Summary
Helsmoortel-Van der Aa syndrome (HVDAS) is a neurodevelopmental condition associated with intellectual disability/developmental delay, autism spectrum disorder, and multiple medical comorbidities. HVDAS is caused by mutations in activity-dependent neuroprotective protein (ADNP). A recent study identified genome-wide DNA methylation changes in 22 individuals with HVDAS, adding to the group of neurodevelopmental disorders with an epigenetic signature. This methylation signature segregated those with HVDAS into two groups based on the location of the mutations. Here, we conducted an independent study on 24 individuals with HVDAS and replicated the existence of the two mutation-dependent episignatures. To probe whether the two distinct episignatures correlate with clinical outcomes, we used deep behavioral and neurobiological data from two prospective cohorts of individuals with a genetic diagnosis of HVDAS. We found limited phenotypic differences between the two HVDAS-affected groups and no evidence that individuals with more widespread methylation changes are more severely affected. Moreover, in spite of the methylation changes, we observed no profound alterations in the blood transcriptome of individuals with HVDAS. Our data warrant caution in harnessing methylation signatures in HVDAS as a tool for clinical stratification, at least with regard to behavioral phenotypes.
Keywords: Helsmoortel-Van der Aa syndrome, ADNP, DNA methylation, autism spectrum disorder, intellectual disability, epigenetic signature, neurodevelopmental disorders, episignature, biomarkers, genotype-phenotype correlations
Main Text
Helsmoortel-Van der Aa syndrome (HVDAS) is an autosomal-dominant neurodevelopmental disorder (NDD) caused by de novo mutations in ADNP (MIM: 615873).1 The syndrome was first described in ten individuals by Helsmoortel and colleagues in 2014.1 The clinical presentation included intellectual disability/developmental delay (ID/DD), autism spectrum disorder (ASD), facial dysmorphisms, hypotonia, and congenital heart disease (CHD).1 A recent study collating clinical information from medical records for 78 participants across 16 countries has further expanded the clinical spectrum and demonstrated high prevalence of CHD, visual problems, and gastrointestinal problems.2
ADNP encodes a multi-functional protein harboring eight zinc fingers, eight low complexity regions, a homeobox domain for the binding to the DNA,3 a binding site for the heterochromatin protein 1 (HP1),4 and a nuclear localization sequence3,5 (Supplemental Notes). In mouse embryonic stem (ES) cells, ADNP interacts with the chromatin remodeler CHD4 and the chromatin architectural protein HP1 to form a complex called ChAHP.6,7 The ChAHP complex binds to specific DNA regions and represses gene expression by locally rendering chromatin inaccessible.7 Additionally, the ChAHP modulates the 3D chromatin architecture by antagonizing chromatin looping at CTCF sites.6 Within the ChAHP, ADNP mediates the binding to specific DNA sites.6,7 The genes transcriptionally repressed by the ChAHP in ES cells control cell-fate decisions.7 In fact, Adnp null (Adnp−/−) ES cells undergo spontaneous differentiation, and neuronal progenitors derived from them express mesodermal markers instead of neural markers.7 In line with the critical role in cell-fate specification, Adnp−/− embryos have impaired tissue specification, especially of the neural tube, and die in utero.8,9 Heterozygous mice, on the other hand, are viable but show developmental delays and behavioral deficits.10, 11, 12, 13 Of note, ADNP has been described to have extra-nuclear functions critical for synaptic formation and maturation.12,14
Three recent studies—two on an overlapping sample of 22 individuals with HVDAS15,16 and a third on an additional ten individuals17—reported a genome-wide DNA methylation signature associated with this syndrome. This observation adds to the body of evidence showing characteristic patterns of methylation changes in the peripheral blood of individuals with NDDs caused by mutations in chromatin factors, including Kabuki (MIM: 147920),15,18,19 Claes-Jensen (MIM: 300534),15,20 Sotos (MIM: 117550),15,21 Coffin-Siris (MIM: 614607, 135900, 618027, 617808, 616938, 614609, and 614608),15,22 CHARGE (MIM: 214800),15,19 and Floating-Harbor syndrome (MIM: 136140).15,23 Importantly, these episignatures are syndrome specific and might have diagnostic value in guiding the interpretation of genetic variants with unclear pathogenicity.15,17,18,24 In addition to the signatures found in genetically-defined NDDs, we recently reported an enrichment of epigenetic changes (epimutations) in individuals diagnosed with NDDs and congenital anomalies,25 ASD,26 and schizophrenia,26 suggesting that sporadic epimutations might contribute to the etiology of these disorders. Unlike in other syndromes,15,18, 19, 20, 21, 22, 23 the methylation changes observed in HVDAS clearly stratify mutations into two classes: class I for individuals with mutations located outside a region between nucleotides 2000 and 2340 of the ADNP coding sequence (GenBank: NM_015339.4) and class II for individuals with mutations between nucleotides 2000 and 2340, including the recurrent mutation p.Tyr719∗ (c.2156dupA, c.2157C>A, or c.2157C>G).16 The two episignatures can be harnessed to predict the syndrome and the specific mutational class,15, 16, 17 but whether they correlate with clinical outcomes or gene-expression changes in blood has not been studied.
To address these questions, we studied 43 individuals with a genetic diagnosis of HVDAS from four cohorts: cohort A, collected as part of the Autism Sequencing Consortium;27,28 cohort R, collected at the Medical Genetics Laboratory of Ospedale San Camillo-Forlanini29 or Ospedale Pediatrico Bambino Gesù, Rome, Italy; cohort S, prospectively assessed at the Seaver Autism Center for Research and Treatment, Icahn School of Medicine at Mount Sinai; and cohort W, prospectively assessed at the University of Washington (Supplemental Notes). Mutations in cohort A were identified through research-based whole-exome sequencing27,28 and validated by Sanger sequencing. Mutations in cohorts R, S, and W were identified and validated in Clinical Laboratory Improvement Amendments (CLIA)-certified laboratories. Participation was approved by the institutional review boards of participating sites. All caregivers provided informed written consent and assent was obtained when appropriate. We analyzed 24 samples (cohorts A, R, and part of S) for methylation analyses, prospective phenotype data from 32 individuals (cohorts S and W) for genotype-phenotype analyses, and 17 samples (part of cohort S) for RNA-seq analyses (Supplemental Notes).
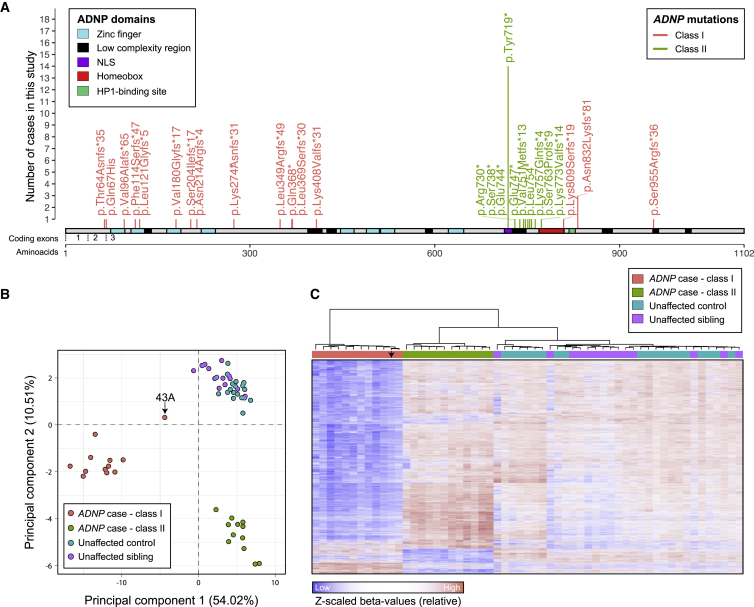
We first performed a genome-wide methylation analysis by using Ilumina EPIC 850K methylation arrays on peripheral blood DNA isolated from 24 individuals with HVDAS, 19 unaffected age-matched controls, and 14 unaffected siblings from cohorts S, A, and R (Supplemental Notes). The 24 HVDAS-affected individuals split evenly into class I and class II mutations (Figure 1A, Table S1). The methylation assays were performed in three batches, each including individuals from multiple cohorts and controls (Table S1, Supplemental Notes). We carefully measured the influence of batch on methylation profiles and modeled batch as a potential confounder (Supplemental Notes, Figure S1). We verified correspondence between nominal and inferred sex and age30 and predicted the fraction of CD4+ T cells, CD8+ T cells, natural killer cells, B lymphocytes, monocytes, and granulocytes in the samples31 (Supplemental Notes). The chronological and inferred epigenetic ages between individuals with class I and class II mutations, unaffected controls, and unaffected siblings were comparable (Supplemental Notes, Figure S2). Subsequently, principal component analysis (PCA) confirmed the clustering of ADNP episignatures into two groups according mutation class (Figure 1B). These two groups were well separated from unaffected age-matched or sibling controls. Specifically, class II mutations were located between nucleotides 2156 and 2317, within the interval defined by Bend and colleagues.16 Eight class II participants harbored the most common variant found in ADNP, p.Tyr719∗ (Figure 1A, Table S1; note that another six participants with p.Tyr719∗ were included in the genotype-phenotype studies). We then performed linear regression using batch, age, sex, predicted blood cell composition, and ADNP mutation status as independent variables; we did this separately for class I and class II mutations. We selected probes associated with disease status at 1% FDR and with ≥10% mean methylation difference between affected individuals and age-matched and unaffected sibling controls (Supplemental Notes).
Figure 1.
Two Distinct Methylation Signatures in Individuals with HVDAS
(A) Lollipop plot of the ADNP mutations in the 42 individuals with point mutations included in the study, including the 24 with methylation data (individual 1S, who carried a deletion, is not shown, Table S1). The amino acid positions are annotated according to ADNP RefSeq protein sequence NP_056154.1. The exon annotation refers to the coding exons in NM_015339.4. The Supplemental Notes contain information on the prediction and re-annotation of linear motifs and structural domains. Mutations in the coding region from nucleotide 2156 to 2317 are classified as class II and are shown in green; mutations outside of this interval are classified as class I and are depicted in pink.
(B) Representation of the two principal dimensions in a principal component analysis of the methylation data from 12 affected individuals in class I, 12 affected individuals in class II, 19 unaffected age-matched controls, and 14 unaffected siblings. The sample indicated by an arrow (43A) harbors the most terminal class I mutation, as shown in (A).
(C) Hierarchical clustering of 12 class I affected individuals, 12 class II affected individuals, 19 unaffected age-matched controls, or 14 unaffected siblings for the 6,448 autosomal CpGs found differentially methylated in individuals with ADNP mutations belonging to class I. The arrow points to data for individual 43A, who carried the most terminal class I mutation, as shown in panel A.
For class I individuals, we identified 6,448 autosomal CpGs that were differentially methylated compared to controls (Figure 1C, Table S2, Figure S3A). Of these differentially methylated sites, 4,143 overlapped with the 5,987 previously identified16 (~69%). Of the 4,143 overlapping CpGs, we observed complete consistency in the direction of effect: 3,974 CpGs hypomethylated in both studies and the remaining hypermethylated in both studies (Figure S3A, Supplemental Notes). The overall difference in methylation between affected individuals and controls for these 4,143 overlapping CpGs was significantly greater compared to unique CpGs from Bend and colleagues16 (Figure S3A). These sites mapped to the gene promoter (defined here as ±2 kb from the transcriptional start sites) and/or gene body (transcription start to transcription end) of 2,802 autosomal RefSeq genes (Table S2). The distribution of these sites on gene promoters or gene body was also comparable to that found by Bend and colleagues16 (Supplemental Notes). Differentially methylated genes in class I were enriched in neuronal and synaptic genes: the top three Gene Ontology (GO) processes at FDR < 0.05 were “neuron cell-cell adhesion” (fold enrichment = 5.59), “receptor localization to synapse” (fold enrichment = 3.83), and “neuronal action potential” (fold enrichment = 3.70). This dataset was also enriched in risk genes for ASD28 (26 genes, p < 0.0001), ID/DD32 (119 genes, p = 4 × 10−4), and CHD (24 genes, p = 0.005) (Supplemental Notes).
For class II individuals, we observed a more modest episignature in line with earlier findings16 with 2,582 differentially methylated autosomal CpGs (Figure 1C, Table S2), 1,007 of which overlapped with the 1,374 previously described16 (~73%) (Figure S3B, Supplemental Notes). Notably, the difference in methylation between affected individuals and controls for these 1,007 overlapping sites was larger than for discordant sites (Figure S3B), as we observed for class I (Figure S3A). Strong and consistent direction of effect was observed for the 1,007 overlapping sites (R2 = 0.91): 771 CpGs were hypermethylated in both studies and the remaining hypomethylated in both studies. Also, the distribution of these sites on gene promoters or gene body was comparable to that found by Bend and colleagues16 (Supplemental Notes). The 1,442 unique RefSeq genes with altered methylation in the promoter and/or gene body were enriched in genes associated with ID/DD32 (69 genes, p = 5 × 10−4) or ASD28 (11 genes, p = 0.013) but not CHD (10 genes, p = 0.13). They were also enriched in neuronal and synaptic genes: the top three GO processes at FDR < 0.05 were “positive regulation of excitatory postsynaptic potential” (fold enrichment = 5.15), “neuromuscular junction development” (fold enrichment = 4.42), and “regulation of synapse assembly” (fold enrichment = 2.95). By selecting clusters comprising three or more differentially methylated CpGs located within 1 kb of each other, we found that class I and class II were associated with 1,398 and 461 differentially methylated regions, respectively (Table S2).
Interestingly, class I and class II episignatures shared 888 differentially methylated probes (1% FDR and with ≥10% mean methylation difference from controls for both classes). However, the direction of change for these CpGs was inversely correlated: there was hypomethylation in class I and hypermethylation in class II (Pearson, R = −0.31, p = 2.2 × 10−16) (Figure S4). This inverse correlation pattern was preserved when we repeated the analyses on the 472 CpG sites shared between the two mutations classes and overlapping with those found by Bend and colleagues16 (Pearson, R = −0.14, p = 0.0019) (Figure S4).
Given the striking divergence between the episignatures in class I and class II (Figures 1B and1C, Figure S4) and the clustering of class II mutations (Figure 1A), we hypothesized that the two types of mutations might differ in their functional impact. Most pathogenic or likely pathogenic mutations in ADNP are nonsense or frameshift mutations. Most mutations, including 40 out of the 43 in this study, fall in exon 5 (coding exon 3) after the last coding exon-exon junction (Figure 1A) and are predicted to escape nonsense-mediated decay (NMD). A previous study has in fact shown that mutant RNAs can be detected in the blood of individuals with HVDAS1 and that mutant ADNP ectopically expressed in HEK293T cells is translated into truncated proteins that can undergo proteasome-mediated degradation or mislocalize in the cytoplasm.5 We therefore asked whether class I and class II mutations located in exon 5 (coding exon 3) differ in NMD escape. To test this hypothesis, we performed Sanger sequencing on the cDNA amplified from RNA isolated from peripheral blood of three individuals with class I mutations [p.Phe114Serfs∗47 (c.339delC), p.Leu349Argfs∗49 (c.1046_1047delTG), and p.Leu369Serfs∗30 (c.1106_1108delTACinsCTGT)] and two individuals with class II mutations [p.Tyr719∗ (c.2157C>A) and p.Glu747∗ (c.2239G>T)] (Table S1, Figure S5A). In all samples examined, we observed bi-allelic expression of ADNP (i.e., both reference and mutant alleles were expressed), confirming that mutant ADNP alleles escape NMD (Figure S5A). We observed no differences between class I and class II mutations, indicating that a differential NMD escape is most likely not the mechanism underlying the divergent episignature. We replicated this observation by examining mutation-mapping reads in genome-wide RNA-seq from 17 individuals with HVDAS (Table S1, Figures S5B–S5D). Notably, all mutations had non-zero RNA-seq read coverage and the ADNP locus was expressed at low abundance (fragments per kilobase million [FPKM], 3.03 ± 0.44). When we compared the overall expression of mutant alleles to the expression of reference alleles, we observed lower expression among the mutant reads on average across all samples (p value for difference in log median expression = 0.013; Figure S5D). Our data, alongside previous evidence,1 indicate that the ADNP pathogenic mutations are not leading to classical haploinsufficiency but rather to the expression of mutant proteins that might or might not gain additional functions. Interestingly, one of our affected participants (1S) harbored a deletion extending from the 5′UTR to the second coding exon, suggesting that class I mutations are more likely resulting from a loss of function. Also, one class I participant showed attenuated genome-wide methylation changes (43A; Figures 1B and 1C). Notably, this individual carries the most distal frameshift mutation (Table S1, Figure 1A). Similarly, Bend and colleagues16 found an individual with a p.Tyr780* (c.2340T>G) mutation having more modest changes compared to other class II mutations. These independent observations suggest that more terminally truncated ADNP mutant proteins might retain partial function associated with an attenuated epigenetic signature. The parsimonious hypothesis that individuals with mild epigenetic signatures (i.e., methylation similar to controls) have milder phenotypic severity remains to be tested.
Genotype-phenotype correlations have begun to emerge in HVDAS. Previous retrospective review of the medical records of 78 participants reported that individuals with p.Tyr719∗ have a higher rate of high pain thresholds and walked significantly later than individuals with other types of ADNP mutations.2 Individuals with p.Tyr719∗ or adjacent variants might also have a higher risk of blepharophimosis.29,33,34 An initial analysis of Bend and colleagues16 on three class I and six class II individuals did not identify differences between the two groups, although the analysis had minimal power. To further probe potential phenotypic differences and relate them to epigenetic changes, we used extensive, prospectively collected clinical data for two cohorts, cohorts S (class I n = 10, class II n = 12) and W (n = 5/class) (Supplemental Notes). Given the profound methylation changes in the blood of individuals with class I mutations, we initially conjectured that individuals with class I mutations would have more severe clinical manifestations. To test this hypothesis, we contrasted data for 70 variables in the cohorts S and W separately and combined them by calculating individual effect sizes and both unadjusted and Bonferroni-corrected p values (Table 1, Table S3). Although we did not perform methylation analysis on cohort W, we classified samples of cohort W in class I and class II on the basis of the mutation they presented, as supported by the 100% accuracy of the two episignatures in predicting class mutation.16 Remarkably, both groups displayed similar frequency of ID, language impairment, ADHD diagnoses, and medical problems. However, we replicated earlier findings2 of a larger delay in first walking independently in class II individuals (class I = 22 ± 2.67 months, class II = 36.33 ± 6.45 months, d = 2.90, adjusted p = 1.4 × 10−5) (Table 1). We also observed some evidence for a higher prevalence and severity of ASD in class II (class I = 4/15, class II = 12/17, d = 1.02) (Table 1). There was also a considerable effect size for rate of self-injurious behavior in class II (d = 0.77), but this failed to reach statistical significance (Table S3). Importantly, these differences also hold up when we considered class II without the 11 individuals with a p.Tyr719∗ variant, indicating that p.Tyr719∗ is not acting as an outlier that skews the results. Overall, the striking divergent methylation patterns in class I and class II individuals did not translate into robust phenotypic differences between the two groups, but there is evidence for subtler associations.
Table 1.
Phenotypic Comparison between the Two HVDAS Groups
| Table 1 | Class I (n = 15) | Class II (n = 17) | Effect | p Value | Adj. p Value |
|---|---|---|---|---|---|
| Sex | 10 females, 5 males | 5 females, 12 males | 0.37 | 0.080 | 1 |
| Age (years) | 7.33 ± 3.33 | 7.18 ± 4.56 | −0.04 | 0.910 | 1 |
| Autism spectrum disorder | 4 | 12 | −0.48 | 0.020 | 1 |
| ASD severity | 4.27 ± 1.83 | 6.4 ± 2.32 | 1.02 | 0.010 | 0.70 |
| Full scale IQ or developmental quotient (DQ) | 36.23 ± 13.82 | 35.72 ± 17.55 | −0.03 | 0.930 | 1 |
| Nonverbal IQ or DQ | 9 | 13 | −0.23 | 0.365 | 1 |
| Verbal IQ or DQ | 53 ± 13.11 | 48.59 ± 12.86 | −0.34 | 0.350 | 1 |
| Motor delays | 14 | 17 | −0.19 | 0.949 | 1 |
| Gait abnormalities | 13 | 15 | −0.27 | 0.464 | 1 |
| First walked independently | 22 ± 2.67 | 36.33 ± 6.45 | 2.90 | 2.1e−07 | 1.4e−5 |
| Language delay | 15 | 17 | 0.00 | 1.000 | 1 |
| Receptive vocabulary (standard score) | 60.5 ± 14.7 | 45.75 ± 20.41 | −0.83 | 0.060 | 1 |
| Expressive vocabulary (standard score) | 53.25 ± 14.9 | 45.14 ± 18.33 | −0.49 | 0.370 | 1 |
| Nonverbal or minimally verbal | 37.68 ± 14.34 | 37.97 ± 19.04 | 0.02 | 0.960 | 1 |
| First phrase | 80 ± 23.51 | 60 ± 15.74 | −1.00 | 0.094 | 1 |
| First single word | 38.71 ± 20.53 | 33.6 ± 12.47 | −0.30 | 0.457 | 1 |
| Intellectual disability | 15 | 16 | 0.00 | 1.000 | 1 |
| Attention deficit/hyperactivity disorder | 6 | 6 | 0.03 | 1.000 | 1 |
| Visual motor integration (standard score) | 58.17 ± 15.99 | 48.7 ± 11.24 | −0.69 | 0.240 | 1 |
| Vision problems | 10 | 16 | −0.35 | 0.126 | 1 |
| Vineland adaptive behavior composite | 35.92 ± 14.53 | 34.33 ± 17.97 | −0.10 | 0.800 | 1 |
| Sensory symptoms | 15 | 17 | 0.00 | 1.000 | 1 |
| High pain threshold | 10 | 15 | −0.3 | 0.252 | 1 |
| Seizures | 3 | 7 | −0.23 | 0.364 | 1 |
| GI problems | 11 | 14 | −0.11 | 0.851 | 1 |
| Hypothyroidism | 4 | 2 | 0.18 | 0.587 | 1 |
| Hypotonia | 11 | 15 | 0.01 | 1.000 | 1 |
| Sleep disturbance | 11 | 12 | −0.02 | 1.000 | 1 |
| Obstructive sleep apnea | 5 | 1 | 0.44 | 0.185 | 1 |
| Congenital heart defect | 7 | 5 | 0.18 | 0.522 | 1 |
| Early tooth eruption | 8 | 7 | 0.27 | 0.388 | 1 |
| Ptosis | 1 | 3 | −0.22 | 0.534 | 1 |
| Hearing problems | 6 | 3 | 0.25 | 0.313 | 1 |
| Neonatal intensive care unit stay at birth | 6 | 8 | −0.07 | 0.964 | 1 |
| Recurrent infections | 6 | 7 | −0.01 | 1.000 | 1 |
| Feeding issues | 3 | 9 | −0.34 | 0.120 | 1 |
| Structural brain changes | 3 | 7 | −0.3 | 0.369 | 1 |
Comparison between clinical measures collected prospectively from 15 class I and 17 class II HVDAS-affected individuals in cohorts S and W. Table S3 presents the analysis of additional measures, broken down by cohort. The Supplemental Notes describe all the measures listed (e.g., ASD severity refers to Comparison Score of the Autism Diagnostic Observation Schedule, 2nd Edition). The Shapiro-Wilk test was used to assess normality of all continuous variables, and either a one-way analysis of variance (ANOVA) or Wilcoxon rank sum test was implemented accordingly. Chi-square tests with Yates correction were used to test discrete variables. We adjusted all tests for multiple testing by using the Bonferroni method to control the false discovery rate. Effect sizes were estimated for discrete measures with the phi coefficient and for continuous measures with Cohen’s d.
We next asked whether changes at the gene expression level could be predicted on the basis of differentially methylated CpGs and also cluster HVDAS-affected individuals from controls. PCA was performed on RNA-seq data from 17 individuals with HVDAS and 19 unaffected siblings via genes harboring differentially methylated CpGs within their respective promoters; no distinct stratification was observed between samples of HVDAS-affected individuals and unaffected siblings (Figure S6A). Subsequently, differential gene expression analysis tested for genes that were over- or under-expressed in affected individuals relative to unaffected siblings and identified two genes associated with class I mutations and nine genes associated with class II mutations under an FDR < 0.05; each had small individual effect sizes (Table S2). A competitive gene-set ranking approach was used to functionally annotate genome-wide trends in gene expression. First, we integrated differentially methylated CpGs from gene promoters and observed a poor consistency between significantly hypermethylated and hypomethylated CpGs relative to genes that are under-expressed and over-expressed in HVDAS-affected individuals, respectively (Figures S6B and S6C). Second, we performed GO enrichment analysis by using our previously identified biological processes found to be significantly enriched for differentially methylated CpGs and confirmed a significant overexpression of genes mapping to gated and ion channel activity (Figure S6D). Notably, there was no significant enrichment for ASD, ID/DD, or CHD genes despite a trend of these genes to be overexpressed in affected individuals. Finally, exploratory GO enrichment analysis was performed and significant enrichment for ephrin, SMAD, and PTK signaling was observed in individuals with class I mutations. Processes related to DNA methylation and replication were enriched in individuals with class II mutations (Figures S6D and S6E). Overall, despite the critical role of ADNP on chromatin accessibility and architecture6,7 and the methylation changes found in individuals with pathogenic ADNP mutations, we did not observe profound alterations in corresponding gene expression profiles.
Here, we replicated and expanded earlier findings15, 16, 17 of DNA methylation changes in HVDAS-affected individuals. The mechanisms leading to these methylation changes are still unknown. Although initial observations suggested an association of ADNP with subunits of the chromatin remodeling complex called SWI/SNF,9 a recent study failed to replicate this finding in mouse ES cells7 and found instead that ADNP interacts with CHD4 and HP1 to form a complex (ChAHP) that restricts local chromatin accessibility7 and controls chromatin looping.6 Defective ChAHP complexes formed by mutant ADNP might potentially change accessibility for DNA methyltransferases and/or demethylases and explain the methylation changes found here. It is still puzzling, however, why mutations affecting residues 719–773 (or 719–78015,16) have a different effect than all other mutations. In fact, even within the groups of 888 differentially methylated CpGs shared between the two groups, the direction of change is opposite: there was more hypomethylation in class I and more hypermethylation in class II. This indicates that the two groups of mutant ADNP affect DNA methylation via different mechanisms. The first important observation is that even frameshift and nonsense mutations do not lead to NMD1 (Figure S4), suggesting that these mutations lead to the expression of truncated proteins. These truncated proteins might act as hypomorphic or gain-of-function alleles. The fact that an individual carrying a deletion extending from the 5′UTR to coding exon 2 clusters with class I mutations (1S) and that the most terminal class I mutation has an attenuated signature [p.Ser955Argfs∗36 (c.2865_2868delTGAG) in 43A] would suggest that this group of mutations might act as hypomorphic alleles. Experiments introducing GFP-tagged ADNP mutants in HEK293T cells have suggested a distinct pattern of expression and localization based on the location of the mutation, dividing the mutations into N-terminal mutations (in the first 412 residues), central mutations (in the 473–719 region), and C-terminal mutations (after 719).5 N-terminal mutants were targeted to proteasomal degradation, expression of central mutations was restricted to the cytoplasm, and expression of C-terminal mutants were expressed in the nucleus but showed reduced co-localization with pericentromeric heterochromatin.5 These data would suggest that N-terminal and C-terminal mutations reduce protein activity, whereas mutations in the central domain are more detrimental. Although our study and Bend et al. (2019)16 do not include mutations in the 473–718 range, the study by Aref-Eshghi et al. (2020)17 includes the p.Ala430Cysfs∗10 (c.1287dupT) and p.Cys627Gly variants, both of which are associated with class I episignature. Overall, the methylation data support a model whereby mutations affecting residues 719–780 (and not residues 473–719) behave differently from all others. Also, independent evidence in mouse ES cells homozygous for p.Tyr718∗ (corresponding to human p.Tyr719∗) show that the mutant ADNP reaches the nucleus.7 Interestingly, this mutant ADNP binds CHD4 and DNA, despite the loss of the homeobox, but cannot interact with HP1.7 Since HP1 mediates the repression activity of the ChAHP complexes, ADNP target genes are overexpressed in the p.Tyr718∗ mutant ES cells.7 Although the analyses from Cappuyins et al.5 and Ostapcuk et al.7 begin to explore the functional impact of different ADNP mutations, relevance to the clinical syndrome is complicated by the use of overexpression systems5 or the use of homozygous targeting.7 Future studies on patient-derived cells, as well as advances in solving 3D structure of ADNP, would be important to better dissect the differential impact of the clinical mutations.
Prior analyses on three class I and six class II individuals failed to detect phenotypic differences between the two groups.16 Our prospectively collected clinical data on 15 class I and 17 class II individuals show that the two mutational groups differ in the mean age of first walking and rates of ASD (Table 1): both were more severe in the class with less profound methylation changes. The lack of correspondence between molecular signatures, including methylation and gene expression, and clinical manifestations caution against making phenotypic inferences on the basis of the blood-based methylation profile. This is important to consider when evaluating the use of these episignatures as biomarkers for patient stratification and/or response to pharmacological agents in clinical trials. In the specific case of HVDAS, these observations are timely as potential experimental therapeutics emerge. In particular, pre-clinical data suggest that an eight-amino-acid ADNP peptide called NAP (NAPVSIPQ) might hold promises for treatment. NAP has been shown to have broad neuroprotective effects in vitro and in vivo,35 and in vivo administration of NAP or its derivatives in a mouse model of HVDAS has been shown to ameliorate synaptic and behavioral defects.10, 11, 12, 13 In conclusion, although the episignatures in HVDAS have proven very valuable for the purpose of complementing clinical genetics and enhancing accuracy of diagnosis, their value as biomarkers for clinical stratification in clinical trials should be carefully evaluated.
Data and Code Availability
The methylation and RNA sequencing data are available at the Gene Expression Omnibus under accession number GEO: GSE152428 (GEO: GSE152427 for the methylation data and GEO: GSE152421 for the RNA sequencing data).
Declaration of Interests
S.D.R., P.M.S., J.D.B., and A.K. are supported by the ADNP Kids Research Foundation. A.K. receives research support from AMO Pharma and is on the scientific advisory board of Ovid Therapeutics. A.K. has also consulted to Takeda, Acadia, and Sema4 in the past year. M.P. has consulted for Servier and Exeltis. C.M. has acted as consultant or participated in the Data Monitoring Committee for Janssen, Servier, Lundbeck, Nuvelution, Angelini, and Otsuka and has received grant support from European Union Funds and Instituto de Salud Carlos III, Spanish Ministry of Economy and Competitiveness. All other authors declare no competing interests.
Acknowledgments
This work was supported by grants from the Beatrice and Samuel A. Seaver Foundation and the ADNP Kids Research Foundation. M.S.B. is a Seaver Foundation Faculty Scholar. S.D.R. is a Fascitelli Research Scholar. The collection of samples in cohort A and some of the molecular and analytical studies were supported by the NIMH (MH111661). The collection and processing of samples, confirmation of diagnoses, and management of data for part of cohort A were also funded by the NIH (R01ES015359, P01ES011269, and UH3OD023365) and the EPA (STAR R829388, R833292, and RD83543201). The work was also supported by the Spanish Ministry of Science and Innovation, Instituto de Salud Carlos III, and CIBERSAM and co-financed by ERDF funds and European Union Seventh Framework Program, the European Union H2020 Program under the Innovative Medicines Initiative 2 Joint Undertaking (grant agreement 777394, project AIMS-2-TRIALS), and Fundación Familia Alonso. The work was partially supported by funding to D.D.M. by Fondazione Marche. Research reported in this paper was supported by the Office of Research Infrastructure of the National Institutes of Health under award number S10OD018522. This work was supported in part through the computational resources and staff expertise provide by Scientific Computing at the Icahn School of Medicine at Mount Sinai. We are grateful to all the families and individuals who participated in this research.
Published: August 5, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.07.003.
Contributor Information
Paige M. Siper, Email: paige.siper@mssm.edu.
Silvia De Rubeis, Email: silvia.derubeis@mssm.edu.
Web Resources
Online Mendelian Inheritance in Man, https://www.omim.org
Supplemental Data
References
- 1.Helsmoortel C., Vulto-van Silfhout A.T., Coe B.P., Vandeweyer G., Rooms L., van den Ende J., Schuurs-Hoeijmakers J.H., Marcelis C.L., Willemsen M.H., Vissers L.E. A SWI/SNF-related autism syndrome caused by de novo mutations in ADNP. Nat. Genet. 2014;46:380–384. doi: 10.1038/ng.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Dijck A., Vulto-van Silfhout A.T., Cappuyns E., van der Werf I.M., Mancini G.M., Tzschach A., Bernier R., Gozes I., Eichler E.E., Romano C., ADNP Consortium Clinical Presentation of a Complex Neurodevelopmental Disorder Caused by Mutations in ADNP. Biol. Psychiatry. 2019;85:287–297. doi: 10.1016/j.biopsych.2018.02.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zamostiano R., Pinhasov A., Gelber E., Steingart R.A., Seroussi E., Giladi E., Bassan M., Wollman Y., Eyre H.J., Mulley J.C. Cloning and characterization of the human activity-dependent neuroprotective protein. J. Biol. Chem. 2001;276:708–714. doi: 10.1074/jbc.M007416200. [DOI] [PubMed] [Google Scholar]
- 4.Mandel S., Rechavi G., Gozes I. Activity-dependent neuroprotective protein (ADNP) differentially interacts with chromatin to regulate genes essential for embryogenesis. Dev. Biol. 2007;303:814–824. doi: 10.1016/j.ydbio.2006.11.039. [DOI] [PubMed] [Google Scholar]
- 5.Cappuyns E., Huyghebaert J., Vandeweyer G., Kooy R.F. Mutations in ADNP affect expression and subcellular localization of the protein. Cell Cycle. 2018;17:1068–1075. doi: 10.1080/15384101.2018.1471313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaaij L.J.T., Mohn F., van der Weide R.H., de Wit E., Buhler M. The ChAHP Complex Counteracts Chromatin Looping at CTCF Sites that Emerged from SINE Expansions in Mouse. Cell. 2019;178:1437–1451.e14. doi: 10.1016/j.cell.2019.08.007. [DOI] [PubMed] [Google Scholar]
- 7.Ostapcuk V., Mohn F., Carl S.H., Basters A., Hess D., Iesmantavicius V., Lampersberger L., Flemr M., Pandey A., Thomä N.H. Activity-dependent neuroprotective protein recruits HP1 and CHD4 to control lineage-specifying genes. Nature. 2018;557:739–743. doi: 10.1038/s41586-018-0153-8. [DOI] [PubMed] [Google Scholar]
- 8.Pinhasov A., Mandel S., Torchinsky A., Giladi E., Pittel Z., Goldsweig A.M., Servoss S.J., Brenneman D.E., Gozes I. Activity-dependent neuroprotective protein: a novel gene essential for brain formation. Brain Res. Dev. Brain Res. 2003;144:83–90. doi: 10.1016/s0165-3806(03)00162-7. [DOI] [PubMed] [Google Scholar]
- 9.Mandel S., Gozes I. Activity-dependent neuroprotective protein constitutes a novel element in the SWI/SNF chromatin remodeling complex. J. Biol. Chem. 2007;282:34448–34456. doi: 10.1074/jbc.M704756200. [DOI] [PubMed] [Google Scholar]
- 10.Vulih-Shultzman I., Pinhasov A., Mandel S., Grigoriadis N., Touloumi O., Pittel Z., Gozes I. Activity-dependent neuroprotective protein snippet NAP reduces tau hyperphosphorylation and enhances learning in a novel transgenic mouse model. J. Pharmacol. Exp. Ther. 2007;323:438–449. doi: 10.1124/jpet.107.129551. [DOI] [PubMed] [Google Scholar]
- 11.Amram N., Hacohen-Kleiman G., Sragovich S., Malishkevich A., Katz J., Touloumi O., Lagoudaki R., Grigoriadis N.C., Giladi E., Yeheskel A. Sexual divergence in microtubule function: the novel intranasal microtubule targeting SKIP normalizes axonal transport and enhances memory. Mol. Psychiatry. 2016;21:1467–1476. doi: 10.1038/mp.2015.208. [DOI] [PubMed] [Google Scholar]
- 12.Hacohen-Kleiman G., Sragovich S., Karmon G., Gao A.Y.L., Grigg I., Pasmanik-Chor M., Le A., Korenková V., McKinney R.A., Gozes I. Activity-dependent neuroprotective protein deficiency models synaptic and developmental phenotypes of autism-like syndrome. J. Clin. Invest. 2018;128:4956–4969. doi: 10.1172/JCI98199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sragovich S., Malishkevich A., Piontkewitz Y., Giladi E., Touloumi O., Lagoudaki R., Grigoriadis N., Gozes I. The autism/neuroprotection-linked ADNP/NAP regulate the excitatory glutamatergic synapse. Transl. Psychiatry. 2019;9:2. doi: 10.1038/s41398-018-0357-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oz S., Kapitansky O., Ivashco-Pachima Y., Malishkevich A., Giladi E., Skalka N., Rosin-Arbesfeld R., Mittelman L., Segev O., Hirsch J.A., Gozes I. The NAP motif of activity-dependent neuroprotective protein (ADNP) regulates dendritic spines through microtubule end binding proteins. Mol. Psychiatry. 2014;19:1115–1124. doi: 10.1038/mp.2014.97. [DOI] [PubMed] [Google Scholar]
- 15.Aref-Eshghi E., Bend E.G., Colaiacovo S., Caudle M., Chakrabarti R., Napier M., Brick L., Brady L., Carere D.A., Levy M.A. Diagnostic Utility of Genome-wide DNA Methylation Testing in Genetically Unsolved Individuals with Suspected Hereditary Conditions. Am. J. Hum. Genet. 2019;104:685–700. doi: 10.1016/j.ajhg.2019.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bend E.G., Aref-Eshghi E., Everman D.B., Rogers R.C., Cathey S.S., Prijoles E.J., Lyons M.J., Davis H., Clarkson K., Gripp K.W. Gene domain-specific DNA methylation episignatures highlight distinct molecular entities of ADNP syndrome. Clin. Epigenetics. 2019;11:64. doi: 10.1186/s13148-019-0658-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aref-Eshghi E., Kerkhof J., Pedro V.P., Barat-Houari M., Ruiz-Pallares N., Andrau J.C., Lacombe D., Van-Gils J., Fergelot P., Dubourg C., Groupe DI France Evaluation of DNA Methylation Episignatures for Diagnosis and Phenotype Correlations in 42 Mendelian Neurodevelopmental Disorders. Am. J. Hum. Genet. 2020;106:356–370. doi: 10.1016/j.ajhg.2020.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aref-Eshghi E., Schenkel L.C., Lin H., Skinner C., Ainsworth P., Paré G., Rodenhiser D., Schwartz C., Sadikovic B. The defining DNA methylation signature of Kabuki syndrome enables functional assessment of genetic variants of unknown clinical significance. Epigenetics. 2017;12:923–933. doi: 10.1080/15592294.2017.1381807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butcher D.T., Cytrynbaum C., Turinsky A.L., Siu M.T., Inbar-Feigenberg M., Mendoza-Londono R., Chitayat D., Walker S., Machado J., Caluseriu O. CHARGE and Kabuki Syndromes: Gene-Specific DNA Methylation Signatures Identify Epigenetic Mechanisms Linking These Clinically Overlapping Conditions. Am. J. Hum. Genet. 2017;100:773–788. doi: 10.1016/j.ajhg.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schenkel L.C., Aref-Eshghi E., Skinner C., Ainsworth P., Lin H., Paré G., Rodenhiser D.I., Schwartz C., Sadikovic B. Peripheral blood epi-signature of Claes-Jensen syndrome enables sensitive and specific identification of patients and healthy carriers with pathogenic mutations in KDM5C. Clin. Epigenetics. 2018;10:21. doi: 10.1186/s13148-018-0453-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choufani S., Cytrynbaum C., Chung B.H., Turinsky A.L., Grafodatskaya D., Chen Y.A., Cohen A.S., Dupuis L., Butcher D.T., Siu M.T. NSD1 mutations generate a genome-wide DNA methylation signature. Nat. Commun. 2015;6:10207. doi: 10.1038/ncomms10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aref-Eshghi E., Bend E.G., Hood R.L., Schenkel L.C., Carere D.A., Chakrabarti R., Nagamani S.C.S., Cheung S.W., Campeau P.M., Prasad C. BAFopathies’ DNA methylation epi-signatures demonstrate diagnostic utility and functional continuum of Coffin-Siris and Nicolaides-Baraitser syndromes. Nat. Commun. 2018;9:4885. doi: 10.1038/s41467-018-07193-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hood R.L., Schenkel L.C., Nikkel S.M., Ainsworth P.J., Pare G., Boycott K.M., Bulman D.E., Sadikovic B. The defining DNA methylation signature of Floating-Harbor Syndrome. Sci. Rep. 2016;6:38803. doi: 10.1038/srep38803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aref-Eshghi E., Rodenhiser D.I., Schenkel L.C., Lin H., Skinner C., Ainsworth P., Paré G., Hood R.L., Bulman D.E., Kernohan K.D., Care4Rare Canada Consortium Genomic DNA Methylation Signatures Enable Concurrent Diagnosis and Clinical Genetic Variant Classification in Neurodevelopmental Syndromes. Am. J. Hum. Genet. 2018;102:156–174. doi: 10.1016/j.ajhg.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barbosa M., Joshi R.S., Garg P., Martin-Trujillo A., Patel N., Jadhav B., Watson C.T., Gibson W., Chetnik K., Tessereau C. Identification of rare de novo epigenetic variations in congenital disorders. Nat. Commun. 2018;9:2064. doi: 10.1038/s41467-018-04540-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garg P., Sharp A.J. Screening for rare epigenetic variations in autism and schizophrenia. Hum. Mutat. 2019;40:952–961. doi: 10.1002/humu.23740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Rubeis S., He X., Goldberg A.P., Poultney C.S., Samocha K., Cicek A.E., Kou Y., Liu L., Fromer M., Walker S., DDD Study. Homozygosity Mapping Collaborative for Autism. UK10K Consortium Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Satterstrom F.K., Kosmicki J.A., Wang J., Breen M.S., De Rubeis S., An J.Y., Peng M., Collins R., Grove J., Klei L. Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell. 2020;180:568–584.e23. doi: 10.1016/j.cell.2019.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pascolini G., Agolini E., Majore S., Novelli A., Grammatico P., Digilio M.C. Helsmoortel-Van der Aa Syndrome as emerging clinical diagnosis in intellectually disabled children with autistic traits and ocular involvement. Eur. J. Paediatr. Neurol. 2018;22:552–557. doi: 10.1016/j.ejpn.2018.01.024. [DOI] [PubMed] [Google Scholar]
- 30.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Houseman E.A., Accomando W.P., Koestler D.C., Christensen B.C., Marsit C.J., Nelson H.H., Wiencke J.K., Kelsey K.T. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deciphering Developmental Disorders S., Deciphering Developmental Disorders Study Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takenouchi T., Miwa T., Sakamoto Y., Sakaguchi Y., Uehara T., Takahashi T., Kosaki K. Further evidence that a blepharophimosis syndrome phenotype is associated with a specific class of mutation in the ADNP gene. Am. J. Med. Genet. A. 2017;173:1631–1634. doi: 10.1002/ajmg.a.38126. [DOI] [PubMed] [Google Scholar]
- 34.Krajewska-Walasek M., Jurkiewicz D., Piekutowska-Abramczuk D., Kucharczyk M., Chrzanowska K.H., Jezela-Stanek A., Ciara E. Additional data on the clinical phenotype of Helsmoortel-Van der Aa syndrome associated with a novel truncating mutation in ADNP gene. Am. J. Med. Genet. A. 2016;170:1647–1650. doi: 10.1002/ajmg.a.37641. [DOI] [PubMed] [Google Scholar]
- 35.Magen I., Gozes I. Microtubule-stabilizing peptides and small molecules protecting axonal transport and brain function: focus on davunetide (NAP) Neuropeptides. 2013;47:489–495. doi: 10.1016/j.npep.2013.10.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The methylation and RNA sequencing data are available at the Gene Expression Omnibus under accession number GEO: GSE152428 (GEO: GSE152427 for the methylation data and GEO: GSE152421 for the RNA sequencing data).