Abstract
Signal transduction through the RAF-MEK-ERK pathway, the first described mitogen-associated protein kinase (MAPK) cascade, mediates multiple cellular processes and participates in early and late developmental programs. Aberrant signaling through this cascade contributes to oncogenesis and underlies the RASopathies, a family of cancer-prone disorders. Here, we report that de novo missense variants in MAPK1, encoding the mitogen-activated protein kinase 1 (i.e., extracellular signal-regulated protein kinase 2, ERK2), cause a neurodevelopmental disease within the RASopathy phenotypic spectrum, reminiscent of Noonan syndrome in some subjects. Pathogenic variants promote increased phosphorylation of the kinase, which enhances translocation to the nucleus and boosts MAPK signaling in vitro and in vivo. Two variant classes are identified, one of which directly disrupts binding to MKP3, a dual-specificity protein phosphatase negatively regulating ERK function. Importantly, signal dysregulation driven by pathogenic MAPK1 variants is stimulus reliant and retains dependence on MEK activity. Our data support a model in which the identified pathogenic variants operate with counteracting effects on MAPK1 function by differentially impacting the ability of the kinase to interact with regulators and substrates, which likely explains the minor role of these variants as driver events contributing to oncogenesis. After nearly 20 years from the discovery of the first gene implicated in Noonan syndrome, PTPN11, the last tier of the MAPK cascade joins the group of genes mutated in RASopathies.
Keywords: ERK2, Noonan syndrome, RASopathies, MAPK cascade, intracellular signaling, RAS signaling, MKP3, RSK, exome sequencing, C. elegans
Introduction
Signaling through the RAF-MEK-ERK pathway, the first described mitogen-associated protein kinase (MAPK) cascade, mediates multiple cellular processes in response to growth factors, cytokines, and hormones.1,2 Due to its nodal role in signal transduction, signal traffic through this cascade is tightly controlled, and enhanced flow through it is a well-characterized event contributing to oncogenesis.3 At the organism level, MAPK signaling participates in early and late developmental programs, including organogenesis, morphology determination, synaptic plasticity, and growth.4, 5, 6 Consistently, germline mutations in genes encoding proteins with a role in the RAS-MAPK signaling cascade or having a functional link to this pathway have been linked causally to the RASopathies, a family of genetic disorders sharing distinctive facial dysmorphisms, short stature, variable cognitive impairment and behavioral problems, and a wide spectrum of cardiac defects as major features.7, 8, 9 These syndromes are also characterized by a variably increased risk for certain childhood malignancies, including rhabdomyosarcoma, neuroblastoma, brain tumors, acute leukemias, and juvenile myelomonocytic leukemia.10, 11, 12 Notably, these discoveries have elucidated a paradigm with two major “classes” of activating pathogenic variants involving proteins of the RAS-MAPK pathway, somatic defects in cancer and germline lesions in RASopathies, that have differential impacts on cell transformation and development.7,13
Noonan syndrome (NS [MIM: PS163950]), the most common and clinically variable disorder among the RASopathies,14 is genetically heterogeneous and is caused by pathogenic variants in more than ten genes encoding proteins with roles in the RAS-MAPK signaling cascade or having functional links to this pathway, with variation in PTPN11 (MIM: 176876),15 SOS1 (MIM: 182530),16,17 RAF1 (MIM: 164760),18,19 RIT1 (MIM: 609591),20 and LZTR1 (MIM: 600574)21,22 accounting for the majority of cases. In about 10%–20% of affected individuals, however, the clinical diagnosis is not molecularly confirmed, suggesting that additional genes are likely implicated in the pathogenesis of this disorder or raising the possibility that other still unrecognized RASopathies (i.e., diseases caused by RAS-MAPK signaling dysregulation) or genocopies (i.e., diseases with significant clinical overlap with NS but caused by distinct molecular mechanisms) may account for these affected individuals.
In this study, we established that de novo missense variants in MAPK1, encoding the mitogen-activated protein kinase 1 (also known as extracellular signal-regulated protein kinase 2, ERK2), underlie a neurodevelopmental disorder within the clinical spectrum of the RASopathies. We characterized the clinical profile of seven unrelated affected individuals and used primary fibroblasts and cell lines ectopically expressing individual mutant alleles to demonstrate that these mutations promote stimulus-dependent gain-of-function (GoF) of the kinase and enhance MAPK signaling in vitro. Knock-in and transgenic mpk-1 lines in Caenorhabditis elegans (C. elegans) were used to demonstrate the activating behavior of the tested amino acid substitutions in vivo. Our in silico and in vitro analyses indicate that the identified variants operate with counteracting effects on MAPK1 function by differentially impacting the ability of the kinase to interact with regulators and substrates and that their GoF retains dependence on MEK activity, which likely explains the minor impact of MAPK1 variation as a driver event in oncogenesis.
Subjects and Methods
Subjects
The study was approved by the local Institutional Ethical Committee of the Ospedale Pediatrico Bambino Gesù, Rome. Subjects 1 and 2 were analyzed in the frame of dedicated research projects focused on RASopathies, while the other individuals were referred for diagnostic genetic testing. Clinical data and DNA specimens were collected, stored, and used following procedures in accordance with the ethical standards of the declaration of Helsinki protocols, with signed informed consents from the participating subjects/families. Explicit permission was obtained to publish the photographs of the subjects as shown in Figure 1.
Figure 1.
Clinical Features of Individuals with De Novo MAPK1 Variants
Facial features of subjects 1 to 7. Note the occurrence of hypertelorism, downslanting palpebral fissures, ptosis, low-set/posteriorly rotated ears with evident antitragus and earlobes with central depression, long philtrum with evident columns, marked upper lip vermilion and everted lower lip, and short/webbed neck. Craniofacial features resemble NS or a related RASopathy in subjects 1, 2, 3, 4, and 6. The evolving phenotype of subject 2 with age is reported in Figure S3.
Exome Sequencing Analyses
Whole-exome sequencing (WES) was performed using DNA samples obtained from leukocytes. A trio-based strategy was used in all cases. Target enrichment kits, sequencing platforms, and WES statistics are reported in Table S1. WES data processing, sequence alignment to GRCh37, and variant filtering and prioritization by allele frequency, predicted functional impact, and inheritance models were performed using the indicated pipeline (UMCUGenetics/IAP; subject 5) or as previously reported.23, 24, 25, 26, 27, 28, 29 WES data output is summarized in Table S2. The de novo origin of the MAPK1 mutations was confirmed by Sanger sequencing in all cases (primer sequences are available on request).
Expression Constructs and Primary Cell Lines
The entire MAPK1 coding sequence was cloned into the pcDNA6/Xpress-HisA eukaryotic expression vector (in vitro studies). Mutations were introduced using primed amplification by polymerase chain reaction (site-directed mutagenesis), as previously described.30 Constructs were subcloned into the pB255 vector (a gift from S. Takagi, University of Nagoya) (in vivo studies). All constructs were Sanger sequenced bidirectionally for their entire open reading frame. Subject 2’s primary fibroblasts were obtained from skin biopsy after signed informed consent, following standard procedures. Human Lenti-X 293T, HEK293T, and fibroblast cells were maintained in DMEM medium supplemented with 10% heat-inactivated FBS, 1% L-glutamine, and antibiotics.
In Vitro Studies
Transient transfections and MAPK/RSK/MCL1 phosphorylation assays were performed as previously reported.24,31,32
Experiments used the following antibodies: mouse monoclonal anti-Xpress (Invitrogen); mouse monoclonal anti-Myc, rabbit polyclonal anti-p44/42 MAPK1/3 (ERK2/1), mouse monoclonal anti-phospho-p44/42 MAPK1/3 (ERK2/1) (Thr202/Tyr204 of Erk1 and Thr185 and Tyr187 of Erk2), rabbit polyclonal anti-MCL1, rabbit polyclonal anti-phospho-MCL1 (Ser159/Thr163), rabbit monoclonal anti-RSK1/RSK2/RSK3 (32D7), rabbit polyclonal anti-phospho-p90RSK (Thr359/Ser363) (Cell Signaling Technology); mouse monoclonal anti-MKP3 and mouse monoclonal anti-GAPDH (Santa Cruz); horseradish peroxidase-conjugated anti-rabbit or anti-mouse (Sigma-Aldrich).
MAPK/RSK/MCL1 phosphorylation assays after treatment with trametinib were performed on transfected HEK293T cells seeded in 6-well plates the day before transfection (70% to 80% confluence) and on subconfluent primary fibroblasts. Cells were serum starved for 16 h and then treated with the MEK1 inhibitor (1.5 ng/mL, 2 h) or left untreated, and then stimulated with EGF (transfected cells: 30 ng/mL, 1 min; primary fibroblasts: 10 ng/mL, 15 min) or left unstimulated.
MAPK1/MEK1 co-IP assays were performed on serum-starved transfected HEK293T cells, while MAPK1/MKP3 co-IP experiments were performed on transfected HEK293T cells and primary fibroblasts serum starved for 16 h, and then stimulated with EGF (transfected cells: 30 ng/mL, 1 min; primary fibroblasts: 10 ng/mL, 15 min), as reported.33
Transactivation reporter assays were performed using Lenti-X 293T cells seeded in 12-well plates the day before transfection. Monolayers were transfected with the following plasmids: pFR-Luc (a reporter plasmid in which luciferase expression is driven by minimal promoters for ELK1), pFA2-Elk1, pRL-TK-Renilla, and vectors expressing wild-type MAPK1 or each of the tested mutants. At 24 h after transfection, cells were serum starved for 18 h and then stimulated with EGF (30 ng/mL) for 5 h or left unstimulated. Normalized luciferase levels were reported as fold activation over vector expressing MAPK1, basally.
For nuclear translocation analyses, Lenti-X 293T cells were seeded at the density of 3 × 103 in 24-well cluster plates onto 12 mm cover glasses. After 24 h of culture in complete medium, cells were transfected with the pcDNA6 constructs. At 24 h after transfection, cells were serum starved for 18 h and then treated with EGF (30 ng/mL, 5 min) or left unstimulated and fixed with 3% paraformaldehyde. Fixed cells were permeabilized with 0.5% Triton X-100 and stained with a mouse monoclonal anti-Xpress followed by goat anti-mouse Alexa Fluor 594 and then with phalloidin antibody conjugated to green-fluorescent Alexa Fluor 488 dye (Invitrogen). After staining, coverslips were extensively rinsed and then mounted on the microscope slide by using Vectashield with DAPI mounting medium (Vector Laboratories). Confocal analysis was performed on a Zeiss LSM 980 with Airyscan2, using the 63× oil objective and excitation spectral laser lines at 405 and 488 nm. Cells stained only with the fluorochrome-conjugated secondary antibodies were used to set up acquisition parameters. Signals from different fluorescent probes were taken in sequential scanning mode.
C. elegans Studies
Culture, maintenance, germline transformation, and genetic crosses were performed using standard techniques.34,35 The Bristol N2 (wild-type animals) and PS21 (let-23(sy1) II; him-5(e1490) V) strains were provided by the Caenorhabditis Genetics Center (CGC, University of Minnesota, Minneapolis, MN). let-23(sy1) is a hypomorphic allele of let-23 (ortholog of the human EGFR gene) harboring a nonsense mutation leading to the lack of the last six amino acids and originally isolated as suppressor of the multivulva phenotype of lin-15(n309) animals.36,37 The engineered change to the endogenous mpk-1 locus was carried out by CRISPR-Cas9 as previously described.38 A total of 20 wild-type animals were injected with a mix containing 750 ng/μL Cas9 (IDT), 700 ng/μL ALT-R CRISPR tracrRNA (IDT), 115 ng/μL dpy-10 crRNA (5′-CTCGTGGTGCCTATGGTAGC-3′), 37.5 ng/μL ssODN dpy-10 (5′-ATAGGCTGTTGGTCTGAAGCCATGTGAAGCTCCGCTACCATAGGCACCGCATGCGGTACGGTTTCCAGTCATTCTCATCTTGCCGTATTGAAGTTCAAGTG-3′), 350 ng/μL mpk-1 crRNA (5′-TCACAAACTGGCTCATCTCC-3′), and 175 ng/μL ssODN mpk-1[D321G] (5′-ATTTCGATTTTCAGTATCGACATCGAGCAAGCATTAGCACATCCTTATTTAGAACAGTATTATGGTCCTGGTGATGAGCCAGTTTGTGAGGAACCATTCACTTTGGA-3′). Worms were then recovered on normal growth medium (NGM) at 20°C. Animals with a roller (rol) or a dumpy (dpy) phenotype were isolated, as well as pools of five wild-type hermaphrodites from jackpot plates (i.e., plates with several rol and dpy animals). To isolate worms harboring the mpk-1 variant, PCR amplification was performed using a single forward primer (5′-GGCTGTGTCGATTTACGTACG-3′) and two reverse primers annealing with the wild-type (5′-GGTGCATTCACATCACAGCTG-3′) or the modified (5′-GCACACAGGTTCGTCACCAGG-3′) sequence. Homozygosity was confirmed by Sanger sequencing. Three independent lines harboring the desired change were generated; two of them were backcrossed three times to the N2 strain to remove any potential off-target mutation and were used for further analyses and crosses. These lines showed an equivalent phenotype and were designated as mpk-1(pan14[D321G]).
The wild-type MAPK1 cDNA and a subset of mutant alleles were subcloned into the pB255 vector to be under the control of the lin-31 promoter (plin-31) and were injected at 100 ng/μL to generate multi-copy extrachromosomal arrays. The pJM371 plasmid (pelt-2::NLS::GFP) (a gift from J.D. McGhee, University of Calgary), driving GFP expression in intestinal cell nuclei, was used as co-injection marker (30 ng/μL). Phenotypic analysis was performed on three independent lines for each construct.
L3/L4 larvae and adult hermaphrodites were scored quantitatively for assessing vulval induction (larvae) and for the presence of multiple ectopic pseudovulvae (multivulva, Muv phenotype) and lack of a vulva (vulvaless, Vul phenotype) (adults). Muv and Vul were scored at a Leica MZ10F dissecting microscope, while vulval induction was evaluated using an Eclipse Ti2-E microscope (Nikon) equipped with DIC optics on live animals mounted on 2% agarose pads containing 10 mM sodium azide as anesthetic. let-23(sy1) hermaphrodites are egg-laying defective (Egl phenotype) due to the lack of a functional vulva, and larvae accumulate and hatch inside the mother resulting in a bag-of-worms phenotype (Bag). Based on that, the prevalence of Vul in let-23(sy1);mpk-1(D321G) and let-23(sy1);plin-31::MAPK1 double mutants was scored blindly at a dissecting microscope by counting the number of eggs retained in the uterus and identifying animals that had become Bag. More specifically, to assess the Egl phenotype, L4 larvae were picked and visually scored after 24 h at 20°C for the accumulation of eggs inside their body, while Bags were later identified by scoring the presence of larvae inside the mother. After each cross, the genotype of individual alleles (mpk-1 and let-23) was confirmed by direct sequencing of the appropriate genomic region. In transgenic experiments, isogenic animals that had lost the transgene were cloned separately and used as controls. p values were calculated using two-tailed Fisher’s exact test.
Structural Analyses
Analyses were performed using the UCSF Chimera package.39 Side chains not solved in the X-ray structures were modeled according to Shapovalov and Dunbrack.40
Results
De Novo Missense Variants in MAPK1 Cause a Neurodevelopmental Disorder within the RASopathy Clinical Spectrum
We performed WES to identify the molecular causes underlying a group of clinically heterogeneous phenotypes with features suggestive of NS or overlapping related RASopathies but without pathogenic variant(s) in previously identified genes. WES data analysis excluded the occurrence of other functionally relevant variants compatible with known Mendelian disorders based on the expected inheritance model and clinical presentation; however, de novo missense changes in MAPK1 were observed in two individuals (c.521C>T [p.Ala174Val], subject 1; c.953A>G [p.Asp318Gly], subject 2; GenBank: NM_002745.4) (Tables S1 and S2). In subject 2, the missense change was confirmed in skin fibroblasts, providing evidence for its germline or early post-zygotic origin. Both variants affected highly conserved residues among MAPK1 orthologs and paralogs (Figure S1), had high CADD scores, and had not previously been reported in gnomAD (Table 1). MAPK1 is highly intolerant to loss-of-function (LoF) variants (pLI 1.0) and highly constrained for missense variation (Z = 3.61),41 further supporting the clinical relevance of this finding. Networking and connection via GeneMatcher42 allowed us to identify five additional unrelated subjects carrying a de novo missense MAPK1 substitution (Table 1). In all subjects, no other clinically associated variant was identified (Tables S1 and S2). All MAPK1 variants were private and affected invariant residues among MAPK1 orthologs and paralogs (Figure S1), and multiple in silico prediction algorithms rated these changes as deleterious or pathogenic (Table 1 and Figure S2). Notably, a subset of the identified variants mapped to the cancer-associated MAPK1 mutation cluster involving Asp321 and Glu322 (COSMIC database). One substitution (c.952G>A [p.Asp318Asn]) affected the same codon as subject 2 (c.953A>G [p.Asp318Gly]), further documenting the non-random distribution of the identified variants. In all samples, no evidence of allelic imbalance was observed (reference versus variant allele ratio between 0.46 and 0.60), strongly suggesting a germline origin of the identified de novo variants.
Table 1.
List of the Identified Pathogenic MAPK1 Missense Variants
| Nucleotide Change | Amino Acid Change | Domain | Mutation Class | Subject | Origin | REVELa | CADD phreda | Metadome dN/dSb | ACMGc |
|---|---|---|---|---|---|---|---|---|---|
| c.221T>A | p.Ile74Asn | active site | 2 | 7 | de novo | 0.82 | 29.0 | 0.21 | pathogenic |
| c.238C>T | p.His80Tyr | DRS | 1 | 3 | de novo | 0.88 | 30.0 | 0.27 | pathogenic |
| c.521C>T | p.Ala174Val | activation lip | 2 | 1 | de novo | 0.45 | 23.9 | 0.10 | pathogenic |
| c.952G>A | p.Asp318Asn | DRS | 1 | 4 | de novo | 0.31 | 28.9 | 0.27 | pathogenic |
| c.953A>G | p.Asp318Gly | DRS | 1 | 2 | de novo | 0.39 | 25.7 | pathogenic | |
| c.964G>C | p.Glu322Gln | DRS | 1 | 6 | de novo | 0.31 | 26.7 | 0.19 | pathogenic |
| c.968C>G | p.Pro323Arg | DRS | 1 | 5 | de novo | 0.73 | 28.2 | 0.18 | pathogenic |
Nucleotide numbering reflects cDNA numbering with 1 corresponding to the A of the ATG translation initiation codon in the MAPK1 reference sequence (GenBank: NM_002745.4). No variants were reported in the public databases ExAC and gnomAD.
All variants were predicted to be “deleterious” by Rare Exome Variant Ensemble Learner (REVEL) and/or Combined Annotation Dependent Depletion (CADD) v.1.3 algorithms.43,44 Scores > 15 (CADDphred) or > 0.5 (REVEL) predict that the sequence change has a significant impact on protein structure and function.
dN/dS metric detects evolutionary pressure in protein-coding regions and genomes and is used to measure genetic tolerance.45 Residues with dN/dS values < 0.53 are considered to map within regions “intolerant” to nonsynonymous variation, those with a ratio < 0.18 are supposed to be within “highly intolerant” regions (see Figure S2).
All changes satisfied the necessary criteria to be classified as pathogenic according to the American College of Medical Genetics criteria (see InterVar in Web Resources).46 c.221T>A: PS2, PS3, PM1, PM2, PP2, PP3. c.238C>T: PS2, PS3, PM1, PM2, PP2, PP3. c.521C>T: PS2, PS3, PM1, PM2, PP2, PP3. c.952G>A: PS2, PS3, PM1, PM2, PP2, PP3. c.953A>G: PS2, PS3, PM1, PM2, PP2, PP3. c.964G>C: PS2, PS3, PM1, PM2, PM5, PP2, PP3. c.968C>G: PS2, PS3, PM1, PM2, PP2, PP3.
The seven subjects with de novo MAPK1 mutations shared a neurodevelopmental disorder, with a phenotype reminiscent of RASopathy in a subset of cases (Figures 1 and S3, Table S3). A clinical history of each individual can be found in the Supplemental Note (Case Reports). While the overall phenotype of these subjects appeared variable in terms of severity, it consistently included developmental delay (DD), intellectual disability (ID), and behavioral problems (e.g., ADHD, anxiety, reduced stress tolerance, aggressive behavior). Postnatally reduced growth was documented in approximately half of affected individuals. Craniofacial anomalies were also common, including hypertelorism (5/7 cases), ptosis (6/7 cases), downslanting palpebral fissures (3/7 cases), low-set/posteriorly rotated ears (6/7 cases), wide nasal bridge (3/7cases), and dental anomalies (4/7 cases), which are observed as major recurrent features in RASopathies. Notably, most subjects shared a distinctive ear morphology with evident antitragus and earlobes with central depression, and a long and deep philtrum, marked upper lip vermilion, and everted lower lip. The craniofacial appearance and the co-occurrence of a short/webbed neck, low posterior hairline, skin features (e.g., multiple lentigines, cafè au lait spots), and reduced growth substantiated a clinical suspicion of NS or a clinically related trait in subjects 1, 2, 3, 4, and 6. The severe phenotype and peculiar facies of subject 6 was suggestive of cardiofaciocutaneous syndrome (MIM: PS115150). Congenital heart defects, consisting of atrial septal defects and mitral valve insufficiency, were reported in four subjects, but none had cardiomyopathy. Hypotonia and EEG anomalies unmasking potential seizure activity were also noted, which was well controlled by pharmacological treatment (subjects 5 and 6). Minor skeletal defects were also a common finding. Bleeding diathesis and lymphedema occurred in single individuals. None of the subjects was diagnosed with cancer.
MAPK1 Missense Variants Are GoF Alleles
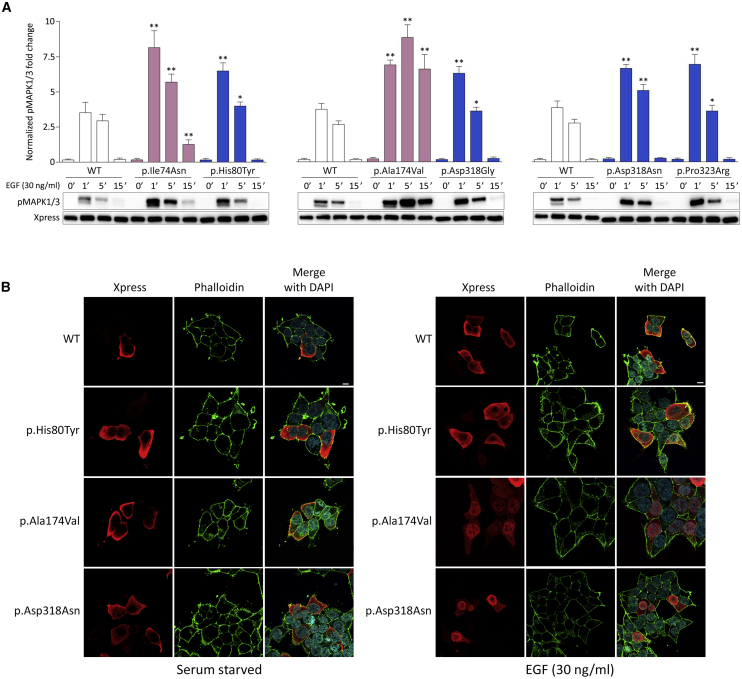
MAPK1 is a ubiquitously expressed protein serine/threonine kinase functioning as terminal tier of the RAS-RAF-MEK-ERK signaling cascade. Its activity requires MEK-mediated phosphorylation of two regulatory phosphorylatable residues, Thr185 and Tyr187, that are located within an activation segment, which favors a conformation of the catalytic domain stabilizing the active site (i.e., DFG-aspartate-in conformation).16 Dephosphorylation of Thr185 and Tyr187 is catalyzed by MAPK phosphatases (MKPs) and is a required step for the inactivation of the kinase.47 The reversible phosphorylated state of the two residues controls the magnitude and duration of activation as well as the nuclear localization of the kinase, which, in turn, determine the nature of the cellular response to stimuli. Due to the clinical overlap of some subjects heterozygous for mutant MAPK1 alleles with NS and the consideration that the latter is caused by upregulated MAPK signaling, we hypothesized a GoF effect of the identified mutations on MAPK1 function. To validate this hypothesis, we assessed the consequences of the disease-associated amino acid substitutions on two key events of MAPK1 function, i.e., its phosphorylation status and translocation to the nucleus. We analyzed the levels of MAPK1 phosphorylation in HEK293T cells transiently transfected with Xpress-tagged MAPK1 constructs encoding each of the p.Ile74Asn, p.His80Tyr, p.Ala174Val, p.Asp318Gly, p.Asp318Asn, and p.Pro323Arg substitutions, under basal conditions and following EGF stimulation, in time-course experiments (Figure 2A). Ectopic expression of all mutants promoted variably boosted stimulus-dependent MAPK1 phosphorylation, compared to cells overexpressing the wild-type protein, suggesting an activating role of each disease-associated variant. The enhanced stimulus-dependent MAPK1 phosphorylation was confirmed in primary skin fibroblasts from subject 2 (Figure S4, left panel). While MAPK1 is located in the cytoplasm basally, the kinase translocates to the nucleus following stimulation, an event that is required to control the function of a wide variety of transcription factors, chromatin modifying enzymes, and structural proteins modulating gene expression.48 MAPK1 nuclear translocation is promoted by phosphorylation of the regulatory Thr185 and Tyr187 residues, which induces a conformational change allowing the release of the kinase from cytoplasmic anchoring proteins, and its nuclear import via importin-mediated transport.49 Confocal laser scanning microscopy analysis was performed in Lenti-X 293T cells transiently expressing Xpress-tagged wild-type and mutant MAPK1 constructs under starved condition and following EGF stimulation. Like the wild-type protein, all disease-associated MAPK1 proteins localized in the cytoplasm basally; however, consistent with their stronger activation following stimulation, they showed a variably more efficient nuclear translocation after treatment with EGF, with amino acid substitutions at residues Ala174, Asp318, and Pro323 displaying the most robust response (Figures 2B and S5).
Figure 2.
Ectopic Expression of Disease-Causing MAPK1 Mutant Alleles Promotes a Variably Boosted Stimulus-Dependent Phosphorylation and Nuclear Translocation of MAPK1
(A) MAPK phosphorylation assays. Representative blots (below) and graphs reporting mean ± SD densitometry values (above) of three independent experiments are shown. Affected residues mapping at the DRS are shown in blue, while those mapping at the activation segment are colored in pink. HEK293T cells were transiently transfected with the indicated Xpress-tagged MAPK1 construct, serum starved (16 h), and stimulated with EGF (30 ng/mL) in time-course experiments or left untreated. Equal amounts of cell lysates were resolved on 10% polyacrylamide gels. Asterisks indicate statistically significant differences in the phosphorylation levels compared to cells overexpressing wild-type MAPK1 at the corresponding experimental points (∗p < 0.05, ∗∗p < 0.01; Student’s t test).
(B) MAPK1 subcellular localization showed by confocal laser scanning microscopy (CLSM) observations. Panels represent central sections. Assays were performed in Lenti-X 293T cells transiently expressing Xpress-tagged wild-type and three representative MAPK1 mutants, basally (left) and after 5 min of EGF stimulation (right). Similarly to the wild type protein, all mutants localized in the cytoplasm basally, whereas they showed a variably more efficient nuclear translocation after EGF stimulation. Fixed cells were stained with an anti-Xpress mouse monoclonal antibody followed by goat anti-mouse Alexa Fluor-594 (red) and an anti-phalloidin antibody conjugated to green-fluorescent Alexa Fluor 488 dye (green). Nuclei are visualized by DAPI staining (blue). Scale bar is 10 μm. Images referred to the complete set of mutants are reported in Figure S5.
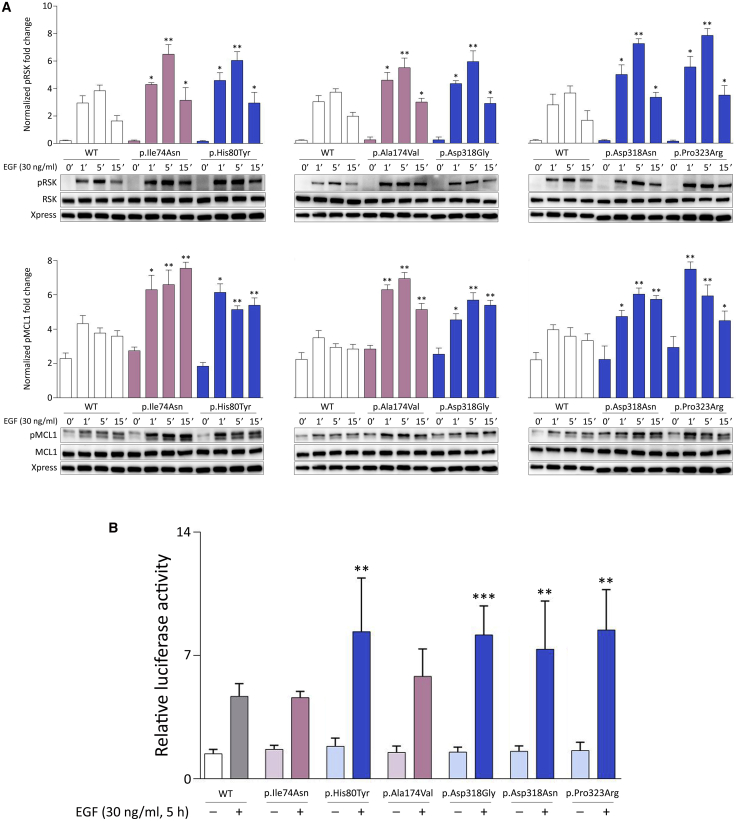
To further confirm the GoF role of the identified variants on protein function, we assessed the consequences of ectopic expression of MAPK1 mutants on the phosphorylation status of two well-known cytoplasmic substrates of the kinase, the p90 ribosomal S6 kinases 1-3 (RSK1 to 3) and BCL2-related pro-survival myeloid cell leukemia 1 (MCL1) protein. Immunoblotting analysis showed that the increased MAPK1 phosphorylation associated with overexpression of each mutant resulted in an enhanced phosphorylation of RSKs and MCL1 (Figure 3A), indicating MAPK signaling upregulation. The enhanced RSK phosphorylation was also confirmed in primary fibroblasts from subject 2 (Figure S4, right panel). To specifically assess the function of individual mutants on nuclear substrates, an ELK1 transactivation assay was performed (Figure 3B). When phosphorylated by MAPK1, ELK1 binds to serum responsive elements of promoters to initiate transcription of target genes.50,51 Lenti-X 293T cells were transfected to co-express either wild-type MAPK1 or each of the six mutants with the C-terminal ERK responsive domain of ELK1 fused to the DNA-binding domain of Gal4, a reporter (i.e., luciferase) under the control of a synthetic promoter with five tandemly arranged GAL4 binding sites, and an internal transfection control (i.e., Renilla luciferase). A variably enhanced induction of the reporter after EGF stimulation was observed in cells expressing most mutants, whereas no difference in the transactivation activity was documented in the unstimulated state. Of note, one mutant (p.Ala174Val) was found to have just a slightly increased transactivation activity, while the activity of the p.Ile74Asn mutant was similar to that of the wild-type protein, suggesting a differential impact of mutations involving the activation segment or regions interacting with it on specific functions of the kinase (see below).
Figure 3.
Disease-Causing MAPK1 Mutations Promote a Variably Enhanced Stimulus-Dependent Activation of the MAPK Signaling Cascade
(A) Overexpression of MAPK1 mutants promote enhanced phosphorylation of RSKs and MCL1, two cytoplasmic substrates of the kinase, as assessed by time-course experiments. Representative blots (below) and graphs reporting mean ± SD densitometry values (above) of three independent experiments are shown. Affected residues mapping at the DRS are shown in blue, while those mapping at the activation segment are colored in pink. HEK293T cells were transiently transfected with the indicated Xpress-tagged MAPK1 construct, serum starved (16 h), and treated with EGF (30 ng/mL) in time-course experiments or left unstimulated. Equal amounts of cell lysates were resolved on 10% polyacrylamide gels. Asterisks indicate statistically significant differences in the phosphorylation levels compared to cells overexpressing wild-type MAPK1 at the corresponding experimental points (∗p < 0.05, ∗∗p < 0.01; Student’s t test).
(B) Luciferase assay in Lenti-X 293T cells. Variably enhanced induction of the reporter after EGF stimulation was observed in cells expressing MAPK1 mutants, whereas no difference in the transactivation activity was documented in the unstimulated state. Elk1-induced expression of luciferase was estimated by measuring luciferase activity in lysates prepared from Lenti-X 293T cells cotransfected with pFR-Luc, pFA2-Elk1, pRL-TK-Renilla, and vectors expressing the wild-type or each of the six MAPK1 mutants. Each value represents luciferase activity in relative light units, which was normalized for Renilla luciferase activity. Three independent experiments were performed, each including duplicate samples. Bars indicate means ± SD. One-way ANOVA (p = 0.0001, R square = 0.8913) followed by Bonferroni’s multiple comparison test (∗∗p < 0.005, ∗∗∗p < 0.001).
In Vivo Assessment of MAPK1 Variants
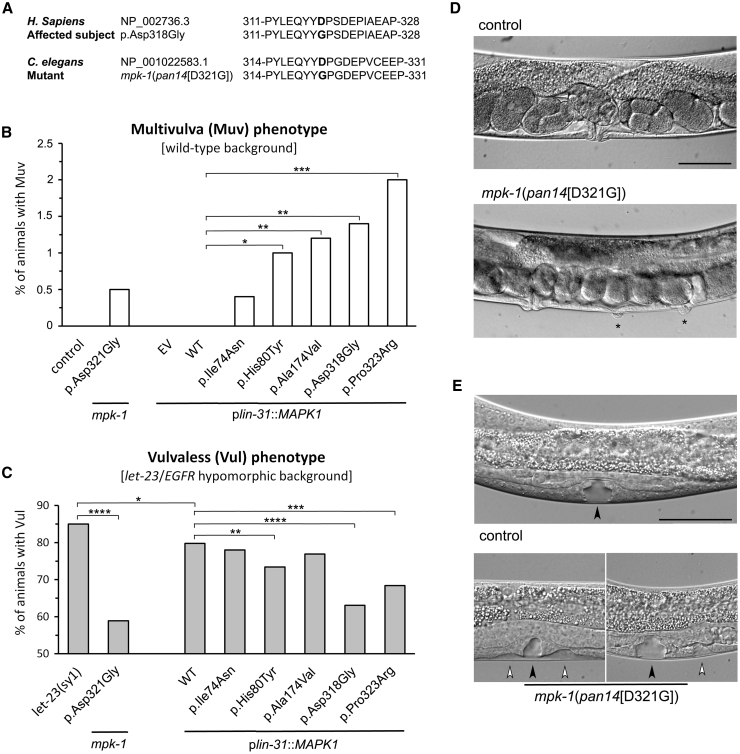
To explore the functional impact of the disease-associated MAPK1 alleles on RAS-MAPK signaling in vivo, we used C. elegans as a model system. The mpk-1/sur-1 gene, ortholog of MAPK1, displays 81% sequence identity and 88% conservation with the human protein (Figure S1). MPK-1 is involved in several LET-60 (RAS)-mediated developmental processes, including vulval cell fate specification.52 Specifically, LET-60 acts via the canonical MAPK cascade (i.e., LIN-45/RAF, MEK-2/MEK, and MPK-1) to promote hermaphrodite vulval development in cooperation with the WNT and LIN-12 (NOTCH) signaling pathways.53 Enhanced LET-60 signaling leads to the formation of multiple ectopic pseudovulvae along the ventral side of the animal as a result of hyperinduction of the six vulval precursor cells (VPCs) (multivulva, Muv phenotype). Conversely, when signal flow through LET-60 is turned off, as in the case of defective upstream signaling elicited by the LET-23 (EGFR) tyrosine kinase receptor, VPCs adopt an alternative cell fate resulting in the absence of any vulval tissue (vulvaless, Vul phenotype). In line with the positive role of MPK-1 in LET-60 signaling, LoF mpk-1 mutations cause a highly penetrant Vul phenotype and suppress the Muv phenotype associated with hyperactive LET-60, while transgenic animals co-expressing GoF mutations of mpk-1 and mek-2 alleles under control of the lin-31 promoter exhibit a high penetrant Muv phenotype.54, 55, 56 Here, we introduced the c.953A>G (p.Asp318Gly) change at the orthologous position of the C. elegans gene by CRISPR-Cas9 to generate mpk-1(pan14[D321G]) knock-in animals (Figure 4A). The resulting nematodes did not show any Vul phenotype, ruling out a LoF/dominant-negative role of the mutation. Conversely, the mutant worms displayed a low penetrant Muv phenotype and partially rescued the Vul phenotype of animals harboring the hypomorphic let-23 (sy1) allele (Figures 4B–4D and Table S4), indicating upregulation of signaling through LET-60 and the MAPK cascade. In line with this evidence, hyperinduction of VPCs was also noted (Figure 4E).
Figure 4.
Consequences of MAPK1 Disease-Causing Variants on C. elegans Vulval Development
(A) Amino acid sequences of wild-type and p.Asp318Gly MAPK1, wild-type MPK-1 (ortholog of MAPK1), and mpk-1(pan14[D321G]) mutant engineered by CRISPR-Cas9. The modified residues are highlighted in bold.
(B) mpk-1(pan14[D321G]) (p.Asp321Gly) knock-in animals displayed a low penetrant multivulva (Muv) phenotype. A variable prevalence of Muv was also observed in worms overexpressing a subset of disease-causing MAPK1 alleles under the control of plin-31, which drives expression principally in vulval precursor cells (VPCs). Asterisks specify significant differences from animals expressing MAPK1WT (∗p < 0.02, ∗∗p < 0.01, ∗∗∗p < 0.0005; two-tailed Fisher’s exact test).
(C) Expression of the mutant alleles ameliorates the vulvaless (Vul) phenotype of animals carrying the hypomorphic let-23 (EGFR) sy1 allele. Asterisks specify significant differences from let-23(sy1) animals (∗p < 0.05, ∗∗∗∗p < 0.00002) or from let-23(sy1) animals expressing MAPK1WT (∗∗p < 0.02, ∗∗∗p < 0.002, ∗∗∗∗p < 0.00002).
(D) Nomarski images show that a normal vulva develops in control adult hermaphrodites (above), whereas a number of multiple ectopic pseudovulvae are observed in mpk-1(pan14[D321G]) animals (below). Asterisks indicate the ectopic pseudovulvae.
(E) Nomarski images of vulval precursor cells (VPCs) at late L3/early L4 larval stages. In control animals, only P6.p detaches from the cuticle generating a single, symmetric invagination (above). This process is altered in a minority of L3/L4 larvae carrying the mpk-1(pan14[D321G]) allele, which show multiple, asymmetric invaginations (below). This phenotype represents the prodromal sign of Muv. Black and white arrowheads point to normal and extra invaginations, respectively. Anterior is to the left and dorsal is up, in all images. Scale bars, 50 μm. EV, empty vector. Number of animals scored are reported in Table S4.
To confirm the GoF behavior of the p.Asp318Gly substitution and extend in vivo our analysis to a larger panel of mutants, transgenic lines overexpressing wild-type MAPK1 or a subset of mutant alleles (those encoding p.Ile74Asn, p.His80Tyr, p.Ala174Val, p.Asp318Gly, and p.Pro323Arg) under the control of plin-31, which drives expression principally in VPCs, were generated and assessed phenotypically. Ectopic expression of the wild-type protein did not elicit any Muv phenotype but slightly ameliorated the Vul phenotype of let-23(sy1) animals (Table S4), indicating that human MAPK1 works appropriately in the context of the C. elegans vulva. Of note, worms overexpressing each of the mutants displayed Muv, although at a low prevalence, and more efficiently rescued the Vul phenotype, with p.Asp318Gly and p.Pro323Arg being the most activating substitutions (Figures 4B and 4C).
Defective Negative MKP3 Regulation Is a Generalizable Mechanism for the GoF Behavior of MAPK1 Variants
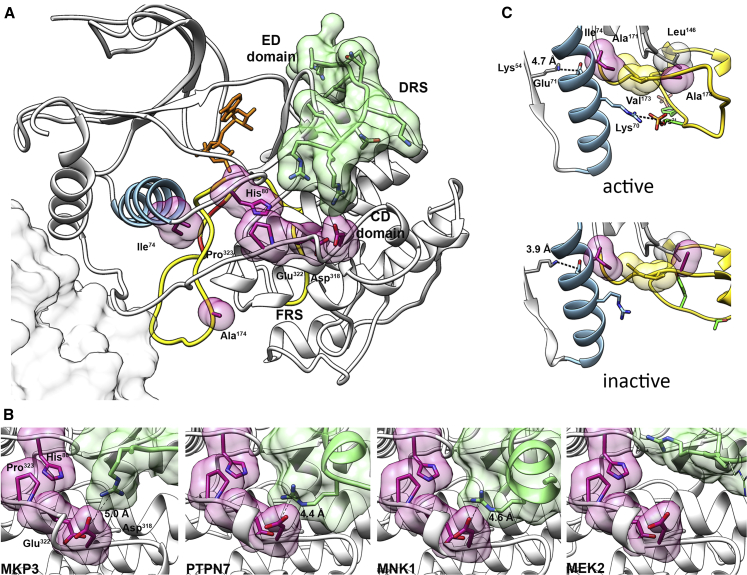
To understand the molecular mechanism(s) by which the identified pathogenic variants promote enhanced MAPK1 function, analysis of the available crystallographic structures of the kinase complexed with regulators and effectors was performed. MAPK1 is characterized by a single kinase domain (residues 25–313), comprising two lobes.57 The small N-terminal lobe is constituted by a five-stranded antiparallel β sheet (1–5) containing residues involved in catalysis, an α helix (helix C) that assumes slightly different orientations in the active and inactive states, and a glycine-rich loop, interacting with the ATP phosphates. The large C-terminal lobe comprises six conserved segments and four short conserved β strands containing most of the catalytic residues. The activation segment, close to the active site, represents the most important regulatory element and encompasses the activation lip, where the phosphorylatable residues Thr185 and Tyr187 are located. Interaction of the kinase with substrates, scaffolding proteins, and positive (MEK1/2) and negative (multiple MKPs) regulators is mediated by two other key regions, the D-site recruitment site (DRS) and F-site recruitment site (FRS).47 Notably, substrates and regulators differentially use those domains to interact with the kinase.58 In particular, the DRS binds to proteins with complementary D-motif linear sequences, and it contains a charged region and a hydrophobic groove. The charged region (termed the common docking [CD] domain) is an anionic patch comprising conserved key acidic residues (i.e., Asp318, Asp321 in MAPK1). The hydrophobic groove (Leu115, Leu121, Leu157, His125, Tyr128) is flanked by a more variable region (ED domain, Thr159, Thr160), important for binding selectivity.58
The affected residues were found to map at two specific regions of the kinase: His80, Asp318, Glu322, and Pro323 are part of or close to the DRS, while Ala174 is located on the activation segment and Ile74 interacts with it (Figure 5A). All residues are distant from the putative dimer interface, or from the FRS, and therefore are not predicted to affect the function of those regions. MAPK1 uses the DRS to bind to positive (i.e., MEK1/2) and negative (e.g., MKP3 and PTPN7) regulators, and a number of substrates (e.g., MNK1) (Figure 5B). MKP3, PTPN7, and MNK1 form a salt bridge interaction with Asp318 and interact with the other affected residues of this region (His80, Glu322, and Pro323). By contrast, the interface is much looser in the case of MEK2. Substitutions affecting residues in this region are thus predicted to affect the DRS affinity for signaling partners, with a weaker effect in the case of MEK2. This analysis is consistent with previous observations indicating that the p.Asp321Asn substitution in X. laevis (equivalent to p.Asp318Asn in the human protein) abolishes binding to MKP3 and MNK1, but only partially reduces association to MEK2.59 Consistent with this, the equivalent substitution in the rat protein, p.Asp316Asn, causes a 10-fold decrease in the affinity for MKP3 and in the catalytic efficiency for the phosphorylation of ELK1 (but not of MBP, another ERK substrate).60 Similarly, the cancer-associated p.Glu322Lys and p.Glu322Val (which are less conservative than the p.Glu322Gln amino acidic change reported here) are refractory to dephosphorylation by MKP3.61
Figure 5.
Location and Functional Impact of Pathogenic Variants
(A) Structure of MAPK1 complexed with a peptide from the D-motif of MKP3 (PDB: 2FYS). Different colors and representations highlight functional important regions. Activation segment, yellow; phosphorylatable residues Thr185 and Tyr187, red; helix C, light blue; mutated residues, semi-transparent violet surfaces; MKP3 D-motif, semi-transparent green surface; putative position of the second monomer in the MAPK1 dimer (according to PDB: 2ERK), gray surface. CD and ED domains, part of the DRS, are indicated. The ATP analog phosphoaminophosphonic acid-adenylate ester, not present in the structure, has been added in the corresponding position from the MAPK1/MNK1 complex (PDB: 2Y9Q) (orange sticks).
(B) Structures of MAPK1 complexes with peptides interacting with the DRS region, corresponding to D-motifs of phosphatases MKP3 (PDB: 2FYS) and PTPN7 (PDB: 2GPH), MAPK1 substrate MNK1 (PDB: 2Y9Q), and kinase MEK2 (PDB: 4H3Q). Representations and colors are used as above. The residues of the interacting peptides forming salt bridges with Asp318 are reported as sticks. In the case of MEK2, no residues form an ion pair with Asp318: the MEK2 residue closest to Asp318 is Arg4, whose side chain was not solved in the crystal structure, indicating that a stable salt bridge between MAPK1-Asp318 and MEK2-Arg4 residues is not present. A putative conformation for Arg4 has been modeled in the structure (thin sticks).
(C) Location and interactions of residues Ile74 and Ala174 in the active and inactive structures of MAPK1 (PDB: 2ERK and 1ERK, respectively). Representations and colors are used as above. Ile74, located on helix C, participates in a hydrophobic cluster comprising activation segment residues Ala171 and Val173, in both protein conformations. This region is critical in the activation mechanism. In the active state, Lys70 forms an ion pair with the phosphorylated Thr185, causing a remodeling of helix C, and the formation of a stronger salt bridge between conserved residues Glu71 and β3 Lys54, which is a prerequisite for activation. Ala174 is in the activation segment and interacts with Leu146, which is in the catalytic region. In all cases, residue numbers correspond to the human sequence.
In contrast, Ala174 is located within the activation segment, i.e., the region that drastically changes its conformation as a consequence of phosphorylation of Thr185 and Tyr187, causing activation of MAPK1 (Figure 5A). In addition, this residue is in contact with the catalytic site (and particularly with the conserved HRD motif) via a hydrophobic interaction with Leu146 (Figure 5C). Substitution with Val is predicted to perturb the allosteric switching of the activation segment. Finally, helix C, where Ile74 is located, has a central role in the activation of MAPK1. It comprises the conserved Glu71, which forms a salt bridge with the phosphate binding residue Lys54, stabilizing its position (Figure 5C).57,62 Phosphorylation of Thr185 and Tyr187 promotes a conformational change of the activation lip that causes multiple interactions between Thr185 and helix C residues. In particular, in the active state, Lys70 and pThr185 form a salt bridge.63 In turn, these interactions cause a remodeling of helix C, strengthening the Glu71-Lys54 ion pair and bringing catalytic residues in the correct alignment.47,63 Residues of helix C, in particular Leu75, have also been implicated in the control of MAPK1 autophosphorylation, through the interaction with the “gatekeeper” residue Gln105.64 Ile74 participates in a hydrophobic cluster comprising the activation segment residues Ala171 and Val173, and it is one helix turn away from conserved Glu71 and Lys70. Its substitution to a polar Asn residue is predicted to perturb the hydrophobic cluster and more in general the interactions of helix C with activation loop and catalytic site residues, which are crucial in the activation process.
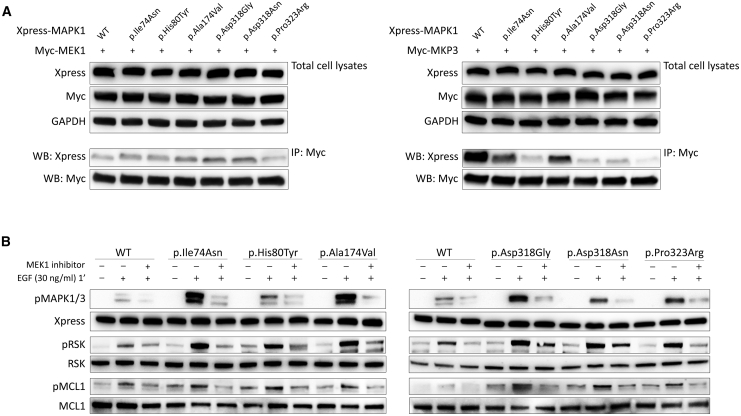
To experimentally validate the hypothesis of a differential impact of the pathogenic MAPK1 amino acid substitutions on the binding properties of the kinase, we performed co-immunoprecipitation assays to assess binding of the mutants to MEK1 and MKP3. Experiments were carried out on lysates collected from HEK293T cells transiently transfected to co-express Xpress-tagged wild-type and mutant MAPK1 proteins together with Myc-tagged MEK1 or MKP3 (Figure 6A). Basally, the co-immunoprecipitated amount of all tested mutants with MEK1 was comparable to that of wild-type MAPK1, demonstrating that mutations do not significantly affect proper binding to the kinase. In contrast, all mutants were less efficiently immunoprecipitated by MKP3 following EGF stimulation compared to the wild-type protein, indicating a variably decreased binding to the phosphatase, with mutants carrying amino acid substitutions at the DRS having the most reduced binding. This finding was confirmed in primary fibroblasts endogenously expressing the p.Asp318Gly variant (Figure S6, left panel). Overall, the data indicate that the activating role of the disease-causing MAPK1 variants is the result, at least in part, of a differential perturbed binding of the kinase with signaling partners.
Figure 6.
Disease-Causing Mutations Impair Binding of MAPK1 with MKP3 but Not Binding to MEK1, and Retain Dependence on MEK Activity in Their Upregulation of MAPK Signaling
(A) Co-immunoprecipitation assays. Lysates from HEK293T cells transiently transfected to express wild-type and mutant Xpress-tagged MAPK1 protein with Myc-MEK1 or Myc-MKP3 were immunoprecipitated with an anti-Myc antibody and assayed by western blotting using the indicated antibodies.
(B) MAPK1 mutation-promoted MAPK signal upregulation retains dependence on MEK activity. MAPK, RSK, and MCL1 phosphorylation assays were performed in transiently transfected HEK293T cells starved for 16 h and stimulated with EGF (30 ng/mL, 1 min) after treatment with the MEK inhibitor trametinib (1.5 ng/mL for 2 h). Blots show a decrease in pMAPK, pRSK and pMCL1 in cells overexpressing wild-type or mutant MAPK1 proteins in presence of trametinib, indicating that the mutation-driven signal upregulation of the MAPK cascade retains dependence on MEK activity.
MAPK1 Variation-Promoted Signaling Upregulation Retains Dependence on MEK Activity
The collected biochemical data indicated that the activating role of MAPK1 variants is stimulus dependent, which raises the possibility that the aberrantly enhanced signaling driven by mutations might be druggable through MEK inhibition. To this goal, assays were performed to assess MAPK1, RSK, and MCL1 phosphorylation following treatment with trametinib, a well-known MEK1 inhibitor,65 on transfected HEK293T cells overexpressing the MAPK1 mutants and primary fibroblasts heterozygous for the p.Asp318Gly change (Figures 6B and S6, right panel). Western blot analysis revealed a significant decrease of MAPK1, RSK, and MCL1 phosphorylation in EGF-stimulated transfected cells and fibroblasts after trametinib treatment, indicating a direct dependency of MAPK1 mutants on MEK1 to drive upregulated signaling through the MAPK cascade.
Discussion
Our findings establish germline MAPK1 missense variants as the cause of a previously unrecognized neurodevelopmental disorder falling within the RASopathy clinical spectrum, with a phenotype reminiscent of NS in a subset of subjects. The non-random distribution of the identified allelic series and our in silico, in vitro, and in vivo data consistently indicate that pathogenic MAPK1 variants cause GoF and promote a boosted signal flow through the MAPK cascade. The collected data suggest that the disease-causing amino acid changes can be subdivided into at least two major classes and offer a mechanistic interpretation for the activating consequences of variants affecting the DRS.
Although two individuals of the present cohort (subjects 1 and 2) were identified among a small cohort of subjects with features suggestive of NS for whom pathogenic variants had not been identified in known RASopathy-associated genes, systematic screening of the entire MAPK1 coding sequence in an additional 267 subjects who had a clinically suspected diagnosis of RASopathy not explained by mutations in the previously identified genes implicated in these disorders did not find other pathogenic variants. This suggests that MAPK1 variation represents a rare event in subjects with NS-related features and that it may be more commonly associated with currently unclassified syndromic DD/ID.
Our collective data indicate that pathogenic MAPK1 variants are activating and can be classified within two mechanistic groups affecting proper binding of the kinase to regulators and effectors or perturbing the mechanism of activation of the kinase. We showed that the defective negative regulation exerted by MKP3 (and possibly other MKPs) represents a generalizable mechanism for the GoF behavior of the identified MAPK1 variants. Notably, substitutions in or close to the DRS can lead to either GoF or LoF61 by impairing proper MAPK1 binding to regulators and substrates. Our structural and experimental analyses support a model in which disease-causing variants operate with counteracting effects on MAPK1 function by differentially impacting the ability of the altered proteins to interact with MEK1, MKP3, and substrates. Remarkably, the evidence that only some intermolecular interactions are impaired by the pathogenic substitutions in the DRS strongly suggests that these lesions produce only a local perturbation of the binding interface, rather than an overall conformational rearrangement of the CD domain. Consistent with the GoF role of the identified MAPK1 variants, no pathogenic amino acid change was found to involve residues at the FRS. This is in line with recent data demonstrating a mutually exclusive distribution of GoF and LoF MAPK1 changes in these domains, with the former limited to DRS residues and the latter involving FRS residues.61
According to the Tissue Cancer Genome Atlas and Cosmic datasets, MAPK1 gene amplification is observed in a significant proportion of ovarian, bladder, lung, and breast cancers (5% to 20% of cases). However, while enhanced MAPK1 activation driven by oncogenic mutations in upstream signal transducers (e.g., PTPN11, HRAS [MIM: 190020], KRAS [MIM: 190070], NRAS [MIM: 164790], and BRAF [MIM: 164757]) is a common theme in cancer, MAPK1 mutations do not represent a major somatic event contributing to oncogenesis. Indeed, they have been reported in a substantially restricted number of tumors and less frequently with respect to other genes coding for signal transducers with role in the RAS-MAPK pathway (COSMIC database). Based on our findings, this is likely linked to the particular behavior of these mutations, which are stimulus dependent and confer only mild and/or context-specific GoF because of their counterbalancing consequences on MAPK1 function. In this proposed model, the disease-causing amino acid substitutions clustering at the DRS significantly affect MKP3 binding and MKP3-mediated MAPK1 dephosphorylation of the regulatory residues, Thr185 and Tyr187. However, since DRS also mediates MAPK1 binding to a number of signaling partners and scaffolding proteins, it is possible that these mutations may also variably impact binding of the kinase to a large number of MAPK1 interacting proteins, including substrates.47,60,66,67 It is therefore reasonable to hypothesize that each of the identified pathogenic variants may variably exert counterbalancing effects on the phosphorylation status of the kinase and its capability to couple its activation to efficient phosphorylation of substrates.
We clinically and molecularly profile a RASopathy caused by activating germline pathogenic variation of MAPK1. After nearly 20 years from the discovery of the first gene implicated in NS, PTPN11,15 the last tier of the MAPK cascade finally joins the group of genes mutated in RASopathies. Our findings further extend the molecular mechanisms by which upregulation of MAPK signaling is linked to human disease and provide a rationale for the differential impact of MAPK1 mutations in development and oncogenesis.
Data and Code Availability
The pathogenic variants identified in this work have been submitted to ClinVar under the following accession numbers: c.221T>A, SCV001337682; c.238C>T, SCV001337683; c.521C>T, SCV001337684; c.952G>A, SCV001337685; c.953A>G, SCV001337686; c.964G>C, SCV001337687; c.968C>G, SCV001337688. WES datasets have not been deposited in a public repository due to privacy and ethical restrictions but are available from the corresponding author on request.
Declaration of Interests
I.M.W. and M.M.M. are employees of GeneDx. S.Á. and M.M.-G. are employees of NimGenetics. All the other authors declare no competing interests.
Acknowledgments
The authors thank the subjects and their families for participating in this study. We are grateful to Shin Takagi (University of Nagoya, Japan) and James D. McGhee (University of Calgary, Canada) for providing the original plasmids used for C. elegans studies, and Serenella Venanzi (Istituto Superiore di Sanità, Rome, Italy) and Rosalba Carrozzo (Ospedale Pediatrico Bambino Gesù, Rome, Italy) for technical assistance. C. elegans strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). This work was supported by grants from EJP-RD (NSEuroNet) to M.Z. and M.T. and by AIRC (IG 21614) and Ministero della Salute (Ricerca Corrente 2019, 2020) to M.T. Funding from the JPB Foundation was provided to W.K.C.
Published: July 27, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.06.018.
Web Resources
ExAC Browser, http://exac.broadinstitute.org/
Gene SeT AnaLysis, http://www.webgestalt.org/option.php
gnomAD Browser, https://gnomad.broadinstitute.org/
InterVar, http://wintervar.wglab.org
OMIM, https://www.omim.org/
RCSB Protein Data Bank, http://www.rcsb.org/pdb/home/home.do
UMCUGenetics/IAP, https://github.com/UMCUGenetics/IAP/tree/v2.2.0
Supplemental Data
References
- 1.Yoon S., Seger R. The extracellular signal-regulated kinase: multiple substrates regulate diverse cellular functions. Growth Factors. 2006;24:21–44. doi: 10.1080/02699050500284218. [DOI] [PubMed] [Google Scholar]
- 2.Kolch W. Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat. Rev. Mol. Cell Biol. 2005;6:827–837. doi: 10.1038/nrm1743. [DOI] [PubMed] [Google Scholar]
- 3.Samatar A.A., Poulikakos P.I. Targeting RAS-ERK signalling in cancer: promises and challenges. Nat. Rev. Drug Discov. 2014;13:928–942. doi: 10.1038/nrd4281. [DOI] [PubMed] [Google Scholar]
- 4.Krens S.F., Spaink H.P., Snaar-Jagalska B.E. Functions of the MAPK family in vertebrate-development. FEBS Lett. 2006;580:4984–4990. doi: 10.1016/j.febslet.2006.08.025. [DOI] [PubMed] [Google Scholar]
- 5.Hayashi S., Ogura Y. ERK signaling dynamics in the morphogenesis and homeostasis of Drosophila. Curr. Opin. Genet. Dev. 2020;63:9–15. doi: 10.1016/j.gde.2020.01.004. [DOI] [PubMed] [Google Scholar]
- 6.Thomas G.M., Huganir R.L. MAPK cascade signalling and synaptic plasticity. Nat. Rev. Neurosci. 2004;5:173–183. doi: 10.1038/nrn1346. [DOI] [PubMed] [Google Scholar]
- 7.Tartaglia M., Gelb B.D. Disorders of dysregulated signal traffic through the RAS-MAPK pathway: phenotypic spectrum and molecular mechanisms. Ann. N Y Acad. Sci. 2010;1214:99–121. doi: 10.1111/j.1749-6632.2010.05790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rauen K.A. The RASopathies. Annu. Rev. Genomics Hum. Genet. 2013;14:355–369. doi: 10.1146/annurev-genom-091212-153523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aoki Y., Niihori T., Inoue S., Matsubara Y. Recent advances in RASopathies. J. Hum. Genet. 2016;61:33–39. doi: 10.1038/jhg.2015.114. [DOI] [PubMed] [Google Scholar]
- 10.Kratz C.P., Franke L., Peters H., Kohlschmidt N., Kazmierczak B., Finckh U., Bier A., Eichhorn B., Blank C., Kraus C. Cancer spectrum and frequency among children with Noonan, Costello, and cardio-facio-cutaneous syndromes. Br. J. Cancer. 2015;112:1392–1397. doi: 10.1038/bjc.2015.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunnett-Kane V., Burkitt-Wright E., Blackhall F.H., Malliri A., Evans D.G., Lindsay C.R. Germline and sporadic cancers driven by the RAS pathway: parallels and contrasts. Ann. Oncol. 2020;31:873–883. doi: 10.1016/j.annonc.2020.03.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Villani A., Greer M.C., Kalish J.M., Nakagawara A., Nathanson K.L., Pajtler K.W., Pfister S.M., Walsh M.F., Wasserman J.D., Zelley K., Kratz C.P. Recommendations for Cancer Surveillance in Individuals with RASopathies and Other Rare Genetic Conditions with Increased Cancer Risk. Clin. Cancer Res. 2017;23:e83–e90. doi: 10.1158/1078-0432.CCR-17-0631. [DOI] [PubMed] [Google Scholar]
- 13.Simanshu D.K., Nissley D.V., McCormick F. RAS proteins and their regulators in human disease. Cell. 2017;170:17–33. doi: 10.1016/j.cell.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roberts A.E., Allanson J.E., Tartaglia M., Gelb B.D. Noonan syndrome. Lancet. 2013;381:333–342. doi: 10.1016/S0140-6736(12)61023-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tartaglia M., Mehler E.L., Goldberg R., Zampino G., Brunner H.G., Kremer H., van der Burgt I., Crosby A.H., Ion A., Jeffery S. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat. Genet. 2001;29:465–468. doi: 10.1038/ng772. [DOI] [PubMed] [Google Scholar]
- 16.Tartaglia M., Pennacchio L.A., Zhao C., Yadav K.K., Fodale V., Sarkozy A., Pandit B., Oishi K., Martinelli S., Schackwitz W. Gain-of-function SOS1 mutations cause a distinctive form of Noonan syndrome. Nat. Genet. 2007;39:75–79. doi: 10.1038/ng1939. [DOI] [PubMed] [Google Scholar]
- 17.Roberts A.E., Araki T., Swanson K.D., Montgomery K.T., Schiripo T.A., Joshi V.A., Li L., Yassin Y., Tamburino A.M., Neel B.G., Kucherlapati R.S. Germline gain-of-function mutations in SOS1 cause Noonan syndrome. Nat. Genet. 2007;39:70–74. doi: 10.1038/ng1926. [DOI] [PubMed] [Google Scholar]
- 18.Pandit B., Sarkozy A., Pennacchio L.A., Carta C., Oishi K., Martinelli S., Pogna E.A., Schackwitz W., Ustaszewska A., Landstrom A. Gain-of-function RAF1 mutations cause Noonan and LEOPARD syndromes with hypertrophic cardiomyopathy. Nat. Genet. 2007;39:1007–1012. doi: 10.1038/ng2073. [DOI] [PubMed] [Google Scholar]
- 19.Razzaque M.A., Nishizawa T., Komoike Y., Yagi H., Furutani M., Amo R., Kamisago M., Momma K., Katayama H., Nakagawa M. Germline gain-of-function mutations in RAF1 cause Noonan syndrome. Nat. Genet. 2007;39:1013–1017. doi: 10.1038/ng2078. [DOI] [PubMed] [Google Scholar]
- 20.Aoki Y., Niihori T., Banjo T., Okamoto N., Mizuno S., Kurosawa K., Ogata T., Takada F., Yano M., Ando T. Gain-of-function mutations in RIT1 cause Noonan syndrome, a RAS/MAPK pathway syndrome. Am. J. Hum. Genet. 2013;93:173–180. doi: 10.1016/j.ajhg.2013.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamamoto G.L., Aguena M., Gos M., Hung C., Pilch J., Fahiminiya S., Abramowicz A., Cristian I., Buscarilli M., Naslavsky M.S. Rare variants in SOS2 and LZTR1 are associated with Noonan syndrome. J. Med. Genet. 2015;52:413–421. doi: 10.1136/jmedgenet-2015-103018. [DOI] [PubMed] [Google Scholar]
- 22.Johnston J.J., van der Smagt J.J., Rosenfeld J.A., Pagnamenta A.T., Alswaid A., Baker E.H., Blair E., Borck G., Brinkmann J., Craigen W., Members of the Undiagnosed Diseases Network Autosomal recessive Noonan syndrome associated with biallelic LZTR1 variants. Genet. Med. 2018;20:1175–1185. doi: 10.1038/gim.2017.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bauer C.K., Calligari P., Radio F.C., Caputo V., Dentici M.L., Falah N., High F., Pantaleoni F., Barresi S., Ciolfi A. Mutations in KCNK4 that affect gating cause a recognizable neurodevelopmental syndrome. Am. J. Hum. Genet. 2018;103:621–630. doi: 10.1016/j.ajhg.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flex E., Martinelli S., Van Dijck A., Ciolfi A., Cecchetti S., Coluzzi E., Pannone L., Andreoli C., Radio F.C., Pizzi S. Aberrant Function of the C-Terminal Tail of HIST1H1E Accelerates Cellular Senescence and Causes Premature Aging. Am. J. Hum. Genet. 2019;105:493–508. doi: 10.1016/j.ajhg.2019.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magini P., Smits D.J., Vandervore L., Schot R., Columbaro M., Kasteleijn E., van der Ent M., Palombo F., Lequin M.H., Dremmen M. Loss of SMPD4 Causes a Developmental Disorder Characterized by Microcephaly and Congenital Arthrogryposis. Am. J. Hum. Genet. 2019;105:689–705. doi: 10.1016/j.ajhg.2019.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thevenon J., Duffourd Y., Masurel-Paulet A., Lefebvre M., Feillet F., El Chehadeh-Djebbar S., St-Onge J., Steinmetz A., Huet F., Chouchane M. Diagnostic odyssey in severe neurodevelopmental disorders: toward clinical whole-exome sequencing as a first-line diagnostic test. Clin. Genet. 2016;89:700–707. doi: 10.1111/cge.12732. [DOI] [PubMed] [Google Scholar]
- 27.Miller C.R., Lee K., Pfau R.B., Reshmi S.C., Corsmeier D.J., Hashimoto S., Dave-Wala A., Jayaraman V., Koboldt D., Matthews T. Disease-associated mosaic variation in clinical exome sequencing: a two-year pediatric tertiary care experience. Cold Spring Harb. Mol. Case Stud. 2020;6:a005231. doi: 10.1101/mcs.a005231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Retterer K., Juusola J., Cho M.T., Vitazka P., Millan F., Gibellini F., Vertino-Bell A., Smaoui N., Neidich J., Monaghan K.G. Clinical application of whole-exome sequencing across clinical indications. Genet. Med. 2016;18:696–704. doi: 10.1038/gim.2015.148. [DOI] [PubMed] [Google Scholar]
- 29.Rodríguez C., Sánchez-Morán I., Álvarez S., Tirado P., Fernández-Mayoralas D.M., Calleja-Pérez B., Almeida Á., Fernández-Jaén A. A novel human Cdh1 mutation impairs anaphase promoting complex/cyclosome activity resulting in microcephaly, psychomotor retardation, and epilepsy. J. Neurochem. 2019;151:103–115. doi: 10.1111/jnc.14828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinelli S., Torreri P., Tinti M., Stella L., Bocchinfuso G., Flex E., Grottesi A., Ceccarini M., Palleschi A., Cesareni G. Diverse driving forces underlie the invariant occurrence of the T42A, E139D, I282V and T468M SHP2 amino acid substitutions causing Noonan and LEOPARD syndromes. Hum. Mol. Genet. 2008;17:2018–2029. doi: 10.1093/hmg/ddn099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Motta M., Chillemi G., Fodale V., Cecchetti S., Coppola S., Stipo S., Cordeddu V., Macioce P., Gelb B.D., Tartaglia M. SHOC2 subcellular shuttling requires the KEKE motif-rich region and N-terminal leucine-rich repeat domain and impacts on ERK signalling. Hum. Mol. Genet. 2016;25:3824–3835. doi: 10.1093/hmg/ddw229. [DOI] [PubMed] [Google Scholar]
- 32.Motta M., Fidan M., Bellacchio E., Pantaleoni F., Schneider-Heieck K., Coppola S., Borck G., Salviati L., Zenker M., Cirstea I.C., Tartaglia M. Dominant Noonan syndrome-causing LZTR1 mutations specifically affect the Kelch domain substrate-recognition surface and enhance RAS-MAPK signaling. Hum. Mol. Genet. 2019;28:1007–1022. doi: 10.1093/hmg/ddy412. [DOI] [PubMed] [Google Scholar]
- 33.Kaboord B., Perr M. Isolation of proteins and protein complexes by immunoprecipitation. Methods Mol. Biol. 2008;424:349–364. doi: 10.1007/978-1-60327-064-9_27. [DOI] [PubMed] [Google Scholar]
- 34.Sulston J.E., Hodgkin J. Methods. In: Wood W.B., editor. The Nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1988. pp. 587–606. [Google Scholar]
- 35.Mello C.C., Kramer J.M., Stinchcomb D., Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aroian R.V., Sternberg P.W. Multiple functions of let-23, a Caenorhabditis elegans receptor tyrosine kinase gene required for vulval induction. Genetics. 1991;128:251–267. doi: 10.1093/genetics/128.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aroian R.V., Lesa G.M., Sternberg P.W. Mutations in the Caenorhabditis elegans let-23 EGFR-like gene define elements important for cell-type specificity and function. EMBO J. 1994;13:360–366. doi: 10.1002/j.1460-2075.1994.tb06269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paix A., Folkmann A., Rasoloson D., Seydoux G. High Efficiency, Homology-Directed Genome Editing in Caenorhabditis elegans Using CRISPR-Cas9 Ribonucleoprotein Complexes. Genetics. 2015;201:47–54. doi: 10.1534/genetics.115.179382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 40.Shapovalov M.V., Dunbrack R.L., Jr. A smoothed backbone-dependent rotamer library for proteins derived from adaptive kernel density estimates and regressions. Structure. 2011;19:844–858. doi: 10.1016/j.str.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ioannidis N.M., Rothstein J.H., Pejaver V., Middha S., McDonnell S.K., Baheti S., Musolf A., Li Q., Holzinger E., Karyadi D. REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. Am. J. Hum. Genet. 2016;99:877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiel L., Venselaar H., Veltman J.A., Vriend G., Gilissen C. Aggregation of population-based genetic variation over protein domain homologues and its potential use in genetic diagnostics. Hum. Mutat. 2017;38:1454–1463. doi: 10.1002/humu.23313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Q., Wang K. InterVar: Clinical interpretation of genetic variants by ACMG-AMP 2015 guidelines. Am. J. Hum. Genet. 2017;100:267–280. doi: 10.1016/j.ajhg.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roskoski R., Jr. ERK1/2 MAP kinases: structure, function, and regulation. Pharmacol. Res. 2012;66:105–143. doi: 10.1016/j.phrs.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 48.Maik-Rachline G., Hacohen-Lev-Ran A., Seger R. Nuclear ERK: Mechanism of Translocation, Substrates, and Role in Cancer. Int. J. Mol. Sci. 2019;20:1194. doi: 10.3390/ijms20051194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Plotnikov A., Chuderland D., Karamansha Y., Livnah O., Seger R. Nuclear ERK translocation is mediated by protein kinase CK2 and accelerated by autophosphorylation. Cell. Physiol. Biochem. 2019;53:366–387. doi: 10.33594/000000144. [DOI] [PubMed] [Google Scholar]
- 50.Marais R., Wynne J., Treisman R. The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell. 1993;73:381–393. doi: 10.1016/0092-8674(93)90237-k. [DOI] [PubMed] [Google Scholar]
- 51.Gille H., Kortenjann M., Thomae O., Moomaw C., Slaughter C., Cobb M.H., Shaw P.E. ERK phosphorylation potentiates Elk-1-mediated ternary complex formation and transactivation. EMBO J. 1995;14:951–962. doi: 10.1002/j.1460-2075.1995.tb07076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sundaram, M.V. (2013). Canonical RTK-Ras-ERK signaling and related alternative pathways, WormBook, ed. The C. elegans Research Community, WormBook, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 53.Sternberg, P.W. (2005). Vulval development, WormBook, ed. The C. elegans Research Community, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 54.Lackner M.R., Kim S.K. Genetic analysis of the Caenorhabditis elegans MAP kinase gene mpk-1. Genetics. 1998;150:103–117. doi: 10.1093/genetics/150.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lackner M.R., Kornfeld K., Miller L.M., Horvitz H.R., Kim S.K. A MAP kinase homolog, mpk-1, is involved in ras-mediated induction of vulval cell fates in Caenorhabditis elegans. Genes Dev. 1994;8:160–173. doi: 10.1101/gad.8.2.160. [DOI] [PubMed] [Google Scholar]
- 56.Wu Y., Han M. Suppression of activated Let-60 ras protein defines a role of Caenorhabditis elegans Sur-1 MAP kinase in vulval differentiation. Genes Dev. 1994;8:147–159. doi: 10.1101/gad.8.2.147. [DOI] [PubMed] [Google Scholar]
- 57.Zhang F., Strand A., Robbins D., Cobb M.H., Goldsmith E.J. Atomic structure of the MAP kinase ERK2 at 2.3 A resolution. Nature. 1994;367:704–711. doi: 10.1038/367704a0. [DOI] [PubMed] [Google Scholar]
- 58.Sammons R.M., Ghose R., Tsai K.Y., Dalby K.N. Targeting ERK beyond the boundaries of the kinase active site in melanoma. Mol. Carcinog. 2019;58:1551–1570. doi: 10.1002/mc.23047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tanoue T., Adachi M., Moriguchi T., Nishida E. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat. Cell Biol. 2000;2:110–116. doi: 10.1038/35000065. [DOI] [PubMed] [Google Scholar]
- 60.Zhang J., Zhou B., Zheng C.F., Zhang Z.Y. A bipartite mechanism for ERK2 recognition by its cognate regulators and substrates. J. Biol. Chem. 2003;278:29901–29912. doi: 10.1074/jbc.M303909200. [DOI] [PubMed] [Google Scholar]
- 61.Brenan L., Andreev A., Cohen O., Pantel S., Kamburov A., Cacchiarelli D., Persky N.S., Zhu C., Bagul M., Goetz E.M. Phenotypic characterization of a comprehensive set of MAPK1/ERK2 missense mutants. Cell Rep. 2016;17:1171–1183. doi: 10.1016/j.celrep.2016.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roskoski R., Jr. A historical overview of protein kinases and their targeted small molecule inhibitors. Pharmacol. Res. 2015;100:1–23. doi: 10.1016/j.phrs.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 63.Canagarajah B.J., Khokhlatchev A., Cobb M.H., Goldsmith E.J. Activation mechanism of the MAP kinase ERK2 by dual phosphorylation. Cell. 1997;90:859–869. doi: 10.1016/s0092-8674(00)80351-7. [DOI] [PubMed] [Google Scholar]
- 64.Emrick M.A., Lee T., Starkey P.J., Mumby M.C., Resing K.A., Ahn N.G. The gatekeeper residue controls autoactivation of ERK2 via a pathway of intramolecular connectivity. Proc. Natl. Acad. Sci. USA. 2006;103:18101–18106. doi: 10.1073/pnas.0608849103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gilmartin A.G., Bleam M.R., Groy A., Moss K.G., Minthorn E.A., Kulkarni S.G., Rominger C.M., Erskine S., Fisher K.E., Yang J. GSK1120212 (JTP-74057) is an inhibitor of MEK activity and activation with favorable pharmacokinetic properties for sustained in vivo pathway inhibition. Clin. Cancer Res. 2011;17:989–1000. doi: 10.1158/1078-0432.CCR-10-2200. [DOI] [PubMed] [Google Scholar]
- 66.Sharrocks A.D., Yang S.H., Galanis A. Docking domains and substrate-specificity determination for MAP kinases. Trends Biochem. Sci. 2000;25:448–453. doi: 10.1016/s0968-0004(00)01627-3. [DOI] [PubMed] [Google Scholar]
- 67.Peti W., Page R. Molecular basis of MAP kinase regulation. Protein Sci. 2013;22:1698–1710. doi: 10.1002/pro.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The pathogenic variants identified in this work have been submitted to ClinVar under the following accession numbers: c.221T>A, SCV001337682; c.238C>T, SCV001337683; c.521C>T, SCV001337684; c.952G>A, SCV001337685; c.953A>G, SCV001337686; c.964G>C, SCV001337687; c.968C>G, SCV001337688. WES datasets have not been deposited in a public repository due to privacy and ethical restrictions but are available from the corresponding author on request.