Graphical abstract
Keywords: SARS-CoV-2, Coronavirus, RNA virus, Respiratory disease, SARS-CoV-2 inhibitors
Abstract
COVID-19 caused by the novel SARS-CoV-2 has been declared a pandemic by the WHO is causing havoc across the entire world. As of May end, about 6 million people have been affected, and 367 166 have died from COVID-19. Recent studies suggest that the SARS-CoV-2 genome shares about 80% similarity with the SARS-CoV-1 while their protein RNA dependent RNA polymerase (RdRp) shares 96% sequence similarity. Remdesivir, an RdRp inhibitor, exhibited potent activity against SARS-CoV-2 in vitro. 3-Chymotrypsin like protease (also known as Mpro) and papain-like protease, have emerged as the potential therapeutic targets for drug discovery against coronaviruses owing to their crucial role in viral entry and host-cell invasion. Crystal structures of therapeutically important SARS-CoV-2 target proteins, namely, RdRp, Mpro, endoribonuclease Nsp15/NendoU and receptor binding domain of CoV-2 spike protein has been resolved, which have facilitated the structure-based design and discovery of new inhibitors. Furthermore, studies have indicated that the spike proteins of SARS-CoV-2 use the Angiotensin Converting Enzyme-2 (ACE-2) receptor for its attachment similar to SARS-CoV-1, which is followed by priming of spike protein by Transmembrane protease serine 2 (TMPRSS2) which can be targeted by a proven inhibitor of TMPRSS2, camostat. The current treatment strategy includes repurposing of existing drugs that were found to be effective against other RNA viruses like SARS, MERS, and Ebola. This review presents a critical analysis of druggable targets of SARS CoV-2, new drug discovery, development, and treatment opportunities for COVID-19.
1. Introduction
Coronavirus disease 2019 (COVID-19) is a pandemic faced by the global community nearly a century after the Spanish flu [1]. The current pandemic is due to a novel beta coronavirus, SARS-CoV-2, taxonomically belonging to the coronaviridae family, other members of which are known to cause respiratory infections in humans [2]. The rapid torrent of COVID-19 infections around the globe at an alarming rate is due to the estimated basic reproductive number R0 value between 1 and 3, predicted to be higher than the SARS-CoV-1, with the main transmission route being respiratory droplets and contact. Based on phylogenetic analysis, the natural host of the virus was found in bats, but to date, there has not been any confirmation regarding the intermediate host [3], [4]. The virus was found to have been first identified in the city of Wuhan, Hubei province China in December 2019, and was declared as a pandemic by the WHO on March 11 and, as of May end, about for 6 million have been affected, and 367 166 have died from COVID-19. [5], [6].
Broadly pyrexia, cough, hemoptysis, diarrhea, dyspnoea, muscle soreness, lymphopenia, dysosmia, and dysgeusia are the symptoms associated with COVID-19, but certain symptoms atypical from other coronavirus infections are to be acknowledged. They include the presence of Gastro-Intestinal distress in some of the cases, and clinical manifestations of lower respiratory tract infection, which are to be addressed [7], [8]. Current diagnostic approaches range from nucleic acid assays- Reverse transcriptase- quantitative Polymerase Chain Reaction (RT-qPCR), Computed Tomography (CT) scans to immunological detection kits [9].
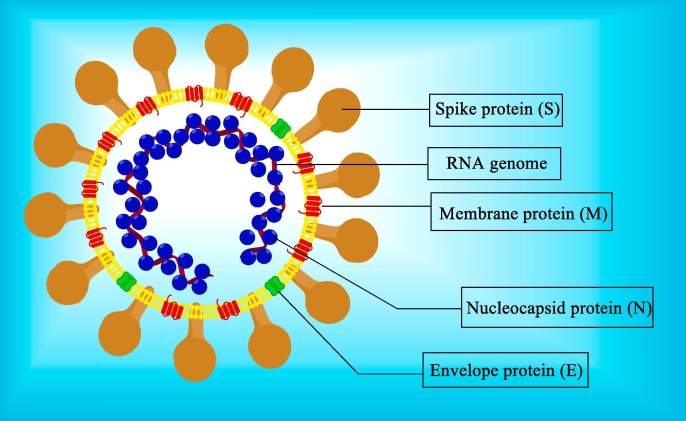
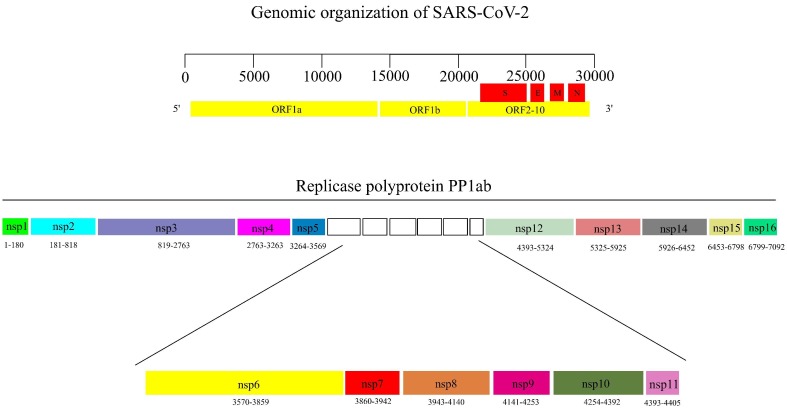
SARS-CoV-2 is an enveloped, single-stranded positive-sense RNA virus. The viral RNA genome contains 29,903 nucleotide bases and has ten open reading frames (ORF). The ORF1ab encodes for the large replicase polyprotein PP1ab, which is cleaved by Papain Like protease (PLpro) and 3-Chymotrypsin like Protease (3CLpro) to non-structural proteins (nsps) 1–16. The structural proteins S, N, E, M, and auxiliary proteins are encoded by ORF2-10 [10], [11] (Fig. 1, Fig. 2 ).
Fig. 1.
Structure of SARS-CoV-2.
Fig. 2.
(A) Genomic organization of SARS-CoV-2. (B) non-structural proteins 1–16. (the numbers below each of the nsps represent the amino acid residues). ORF: Open reading frame, S: spike protein, E: Envelope protein, N: Nucleocapsid protein, M: Membrane protein, nsp: non-structural protein.
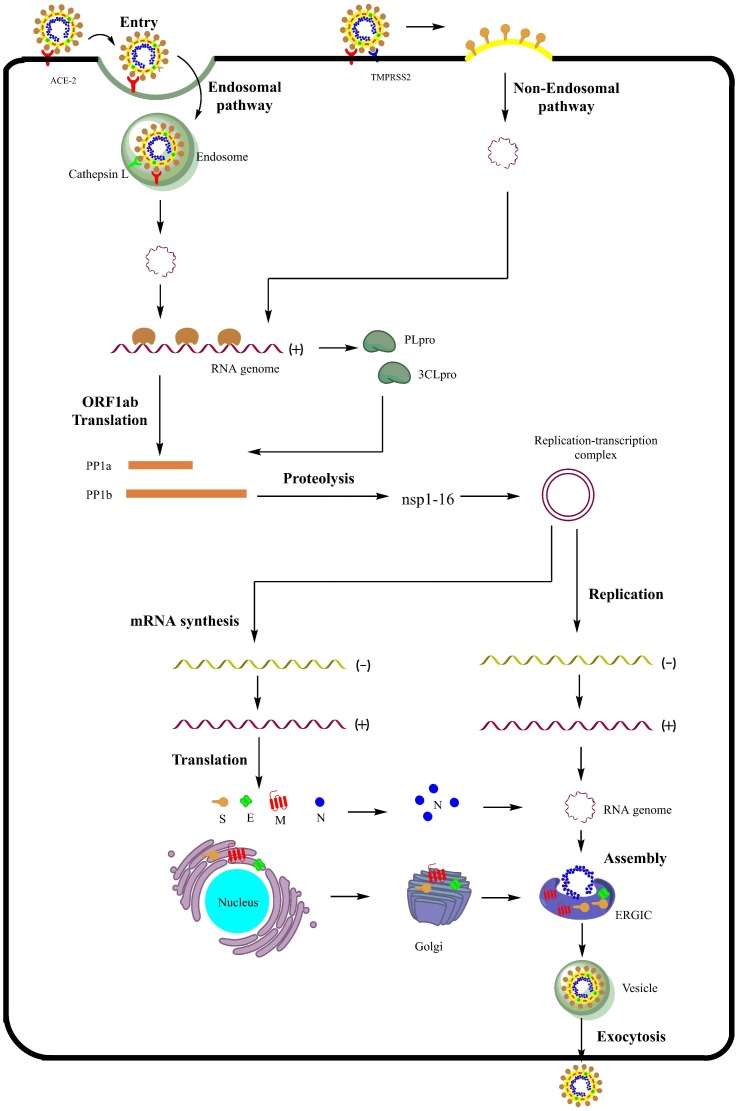
Entry of the virus into the host’s cells is aided by the attachment of the Spike protein (S) onto the host’s Angiotensin Converting Enzyme-2 (ACE-2) receptor resulting in conformational changes that lead to the fusion of viral and cell membranes. Inside the endocytic vesicle, pH-dependent activation of cathepsin L leads to priming of the spike proteins that result in the release of the viral RNA genome into the cytoplasm (endocytic pathway). Alternatively, Transmembrane Protease Serine type 2 (TMPRSS2) is also involved in priming of the spike proteins at the cell surface, enabling entry of the viral genome (non-endocytic pathway). Once inside the cytoplasm, the ORF1ab fragment of the viral RNA genome is translated into the replicase polyprotein PP1ab, which is acted upon by viral enzymes like PLpro and 3CLpro (Mpro) to produce nsps 1–16 including RdRp and helicase that play a crucial role in the replication of the virus. The nsps assemble to form the replication-transcription complex. The positive strand of RNA genome is transcribed to produce a negative-strand template for the synthesis of the new viral RNA genome. The transcribed mRNAs are translated to produce structural proteins S, M, E, and N. The viral RNA genome along with the N protein interacts with other structural proteins, and the assembled virions are delivered outside of the cell via exocytosis (Fig. 3 ).
Fig. 3.
The life cycle of SARS-CoV-2. ACE-2: Angiotensin converting enzyme-2, TMPRSS2: Transmembrane protease serine 2, PLpro: Papain-like protease, 3-CLpro: 3-Chymotrypsin-like protease, ORF1ab: Open reading frame 1ab, PP1ab: Polyprotein 1ab, nsp: non-structural protein, S: spike protein, E: Envelope protein, N: Nucleocapsid protein, M: Membrane protein, ERGIC: Endoplasmic reticulum-Golgi intermediate compartment.
1.1. Approaches to anti-coronavirus drug discovery
In general, three approaches were followed for the discovery and treatment options for the pathogenic coronaviruses- SARS and MERS [12]. Similar approaches were also being followed for the current pathogenic SARS-CoV-2. The first approach is the testing of broad-spectrum antiviral agents (mostly drugs) that have been used or previously tested for other viral infections by using standardized assays that measures the effects of these compounds on virus yields, infection rates, virus entry, and plaque formation. Some of the drugs identified via this approach during the previous two epidemics- SARS and MERS include ribavirin, Interferon-α, β, and γ [13], [14], [15]. A similar approach for the current pandemic has also identified antiviral agents like remdesivir, lopinavir/ritonavir, favipiravir, etc. A significant advantage of this approach is that the pharmacodynamic, pharmacokinetic, toxicity, dosing, etc. of the drugs will be readily available, thus reducing significant time in the drug discovery process. The second approach to the anti-coronavirus drug discovery involves the in-silico screening of chemical libraries that contains a large number of compounds. Rapid, high-throughput virtual screening (HTVS) of a large number of compounds is possible via this approach, which can be further validated by suitable antiviral assays. Drugs or investigational molecules belonging to different categories have been identified for the past epidemics- SARS and MERS as well for the current pandemic [15], [16], [17], [18], [19]. The major con associated with this approach is that, although the identified compounds exhibit antiviral activity, they are associated with several side effects. The third approach is the de-novo design and development of novel inhibitors based on the biophysical and genomic understanding of the individual coronaviruses. Examples include the development of antiviral peptides targeting the spike proteins, development of novel inhibitors targeting different viral enzymes, development of inhibitors targeting host cellular proteases., development of monoclonal antibody (MAbs). Theoretically, this approach will lead to potent compounds, but the development of these compounds usually takes years, as they undergo the entire drug discovery process.
2. Druggable molecular targets of SARS-CoV-2
2.1. Spike proteins of novel SARS-CoV-2
The entry of SARS-CoV-2 depends upon the binding of its spike protein to the cell surface receptor. The spike proteins of SARS-CoV-2 are 1273 amino acids long and structurally consists of two domains- the N-terminal S1 domain for binding to the cellular receptor and C-terminal S2 domain for fusion with the cell membrane [20]. The S1 subunit (aa 14-685) encompasses an N-terminal domain (aa 14-305) and a receptor-binding domain (RBD) (aa 319-541) [21], [22]. The RBD consists of core and external sub-domains responsible for forming trimer particle and for interaction with the receptor, respectively [23], [24]. The S2 sub-domain encompasses Heptad repeat 1 (HR1), Heptad repeat 2 (HR2), Fusion peptide (FP), and transmembrane domain. After binding of the S1 domain of spike protein to the host receptor, conformational changes take place in the S2 domain. The heptad repeat domains, HR1 and HR2, interact with themselves to form a six-helix bundle fusion core bringing about the close association of cellular and viral membranes for fusion [22]. Xia et al. designed novel peptides based on the HR1 and HR2 domains as fusion inhibitors [22] similar to the approach that was undertaken against SARS-CoV and MERS-CoV [25], [26], [27]. The two peptides designed were designated as 2019-nCoV-HR1P and 2019-nCoV-HR2P. 2019-nCoV-HR2P was shown to potently inhibit fusion in 2019-nCoV spike mediated cell-cell fusion assay with a half-maximal inhibitory concentration (IC50) value of 0.18 µM. EK1, reported previously as a pan CoV fusion inhibitor targeting the HR1 domain [27], also showed potent fusion inhibition (IC50 = 0.19 µM). Furthermore, 2019-nCoV-HR2P and EK1 were also found to effectively inhibit 2019-nCoV pseudovirus infection in 293 T cells with IC50 values of 0.98 µM and 2.38 µM, respectively (Fig. 4 ).
Fig. 4.
Peptide viral entry inhibitors.
Following the initial findings, Xia et al. and others designed a series of derivatives based on EK-1 [28]. EK-1 was covalently linked with cholesterol (designated as EK1C) and palmitic (designated as EK1P) at their C-terminus end with a polyethylene glycol (PEG) spacer (Fig. 4). Both the peptides exhibited potent fusion inhibition in SARS-CoV-2 mediated cell-cell fusion (IC50 = 48.1 and 69.2 nM for EK1C and EK1P, respectively). Based on the inhibitory potential of EK1C, further derivatives were designed with glycine / serine-based linkers (GSG) and PEG-based spacers. The peptides were designated as EK1C1-EK1C7 (Table 1 ). The presence of the linker/spacer and length of the spacer influenced the activity, especially increased the activity of these lipopeptides. The most potent lipopeptide, EK1C4, contained the optimal five residues linker/spacer GSGSG-PGE4. EK1C4 also exhibited potent activity against SARS-CoV-2 pseudovirus infection and live SARS-CoV-2 infection in vitro with IC50 values of 15.8 nM and 36.5 nM, respectively. Furthermore, in vivo studies were conducted to evaluate the prophylactic and post-infection potential of EK1C4 on the HCoV-OC43 infection mouse model. The studies revealed that EK1C4 was effective prophylactically up to 12 h before challenging with the infection at a dose of 0.5 mg/kg intranasally, suggesting good stability and half-life of the peptide. EK1C4 also afforded protection to mice when administered at 0.5 h post-infection [28]. Considering these results together, EK1C4 has all the potential to be a drug candidate for treating COVID-19.
Table 1.
Derivatives of EK1C.
| DESIGNATION | PEPTIDE | LINKER | LIPID | INHIBITION OF SARS-CoV-2 MEDIATED CELL-CELL FUSION (IC50 in nM) | INHIBITION OF SARS-CoV-2 PSEUDOVIRUS INFECTION (IC50 in nM) |
|---|---|---|---|---|---|
| EK1C1 | EK1 | CHOLESTEROL | 56.8 | 480.3 | |
| EK1C2 | EK1 | GSG | CHOLESTEROL | 48.2 | 418.6 |
| EK1C3 | EK1 | GSG-PGE4 | CHOLESTEROL | 10.6 | 86.8 |
| EK1C4 | EK1 | GSGSG-PGE4 | CHOLESTEROL | 1.3 | 15.8 |
| EK1C5 | EK1 | GSGSG-PGE8 | CHOLESTEROL | 3.1 | 31.3 |
| EK1C6 | EK1 | GSGSG-PGE12 | CHOLESTEROL | 3.9 | 77.4 |
| EK1C7 | EK1 | GSGSG-PEG24 | CHOLESTEROL | 3.9 | 84.4 |
Similarly, Zhang et al. designed and synthesized two peptides streptavidin-binding peptide-1 (SBP-1) and streptavidin-binding peptide-2 (SBP-2) based on the sequence of ACE-2 alpha-1 helix peptidase domain targeting RBD of the SARS-CoV-2 Spike protein in an attempt to disrupt the interaction between SARS-CoV-2- RBD and ACE-2 receptors (Fig. 5 ) [29]. Although SBP-2 did not show any affinity towards SARS-CoV-2 RBD, SBP-1 was found to potently bind to the SARS-CoV-2 RBD with a KD value of 47 nM, which was comparable to the binding of ACE-2 to SARS-CoV-2 RBD (KD = 14.7 nM).
Fig. 5.
Peptide viral entry inhibitors.
2.2. Extracellular proteases
Entry and invasion of SARS-CoVs into the host cells are facilitated through the concerted action of membrane proteases. These proteases have been confirmed as valuable therapeutic targets, and the inhibitors have been identified as promising drug leads against SARS-CoV-2.
2.2.1. Angiotensin converting enzyme-2 (ACE-2)
Coronaviruses make use of the host receptors as a doorway for entry into the host cell. Spike proteins of MERS-CoV use human dipeptidyl peptidase-4 (DPP-4) enzymes as its receptor for attachment [30] while SARS-CoV uses human angiotensin converting enzyme-2 (ACE-2) as a gateway for entry into the host cell [31]. Recent studies have shown that spike proteins of SARS-CoV-2 bind to the human ACE-2 for entry into the host cell, thus making ACE-2 a druggable target for COVID-19 [32], [33]. However, the simplicity ends there. ACE-2, apart from being used as a receptor for cellular entry by SARS coronaviruses, has several protective functions. Angiotensin I is synthesized from angiotensinogen, a process catalyzed by renin. Angiotensin I then serve as a substrate for the two enzymes Angiotensin converting enzyme-1 (ACE-1) and Angiotensin converting enzyme-1 (ACE-2). Angiotensin I is converted to an octapeptide angiotensin II by ACE-1 and a heptapeptide Angiotensin 1-7 by ACE-2. Angiotensin II and Angiotensin 1-7 have opposing effects on the vascular dynamics, the former being vasoconstrictive in nature and latter being vasodilative in nature, mediated by binding to their receptor Angiotensin-1 (AT-1) and Mas, respectively. Thus ACE-2/Ang 1-7/Mas receptor complex serves as the suppressive axis of the classical renin-angiotensin system (RAS) [34]. Chronic administration of Angiotensin Receptor Blockers (ARBs) leads to the up-regulation of ACE-2 [35], [36]. Both SARS-CoV and SARS-CoV-2 use ACE-2 as the receptor for binding, and hence it might sound absurd to suggest ARBs for SARS patients. Studies have shown downregulation in ACE-2 levels following viral attachment and fusion. This leads to the elevation of Angiotensin I levels due to the enhanced action of ACE-1 and repressed action of ACE-2, resulting in catastrophic changes in lung physiology mediated via AT-1 receptors [37]. Therefore, treating COVID-19 patients with ARBs, although paradoxical, may do more good than harm to the lungs. However, it is too early to recommend such therapy for COVID-19 due to its complexity. Currently, no molecule in the clinical or preclinical trial is known to inhibit ACE-2. The drug discovery directed against it for treating COVID-19 may not be a fruitful approach considering its protective effects on the lungs. However, a rational approach would be to use a recombinant form of soluble ACE-2 that can act as a decoy receptor and bind to the spike proteins of SARS-CoV-2 to slow its entry into the host cell as well as to afford protection to the lungs [38]. Ou et al. demonstrated that the entry of SARS-CoV-2 into 293/hACE2 cells was significantly suppressed by the pre-incubation of the pseudoviruses with a soluble form of hACE2 at 10 and 50 µg/ml [39].
2.2.2. Transmembrane protease serine 2 (TMPRSS2)
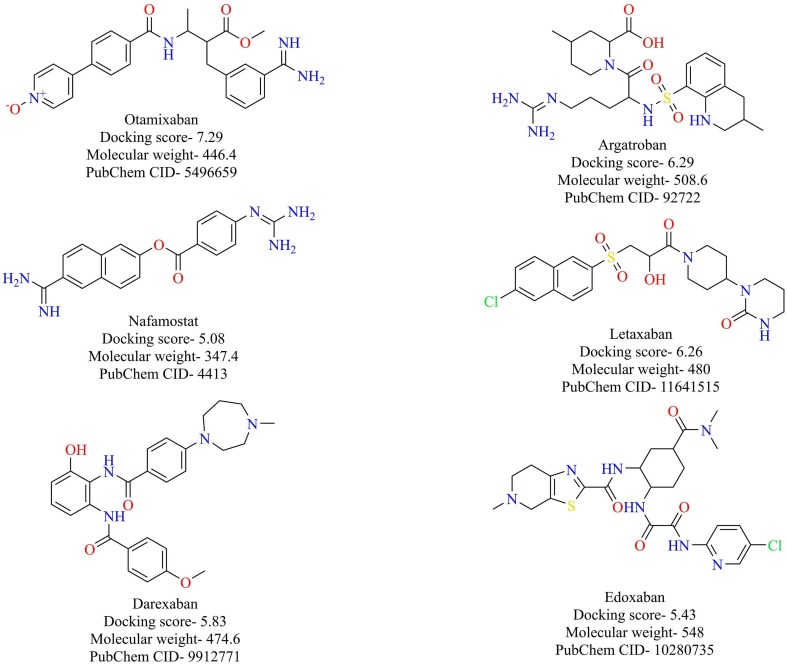
As previously described, spike proteins of SARS-CoV-2 bind to the ACE-2 receptor in order to enter into the host cell. However, other players are also associated with the entry of the virus into the host cell. Transmembrane protease serine 2 (TMPRSS2) is a host cellular protease encoded by the TMPRSS2 gene. TMPRSS2 is predominantly localized in the cell membrane of the lung’s epithelial cells and aids in priming the spike proteins, a process that results in the fusion of viral and cellular membranes. Previous results have shown that the priming of spike proteins of SARS-CoV by TMPRSS2 is crucial for the entry of the virus into the host cells [40], [41]. Recently, it was shown that entry of the novel SARS-CoV-2 also depends upon the priming of its spike proteins by TMPRSS2 and was blocked by a clinically known inhibitor of TMRSS2, camostat [33]. Hence, TMPRSS2 based therapeutics could prove to be useful armory in the war against COVID-19. Recently, Rensi et al. developed seven protein models of TMPRSS2 based on homology modeling and screened them against the clinically known serine protease inhibitors [42]. The top six molecules (anticoagulants with significant and potentially dangerous clinical effects and side effects), along with their average docking scores (kcal/mol), are given in Fig. 6 . An advantage of screening clinical molecules is that the safety and adverse effects of these molecules would have been already established, and priority thus would be to prove their efficacy. Further, in vitro studies of these molecules are currently underway to validate the predictions.
Fig. 6.
Structure of reported TMPRSS2 inhibitors.
2.2.3. Furin
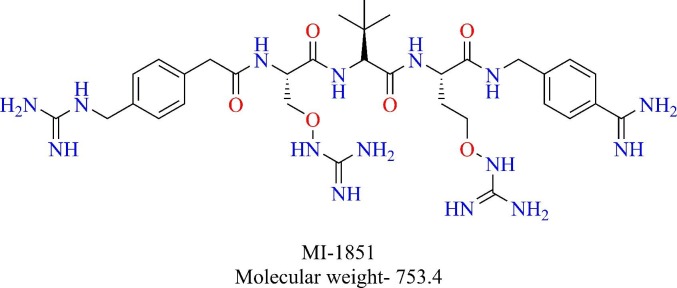
Furin belonging to the family of proprotein convertases, an enzyme that is responsible for activating precursor proteins like hormones, growth factors, cell surface receptors [48]. Furin selectively cleaves the proteins at multi-basic motifs of the sequence R-X-K/R-R↓ [49]. Furin is also known to activate fusion proteins of viruses like Human immunodeficiency virus (HIV), Ebola, highly pathogenic avian influenza A virus (HPAIV) as well as human coronaviruses like MERS and HCoV-OC43 [49], [50], [51], [52]. However, no such activation is reported for SARS-CoV. Recently, the genetic sequence analysis of SARS-CoV-2 S protein identified a polybasic cleavage site that may be accessible for proteases like furin, making it distinct from SARS-CoV [53], [54], [55]. Four amino acid sequence (RRAR) was found to be present in the S1/S2 site of SARS-CoV-2 S protein, thus corresponding to a canonical furin cleavage site. Hence, furin can also prime SARS-CoV-2 S proteins and aid in the entry of the virus. Bestle et al. and others demonstrated that furin activates SARS-CoV-2 S protein [56]. Furin was shown to cleave the S1/S2 site in the S protein of SARS-CoV-2 in HEK293 cells, which was decreased in the presence of furin inhibitor MI-1851 (Fig. 7 ). They also demonstrated that TMPRSS2 cleaves the S2′ site of SARS-CoV-2 S protein. Inhibition of either of these enzymes was shown to inhibit the replication of the virus in Calu-3 cells while inhibition of cathepsin L did not have any effect on the replication indicating that furin and TMPRRS2 are required for S protein activation while cathepsin L may or may not have a role in the activation of S protein in Calu-3 cells [56]. Thus, furin is essential for the multiplication of SARS-CoV-2 in airway epithelial cells and hence provides a favorable target of further exploration.
Fig. 7.
Structure of furin inhibitor- MI-1851.
2.3. Intracellular proteases
After the virus enters into the host cells, some of the critical processes regarding endosomal proliferation and assembly of viral proteins require the processing of these proteins through intracellular proteases. These processes are essential for the proliferation of the virus. Disruption of these process through target protease inhibitors breaks the cyclic of virus proliferation and have been suggested as the antiviral drug targets.
2.3.1. Cathepsin L
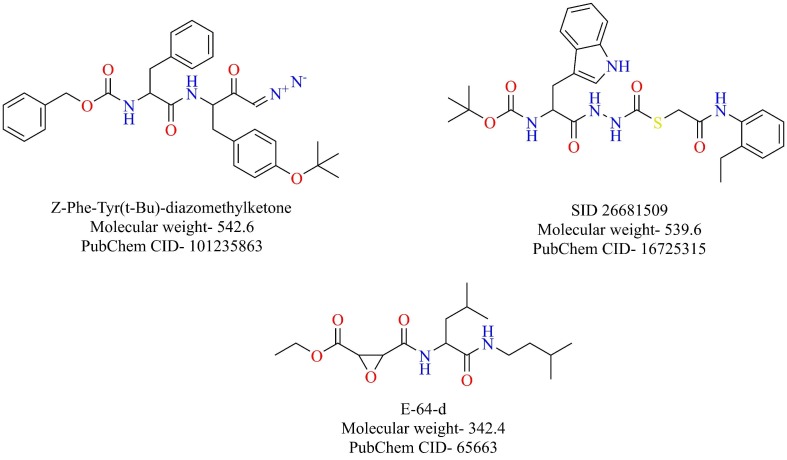
Cathepsins are cysteine proteases that play a crucial role in protein catabolism in the endosomes and lysosomes. The ubiquitously expressed cathepsin L plays a fundamental part in the entry of the SARS-CoV into the host cells [43], [44]. Once the virus enters the endosomes after binding onto the ACE-2 receptor, the spike proteins of SARS-CoV are activated by the pH-dependent cathepsin L, which results in the fusion of virus and endosomal membranes releasing the genetic material of virus into the cytoplasm [45]. SID 26681509, Z-Phe-Tyr(t-Bu)-diazomethylketone, and E-64-d are some of the well-known cathepsin L inhibitors (Fig. 8 ). Teicoplanin, a glycopeptide antibiotic, was also shown to inhibit the entry of SARS-CoV and MERS-CoV by blocking cathepsin L [46]. It was also shown to inhibit the entry of SARS-CoV-2 pseudoviruses [47]. Ou et al. demonstrated that cathepsin L is specifically required for the entry of SARS-CoV-2 S pseudovirus into 293/hACE2 cells. Broad-spectrum cathepsin inhibitor, E-64-d, reduced the SARS-CoV-2 S pseudovirus entry by 92.5% and cathepsin L inhibitor, SID 26681509, reduced the entry by about 76% while cathepsin B inhibitor, (CA-074), had no significant effect on the virus entry [39]. Taken together, the priming of spike proteins of SARS-CoV-2 is dependent on cathepsin L in the endosomes, making it an attractive therapeutic target for further exploration.
Fig. 8.
Structure of reported cathepsin L inhibitors.
2.4. RNA dependent RNA Polymerase (RdRp)
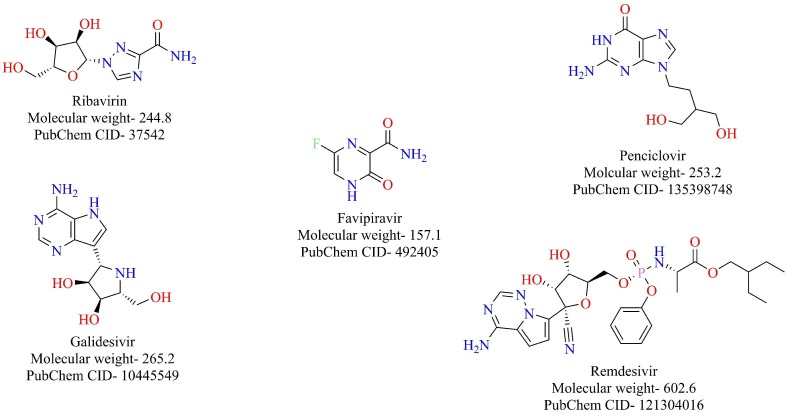
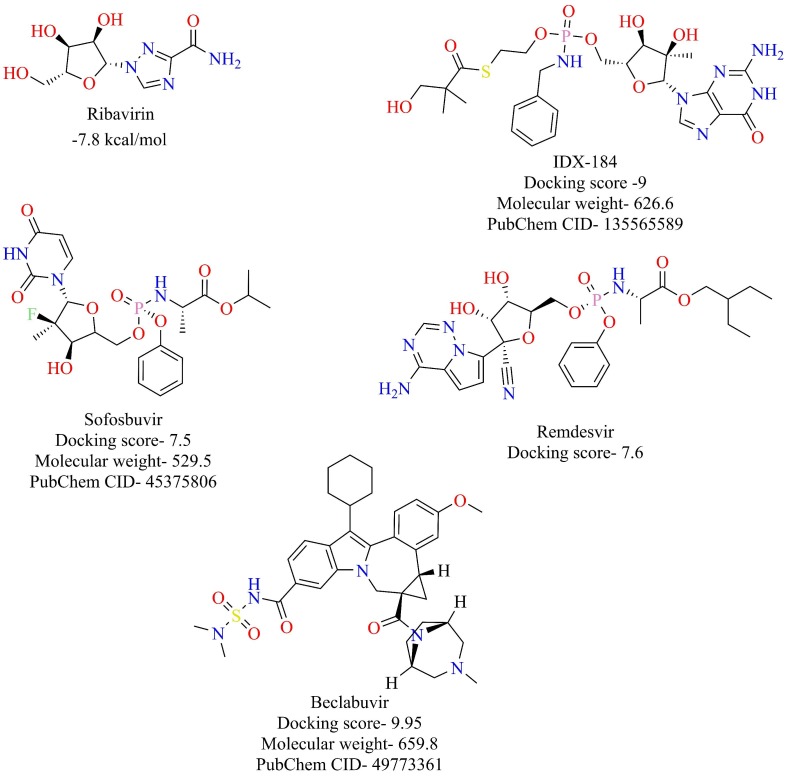
RNA dependent RNA Polymerase (RdRp) is an enzyme that is responsible for the replication of RNA from an RNA template. RdRp is one of the nsp (nsp-12) that play an essential role in the life cycle of RNA viruses like hepatitis C virus, Zika virus, and coronaviruses [57], [58]. In SARS-Cov-1 RdRp (nsp-12) functions as a tripartite polymerase complex with nsp-7 and nsp-8. The nsp7 and nsp8 activate and confer processivity to the RNA-synthesizing activity of nsp12. This tripartite polymerase complex further associates with nsp-14, which confers proofreading exonuclease function [59]. Remarkably, RdRp of SARS-CoV and SARS-CoV-2 share 96% sequence similarity [60]. Therefore, it is very likely that the agents that target RdRp of SARS-CoV might also target the RdRp of SARS-CoV-2. Some of the compounds that target RdRp of SARS-CoV include favipiravir, ribavirin, penciclovir, galidesivir, remdesivir, among others (Fig. 9 ) [61]. Recently, ribavirin, favipiravir, penciclovir, and remdesivir were shown to inhibit SARS-CoV-2 in vitro, probably due to their inhibitory activity towards RdRp [62]. Several computer-aided molecular modeling studies have been done to find potential candidates that target RdRp of SARS-CoV-2. Elfiky et al. evaluated the effectiveness of antiviral agents to bind to SARS-CoV-2 RdRp through a combination of homology modeling and molecular modeling studies [63]. The structures of the antiviral agents, along with their docking scores in kcal/mol, are given in Fig. 10 . Datta et al. constructed a model of SARS-CoV-2 RdRp using sequence alignment and homology modeling and screened the model against antiviral drugs. Beclabuvir, an HCV RdRp inhibitor, was shown to bind effectively to SARS-CoV-2 with a docking score of −9.95 kcal/mol and inhibition constant of 51.03 nM [64]. Recently, the structure of RdRp of SARS-CoV-2 was resolved in complex with nsp-7 and nsp-8 by the cryo-EM [65]. The overall architecture of the SARS CoV-2 nsp12/nsp7/nsp8 complex was observed to be similar to the RdRp complex of SARS-CoV-1. Binding of remdesivir diphosphate, the RdRp inhibitor under clinical trials for the treatment of COVID-19, to SARS CoV-2 nsp12, was investigated based on modeling with sofosbuvir bound to hepatitis C virus (HCV) ns5b. The nsp12 of the SARS CoV-2 virus showed the highest similarity with the Apostate of ns5b. Sofosbuvir, a nucleoside prodrug, targets HCV ns5b polymerase and selectively binding catalytic site of the HCV polymerase. Sofosbuvir has been approved for the treatment of chronic HCV infection [66]. The catalytic site of SARS CoV-2 nsp13 shows remarkable structural similarity to HCV ns5b polymerase. The antiviral RdRp inhibitors (Fig. 9) are likely to target SARS CoV-2 RdRp and are excellent drug candidates for the treatment of COVID-19.
Fig. 9.
Antiviral drugs targeting RdRp functions.
Fig. 10.
Structure of reported RdRp inhibitors along with their docking scores.
2.5. 3-Chymotrypsin like protease or main protease (3CLpro or Mpro)
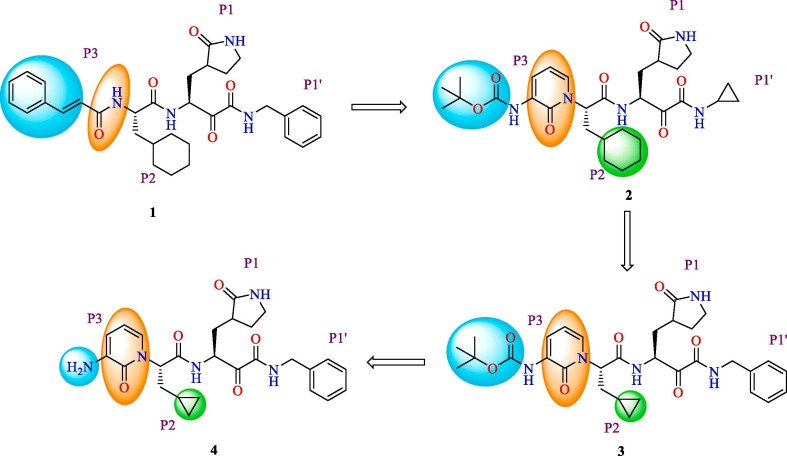
3-Chymotrypsin like Protease or Main protease (Mpro) is one of the two crucial proteolytic enzymes that help in cleaving the replicase polyprotein 1ab in SARS-CoV-2. The Mpro, which is also known as non-structural protein 5 (nsp5), is 306 amino acid long and is translated from the Orf1ab of viral RNA genome[67]. Mpro is known to cleave the replicase polyprotein at 11 specific sites, the recognition sites being Leu-Gln↓(Ser, Ala, Gly), to release 12 nsps (nsp4, nsp6-16) that are essential for viral replication as well as viral assembly [10], [67]. Structurally, Mpro is a dimer, and each monomer has three domains- domain I, which includes residues 8-101, domain II which includes residues 102-184 and domain III, which includes residues 201-303. The domains II and III are connected through the loop region that includes 185-200 residues. The substrate-binding site containing the catalytic dyad (Cys145-His41) is located between domain I and domain II [19]. The functional importance of the enzyme in the life cycle of SARS-CoV-2 and the fact that no human proteases possess similar cleavage recognition site makes Mpro appealing target for drug discovery. Zhang et al. reported the optimization of α-ketamides for activity against the Mpro of SARS-CoV-2 [68] (Fig. 11 ). The lead compound 1 was found to be active against the SARS-CoV Mpro in picomolar concentration [69]. However, short half-life in plasma and unfavorable pharmacokinetic profile of compound 1 prevented further development and study. In order to improve its pharmacokinetic profile, the amide linkage between P2-P3 was replaced with a pyridine ring for improved metabolic stability, and the cinnamoyl moiety was replaced with a butyloxycarbonyl (BOC) group for increased solubility and reduced plasma protein binding. However, the better pharmacokinetic profile met with a decreased activity against the Mpro of SARS-CoV-2 of compound 2 (IC50 = 2.39 µM) when compared with compound 1 (IC50 = 0.18 µM). In the next modification, P2 cyclohexyl moiety was replaced with a smaller cyclopropyl moiety in order to improve the antiviral activity. The resulting compound 3 inhibits Mpro of SARS-CoV-2 with an IC50 of 0.67 µM. Further, compound 3 effectively inhibited the replication of SARS-CoV-2 in Calu3 cells, while compound 4 formed by the removal of the BOC group was almost inactive, emphasizing that the BOC group is indispensable. The X-ray crystal structure of compound 3 with SARS-CoV-2 Mpro is depicted in Fig. 12 . Compound 3 also had a favorable pharmacokinetic profile in vivo, suggesting it as an ideal lead for further development.
Fig. 11.
Lead optimization of α-ketamides as Mpro inhibitor (Color representation indicates groups that are modified during the lead optimization of compound 1).
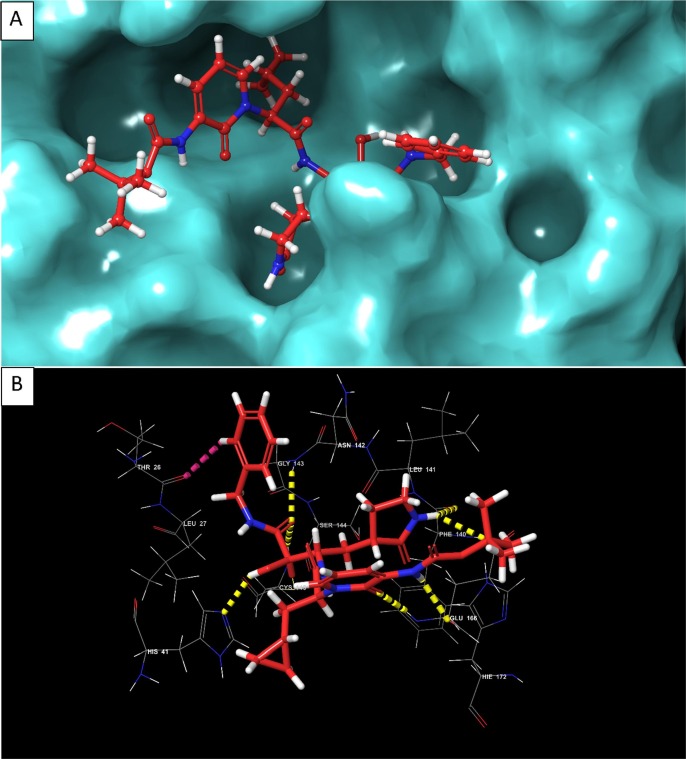
Fig. 12.
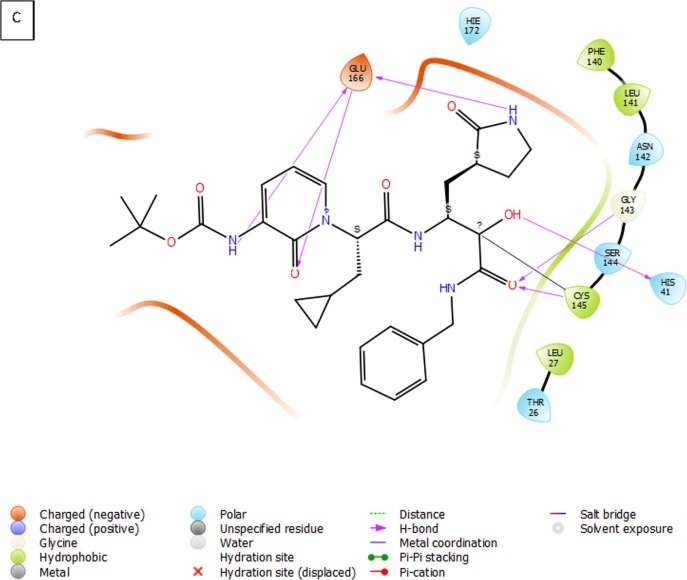
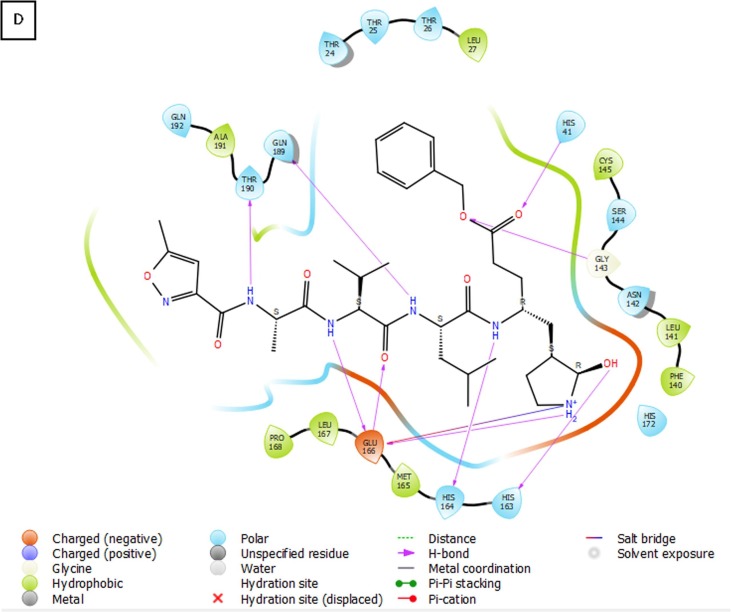
(A) 3D view of compound 3 inside the binding pocket of SARS-CoV-2 Mpro (B) 3D interaction diagram of compound 3 with active site residues of the target protein (C) 2D interaction diagram of compound 3 with active site residues of the target protein (PDB ID- 6Y2F) (Visualized using Maestro visualizer) [70].
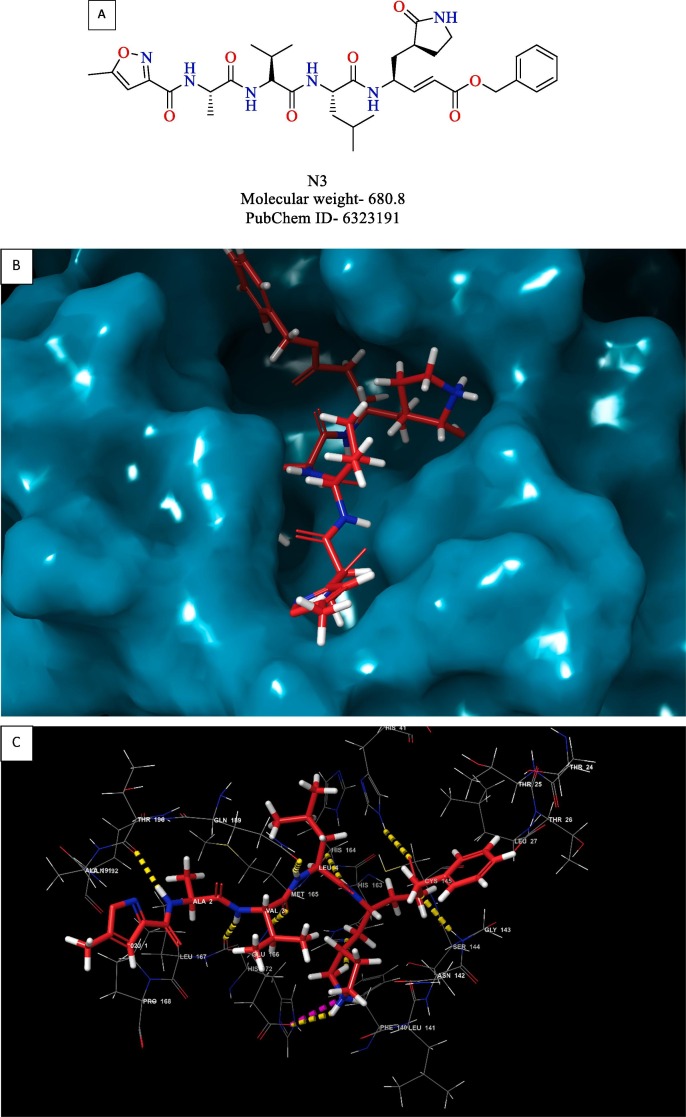
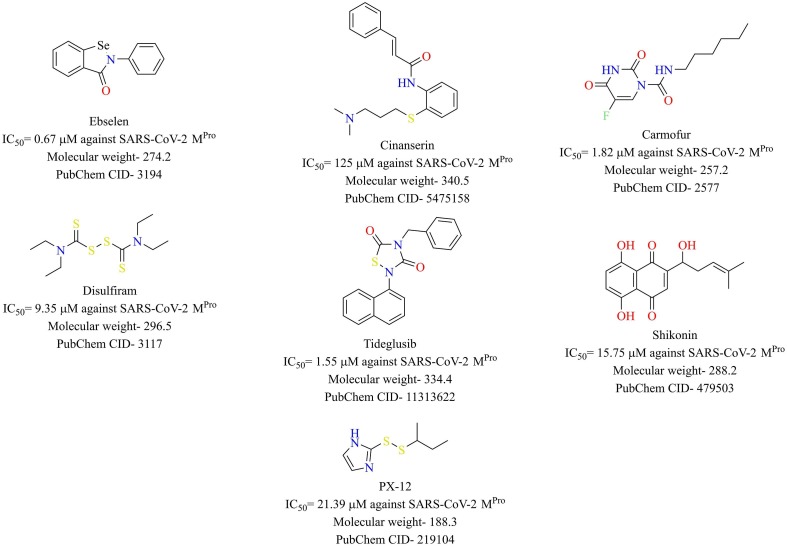
Jin et al. developed a strategy encompassing structure-based drug design, virtual screening, and high throughput screening to repurpose existing drugs or agents to target Mpro of SARS-CoV-2 [19]. N3 (Fig. 13 ) was designed based on computer-aided drug design to target the Mpro of SARS-CoV and MERS-CoV [71], [72]. Since Mpro is highly conserved among coronaviruses, compound N3 was also tested against Mpro of SARS-CoV-2, and it was found to be a potent inhibitor of Mpro. Enzyme kinetics studies revealed compound N3 as an irreversible inhibitor. The X-ray crystal structure of compound N3 in the binding pocket of SARS-CoV-2 Mpro is shown in Fig. 13. Based on the crystal structure of SARS-CoV-2 Mpro with N3, a virtual screening study identified cinanserin, a serotonin antagonist, as a promising antiviral candidate. Cinanserin exhibited activity against SARS-CoV-2 Mpro in vitro (IC50 = 125 µM). A high-throughput screening study consisting of approved drugs, natural products, and clinical drug candidates was performed to identify hit compounds against SARS-CoV-2 Mpro. The top six compounds, along with their IC50 values, are depicted in Fig. 14 . These compounds were evaluated for the potential to inhibit the replication of SARS-CoV-2 in Vero E6 cells. Ebselen (also called PZ 51, DR3305, and SPI-1005, is a synthetic organoselenium drug molecule with anti-inflammatory, antioxidant and cytoprotective activity) and N3 with half-maximal Effective Concentration (EC50) values of 4.67 and 16.77 µM, respectively were found to be the most potent compounds. Ebselen has low cytotoxicity, and its safety has been evaluated in a previous clinical trial [73], [74], making it an ideal candidate for further development.
Fig. 13.
(A) Structure of compound N3 (B) 3D view of compound N3 inside the binding pocket of SARS-CoV-2 Mpro (C) 3D interaction diagram of compound N3 with active site residues of the target protein (D) 2D interaction diagram compound N3 with active site residues of the target protein (PDB ID- 6LU7) (Visualized using Maestro visualizer) [70].
Fig. 14.
Structure of reported Mpro inhibitors.
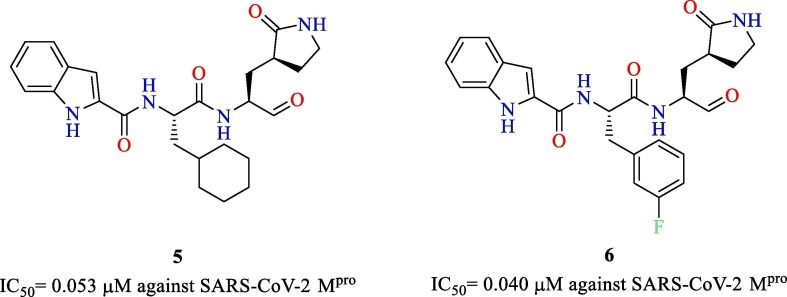
Dai et al. reported the synthesis and evaluation of two peptidomimetic aldehydes targeting the Mpro of SARS-CoV-2 [75]. The two peptidomimetic aldehydes 5 and 6 potently inhibited SARS-CoV-2 Mpro with IC50 values of 0.053 µM and 0.040 µM, respectively (Fig. 15 ). The crystal structures of SARS-CoV-2 Mpro with both the inhibitors were resolved and were found to bind with the substrate-binding site of the protein and showed vital interactions with the active site residues. Compounds 5 and 6 were also shown to possess potent antiviral activity against SARS-CoV-2 in Vero E6 cells with EC50 values of 0.42 and 0.33 µM, respectively. Furthermore, in vivo pharmacokinetic and toxicity studies were conducted to substantiate these compounds as promising candidates for COVID-19. Both the compounds exhibited good pharmacokinetic properties, and compound 5 exhibited low toxicity in vivo.
Fig. 15.
Peptidomimetic aldehydes 5 and 6 targeting SARS-CoV-2 Mpro.
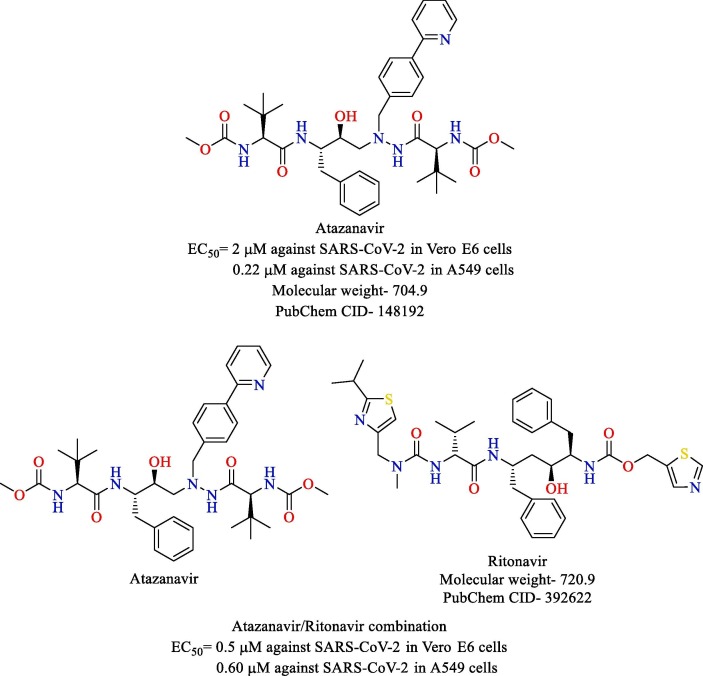
Fintelman-Rodrigues and co-workers shed light on the promising potential of atazanavir and atazanavir/ritonavir combination for the treatment of COVID-19 [76]. Through a combination of molecular docking and molecular dynamics, atazanavir was shown to bind to the active site of SARS-CoV-2 Mpro. It was also shown to inhibit the enzyme through zymographic studies. Atazanavir and atazanavir/ritonavir combination were evaluated for their inhibitory potential against SARS-CoV-2 in Vero E6 cells and human epithelial pulmonary cell lines (A549). The solo treatment, as well as the combination treatments, was found to inhibit SARS-CoV-2 in both the cell lines potently. The EC50 values of atazanavir and atazanavir/ritonavir combination against SARS-CoV-2 in Vero E6 cells and A549 cells are shown in Fig. 16 . Furthermore, atazanavir and atazanavir/ritonavir also prevented pro-inflammatory cytokine production in monocytes infected with SARS-CoV-2, as confirmed by the low levels of lactate dehydrogenase (LDH) and Interleukin-6 (IL-6).
Fig. 16.
Structure of atazanavir and ritonavir.
2.6. Papain like protease (PLpro)
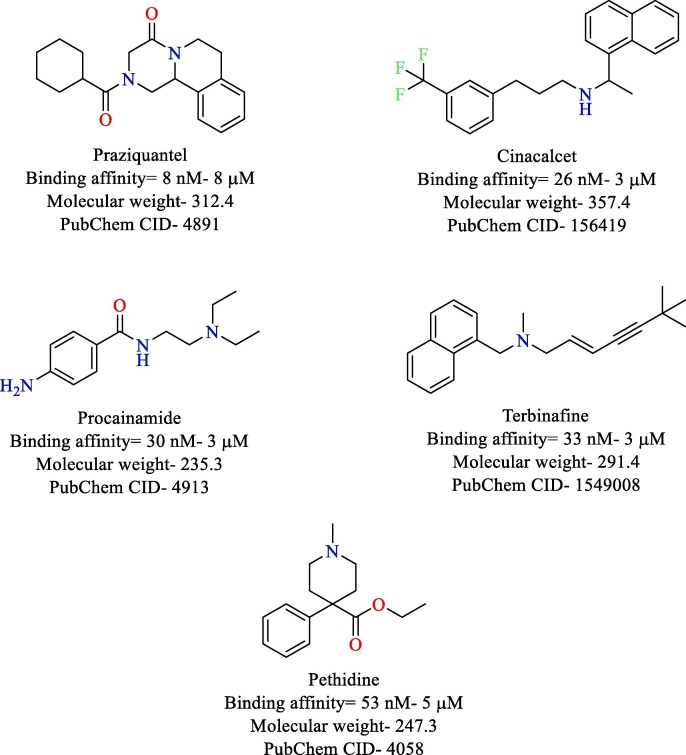
Papain like protease is the other proteolytic enzyme that helps in processing of the replicase polyprotein. PLpro is known to cleave the replicase polyprotein at three sites releasing the nsp-1, nsp-2, and nsp-3, which are essential for viral replication [10]. Apart from its proteolytic function, PLpro is also reported to possess de-ubiquitinating property [77]. It also plays a central role in countering the strength of host immune response to viral infection by shutting down crucial pathways [78], [79], [80]. Currently, no in vitro studies have been performed against PLpro of SARS-CoV-2. However, computational studies have been performed which could provide a direction for further exploitation of PLpro. Wu et al. constructed a homology modeling of SARS-CoV-2 PLpro and screened it virtually against the ZINC drug database and an in-house database of Chinese medicine and natural products. The virtual screening study identified ribavirin, valganciclovir, β-thymidine, and natural products Platycodin D, Chrysin, and Neohesperidin as potential leads with high binding affinity with SARS Cov-2 PLpro [10]. Arya et al. also constructed a homology model of SARS-CoV-2 PLpro and screened it against a library of 2525 FDA approved drugs. Praziquantel (anti-parasitic), cinacalcet (anti-hyperparathyroidism), procainamide (anti-arrhythmic), terbinafine (antifungal) and pethidine (opioid analgesic) were identified with potent binding affinity with SARS-CoV-2 PLpro [81] (Fig. 17 ). No major adverse effects have been reported for praziquantel and cinacalcet. However, procainamide, terbinafine, and pethidine have major adverse effects like cardiac abnormalities, hepatic failure, and respiratory depression, respectively [82], [83], [84].
Fig. 17.
Structure of top 5 compounds reported as PLpro inhibitor.
3. Treatment opportunities for COVID-19
3.1. Antivirals
3.1.1. Remdesivir
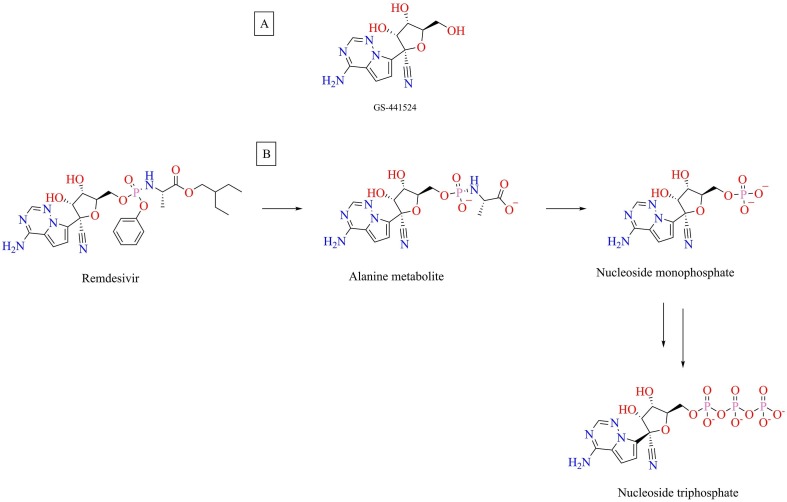
Remdesivir, a prodrug of adenosine analog GS-441524 (Fig. 18 ), is an investigational antiviral molecule that was developed by Gilead Sciences for the treatment of the Ebola virus [85]. Remdesivir is in the frontline in ongoing clinical trials for COVID-19. Remdesivir is a direct-acting antiviral drug, which stops replication of SARS-CoV-2 in host-cells [86]. Remdesivir was also found to exhibit antiviral activity against other flavo viruses like Marburg virus, Pneumo viruses like respiratory syncytial virus, and paramyxoviruses like mumps, measles, Nipah virus, and Hendra virus [87]. A recent study reported prophylactic as well as the therapeutic potential of remdesivir in the rhesus macaque model of MERS-CoV infection [88]. Remdesivir potently inhibited the replication of SARS-CoV and MERS-CoV in human airway epithelial cell cultures with IC50 values of 0.069 and 0.074 µM, respectively [89]. In addition, it was also effective against zoonotic coronaviruses [89], [90]. Recently, remdesivir was shown to exhibit potent activity against the novel coronavirus SARS-CoV-2 in vitro. Remdesivir was effective in blocking the viral infection in Vero E6 cells infected with nCoV-2019BetaCoV/Wuhan/WIV04/2019 (EC50 = 0.77 µM) [62]. Remdesivir was also shown to afford clinical benefits in rhesus macaques infected with SARS-CoV-2 [91]. The preliminary results of a double-blinded, randomized, placebo-controlled study evaluating the effectiveness of intravenous remdesivir for the treatment of COVID-19 was recently published. The study findings revealed that remdesivir was superior to placebo in reducing the time to recovery in adults hospitalized with COVID-19 with a median recovery time of 11 days as compared to 15 days for the placebo-treated group [92]. Recently, Gilead Sciences announced that a 5-day treatment course of remdesivir was associated with significantly greater clinical improvement in hospitalized patients with moderate COVID-19 when compared with standard care in a Phase-3 clinical study, further demonstrating the importance of remdesivir in the battle against COVID-19 [93]. The adverse effects that have been reported due to remdesivir administration include hypersensitivity- infusion-related and anaphylaxis and elevation in liver transaminases [94]. In the USA, the Food and Drug Administration (FDA) has granted remdesivir an Emergency Use Authorization (EUA) for the management of hospitalized patients with severe COVID-19.
Fig. 18.
(A) Structure of GS-441524 (B) Metabolic activation of remdesivir.
3.1.1.1. Mechanism of action
Remdesivir being a nucleoside analog inhibits the viral genome replication process by targeting the RdRp enzyme. After host-dependent conversion to its active form- nucleoside triphosphate (NTP) (Fig. 18), it incorporates into the RNA strand after fending off Adenosine triphosphate (ATP), the natural nucleotide incorporated in this process resulting in premature termination of RNA synthesis [85], [86], [95]
3.1.2. Lopinavir/ritonavir combination
Lopinavir and ritonavir are two structurally related protease inhibitors that were developed for the treatment of (HIV). Lopinavir exhibited higher selectivity, about 10-fold against both the wild and mutant strains of HIV-1 protease enzyme in vitro than ritonavir. However, it lacked the same potency in vivo due to metabolic inactivation by cytochrome P-450 (CYP) enzymes [96]. Ritonavir inhibits cytochrome P-450-3A4 (CYP3A4) at sub-therapeutic levels and hence is always co-administered with lopinavir to prevent its metabolic degradation [97]. Lopinavir was found to inhibit both SARS-CoV and MERS-CoV replication in vitro (EC50 = 17.1 and 8 µM against SARS-CoV and MERS-CoV, respectively). The combination of lopinavir/ritonavir was found to be effective in the animal model as well [98], [99]. It was proposed that the 3CLpro inhibition capacity of lopinavir/ritonavir partly contributes to their anti-CoV activity [100]. It was postulated that the combination could be effective against SARS-CoV-2. A study on analysis of molecular complexation between lopinavir and ritonavir with SARS-CoV-2 3CLpro suggested interaction of these drugs with residues at the active site of the enzyme [101]. There are 39 ongoing studies registered in ClinicalTrial.gov on clinical evaluation of lopinavir/ritonavir for the treatment of COVID-19. The results posted on a recently conducted clinical trial, Lopinavir Trial for Suppression of SARS-Cov-2 in China (LOTUS China), revealed that the combination of lopinavir/ritonavir offered no benefits over the standard care in hospitalized adult COVID-19 patients [102]. Considering the dynamic situation and differential pathologies of COVID-19 in different areas, it is important to wait for reports and outcomes of other ongoing clinical trials before drawing any conclusion on the therapeutic utility of this combination for COVID-19.
3.1.3. Favipiravir
Favipiravir is an antiviral drug approved for the treatment of the influenza virus. Favipiravir is a prodrug that is metabolically activated by the human hypoxanthine-guanine phosphoribosyltransferase (HGPRT) to its active metabolite-favipiravir-ribofuranosyl-5′-triphosphate (favipiravir-RTP) [103]. Favipiravir is an RdRp inhibitor that has shown promise as a therapeutic intervention for the Ebola virus [104], [105], [106], [107]. Favipiravir was shown to inhibit the replication of SARS-CoV-2 in vitro (EC50 = 61.88 µM) [62]. An open-label non-randomized control study was undertaken to compare the clinical effectiveness of favipiravir and lopinavir/ritonavir as a treatment strategy for COVID-19. The study findings revealed that favipiravir had better treatment outcomes on COVID-19 in terms of viral clearance and disease progression [108]. A clear understanding of clinical pharmacokinetics and pharmacodynamics profiles of favipiravir based on its previous clinical trials for influenza treatment [109] may be useful for determining the appropriate doses and course of treatment for COVID-19.
3.1.4. Ribavirin
Ribavirin, a guanosine analog, has been used for the treatment of RNA viruses like hepatitis and respiratory syncytial virus. It interferes with the viral RNA synthesis and viral mRNA capping after undergoing metabolic activation to its active metabolite [110]. Past studies have highlighted the combination of ribavirin and interferon therapy as an effective treatment strategy for SARS-CoV and MERS-CoV [111], [112], [113]. Recently, Wang et al. highlighted the potential of ribavirin to inhibit SARS-CoV-2 in vitro in Vero E6 cells (EC50 = 109.50 µM). Clinical trials are underway to evaluate the efficacy of ribavirin in the treatment of COVID-19 [62].
3.1.5. Other antiviral drugs
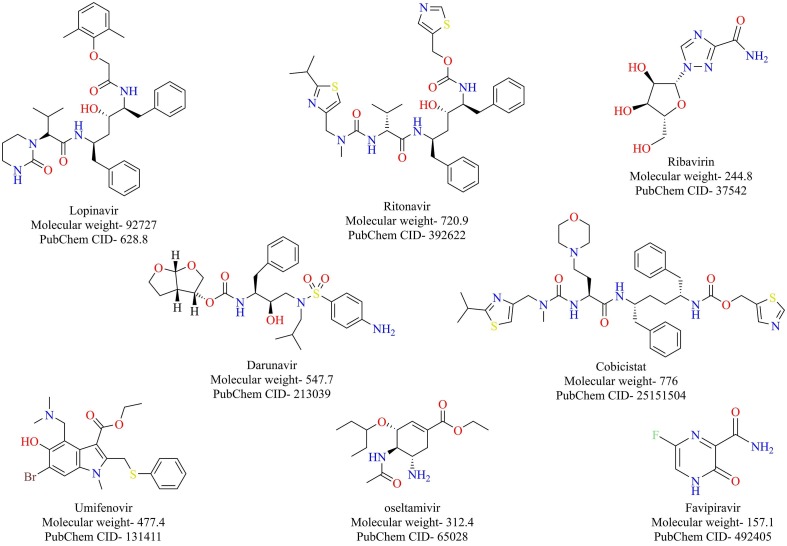
Other antivirals that are currently being tested include umifenovir, oseltamivir, darunavir/cobicistat combination (Fig. 19 ). Umifenovir is used for the treatment of influenza in Russia and China. Umifenovir exhibits its action by preventing the fusion of the virus with the cell membrane blocking its entry into the host cell [114]. Oseltamivir, also used for the treatment of influenza viruses, acts as a neuraminidase inhibitor, preventing the release of new virus particles [115]. Darunavir is a protease inhibitor approved for the treatment of HIV, and cobicistat prevents the catabolism of darunavir [116]. Clinical trials are underway to evaluate the efficacy of these drugs for treating COVID-19.
Fig. 19.
Antiviral drugs currently being repurposed for treating COVID-19.
3.2. Antibiotics
3.2.1. Teicoplanin
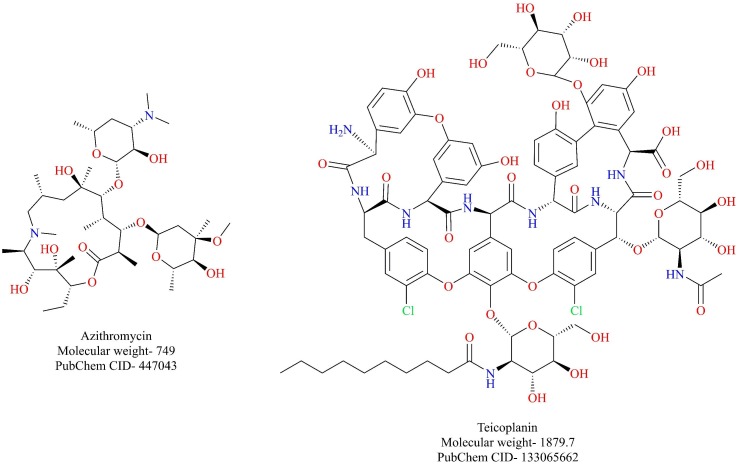
Teicoplanin belonging to the class of glycopeptide antibiotic is used for the treatment of gram-positive bacterial infections. The previous report suggested that teicoplanin and its derivatives were capable of blocking the viral entry of the Ebola virus, SARS-CoV, and MERS-CoV into the cell by blocking cathepsin L [46]. A recent report by the same research group suggests that teicoplanin potently inhibits the entry of SARS-CoV-2-S-pseudoviruses by blocking cathepsin L. (IC50 = 1.66 µM) [47]. Given its antibacterial prowess, teicoplanin could be considered as a dual inhibitor of the SARS-CoV-2 and its associated bacterial co-infections. However, further evidence is required to support such a claim.
3.2.2. Azithromycin
Azithromycin is a broad-spectrum antibiotic that is used for the management of several bacterial infections (Fig. 20 ). A recently conducted clinical trial highlighted the importance of azithromycin for the management of SARS-CoV-2 infection [117]. A total of 36 patients were enrolled for the study, out of which 20 patients received chloroquine, and 16 patients served as control. Among the chloroquine treated patients, six patients received azithromycin as a precautionary measure against bacterial superinfection. At the end of day 6, 100% of the patients treated with chloroquine and azithromycin were cured off the virus while only 57.1% of the patients were cured who received chloroquine alone (negative PCR results). These outcomes should be further dwelled on to consider azithromycin as viable adjuvant therapy for SARS-CoV-2 infection. Fascinatingly, azithromycin has been reported to be active against other RNA viruses like Ebola and Zika virus in vitro [118], [119]. Further studies might shed light on its potential against SARS-CoV-2.
Fig. 20.
Antibiotics that are being explored for the treatment of COVID-19.
3.2.3. Carrimycin
Carrimycin, a macrolide antibiotic, is under clinical investigation (NCT04286503) to evaluate its efficacy in treating patients with COVID-19. Carrimycin has been patented for the treatment of mycobacterial infections (European patent EP 3384915 A1). However, after a thorough search, no information could be obtained regarding the structure or mechanism of action of this drug.
3.3. Antiprotozoal drugs
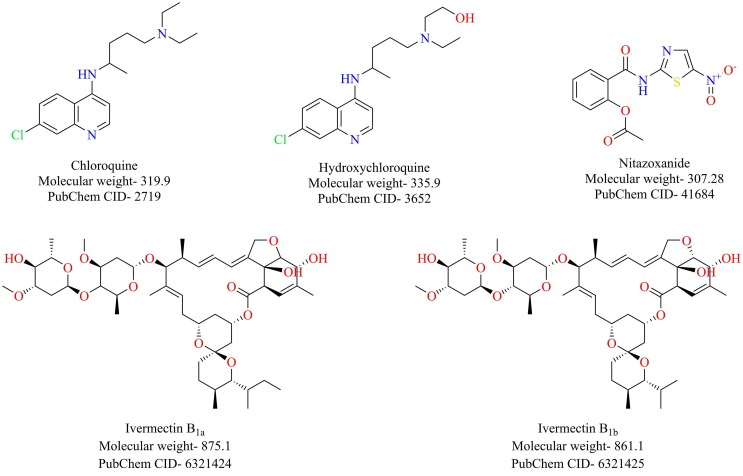
3.3.1. Chloroquine and hydroxychloroquine
Chloroquine and hydroxychloroquine belonging to the class of 4-aminoquinolines are primarily used as antimalarials. Structurally, hydroxychloroquine differs from chloroquine by the presence of one extra hydroxyl group. Besides its antimalarial prowess, chloroquine has established itself as a broad-spectrum antiviral agent exhibiting potent activity against both DNA and RNA viruses [120], [121], [122], [123]. Concerning coronaviruses, chloroquine has been reported to be potent against the SARS-CoV-1 [124]. A recent report by Wang et al. shed light on the antiviral prowess of chloroquine against the SARS-CoV-2. Chloroquine effectively blocked the viral infection with a high selectivity index (EC50 = 1.13 µM) [62]. Gao et al., in a recent publication, disclosed that chloroquine treatment offered superior efficacy than control treatment in a trial that involved a hundred infected patients [125].
Now answering the elephant in the room- Can hydroxychloroquine also be used for the treatment of Covid-19? A recent report demonstrated that hydroxychloroquine is also effective against the SARS-CoV-2, and a study also highlighted that hydroxychloroquine is more potent against SARS-CoV-2 than chloroquine [126], [127]. More arguments favor the use of hydroxychloroquine over chloroquine. Firstly, the presence of an extra hydroxyl group makes it more hydrophilic and hence faster clearance lessening the risk of retinal toxicity than chloroquine. Secondly, hydroxychloroquine is a safer option than chloroquine with respect to therapeutic window and safety margins. Thirdly, owing to its proven immunomodulatory properties, hydroxychloroquine can act as an ideal treatment intervention in severely ill cases where cytokine storm is a significant issue to deal with COVID-19 [128].
Recently, a controversial observational study on the effectiveness of chloroquine or hydroxychloroquine with or without a macrolide for the treatment of COVID-19 was published. The study comprising data from over 600 hospitals in six continents concluded that chloroquine or hydroxychloroquine treatment afforded no protection to COVID-19 patients and was associated with an increased incidence of ventricular arrhythmias. However, serious questions were raised on the validity of the data published in the study. The study was later retracted from the journal as independent auditing of the data was not possible [129]. Reports from clinical trials like solidarity trials, RECOVERY trials have demonstrated that hydroxychloroquine offers no benefit to COVID-19 patients [130], [131]. USFDA has also withdrawn its EUA status given to chloroquine and hydroxychloroquine in view of the recent developments [132].
3.3.1.1. Mechanism of action of chloroquine
Chloroquine is claimed to act via two mechanisms. Chloroquine, in its unprotonated state, spontaneously moves across the cell membranes and gets accumulated in highly acidic vesicles like lysosomes and endosomes and organelles like Golgi, where it gets ionized and raises their pH thereby inactivating several proteolytic enzymes. Evidence suggests that viral entry, replication, and infection requires an acidic environment in the endosome-lysosomes and the catalyzing efficiency of endosomal-lysosomal enzymes. By raising the pH of the vesicles, chloroquine can effectively inhibit the entry and replication process [133].
Chloroquine indirectly interferes with the interaction between the SARS-CoV-1 spike proteins and ACE-2 receptors. Interactions between the glycosylated form of ACE-2 receptor and SARS-CoV-1 S protein is necessary for the viral entry into the cell. Chloroquine impairs the Golgi mediated N-terminal glycosylation of ACE-2, which results in decreased affinity to SARS-CoV-1 S proteins. Hence weak interaction ensues, and the viral entry is prevented [124].
3.3.2. Nitazoxanide
Nitazoxanide, a thiazolide class of drugs, is an antiprotozoal agent used for the treatment of various protozoal and helminthic infections [134]. Apart from its antiprotozoal potential, nitazoxanide has established itself as a broad-spectrum antiviral agent showing potency against both the DNA and RNA viruses [135]. Recently, nitazoxanide was shown to effectively inhibit the SARS-CoV-2 in Vero E6 cells (EC50 = 2.12 µM) [62]. Considering its antiviral property along with its anti-inflammatory potential [136], nitazoxanide could act as a possible therapeutic intervention for COVID-19. However, further studies must be conducted to warrant such a claim.
3.3.3. Ivermectin
Ivermectin (Fig. 21 ) is a broad-spectrum anti-parasitic agent [137]. It has also been reported to possess antiviral activity against viruses such as HIV and dengue [138]. Ivermectin was shown to inhibit the replication of SARS-CoV-2 in vitro [139]. A single treatment of ivermectin at the dose of 5 µM resulted in an about 5000-fold reduction in viral titers following 48 h of ivermectin treatment. The safety profile of ivermectin is well established, and hence ivermectin can be potentially repurposed against COVID-19.
Fig. 21.
Antiprotozoal drugs that are being repurposed for treatment of COVID-19.
3.4. Fusion inhibitors
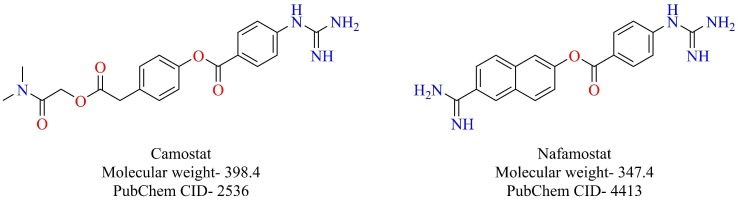
3.4.1. Camostat
As previously mentioned, SARS-CoV-2 also uses the ACE-2 receptor for its attachment and TMPRSS2 for S-protein priming [33]. Camostat, a serine protease inhibitor that has been approved in Japan for the treatment of pancreatitis, was shown to block the cellular entry of SARS-CoV-2 by inhibiting TMPRSS2 [33]. Having already established its safety profile, camostat might prove to be a valuable drug in the arsenal, which can be repurposed to combat COVID-19. Camostat is currently under clinical trials (NCT04338906, NCT04321096) to prove its efficacy for the treatment of COVID-19.
3.4.2. Nafamostat
Following the footsteps of camostat is nafamostat (Fig. 22 ), a serine protease inhibitor used as an anticoagulant [140]. Nafamostat has been shown to inhibit the entry of MERS-CoV by inhibiting cellular TMPRSS2 [141]. Recently, nafamostat was also shown to inhibit the novel SARS-CoV-2 in vitro (EC50 = 22.50 µM) [62]. Scientists from Research Center for Asian Infectious Diseases of the Institute of Medical Science, the University of Tokyo, have disclosed that nafamostat prevents the entry of novel SARS-CoV-2 by potently binding to the transmembrane protease TMPRSS2. Nafamostat was able to achieve the fusion inhibition at less than one-tenth of the concentration required by camostat [142]. Nafamostat can also be repurposed for therapeutic utility against COVID-19.
Fig. 22.
Fusion inhibitors- camostat and nafamostat.
3.5. Immunomodulators and anti-inflammatory drugs
A coordinated effort by the host’s immune system is essential for battle against SARS-CoV-2. However, a dysregulated immune response has been reported in patients with COVID-19. Patients who were admitted to ICU on account of severe COVID-19 were reported to possess higher levels of IL-7, IL-2, IL-10, Granulocyte colony-stimulating factor (GCSF), Monocyte chemoattractant protein-1 (MCP-1), Tumor necrosis factor- α (TNF-α) compared with non-ICU patients suggesting a possible cytokine storm that ultimately culminates in Acute Respiratory Distress Syndrome (ARDS) [143]. Therefore, corticosteroids can be considered as an adjunct treatment option. However, currently, corticosteroid therapy is a topic of debate. Recently, Russel et al. suggested that there is no clinical evidence to support the use of corticosteroids in COVID-19 patients [144]. Notably, in a retrospective study, it was demonstrated that early, low-dose treatment intervention with corticosteroids was associated with quicker improvement of clinical symptoms [145]. Therefore, the use of corticosteroids for patients with COVID-19 is still under debate, and additional evidence is needed to be considered as a treatment intervention. Currently, a clinical trial is underway to evaluate the efficacy of dexamethasone in patients with ARDS secondary to SARS-CoV-2 infection (NCT04325061). Zhou et al. demonstrated that inflammatory and pro-inflammatory mediators like Granulocyte-macrophage colony-stimulating factor (GM-CSF) and IL-6 are highly expressed following viral infection [146]. Therefore, blocking GM-CSF and IL-6 receptors can reduce the inflammatory response. In accordance with this, Mavrilimumab, a monoclonal antibody targeting the GM-CSF receptor, is currently under clinical trials (NCT04337216) to evaluate its efficacy. Clinical trials are also underway to evaluate the safety and efficacy of IL-6 receptor blockers like Sarilumab (NCT04315298), Tocilizumab (NCT04320615, NCT04315480) and Siltuximab (NCT04329650) for treating COVID-19. Baricitinib has been proposed as a potential drug for the treatment of COVID-19 [147]. AP-2 associated protein kinase 1 (AAK1) is a promoter of endocytosis. Baricitinib is known to bind and inhibit AAK1 potently even at therapeutic doses (2 mg or 4 mg) and hence has the potential to prevent the virus entry. Moreover, it is also a known inhibitor of Janus kinases (JAK1 and JAK2), essential enzymes that are involved in signal transduction initiated by cytokines [147]. Therefore, inhibiting JAK enzymes contributes to its anti-inflammatory properties. However, inhibition of JAK1 and JAK2 by baricitinib appears to be a double-edged sword with respect to the treatment of COVID-19 [148]. Interferons are essential soldiers deployed by host’s immune system in response to virus entry. Interferons mediate their antiviral effects via the JAK-STAT pathway. Hence, inhibiting JAK enzymes by baricitinib can inhibit interferon mediated antiviral response and might aid in the replication of SARS-CoV-2 [148]. Currently, baricitinib is under clinical investigation (NCT04321993) as a potential drug for treating SARS-CoV-2 infection.
3.6. Traditional Chinese Medicine (TCM) and natural products
Traditional Chinese Medicine (TCM) has played a substantial role in the treatment and prevention of past epidemics. Past clinical studies have underlined the importance of TCM in the treatment and prevention of SARS and H1N1 influenza [149], [150]. Since there is a high correlation between SARS-CoV and SARS-CoV-2, it was proposed that TCM can also be beneficial for the management of COVID-19 [151]. National Health Commission (NHC) of the People’s Republic of China has recommended the use of TCM for Covid-19 since the fourth version of Diagnosis and Treatment Protocol for Novel Coronavirus Pneumonia [152]. The TCM recommended in the current version (seventh trial version) of Diagnosis and Treatment Protocol for Novel Coronavirus Pneumonia are highlighted in Table 2 [153].
Table 2.
Chinese patent medicines recommended under the diagnosis and treatment protocol for novel Coronavirus Pneumonia (Trial version seven) [153].
| Disease stage | Clinical manifestation | Recommended Chinese patent medicine |
|---|---|---|
| Medical observation | Fatigue and Gastrointestinal discomfort | HuoxiangZhengqi capsules |
| Fatigue and fever | Jinhua Qinggan granules, LianhuaQingwen capsules (granules), ShufengJiedu capsules (granules), FangfengTongsheng pills (granules) | |
| Clinical treatment (confirmed cases) | Severe cases | Xiyanping injection, Xuebijing injection, Reduning injection, Tanreqing injection, Xingnaojing injection |
| Critical cases | Xuebijing injection, Reduning injection, Tanreqing injection, Xingnaojing injection, Shenfu injection, Shengmai injection, Shenmai injection |
Ni et al. reported the treatment of 3 COVID-19 patients in Wuhan with Shuanghuanglian oral liquid (SHL). SHL is a patented TCM that is used for the treatment of cough, cold, and fever. The symptoms of all three patients improved after the consumption of SHL, and the patients finally recovered without any side effects [154]. Currently, it is under clinical investigation (ChiCTR2000029605). Lianhuaqingwen (LH), a TCM formula consisting of 13 herbs, was found to exhibit antiviral activity against the SARS-CoV-2 in vitro [155]. LH inhibited the replication of SARS-CoV-2 with an IC50 value of 411.2 µg/ml. Electron microscopic studies revealed that LH caused viral deformities. Furthermore, LH was also shown to suppress the levels of inflammatory cytokines like TNF-α and IL-6 [155]. Though TCM seems to be a treatment strategy for COVID-19, the safety aspects of TCM needs further warranting. Recently, Gray et al. addressed the potentially harmful effects of TCMs [156]. Although TCMs were found to exhibit potential beneficial effects in clinical trials conducted for SARS-CoV, most of these studies were poorly designed [152], [157]. Hence, double-blinded, randomized, placebo-controlled clinical trials should be conducted to prove safety as well as efficacy aspects of TCMs.
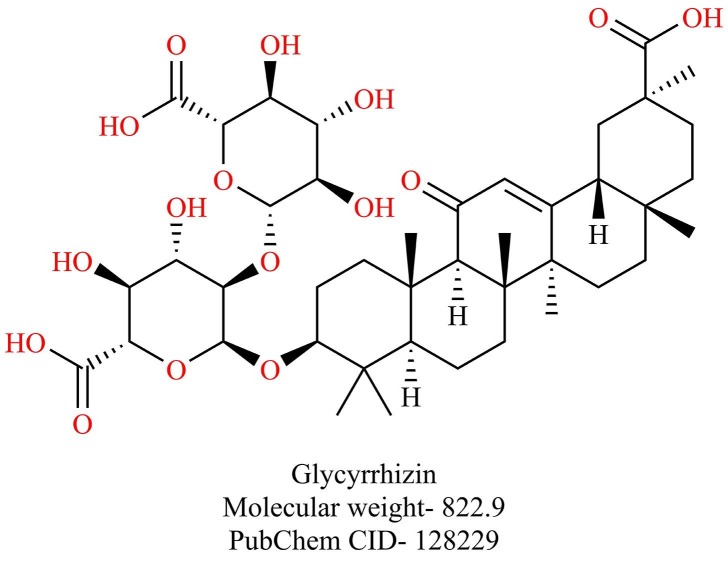
Chen et al. identified five natural compounds- baicalin, hesperetin, scutellarin, nicotianamine, and glycyrrhizin that could potently bind to ACE-2 by molecular docking studies [158]. Incidentally, glycyrrhizin (Fig. 23 ), a triterpene saponin found in liqourice roots, has been previously reported to inhibit the replication of SARS-CoV [159], [160]. A derivative of glycyrrhizin with 2-acetamido-β-d-glucopyranosyl amine into the glycoside chain showed 10-fold increased activity in vitro against SARS-CoV. An amine derivative of glycyrrhizin and the conjugates of glycyrrhizin with two amino acid residues and a free 30-COOH function showed 70-fold increased activity against SARS-CoV. However, these modifications also increased cytotoxicity [161]. Since the safety and toxicity profiles of glycyrrhizin are well established, further studies are needed to establish the efficacy of glycyrrhizin against SARS-CoV-2.
Fig. 23.
Structure of Glycyrrhizin.
4. Conclusion and future perspectives
Extensive efforts are being made across the whole globe to find potential vaccines and therapeutic agents against COVID-19. However, until now, no vaccine or therapeutic agent has shown the results required for the approval for the treatment of COVID-19 or protection against SARS-CoV-2 infections. This review summarizes the potential druggable targets of SARS-CoV-2 for the discovery and development of novel therapeutics against COVID-19 based on the current understanding of molecular structures and functions of the targets as well as an understanding of the pathogenesis of the disease. The treatment approaches that are currently being explored have also been highlighted. The drugs that are currently being explored for the treatment of COVID-19 is summarized in Table 3 .
Table 3.
Summary of the Drugs/investigational agents that are currently being explored for the treatment of COVID-19.
| Compound name | Category | Potency against SARS-CoV-2 (in vitro EC50 or IC50 values in µM) | Mechanism of action | Major Adverse effects | Status | Reference |
|---|---|---|---|---|---|---|
| Remdesivir | Antiviral | 0.77 | RdRp inhibitor | hypersensitivity- infusion-related and anaphylaxis, elevation in liver transaminases | Phase 3 clinical trial (NCT04292899), EAU in the USA | [62], [94] |
| Lopinavir | Antiviral | 26.63 | Mpro inhibitor | Serious cardiotoxicity, lactic acidosis, acute renal failure reported in preterm neonates | Phase 2 clinical trial (NCT04330690) | [162], [163] |
| Favipiravir | Antiviral | 61.8 | RdRp inhibitor | Teratogenic | Phase 3 clinical trials (NCT04336904) | [62], [164] |
| Ribavirin | Antiviral | 106.5 | RdRp inhibitor | Teratogenic, hemolytic anemia | Phase 3 clinical trials (NCT04392427) | [62], [165] |
| Umifenovir | Antiviral | 4.11 | Fusion inhibitor | No major adverse effects have been reported. | Phase 4 clinical trials (NCT04252885) | [166] |
| Oseltamivir | Antiviral | – | – | Steven-Johnson syndrome, anaphylactic reactions, cardiac arrhythmia, seizures have been reported | Phase 3 clinical trials (NCT04338698) | [167] |
| Darunavir | Antiviral | – | Mpro inhibitor | Liver injury, severe skin reactions like erythema multiforme and Steven-Johnson syndrome | Phase 3 clinical trial (NCT04252274) | [168] |
| Teicoplanin | Antibiotic | 1.66 | Cathepsin L inhibitor | Not much information is available on the adverse effects associated with teicoplanin | – | [47] |
| Azithromycin | Antibiotic | – | – | QT interval prolongation, severe hypersensitivity reactions have been reported | Phase 3 clinical trial (NCT04332107) | [169] |
| Carrimycin | Antibiotic | – | – | Not much clinical data is available on Carrimycin | Phase 4 clinical trial (NCT04286503) | |
| Chloroquine | Antiprotozoal | 1.13 | Raises the pH in the endocytic vesicles, interferes with terminal glycosylation of ACE-2 | Prolongation of QT interval, hypoglycemia, neuropsychiatric effects | Phase 3 clinical trial (NCT04303507) | [62], [170] |
| Hydroxychloroquine | Antiprotozoal | 0.72 | Raises the pH in the endocytic vesicles, interferes with terminal glycosylation of ACE-2 | Prolongation of QT interval, hypoglycemia, neuropsychiatric effects | Phase 3 clinical trial (NCT04340544) | [127], [170] |
| Nitazoxanide | Antiprotozoal | 2.12 | – | No major adverse effects have been reported | Phase 2 clinical trial (NCT04423861) | [62] |
| Ivermectin | Antiprotozoal | ~2 | – | No major adverse effects reported | Phase 2 clinical trial (NCT04438850) | [139] |
| Camostat | Antifibrinolytic | – | TMPRSS2 inhibitor | Not much information is available on the adverse events | Phase 2 clinical trial (NCT04374019) | |
| Nafamostat | Anticoagulant | 22.5 | TMPRSS2 inhibitor | Hyperkalemia, agranulocytosis, anaphylaxis | Phase 2 clinical trial (NCT04418128) | [62], [171] |
Infectious diseases resulting from novel pathogens will continue to be a global health concern, further epitomized by COVID-19. The world is still unprepared for such epidemics/pandemics despite former coronaviruses infections SARS-CoV and MERS-CoV. Although vaccines are the only way to prevent/eradicate SARS-CoV-2, at least a year is needed to develop vaccines with desired protection and address their safety issues before they can be deployed for large scale vaccination campaigns. Given the lengthy process associated with drug development, repurposing of approved drugs appears to be an appropriate solution to answer the short-term goals for the management of COVID-19. Only the efficacy of these drugs needs to be established against COVID-19 since their safety profiles are already well established. Many clinical studies are currently underway to repurpose existing drugs for COVID-19 management.
The long-term goals for management of COVID-19 should be directed against the development of novel inhibitors aimed at any of the druggable targets discussed in this review. The starting point of such a journey would be to modify the agents that have been shown to be promising against other coronaviruses like SARS-CoV, as demonstrated by Xia et al. and Zhang et al. in their respective studies [28], [68]. The main protease of SARS-CoV-2 appears to be an attractive target for the discovery and development of novel drug candidates because of its importance in virus replication and the fact that its active site is highly conserved among coronaviruses. Peptidomimetics targeting the entry of the virus and small peptide molecules targeting Mpro have shown the potential to combat SARS-CoV-2. However, questions regarding their stability inside the body, selectivity against the target protease, and their delivery to the target site should be addressed. Structures and functions of RdRp have been extensively investigated for RNA viruses. Considerable efforts have also been made, and several lead compounds have been identified targeting specific RdRp functions through extensive structure-activity-relationship studies. A few of them have been advanced to the clinical use and are clinical evaluation. Current repertoire of lead RdRp inhibitors offer unparalleled starting point for optimization of their efficacy against SARS CoV-2. The inhibitors of RdRp may have low propensity to development of resistance, due to requirement of high fidelity for CoV RdRp functions.
Well-coordinated efforts between various disciplines are required to contain this global pandemic. Clinically proven therapeutics and vaccines are the needs of the hour to curb the growth of this pandemic and ensure global safety.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Authors gratefully acknowledge BITS-Pilani for providing the necessary facilities to do this work. This work was carried out under grants from the Department of Biotechnology Indo-Spain, New Delhi. (Ref. No: BT/IN/Spain/39/SM/2017-2018) and Ministerio de Economía y Competitividad (MINECO, AEI, FEDER, UE) MINECO: AGL2016-79813-C2-1R. BLT is supported by the collaborative Antiviral Drug Discovery and Development Center (AD3C) led by the School of Medicine University of Alabama Birmingham (UAB) (https://www.uab.edu/medicine/ad3c/). The AD3C is one of the Centers of Excellence for Translational Research from the National Institute of Allergy and Infectious Diseases (NIAID), funded by grant U19 AI142759.
References
- 1.Shi Y., Wang Y., Shao C., Huang J., Gan J., Huang X., Bucci E., Piacentini M., Ippolito G., Melino G. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 2020 doi: 10.1038/s41418-020-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shereen M.A., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Y., Gayle A.A., Wilder-Smith A., Rocklöv J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J. Travel Med. 2020;27 doi: 10.1093/jtm/taaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park S.E. Epidemiology, virology, and clinical features of severe acute respiratory syndrome -coronavirus-2 (SARS-CoV-2; Coronavirus Disease-19) Clin. Exp. Pediatr. 2020;63:119–124. doi: 10.3345/cep.2020.00493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adhikari S.P., Meng S., Wu Y.-J., Mao Y.-P., Ye R.-X., Wang Q.-Z., Sun C., Sylvia S., Rozelle S., Raat H., Zhou H. Epidemiology, causes, clinical manifestation and diagnosis, prevention and control of coronavirus disease (COVID-19) during the early outbreak period: a scoping review. Infect. Dis. Poverty. 2020;9:29. doi: 10.1186/s40249-020-00646-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.COVID-19 situation reports, (n.d.). https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/ (accessed April 29, 2020).
- 7.Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020;102433 doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keyhan S.O., Fallahi H.R., Cheshmi B. Dysosmia and dysgeusia due to the 2019 Novel Coronavirus; a hypothesis that needs further investigation. Maxillofac. Plast. Reconstr. Surg. 2020;42:9. doi: 10.1186/s40902-020-00254-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X., Geng M., Peng Y., Meng L., Lu S. Molecular immune pathogenesis and diagnosis of COVID-19. J. Pharm. Anal. 2020 doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., Zheng M., Chen L., Li H. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020 doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.F. Wu, S. Zhao, B. Yu, Y.-M. Chen, W. Wang, Z.-G. Song, Y. Hu, Z.-W. Tao, J.-H. Tian, Y.-Y. Pei, M.-L. Yuan, Y.-L. Zhang, F.-H. Dai, Y. Liu, Q.-M. Wang, J.-J. Zheng, L. Xu, E.C. Holmes, Y.-Z. Zhang, A new coronavirus associated with human respiratory disease in China Supplementary information, (n.d.). doi:10.1038/s41586-020-2008-3.
- 12.Zumla A., Chan J.F.W., Azhar E.I., Hui D.S.C., Yuen K.Y. Coronaviruses-drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cinatl J., Morgenstern B., Bauer G., Chandra P., Rabenau H., Doerr H.W. Treatment of SARS with human interferons. Lancet. 2003;362:293–294. doi: 10.1016/S0140-6736(03)13973-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falzarano D., De Wit E., Martellaro C., Callison J., Munster V.J., Feldmann H. Inhibition of novel β coronavirus replication by a combination of interferon-α2b and ribavirin. Sci. Rep. 2013;3 doi: 10.1038/srep01686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan J.F.W., Chan K.H., Kao R.Y.T., To K.K.W., Zheng B.J., Li C.P.Y., Li P.T.W., Dai J., Mok F.K.Y., Chen H., Hayden F.G., Yuen K.Y. Broad-spectrum antivirals for the emerging Middle East respiratory syndrome coronavirus. J. Infect. 2013;67:606–616. doi: 10.1016/j.jinf.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elshabrawy H.A., Fan J., Haddad C.S., Ratia K., Broder C.C., Caffrey M., Prabhakar B.S. Identification of a broad-spectrum antiviral small molecule against severe acute respiratory syndrome coronavirus and ebola, hendra, and nipah viruses by using a novel high-throughput screening assay. Am Soc Microbiol. 2014 doi: 10.1128/JVI.03050-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dyall J., Coleman C.M., Hart B.J., Venkataraman T., Holbrook M.R., Kindrachuk J., Johnson R.F., Olinger G.G., Jahrling P.B., Laidlaw M., Johansen L.M., Lear-Rooney C.M., Glass P.J., Hensley L.E., Frieman M.B. Repurposing of clinically developed drugs for treatment of middle east respiratory syndrome coronavirus infection. Am Soc Microbiol. 2014 doi: 10.1128/AAC.03036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Huettenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.P., Liu Y.F., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O’Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu D., Wang H.Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d’Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., García-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020:1–13. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from COVID-19 virus and discovery of its inhibitors. Nature. 2020 doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 20.D. Wrapp, N. Wang, K.S. Corbett, J.A. Goldsmith, C.L. Hsieh, O. Abiona, B.S. Graham, J.S. McLellan, Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation, Science (80-.). 367 (2020) 1260–1263. doi:10.1126/science.aax0902. [DOI] [PMC free article] [PubMed]
- 21.Liu Z., Xiao X., Wei X., Li J., Yang J., Tan H., Zhu J., Zhang Q., Wu J., Liu L. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J. Med. Virol. 2020 doi: 10.1002/jmv.25726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xia S., Zhu Y., Liu M., Lan Q., Xu W., Wu Y., Ying T., Liu S., Shi Z., Jiang S., Lu L. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell. Mol. Immunol. 2020:1–3. doi: 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu G., Hu Y., Wang Q., Qi J., Gao F., Li Y., Zhang Y., Zhang W., Yuan Y., Bao J., Zhang B., Shi Y., Yan J., Gao G.F. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013;500:227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.F. Li, W. Li, M. Farzan, S.C. Harrison, Structural biology: Structure of SARS coronavirus spike receptor-binding domain complexed with receptor, Science (80-.). 309 (2005) 1864–1868. doi:10.1126/science.1116480. [DOI] [PubMed]
- 25.Liu S., Xiao G., Chen Y., He Y., Niu J., Escalante C.R., Xiong H., Farmar J., Debnath A.K., Tien P., Jiang S. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: Implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet. 2004;363:938–947. doi: 10.1016/S0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu L., Liu Q., Zhu Y., Chan K.H., Qin L., Li Y., Wang Q., Chan J.F.W., Du L., Yu F., Ma C., Ye S., Yuen K.Y., Zhang R., Jiang S. Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor. Nat. Commun. 2014;5 doi: 10.1038/ncomms4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia S., Yan L., Xu W., Agrawal A.S., Algaissi A., Tseng C.T.K., Wang Q., Du L., Tan W., Wilson I.A., Jiang S., Yang B., Lu L. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci. Adv. 2019;5 doi: 10.1126/sciadv.aav4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Qi F., Bao L., Du L., Liu S., Qin C., Sun F., Shi Z., Zhu Y., Jiang S., Lu L. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.G. Zhang, S. Pomplun, A.R. Loftis, A. Loas, B.L. Pentelute, The first-in-class peptide binder to the SARS-CoV-2 spike protein, BioRxiv. (2020) 2020.03.19.999318. doi:10.1101/2020.03.19.999318.
- 30.Raj V.S., Mou H., Smits S.L., Dekkers D.H.W., Müller M.A., Dijkman R., Muth D., Demmers J.A.A., Zaki A., Fouchier R.A.M., Thiel V., Drosten C., Rottier P.J.M., Osterhaus A.D.M.E., Bosch B.J., Haagmans B.L. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuhn J.H., Li W., Choe H., Farzan M. Angiotensin-converting enzyme 2: A functional receptor for SARS coronavirus. Cell. Mol. Life Sci. 2004;61:2738–2743. doi: 10.1007/s00018-004-4242-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou P., Lou Yang X., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Di Jiang R., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020:1–10. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paz Ocaranza M., Riquelme J.A., García L., Jalil J.E., Chiong M., Santos R.A.S., Lavandero S. Counter-regulatory renin–angiotensin system in cardiovascular disease. Nat. Rev. Cardiol. 2020;17:116–129. doi: 10.1038/s41569-019-0244-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klimas J., Olvedy M., Ochodnicka-Mackovicova K., Kruzliak P., Cacanyiova S., Kristek F., Krenek P., Ochodnicky P. Perinatally administered losartan augments renal ACE2 expression but not cardiac or renal Mas receptor in spontaneously hypertensive rats. J. Cell Mol. Med. 2015;19:1965–1974. doi: 10.1111/jcmm.12573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Furuhashi M., Moniwa N., Mita T., Fuseya T., Ishimura S., Ohno K., Shibata S., Tanaka M., Watanabe Y., Akasaka H., Ohnishi H., Yoshida H., Takizawa H., Saitoh S., Ura N., Shimamoto K., Miura T. Urinary angiotensin-converting enzyme 2 in hypertensive patients may be increased by olmesartan, an angiotensin II receptor blocker. Am. J. Hypertens. 2015;28:15–21. doi: 10.1093/ajh/hpu086. [DOI] [PubMed] [Google Scholar]
- 37.Gurwitz D. Angiotensin receptor blockers as tentative SARS-CoV-2 therapeutics. Drug Dev. Res. 2020:2–5. doi: 10.1002/ddr.21656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang H., Penninger J.M., Li Y., Zhong N., Slutsky A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;2 doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., Jin Q., Wang J., Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 2010;84:12658–12664. doi: 10.1128/jvi.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Glowacka I., Bertram S., Muller M.A., Allen P., Soilleux E., Pfefferle S., Steffen I., Tsegaye T.S., He Y., Gnirss K., Niemeyer D., Schneider H., Drosten C., Pohlmann S. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011;85:4122–4134. doi: 10.1128/jvi.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.S. Rensi, R. B Altman, T. Liu, Y.-C. Lo, G. McInnes, alex derry, alli keys, Homology Modeling of TMPRSS2 Yields Candidate Drugs That May Inhibit Entry of SARS-CoV-2 into Human Cells, ChemRxiv. (n.d.). doi:10.26434/chemrxiv.12009582.
- 43.Simmons G., Gosalia D.N., Rennekamp A.J., Reeves J.D., Diamond S.L., Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. USA. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang I.C., Bosch B.J., Li F., Li W., Kyoung H.L., Ghiran S., Vasilieva N., Dermody T.S., Harrison S.C., Dormitzer P.R., Farzan M., Rottier P.J.M., Choe H. SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells. J. Biol. Chem. 2006;281:3198–3203. doi: 10.1074/jbc.M508381200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simmons G., Zmora P., Gierer S., Heurich A., Pöhlmann S. Proteolytic activation of the SARS-coronavirus spike protein: Cutting enzymes at the cutting edge of antiviral research. Antiviral Res. 2013;100:605–614. doi: 10.1016/j.antiviral.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou N., Pan T., Zhang J., Li Q., Zhang X., Bai C., Huang F., Peng T., Zhang J., Liu C., Tao L., Zhang H. Glycopeptide antibiotics potently inhibit cathepsin l in the late endosome/lysosome and block the entry of ebola virus, middle east respiratory syndrome coronavirus (MERS-CoV), and severe acute respiratory syndrome coronavirus (SARS-CoV) J. Biol. Chem. 2016;291:9218–9232. doi: 10.1074/jbc.M116.716100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.J. Zhang, X. Ma, F. Yu, J. Liu, F. Zou, T. Pan, H. Zhang, Teicoplanin potently blocks the cell entry of 2019-nCoV, BioRxiv. (2020) 2020.02.05.935387. doi:10.1101/2020.02.05.935387.
- 48.Seidah N.G., Chretien M. Proprotein and prohormone convertases: A family of subtilases generating diverse bioactive polypeptides. Brain Res. 1999;848:45–62. doi: 10.1016/S0006-8993(99)01909-5. [DOI] [PubMed] [Google Scholar]
- 49.W. Garten, Characterization of proprotein convertases and their involvement in virus propagation, in: Act. Viruses by Host Proteases, Springer International Publishing, 2018: pp. 205–248. doi:10.1007/978-3-319-75474-1_9.
- 50.Moulard M., Decroly E. Maturation of HIV envelope glycoprotein precursors by cellular endoproteases. Biochim. Biophys. Acta - Rev. Biomembr. 2000;1469:121–132. doi: 10.1016/S0304-4157(00)00014-9. [DOI] [PubMed] [Google Scholar]
- 51.Mille J.K., Whittaker G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. USA. 2014;111:15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Le Coupanec A., Desforges M., Meessen-Pinard M., Dubé M., Day R., Seidah N.G., Talbot P.J. Cleavage of a neuroinvasive human respiratory virus spike glycoprotein by proprotein convertases modulates neurovirulence and virus spread within the central nervous system. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176 doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.D. Bestle, M.R. Heindl, H. Limburg, T.V.L. Van, O. Pilgram, H. Moulton, D.A. Stein, K. Hardes, M. Eickmann, O. Dolnik, C. Rohde, S. Becker, H.-D. Klenk, W. Garten, T. Steinmetzer, E. Böttcher-Friebertshäuser, TMPRSS2 and furin are both essential for proteolytic activation and spread of SARS-CoV-2 in human airway epithelial cells and provide promising drug targets, BioRxiv. (2020) 2020.04.15.042085. doi:10.1101/2020.04.15.042085.
- 57.Elfiky A.A. Zika viral polymerase inhibition using anti-HCV drugs both in market and under clinical trials. J. Med. Virol. 2016;88:2044–2051. doi: 10.1002/jmv.24678. [DOI] [PubMed] [Google Scholar]
- 58.Gordon C.J., Tchesnokov E.P., Feng J.Y., Porter D.P., Gotte M. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 2020 doi: 10.1074/jbc.ac120.013056. jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Subissi L., Posthuma C.C., Collet A., Zevenhoven-Dobbe J.C., Gorbalenya A.E., Decroly E., Snijder E.J., Canard B., Imbert I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. USA. 2014;111:E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morse J.S., Lalonde T., Xu S., Liu W.R. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. ChemBioChem. 2020;21:730–738. doi: 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zumla A., Chan J.F.W., Azhar E.I., Hui D.S.C., Yuen K.Y. Coronaviruses-drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Elfiky A.A. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020;248 doi: 10.1016/j.lfs.2020.117477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.K. Dutta, S. Shityakov, O. Morozova, I. Khalifa, J. Zhang, A. Panda, C. Ghosh, Beclabuvir can Inhibit the RNA-dependent RNA Polymerase of Newly Emerged Novel Coronavirus (SARS-CoV-2), (2020). doi:10.20944/PREPRINTS202003.0395.V1.
- 65.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., Zhu C., Hu T., Hua T., Zhang B., Yang X., Li J., Yang H., Liu Z., Xu W., Guddat L.W., Wang Q., Lou Z., Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020 doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Heo Y.A., Deeks E.D. Sofosbuvir/velpatasvir/voxilaprevir: a review in chronic hepatitis C. Drugs. 2018;78:577–587. doi: 10.1007/s40265-018-0895-5. [DOI] [PubMed] [Google Scholar]
- 67.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.L. Zhang, D. Lin, X. Sun, U. Curth, C. Drosten, L. Sauerhering, S. Becker, K. Rox, R. Hilgenfeld, Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors, Science (80-.). (2020) eabb3405. doi:10.1126/science.abb3405. [DOI] [PMC free article] [PubMed]
- 69.Zhang L., Lin D., Kusov Y., Nian Y., Ma Q., Wang J., Von Brunn A., Leyssen P., Lanko K., Neyts J., De Wilde A., Snijder E.J., Liu H., Hilgenfeld R. α-Ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication: structure-based design, synthesis, and activity assessment. J. Med. Chem. 2020 doi: 10.1021/acs.jmedchem.9b01828. [DOI] [PubMed] [Google Scholar]
- 70.Schrödinger Release 2020-1: Maestro, Schrödinger, LLC, New York, NY, 2020, (n.d.).
- 71.Yang H., Xie W., Xue X., Yang K., Ma J., Liang W., Zhao Q., Zhou Z., Pei D., Ziebuhr J., Hilgenfeld R., Kwok Y.Y., Wong L., Gao G., Chen S., Chen Z., Ma D., Bartlam M., Rao Z. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3 doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xue X., Yu H., Yang H., Xue F., Wu Z., Shen W., Li J., Zhou Z., Ding Y., Zhao Q., Zhang X.C., Liao M., Bartlam M., Rao Z. Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design. J. Virol. 2008;82:2515–2527. doi: 10.1128/jvi.02114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.E. Lynch, J. Kil, Development of ebselen, a glutathione peroxidase mimic, for the prevention and treatment of noise-induced hearing loss, in: Semin. Hear., © Thieme Medical Publishers, 2009: pp. 47–55. doi:10.1055/s-0028-1111106.
- 74.Kil J., Lobarinas E., Spankovich C., Griffiths S.K., Antonelli P.J., Lynch E.D., Le Prell C.G. Safety and efficacy of ebselen for the prevention of noise-induced hearing loss: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2017;390:969–979. doi: 10.1016/S0140-6736(17)31791-9. [DOI] [PubMed] [Google Scholar]
- 75.W. Dai, B. Zhang, X.-M. Jiang, H. Su, J. Li, Y. Zhao, X. Xie, Z. Jin, J. Peng, F. Liu, C. Li, Y. Li, F. Bai, H. Wang, X. Cheng, X. Cen, S. Hu, X. Yang, J. Wang, X. Liu, G. Xiao, H. Jiang, Z. Rao, L. Zhang, Y. Xu, H. Yang, H. Liu, Structure-Based Design, Synthesis and Biological Evaluation of Peptidomimetic Aldehydes as a Novel Series of Antiviral Drug Candidates Targeting the SARS-CoV-2 Main Protease, BioRxiv. (2020) 2020.03.25.996348. doi:10.1101/2020.03.25.996348.
- 76.N. Fintelman-Rodrigues, C.Q. Sacramento, C.R. Lima, F.S. da Silva, A. Ferreira, M. Mattos, C.S. de Freitas, V.C. Soares, S. da S.G. Dias, J.R. Temerozo, M. Miranda, A.R. Matos, F.A. Bozza, N. Carels, C.R. Alves, M.M. Siqueira, P.T. Bozza, T.M.L. Souza, Atazanavir inhibits SARS-CoV-2 replication and pro-inflammatory cytokine production, BioRxiv. (2020) 2020.04.04.020925. doi:10.1101/2020.04.04.020925.
- 77.Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005;79:15189–15198. doi: 10.1128/jvi.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yuan L., Chen Z., Song S., Wang S., Tian C., Xing G., Chen X., Xiao Z.X., He F., Zhang L. P53 degradation by a coronavirus papain-like protease suppresses type I interferon signaling. J. Biol. Chem. 2015;290:3172–3182. doi: 10.1074/jbc.M114.619890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen X., Yang X., Zheng Y., Yang Y., Xing Y., Chen Z. SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING-TRAF3-TBK1 complex. Protein Cell. 2014;5:369–381. doi: 10.1007/s13238-014-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li S.W., Wang C.Y., Jou Y.J., Huang S.H., Hsiao L.H., Wan L., Lin Y.J., Kung S.H., Lin C.W. SARS coronavirus papain-like protease inhibits the TLR7 signaling pathway through removing Lys63-linked polyubiquitination of TRAF3 and TRAF6. Int. J. Mol. Sci. 2016;17:678. doi: 10.3390/ijms17050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Arya R., Das A., Prashar V., Kumar M. Potential inhibitors against papain-like protease of novel coronavirus (SARS-CoV-2) from FDA approved drugs. Chemrxiv. Org. 2020 doi: 10.26434/chemrxiv.11860011.v2. [DOI] [Google Scholar]
- 82.Procainamide | C13H21N3O - PubChem, (n.d.). https://pubchem.ncbi.nlm.nih.gov/compound/Procainamide (accessed June 22, 2020).
- 83.terbinafine | C21H25N - PubChem, (n.d.). https://pubchem.ncbi.nlm.nih.gov/compound/Terbinafine (accessed June 22, 2020).
- 84.Meperidine | C15H21NO2 - PubChem, (n.d.). https://pubchem.ncbi.nlm.nih.gov/compound/Meperidine (accessed June 22, 2020).
- 85.Warren T.K., Jordan R., Lo M.K., Ray A.S., Mackman R.L., Soloveva V., Siegel D., Perron M., Bannister R., Hui H.C., Larson N., Strickley R., Wells J., Stuthman K.S., Van Tongeren S.A., Garza N.L., Donnelly G., Shurtleff A.C., Retterer C.J., Gharaibeh D., Zamani R., Kenny T., Eaton B.P., Grimes E., Welch L.S., Gomba L., Wilhelmsen C.L., Nichols D.K., Nuss J.E., Nagle E.R., Kugelman J.R., Palacios G., Doerffler E., Neville S., Carra E., Clarke M.O., Zhang L., Lew W., Ross B., Wang Q., Chun K., Wolfe L., Babusis D., Park Y., Stray K.M., Trancheva I., Feng J.Y., Barauskas O., Xu Y., Wong P., Braun M.R., Flint M., McMullan L.K., Chen S.S., Fearns R., Swaminathan S., Mayers D.L., Spiropoulou C.F., Lee W.A., Nichol S.T., Cihlar T., Bavari S. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gordon C.J., Tchesnokov E.P., Woolner E., Perry J.K., Feng J.Y., Porter D.P., Gotte M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020 doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lo M.K., Jordan R., Arvey A., Sudhamsu J., Shrivastava-Ranjan P., Hotard A.L., Flint M., McMullan L.K., Siegel D., Clarke M.O., Mackman R.L., Hui H.C., Perron M., Ray A.S., Cihlar T., Nichol S.T., Spiropoulou C.F. GS-5734 and its parent nucleoside analog inhibit Filo-, Pneumo-, and Paramyxoviruses. Sci. Rep. 2017;7 doi: 10.1038/srep43395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.de Wit E., Feldmann F., Cronin J., Jordan R., Okumura A., Thomas T., Scott D., Cihlar T., Feldmann H. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc. Natl. Acad. Sci. USA. 2020;117:6771–6776. doi: 10.1073/pnas.1922083117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sheahan T.P., Sims A.C., Graham R.L., Menachery V.D., Gralinski L.E., Case J.B., Leist S.R., Pyrc K., Feng J.Y., Trantcheva I., Bannister R., Park Y., Babusis D., Clarke M.O., MacKman R.L., Spahn J.E., Palmiotti C.A., Siegel D., Ray A.S., Cihlar T., Jordan R., Denison M.R., Baric R.S. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Brown A.J., Won J.J., Graham R.L., Dinnon K.H., Sims A.C., Feng J.Y., Cihlar T., Denison M.R., Baric R.S., Sheahan T.P. Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic deltacoronaviruses with a highly divergent RNA dependent RNA polymerase. Antiviral Res. 2019;169 doi: 10.1016/j.antiviral.2019.104541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Williamson B.N., Feldmann F., Schwarz B., Meade-White K., Porter D.P., Schulz J., van Doremalen N., Leighton I., Yinda C.K., Pérez-Pérez L., Okumura A., Lovaglio J., Hanley P.W., Saturday G., Bosio C.M., Anzick S., Barbian K., Cihlar T., Martens C., Scott D.P., Munster V.J., de Wit E. Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2. Nature. 2020 doi: 10.1038/s41586-020-2423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., Lopez de Castilla D., Finberg R.W., Dierberg K., Tapson V., Hsieh L., Patterson T.F., Paredes R., Sweeney D.A., Short W.R., Touloumi G., Lye D.C., Ohmagari N., Oh M., Ruiz-Palacios G.M., Benfield T., Fätkenheuer G., Kortepeter M.G., Atmar R.L., Creech C.B., Lundgren J., Babiker A.G., Pett S., Neaton J.D., Burgess T.H., Bonnett T., Green M., Makowski M., Osinusi A., Nayak S., Lane H.C. Remdesivir for the treatment of covid-19 — preliminary report. N. Engl. J. Med. 2020 doi: 10.1056/nejmoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.D. Maffei, Gilead announces results from phase 3 trial of investigational antiviral Remdesivir in patients with Severe COVID-19, Gilead Sci. Inc. (2020) 1. https://www.gilead.com/news-and-press/press-room/press-releases/2020/6/gilead-announces-results-from-phase-3-trial-of-remdesivir-in-patients-with-moderate-covid-19 (accessed June 20, 2020).
- 94.FDA, Fact sheet for health care providers emergency use authorization (EUA) of remdesivir (GS-5734™), Food Drug Adm. (2020) 1–36. https://www.fda.gov/emergency (accessed June 22, 2020).
- 95.C.J. Gordon, E.P. Tchesnokov, J.Y. Feng, D.P. Porter, M. Gotte, The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus., J. Biol. Chem. (2020). doi:10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed]
- 96.Cvetkovic R.S., Goa K.L. Lopinavir/ritonavir: A review of its use in the management of HIV infection. Drugs. 2003;63:769–802. doi: 10.2165/00003495-200363080-00004. [DOI] [PubMed] [Google Scholar]
- 97.Zeldin R.K., Petruschke R.A. Pharmacological and therapeutic properties of ritonavir-boosted protease inhibitor therapy in HIV-infected patients. J. Antimicrob. Chemother. 2004;53:4–9. doi: 10.1093/jac/dkh029. [DOI] [PubMed] [Google Scholar]
- 98.De Wilde A.H., Jochmans D., Posthuma C.C., Zevenhoven-Dobbe J.C., Van Nieuwkoop S., Bestebroer T.M., Van Den Hoogen B.G., Neyts J., Snijder E.J. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob. Agents Chemother. 2014;58:4875–4884. doi: 10.1128/AAC.03011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chan J.F.-W., Yao Y., Yeung M.-L., Deng W., Bao L., Jia L., Li F., Xiao C., Gao H., Yu P., Cai J.-P., Chu H., Zhou J., Chen H., Qin C., Yuen K.-Y. Treatment with lopinavir/ritonavir or interferon-β1b improves outcome of MERS-CoV infection in a nonhuman primate model of common marmoset. J. Infect. Dis. 2015;212:1904–1913. doi: 10.1093/infdis/jiv392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nukoolkarn V., Lee V.S., Malaisree M., Aruksakulwong O., Hannongbua S. Molecular dynamic simulations analysis of ritronavir and lopinavir as SARS-CoV 3CLpro inhibitors. J. Theor. Biol. 2008;254:861–867. doi: 10.1016/j.jtbi.2008.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nutho B., Mahalapbutr P., Hengphasatporn K., Pattaranggoon N.C., Simanon N., Shigeta Y., Hannongbua S., Rungrotmongkol T. Why are lopinavir and ritonavir effective against the newly emerged Coronavirus 2019?: Atomistic insights into the inhibitory mechanisms. Biochemistry. 2020 doi: 10.1021/acs.biochem.0c00160. [DOI] [PubMed] [Google Scholar]
- 102.B. Cao, Y. Wang, D. Wen, W. Liu, J. Wang, G. Fan, L. Ruan, B. Song, Y. Cai, M. Wei, X. Li, J. Xia, N. Chen, J. Xiang, T. Yu, T. Bai, X. Xie, L. Zhang, C. Li, Y. Yuan, H. Chen, H. Li, H. Huang, S. Tu, F. Gong, Y. Liu, Y. Wei, C. Dong, F. Zhou, X. Gu, J. Xu, Z. Liu, Y. Zhang, H. Li, L. Shang, K. Wang, K. Li, X. Zhou, X. Dong, Z. Qu, S. Lu, X. Hu, S. Ruan, S. Luo, J. Wu, L. Peng, F. Cheng, L. Pan, J. Zou, C. Jia, J. Wang, X. Liu, S. Wang, X. Wu, Q. Ge, J. He, H. Zhan, F. Qiu, L. Guo, C. Huang, T. Jaki, F.G. Hayden, P.W. Horby, D. Zhang, C. Wang, A trial of lopinavir-ritonavir in adults hospitalized with severe covid-19, N. Engl. J. Med. (2020) 1–13. doi:10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed]
- 103.Naesens L., Guddat L.W., Keough D.T., Van Kuilenburg A.B.P., Meijer J., Vande Voorde J., Balzarini J. Role of human hypoxanthine guanine phosphoribosyltransferase in activation of the antiviral agent T-705 (favipiravir) Mol. Pharmacol. 2013;84:615–629. doi: 10.1124/mol.113.087247. [DOI] [PubMed] [Google Scholar]
- 104.Oestereich L., Lüdtke A., Wurr S., Rieger T., Muñoz-Fontela C., Günther S. Successful treatment of advanced Ebola virus infection with T-705 (favipiravir) in a small animal model. Antiviral Res. 2014;105:17–21. doi: 10.1016/j.antiviral.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 105.Madelain V., Oestereich L., Graw F., Nguyen T.H.T., De Lamballerie X., Mentré F., Günther S., Guedj J. Ebola virus dynamics in mice treated with favipiravir. Antiviral Res. 2015;123:70–77. doi: 10.1016/j.antiviral.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 106.Bouazza N., Treluyer J.M., Foissac F., Mentré F., Taburet A.M., Guedj J., Anglaret X., De Lamballerie X., Keïta S., Malvy D., Frange P. Favipiravir for children with Ebola. Lancet. 2015;385:603–604. doi: 10.1016/S0140-6736(15)60232-X. [DOI] [PubMed] [Google Scholar]
- 107.D. Sissoko, C. Laouenan, E. Folkesson, A.-B. M’Lebing, A.-H. Beavogui, S. Baize, A.-M. Camara, P. Maes, S. Shepherd, C. Danel, S. Carazo, M.N. Conde, J.-L. Gala, G. Colin, H. Savini, J.A. Bore, F. Le Marcis, F.R. Koundouno, F. Petitjean, M.-C. Lamah, S. Diederich, A. Tounkara, G. Poelart, E. Berbain, J.-M. Dindart, S. Duraffour, A. Lefevre, T. Leno, O. Peyrouset, L. Irenge, N. Bangoura, R. Palich, J. Hinzmann, A. Kraus, T.S. Barry, S. Berette, A. Bongono, M.S. Camara, V. Chanfreau Munoz, L. Doumbouya, Souley Harouna, P.M. Kighoma, F.R. Koundouno, Réné Lolamou, C.M. Loua, V. Massala, K. Moumouni, C. Provost, N. Samake, C. Sekou, A. Soumah, I. Arnould, M.S. Komano, L. Gustin, C. Berutto, D. Camara, F.S. Camara, J. Colpaert, L. Delamou, L. Jansson, E. Kourouma, M. Loua, K. Malme, E. Manfrin, A. Maomou, A. Milinouno, S. Ombelet, A.Y. Sidiboun, I. Verreckt, P. Yombouno, A. Bocquin, C. Carbonnelle, T. Carmoi, P. Frange, S. Mely, V.-K. Nguyen, D. Pannetier, A.-M. Taburet, J.-M. Treluyer, J. Kolie, R. Moh, M.C. Gonzalez, E. Kuisma, B. Liedigk, D. Ngabo, M. Rudolf, R. Thom, R. Kerber, M. Gabriel, A. Di Caro, R. Wölfel, J. Badir, M. Bentahir, Y. Deccache, C. Dumont, J.-F. Durant, K. El Bakkouri, M. Gasasira Uwamahoro, B. Smits, N. Toufik, S. Van Cauwenberghe, K. Ezzedine, E. Dortenzio, L. Pizarro, A. Etienne, J. Guedj, A. Fizet, E. Barte de Sainte Fare, B. Murgue, T. Tran-Minh, C. Rapp, P. Piguet, M. Poncin, B. Draguez, T. Allaford Duverger, S. Barbe, G. Baret, I. Defourny, M. Carroll, H. Raoul, A. Augier, S.P. Eholie, Y. Yazdanpanah, C. Levy-Marchal, A. Antierrens, M. Van Herp, S. Günther, X. de Lamballerie, S. Keïta, F. Mentre, X. Anglaret, D. Malvy, Experimental Treatment with Favipiravir for Ebola Virus Disease (the JIKI Trial): A Historically Controlled, Single-Arm Proof-of-Concept Trial in Guinea, PLOS Med. 13 (2016) e1001967. doi: 10.1371/journal.pmed.1001967. [DOI] [PMC free article] [PubMed]
- 108.Cai Q., Yang M., Liu D., Chen J., Shu D., Xia J., Liao X., Gu Y., Cai Q., Yang Y., Shen C., Li X., Peng L., Huang D., Zhang J., Zhang S., Wang F., Liu J., Chen L., Chen S., Wang Z., Zhang Z., Cao R., Zhong W., Liu Y., Liu L. Experimental treatment with favipiravir for COVID-19: an open-label control study. Engineering. 2020 doi: 10.1016/j.eng.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Du Y.-X., Chen X.-P. Favipiravir: pharmacokinetics and concerns about clinical trials for 2019-nCoV infection. Clin. Pharmacol. Ther. 2020 doi: 10.1002/cpt.1844. [DOI] [PubMed] [Google Scholar]
- 110.Graci J.D., Cameron C.E. Mechanisms of action of ribavirin against distinct viruses. Rev. Med. Virol. 2006;16:37–48. doi: 10.1002/rmv.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Falzarano D., De Wit E., Rasmussen A.L., Feldmann F., Okumura A., Scott D.P., Brining D., Bushmaker T., Martellaro C., Baseler L., Benecke A.G., Katze M.G., Munster V.J., Feldmann H. Treatment with interferon-α2b and ribavirin improves outcome in MERS-CoV-infected rhesus macaques. Nat. Med. 2013;19:1313–1317. doi: 10.1038/nm.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Omrani A.S., Saad M.M., Baig K., Bahloul A., Abdul-Matin M., Alaidaroos A.Y., Almakhlafi G.A., Albarrak M.M., Memish Z.A., Albarrak A.M. Ribavirin and interferon alfa-2a for severe Middle East respiratory syndrome coronavirus infection: A retrospective cohort study. Lancet Infect. Dis. 2014;14:1090–1095. doi: 10.1016/S1473-3099(14)70920-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Arabi Y.M., Shalhoub S., Mandourah Y., Al-Hameed F., Al-Omari A., Al Qasim E., Jose J., Alraddadi B., Almotairi A., Al Khatib K., Abdulmomen A., Qushmaq I., Sindi A.A., Mady A., Solaiman O., Al-Raddadi R., Maghrabi K., Ragab A., Al Mekhlafi G.A., Balkhy H.H., Al Harthy A., Kharaba A., Gramish J.A., Al-Aithan A.M., Al-Dawood A., Merson L., Hayden F.G., Fowler R. Ribavirin and interferon therapy for critically Ill patients with middle east respiratory syndrome: a multicenter observational study. Clin. Infect. Dis. 2019 doi: 10.1093/cid/ciz544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Blaising J., Polyak S.J., Pécheur E.I. Arbidol as a broad-spectrum antiviral: an update. Antiviral Res. 2014;107:84–94. doi: 10.1016/j.antiviral.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bardsley-Elliot A., Noble S. Oseltamivir. Drugs. 1999;58:851–860. doi: 10.2165/00003495-199958050-00007. [DOI] [PubMed] [Google Scholar]
- 116.Capetti A., Cossu M.V., Rizzardini G. Darunavir/cobicistat for the treatment of HIV-1: A new era for compact drugs with high genetic barrier to resistance. Expert Opin. Pharmacother. 2015;16:2689–2702. doi: 10.1517/14656566.2015.1109632. [DOI] [PubMed] [Google Scholar]
- 117.Gautret P., Lagier J.-C., Parola P., Hoang V.T., Meddeb L., Mailhe M., Doudier B., Courjon J., Giordanengo V., Vieira V.E., Dupont T., Honoré S., Colson P., Chabrière E., La Scola B., Rolain J.-M., Brouqui P., Raoult D. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bosseboeuf E., Aubry M., Nhan T., de Pina J.J., Rolain J.M., Raoult D., Musso D. Azithromycin Inhibits the Replication of Zika Virus. J. Antivir. Antiretrovir. 2018;10 doi: 10.4172/1948-5964.1000173. [DOI] [Google Scholar]
- 119.Madrid P.B., Panchal R.G., Warren T.K., Shurtleff A.C., Endsley A.N., Green C.E., Kolokoltsov A., Davey R., Manger I.D., Gilfillan L., Bavari S., Tanga M.J. Evaluation of ebola virus inhibitors for drug repurposing. ACS Infect. Dis. 2016;1:317–326. doi: 10.1021/acsinfecdis.5b00030. [DOI] [PubMed] [Google Scholar]
- 120.Delvecchio R., Higa L.M., Pezzuto P., Valadão A.L., Garcez P.P., Monteiro F.L., Loiola E.C., Dias A.A., Silva F.J.M., Aliota M.T., Caine E.A., Osorio J.E., Bellio M., O’Connor D.H., Rehen S., De Aguiar R.S., Savarino A., Campanati L., Tanuri A. Chloroquine, an endocytosis blocking agent, inhibits zika virus infection in different cell models. Viruses. 2016;8 doi: 10.3390/v8120322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Freiberg A.N., Worthy M.N., Lee B., Holbrook M.R. Combined chloroquine and ribavirin treatment does not prevent death in a hamster model of Nipah and Hendra virus infection. J. Gen. Virol. 2010;91:765–772. doi: 10.1099/vir.0.017269-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.E.A. Kouroumalis, J. Koskinas, Treatment of chronic active hepatitis B (CAH B) with chloroquine: a preliminary report, Ann. Acad. Med. Singapore 15 (1986) 149–152. http://www.ncbi.nlm.nih.gov/pubmed/3752892 (accessed March 31, 2020). [PubMed]
- 123.Koyama A.H., Uchida T. Inhibition of multiplication of herpes simplex virus type 1 by ammonium chloride and chloroquine. Virology. 1984;138:332–335. doi: 10.1016/0042-6822(84)90356-8. [DOI] [PubMed] [Google Scholar]
- 124.Vincent M.J., Bergeron E., Benjannet S., Erickson B.R., Rollin P.E., Ksiazek T.G., Seidah N.G., Nichol S.T. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2005;2 doi: 10.1186/1743-422X-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gao J., Tian Z., Yang X. Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci. Trends. 2020;14:72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- 126.Liu J., Cao R., Xu M., Wang X., Zhang H., Hu H., Li Y., Hu Z., Zhong W., Wang M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020;6:16. doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yao X., Ye F., Zhang M., Cui C., Huang B., Niu P., Liu X., Zhao L., Dong E., Song C., Zhan S., Lu R., Li H., Tan W., Liu D. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Singh A.K., Singh A., Shaikh A., Singh R., Misra A. Chloroquine and hydroxychloroquine in the treatment of COVID-19 with or without diabetes: A systematic search and a narrative review with a special reference to India and other developing countries. Diabetes Metab. Syndr. Clin. Res. Rev. 2020 doi: 10.1016/j.dsx.2020.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mehra M.R., Desai S.S., Ruschitzka F., Patel A.N. Hydroxychloroquine or chloroquine with or without a macrolide for treatment of COVID-19: a multinational registry analysis. Lancet. 2020 doi: 10.1016/S0140-6736(20)31180-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 130.Chief Investigators of the RECOVERY Trial, No clinical benefit from use of hydroxychloroquine in hospitalised patients with COVID-19, Recover. Trial. (2020). https://www.recoverytrial.net/news/statement-from-the-chief-investigators-of-the-randomised-evaluation-of-covid-19-therapy-recovery-trial-on-hydroxychloroquine-5-june-2020-no-clinical-benefit-from-use-of-hydroxychloroquine-in-hospitalised-patients-with-co (accessed June 20, 2020).
- 131.“Solidarity” clinical trial for COVID-19 treatments, (n.d.). https://www.who.int/emergencies/diseases/novel-coronavirus-2019/global-research-on-novel-coronavirus-2019-ncov/solidarity-clinical-trial-for-covid-19-treatments (accessed June 20, 2020).
- 132.Coronavirus (COVID-19) Update: FDA Revokes Emergency Use Authorization for Chloroquine and Hydroxychloroquine | FDA, (n.d.). https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-revokes-emergency-use-authorization-chloroquine-and (accessed June 20, 2020).
- 133.Al-Bari M.A.A. Targeting endosomal acidification by chloroquine analogs as a promising strategy for the treatment of emerging viral diseases. Pharmacol. Res. Perspect. 2017;5 doi: 10.1002/prp2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Fox L.M., Saravolatz L.D. Nitazoxanide: a new thiazolide antiparasitic agent. Clin. Infect. Dis. 2005;40:1173–1180. doi: 10.1086/428839. [DOI] [PubMed] [Google Scholar]
- 135.Rossignol J.F. Nitazoxanide: A first-in-class broad-spectrum antiviral agent. Antiviral Res. 2014;110:94–103. doi: 10.1016/j.antiviral.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rossignol J.F. Nitazoxanide, a new drug candidate for the treatment of Middle East respiratory syndrome coronavirus. J. Infect. Public Health. 2016;9:227–230. doi: 10.1016/j.jiph.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.A.G. Canga, A.M.S. Prieto, M.J. Diez Liébana, N.F. Martínez, M. Sierra Vega, J.J. García Vieitez, The pharmacokinetics and interactions of ivermectin in humans - A mini-review, AAPS J. 10 (2008) 42–46. doi:10.1208/s12248-007-9000-9. [DOI] [PMC free article] [PubMed]
- 138.Wagstaff K.M., Sivakumaran H., Heaton S.M., Harrich D., Jans D.A. Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem. J. 2012;443:851–856. doi: 10.1042/BJ20120150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Caly L., Druce J.D., Catton M.G., Jans D.A., Wagstaff K.M. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res. 2020;178 doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Al-Horani R.A., Desai U.R. Recent advances on plasmin inhibitors for the treatment of fibrinolysis-related disorders. Med. Res. Rev. 2014;34:1168–1216. doi: 10.1002/med.21315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yamamoto M., Matsuyama S., Li X., Takeda M., Kawaguchi Y., Inoue J.I., Matsuda Z. Identification of nafamostat as a potent inhibitor of middle east respiratory syndrome Coronavirus s protein-mediated membrane fusion using the split-protein-based cell-cell fusion assay. Antimicrob. Agents Chemother. 2016;60:6532–6539. doi: 10.1128/AAC.01043-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Identification of an existing Japanese pancreatitis drug, Nafamostat, which is expected to prevent the transmission of new coronavirus infection (COVID-19)|THE INSTITUTE OF MEDICAL SCIENCE, THE UNIVERSITY OF TOKYO, (n.d.). https://www.ims.u-tokyo.ac.jp/imsut/en/about/press/page_00002.html (accessed April 5, 2020).
- 143.D. Tian, C. Qin, C. Qin, L. Zhou, Z. Hu, S. Zhang, S. Yang, Y. Tao, C. Xie, W. Wang, K. Shang, The Lancet Infectious Diseases Dysregulation of immune response in patients with COVID-19 in Wuhan , China, (2020). [DOI] [PMC free article] [PubMed]
- 144.Russell C.D., Millar J.E., Baillie J.K. Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. Lancet. 2020;395:473–475. doi: 10.1016/S0140-6736(20)30317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Y. Wang, W. Jiang, Q. He, C. Wang, B. Wang, P. Zhou, N. Dong, Q. Tong, Early, low-dose and short-term application of corticosteroid treatment in patients with severe COVID-19 pneumonia: single-center experience from Wuhan, China, MedRxiv. (2020) 2020.03.06.20032342. doi:10.1101/2020.03.06.20032342.
- 146.Y. Zhou, B. Fu, X. Zheng, D. Wang, C. Zhao, Y. Qi, R. Sun, Z. Tian, X. Xu, H. Wei, Aberrant pathogenic GM-CSF+ T cells and inflammatory CD14+CD16+ monocytes in severe pulmonary syndrome patients of a new coronavirus, BioRxiv. (2020) 2020.02.12.945576. doi:10.1101/2020.02.12.945576.
- 147.Richardson P., Griffin I., Tucker C., Smith D., Oechsle O., Phelan A., Stebbing J. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet. 2020;395:e30–e31. doi: 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Favalli E.G., Biggioggero M., Maioli G., Caporali R. Baricitinib for COVID-19: a suitable treatment? Lancet Infect. Dis. 2020 doi: 10.1016/s1473-3099(20)30262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.H. Luo, Q. ling Tang, Y. xi Shang, S. bing Liang, M. Yang, N. Robinson, J. ping Liu, Can Chinese Medicine Be Used for Prevention of Corona Virus Disease 2019 (COVID-19)? A Review of Historical Classics, Research Evidence and Current Prevention Programs, Chin. J. Integr. Med. 26 (2020) 243–250. doi:10.1007/s11655-020-3192-6. [DOI] [PMC free article] [PubMed]
- 150.Chen Z., Nakamura T. Statistical evidence for the usefulness of Chinese medicine in the treatment of SARS. Phyther. Res. 2004;18:592–594. doi: 10.1002/ptr.1485. [DOI] [PubMed] [Google Scholar]
- 151.J. ling Ren, A.H. Zhang, X.J. Wang, Traditional Chinese medicine for COVID-19 treatment, Pharmacol. Res. 155 (2020) 104743. doi:10.1016/j.phrs.2020.104743. [DOI] [PMC free article] [PubMed]
- 152.Yang Y., Islam M.S., Wang J., Li Y., Chen X. Traditional Chinese medicine in the treatment of patients infected with 2019-new coronavirus (SARS-CoV-2): a review and perspective. Int. J. Biol. Sci. 2020;16:1708–1717. doi: 10.7150/ijbs.45538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Diagnosis and Treatment Protocol for Novel Coronavirus Pneumonia (Trial Version 6, Revised), n.d. http://en.nhc.gov.cn/2020-03/29/c_78469.htm (accessed April 15, 2020). [DOI] [PMC free article] [PubMed]
- 154.Ni L., Zhou L., Zhou M., Zhao J., Wang D.W. Combination of western medicine and Chinese traditional patent medicine in treating a family case of COVID-19 in Wuhan. Front. Med. 2020 doi: 10.1007/s11684-020-0757-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Runfeng L., Yunlong H., Jicheng H., Weiqi P., Qinhai M., Yongxia S., Chufang L., Jin Z., Zhenhua J., Haiming J., Kui Z., Shuxiang H., Jun D., Xiaobo L., Xiaotao H., Lin W., Nanshan Z., Zifeng Y. Lianhuaqingwen exerts anti-viral and anti-inflammatory activity against novel coronavirus (SARS-CoV-2) Pharmacol. Res. 2020 doi: 10.1016/j.phrs.2020.104761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Gray P.E., Belessis Y. The use of Traditional Chinese Medicines to treat SARS-CoV-2 may cause more harm than good. Pharmacol. Res. 2020 doi: 10.1016/j.phrs.2020.104776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Liu J., Manheimer E., Shi Y., Gluud C. Chinese herbal medicine for severe acute respiratory syndrome: A systematic review and meta-analysis. J. Altern. Complement. Med. 2004;10:1041–1051. doi: 10.1089/acm.2004.10.1041. [DOI] [PubMed] [Google Scholar]
- 158.H. Chen, Q. Du, Potential natural compounds for preventing SARS-CoV-2 (2019-nCoV) infection, Preprints. (2020) 2020010358. doi:10.20944/preprints202001.0358.v3.
- 159.Cinatl J., Morgenstern B., Bauer G., Chandra P., Rabenau H., Doerr H.W. Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet. 2003;361:2045–2046. doi: 10.1016/S0140-6736(03)13615-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Chen F., Chan K.H., Jiang Y., Kao R.Y.T., Lu H.T., Fan K.W., Cheng V.C.C., Tsui W.H.W., Hung I.F.N., Lee T.S.W., Guan Y., Peiris J.S.M., Yuen K.Y. In vitro susceptibility of 10 clinical isolates of SARS coronavirus to selected antiviral compounds. J. Clin. Virol. 2004;31:69–75. doi: 10.1016/j.jcv.2004.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hoever G., Baltina L., Michaelis M., Kondratenko R., Baltina L., Tolstikov G.A., Doerr H.W., Cinatl J. Antiviral activity of glycyrrhizic acid derivatives against SARS-coronavirus. J. Med. Chem. 2005;48:1256–1259. doi: 10.1021/jm0493008. [DOI] [PubMed] [Google Scholar]
- 162.Choy K.T., Wong A.Y.L., Kaewpreedee P., Sia S.F., Chen D., Hui K.P.Y., Chu D.K.W., Chan M.C.W., Cheung P.P.H., Huang X., Peiris M., Yen H.L. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antiviral Res. 2020;178 doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lopinavir | C37H48N4O5 - PubChem, (n.d.). https://pubchem.ncbi.nlm.nih.gov/compound/Lopinavir (accessed June 22, 2020).
- 164.Favipiravir - DrugBank, (n.d.). https://www.drugbank.ca/drugs/DB12466 (accessed June 22, 2020).
- 165.ribavirin | C8H12N4O5 - PubChem, (2017). https://pubchem.ncbi.nlm.nih.gov/compound/Ribavirin (accessed June 22, 2020).
- 166.Wang X., Cao R., Zhang H., Liu J., Xu M., Hu H., Li Y., Zhao L., Li W., Sun X., Yang X., Shi Z., Deng F., Hu Z., Zhong W., Wang M. The anti-influenza virus drug, arbidol is an efficient inhibitor of SARS-CoV-2 in vitro. Cell Discov. 2020;6:1–5. doi: 10.1038/s41421-020-0169-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.C. Summary, Oseltamivir _ C16H28N2O4 - PubChem, 18 (2020). https://pubchem.ncbi.nlm.nih.gov/compound/Oseltamivir#section=Pharmacology (accessed June 22, 2020).
- 168.Darunavir _ C27H37N3O7S - PubChem, (n.d.). https://pubchem.ncbi.nlm.nih.gov/compound/Darunavir#section=Drug-Warnings (accessed June 22, 2020).
- 169.F.S. Structures, Azithromycin _ C38H72N2O12 - PubChem, (2020). https://pubchem.ncbi.nlm.nih.gov/compound/Azithromycin (accessed June 22, 2020).
- 170.Juurlink D.N. Safety considerations with chloroquine, hydroxychloroquine and azithromycin in the management of SARS-CoV-2 infection. CMAJ. 2020;192:E450–E453. doi: 10.1503/cmaj.200528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Nafamostat | C19H17N5O2 - PubChem, (n.d.). https://pubchem.ncbi.nlm.nih.gov/compound/Nafamostat (accessed June 22, 2020).