Summary
KAT5 encodes an essential lysine acetyltransferase, previously called TIP60, which is involved in regulating gene expression, DNA repair, chromatin remodeling, apoptosis, and cell proliferation; but it remains unclear whether variants in this gene cause a genetic disease. Here, we study three individuals with heterozygous de novo missense variants in KAT5 that affect normally invariant residues, with one at the chromodomain (p.Arg53His) and two at or near the acetyl-CoA binding site (p.Cys369Ser and p.Ser413Ala). All three individuals have cerebral malformations, seizures, global developmental delay or intellectual disability, and severe sleep disturbance. Progressive cerebellar atrophy was also noted. Histone acetylation assays with purified variant KAT5 demonstrated that the variants decrease or abolish the ability of the resulting NuA4/TIP60 multi-subunit complexes to acetylate the histone H4 tail in chromatin. Transcriptomic analysis in affected individual fibroblasts showed deregulation of multiple genes that control development. Moreover, there was also upregulated expression of PER1 (a key gene involved in circadian control) in agreement with sleep anomalies in all of the individuals. In conclusion, dominant missense KAT5 variants cause histone acetylation deficiency with transcriptional dysregulation of multiples genes, thereby leading to a neurodevelopmental syndrome with sleep disturbance, cerebellar atrophy, and facial dysmorphisms, and suggesting a recognizable syndrome.
Keywords: KAT5, TIP60, brain malformations, epilepsy, intellectual disability, sleep disturbance, epigenetic, histone acetyltransferase, histone
Introduction
Epigenetic regulation by histone acetylation is essential for proper development, and its role in human genetic diseases is increasingly being recognized. Notably, variants in lysine acetyltransferase genes, such as KAT6A (MIM: 601408) and KAT6B (MIM: 605880), have been identified in individuals with neurodevelopmental disorders characterized by intellectual disability and malformations.1,2 KAT5 (MIM: 601409) (a.k.a. TIP60) variants have not yet been associated with a syndrome. KAT5 can act as a haploinsufficient tumor suppressor gene, and it encodes an essential lysine acetyltransferase involved in gene expression, DNA repair, chromatin remodeling, apoptosis, and cell proliferation.3,4 It is part of a large, multi-protein complex named NuA4 (also known as the TIP60/p400 complex), which includes TRRAP, EP400, and ING3 among other proteins.5 Local recruitment of the NuA4 complex and KAT5-mediated acetylation of conserved lysine residues on histones H4 and H2A(.Z/.X) are linked to transcription activation as well as repair of DNA double-strand breaks, in part through chromatin relaxation but also through signaling and/or crosstalk with other chromatin-binding factors.6,7 KAT5 can also directly acetylate non-histone proteins such as ATM in DNA damage response, p53 at lysine 120 in apoptosis, and other mitotic regulators that impact cell cycle control. KAT5-dependent acetylation of specific transcription factors can also lead to transcription activation or repression.8, 9, 10 The NuA4/TIP60 complex is essential for stem cell maintenance and renewal,11 and recent work revealed that KAT5 may play a role in epithelial-mesenchymal transition induction12; all of these are key processes in the developing embryo. Finally, KAT5 contributes to genome integrity by maintaining accurate chromosome alignment and segregation.13 KAT5 depletion was shown to impair the chromosomal segregation during mitosis and to result in polyploidy.14
We studied three individuals with de novo heterozygous missense variants in KAT5 that affect normally invariant residues. All three individuals have short stature, cerebral malformations, seizures, and global developmental delay or intellectual disability along with a significant speech disorder and a severe sleep disorder.
In order to understand the molecular mechanisms underlying the phenotype in these individuals with KAT5 variants, we engineered cell lines to purify native NuA4 complexes that contain the variant catalytic subunit so that we could determine the possible effects on complex assembly/protein interactions and acetylation of chromatin substrates, and we performed transcriptomic analyses in primary cells to determine possible target genes implicated in the pathology.
Material and Methods
Recruitment and Sequencing
Apart from the published individual, other individuals were recruited through GeneMatcher.15 Information was obtained from each clinical team. Exome sequencing (ES) was performed by the National Institutes of Health (NIH) Intra-mural Sequencing Center (NISC) for individual 1 (complete method described by Berger et al.16). ES was performed on a research basis for individual 2 at University of Geneva Medical School and Geneva University Hospitals. ES was performed in a commercial laboratory for individual 3. Informed consent to publish clinical information and photographs was obtained from the parents of the individuals reported in this article. For each individual, the procedures followed were in accordance with the ethical standards of the responsible committees on human experimentation.
Cell Culture and Transfection
K562 cells were obtained from the American Type Culture Collection (ATCC) and maintained at 37°C under 5% CO2 in RPMI medium supplemented with 10% newborn calf serum (Wisent) and GlutaMAX (Thermo Fisher). When cultivated in spinner flasks, 25 mM HEPES-NaOH (pH 7.4) was added. Cells were transfected using Lipofectamine 2000 (Thermo Fisher) per the manufacturer’s instructions.
Generation of Stable Cell Lines Producing Tagged KAT5 Variants and Affinity Purification of NuA4/TIP60 Complexes
KAT5 (461aa isoform) was cloned into the AAVS1_Puro_PGK1_3xFLAG_Twin_Strep plasmid (addgene #68375), and the variants found in each individual were introduced via site-directed mutagenesis. Generation of K562 cells that expressed either wild-type (WT) or variant-tagged KAT5 was performed through break-induced recombination and/or insertion at the AAVS1 locus (MIM: 102699) as described.17
Nuclear cell extracts were prepared from 3.10ˆ9 cells and used to perform tandem affinity purification as described.18 In brief, nuclear extracts were adjusted to 0.1% Tween-20, and ultracentrifuged at 100,000 g for 1 h. Extracts were precleared with 250 μl Sepharose CL-6B (Sigma), then 250 μl anti-FLAG M2 affinity resin (Sigma) was added for 2 h at 4°C. The beads were then washed in Poly-Prep columns (Bio-Rad) with 40 column volumes (CV) of buffer #1 (20 mM HEPES-KOH [pH 7.9], 10% glycerol, 300 mM KCl, 0.1% Tween 20, 1 mM DTT, 1 mM PMSF, 2 μg/mL Leupeptin, 5 μg Aprotinin, 2 μg/mL Pepstatin, 10 mM Na-butyrate, 10 mM β-glycerophosphate, 100 μM Sodium Orthovanadate, 5 mM N-Ethylmaleimide, 2 mM Ortho-Phenanthroline) followed by 40 CV of buffer #2 (20 mM HEPES-KOH [pH 7.9], 10% glycerol, 150 mM KCl, 0.1% Tween 20, 1mMDTT, 1 mM PMSF, 2 μg/mL Leupeptin, 5 μg Aprotinin, 2 μg/mL Pepstatin, 10 mM Na-butyrate, 10 mM β-glycerophosphate, 100 μM Sodium Orthovanadate, 5 mM N-Ethylmaleimide, 2 mM Ortho-Phenanthroline). Complexes were eluted in two fractions with 2.5 CV of buffer #2 supplemented with 200 ug/mL 3xFLAG peptide (Sigma) for 1 h at 4°C. Typically, 15 ul of the first elution (3% of total) was loaded on NuPAGE 4%–12% Bis-Tris gels (Invitrogen) and analyzed via silver staining.
In Vitro HAT Assays
1 μg of core histones (CH) or short oligonucleosomes (SON) was incubated with affinity-purified NuA4/TIP60 complexes harboring the different KAT5 variants and 3H-labeled acetyl-CoA (0,1 μCi, Perkin-Elmer) in HAT buffer (50 mM Tris-HCl ph8.0, 50 mM KCl, 5% glycerol, 0,1 mM EDTA, 1 mM DTT, 1 mM PMSF, 10 mM Sodium Butyrate) for 30 min at 30°C. Half of the reaction was spotted on P81 filter paper, washed, and analyzed via liquid scintillation. The other half was loaded on SDS-PAGE 15% gels. Gels were Coomassie-stained to ensure homogeneous loading, then destained, fluorographed using EN3HANCE (Perkin-Elmer), dried, and exposed at −80°C. The amounts of purified TIP60/NuA4 complex used in the reactions were normalized between samples based on 3H counts on CH and Flag-KAT5 signal measured via immunoblot. All reactions were done in triplicates, and the assay was performed two times.
RNaseq Methods
RNA libraries were prepared from low-passage fibroblasts from individuals 2 and 3 through the use of the Illumina RNA Truseq V2 and Truseq mRNA stranded kits, respectively. The libraries were then sequenced on an Illumina HiSeq4000 sequencer at 2 × 100bp and 1 × 100bp read lengths, respectively. Two and four healthy control low-passage fibroblasts were also sequenced from Truseq V2 and Truseq mRNA stranded libraries at 2 × 100bp and 1 × 100bp read lengths, respectively. Transcriptomics analyses were performed as previously described.19 Common differentially expressed genes were selected by using the DESeq2 R package to compare the affected individuals to the respective healthy controls with thresholds at |Log2FC| > 0.5, 5% FDR, and adjusted p value < 0.05.
RT-qPCR
Total RNA from affected individuals and three new controls was isolated from low-passage fibroblasts through the use of the PureLink RNA mini kit (Life Technologies). Controls were male children of White, African American, and Asian backgrounds. Equal amounts of RNA were used to synthesize cDNA through the use of the qScript cDNA synthesis kit (Quanta Biosciences). cDNA was quantified through the use of PowerUp SYBR green Master Mix (Applied Biosystems) on a LightCycler® 96 system (Roche) using primers listed in Table S3. Amplicons were resolved by using agarose gel to determine the size. Relative gene expression levels were analyzed via 2−ΔΔCT method with β-actin used as the reference gene. Statistical significance was determined via two-way ANOVA with Dunnett’s multiple comparisons test. Variation was reported as standard deviation (SD).
Results
Clinical Descriptions
Individual 1 is a 30-year-old female with intellectual disability who has been reported by Berger et al. in a Smith-Magenis Syndrome (SMS)-like cohort.16 She presented with behavioral problems with perseverative speech, poor language function, and sleep disorder. At age 10, she had disruptive behaviors and a diagnosis of attention deficit disorder. At age 29, she had an IQ of 40 with expressive language at the 8-year-old level and receptive language at 4-to-5-year-old skill level. Her head circumference is 55cm (73rd centile). She also presents with adult-onset seizures, severe myopia, hyperacusis, kyphoscoliosis, brachydactyly, and frequent urinary tract infections. Her facial dysmorphisms include a round face with a flat facial profile, prognathism, down-slanting corners of the mouth, low-set ears, depressed nasal bridge, and almond-shaped eyes. She has partial agenesis of the corpus callosum. She developed secondary amenorrhea at 29 years of age. Sleep problems present since early childhood included early sleep offset, nighttime awakenings (1–2), and increased daytime naps. In adulthood, sleep diaries document 24 h sleep cycle characterized by early morning awakening (between 05:30–06:30), bedtime at 20:30, two daytime naps (09:30–10:30 and 13:00–14:30), and nocturnal awakenings (~30 min long) after sleep onset, usually occurring at 23:00 and 01:00. Increased daytime salivary melatonin level was documented at 11:15 (mean 46 pg/mL for two samples), which is consistent with the inverted circadian melatonin profile observed in SMS.20 ES identified a de novo missense variant c.158G>A (p.Arg53His) in KAT5 (RefSeq accession number NM_006388.3). No other variants met the filtering exome criteria.
Individual 2 is a 13-year-old male with intellectual disability and multiple malformations. He was born at 38 weeks with a unilateral cleft lip and palate. At 12 years of age, he is nonverbal. and a cognitive evaluation documented an IQ of 20–30. He has disruptive behavior with hyperactivity and multiple stereotypies. He suffers from generalized tonic-clonic seizures and has severe sleep disorder (with sleep onset delay and night waking). His head circumference was 50 cm (1st centile, −2.6 SD). Facial dysmorphisms include prognathism, lateral thinning of the eyebrows, macrostomia, thick lower lip, and bulbous and asymmetric nose. He also has bilateral single palmar creases and fifth finger clinodactyly, as well as unilateral cryptorchidism. Horseshoe kidney and bilateral vesico-ureteral reflux were diagnosed during childhood. Brain MRI showed global progressive cerebellar atrophy (vermis more than hemispheres), dysgenesis of corpus callosum (short, thickened, and hypoplasia of rostrum and splenium), and a small anterior pituitary gland. He suffers from growth hormone (GH) deficiency diagnosed at the age of 2 years and for which he is treated with GH injections. ES identified a de novo missense variant c.1105T>A (p.Cys369Ser) in KAT5 (RefSeq NM_006388.3).
Individual 3 is a 2-year-old male with developmental delay and multiple malformations. At 16 months of age, he presented with short stature and congenital microcephaly, height 71.1 cm (<1st centile, −2.9 SD), weight 11.2 kg (48th centile), and head circumference 44.5 cm (<1st centile, −2.2 SD). He has severe developmental delay with disruptive behaviors and an important sleep disorder (night waking and sleep onset delay which was improved by nighttime clonidine, which was prescribed because daytime clonidine caused daytime sleepiness). He suffers from generalized myoclonic seizures. He has a perimembranous ventricular septal defect and a dysplastic pulmonary valve with supravalvular and valvular stenosis. He also has a high-arched palate with a submucous cleft. His genitourinary anomalies consist of hypospadias and bilateral cryptorchidism. His facial dysmorphisms include a round face with a flat facial profile, epiblepharon, epicanthal folds, down-slanting corners of the mouth, and upturned nose with depressed nasal bridge. He also has bilateral fifth finger clinodactyly. His medication includes risperidone and clonidine. Brain MRI showed polymicrogyria of right sylvian fissure, cystic dilation of 4th ventricle, and inferior cerebellar vermis atrophy. ES identified a de novo missense variant c.1237T>G (p.Ser413Ala) in KAT5 (RefSeq NM_006388.3).
Additional clinical information is available for these three individuals in Tables S1 and S2 (for comparison of sleep disorder characteristics).
Analysis of the Variants
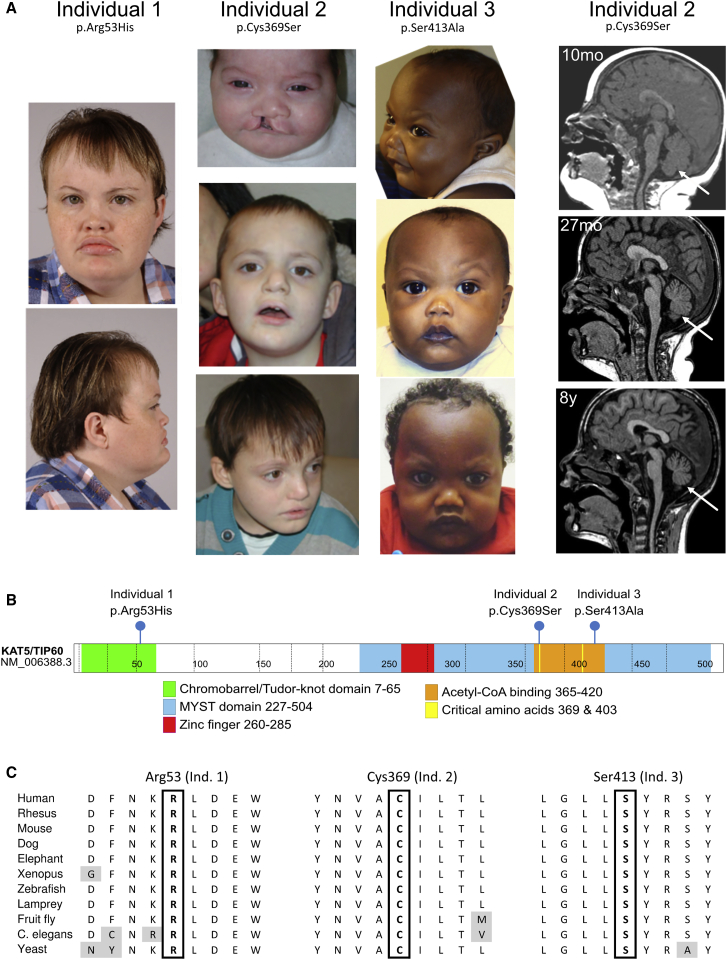
We named the variants through the use of isoform NM_006388.3 (513 amino acids) because it is highly expressed and is the canonical isoform in Uniprot.21 However, the longest isoform is NM_182710.2 (546 amino acids), and a commonly studied isoform is NM_182709.2 (also known as PLIP, 461 amino acids). All three variants are absent from the Genome Aggregation Database (gnomAD),22 and this absence indicates that these variants are not present in the more than 100,000 individuals from population genetic studies included in this database. As shown in Figure 1B, the p.Arg53His variant is in the chromodomain, following an acetylated lysine residue. In addition to potentially affecting KAT5’s ability to interact with histones via its chromodomain, the p.Arg53His variant may disrupt the protein’s structure and thus stability (as suggested by STRUM analysis,23 and by the protein yields in K562 cell extracts shown in Figure S1). The p.Cys369Ser variant is near the Acetyl-CoA binding domain, and this residue has been shown to be critical for the catalysis of yeast Esa1 (KAT5 ortholog) and other MYST-family acetyltransferases,24 as well as for KAT5 autoacetylation.25 Finally, the p.Ser413Ala variant is in the Acetyl-CoA binding domain. All residues are invariant throughout evolution, and in fact, they are conserved down to yeast Esa1 (Figure 1C). An analysis of the 3D structure of KAT5 (Figure 2A) suggests that the p.Cys369Ser and p.Ser413Ala variants may alter the interaction of the protein with Acetyl-CoA. This was also suggested by 3D mutation impact analysis using HOPE and VarSite.26,27
Figure 1.
Clinical Images and Variant Details
(A) Photographs of the three individuals, showing shared facial dysmorphisms. Individual 1 and individual 3 have round faces, flat facial profiles, down-slanting corners of their mouths, and depressed nasal bridges. Individual 1 and individual 2 have prognathism. The images on the right are sagittal MRI images for individual 2 at the indicated ages, showing progressive cerebellar atrophy (arrow).
(B) Variant location in functional domains of the KAT5 protein.
(C) Affected amino acids are invariant between different species.
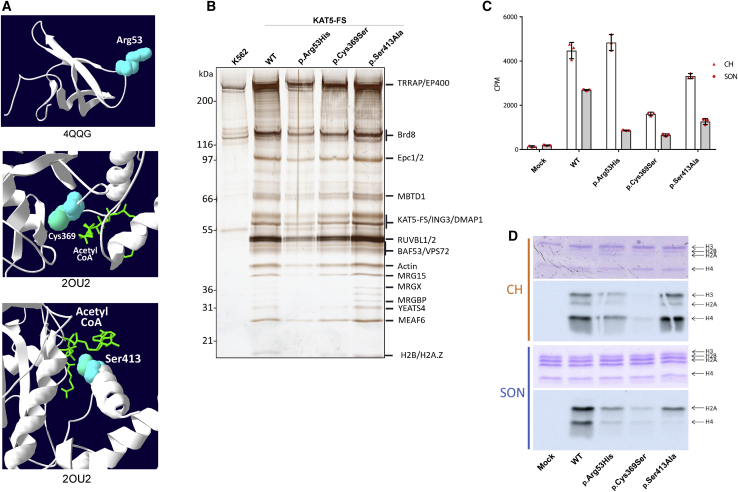
Figure 2.
Functional Impact of KAT5 Variants on the Native NuA4/TIP60 Acetyltransferase Complex
(A) Predicted variant location in 3D reconstruction of KAT5 protein. Annotations below the images refer to the RCSB PBD (Research Collaboratory for Structural Bioinformatics Protein Data Bank) structure IDs.
(B) Variant KAT5 proteins assemble in normal NuA4/TIP60 complexes. WT and variant KAT5 proteins were fractionated from nuclear extracts via tandem affinity purification. Purified fractions were loaded on gel and stained with silver. Bone fide NuA4/TIP60 subunits are identified on the right. Note: the isoform used for experiments was the commonly used 461-amino-acid isoform (RefSeq NM_182709.2), and thus variants are at positions 53, 317, and 361 in that protein, but for all figures, they were identified with the canonical isoform RefSeq NM_006388.3 numbering for consistency with the rest of the manuscript.
(C) In vitro histone acetylation assay performed with purified native WT and variant complexes. The graph shows the scintillation counts of the liquid assays with 3H-Acetyl-CoA with free core histones (CH) or native short oligonucleosomes (SON). Error bars represent standard deviations of technical replicates.
(D) Fluorograph of in vitro histone acetylation assays with native WT and variant complexes. Protein gels were treated with En3Hance, dried, and exposed on film to assess 3H-labeled protein bands and/or acetylation in order to visualize the effect on specific histones (lower panels). Coomassie stained gels are shown to control relative substrate amounts in the reactions (upper panels).
Expected versus observed counting of single-nucleotide changes in gnomAD show that KAT5 is only moderately intolerant to loss-of-function (LoF) variants (pLI score [probability of being loss of function intolerant] 0.09; observed/expected [o/e] ratio 0.26 [90% confidence interval (CI): 0.15–0.47]).22 Moreover, fewer missense variants were observed than were expected (o/e ratio 0.44 [90% CI: 0.39-0.51] with a Z score of 3.61) (gnomAD v2.1.1). Regarding other assessments of KAT5 as a gene potentially associated with a dominant disease, the %HI score (haploinsufficiency score from DECIPHER) is 4.47%. %HI scores below 10% indicate that a gene is more likely to be deleterious if haploinsufficient.28 The KAT5 P(AD) score is 0.996 (probability for a gene to carry dominant mutations from the DOMINO website, accessed June 2, 2020).3 A P(AD) score of 0.95 is highly associated with autosomal dominant inheritance through haploinsufficiency, gain of function, or dominant-negative effects.29 An analysis of the affected residues performed through the use of Metadome and the MTR Gene Viewer suggested that all affected residues are intolerant to variations.30,31 Moreover, most pathogenicity prediction tools we used considered the variants to be likely pathogenic. That was the case for DANN,32 DEOGEN2, EIGEN, FATHMM-MKL, M-CAP, MutationAssessor, MutationTaster, and SIFT (scores from the dbNSFP33 database except for DANN and analyzed through the Varsome website34). The variants had CADD scores of 32, 27, and 26 respectively (scores 20 or above indicate that they are among the 1% most likely pathogenic variants in the genome).35 The variants were also considered to be deleterious according to results from Rhapsody and MutPred2.36,37
Purification and Biochemical Analysis of KAT5 Variants
In order to determine the effect of the variants on KAT5 protein interactome and enzymatic activity, we used genome editing to introduce WT and mutant KAT5 cDNAs at the safe harbor AAVS1 locus in human K562 cells.17 Equivalent accumulation of the C-terminally tagged proteins (3×Flag-2×Strep) was measured and clones were selected (Figure S2). Production in these clones is near physiological levels compared to endogenous KAT5. Native NuA4/TIP60 complexes were then obtained via tandem affinity purification.18 Analysis of the purified fractions through the use of protein gel, immunoblotting, and mass spectrometry showed that WT and variant KAT5 normally assemble into full stoichiometric NuA4/TIP60 complexes (Figure 2B). Then, the histone acetyltransferase (HAT) activity of the different complexes was measured in vitro with 3H-Acetyl-CoA using free CH or native chromatin (SON) as substrates. All variants displayed impaired HAT activity to varying degrees compared to WT KAT5 (Figures 2C–D). The p.Cys369Ser variant showed the most dramatic effect, being unable to acetylate both free histones and chromatin, as expected based on its localization in the catalytic site. On the other hand, the complexes containing KAT5 p.Arg53His and p.Ser413Ala variants are mostly defective in their HAT activity toward chromatin, not free histones (Figure 2C). Strikingly, as shown through the use of gel fluorography (Figure 2D), this defect is clearly more specific toward nucleosomal histone H4 tail acetylation, whereas H2A acetylation is still detected. Altogether, these data clearly demonstrate that the de novo variants detected in the individuals described above cripple the lysine acetyltransferase activity of KAT5, leading to partial loss of function in vivo and impairing the ability of the NuA4/TIP60 complex to properly acetylate its targets in a chromatin context. Based on KAT5’s critical role in genome expression and maintenance, control of cell proliferation, and development, these variants are likely implicated in the neurodevelopmental defects seen in these individuals.
Transcriptomic Analyses
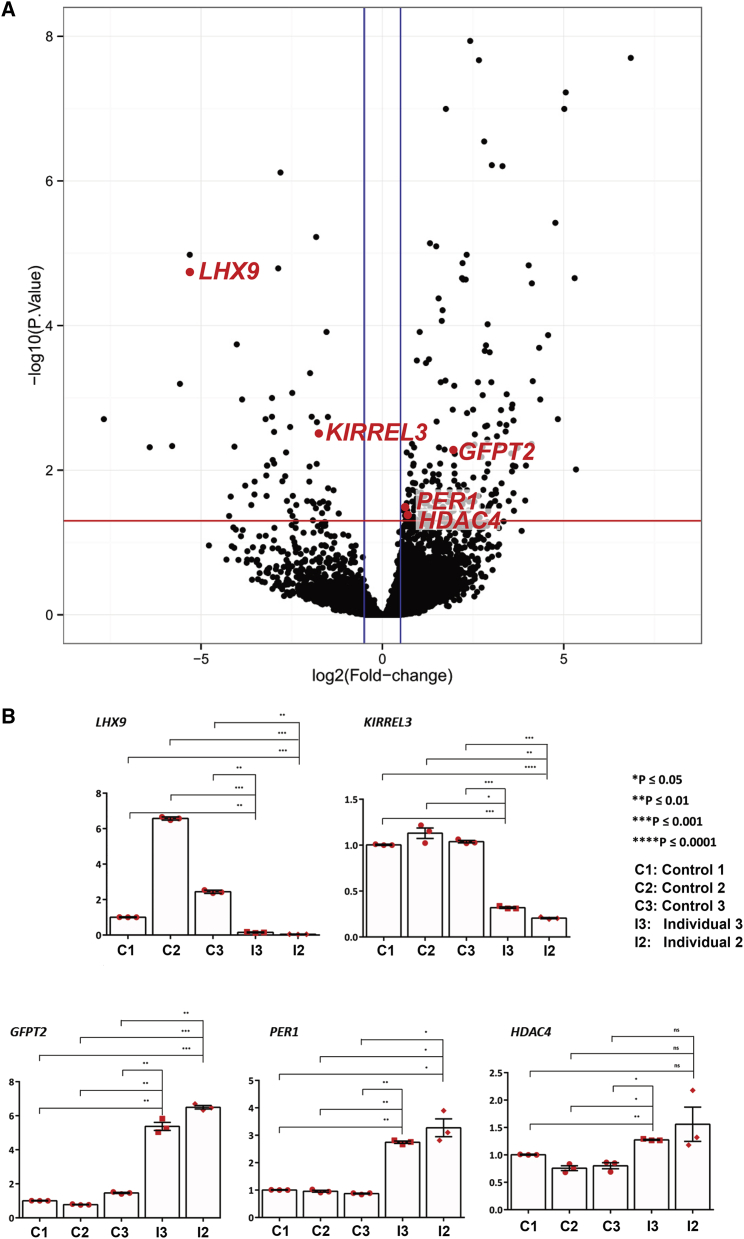
We performed an analysis of dysregulated genes in primary human fibroblasts from individuals 2 and 3, as described previously.19 From the RNaseq data (cutoff Log2FC of >0.5 or <−0.5, p £ 0.05 [Figure 3A]), we selected genes which were involved in development, neuronal function, and circadian rhythm control, and we performed qPCR analysis with additional controls (Figure 3B and Figure S2). Genes which showed consistent downregulations were LHX9 (MIM: 606066) and KIRREL3 (MIM: 607761). Genes which showed consistent upregulations were GFPT2 (MIM: 603865), PER1 (MIM: 602260), and HDAC4 (MIM: 605314).
Figure 3.
RNaseq Was Performed on Fibroblasts from Individuals 2 and 3 and Six Healthy Controls
(A) Volcano plot showing common DEGs (differentially expressed genes) of individuals 2 and 3. Significant DEGs. The red line indicates a −log10 (adjusted p value) of 1.3 (padj of 0.05); and the blue line a Log2 Fold Change of −0.05 and 0.05. Significant DEGs shown in panel B are represented by red dots.
(B) Reverse transcriptase-qPCR analysis of specific genes deregulated in fibroblasts from three new controls and from individuals 2 and 3. β-actin was used as the reference gene. Triplicates were used. Error bars represent standard deviation. p values were generated through the use of two-way ANOVA with Dunnett’s multiple comparisons test.
Discussion
As shown in Table 1, Table S1, and Figure 1A, the three affected individuals share many features. They are moderately short (−1.95 SD, −2.1 SD, and −2.9 SD) and two have microcephaly (−2.6 SD and −2.2 SD). They have severe developmental delay or moderate-to-severe intellectual disability. All three individuals present with disruptive behavior and have severe sleep disorders. All have night waking, and individuals 2 and 3 have sleep onset delay (improved by clonidine for individual 3), and individual 1 has daytime sleepiness. Individuals 1 and 2 have anomalies of the corpus callosum, individuals 2 and 3 have cerebellar atrophy, and individual 3 has focal polymicrogyria. Facial dysmorphisms, each present in at least two individuals, include a round face with a flat facial profile, a depressed nasal bridge, downturned corners of the mouth, and prognathism. Individual 2 has a cleft lip and palate, and individual 3 has a submucous cleft palate. All three individuals have genitourinary anomalies, including cryptorchidism, hypospadias, horseshoe kidney, and vesico-ureteral reflux. Although there is some clinical overlap between the individuals we studied and individuals with SMS, such as sleep disturbances and some facial dysmorphisms, there are also several differences, such as the seizures and genitourinary anomalies seen in all individuals here, which are present in only a minority of individuals with SMS. Progressive cerebellar atrophy and CNS malformations are not observed in SMS, whereas dental anomalies and broad hands are common in SMS and are not observed here, among other differences.
Table 1.
Main Clinical Features
| Individuals | 1 | 2 | 3 |
|---|---|---|---|
| KAT5 variants (RefSeq NM_006388.3) | c.158G>A (p.Arg53His) | c.1105T>A (p.Cys369Ser) | c.1237T>G (p.Ser413Ala) |
| Chromosomal positions (hg19) | Chr11:65480402G>A | Chr11:65484393T>A | Chr11:65486132T>G |
| Age and gender | 29-year-old female | 13-year-old male | 18-month-old male |
| Microcephaly | - | + | + |
| Developmental delay or intellectual disability | +, IQ 40 | +, IQ 20–30 | +, severe |
| Behavioral issues | ADHD, sleep disorder, disruptive behavior | ADHD, severe sleep disorder, multiple stereotypies and disruptive behavior | behavioral difficulties with tantrums and head banging |
| Seizures | + | + | + |
| Cerebral malformations | partial agenesis of the corpus callosum | corpus callosum dysgenesis, cerebellar atrophy | focal polymicrogyria, cerebellar atrophy |
| Urogenital anomalies | recurrent urinary tract infections | horseshoe kidney, vesico-ureteral reflux, cryptorchidism | hypospadias, cryptorchidism |
| Congenital heart defect | - | - | VSD, dysplastic pulmonary valve |
| Orofacial malformations | - | unilateral cleft lip and palate | submucous cleft palate |
| Ocular anomaly | severe myopia | strabismus and hypermetropia | epiblepharon |
| Dysmorphisms | round face, flat facial profile; down-slanting corners of mouth; depressed nasal bridge; prognathism; low-set ears; almond-shaped eyes | lateral thinning of eyebrows, macrostomia, bulbous and asymmetric nose, thick lower lip, prominent chin | round face, flat facial profile; down-slanting corners of mouth |
KAT5 variants cause histone acetylation deficiency and gene expression deregulation, and thereby lead to a neurodevelopmental syndrome with facial dysmorphisms, various malformations, and sleep disturbances. Berger et al.16 had already reported individual 1 in 2017 and had then proposed KAT5 as a candidate gene to explain the phenotype. The addition of two new individuals with overlapping phenotypes and having a KAT5 variant confirms the involvement of KAT5 in human diseases. The observed deficient histone acetylation by biochemical assays using native KAT5 complexes suggests an LoF mechanism during development. Mice heterozygous for a knockout Kat5 allele have normal development, growth, and fertility in the literature.3,38,39 Heterozygous mice phenotyped by the International Mouse Phenotyping Consortium (IMPC) are also essentially normal (IMPC website accessed June 3, 2020).40 Homozygous knockout mice are embryonic lethal both in the literature and the IMPC study. It is possible that in humans, haploinsufficiency for KAT5 does not lead to a syndrome, and it is even likely given that over 10 high-confidence LoF variants are found in gnomAD, but that missense variants abrogating KAT5 activity might cause a dominant deleterious effect. Moreover, regulatory mechanisms may lead to near-normal KAT5 protein levels in case of haploinsufficiency. This was observed in adipose tissue of Kat5 haploinsufficient mice, in which Kat5 mRNA was reduced to 50%, but protein levels were normal, and similar observations were also made in different tissues in other studies.39,41,42 We hypothesize that NuA4 complexes with inactive KAT5 have widespread epigenetic consequences (as suggested by our transcriptomic studies), whereas the presence of slightly fewer NuA4 complexes does not, but future studies, ideally in vivo, will be required to assess this hypothesis.
LHX9 and KIRREL3 were consistently downregulated in primary fibroblasts. LHX9 is important for thalamic neuronal differentiation.43 Knockout mice have profound hypersomnolence, likely because Lhx9 may be important for specification or survival of a subset of hypocretin-containing neurons of the hypothalamus that are essential for the normal regulation of sleep.44 LHX9 is downregulated in individuals with Pallister-Kallian syndrome, a neurodevelopmental disorder.45 KIRREL3 is an IgSF-adhesion molecule implicated in synapse formation, synaptic transmission, and ultrastructure. It regulates mossy fiber synapse development in the hippocampus46 and has been implicated in neurodevelopmental disorders.47,48
GFPT2, PER1, and HDAC4 were consistently upregulated in primary fibroblasts. GFPT2 controls the flux of glucose into the hexosamine pathway involved in protein glycosylation. An individual with severe intellectual disability was reported with a de novo missense variant in this gene.49 PER1 is a key component of the circadian clock and acts as a transcriptional repressor.50 HDAC4 is a histone deacetylase which binds promoters through transcription factors MEF2C and MEF2D and represses transcription. Its deletion causes cognitive and behavioral issues often associated with brachydactyly.51 Interestingly, genome-wide location analysis of the NuA4/TIP60 complex in human K562 cells previously reported its presence on the PER1 and HDAC4 genes (see Figure S3).7
Importantly, KAT5 has been shown to be critical for learning and memory in Drosophila52, 53, 54 and has also been shown to control sleep in Drosophila by regulating axonal growth in pacemaker cells.55 In addition, mammalian KAT5 has recently been reported to be an important regulator of the circadian clock cycle through direct action at gene promoters and BMAL1 acetylation.56 Moreover, another HAT, ELP3, has also been associated with sleep anomalies in Drosophila,57 and sleep deprivation induces the expression of Hdac2 in rat hippocampi.58 The role of epigenetics in the regulation of sleep has been reviewed by Quershi and Mehler in 2014.59 Sleep disturbances not associated with sleep apnea are also seen in diseases caused by variants in epigenetic regulators. Dominant variants in or deletions of the histone deacetylase HDAC4 have been implicated in the pathophysiology of chromosome 2q37 deletion syndrome (MIM: 600430), in which there is a sleep disturbance, and lead to reduced expression of RAI1 (MIM: 607642), a gene for which variants cause the overlapping SMS (MIM: 182290).60 SMS due to deletions of 17p11.2 or RAI1 variants is associated with a recognized circadian sleep disorder characterized by an advanced sleep phase and inverted melatonin secretion profile.61,62 Autosomal dominant mental retardation type 1 (MIM: 156200) is caused by variants in MBD5 (MIM: 611472) that encode Methyl-CpG-binding domain protein 5, which is part of a polycomb repressive complex that deubiquitinates a lysine of histone H2A. Interestingly, disturbed PER1 levels were noted with both MBD5 mutations and with SMS.63,64 Diseases caused by mutations in other epigenetic regulators are associated with sleep disturbances (KDM5B [MIM: 605393],65 MECP2 [MIM: 300005],66 EHMT1 [MIM: 607001], KMT2C [MIM: 606833], and HDAC8 [MIM: 300269]67), as well as several other genetic diseases.68, 69, 70, 71, 72
Other HATs associated with Mendelian disorders are KAT6A and KAT6B. KAT6A variants cause autosomal dominant mental retardation 32 (MIM: 616268), and overlapping features with the syndrome described here include intellectual disability, microcephaly, epilepsy, and sleep disturbances.73 KAT6B variants cause Genitopatellar syndrome (MIM: 606170) and Say-Barber-Biesecker-Young-Simpson (SBBYS) syndrome (MIM: 603736). Overlapping features with both of these syndromes include intellectual disability, microcephaly, and genital anomalies. Specifically, corpus callosum anomalies (for Genitopatellar syndrome) and cleft palate (for SBBYS syndrome) are overlapping features with the syndrome described here.74
It will be interesting in the future to determine whether similar pathways are dysregulated in neuronal models of the various epigenetic disorders associated with sleep disturbances mentioned above, as this could perhaps lead to the development of better targeted therapies for such symptoms.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We thank all affected individuals and family members for their participation in this study. Work was supported by grants from the Canadian Institutes of Health Research (CIHR) to J.C. (FDN-143314). P.M.C. is supported by salary awards from the CIHR and the Fonds de Recherche Québec—Santé. J.C. holds the Canada Research Chair in Chromatin Biology and Molecular Epigenetics. S.E.A.’s laboratory was supported by a ChildCare Foundation grant and a Swiss National Science Foundation (SNSF) grant (163180).
Published: August 20, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.08.002.
Data and Code Availability
RNaseq data are available on the National Center for Biotechnology Information (NCBI)’s Gene Expression Omnibus with accession number GSE154199.
Web Resources
DECIPHER, https://decipher.sanger.ac.uk/
DOMINO, https://wwwfbm.unil.ch/domino/
ExAC Browser, http://exac.broadinstitute.org/
Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo/
OMIM, https://www.omim.org
Philippe Campeau’s laboratory, http://pcampeaulab.org
Supplemental Data
References
- 1.Arboleda V.A., Lee H., Dorrani N., Zadeh N., Willis M., Macmurdo C.F., Manning M.A., Kwan A., Hudgins L., Barthelemy F., UCLA Clinical Genomics Center De novo nonsense mutations in KAT6A, a lysine acetyl-transferase gene, cause a syndrome including microcephaly and global developmental delay. Am. J. Hum. Genet. 2015;96:498–506. doi: 10.1016/j.ajhg.2015.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campeau P.M., Kim J.C., Lu J.T., Schwartzentruber J.A., Abdul-Rahman O.A., Schlaubitz S., Murdock D.M., Jiang M.M., Lammer E.J., Enns G.M. Mutations in KAT6B, encoding a histone acetyltransferase, cause Genitopatellar syndrome. Am. J. Hum. Genet. 2012;90:282–289. doi: 10.1016/j.ajhg.2011.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gorrini C., Squatrito M., Luise C., Syed N., Perna D., Wark L., Martinato F., Sardella D., Verrecchia A., Bennett S. Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature. 2007;448:1063–1067. doi: 10.1038/nature06055. [DOI] [PubMed] [Google Scholar]
- 4.Avvakumov N., Côté J. The MYST family of histone acetyltransferases and their intimate links to cancer. Oncogene. 2007;26:5395–5407. doi: 10.1038/sj.onc.1210608. [DOI] [PubMed] [Google Scholar]
- 5.Doyon Y., Côté J. The highly conserved and multifunctional NuA4 HAT complex. Curr. Opin. Genet. Dev. 2004;14:147–154. doi: 10.1016/j.gde.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 6.Steunou A.-L., Rossetto D., Côté J. Fundamentals of chromatin. Springer; 2014. Regulating chromatin by histone acetylation; pp. 147–212. [DOI] [Google Scholar]
- 7.Jacquet K., Fradet-Turcotte A., Avvakumov N., Lambert J.P., Roques C., Pandita R.K., Paquet E., Herst P., Gingras A.C., Pandita T.K. The TIP60 Complex Regulates Bivalent Chromatin Recognition by 53BP1 through Direct H4K20me Binding and H2AK15 Acetylation. Mol. Cell. 2016;62:409–421. doi: 10.1016/j.molcel.2016.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao H., Chung J., Kao H.Y., Yang Y.C. Tip60 is a co-repressor for STAT3. J. Biol. Chem. 2003;278:11197–11204. doi: 10.1074/jbc.M210816200. [DOI] [PubMed] [Google Scholar]
- 9.Bararia D., Trivedi A.K., Zada A.A., Greif P.A., Mulaw M.A., Christopeit M., Hiddemann W., Bohlander S.K., Behre G. Proteomic identification of the MYST domain histone acetyltransferase TIP60 (HTATIP) as a co-activator of the myeloid transcription factor C/EBPalpha. Leukemia. 2008;22:800–807. doi: 10.1038/sj.leu.2405101. [DOI] [PubMed] [Google Scholar]
- 10.Huang J., Stewart A., Maity B., Hagen J., Fagan R.L., Yang J., Quelle D.E., Brenner C., Fisher R.A. RGS6 suppresses Ras-induced cellular transformation by facilitating Tip60-mediated Dnmt1 degradation and promoting apoptosis. Oncogene. 2014;33:3604–3611. doi: 10.1038/onc.2013.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fazzio T.G., Huff J.T., Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134:162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fukagawa A., Ishii H., Miyazawa K., Saitoh M. δEF1 associates with DNMT1 and maintains DNA methylation of the E-cadherin promoter in breast cancer cells. Cancer Med. 2015;4:125–135. doi: 10.1002/cam4.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mo F., Zhuang X., Liu X., Yao P.Y., Qin B., Su Z., Zang J., Wang Z., Zhang J., Dou Z. Acetylation of Aurora B by TIP60 ensures accurate chromosomal segregation. Nat. Chem. Biol. 2016;12:226–232. doi: 10.1038/nchembio.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grézy A., Chevillard-Briet M., Trouche D., Escaffit F. Control of genetic stability by a new heterochromatin compaction pathway involving the Tip60 histone acetyltransferase. Mol. Biol. Cell. 2016;27:599–607. doi: 10.1091/mbc.E15-05-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berger S.I., Ciccone C., Simon K.L., Malicdan M.C., Vilboux T., Billington C., Fischer R., Introne W.J., Gropman A., Blancato J.K., NISC Comparative Sequencing Program Exome analysis of Smith-Magenis-like syndrome cohort identifies de novo likely pathogenic variants. Hum. Genet. 2017;136:409–420. doi: 10.1007/s00439-017-1767-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dalvai M., Loehr J., Jacquet K., Huard C.C., Roques C., Herst P., Côté J., Doyon Y. A Scalable Genome-Editing-Based Approach for Mapping Multiprotein Complexes in Human Cells. Cell Rep. 2015;13:621–633. doi: 10.1016/j.celrep.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 18.Doyon Y., Côté J. Preparation and Analysis of Native Chromatin-Modifying Complexes. Methods Enzymol. 2016;573:303–318. doi: 10.1016/bs.mie.2016.01.017. [DOI] [PubMed] [Google Scholar]
- 19.Cogné B., Ehresmann S., Beauregard-Lacroix E., Rousseau J., Besnard T., Garcia T., Petrovski S., Avni S., McWalter K., Blackburn P.R., CAUSES Study. Deciphering Developmental Disorders study Missense Variants in the Histone Acetyltransferase Complex Component Gene TRRAP Cause Autism and Syndromic Intellectual Disability. Am. J. Hum. Genet. 2019;104:530–541. doi: 10.1016/j.ajhg.2019.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chik C.L., Rollag M.D., Duncan W.C., Smith A.C. Diagnostic utility of daytime salivary melatonin levels in Smith-Magenis syndrome. Am. J. Med. Genet. A. 2010;152A:96–101. doi: 10.1002/ajmg.a.33158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.UniProt Consortium UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47(D1):D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quan L., Lv Q., Zhang Y. STRUM: structure-based prediction of protein stability changes upon single-point mutation. Bioinformatics. 2016;32:2936–2946. doi: 10.1093/bioinformatics/btw361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan Y., Harper S., Speicher D.W., Marmorstein R. The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate. Nat. Struct. Biol. 2002;9:862–869. doi: 10.1038/nsb849. [DOI] [PubMed] [Google Scholar]
- 25.Yang C., Wu J., Zheng Y.G. Function of the active site lysine autoacetylation in Tip60 catalysis. PLoS ONE. 2012;7:e32886. doi: 10.1371/journal.pone.0032886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Venselaar H., Te Beek T.A., Kuipers R.K., Hekkelman M.L., Vriend G. Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC Bioinformatics. 2010;11:548. doi: 10.1186/1471-2105-11-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laskowski R.A., Stephenson J.D., Sillitoe I., Orengo C.A., Thornton J.M. VarSite: Disease variants and protein structure. Protein Sci. 2020;29:111–119. doi: 10.1002/pro.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang N., Lee I., Marcotte E.M., Hurles M.E. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 2010;6:e1001154. doi: 10.1371/journal.pgen.1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quinodoz M., Royer-Bertrand B., Cisarova K., Di Gioia S.A., Superti-Furga A., Rivolta C. DOMINO: Using Machine Learning to Predict Genes Associated with Dominant Disorders. Am. J. Hum. Genet. 2017;101:623–629. doi: 10.1016/j.ajhg.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wiel L., Baakman C., Gilissen D., Veltman J.A., Vriend G., Gilissen C. MetaDome: Pathogenicity analysis of genetic variants through aggregation of homologous human protein domains. Hum. Mutat. 2019;40:1030–1038. doi: 10.1002/humu.23798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Traynelis J., Silk M., Wang Q., Berkovic S.F., Liu L., Ascher D.B., Balding D.J., Petrovski S. Optimizing genomic medicine in epilepsy through a gene-customized approach to missense variant interpretation. Genome Res. 2017;27:1715–1729. doi: 10.1101/gr.226589.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quang D., Chen Y., Xie X. DANN: a deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics. 2015;31:761–763. doi: 10.1093/bioinformatics/btu703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X., Wu C., Li C., Boerwinkle E. dbNSFP v3.0: A One-Stop Database of Functional Predictions and Annotations for Human Nonsynonymous and Splice-Site SNVs. Hum. Mutat. 2016;37:235–241. doi: 10.1002/humu.22932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kopanos C., Tsiolkas V., Kouris A., Chapple C.E., Albarca Aguilera M., Meyer R., Massouras A. VarSome: the human genomic variant search engine. Bioinformatics. 2019;35:1978–1980. doi: 10.1093/bioinformatics/bty897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rentzsch P., Witten D., Cooper G.M., Shendure J., Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47(D1):D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ponzoni L., Peñaherrera D.A., Oltvai Z.N., Bahar I. Rhapsody: predicting the pathogenicity of human missense variants. Bioinformatics. 2020;36:3084–3092. doi: 10.1093/bioinformatics/btaa127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pejaver V., Urresti J., Lugo-Martinez J., Pagel K.A., Lin G.N., Nam H.-J., Mort M., Cooper D.N., Sebat J., Iakoucheva L.M. MutPred2: inferring the molecular and phenotypic impact of amino acid variants. bioRxiv. 2017 doi: 10.1101/134981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu Y., Fisher J.B., Koprowski S., McAllister D., Kim M.S., Lough J. Homozygous disruption of the Tip60 gene causes early embryonic lethality. Dev. Dyn. 2009;238:2912–2921. doi: 10.1002/dvdy.22110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao Y., Hamers N., Rakhshandehroo M., Berger R., Lough J., Kalkhoven E. Allele compensation in tip60+/- mice rescues white adipose tissue function in vivo. PLoS ONE. 2014;9:e98343. doi: 10.1371/journal.pone.0098343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dickinson M.E., Flenniken A.M., Ji X., Teboul L., Wong M.D., White J.K., Meehan T.F., Weninger W.J., Westerberg H., Adissu H., International Mouse Phenotyping Consortium. Jackson Laboratory. Infrastructure Nationale PHENOMIN, Institut Clinique de la Souris (ICS) Charles River Laboratories. MRC Harwell. Toronto Centre for Phenogenomics. Wellcome Trust Sanger Institute. RIKEN BioResource Center High-throughput discovery of novel developmental phenotypes. Nature. 2016;537:508–514. doi: 10.1038/nature19356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fisher J.B., Kim M.S., Blinka S., Ge Z.D., Wan T., Duris C., Christian D., Twaroski K., North P., Auchampach J., Lough J. Stress-induced cell-cycle activation in Tip60 haploinsufficient adult cardiomyocytes. PLoS ONE. 2012;7:e31569. doi: 10.1371/journal.pone.0031569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gehrking K.M., Andresen J.M., Duvick L., Lough J., Zoghbi H.Y., Orr H.T. Partial loss of Tip60 slows mid-stage neurodegeneration in a spinocerebellar ataxia type 1 (SCA1) mouse model. Hum. Mol. Genet. 2011;20:2204–2212. doi: 10.1093/hmg/ddr108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peukert D., Weber S., Lumsden A., Scholpp S. Lhx2 and Lhx9 determine neuronal differentiation and compartition in the caudal forebrain by regulating Wnt signaling. PLoS Biol. 2011;9:e1001218. doi: 10.1371/journal.pbio.1001218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dalal J., Roh J.H., Maloney S.E., Akuffo A., Shah S., Yuan H., Wamsley B., Jones W.B., de Guzman Strong C., Gray P.A. Translational profiling of hypocretin neurons identifies candidate molecules for sleep regulation. Genes Dev. 2013;27:565–578. doi: 10.1101/gad.207654.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaur M., Izumi K., Wilkens A.B., Chatfield K.C., Spinner N.B., Conlin L.K., Zhang Z., Krantz I.D. Genome-wide expression analysis in fibroblast cell lines from probands with Pallister Killian syndrome. PLoS ONE. 2014;9:e108853. doi: 10.1371/journal.pone.0108853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin E.A., Muralidhar S., Wang Z., Cervantes D.C., Basu R., Taylor M.R., Hunter J., Cutforth T., Wilke S.A., Ghosh A., Williams M.E. The intellectual disability gene Kirrel3 regulates target-specific mossy fiber synapse development in the hippocampus. eLife. 2015;4:e09395. doi: 10.7554/eLife.09395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kalsner L., Twachtman-Bassett J., Tokarski K., Stanley C., Dumont-Mathieu T., Cotney J., Chamberlain S. Genetic testing including targeted gene panel in a diverse clinical population of children with autism spectrum disorder: Findings and implications. Mol. Genet. Genomic Med. 2018;6:171–185. doi: 10.1002/mgg3.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Conrad S., Demurger F., Moradkhani K., Pichon O., Le Caignec C., Pascal C., Thomas C., Bayart S., Perlat A., Dubourg C. 11q24.2q24.3 microdeletion in two families presenting features of Jacobsen syndrome, without intellectual disability: Role of FLI1, ETS1, and SENCR long noncoding RNA. Am. J. Med. Genet. A. 2019;179:993–1000. doi: 10.1002/ajmg.a.61113. [DOI] [PubMed] [Google Scholar]
- 49.Gilissen C., Hehir-Kwa J.Y., Thung D.T., van de Vorst M., van Bon B.W.M., Willemsen M.H., Kwint M., Janssen I.M., Hoischen A., Schenck A. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–347. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
- 50.Franken P., Dijk D.J. Circadian clock genes and sleep homeostasis. Eur. J. Neurosci. 2009;29:1820–1829. doi: 10.1111/j.1460-9568.2009.06723.x. [DOI] [PubMed] [Google Scholar]
- 51.Le T.N., Williams S.R., Alaimo J.T., Elsea S.H. Genotype and phenotype correlation in 103 individuals with 2q37 deletion syndrome reveals incomplete penetrance and supports HDAC4 as the primary genetic contributor. Am. J. Med. Genet. A. 2019;179:782–791. doi: 10.1002/ajmg.a.61089. [DOI] [PubMed] [Google Scholar]
- 52.Schmidt R.L., Sheeley S.L. Mating and memory: an educational primer for use with “epigenetic control of learning and memory in Drosophila by Tip60 HAT action”. Genetics. 2015;200:21–28. doi: 10.1534/genetics.115.176313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu S., Wilf R., Menon T., Panikker P., Sarthi J., Elefant F. Epigenetic control of learning and memory in Drosophila by Tip60 HAT action. Genetics. 2014;198:1571–1586. doi: 10.1534/genetics.114.171660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu S., Elefant F. Tip off the HAT- Epigenetic control of learning and memory by Drosophila Tip60. Fly (Austin) 2015;9:22–28. doi: 10.1080/19336934.2015.1080887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pirooznia S.K., Chiu K., Chan M.T., Zimmerman J.E., Elefant F. Epigenetic regulation of axonal growth of Drosophila pacemaker cells by histone acetyltransferase tip60 controls sleep. Genetics. 2012;192:1327–1345. doi: 10.1534/genetics.112.144667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Petkau N., Budak H., Zhou X., Oster H., Eichele G. Acetylation of BMAL1 by TIP60 controls BRD4-P-TEFb recruitment to circadian promoters. eLife. 2019;8:e43235. doi: 10.7554/eLife.43235. 10.7554/eLife.43235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singh N., Lorbeck M.T., Zervos A., Zimmerman J., Elefant F. The histone acetyltransferase Elp3 plays in active role in the control of synaptic bouton expansion and sleep in Drosophila. J. Neurochem. 2010;115:493–504. doi: 10.1111/j.1471-4159.2010.06892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Duan R., Liu X., Wang T., Wu L., Gao X., Zhang Z. Histone Acetylation Regulation in Sleep Deprivation-Induced Spatial Memory Impairment. Neurochem. Res. 2016;41:2223–2232. doi: 10.1007/s11064-016-1937-6. [DOI] [PubMed] [Google Scholar]
- 59.Qureshi I.A., Mehler M.F. Epigenetics of sleep and chronobiology. Curr. Neurol. Neurosci. Rep. 2014;14:432. doi: 10.1007/s11910-013-0432-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Williams S.R., Aldred M.A., Der Kaloustian V.M., Halal F., Gowans G., McLeod D.R., Zondag S., Toriello H.V., Magenis R.E., Elsea S.H. Haploinsufficiency of HDAC4 causes brachydactyly mental retardation syndrome, with brachydactyly type E, developmental delays, and behavioral problems. Am. J. Hum. Genet. 2010;87:219–228. doi: 10.1016/j.ajhg.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.De Leersnyder H., De Blois M.C., Claustrat B., Romana S., Albrecht U., Von Kleist-Retzow J.C., Delobel B., Viot G., Lyonnet S., Vekemans M., Munnich A. Inversion of the circadian rhythm of melatonin in the Smith-Magenis syndrome. J. Pediatr. 2001;139:111–116. doi: 10.1067/mpd.2001.115018. [DOI] [PubMed] [Google Scholar]
- 62.Smith A.C.M., Morse R.S., Introne W., Duncan W.C., Jr. Twenty-four-hour motor activity and body temperature patterns suggest altered central circadian timekeeping in Smith-Magenis syndrome, a neurodevelopmental disorder. Am. J. Med. Genet. A. 2019;179:224–236. doi: 10.1002/ajmg.a.61003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mullegama S.V., Pugliesi L., Burns B., Shah Z., Tahir R., Gu Y., Nelson D.L., Elsea S.H. MBD5 haploinsufficiency is associated with sleep disturbance and disrupts circadian pathways common to Smith-Magenis and fragile X syndromes. Eur. J. Hum. Genet. 2015;23:781–789. doi: 10.1038/ejhg.2014.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nováková M., Nevsímalová S., Príhodová I., Sládek M., Sumová A. Alteration of the circadian clock in children with Smith-Magenis syndrome. J. Clin. Endocrinol. Metab. 2012;97:E312–E318. doi: 10.1210/jc.2011-2750. [DOI] [PubMed] [Google Scholar]
- 65.Martin H.C., Jones W.D., McIntyre R., Sanchez-Andrade G., Sanderson M., Stephenson J.D., Jones C.P., Handsaker J., Gallone G., Bruntraeger M., Deciphering Developmental Disorders Study Quantifying the contribution of recessive coding variation to developmental disorders. Science. 2018;362:1161–1164. doi: 10.1126/science.aar6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Della Ragione F., Vacca M., Fioriniello S., Pepe G., D’Esposito M. MECP2, a multi-talented modulator of chromatin architecture. Brief. Funct. Genomics. 2016;15:420–431. doi: 10.1093/bfgp/elw023. [DOI] [PubMed] [Google Scholar]
- 67.Deardorff M.A., Bando M., Nakato R., Watrin E., Itoh T., Minamino M., Saitoh K., Komata M., Katou Y., Clark D. HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature. 2012;489:313–317. doi: 10.1038/nature11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tietze A.L., Blankenburg M., Hechler T., Michel E., Koh M., Schlüter B., Zernikow B. Sleep disturbances in children with multiple disabilities. Sleep Med. Rev. 2012;16:117–127. doi: 10.1016/j.smrv.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 69.Gadoth N., Oksenberg A. Sleep and sleep disorders in rare hereditary diseases: a reminder for the pediatrician, pediatric and adult neurologist, general practitioner, and sleep specialist. Front. Neurol. 2014;5:133. doi: 10.3389/fneur.2014.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen L., Mullegama S.V., Alaimo J.T., Elsea S.H. Smith-Magenis syndrome and its circadian influence on development, behavior, and obesity - own experience. Dev Period Med. 2015;19:149–156. [PubMed] [Google Scholar]
- 71.Trickett J., Heald M., Oliver C., Richards C. A cross-syndrome cohort comparison of sleep disturbance in children with Smith-Magenis syndrome, Angelman syndrome, autism spectrum disorder and tuberous sclerosis complex. J. Neurodev. Disord. 2018;10:9. doi: 10.1186/s11689-018-9226-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ansar M., Paracha S.A., Serretti A., Sarwar M.T., Khan J., Ranza E., Falconnet E., Iwaszkiewicz J., Shah S.F., Qaisar A.A. Biallelic variants in FBXL3 cause intellectual disability, delayed motor development and short stature. Hum. Mol. Genet. 2019;28:972–979. doi: 10.1093/hmg/ddy406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kennedy J., Goudie D., Blair E., Chandler K., Joss S., McKay V., Green A., Armstrong R., Lees M., Kamien B., DDD Study KAT6A Syndrome: genotype-phenotype correlation in 76 patients with pathogenic KAT6A variants. Genet. Med. 2019;21:850–860. doi: 10.1038/s41436-018-0259-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Campeau P.M., Lu J.T., Dawson B.C., Fokkema I.F., Robertson S.P., Gibbs R.A., Lee B.H. The KAT6B-related disorders genitopatellar syndrome and Ohdo/SBBYS syndrome have distinct clinical features reflecting distinct molecular mechanisms. Hum. Mutat. 2012;33:1520–1525. doi: 10.1002/humu.22141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNaseq data are available on the National Center for Biotechnology Information (NCBI)’s Gene Expression Omnibus with accession number GSE154199.