
- Tsai J.-J., Liu W.-L., Lin P.-C., Huang B.-Y., Tsai C.-Y., Lee P.-Y.A., Tsai Y.-L., Chou P.-H., Chung S., Liu L.-T., Chen C.-H. PLoS One. 2019;14 [Google Scholar]
- Tsaloglou M.N., Bahi M.M., Waugh E.M., Morgan H., Mowlem M. Anal. Methods. 2011;3:2127–2133. [Google Scholar]
- Valasevich N., Schneider B. J. Phytopathol. 2017;165:762–770. [Google Scholar]
- Van Heirstraeten L., Spang P., Schwind C., Drese K.S., Ritzi-Lehnert M., Nieto B., Camps M., Landgraf B., Guasch F., Corbera A.H., Samitier J., Goossens H., Malhotra-Kumar S., Roeser T. Lab Chip. 2014;14:1519–1526. doi: 10.1039/c3lc51339d. [DOI] [PubMed] [Google Scholar]
- Walker F.M., Hsieh K. Biosensors. 2019;9:117. doi: 10.3390/bios9040117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker H.K., Hall W.D., Hurst J.W. third ed. Butterworths; 1990. Clinical Methods: the History, Physical, and Laboratory Examinations. [PubMed] [Google Scholar]
- Wand N.I.V., Bonney L.C., Watson R.J., Graham V., Hewson R. J. Gen. Virol. 2018;99:1012–1026. doi: 10.1099/jgv.0.001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.H., Lien K.Y., Wu J.J., Lee G. Bin. Lab Chip. 2011;11:1521–1531. doi: 10.1039/c0lc00430h. [DOI] [PubMed] [Google Scholar]
- Wang H., Chen H.-W., Hupert M.L., Chen P.-C., Datta P., Pittman T.L., Goettert J., Murphy M.C., Williams D., Barany F., Soper S.A. Angew. Chem. Int. Ed. 2012;51:4349–4353. doi: 10.1002/anie.201200732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Qi M., Cutler A.J. Nucleic Acids Res. 1993;21:4153–4154. doi: 10.1093/nar/21.17.4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Hua, Ma Z., Qin J., Shen Z., Liu Q., Chen X., Wang Hengliang, An Z., Liu W., Li M. Biosens. Bioelectron. 2019;126:373–380. doi: 10.1016/j.bios.2018.11.011. [DOI] [PubMed] [Google Scholar]
- Wang L.-X., Fu J.-J., Zhou Y., Chen G., Fang C., Lu Z.S., Yu L. Talanta. 2020;208 doi: 10.1016/j.talanta.2019.120407. [DOI] [PubMed] [Google Scholar]
- Wang R., Zhao R., Li Y., Kong W., Guo X., Yang Y., Wu F., Liu W., Song H., Hao R. Lab Chip. 2018;18:3507–3515. doi: 10.1039/c8lc00841h. [DOI] [PubMed] [Google Scholar]
- Wang X., Yin F., Bi Y., Cheng G., Li J., Hou L., Li Y., Yang B., Liu W., Yang L. J. Virol. Methods. 2016;238:86–93. doi: 10.1016/j.jviromet.2016.10.010. [DOI] [PubMed] [Google Scholar]
- Wu J., Kodzius R., Cao W., Wen W. Microchim. Acta. 2014;181:1611–1631. [Google Scholar]
- Wu L.T., Thomas I., Curran M.D., Ellis J.S., Parmar S., Goel N., Sharma P.I., Allain J.P., Lee H.H. J. Clin. Microbiol. 2013;51:3031–3038. doi: 10.1128/JCM.00740-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H., Zhu Y., Zhang Y., Wang L., Chen J., Lu Y., Xu Y., Xing W. Sci. Rep. 2017;7:1–11. doi: 10.1038/s41598-017-01415-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Chen Z., Cao X., Li Z., Stavrakis S., Choo J., DeMello A.J., Howes P.D., He N. Anal. Bioanal. Chem. 2018;410:7019–7030. doi: 10.1007/s00216-018-1335-9. [DOI] [PubMed] [Google Scholar]
- Yang Z., Xu G., Reboud J., Ali S.A., Kaur G., McGiven J., Boby N., Gupta P.K., Chaudhuri P., Cooper J.M. ACS Sens. 2018;3:403–409. doi: 10.1021/acssensors.7b00825. [DOI] [PubMed] [Google Scholar]
- Yaren O., Alto B.W., Gangodkar P.V., Ranade S.R., Patil K.N., Bradley K.M., Yang Z., Phadke N., Benner S.A. BMC Infect. Dis. 2017;17:293. doi: 10.1186/s12879-017-2382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X., Xu J., Lu L., Li X., Fang X., Kong J. Anal. Chim. Acta. 2018;1018:78–85. doi: 10.1016/j.aca.2018.02.068. [DOI] [PubMed] [Google Scholar]
- Yeh E.-C., Fu C.-C., Hu L., Thakur R., Feng J., Lee L.P. Sci. Adv. 2017;3 doi: 10.1126/sciadv.1501645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J., Hu J., Sun J., Wang B., Mu Y. Analyst. 2019;144:7032–7040. doi: 10.1039/c9an01067j. [DOI] [PubMed] [Google Scholar]
- Yin J., Suo Y., Zou Z., Sun J., Zhang S., Wang B., Xu Y., Darland D., Zhao J.X., Mu Y. Lab Chip. 2019;19:2769–2785. doi: 10.1039/c9lc00389d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z., Ramshani Z., Waggoner J.J., Pinsky B.A., Senapati S., Chang H.-C. Sensor. Actuator. B Chem. 2020;310 [Google Scholar]
- Yoo S.M., Lee S.Y. Trends Biotechnol. 2016;34:7–25. doi: 10.1016/j.tibtech.2015.09.012. [DOI] [PubMed] [Google Scholar]
- Yoon J., Yoon Y.-J.J., Lee T.Y., Park M.K., Chung J., Shin Y., Kyoung Park M., Chung J., Shin Y., Park M.K., Chung J., Shin Y. Sensor. Actuator. B. 2018;255:1491–1499. [Google Scholar]
- Zhang J.J., Su X., Xu J., Wang J., Zeng J., Li C., Chen W., Li T., Min X., Zhang D., Zhang S., Ge S., Zhang J.J., Xia N. Biomicrofluidics. 2019;13:34102. doi: 10.1063/1.5088552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Cai Q., Wiederkehr R.S., Fauvart M., Fiorini P., Majeed B., Tsukuda M., Matsuno T., Stakenborg T. Lab Chip. 2016;16:4012–4019. doi: 10.1039/c6lc01046f. [DOI] [PubMed] [Google Scholar]
- Zhang L., Zhang Y., Wang C., Feng Q., Fan F., Zhang G., Kang X., Qin X., Sun J., Li Y., Jiang X. Anal. Chem. 2014;86:10461–10466. doi: 10.1021/ac503072a. [DOI] [PubMed] [Google Scholar]
- Zhang M., Ye J., He J., Zhang F., Ping J., Qian C., Wu J. Anal. Chim. Acta. 2020;1099:1–15. doi: 10.1016/j.aca.2019.11.056. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Park S., Yang S., Wang T.H. Biomed. Microdevices. 2010;12:1043–1049. doi: 10.1007/s10544-010-9458-6. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhang L., Sun J., Liu Y., Ma X., Cui S., Ma L., Xi J.J., Jiang X. Anal. Chem. 2014;86:7057–7062. doi: 10.1021/ac5014332. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhang Ying, Xie K. Mol. Breed. 2020;40:11. [Google Scholar]
- Zhao F., Lee E.Y., Noh G.S., Shin J., Liu H., Qiao Z., Shin Y. Sci. Rep. 2019;9:1–12. doi: 10.1038/s41598-019-52922-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Hu A., Cui J., Yang K., Zhu X., Liu Y., Deng G., Zhu L. Micromachines. 2019;10:537. doi: 10.3390/mi10080537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Zhao J., Hu A., Pan J., Deng G., Hua C., Zhu C., Liu Y., Yang K., Zhu L. Micromachines. 2020;11:186. doi: 10.3390/mi11020186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang B., Gan W., Wang S., Han J., Xiang G., Li C.X., Sun J., Liu P. Anal. Chem. 2015;87:1202–1209. doi: 10.1021/ac5039303. [DOI] [PubMed] [Google Scholar]
- Zou Y., Mason M.G., Wang Y., Wee E., Turni C., Blackall P.J., Trau M., Botella J.R. PLoS Biol. 2017;15 doi: 10.1371/journal.pbio.2003916. [DOI] [PMC free article] [PubMed] [Google Scholar]