Abstract
Bacillus velezensis 83 was isolated from mango tree phyllosphere of orchards located in El Rosario, Sinaloa, México. The assessment of this strain as BCA (biological control agent), as well as PGPB (plant growth-promoting bacteria), were demonstrated through in vivo and in vitro assays. In vivo assays showed that B. velezensis 83 was able to control anthracnose (Kent mangoes) as efficiently as chemical treatment with Captan 50 PH™ or Cupravit hidro™. The inoculation of B. velezensis 83 to the roots of maize seedlings yielded an increase of 12% in height and 45% of root biomass, as compared with uninoculated seedlings. In vitro co-culture assays showed that B. velezensis 83 promoted Arabidopsis thaliana growth (root and shoot biomass) while, under the same experimental conditions, B. velezensis FZB42 (reference strain) had a suppressive effect on plant growth. In order to characterize the isolated strain, the complete genome sequence of B. velezensis 83 is reported. Its circular genome consists of 3,997,902 bp coding to 3949 predicted genes. The assembly and annotation of this genome revealed gene clusters related with plant-bacteria interaction and sporulation, as well as ten secondary metabolites biosynthetic gene clusters implicated in the biological control of phytopathogens. Despite the high genomic identity (> 98%) between B. velezensis 83 and B. velezensis FZB42, they are phenotypically different. Indeed, in vitro production of compounds such as surfactin and bacillomycin D (biocontrol activity) and γ-PGA (biofilm component) is significantly different between both strains. 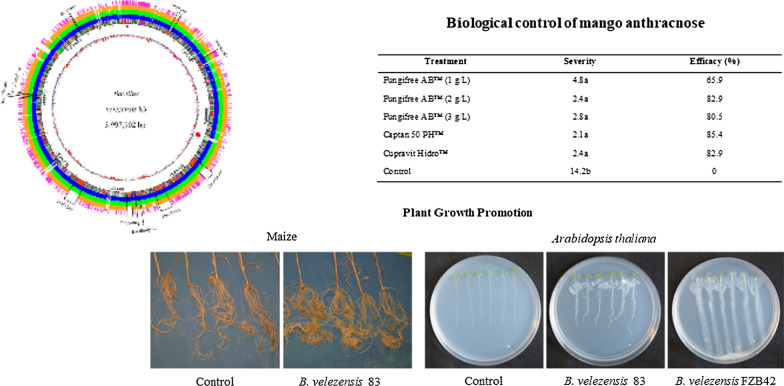
Keywords: Bacillus velezensis 83, Genome sequencing, Biological control agent, PGPB, Secondary metabolites production
Keypoints
B. velezensis 83 is a biological control agent and plant growth-promoting bacteria. Phylogenomic analysis allowed to reclassify B. subtilis 83 as B. velezensis 83. B. velezensis 83 has genes to establish a rhizosphere plant-bacteria interaction.
Introduction
Bacillus is a bacterial genus which comprises several species that establish, directly or indirectly, beneficial relations with plants (Olanrewaju et al. 2017). By direct mechanisms some Bacillus strains promote plant growth, by improving the acquisition of nutrients such as nitrogen and phosphate (biofertilization), or through the production of phytohormones (biostimulants) such as IAA (indol acetic acid), enzymes such as ACC deaminase (1-amyclocyclopropane-1-carboxylate deaminase) or volatile organic compounds (VOC) such as 2,3-butanediol and acetoin (Xu et al. 2014; Hanif et al. 2015; Asari et al. 2016, 2017; Vejan et al. 2016). On the other hand, through indirect mechanisms (production of antimicrobial compounds), they naturally exercise biological control over several phytopathogenic bacteria such as Pseudomonas syringae, Agrobacterium tumefaciens, Xanthomonas campestris, Xanthomonas oxonopodis, Erwinia amylora and fungal pathogens as Botrytis cinerea, Fusarium oxysporum, Colletotrichum gloeosporioides, Rhizoctonia solani and Penicillium expansum (Fira et al. 2018). Bacillus spp. biological control mechanisms include competition for nutrient and space, antibiosis or induced systemic resistance (ISR), which alone or together can reduce incidence and/or severity of plant diseases (Kloepper et al. 2004; Ongena and Jacques 2007; Choudhary and Johri 2009; Mongkolthanaruk 2012; Fan et al. 2018).
Genome sequencing strategies complemented with the analysis of secondary metabolite gene cluster profiles of several Bacillus strains have been helpful in order to identify potential biological control agents (BCA) or plant growth-promoting bacteria (PGPB); but also to estimate their disease prevention and benefical plant interaction potential (Borriss et al. 2011; Niazi et al. 2014; Palazzini et al. 2016; Cai et al. 2017; Belbahri et al. 2017; Fan et al. 2018; Chen et al. 2019). As part of the operational group Bacillus amyloliquefaciens (B. amyloliquefaciens, Bacillus siamensis and Bacillus velezensis), B. velezensis species has been recognized as a plant-associated bacteria due to the presence of nine gene clusters encoding enzymes involved in the nonribosomal synthesis of lipopeptides (surfactin, bacillomicyn D, fengicyn) and polyketides (macrolactin, bacillaene, difficidin), a siderophore (bacillibactin), a dipeptide (bacilysin) and a putative peptide with nematicide activity (Chen et al. 2007; Fan et al. 2017; Rabbee et al. 2019). Other genes involved in carbohydrate metabolism and plant cell wall degradation, such as amylase E (amyE), cellulase (bglC), xylanase (xynACD), galactokinase (gal1), and betaglucanase (bglCS) are also exclusively present in Bacillus strains associated to plants (Borriss et al. 2011). B. velezensis strains can show high genomic homology with phenotypic similarities, or differences, such as nutritional requirements, ability to colonize plants or production of antimicrobial metabolites (therefore antagonistic in vitro and in vivo activity) because they respond differently to stimuli from the habitat to which they are exposed. For example, the strains B. velezensis UCMB5044 (a oligotrophic PGPR isolated from desert soils), B. velezensis UCMB5113 and B. velezensis At1 (both strain plant endophytes) and B. velezensis UCMB55007 (a copiotroph isolated from calf gut) share 99% of genetic homology. However, their gene expression in response to root maize exudates were significantly different because they possess different DNA methylation patterns (Reva et al. 2019).
B. velezensis 83, is an aerobic, gram positive, rod shaped and spore forming bacteria isolated by our research team from the mango tree phyllosphere of orchards located in El Rosario, Sinaloa, México. This strain is the biological component of Fungifree AB™ marketed in México since 2012, as a foliar fungicide, very effective for the biocontrol of five different genera of phytopathogenic fungi (Colletotrichum, Erysiphe, Botrytis, Sphaerotheca and Leveillula) in crops of high agricultural importance such as mango, avocado, papaya, citrus, tomato, blueberry, blackberry, raspberry, zucchini, melon, cucumber, watermelon and others (Galindo et al. 2013). Here, we reported in vivo assays to demonstrate the activity of this strain as BCA (mango) as well as PGPB (maize). We have shown that B. velezensis 83 is an efficient strain for controlling mango anthracnose as well as a potent plant growth promotor agent in maize and A. thaliana. In addition, in vitro production by this strain of key compounds such as surfactin, bacillomycin D (biocontrol activity), acetoine and 2,3-butanediol, and particularly γ-PGA (biofilm component), was characterized, and helped to understand the mechanisms by which this strain exerts its action as a biofungicide and plant growth promotor. The genome sequence of B. velezensis 83, including genome assembly and annotation is also reported.
B. velezensis FZB42 was used as a reference for plant growth promotion and biological control strain (Fan et al. 2018). We show a comparison of metabolic and biological activities between B. velezensis 83 and B. velezensis FZB42 (> 98% genome identity) under in vitro conditions. However, as shown in this paper, a high genomic homology with a well characterized BCA strain is only the first step to fully characterize a newly isolate as BCA. The strain characterized in this work (B. velezensis 83), exhibited unique phenotypical traits in terms of biocontrol and growth promoting activities when compared with B. velezensis FZB42.
Materials and methods
Bacteria
B. velezensis 83 strain was deposited at Belgian Co-ordinated Collection of Micro-organisms (BCCM) by our research team under the accession number LMG S-30921. B. velezensis 83 was grown in 500 mL shake flasks containing 50 mL of Luria Bertani (LB) medium and incubated at 29 °C and 200 rpm for 12 h. Genomic DNA was isolated using a commercial kit (Quiagen). For the biocontrol and plant growth promotion assays, B. velezensis 83 was used as powder commercial formulation (Fungifree AB™ obtained from Agro&Biotecnia S. de R.L. de C.V.). B. velezensis FZB42 (BGSC 10A6, DSM23117) was kindly donated by Nord Reet UG Greifswald (Germany).
Biological control and PGPB assays
A trial for biological control of mango anthracnose caused by C. gloeosporioides was done in a mango production orchard in El Rosario, Sinaloa, México. An experimental randomized block design was established with six treatments and four repetitions. The experimental unit was a mango Kent tree with more than 10 years old. In the experimental design, three B. velezensis 83 treatments (Fungifree AB™ in 1, 2 or 3 g/L), two chemical treatments (Captan 50™ in 300 g/L, and Cupravit hidro™ in 400 g /L) and a control (without treatment) were included. The treatments were applied to the foliage with a motorized-spray backpack. Four liters of the corresponding treatment were applied once a month in each tree from the beginning of flowering to the harvest of the fruits (six applications in total). Twelve fruits were taken from each experimental unit and stored (under commercial conditions) for 21 days, then the severity of anthracnose at postharvest was evaluated. The evaluation of severity and control efficacy obtained with each treatment was done using a visual hedonic scale, where: (0) healthy fruit, (1) < 2 mm spots, (2) < 5%, (3) < 12.5%, (4) < 25%, (5) < 50% and (6) > 50% of affected area in the mango fruit. The data collected were transformed to obtain the percentage of severity by means of the Eq. 1. (Townsend and Heuberger 1943):
| 1 |
where:
P = severity (%).
n = number of samples per category.
v = numerical value of the category.
N = total number of samples.
C = highest category.
After that, the control efficacy of each treatment was calculated by Eq. 2. (Abbott, 1925):
| 2 |
where:
IT = severity (%) in the control.
it = severity (%) in the treatment.
Plant growth promoting assays
Maize growth promotion study was carried out under greenhouse conditions. First of all, the seeds were disinfected by immersion in 1% NaClO aqueous solution for 1 min followed by three rinses with sterile distilled water. B. velezensis 83 was tested with two different treatments: 1) as a seed treatment or 2) as root treatment of seedlings. For seed treatment, the seeds were immersed in Fungifree AB™ aqueous solution (using 10 g/kg seed) during 2 h then sown in sterile substrate (peat moss: black earth, 3:1). For root treatment, Fungifree AB™ (at a dose equivalent to 2.5 kg/ha) was applied to each pot seedling at 10 and 22 days after sowing (DAS). For each treatment, 30 seeds or seedlings were used. For the control treatment, only distilled water was applied and the same quantity of seeds or seedlings was used. The evaluation of length shoot, dry weight (DW) of shoot and root was carried out with a sample of 15 plants at 37 DAS.
Another assay to show the plant growth promoting effects caused by B. velezensis 83 was done with in vitro co-cultures with A. thaliana (Col-0) seedlings and comparing with those effects caused by B. velezensis FZB42 and against a control (uninoculated plants). The seeds were disinfected and sown in aseptic conditions to obtain seedlings (Barrera-Ortiz et al. 2018). A. thaliana seedlings of 4 days after germination (DAG) were transferred to fresh agar plates with 0.2 × MS media inoculated with an aliquot of 10 µL (1 × 105 cfu/mL) from an overnight culture of B. velezensis 83 or B. velezensis FZB42 (Fan et al. 2018). Six seedlings in each plate were carefully placed over the bacterial stria and whose shoots were approximately 1 cm from the bacterial inoculum. For Col-0 seedlings, the plates were incubated for an additional 6-day period and the root and primary root length, lateral root length (of the longest lateral root in each seedling) and shoot diameter were measured with a ruler, while lateral roots in the primary root and leaves in the shoot were recorded using a stereomicroscope Olympus SZ40 (Olympus Iberia S.A.U, Barcelona, España) at a 10X magnification. Lateral root density was calculated dividing lateral root number between the primary root length, and those parameters were obtained of 18 individuals for each treatment. Plant biomass, total fresh weight, shoot fresh weight and root fresh weight of 6 seedlings grown on the same plate were measured with an analytical balance Ohaus PA224 (Ohaus Corporation, Newjersey, USA), and three plates of each treatment were evaluated.
Secondary metabolites production
For secondary metabolites production, B. velezensis 83 and B. velezensis FZB42 were grown in liquid batch cultures using a mineral medium with the following composition (in g/L): glucose 30.0, (NH4)2SO4 6.0; K2HPO4 7.98; KH2PO4 9.6; MgSO4·7H2O 0.4; CaCl2 0.1; FeSO4·7H2O 0.08; MnCl2·4H2O 0.019. The batch cultures of each strain were carried out inoculating 5 mL of an overnight culture in YPG medium (2 × 109 cfu/mL) in 500 mL shaken flasks with 100 mL of working volume and incubated at 30 °C and 200 rpm for 48 h. Cultures were conducted at initial pH of 6.8 adjusted with NaOH before sterilization. B. velezensis FZB42 liquid batch culture was carried out under the same experimental conditions. Cell concentration (cell/mL) was determined using a Neubauer chamber.
Glucose, acetoin and 2,3-butanediol concentrations
A 1 mL sample was collected from the shaken flask and bacterial cells were removed from the medium by centrifugation (10,000 × g, 15 min) and filtration through a 0.2 µm membrane (hydrophilic and nonpyrogenic, Sartorious AG, Goettingen, Germany). Glucose, acetoin and butanediol in the supernatant were analyzed by high resolution reverse phase high-performance liquid chromatography (HPLC) using methodology reported previously (Cristiano-Fajardo et al. 2019) with minor modifications. Briefly, 20 µl of the sample was loaded to an Aminex HPX-87H column (7.8 × 300 mm; Bio-Rad Laboratories Inc., California, USA) and separated by using a Waters 2695 HPLC system (Waters Corporation, Massachusetts, USA). Acetoin was determined by absorbance at 210 nm and glucose and butanediol by refraction index. H2SO4 5 mM was used as mobile phase with a flow of 0.6 mL/min. Column temperature was adjusted at 50 °C. Pure glucose, acetoin and butanediol (Sigma Chemical, St. Louis, MO, USA) were used as standards.
Surfactin and bacillomycin concentration
A 1 mL sample was collected from the shaken flasks and bacterial cells were removed from the medium as described above. The supernatant was loaded to a ZorbaxSB-C18 column (4.6 mm × 150 mm; Agilent Technologies, Palo Alto, CA, USA) and separated by HPLC by using a Waters 2695 HPLC system. The mobile phase was composed by 0.1% trifluoroacetic acid (TFA) in water (phase A), and 0.1% TFA in acetonitrile (phase B). Samples were eluted for 14 min using 40% of phase B at 32 °C with a flow rate of 0.3 mL/min, followed with a linear gradient of 40–85% for 1 min and maintained at 85% for another 36 min; finally a new linear gradient of 85–40% for 1 min, continued at 40% for 8 min. The elution pattern was monitored by determining absorbance at 205 nm. Pure surfactin and iturin (Sigma Chemical, St. Louis, MO, USA) were used as standards.
Poly-γ-glutamic acid concentration and mean molecular weight
Concentration of γ-PGA and mean molecular weight (MW) in the supernatant was determined by gel permeation chromatography (GPC) using methodology reported previously (Cristiano-Fajardo et al. 2019). Summarizing, 100 µl of cell-free supernatant was loaded into serial connected Ultrahydrogel columns (UG500/Linear; Waters Corporation, Massachusetts, USA), using a Waters 2695 HPLC system. γ-PGA was eluted with 0.1 M NaNO3 using a flow rate of 0.8 mL/min, columns temperature was adjusted at 38 °C. γ-PGA was detected with a refractive index detector (Waters 2414, USA). Pure γ-PGA (Sigma-Aldrich Inc., Missouri, USA) solutions were used as standards for polymer concentration calculation and poly(ethylene oxide) (Waters Corporation, Massachusetts, USA) with MWs in the range of 24–933 kDa were used as standards for mean MW.
Genome sequencing, assembly an annotation
Genomic DNA from B. velezensis 83 was sent to MOgene Genome Sequencing Services (Mogene LC; St. Louis, MO, USA) with the following requests: libraries of 350 bp fragment size and paired end sequencing (2 × 250 bp) in MiSeq sequencer. The total number of reads was about ~ 7,000,000 paired reads that represents a genome coverture ~ 400X. Genome assembly was performed as described previously in Pérez-Carrascal et al. (2016). Briefly, a combined de novo and reference-based assembly was obtained with the Spades Genome Assembler (SPAdes; Bankevich et al. 2012). First, the SPAdes contigs were aligned, using NUCmer (Kurtz et al. 2004), to the complete genome of B. velezensis YAU-B9601-Y2 the closest phylogenetic relative to B. velezensis 83 (Hao et al. 2012). Second, the Illumina reads were mapped onto the contigs assembly, oriented according the 5´-3´directions of their pair-ends, and joined manually with Consed (Gordon et al. 2013). At the end, we obtained a single contig representing a closed circular chromosome 3,997,902 bp length. The protein-coding regions (ORFS, open reading frames) were predicted with Glimmer 3.02 (Delcher et al. 2007). The ORFs model was uploaded into Artemis 12.0 (Carver et al. 2012) to make ORFs frame rectifications and register manual annotations. Functional descriptions of genes were obtained by BlastX searches of the complete set of ORFs against the non-redundant database of the GenBank (Benson et al. 2008). A best-blast hits table was created with the parameters of the percentage of identity and similarity, coverture of the ORFs, coordinates of the matches, and the annotated function of the protein. Additional comparisons with the Conserved Domain Database (CDD) of the GenBank (Marchler-Bauer et al. 2015), Interpro (Mitchell et al. 2015), and IS-database (Siguier et al. 2006), contributed to confirm the GenBank-based annotations and to solve controversial cases. COG annotation was done using the NCBI COGs database (Tatusov et al. 2000) with BlastP comparisons (https://blast.ncbi.nlm.nih.gov) with the minimal similarity of 30% and e-value < 1 X10−6. The whole genome sequence of B. velezensis 83 was deposited in GenBank under the accession number CP034203.
Genome analysis
Genome analysis was performed following the standard methodology described in González et al. (2019). In short, the pangenome model was obtained with the Bacterial Pangenome Analysis program (BPGA), by setting the USEARCH clustering algorithm to the default values (the minimal identity of 50% and 20 combinations) (Chaudhari et al. 2016). The phylogenetic tree was done in MEGA-6 evolutionary analysis software (Tamura et al. 2013) by Maximum Likelihood (ML) method using the JTT matrix (Jones et al. 1992) and bootstrap of 1000 replicates.
The accessory genome obtained with BPGA, was represented in a heatmap of presence and absence of genes and schematized with heatmap.2 (R´s gplots package; https://rdrr.io/cran/gplots). Genome comparisons were illustrated by circular maps obtained with GenVision of DNASTAR (Lasergene Core Suite (https://www.snapgene.com). Whole-genome comparisons were performed with selected Bacillus strains using Average Nucleotide Identity (ANI) calculated with JSpecies (Richter et al. 2016), and MUMmer 3.06 (Kurtz et al. 2004). Prophage searches were done with the Phaster program (Arndt et al. 2016).
Statistical analysis
All data were analyzed by one-way analysis of variance (ANOVA) and Tukey comparison procedure assuming equal variances using Minitab™ 17 Statistical Software (Minitab, LLC, Pennsylvania, USA).
Results
B. velezensis 83 is a biological control agent and plant growth-promoting bacteria
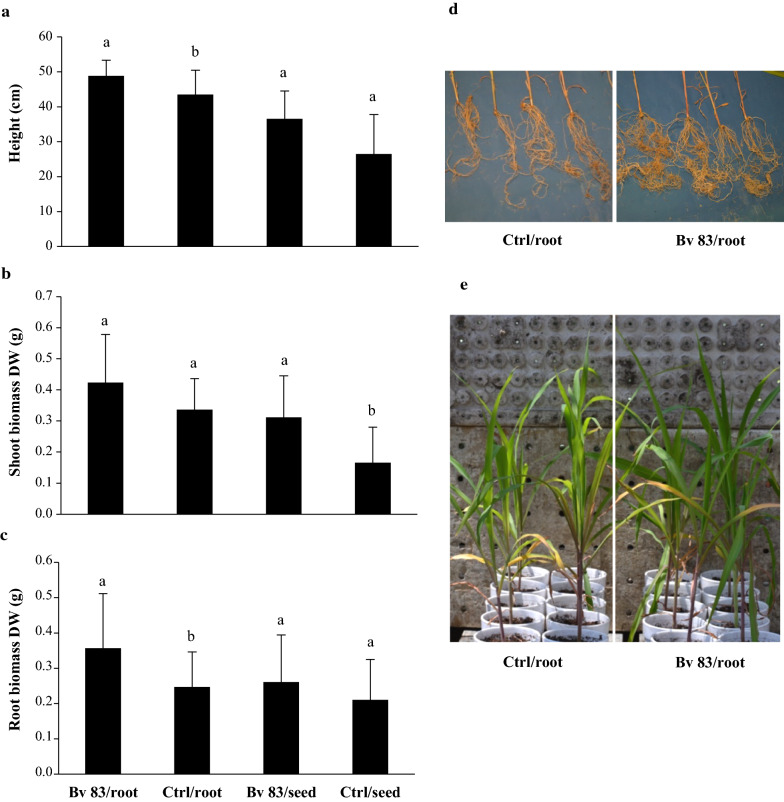
Mango anthracnose severity was reduced in trees treated with B. velezensis 83 at three different doses, and fruit damage was similarly to those fruits harvested from trees with chemical conventional treatments (Captan 50 PH™ or Cupravit hydro™). The highest anthracnose severity in Kent mango fruits was found in the control treatment (Table 1). In maize, the application of B. velezensis 83 to the root caused an increase of 12% in height (Fig. 1a) of the seedlings, however, it has no effect on their shoot biomass (Fig. 1b) but an increase of 45% of the root biomass (Fig. 1c) of the seedlings was observed with respect to the uninoculated control. When the treatment was applied to seeds, the growth-promotion capacity of B. velezensis 83 was also evident. With respect to the control, the application of B. velezensis 83 caused an increase of 38% of the height and 88% of the shoot biomass, even if no effect on the root biomass was observed.
Table 1.
Severity and biocontrol efficacy of B. velezensis 83 in mango ocharchs
| Treatment | Severity | Efficacy (%) |
|---|---|---|
| Fungifree AB™ (1 g/L) | 4.8a | 65.9 |
| Fungifree AB™ (2 g/L) | 2.4a | 82.9 |
| Fungifree AB™ (3 g/L) | 2.8a | 80.5 |
| Captan 50 PH™ | 2.1a | 85.4 |
| Cupravit Hidro™ | 2.4a | 82.9 |
| Control | 14.2b | 0 |
Different letters indicate significant differences among the treatments at P ≤ 0.05
Fig. 1.
Growth-promoting effect of B. velezensis 83 on maize seedlings. a Height (cm) of the seedlings, b Shoot biomass (g) dry weight (DW) of the seedlings, c Root biomass (g) dry weight (DW) of the seedlings. Bv 83/root: Fungifree AB™ treatment to the root, Ctrl/root: without treatment to the root, Bv 83/seed: Fungifree AB™ treatment to the seed, Ctrl/seed: without treatment to the seed. d Aspect of the root of seedlings of Ctrl/root and Bv 83/root treatment. e Aspect of the shoot of seedlings of Ctrl/root and Bv 83/root treatment. N = 15. Different letters indicate statistically significant differences among treatments at P ≤ 0.05
Genomic and pangenomic features of B. velezensis 83
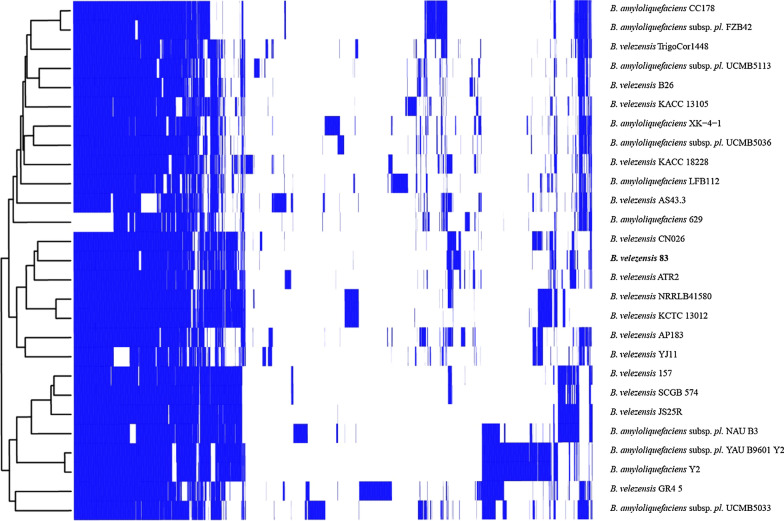
The complete genome of B. velezensis 83 consists of 3,997,902 bp with an average G + C content of 46%, 7 copies of the rRNAs operon (16S, 23S and 5S RNA) and 68 tRNA genes. The genome was predicted to encode 3752 coding sequences (CDS) of which 3255 were functionally annotated, whereas 497 were hypothetical. A total of 2892 CDS were assigned to COGs (cluster of orthologous groups). Functional classes defined by COGs indicate that B. velezensis 83 harbor a high proportion of proteins involved in carbohydrate (COG G) and amino acids transport and metabolism (COG E), as well as in transcription (COG K). The pangenome model of 27 selected strains of B. velezensis (including B. velezensis 83) and B. amyloliquefaciens strains approaches an asymptote (Additional file 1. Fig. S1) indicating, as in other Bacillus species, that B. velezensis has a closed pangenome structure and limited variation in gene content. The pangenome size consist of 5263 gene families, of which 2683 belong to the core genome, 1928 gene families in the accessory component, and a total of 652 unique gene families unevenly distributed in the 27 B. velezensis strains. Particularly, only 12 genes were unique in B. velezensis 83, most of them were hypothetical. The accessory genome described by a heatmap of presence/absence of genes indicates that although B. velezensis belongs to a clade with very related strains, each one still has some genetic differences that make each strain unique (Fig. 2).
Fig. 2.
Accessory genome profile of B. velezensis. The heat-map indicates the presence (blue) and absence (white) of the accessory genes in 27 B. velezensis strains obtained from the BPGA pangenome model (Chaudhari et al. 2016). B. velezensis 83 is in bold letter
Taxonomic affiliation of strain 83
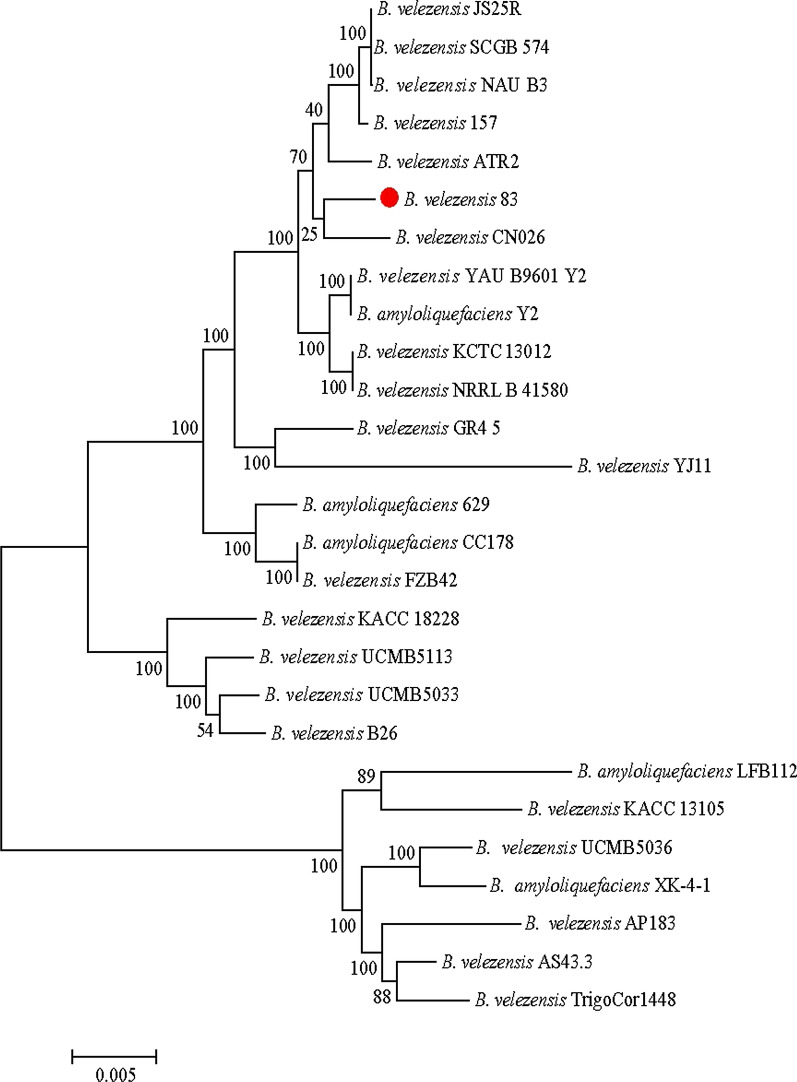
Based on phylogenetic analysis of the 16S ribosomal genes, Bacillus strain 83 was initially classified as Bacillus subtilis (Galindo et al. 2013). To review this classification in the light of genome sequence presented here, we performed pairwise whole-genome comparisons between this strain and other Bacillus species using the Average Nucleotide Identity (ANI) (Richter et al. 2016). The results indicate that B. subtilis 83 is closely related to B. velezensis (ANIm > 97%, Additional file 1. Fig. S2). To support this observation, we did a phylogenomic analysis using 20 core housekeeping genes determined from the pangenome analysis of 27 B. velezensis strains (including strain 83). The phylogenetic tree, located the B. subtilis 83 together strains of B. velezensis, Bacillus methylotrophicus, and B. amyloliquefaciens subsp. plantarum, into a very related clade forming the recently recognized species called B. velezensis (Fig. 3) (Dunlap et al. 2015; Fan et al. 2017). Although B. velezensis and B. amyloliquefaciens are known plant growth-promoting bacteria, strains of the former are commonly associated with plants while the latter is constituted by soil-borne strains (Fan et al. 2017). These results support place B. subtilis strain 83 within B. velezensis species and allow us to correct its previous wrong classification.
Fig. 3.
Taxonomic affiliation of B. velezensis 83. The phylogenomic tree was performed with 20 concatenated genes of the core families determined with BPGA (Chaudhari et al. 2016; see methods). Bacillus strains were selected from the phylogenomic neighbor joining tree reported in Dunlap et al. (2015). The tree is the result of 1000 replicates using the maximum likelihood method (ML) in MEGA program (Tamura et al. 2013). Red dot indicates the position of B. velezensis 83
Genes related to biological control and sporulation
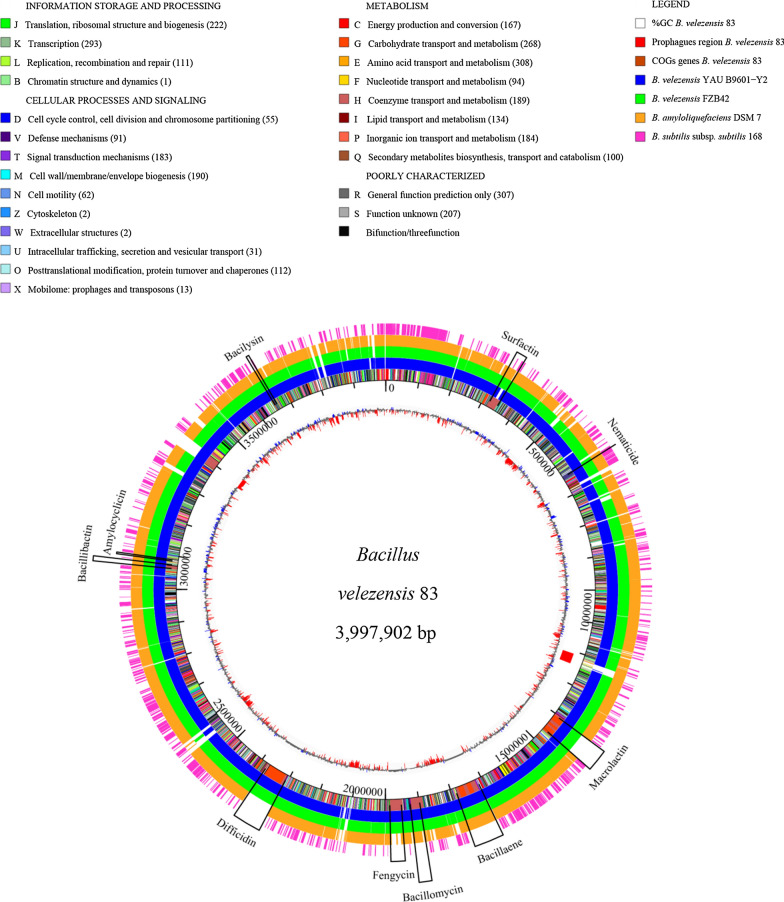
Genomic comparisons of B. velezensis 83 with the strains B. velezensis FZB42, B. velezensis YAUB601-Y2, B. amyloliquefaciens DMS7 and B. subtilis 168, showed high conservation and collinearity in the chromosomal sequence (Fig. 4). The genome of B. velezensis 83 harbors ten gene clusters dedicated to the synthesis of biocontrol metabolites (Table 2) with high genomic homology with B. velezensis FZB42 (Additional file 1. Table S1), covering about 8.2% of its genome. The genome of B. velezensis 83 contains five Non-Ribosomal Peptide Synthetases (NRPS) and three Polyketide Synthases (PKS) gene clusters. The sfp gene coding for the 4′-phosphopantetheinyl transferase responsible for the conversion of the apo-ACP domains of PKS and NRPS to their active holo-forms, was also identified in this strain together with the regulatory gene yczE. The predicted products of these NRPS gene clusters are the lipopeptides surfactin (srfA), bacillomycin (bmy) and fengycin (fen), the siderophore bacillibactin (dhb) and the dipeptide bacilysin (bac). In the PKS gene cluster, the genes encoding for macrolactin (mln), bacillaene (bae) and difficidin (dfn) were identified. In addition, a gene cluster probably involved in the production of the lantibiotic amylocyclicin, and the pur gene cluster for synthesis of a nematicide compound were located (Xia et al. 2011). However, B. velezensis 83 showed some incomplete gene clusters. For instance, the cluster for subtilin (spa) synthesis presents only five genes (spaEFGRK) out of ten reported; lacking those genes involved in the synthesis and transport of subtilin (Stein et al. 2002). The gene cluster for mersacidin (mrs) synthesis was also partially present, as only five genes (mrsK2, mrsR2, mrsFGE) out of ten reported were found; the genes for synthesis, modification and export of mersacidin are absent, keeping only those for regulation and immunity for this antibiotic (Schmitz et al. 2006). The results indicate that B. velezensis 83 may produce a wide repertoire of metabolites with biocontrol properties. The sporulation capacity of Bacillus has been exploited by the biopesticide industry to the preparation of powder formulations because spores are easier to handle and store, presenting a longer shelf life compared to liquid preparations. The sporulation process of Bacillus strains is carried out through the Spo0A pathway (Romero 2013; Liaqat et al. 2013; Yan et al. 2016) and the lack of some genes for the signaling cascade can affect the sporulation process (Branda et al. 2001; Yan et al. 2016). As predictable, in the genome of B. velezensis 83, all the genes involved in the different stages of the sporulation process (Additional file 1. Table S2), described by other authors (Romero 2013; López and Kolter 2010; Tan and Ramamurthi 2013) were found. Ten genes coding for Rap (response regulator aspartate phosphatase) proteins (Rap A1,-A2,-B,-C,-D,-F,-H,-J,-K1 and -K2) and three genes coding for Phr peptides (PhrA,-C-K) were identified. The interaction between Rap protein-Phr peptide results in a regulatory system which function is being the communication bridge between sporulation and competence process in B. subtilis (Schultz et al. 2009), the competence is a physiological state through the cell uptakes of exogenous DNA through which genomic diversity and evolution is generated (Brito et al. 2018).
Fig. 4.
Graphical representation of B. velezensis 83 genome. From the innermost circle to the outermost: average GC (%) content profile, prophage region (red), the complete genome of B. velezensis 83. Color bars in the circle indicate the corresponding COG classification showed in the upper part of the scheme, the number of protein identified for each functional class is shown in (parentheses); subsequent circles indicate conserved regions with the genome of B. velezensis YAUB9601-Y2 (blue), B. velezensis FZB42 (green), B amyloliquefaciens DSM7 (orange), Bacillus subsp. subtilis (purple). The distribution of metabolite gene clusters is indicated by black rectangles and the cluster name
Table 2.
Genes involved in synthesis of biocontrol metabolites in B. velezensis 83
| Metabolite | Gene cluster | Size (kb) | Position |
|---|---|---|---|
| Surfactin | srfAABCD | 26.1 | 320,917–347,075 |
| Macrolactin | mlnABCDEFGHI | 53.2 | 1,404,749–1,458,017 |
| Bacillaene | baeBCDE,acpK,bacGHIJLMNRS | 71.7 | 1,714,735–1,787,205 |
| Bacillomycin | bmyCBAD | 36.9 | 1,890,815–1,928,064 |
| Fengycin | fenEDCBA | 37.6 | 1,950,976–1,988,645 |
| Difficidin | dfnMLKJIHGFEDCBXYA | 69.1 | 2,308,397–2,377,919 |
| Amylocyclicin | acnFEDCAB | 4.0 | 3,085,092–3,089,264 |
| Bacillibactin | dhbFBECA | 11.7 | 3,061,830–3,073,566 |
| Bacilysin | bacEDCBA | 4.7 | 3,661,180–3,665,866 |
| Nematicide | pur(EKBCSQLFMNHD) | 12.7 | 638,387–651,184 |
Genes related to plant-bacteria interaction
B. velezensis 83 genome harbors genes involved in different process of plant-bacteria interaction (Additional file 1. Table S3). For biofilm formation, the operon for exopolysaccharide (epsA-O) synthesis and the operon yqxM-sipW-tasA for TasA protein fibers synthesis, are present (Al-Ali et al. 2018). Besides, genes coding for other biofilm components (Marvasi et al. 2010), such as the γ-polyglutamic acid polymer (pgdS, pgsEACB), the levansucrase (sacB) enzyme, and the proteases bacillopeptidase F (bpr), glutamyl endopeptidase protein (mpr), epr subtilisin family serine protease protein (epr), bacillolysin (npr) and the extracellular serine protease protein (vpr), were all found. Several genes exclusively present in Bacillus strains associated to plants were also found in B. velezensis 83 genome. That genes are involved in carbohydrate metabolism and plant cell wall degradation as described by Borriss et al. (2011), like amylase E (amyE), cellulase (bglC), xylanase (xynACD), galactokinase (gal1), and betaglucanase (bglCS). To assess the potential of B. velezensis 83 as plant growth-promoting bacteria, we also looked for genes already reported to promote the plant growth (Belbahri et al. 2017). Three different pathways described (Idris et al. 2007) for synthesis of the auxin indole acetic acid (IAA) were found: 1) the one of indole-3-pyruvate (IPyA) dependent of tryptophan transaminase gene clustering (patB, YclC, YclB, DhaS) products, 2) that of indole-3-acetonitrile (IAN) in which the nitrilase gene (yhcX) product acts, and 3) an uncharacterized IAA biosynthesis pathway in which the product of the gene acetyltransferase (ysnE) participates. The gene coding the protein involved in auxin excretion (ywkB) was also found. In addition, the cluster (alsDSR) encoding a α-acetolactate synthase for the synthesis of acetoin and 2,3-butanediol was present. Interestingly, even though B. velezensis 83 was isolated from mango tree phyllosphere, the strain has all the necessary genes to establish a rhizosphere plant-bacteria interaction. Therefore, in addition to control foliar phytopathogens, B. velezensis 83 seems to have the potential to be a biological control agent for root phytopathogens, probably eliciting ISR and/or promoting root growth.
Phenotypic differences between B. velezensis 83 and B. velezensis FZB42.
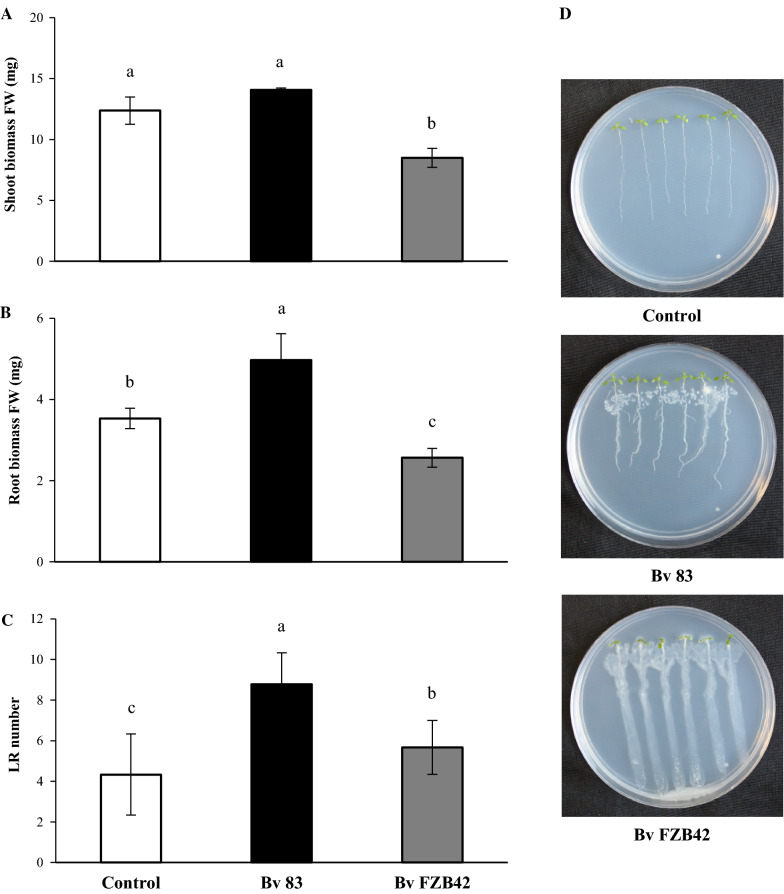
B. velezensis 83 and B. velezensis FZB42 (reference strain) had > 98% of identity (Additional file 1. Fig. S2). Through an in vitro co-culture assay with A. thaliana we showed the plant growth promotion effect of B. velezensis 83, it was compared with that caused by B. velezensis FZB42 and also both strains were compared against a control (uninoculated seedlings) (Fig. 5). After two days, B. velezensis 83 promoted the growth of A. thaliana, in contrast to the inoculation with B. velezensis FZB42, which had a suppressive effect over plant growth. There were no significant effects in terms of shoot biomass of A. thaliana caused by B. velezensis 83 inoculation (Fig. 5a); however, the main effect was observed in the root biomass (Fig. 5b). B. velezensis 83 increased significantly the lateral root number (102% more than the control) (Fig. 5c). This was similar to data reported for other Bacillus strains (Ryu et al. 2004, 2005; López-Bucio et al. 2007; Niazi et al. 2014; Asari et al. 2016, 2017; Verbon and Liberman, 2016; Islam et al. 2016; Kuan et al. 2016). Surprisingly, seedlings inoculated with B. velezensis FZB42 had only 69% and 73% shoot and root biomass (FW), respectively, as compared with the control. Furthermore, there were significant differences in other parameters of plant growth caused by both strains in A. thaliana: the primary root length, root density, lateral root length, shoot number, shoot diameter and total biomass were increased when using B. velezensis 83 but decreased when using B. velezensis FZB42 (Additional file 1. Fig. S3). Something characteristic of the inoculation of each strain was the bacterial pattern of seedling colonization. It was observed that B. velezensis 83 colonized forming a well defined and robust biofilm following the contour and development of the roots of each seedling, while B. velezensis FZB42 had an exacerbated growth and colonized extending its biofilm beyond the root and also invading the seedling shoot (Fig. 5d).
Fig. 5.
B. velezensis 83 and B. velezensis FZB42 growth-promoting effect on A. thaliana seedlings. a Shoot biomass (mg) fresh weight (FW). b Root biomass (mg) fresh weight (FW), c Lateral root (LR) number. N = 18. Different letters indicate statistically significant differences among treatments at P ≤ 0.05. d Aspect of control (not inoculated seedlings), B. velezensis 83 (Bv 83) and B. velezensis FZB42 (Bv FZB42) inoculated seedlings
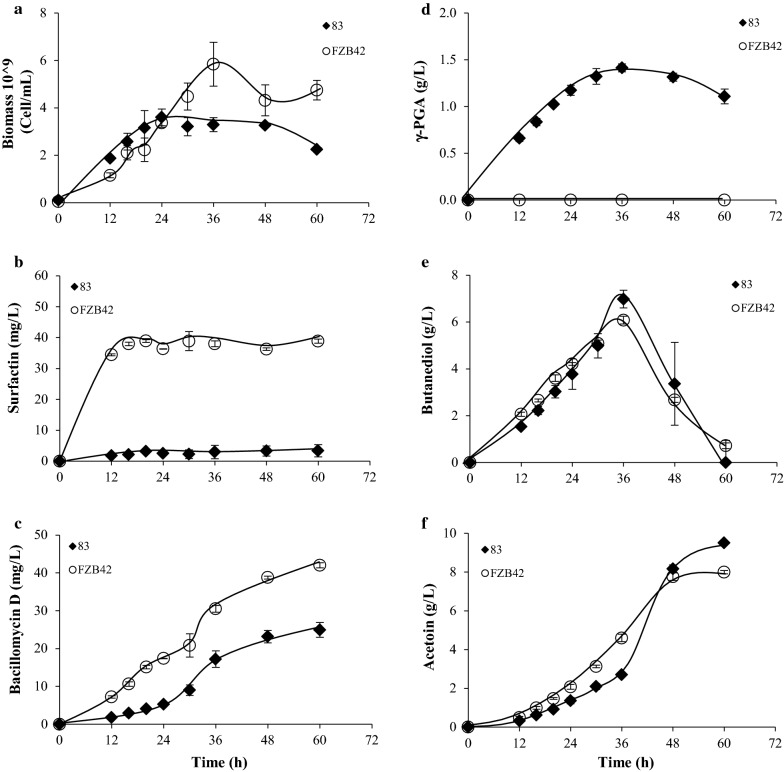
To test the functionality of some B. velezensis 83 gene clusters coding for synthesis of secondary metabolites related with biological control and plant interaction, several in vitro experiments were carried out (Fig. 6). With the culture conditions used in this study, B. velezensis 83 reached a maximum biomass of 3.3 × 109 cell/mL (Fig. 6a), the stationary growth phase started after glucose and acetate were depleted in the medium (Additional file 1. Fig. S4). Surfactin and bacillomycin production was detected since 12 h of culture. B. velezensis 83 produced up to 3.4 mg/L of surfactin (Fig. 6b) and 23.2 mg/L of bacillomycin (Fig. 6c). The γ-PGA was produced between the 12th and 36th h of cultivation, the maximum γ-PGA concentration was 1.4 g/L (Fig. 6d) and it was associated with the increase of apparent viscosity (3.9–4.9 cp) in the media. The γ-PGA MW was 1.8–2.0 MDa, but this compound was consumed after glucose depletion in the culture media (Additional file 1. Fig. S4). The production of 2,3-butanediol and acetoin were associated with biomass growth and glucose metabolism; nevertheless, 2,3-butanediol was consumed when glucose was depleted in the media. The maximum 2,3-butanediol concentration was 6.97 g/L (Fig. 6e) and acetoin was 9.51 g/L (Fig. 6f). On the other hand, the strain B. velezensis FZB42 showed a similar behavior in glucose consumption, as well as acetate, 2,3-butanediol and acetoin production. Nevertheless, the main differences observed between B. velezensis 83 and B. velezensis FZB42 strains were the maximum growth, γ-PGA as well as lipopeptides production. With the culture conditions used in this work, B. velezensis FZB42 did not produce γ-PGA but instead produced more biomass (5.8 × 109 cell/mL), and more surfactin (34.5 -39 mg/L since 12 h of culture time) and bacillomycin, than B. velezensis 83. We know that the gene cluster (pgdS, pgsEACB) involved in the synthesis γ-PGA are present in both strains B. velezensis 83 and B. velezensis FZB42 and they are of high identity (98%). Nevertheless, is known that B. velezensis 83 is a glutamic acid independent γ-PGA producing strain (Cristiano-Fajardo et al. 2019) as other few Bacillus strains (B. subtilis C10, Bacillus licheniformis A13, B. licheniformis TISTR 1010, B. methylotrophicus, B. subtilis NX2, B. amyloliquefaciens LL3) which only need glucose and NH4Cl as carbon and nitrogen sources, respectively, for γ-PGA synthesis (Cao et al. 2011; Hsueh et al. 2017; Sha et al. 2019). In contrast, lack of γ-PGA production by B. velezensis FZB42 could be explained due to a glutamate dependent mechanism. Therefore, B. velezensis 83 in addition to being a biological control agent and plant growth-promoting bacteria, is a new B. velezensis reported in the list of glutamate-independent γ-PGA producer strains reported recently (Sirisansaneeyakul et al. 2017).
Fig. 6.
B. velezensis 83 vs B. velezensis FZB42 secondary metabolites production in liquid culture. a Biomass (cell/mL), b surfactin (mg/L), c bacillomycin (mg/L), d γ-PGA (g/L), e 2,3-butanediol (g/L) and f acetoin (g/L)
A BlastP analysis (70% coverage with 50% identity) between B. velezensis 83 and B. velezensis FZB42 genomes shown that these two strains share 3475 core genes. In B. velezensis 83 genome there were 371 unique genes, 57% of them were genes with known function, 40% hypothetical proteins, 3% phage sequences and 1% kinase genes, while in B. velezensis FZB42 genome there were 262 unique genes. In B. velezensis 83 the cysTWA gene cluster (sulfate permease) and the sbp gene (sulfate ABC transporter substrate-binding protein) were found, these genes provide the ability to assimilating sulfite as well as thiosulfate to bacteria as sulfur sources (Guédon and Martin-Verstraete, 2006). Furthermore, cysTWA, sbp and cysP (thiosulfate transporter) genes complete the synthesis pathway of amino acid cysteine which has only been reported for the B. subtilis BD170 strain and other bacteria such as Escherichia coli and Salmonella typhimurium (Mansilla and de Mendoza, 1997). Cysteine is an amino acid present in antimicrobial peptides of ribosomal synthesis as bacteriocins (Abriouel et al. 2011) and other peptides as subtilisin (Graycar et al. 2013), surprisingly subtilin and mersacidin gene cluster were incomplete and only three genes for subtilisin (serine proteases: apr, aprx and epr) were found in B. velezensis 83 genome. Some Bacillus spp. strains also produce sulfure containing VOC such as carbon disulfide, dimethyltrisulfide and thiophene, which has been reported with antifungal activity (Caulier et al. 2019). Other relevant genes such as quorum-sensing (QS) pheromone comX, rap proteins (rapK1, rapK2 and rapH) and phrK peptide were identified as unique genes. In B. subtilis group ComX pheromone determines the pherotype and that the social communication cell–cell to stablish community structures within and between biofilms (Kalamara et al. 2018). ComX is the first signal to activate the transcription factor ComA which regulates the expression of several genes involved in the sporulation-competence signal transduction network (Schultz et al. 2009) as well as the production of surfactin, exoproteases and extracellular matrix for biofilm formation (Kalamara et al. 2018). ComX also could be different for strains of the same specie as reported for Bacillus spp. strains isolated from rhizoplane of tomato plants, which also showed different growth promotion effect over the plants (Oslizlo et al. 2015). There are two groups of Rap proteins, those that show phosphatase activity (RapA, RapB, RapE, RapH and RapJ) over effector response regulators (RR) and those that exert their function blocking the DNA-binding activity (RapC, RapF, RapG, RapH, and RapK) of their RR target, each Rap protein is specifically inhibited by a Phr peptide (Gallego del Sol and Marina, 2013). RapH protein negatively regulates srfA expression in B. velezensis MT45 and B. velezensis DSM7 (Zhi et al. 2017) and RapK-PhrK regulates the expression of several genes known to be activated directly by ComA (competence-sporulation genes) and indirectly by Spo0A (sporulation-biofilm genes) (Auchtung et al. 2006).
Discussion
B. velezensis 83 is an efficient biocontrol agent of mango anthracnose and plant growth-promoting bacteria of maize. México is one of the biggest worldwide mango exporters, for this reason Mexican mango orchards are worked carefully to obtain fruit with phytosanitary quality for its competitiveness in the international market. This means that mangoes pests (like fruit fly) and diseases (like anthracnose) must be absent at the harvest day as well as during the time of storage previous to its commercialization at the final export destination (21 days for boat transport from México to Japan). When chemical products are used for the control of anthracnose, it is important to ensure that the maximum residue limits (MRLs) of pesticides are under the maximal concentration allowed by international standard. Therefore, the relevance of biological control with B. velezensis 83 lies in the fact that it is an innocuous product for human consumption, whose application is efficient for the control of anthracnose in mango Kent (between 65 and 80%) such as treatments with agrochemicals (Captan 50 PH™ or Cupravit hydro™). B. velezensis 83 produces, in vitro, bacillomycin D, a lipopeptide that inhibits both spore germination and mycelial growth, propidium iodide staining of spores and mycelium showed that bacillomycin D caused damage to the cell membranes of C. gloeosporioides, affecting directly its viability (Luna-Bulbarela et al. 2018). Interestingly, B. velezensis 83 genome harbors three gene clusters coding synthases dedicated to the production of antifungal compounds, including the lipopeptide fengycin, the siderophore bacillibactin and the dipeptide bacilysin, which could contribute to its ability for the biocontrol of mango trees anthracnose. Other Bacillus strains have similar antimicrobial compounds production and biocontrol capacity. For example, a 70% of control for crown rot causing pathogens (Lasiodiplodia theobromae, Thielaviopsis paradoxa, Colletotrichum musae and Fusarium verticillioides) was obtained in postharvest banana fruits inoculated with B. amyloliquefaciens DGA14 (108 cfu/mL) (Alvindia, 2013). B. amyloliquefaciens GYL4 (104–108 cfu/mL) displayed between 46–93% of control efficacy on anthracnose of cucumber (Kim et al. 2016), B. velezensis RC 218 (104–106 cfu/mL) (Palazzini et al. 2016) and B. velezensis TrigoCor1448 (> 108 cfu/mL) (Crane et al. 2013) were able to control the Fusarium head blight in wheat.
B. velezensis 83 as plant growth-promoting bacteria had a different effect on the growth of the maize seedlings development depending on the phenological state of the maize, application to root seedling promoted the root development, in contrast application in seed state promoted the shoot development. Traits as phosphate solubilization, auxin and VOC (as 2,3-butanediol and acetoin) production by B. velezensis 83 could be involved in the growth promotion of maize as has been reported for other strains (Bentes et al. 2019; Cui et al. 2019). Plant hormone homeostasis is affected by PGPR production of auxins, ethylene, gibberellins, abscisic acid, salicylic acid, jasmonic acid, and different effects are observed on shoot or root system, being auxin the main hormone which regulates the plant growth and development (Tsukanova et al. 2017). PGPR production of auxins promotes plant-PGPR interaction, activates jasmonic acid dependent plant resistance, affects the expression of genes involved in auxin synthesis and transport (influx and efflux carriers), the PGPR colonization site affect the auxin gradient in the plant and also the bacterial VOC affect the auxin genes expression (Tsukanova et al. 2017). However, it has been reported that auxin has a complex crosstalk network (involving gene expression, signal transduction, and metabolic conversion process) with cytokinin and ethylene to coordinate the root development in A. thaliana (Liu et al. 2017a, b), thus the PGPR could affects several physiological processes in the plant at the same time.
In order to show the plant growth promotion effect, B. velezensis 83 was used in the in vitro assays with the model plant A. thaliana using 105 cfu/mL for in vitro inoculation, because a higher concentration of B. velezensis 83 cells (> 107 cfu/mL) had a growth suppressive effect on A. thaliana (data not shown). On the other hand, B. velezensis FZB42 in 103 cfu/mL concentration of bacteria did not show sufficient growth in the stria to generate reproducible results on the A. thaliana seedlings (data not shown). The plant growth promotion activity by B. velezensis FZB42 on Lemna minor plantlets (grown in 48-well microtiter plates) was dependent on the concentration of bacterial culture filtrate (%) or the quantity of growing bacterial cells (cfu) added (Idris et al. 2007). The higgest stimulatory effect on L. minor plantlets was using 2 × 105 cfu of B. velezensis FZB42, the result was attributed to the IAA production by this strain. Also, the authors reported that the lowest dilution of bacterial culture filtrates (0.1%) had a stimulatory effect on the seedling growth, but the highest concentration of bacterial growing cells (1 × 107 cfu) added had a negative effect on plantlets growth (Idris et al. 2007). B. velezensis FZB42 (former B. amyloliquefaciens FZB42) inoculation (106 cfu/mL) promoted the plant growth of A. thaliana under non-stress and saline conditions, the strain enhanced a 28.3% and 27.2% the shoot biomass dry weight at 0 and 100 mM NaCl compared with non-inoculated seedlings, respectively, nevertheless the effect on the root biomass dry weight was not considered in their evaluation. The inoculation of B. velezensis FZB42 increased the expression of auxin and photosynthesis related genes on A. thaliana (Liu et al. 2017a, b).
This study showed the capability B. velezensis 83 to synthetize in vitro antimicrobial compounds such as bacillomycin D, ISR elicitor compounds as surfactin, growth promotion compounds such as acetoin and 2,3-butanediol, and also the biopolymer γ-PGA (a biofilm component). The in vitro results of secondary metabolites production and B. velezensis 83-A. thaliana co-cultures assays suggest that the capacity of B. velezensis 83 to synthesize high concentration of γ-PGA and low concentration of surfactin (in contrast with B. velezensis FZB42) which contributed significantly to the pattern of biofilm formation on the root and resulted in a positive effect on seedling growth. The lipopeptides and polyketides compounds have the fundamental function of inhibiting plant pathogens (Mongkolthanaruk 2012); on the other hand, surfactin is a metabolite known for its antimicrobial activity, elicitor of the plant immune response and biofilm promoter (Ongena and Jacques 2007; Jourdan et al. 2009). Some components of the culture medium such as carbon, nitrogen and metal ion sources, were determinant of the level of production of antifungal activity of Bacillus sp. BH072 against Aspergillus niger, B. cinerea and F. oxysporum (Zhao et al. 2014). The γ-PGA could improve the abilities of B. amyloliquefaciens C06 cells to attach to smoot surfaces, to form biofilm and colonies, as well as to swarm on semisolid surface in vitro, is critical for increasing the robustness and complex morphology of the colony biofilm (Yu et al. 2016). B. subtilis γ-PGA producer cells had a higher root colonization efficiency (cfu of root-associated B. subtilis cells per gram of collected soil) than γ-PGA non-producers B. subtilis cells. Indeed, γ-PGA production improves colonization efficiency of B. amyloliquefaciens C06 on apple surface (Liu et al. 2010). In terms of VOC production, 2,3-butanediol and acetoin of B. subtilis GB03 and B. amyloliquefaciens IN937a, can reduce infection severity of A. thaliana seedlings by Erwinia carotovora subsp. carotovora strain SCC1 (Ryu et al. 2004). These VOCs also trigger plant growth promotion in A. thaliana (Farag et al. 2013). Several VOC structures produced by B. amyloliquefaciens UCMB5113 were identified by GC–MS analysis, different VOC profiles were found depending on the growth medium composition (Asari et al. 2016). On the other hand, acetoin and butanediol activated the abscisic and salicylic acid signaling networks in A. thaliana and Nicotiana benthamiana (tobacco), stimulating the production of nitric oxide and hydrogen peroxide during stomatal closure, which may prevent the plant from microbial infection (Wu et al. 2018).
Phenotypic differences between strains of high genetic homology may be caused by differences in the methylation patterns of the genes as a result the presence of different restriction-modification genes between the strains. For example, the type I restriction-modification DNA-methyltransferases hsdMSR was found in B. velezensis UCMB5113 and At1 strains and methyltransferase BamHIM-restriction enzyme BamHI complex, was uniquely identified in B. velezensis UCMB5007 and UCMB5044 strains. These methyltransferases can interfere in the sequences of the promoters in the genes (i.e. in the transcriptional regulator SigA which in turn regulates other sigma factors) which affects their expression and causes phenotypic differences between the strains when faced with environmental stimuli (Reva et al. 2019). In terms of gene expression, the B. amyloliquefaciens LL3 strain is a model because it is a glutamate-independent producer of γ-PGA and several mutants have been constructed. The mutant B. amyloliquefaciens LL3 ΔpgsBCA (non-producer of γ-PGA) showed a significant increase in the expression levels of the genes for synthesis of the antibiotics bacillaene, surfactin, iturin and fengicin (Gao et al. 2017). These authors suggest that the synthesis of these four antibiotics directly or indirectly affects the synthesis of γ-PGA and vice versa, in particular the synthesis of iturin and γ-PGA depends on the same substrate (glutamates or glutamines) and on the acetyl-coA of the TCA that is part of the synthesis machinery of both compounds. Also, the mutant B. amyloliquefaciens LL3 Δsrf (non-producer of surfactin), shown defects in biofilm formation and swarming, but increased γ-PGA production by ~ 24% more than B. amyloliquefaciens LL3 wild strain (Gao et al. 2017). B. amyloliquefaciens LL3 has the complete pathway for synthesis of iturin, however, the lipopeptide could not be detected in the culture medium also with the γ-PGA synthetase knockout strain NK-∆LP (non-producer γ-PGA), but B. amyloliquefaciens C2LP (derived from NK-∆LP) showed a considerable increase in the expression levels of the iturin synthesis genes due to the insertion of a constitutive promoter (C2up) in the iturin operon (Dang et al. 2019). B. amyloliquefaciens MT45 was able to achieve a tenfold surfactin production in comparison to the very close-related strain B. amyloliquefaciens DSM17 (Zhi et al 2017). The high production of surfactin in B. amyloliquefaciens MT45 was due to factors such as: a) the amount of ABC transporter proteins in the cell (as this contributes to detoxification of surfactin and to the assimilation of glutamate); b) the overexpression of genes related to nitrogen metabolism (important for amino acid synthesis); and c) changes in central carbon metabolism and lipid synthesis. B. amyloliquefaciens MT45 has eight encoding genes ABC transporters and two resistance-related genes and has been suggested that they favor the production of surfactin because they provide resistance to the cells against the antibiotic. On other hand, it has been shown that ~ 23% of the B. amyloliquefaciens FZB42 genes that showed changes in expression levels in the presence of maize plant root exudates (Fan et al. 2012), were genes of unknown function of which nineteen genes were unique to the strain and 15% were hypothetical or putatively functioning proteins. Therefore it is probable that the differences observed between B. velezensis 83 and B. velezensis FZB42 in this work about the production of surfactin, bacillomycin, γ-PGA, as well as in the formation of biofilm, are due to differences in gene regulation and expression related to the srf, bmy and γ-PGA gene clusters and also of unique genes as has been showed for other Bacillus spp. strains.
In conclusion, B. velezensis 83 is an efficient biocontrol agent of mango anthracnose and plant growth-promoting bacteria of maize and A. thaliana. B. subtilis 83 was reclassified as B. velezensis 83 according to a phylogenomic analysis. B. velezensis 83 genome harbors the genes to produce numerous secondary metabolites that are determinant for plant-bacteria interaction, sporulation, biocontrol and PGPB activity. B. velezensis 83 strain has all the necessary genes to establish a rhizosphere plant-bacteria interaction. Therefore, in addition to control foliar phytopathogens, B. velezensis 83 has the potential to be a biological control agent for root phytopathogens, probably eliciting ISR and promoting root growth. Our results are evidence that the B. velezensis 83 and B. velezensis FZB42 strains are phenotypically different, despite the fact that they have high genetic identity (> 98%), and it allows us to highlight the importance of the complementarity of the genomic sequencing with in vitro test and plant assays to identify B. velezensis 83 as a new effective strain for biological control, plant growth promotion and glutamate-independent γ-PGA production.
Supplementary information
Additional file 1: Figure S1. The pangenome model of 27 selected strains of B. velezensis (including B. velezensis 83) and B. amyloliquefaciens strains. Figure S2. Whole-comparison of species and strains of Bacillus. Pairwise ANIm was calculated using JSpecies (Richter et al. 2016). Figure S3. B. velezensis 83 and B. velezensis FZB42 growth-promoting effect on A. thaliana seedlings. Figure S4. B. velezensis 83 vs B. velezensis FZB42 secondary metabolites production in liquid culture. Table S1. Comparative BlastP (70% coverage and 50% identity) analysis of secondary metabolites genes of B. velezensis 83 and B. velezensis FZB42. Table S2. Genes involved in bacterium sporulation. Table S3. Genes involved in plant-bacterium interactions.
Acknowledgements
The authors thank to Celia Flores Ocampo for the excellent technical assistance to realize the chromatographic analysis of secondary metabolites produced by B. velezensis 83.
Authors’ contributions
All authors contributed to the study conception and design research. KB, SC, SB, MM conducted experiments. PB, RS and KB analyzed genomic data. KB wrote the original draft of manuscript. All authors reviewed and edited the manuscript. All authors read and approved the manuscript.
Funding
This work was supported by grants from the Consejo Nacional de Ciencia y Tecnología (CONACyT 247473 and CB-251848) and by UNAM-DGAPA-PAPIIT (grants IN206715, IG200618). KABR & SACF acknowledge CONACyT for the PhD scholarships 361862 & 290773, respectively.
Availability of data and materials
The datasets supporting the conclusions of this article are included withing the article and its additional files.
Ethics approval and consent to participate
This article does not contain any studies with human participants or animals performed by any of the authors.
Competing interest
The authors declare that they have no competing interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13568-020-01101-8.
References
- Abbott WS. A method of computing the effectiveness of an insecticide. J Econ Entomol. 1925;18:265–267. doi: 10.1093/jee/18.2.265a. [DOI] [Google Scholar]
- Abriouel H, Franz CM, Omar NB, Gálvez A. Diversity and applications of Bacillus bacteriocins. FEMS Microbiol Rev. 2011;35:201–232. doi: 10.1111/j.1574-6976.2010.00244.x. [DOI] [PubMed] [Google Scholar]
- Al-Ali A, Deravel J, Krier F, Béchet M, Ongena M, Jacques P. Biofilm formation is determinant in tomato rhizosphere colonization by Bacillus velezensis FZB42. Environ Sci Pollut Res. 2018;25:29910–29920. doi: 10.1007/s11356-017-0469-1. [DOI] [PubMed] [Google Scholar]
- Alvindia DG. Improving control of crown rot disease and quality of pesticide-free banana fruit by combining Bacillus amyloliquefaciens DGA14 and hot water treatment. Eur J Plant Pathol. 2013;136:183. doi: 10.1007/s10658-012-0154-7. [DOI] [Google Scholar]
- Arndt D, Grant J, Marcu A, Sajed T, Pon A, Liang Y, Wishart DS. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asari S, Matzén S, Petersen MA, Bejai S, Meijer J. Multiple effects of Bacillus amyloliquefaciens volatile compounds: plant growth promotion and growth inhibition of phytopathogens. FEMS Microbiol. Ecol. 2016;92:fiw070. doi: 10.1093/femsec/fiw070. [DOI] [PubMed] [Google Scholar]
- Asari S, Tarkowská D, Rolčík J, Novák O, Palmero DV, Bejai S, Meijer J. Analysis of plant growth-promoting properties of Bacillus amyloliquefaciens UCMB5113 using Arabidopsis thaliana as host plant. Planta. 2017;245:15–30. doi: 10.1007/s00425-016-2580-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auchtung JM, Lee CA, Grossman AD. Modulation of the ComA-dependent quorum response in Bacillus subtilis by multiple Rap proteins and Phr peptides. J Bacteriol. 2006;188(14):5273–5285. doi: 10.1128/JB.00300-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrera-Ortiz S, Garnica-Vergara A, Esparza-Reynoso S, García-Cárdenas E, Raya-González J, Ruiz-Herrera LF, López-Bucio J. Jasmonic acid-ethylene crosstalk via ETHYLENE INSENSITIVE 2 reprograms Arabidopsis root system architecture through nitric oxide accumulation. J Plant Growth Regul. 2018;37:438–451. doi: 10.1007/s00344-017-9741-3. [DOI] [Google Scholar]
- Belbahri L, Chenari Bouket A, Rekik I, Alenezi FN, Vallat A, Luptakova L, Petrovova E, Oszako T, Cherrad S, Vacher S, Rateb ME. Comparative genomics of Bacillus amyloliquefaciens strains reveals a core genome with traits for habitat adaptation and a secondary metabolites rich accessory genome. Front Microbiol. 2017;8:1438–1438. doi: 10.3389/fmicb.2017.01438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2008;37:D26–D31. doi: 10.1093/nar/gkn723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentes L, Mendes R, Cid E. Promotion of maize growth using endophytic bacteria under greenhouse and field conditions. Australian J Crop Sci. 2019;13(12):2067–2074. doi: 10.21475/ajcs.19.13.12.p2077. [DOI] [Google Scholar]
- Borriss R, Chen X, Rueckert C, Blom J, Becke A, Baumgarth B, Fan B, Pukall R, Schumann P, Spröer C, Junge H, Vater J, Pühler A, Klenk H. Int J Syst Evol Microbiol. 2011;61:1786–1801. doi: 10.1099/ijs.0.023267-0. [DOI] [PubMed] [Google Scholar]
- Branda SS, González-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci USA. 2001;98(20):11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito PH, Chevreux B, Serra CR, Schyns G, Henriques AO, Pereira-Leal JB. Genetic competence drives genome diversity in Bacillus subtilis. Genome Biol Evol. 2018;10(1):108–124. doi: 10.1093/gbe/evx270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics (Oxford, England) 2012;28:464–469. doi: 10.1093/bioinformatics/btr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai XC, Liu CH, Wang BT, Xue YR. Genomic and metabolic traits endow Bacillus velezensis CC09 with a potential biocontrol agent in control of wheat powdery mildew disease. Microbiol Res. 2017;196:89–94. doi: 10.1016/j.micres.2016.12.007. [DOI] [PubMed] [Google Scholar]
- Cao M, Geng W, Liu L, Song C, Xie H, Guo W, Jin Y, Wang S. Glutamic acid independent production of poly-γ-glutamic acid by Bacillus amyloliquefaciens LL3 and cloning of pgsBCA genes. Bioresource Technol. 2011;102:4251–4257. doi: 10.1016/j.biortech.2010.12.065. [DOI] [PubMed] [Google Scholar]
- Caulier S, Nannan C, Gillis A, Licciardi F, Bragard C, Mahillon J. Overview of the antimicrobial compounds produced by members of the Bacillus subtilis group. Front Microbiol. 2019 doi: 10.3389/fmicb.2019.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhari NM, Gupta VK, Dutta C. BPGA- an ultra-fast pan-genome analysis pipeline. Sci Rep. 2016;6:24373–24373. doi: 10.1038/srep24373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XH, Koumoutsi A, Scholz R, Eisenreich A, Schneider K, Heinemeyer I, Morgenstern B, Voss B, Hess WR, Reva O, Junge H, Voigt B, Jungblut PR, Vater J, Süssmuth R, Liesegang H, Strittmatter A, Gottschalk G, Borriss R. Comparative analysis of the complete genome sequence of the plant growth-promoting bacterium Bacillus amyloliquefaciens FZB42. Nature Biotechnol. 2007;25(9):1007–1014. doi: 10.1038/nbt1325. [DOI] [PubMed] [Google Scholar]
- Chen L, Shi H, Heng J, Wang D, Bian K. Antimicrobial, plant growth-promoting and genomic properties of the peanut endophyte Bacillus velezensis LDO2. Microbiol Res. 2019;218:41–48. doi: 10.1016/j.micres.2018.10.002. [DOI] [PubMed] [Google Scholar]
- Choudhary DK, Johri BN. Interactions of Bacillus spp. and plants—with special reference to induced systemic resistance ISR. Microbiol Res. 2009;164:493–513. doi: 10.1016/j.micres.2008.08.007. [DOI] [PubMed] [Google Scholar]
- Crane JM, Gibson DM, Vaughan RH, Bergstrom GC. Iturin levels on wheat spikes linked to biological control of Fusarium head blight by Bacillus amyloliquefaciens. Phytopathology. 2013;103:146–155. doi: 10.1094/PHYTO-07-12-0154-R. [DOI] [PubMed] [Google Scholar]
- Cristiano-Fajardo SA, Flores C, Flores N, Tinoco-Valencia R, Serrano-Carreón L, Galindo E. Glucose limitation and glucose uptake rate determines metabolite production and sporulation in high cell density continuous cultures of Bacillus amyloliquefaciens 83. J Biotechnol. 2019;299:57–65. doi: 10.1016/j.jbiotec.2019.04.027. [DOI] [PubMed] [Google Scholar]
- Cui W, He P, Munir S, He P, Li X, Li Y, Wu J, Wu Y, Yang L, He P, He Y. Efficacy of plant growth promoting bacteria Bacillus amyloliquefaciens B9601–Y2 for biocontrol of southern corn leaf blight. Biol Control. 2019;139:104080. doi: 10.1016/j.biocontrol.2019.104080. [DOI] [Google Scholar]
- Dang Y, Zhao F, Liu X, Fan X, Huang R, Gao W, Wang S, Yang C. Enhanced production of antifungal lipopeptide iturin A by Bacillus amyloliquefaciens LL3 through metabolic engineering and culture conditions optimization. Microb Cell Fact. 2019;18:68. doi: 10.1186/s12934-019-1121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics (Oxford, England) 2007;23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap CA, Kim SJ, Kwon SW, Rooney AP. Phylogenomic analysis shows that Bacillus amyloliquefaciens subsp. plantarum is a later heterotypic synonym of Bacillus methylotrophicus. Int J Syst Evol Microbiol. 2015;65:2104–2109. doi: 10.1099/ijs.0.000226. [DOI] [PubMed] [Google Scholar]
- Fan B, Carvalhais LC, Becker A, Fedoseyenko D, von Wirén N, Borriss R. Transcriptomic profiling of Bacillus amyloliquefaciens FZB42 in response to maize root exudates. BMC Microbiol. 2012;12:116. doi: 10.1186/1471-2180-12-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan B, Blom J, Klenk HP, Borriss R. Bacillus amyloliquefaciens, Bacillus velezensis, and Bacillus siamensis form an “operational group B. amyloliquefaciens” within the B. subtilis species complex. Front Microbiol. 2017;8:1–15. doi: 10.3389/fmicb.2017.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan B, Wang C, Song X, Ding X, Wu L, Wu H, Gao X, Borriss R. Bacillus velezensis FZB42 in 2018: The gram-positive model strain for plant growth promotion and biocontrol. Front Microbiol. 2018;9:1–14. doi: 10.3389/fmicb.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farag M, Zhang H, Ryu CM. Dynamic chemical communication between plants and bacteria through airborne signals: induced resistance by bacterial volatiles. J Chem Ecol. 2013;39:1007–1018. doi: 10.1007/s10886-013-0317-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fira D, Dimkić I, Berić T, Lozo J, Stanković S. Biological control of plant pathogens by Bacillus species. J Biotechnol. 2018;285:44–55. doi: 10.1016/j.jbiotec.2018.07.044. [DOI] [PubMed] [Google Scholar]
- Galindo E, Serrano-Carreón L, Gutiérrez C, Allende R, Balderas K, Patiño M, Trejo M, Wong M, Rayo E, Isauro D, Jurado C. The challenges of introducing a new biofungicide to the market: A case study. Elect J Biotechnol. 2013;16:5–5. doi: 10.2225/vol16-issue3-fulltext-6. [DOI] [Google Scholar]
- Gallego del Sol F, Marina A. Structural basis of Rap phosphatase Inhibition by Phr peptides. PLoS Biol. 2013;11(3):e1001511. doi: 10.1371/journal.pbio.1001511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W, Liu F, Zhang W, Quan Y, Dang Y, Feng J, Gu Y, Wang S, Song C, Yang C. Mutations in genes encoding antibiotic substances increase the synthesis of poly-γ-glutamic acid in Bacillus amyloliquefaciens LL3. MicrobiologyOpen. 2017;6(1):e00398. doi: 10.1002/mbo3.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guédon E, Martin-Verstraete I. Cysteine metabolism and its regulation in bacteria. In: Wendisch VF, editor. Amino acid biosynthesis ~ pathways, regulation and metabolic engineering. Microbiol monograph. Berlin: Springer; 2006. [Google Scholar]
- González V, Santamaría RI, Bustos P, Pérez-Carrascal OM, Vinuesa P, Juárez S, Martínez-Flores I, Cevallos MÁ, Brom S, Martínez-Romero E, Romero D. Phylogenomic Rhizobium species are structured by a continuum of diversity and genomic clusters. Front Microbiol. 2019;10(910):1–15. doi: 10.3389/fmicb.2019.00910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P. Consed: a graphical editor for next-generation sequencing. Bioinformatics. 2013;29:2936–2937. doi: 10.1093/bioinformatics/btt515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graycar TP, Bott RR, Power SD, Estell DA. Chapter 693—Subtilisins. In: Rawlings ND, Salvesen G, editors. Handbook of proteolytic enzymes. 3. Cambridge: Academic Press; 2013. pp. 3148–3155. [Google Scholar]
- Hanif K, Hameed S, Imran A, Naqqash T, Shahid M, Van Elsas JD. Isolation and characterization of a β-propeller gene containing phosphobacterium Bacillus subtilis strain KPS-11 for growth promotion of potato Solanum tuberosum L. Front Microbiol. 2015;6:583. doi: 10.3389/fmicb.2015.00583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao K, He P, Blom J, Rueckert C, Mao Z, Wu Y, He Y, Borriss R. The genome of plant growth-promoting Bacillus amyloliquefaciens subsp. plantarum strain YAU B9601–Y2 contains a gene cluster for mersacidin synthesis. J Bacteriol. 2012;194:3264–3265. doi: 10.1128/jb.00545-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh YH, Huang KY, Kunene SC, Lee TY. Poly-γ-glutamic acid synthesis, gene regulation, phylogenetic relationships, and role in fermentation. Int J Mol Sci. 2017;18(12):1–20. doi: 10.3390/ijms18122644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idris EE, Iglesias DJ, Talon M, Borriss R. Tryptophan-dependent production of indole-3-acetic acid (IAA) affects level of plant growth promotion by Bacillus amyloliquefaciens FZB42. Mol Plant-Microbe Interact. 2007;20(6):619–626. doi: 10.1094/mpmi-20-6-0619. [DOI] [PubMed] [Google Scholar]
- Islam S, Akanda AM, Prova A, Islam MT, Hossain MM. Isolation and identification of plant growth promoting rhizobacteria from cucumber rhizosphere and their effect on plant growth promotion and disease suppression. Front Microbiol. 2016;6:1360–1360. doi: 10.3389/fmicb.2015.01360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8(3):275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Jourdan E, Henry G, Duby F, Dommes J, Barthélemy JP, Thonart P, Ongena M. Insights into the defense-related events occurring in plant cells following perception of surfactin-type lipopeptide from Bacillus subtilis. Mol Plant-Microbe Interact. 2009;22:456–468. doi: 10.1094/MPMI-22-4-0456. [DOI] [PubMed] [Google Scholar]
- Kalamara M, Spacapan M, Mandic-Mulec I, Stanley-Wall NR. Social behaviours by Bacillus subtilis: quorum sensing, kin discrimination and beyond. Mol Microbiol. 2018;110(6):863–878. doi: 10.1111/mmi.14127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JD, Jeon BJ, Han JW, Park MY, Kang SA, Kim BS. Evaluation of the endophytic nature of Bacillus amyloliquefaciens strain GYL4 and its efficacy in the control of anthracnose. Pest Manag Sci. 2016;72:1529–1536. doi: 10.1002/ps.4181. [DOI] [PubMed] [Google Scholar]
- Kloepper JW, Ryu CM, Zhang S. Induced systemic resistance and promotion of plant growth by Bacillus spp. Phytopathology. 2004;94:1259–1266. doi: 10.1094/phyto.2004.94.11.1259. [DOI] [PubMed] [Google Scholar]
- Kuan KB, Othman R, Abdul RK, Shamsuddin ZH. Plant growth-promoting rhizobacteria inoculation to enhance vegetative growth, nitrogen fixation and nitrogen remobilisation of maize under greenhouse conditions. PLoS ONE. 2016;11:e0152478–e0152478. doi: 10.1371/journal.pone.0152478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaqat I, Ahmed SI, Jahan N. Biofilm formation and sporulation in Bacillus subtilis. Int J Microbiol Res Rev. 2013;4:061–067. [Google Scholar]
- Liu J, He D, Li XZ, Gao S, Wu H, Liu W, Gao X, Zhou T. γ-polyglutamic acid (γ-PGA) produced by Bacillus amyloliquefaciens C06 promoting its colonization on fruit surface. Int J Food Microbiol. 2010;142:190–197. doi: 10.1016/j.ijfoodmicro.2010.06.023. [DOI] [PubMed] [Google Scholar]
- Liu J, Moore S, Chen C, Lindsey K. Crosstalk complexities between auxin, cytokinin, and ethylene in Arabidopsis root development: from experiments to systems Modeling, and back again. Mol Plant. 2017;10(12):1480–1496. doi: 10.1016/j.molp.2017.11.002. [DOI] [PubMed] [Google Scholar]
- Liu S, Hao H, Lu X, Zhao X, Wang Y, Zhang Y, Xie Z, Wang R. Transcriptome profiling of genes involved in induced systemic salt tolerance conferred by Bacillus amyloliquefaciens FZB42 in Arabidopsis thaliana. Sci Rep. 2017;7(1):10795. doi: 10.1038/s41598-017-11308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López D, Kolter R. Extracellular signals that define distinct and coexisting cell fates in Bacillus subtilis. FEMS Microbiol Rev. 2010;34:134–149. doi: 10.1111/j.1574-6976.2009.00199.x. [DOI] [PubMed] [Google Scholar]
- López-Bucio J, Campos-Cuevas JC, Hernández-Calderón E, Velásquez-Becerra C, Farías-Rodríguez R, Macías-Rodríguez LI, Valencia-Cantero E. Bacillus megaterium rhizobacteria promote growth and alter root-system architecture through an auxin- and ethylene-independent signaling mechanism in Arabidopsis thaliana. Mol Plant-Microbe Inter. 2007;20:207–217. doi: 10.1094/MPMI-20-2-0207. [DOI] [PubMed] [Google Scholar]
- Luna-Bulbarela A, Tinoco-Valencia R, Corzo G, Kazuma K, Konno K, Galindo E, Serrano-Carreón L. Effects of bacillomycin D homologues produced by Bacillus amyloliquefaciens 83 on growth and viability of Colletotrichum gloeosporioides at different physiological stages. Biol Control. 2018;127:145–154. doi: 10.1016/j.biocontrol.2018.08.004. [DOI] [Google Scholar]
- Mansilla MC, Mendoza D. L-Cysteine biosynthesis in Bacillus subtilis: identification, sequencing, and functional characterization of the gene coding for phosphoadenylylsulfate sulfotransferase. J Bacteriol. 1997;179(3):976–981. doi: 10.1128/2Fjb.179.3.976-981.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Derbyshire M, Gonzales N, Lu S, Chitsaz F, Geer L, Geer C, He J, Gwadz M, Hurwitz D, Lanczycki C, Lu F, Marchler G, Song J, Thanki N, Wang Z, Yamashita R, Zhang D, Zheng C, Bryant S. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015;43:D222–D232. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvasi M, Visscher PT, Martinez LC. Exopolymeric substances (EPS) from Bacillus subtilis: polymers and genes encoding their synthesis. FEMS Microbiol Lett. 2010;313:1–9. doi: 10.1111/j.1574-6968.2010.02085.x. [DOI] [PubMed] [Google Scholar]
- Mitchell A, Chang HY, Daugherty L, Fraser M, Hunter S, Lopez R, Finn RD. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 2015;43:D213–D221. doi: 10.1093/nar/gku1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongkolthanaruk W. Classification of Bacillus beneficial substances related to plants, humans and animals. J Microbiol Biotechnol. 2012;22:1597–1604. doi: 10.4014/jmb.1204.04013. [DOI] [PubMed] [Google Scholar]
- Niazi A, Manzoor S, Asari S, Bejai S, Meijer J, Bongcam-Rudloff E. Genome analysis of Bacillus amyloliquefaciens Subsp. plantarum UCMB5113: a rhizobacterium that improves plant growth and stress management. PLoS ONE. 2014;9:e104651. doi: 10.1371/journal.pone.0104651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olanrewaju OS, Glick BR, Babalola OO. Mechanisms of action of plant growth promoting bacteria. W J Microbiol Biotechnol. 2017;33:197. doi: 10.1007/s11274-017-2364-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oslizlo A, Stefanic P, Vatovec S, Beigot Glaser S, Rupnik M, Mandic-Mulec I. Exploring ComQXPA quorum-sensing diversity and biocontrol potential of Bacillus spp. isolates from tomato rhizoplane. Microb Biotechnol. 2015;8(3):527–540. doi: 10.1111/1751-7915.12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongena M, Jacques P. Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 2007;16:115–125. doi: 10.1016/j.tim.2007.12.009. [DOI] [PubMed] [Google Scholar]
- Palazzini JM, Dunlap CA, Bowman MJ, Chulze SN. Bacillus velezensis RC 218 as a biocontrol agent to reduce Fusarium head blight and deoxynivalenol accumulation: Genome sequencing and secondary metabolite cluster profiles. Microbiol Res. 2016;192:30–36. doi: 10.1016/j.micres.2016.06.002. [DOI] [PubMed] [Google Scholar]
- Pérez-Carrascal OM, VanInsberghe D, Juárez S, Polz MF, Vinuesa P, González V. Population genomics of the symbiotic plasmids of sympatric nitrogen-fixing Rhizobium species associated with Phaseolus vulgaris. Environ Microbiol. 2016;18:2660–2676. doi: 10.1111/1462-2920.13415. [DOI] [PubMed] [Google Scholar]
- Rabbee MF, Ali MS, Choi J, Hwang BS, Jeong SC, Baek KH. Bacillus velezensis: a valuable member of bioactive molecules within plant microbiomes. Molecules. 2019;24:1046. doi: 10.3390/molecules24061046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reva ON, Swanevelder DZH, Mwita LA, Mwakilili AD, Muzondiwa D, Joubert M, Chan WY, Lutz S, Ahrens CH, Avdeeva LV, Kharkhota MA, Tibuhwa D, Lyantagaye S, Vater J, Borriss R, Meijer J. Genetic, epigenetic and phenotypic diversity of four Bacillus velezensis strains used for plant protection or as probiotics. Front Microbiol. 2019;10:2610. doi: 10.3389/fmicb.2019.02610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M, Rosselló-Móra R, Glöckner F, Peplies J. JSpecies WS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics. 2016;32(6):929–931. doi: 10.1093/bioinformatics/btv681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero D. Bacterial determinants of the social behavior of Bacillus subtilis. Res Microbiol. 2013;164:788–798. doi: 10.1016/j.resmic.2013.06.004. [DOI] [PubMed] [Google Scholar]
- Ryu CM, Farag MA, Hu CH, Reddy MS, Kloepper JW, Paré PW. Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol. 2004;134:1017–1026. doi: 10.1104/pp.103.026583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu CM, Hu CH, Locy RD, Kloepper JW. Study of mechanisms for plant growth promotion elicited by rhizobacteria in Arabidopsis thaliana. Plant Soil. 2005;268:285–292. doi: 10.1007/s11104-004-0301-9. [DOI] [Google Scholar]
- Sha Y, Sun T, Qiu Y, Zhu Y, Zhan Y, Zhang Y, Xu Z, Li S, Feng X, Xu H. Investigation of glutamate dependence mechanism for poly-γ-glutamic acid production in Bacillus subtilis on the basis of transcriptome analysis. J Agric Food Chem. 2019;67:6263–6274. doi: 10.1021/acs.jafc.9b01755. [DOI] [PubMed] [Google Scholar]
- Schmitz S, Hoffmann A, Szekat C, Rudd B, Bierbaum G. The lantibiotic mersacidin is an autoinducing peptide. Appl Environ Microbiol. 2006;72:7270–7277. doi: 10.1128/aem.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz D, Wolynes PG, Ben Jacob E, Onuchic JN. Deciding fate in adverse times: sporulation and competence in Bacillus subtilis. Proc Natl Acad Sci USA. 2009;106(50):21027–21034. doi: 10.1073/pnas.0912185106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siguier P, Filée J, Chandler M. Insertion sequences in prokaryotic genomes. Curr Opin Microbiol. 2006;9:526–531. doi: 10.1016/j.mib.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Sirisansaneeyakul S, Cao M, Kongklom N, Chuensangjun C, Zhongping S, Chisti Y. Microbila production of poly-γ-glutamic acid. World J Microbiol Biotechnol. 2017;33:173. doi: 10.1007/s11274-017-2338-y. [DOI] [PubMed] [Google Scholar]
- Stein T, Borchert S, Kiesau P, Heinzmann S, Klöss S, Klein C, Helfrich M, Entian KD. Dual control of subtilin biosynthesis and immunity in Bacillus subtilis. Mol Microbiol. 2002;44:403–416. doi: 10.1046/j.1365-2958.2002.02869. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6. Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan IS, Ramamurthi KS. Spore formation in Bacillus subtilis. Environ Microbial Rep. 2013;6:212–225. doi: 10.1111/2F1758-2229.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28(1):33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend GR, Heuberger JW. Methods for estimating losses caused by diseases in fungicides experiments. Plant Dis Reporter. 1943;27:340–343. [Google Scholar]
- Tsukanova KA, Chebotar VK, Meyer JJM, Bibikova TN. Effect of plant growth-promoting rhizobacteria on plant hormone homeostasis. S Afr J Bot. 2017;113:91–102. doi: 10.1016/j.sajb.2017.07.007. [DOI] [Google Scholar]
- Vejan P, Abdullah R, Khadiran T, Ismail S, Nasrulhaq BA. Role of plant growth promoting rhizobacteria in agricultural sustainability—A Review. Molecules. 2016;21:573. doi: 10.3390/molecules21050573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbon EH, Liberman LM. Beneficial microbes affect endogenous mechanisms controlling root development. Trends Plant Sci. 2016;21:218–229. doi: 10.1016/j.tplants.2016.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Ma L, Li X, Gao X, Borriss R, Wu Z. Acetoin and 2,3-butanediol from Bacillus amyloliquefaciens induce stomatal closure in Arabidopsis thaliana and Nicotiana benthamiana. J Exp Bot. 2018;69:5625–5635. doi: 10.1093/jxb/ery326. [DOI] [PubMed] [Google Scholar]
- Xia Y, Xie S, Ma X, Wu H, Wang X, Gao X. The purL gene of Bacillus subtilis is associated with nematicidal activity. FEMS Microbiol Lett. 2011;322:99–107. doi: 10.1111/j.1574-6968.2011.02336.x. [DOI] [PubMed] [Google Scholar]
- Xu M, Sheng J, Chen L, Men Y, Gan L, Guo S, Shen L. Bacterial community compositions of tomato Lycopersicum esculentum Mill. seeds and plant growth promoting activity of ACC deaminase producing Bacillus subtilis HYT-12–1 on tomato seedlings. W J Microbiol Biotechnol. 2014;30:835–845. doi: 10.1007/s11274-013-1486-y. [DOI] [PubMed] [Google Scholar]
- Yan F, Yu Y, Wang L, Luo Y, Guo JH, Chai Y. The comER gene plays an important role in biofilm formation and sporulation in both Bacillus subtilis and Bacillus cereus. Front Microbiol. 2016;7:1025. doi: 10.3389/fmicb.2016.01025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Yan F, Chen Y, Jin C, Guo JH, Chai Y. Poly-γ-glutamic acids contribute to biofilm formation and plant root colonization in selected environmental isolates of Bacillus subtilis. Front Microbiol. 2016;7:1811. doi: 10.3389/fmicb.2016.01811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Han Y, Tan XQ, Wang J, Zhou ZJ. Optimization of antifungal lipopeptide production from Bacillus sp. BH072 by response surface methodology. J Microbiol. 2014;52:324–332. doi: 10.1007/s12275-014-3354-3. [DOI] [PubMed] [Google Scholar]
- Zhi Y, Wu Q, Xu Y. Genome and transcriptome analysis of surfactin biosynthesis in Bacillus amyloliquefaciens MT45. Sci Rep. 2017;7:40976. doi: 10.1038/srep40976. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. The pangenome model of 27 selected strains of B. velezensis (including B. velezensis 83) and B. amyloliquefaciens strains. Figure S2. Whole-comparison of species and strains of Bacillus. Pairwise ANIm was calculated using JSpecies (Richter et al. 2016). Figure S3. B. velezensis 83 and B. velezensis FZB42 growth-promoting effect on A. thaliana seedlings. Figure S4. B. velezensis 83 vs B. velezensis FZB42 secondary metabolites production in liquid culture. Table S1. Comparative BlastP (70% coverage and 50% identity) analysis of secondary metabolites genes of B. velezensis 83 and B. velezensis FZB42. Table S2. Genes involved in bacterium sporulation. Table S3. Genes involved in plant-bacterium interactions.
Data Availability Statement
The datasets supporting the conclusions of this article are included withing the article and its additional files.