Abstract
RNA polymerase II interacts with various other complexes and factors to ensure correct initiation, elongation, and termination of mRNA transcription. One of these proteins is SR-related CTD-associated factor 4 (SCAF4), which is important for correct usage of polyA sites for mRNA termination. Using exome sequencing and international matchmaking, we identified nine likely pathogenic germline variants in SCAF4 including two splice-site and seven truncating variants, all residing in the N-terminal two thirds of the protein. Eight of these variants occurred de novo, and one was inherited. Affected individuals demonstrated a variable neurodevelopmental disorder characterized by mild intellectual disability, seizures, behavioral abnormalities, and various skeletal and structural anomalies. Paired-end RNA sequencing on blood lymphocytes of SCAF4-deficient individuals revealed a broad deregulation of more than 9,000 genes and significant differential splicing of more than 2,900 genes, indicating an important role of SCAF4 in mRNA processing. Knockdown of the SCAF4 ortholog CG4266 in the model organism Drosophila melanogaster resulted in impaired locomotor function, learning, and short-term memory. Furthermore, we observed an increased number of active zones in larval neuromuscular junctions, representing large glutamatergic synapses. These observations indicate a role of CG4266 in nervous system development and function and support the implication of SCAF4 in neurodevelopmental phenotypes. In summary, our data show that heterozygous, likely gene-disrupting variants in SCAF4 are causative for a variable neurodevelopmental disorder associated with impaired mRNA processing.
Keywords: SCAF4, neurodevelopmental disorder, mRNA processing, intellectual disability, seizures, epilepsy
Main Text
The regulation of protein-coding gene transcription is a highly complex process that is crucial for proper gene expression. RNA polymerase II (RNAPII) interacts with many other complexes and factors to ensure accurate pre-initiation, initiation, elongation, and termination of mRNA transcription (reviewed in Cramer,1 Orphanides and Reinberg,2 Kornberg,3 and Lee and Young4). Mutations in several genes and proteins involved in mRNA processing have been implicated in human diseases. For example, heterozygous pathogenic variants in POLR2A (MIM: 180660), which encodes the largest subunit of RNAPII, have recently been identified as the cause of a neurodevelopmental disorder (NDD) with hypotonia and variable intellectual and behavioral anomalies (NEDHIB [MIM: 618603]).5 Similarly, pathogenic variants in several subunits of the transcription factor IID complex, which plays a key role in transcriptional initiation,6,7 have been implicated in NDDs. These include pathogenic variants in TATA binding protein associated factors, e.g., an X-linked NDD (MRXS33 [MIM: 300966]) is caused by variants in TAF1 (MIM: 313650) and an autosomal-recessive NDD (MRT60 [MIM: 617432]) by variants in TAF13 (MIM: 600774).8,9 Furthermore, bi-allelic variants in two subunits of the RNAPII interacting Integrator complex, INTS1 (MIM: 611345) and INTS8 (MIM: 611351), have been shown to be associated with NDDs (NDCAGF [MIM: 618571] and NEDCHS [MIM: 618572]) and lead to both altered splicing patterns and differential gene expression in cells derived from affected individuals.10
Using trio exome sequencing on an Illumina HiSeq 2500 platform and data analysis with an in-house pipeline as described previously,11 we identified the de novo splice-site variant c.321+1G>T in SCAF4 (MIM: 616023) (GenBank: NM_020706.2) in an individual with mild intellectual disability (ID) and seizures. SCAF4 consists of 20 exons and encodes SR (serine and arginine)-related CTD (C-terminal domain)-associated factor 4, consisting of 1,147 amino acids and containing an N-terminal conserved CTD-interacting domain and an RNA recognition motif (Figures 1A and 1B).13 SCAF4 interacts with the C-terminal domain of the largest subunit of RNAPII, and together with SCAF8, is required for correct polyA site selection and mRNA termination.13,14 So far, SCAF4 has not been implicated in NDDs or other diseases. According to gnomAD15 constraint scores, SCAF4 is intolerant toward loss-of-function variants (pLI = 1, o/e = 0.03 (0.01–0.11)), thus supporting a pathogenic relevance of the identified splice-site variant.
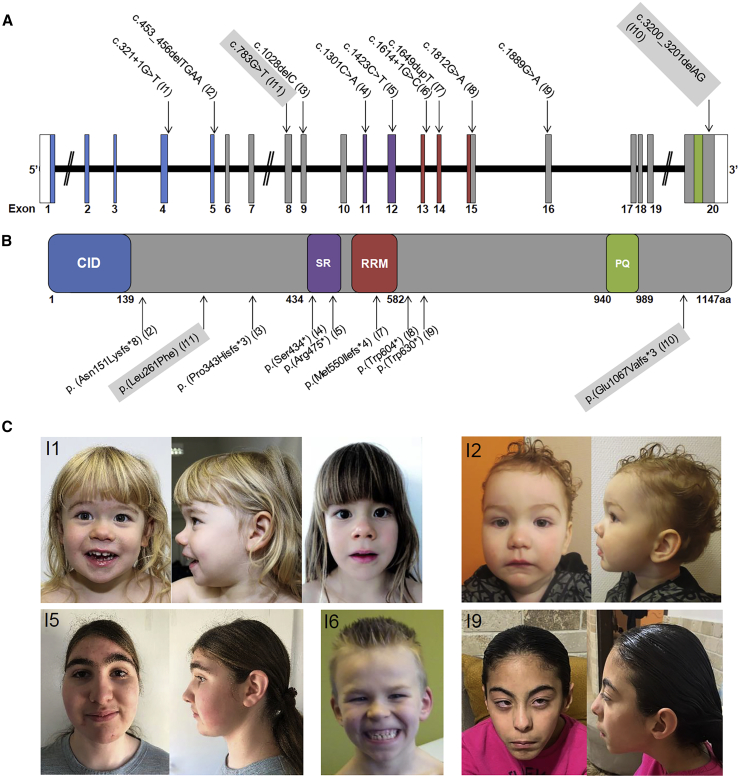
Figure 1.
SCAF4 Variants Observed in Individuals with NDDs
(A) Schematic drawing of SCAF4 (longest isoform GenBank: NM_020706.2) with identified variants. Non-coding exonic regions are displayed in white, coding exons in gray, and encoded domains (according to Ensembl12) in color.
(B) Schematic drawing of the SCAF4 protein. One missense and eight likely gene-disrupting variants (excluding two splice-site variants) are displayed below the scheme. Variants of unknown significance are shaded in gray.
(C) Clinical pictures of individuals with pathogenic variants in SCAF4 (I1 at age 3 and 5 years; I2 at age 20 months; I5 at age 13 years; I6 at age 6.5 years; I9 at age 9 years). Note common facial features such as epicanthus, a deep nasal bridge, bulbous nasal tip, and deep philtrum, particularly in I1 and I2.
Abbreviations: CID, conserved CTD-interacting domain; SR, Ser/Arg-rich domain; RRM, RNA recognition motif; PQ, Pro/Gln-rich domain.
We used GeneMatcher16 to identify eight other unrelated individuals with NDDs and likely pathogenic variants in SCAF4 and two individuals with variants of unknown significance (Table 1, Table S1). Variants were identified through routine diagnostic testing or in a research setting approved by the ethical review boards of the respective institutions (Table S1). Informed consent was obtained from the parents or legal guardians of the affected individuals.
Table 1.
Clinical Details
| Individual |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gender | f | m | m | m | f | m | m | m | f | m | m |
| Age | 3y 3mo | 20mo | 16y | 18y | 13y | 6y 6mo | 6y 5mo | 11y | 4.5y | 4y | 10y |
| Variant cDNA | c.321+1G>T | c.453_456delTGAA | c.1028delC | c.1301C>A | c.1423C>T | c.1614+1G>C | c.1649dupT | c.1812G>A | c. 1889G>A | c.3200_3201delAG | c.783G>T |
| Variant RNA/protein | r.spl? | p.Asn151Lysfs∗8 | p.Pro343Hisfs∗3 | p.Ser434∗ | p.Arg475∗ | r.spl? | p.Met550Ilefs∗4 | p.Trp604∗ | p.Trp630∗ | p.Glu1067Valfs∗3 | p.Leu261Phe |
| De novo | yes | yes | yes | yes | yes | yes | yes | maternal | yes | unknown (also in affected sister) | yes |
| Height cm/SD | 101 / 0.66 | 84.5 / −0.28 | 162.6 / −1.6 | 165.5 / −1.51 | 157.5 / −0.41 | 126.5 / 1 | 114 / −0.22 (5y) | 136 / −1.69 | 125 / −1.24 (8.5y) | N/A | 142 / −0.25 |
| Weight kg/SD | 17.2 / 1.04 | 12.5 / +0.10 | 67.5 / 0.40 | 56.9 / −1.25 | 44.4 / −0.61 | 24.9 / 0 | 26 / 1.56 (5y) | 38.2 / −0.14 | 21 / 1.19 | N/A | 62.7 / 2.39 |
| OFC cm/SD | 49.2 / −0.38 | 47.5 / −0.8 | N/A | 59 / 1.42 | 51.8 / −1.9 | 52.8 / 0 | N/A | 55 / 0.54 | 49.2 / −0.92 | N/A | 57.6 / 2.77 |
| DD/ID | mild-moderate (SON-IQ 1x67, 1x50) | mild | mild | severe | mild | mild (tIQ 91 WISC-V) | language delay | learning diff. | mild-moderate (DQ 56) | moderate | yes |
| Walking | 14mo | 17mo | 14mo | 4–5y | 14mo | 12mo | 18mo | 14–15mo | 29mo | N/A | 15mo |
| First words | 12mo | 18mo | 2.5y | 5y | 14mo | 16mo | 3y | 2–3y | 3.7y | not yet | 3–4y |
| Speech | 3y 3mo: 30 words | N/A | 3y: speech therapy | 10 words | >2y: 2-word comb. | delay | N/A | 11y: 20 words | simple sentences | 4y: no speech | N/A |
| Regression | stagnation with seiz. | no | no | N/A | possibly in speech | no | no | no | yes (10–12mo) | N/A | possibly |
| Seizures, onset age | 4x, 27–30mo | no | no | intractable (myoclonic/tonic), 10mo | myoclonus, 12y | no | myoclonic astatic epilepsy, 4y | intractable, 18mo | no | N/A | yes, 2y |
| MRI anom. | subcort., periventr. hypomyelination | N/D | N/D (normal US) | possible focal cortical dysplasia, volume loss | normal | N/D | nonspecific FLAIR hyperintensities frontal subcortical area left > right | white matter changes, cerebellar atrophy | PCH, HCC | N/A | HCC |
| Muscular hypotonia | yes | yes | no | yes | no | N/R | N/R | hypertonia | yes | N/A | yes |
| Behavioral anom. | autistic features | N/A | autistic features in infancy | autism, aggression, hyperactivity | autism | autism, hyperactivity, aggression | no | self-injurous, aggression | aggression | autism, hyperactivity | disruptive behavior, autism |
| Cardiac defect | N/D | murmur, US normal | VSD, bicuspid aortic valve, hypoplastic aortic arch, dilated cardiomyopathy | N/A | VSD (self-resolved) | ND | N/R | N/R | VSD (self-resolved), PFO | N/A | N/R |
| Renal ano. | N/D | N/D | unilateral agenesis | multicystic kidneys | unilateral hydronephrosis | no | N/R | N/R | multicystic kidneys | N/A | N/R |
| Urogenital anom. | no | no | cryptorchidism | cryptorchidism | no | no | N/R | N/R | inguinal hernia | N/A | N/R |
| GI anomalies | no | no | TE fistula, imperforate anus | s/p pyloroplasty, nissen fundoplication, G-tube | none | no | N/R | N/R | no | N/A | N/R |
| Skeletal anom. | no | no | sacrum segmentation anom., brachydactyly | kyphosis, scoliosis | lordosis, hallux valgus, toe syndactyly II/III | kyphosis | N/R | bilateral ankle rotation, pectus excavatum | antevertion of femur | N/A | scoliosis, pronation of feet |
| Other | none | none | tethered cord, hypothyroidism | sleep apnea, chronic lung disease, bronchopulmonary dysplasia, Sotos syndrome | premature adrenarche | none | none | delayed teeth eruption | none | none | none |
f, female; m, male; y, years; mo, months; SD, standard deviation; OFC, occipito-frontal head circumference; DD, developmental delay; ID, intellectual disability; N/A, not available or not applicable; N/D, not done; N/R, not reported; anom., anomaly; HCC, hypoplasia of corpus callosum; VSD, ventricle septum defect; PFO, persistent foramen ovale; PCH, pontocerebellar hypoplasia; TE, tracheesophageal; US, ultrasound.
The nine variants that were considered likely pathogenic included two splice-site and seven truncating variants (Figures 1A and 1B). None of them were present in gnomAD.15 Eight occurred de novo, and one nonsense variant c.1812G>A (p.Trp604∗) was inherited from a healthy mother. gnomAD15 contains 14 presumably truncating variants in SCAF4, of which 10 are located in the last exon and probably are benign due to escaping nonsense-mediated mRNA decay. In contrast, the two splice-site and seven truncating variants in our cohort are predicted to affect the N-terminal two thirds of the protein, and therefore likely trigger nonsense-mediated mRNA-decay or produce a severely truncated protein. Nonsense-mediated mRNA decay was indicated by reverse transcription PCR (RT-PCR) on cDNA/RNA from a PaxGene (PreAnalytiX, BD and QIAGEN) blood sample of I4 with the c.1301C>A (p.Ser434∗) variant (Figure S1A). In contrast, the mutant c.1889G>A (p.Trp630∗) allele in I9 was still equally visible. However, as RNA sequencing (see below) indicated reduced SCAF4 expression also in this individual (Figure S1B, Table S2), a later decay process might occur. Furthermore, we confirmed aberrant splicing for the splice-site variant in I1 by showing several aberrant transcripts with complete or partial loss of exons 3, 4, or 5 (Figures S2A and S2B). RNA sequencing in I1 additionally revealed another isoform with intron 4 retained (Figures S2C and S2D). For all truncating variants in the N-terminal part of SCAF4, a general loss-of-function mechanism or haploinsufficiency is therefore likely.
The Decipher database17,18 contains several deletions including SCAF4, but all encompass a large number of additional genes, thereby preventing specific deductions regarding SCAF4 haploinsufficiency in these individuals.
Additionally, we identified two variants of unknown significance. The frameshifting variant c.3200_3201del (p.Glu1067Valfs∗3) segregated in two siblings with neurodevelopmental phenotypes. Parents were not available for testing. However, it is located in the C-terminal part of the gene/protein, where most of the truncating variants in gnomAD reside. The de novo missense variant c.783G>T (p.Leu261Phe) was identified in an individual with a consistent NDD phenotype. Though SCAF4 is not particularly intolerant toward missense variants (gnomAD constraint scores: z = 1.94, o/e = 0.79 (0.73–0.85)) and though this variant is not located in any of the known functional domains, there is some evidence pointing to a possible pathogenic relevance. This variant is not present in gnomAD, affects a highly conserved amino acid (Figure S3A), is predicted to be deleterious by Mutation Taster,19 PP2,20 SIFT,21 and M-CAP22 (Figure S3B), and resulted in reduced SCAF4 expression in RNA sequencing in I11, while mutant and wild-type alleles were equally visible in RT-PCR (Figure S1C).
The clinical features of the affected individuals are summarized in Table 1. For phenotypic delineation, we excluded I10 and I11 due to their unclear variant status and I4 who additionally has Sotos syndrome (SOTOS1 [MIM: 117550]) due to a pathogenic variant in NSD1 (MIM: 606681), confounding the clinical picture. The remaining eight individuals all had developmental delay and intellectual disability, mostly in the mild range. Speech was more severely affected than motor development. While age of walking was within a normal range of 12–18 months in seven individuals, only two individuals had normal speech development (first words before 15 months). Speech was severely delayed (first words after 2 years) in four individuals. Developmental stagnation co-occurring with seizures was reported in one and possible developmental regression in two individuals. Behavioral anomalies were reported in five individuals (63%) and included autistic features, hyperactivity, and aggressive behavior. Seizures occurred in four individuals (50%) and included myoclonic seizures in two and intractable seizures in one. Brain MRIs were performed in five individuals showing nonspecific white matter anomalies in three of them. I9 was diagnosed with pontocerebellar hypoplasia and a thin corpus callosum. Variable other features included renal (38%), cardiac (38%), or skeletal (63%) anomalies. I3 presented with multiple malformations and anomalies. Minor but rather unspecific facial dysmorphism were noted in most of the individuals. Shared facial features in two individuals (I1, I2) were epicanthus, a flat nasal bridge, a bulbous nasal tip, and a deep philtrum (Figure 1C).
Taken together, likely gene-disruptive variants in SCAF4 are associated with a variable neurodevelopmental phenotype with predominantly mild developmental delay and intellectual disability and frequently with seizures and behavioral anomalies. As increasingly observed for other genes associated with mild NDDs, putatively pathogenic, autosomal-dominant variants in SCAF4 may be inherited from a mildly affected or presumably healthy parent, as observed for I8.
SCAF4 interacts with RNAPII. Interestingly, also variants in POLR2A, encoding the largest subunit of RNAPII, have been identified to cause a neurodevelopmental disorder, possibly due to a dominant-negative effect on RNA transcription.5 While variable developmental delay, behavioral abnormalities, and white matter anomalies in brain imaging overlap between individuals with either SCAF4 or POLR2A variants, the POLR2A-associated phenotype appears to be more severe with profound muscular hypotonia as a prominent feature.5
In the context of its interaction with RNAPII, an important role for SCAF4 in mRNA polyA recognition and mRNA termination was characterized recently.14 CRISPR-Cas9-mediated knockout of SCAF4 in HEK293 cells resulted in increased transcriptional read-through or in the usage of alternative last exons. This, in turn, was visible as an increase in the number of genes with an elongated 3′ sequence over genes with a truncated one.14
To investigate whether SCAF4 defects also result in incorrect mRNA transcription termination and mRNA processing in cells from affected individuals, we performed paired-end RNA sequencing on RNA from peripheral blood, collected and extracted with the PaxGene system (PreAnalytiX, BD and QIAGEN), of four individuals with SCAF4 variants (I1, I4, I9, I11) and eight healthy control subjects. In the affected individuals, we then determined deregulated genes, performed principal component analysis (PCA) using DESeq2 package v.1.24.0,23 and determined differentially used isoforms using salmon v.1.0.24,25 More details are available in Supplemental Material and Methods. Of note, this approach detects only known and annotated isoforms and indicates shifts in their respective usage.
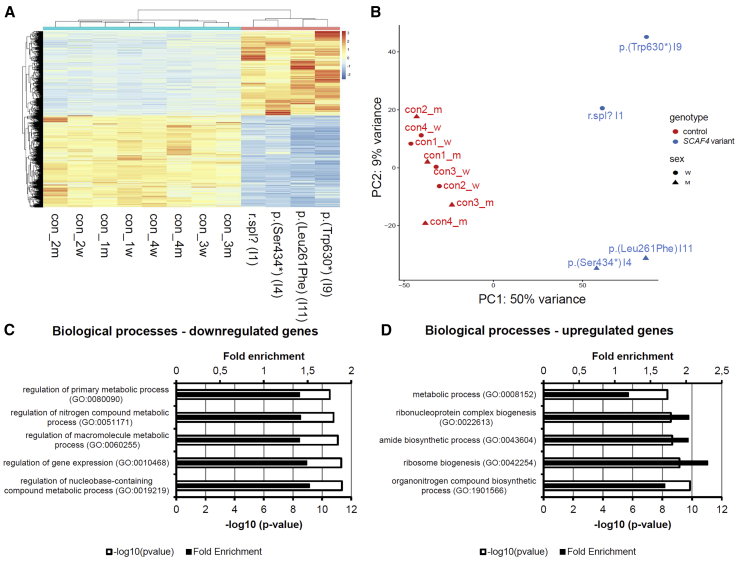
We found that a very large number of genes (n = 9,038) were differentially expressed between the groups of four affected individuals and the eight control subjects, with an adjusted p value < 0.01 (FDR corrected). Of those, 4,328 were upregulated and 4,710 were downregulated (Table S2). Affected individuals and control subjects readily clustered within their group (Figures 2A and 2B), including I11 with the missense variant, thus pointing to a similar loss-of-function mechanism as for the truncating variants and supporting its pathogenic relevance. However, additional data as well as direct comparison to expression profiles of individuals with other NDDs would be required to draw any further conclusions on the possible pathogenicity of this or other missense variants in SCAF4.
Figure 2.
Transcriptome Analysis in Four Individuals with Variants in SCAF4
(A) Heatmap displaying 9,038 differentially expressed genes between four affected individuals and eight healthy control subjects and depicting the expression similarity between individuals with adjusted p value < 0.01. A total of 4,710 genes were downregulated and 4,328 genes were upregulated in the affected individuals. Heatmap was created using the pheatmap package v.1.0.12 (see Web Resources) applying the standard settings (complete linkage method for hierarchical clustering for both columns and rows). Expression values were normalized, and genes were sorted by adjusted p value. Blue, downregulated; red, upregulated.
(B) Principal component analysis (PCA) of affected individuals and control subjects separated by genotype and sex. Affected individuals and control subjects readily clustered within their group. PCA plot was created using the DESeq2 package version 1.24.0.23
(C and D) Gene Ontology (GO) term analysis of (C) downregulated and (D) upregulated genes depicting the top five GO terms for biological processes.
Enrichment of Gene Ontology (GO) terms as well as Reactome Pathways26 among down- and upregulated genes were analyzed using the PANTHER v.14.027 enrichment tool from Gene Ontology28,29 with the following settings: test type: “Fisher’s Exact;” correction: “Bonferroni correction for multiple testing.” A list of all expressed genes within control subjects and the affected individuals with a base mean > 2 was used as background. In this analysis, we found that downregulated genes were enriched for GO terms that included regulation of gene expression and regulation of nucleobase-containing compound metabolic processes (Figure 2C). Furthermore, they were enriched for Reactome pathways that included translation, gene expression, and RNA Polymerase II Transcription (Table S2). Upregulated genes were enriched for GO terms like ribosome biogenesis and organonitrogen compound biosynthetic processes (Figure 2D), as well as for Reactome pathways like translation, metabolism of RNA, and rRNA processing (Table S2).
In addition to broad transcriptional deregulation, we also detected differential splicing of 2,942 genes (adjusted p value < 0.05) between affected individuals and control subjects, supporting the role of SCAF4 in mRNA processing. We selected two alternatively spliced genes and confirmed a shift in the usage of alternative transcripts using RT-PCR with primers flanking differentially spliced regions (Figure S4).
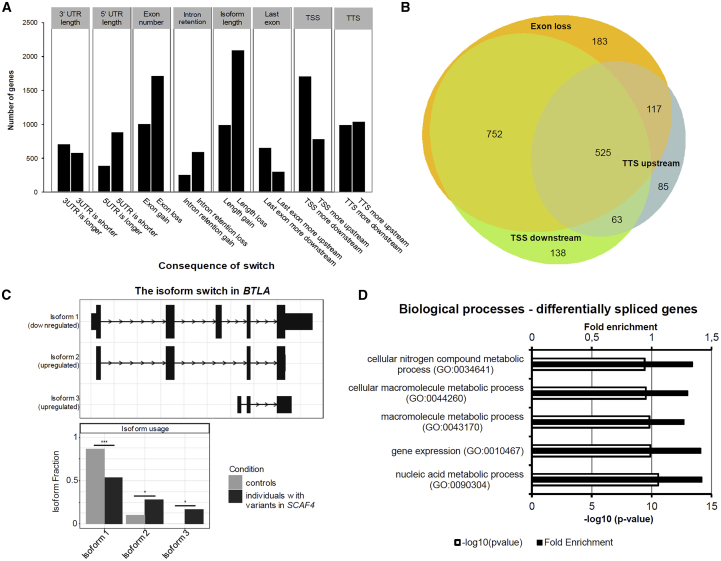
The most prominent splicing effect observed was the increased fraction of short transcripts in more than 70% of all events (2,095 out of 2,942 total) (Figure 3A). This was mainly due to usage of alternative down-stream transcription start sites (TSSs) (70%) and/or alternative up-stream transcription termination sites (TTSs) (37%) and/or exon loss (75%) (Figure 3B). Interestingly, for most of the transcripts, truncation was caused by a combination of two or more of these alterations with all three modifications occurring simultaneously in 17.8% of all altered and in 25.1% of shortened transcripts. As an example, the isoform switch of BTLA (MIM: 607925) is shown. Here, two shorter isoforms due to usage of downstream TSS, upstream TTS, and exon loss have an increased expression level (Figure 3C).
Figure 3.
Impact of SCAF4 Variants on mRNA Processing and Splicing
(A) mRNA isoform expression changes of four individuals with variants in SCAF4 detected by IsoformSwitchAnalyzeR30, 31, 32 analysis of RNA-sequencing data. Most frequent changes include length loss, exon loss, and usage of an alternative transcription start site (TSS) more downstream.
(B) Venn diagram depicting the most prominent causes for a length loss of isoforms. Truncation results from a combination of exon loss, alternative down-stream transcription start sites (TSS downstream), and alternative up-stream transcription termination sites (TTS upstream). Venn diagram was created using the eulerr package version 6.0.0 (see Web Resources).
(C) Example of an alternative spliced gene (BTLA). All expressed transcripts are shown in the upper panel. The isoform usage is shown in the lower panel. In individuals with variants in SCAF4, the two shorter isoforms with downstream TSSs, upstream TTSs, and exon loss showed increased expression while the longest isoform 1 is used less.
(D) Gene Ontology (GO) term analysis of differentially spliced genes depicting the top five GO terms for biological processes.
Abbreviations: TSS, transcription start site; TTS, transcription termination site. Asterisks indicate statistical significance (∗p < 0.05, ∗∗∗p < 0.001).
Similarly to the observations from differentially expressed genes, differentially spliced genes were enriched for GO terms like nucleic acid metabolic processes and gene expression as well as for Reactome pathways like chromatin modifying enzymes, chromatin expression, and gene expression (transcription) (Figure 3D, Table S2).
We compared the differentially spliced genes in individuals with variants in SCAF4 with the 565 differentially spliced genes after SCAF4 knockout in HEK293 cells14 and found an overlap of 25.7% (145 out of 565). A total of 66 (45.5%) of these genes used an alternative last exon (ALE). As various cell types usually have unique transcriptome profiles, usage of HEK293 versus human blood cells might contribute to discrepancies between differentially spliced genes. Also, complete SCAF4 knockout in HEK293 cells versus a heterozygous variant in affected individuals could play a role, since the remaining allele might still cover some of the SCAF4 function. Nevertheless, the overlap between differentially expressed genes in HEK293 and blood cells is larger than expected by chance (95 out of 565) as determined in a hypergeometric test (p value < 0.001), and the general consequence of altered mRNA termination is similar in both systems. Furthermore, we frequently found usage of alternative transcription start sites highlighting the potential that SCAF4 is involved not only in correct mRNA termination but also in TSS recognition. Such a function could directly contribute to the large number of changes in gene expression.
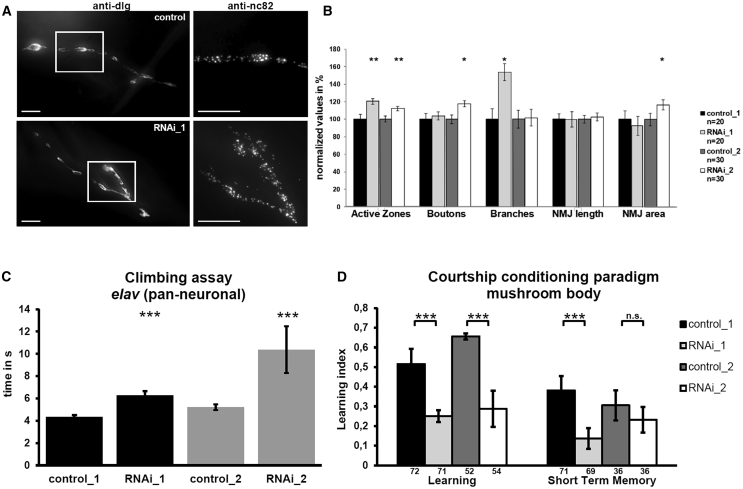
To find more evidence that SCAF4 truncation or haploinsufficiency causes NDDs, we utilized Drosophila melanogaster as a model organism and investigated the effect of knockdown of CG4266, the ortholog of both human SCAF4 and SCAF8 (MIM: 616024), with regard to nervous system development and function. We obtained two CG4266 UAS-RNAi lines (RNAi_1, BL#55354 and RNAi_2, VDRC 26472) from the Transgenic RNAi Project33 from the Bloomington Stock Center (BL) and from the Vienna Drosophila Resource Center (VDRC), respectively. Ubiquitous or tissue-specific knockdown was induced using the UAS/Gal4 system34 with different promoter lines (Table S3). By real-time quantitative PCR we confirmed reduced CG4266 expression to approximately 60%–70% upon ubiquitous knockdown (Figure S5). We first investigated development and morphology of larval neuromuscular junctions (NMJ), an established model for vertebrate glutamatergic synapses,35,36 upon pan-neuronal knock-down. Specifically, we dissected stage 3 larvae and stained NMJs in larval muscle 4 as described previously,37 to assess parameters such as NMJ length and area, number of synaptic boutons, and the number of active zones where neurotransmitters are released (Supplemental Material and Methods, Figure 4A). Pan-neuronal knockdown of CG4266 with either of the two RNAi lines resulted in an increased number of active zones, while the number of branches or the number of boutons was elevated only in one of the two lines, respectively (Figures 4A and 4B). These findings suggest a role of CG4266 in synapse development and morphology.
Figure 4.
Impact of CG4266 Knockdown on Drosophila Nervous System
(A) Representative pictures of neuromuscular junctions (NMJs) from L3 control (upper panel) and pan-neuronal CG4266 knockdown (lower panel) larvae. The white-framed box indicates the cutout on the right side. NMJ area and length as well as the number of synaptic boutons, islands, and branches were determined. In the RNAi_1 larva, an increased number of AZs can be observed in comparison to control larva. Scale bars represent 10 μm.
(B) Quantification revealed a significant increase in the number of AZs upon CG4266 knockdown with two RNAi fly lines using the elav-x;dicer II driver (BL#25750).
(C) Flies with knockdown of CG4266 in all neurons (elav-Gal4/Cyo, BL#8765) showed significant locomotor impairment in the climbing assay, as measured by the amount of time that 70% of flies in a vial needed to crawl up 8.8 cm after being tapped down. Data represent the mean from a minimum of 170 flies tested per genotype.
(D) In the courtship conditioning paradigm, both CG4266 RNAi lines showed significant impairment of learning upon knockdown with a mushroom-body-specific driver line (UAS-Dcr-2;247-Gal4). The short-term memory was reduced significantly in RNAi line 1 with RNAi line 2 showing the same tendency. Graphs display number of animals below the columns per genotype. Differences between learning indices of control and mutant flies were statistically compared by a randomization test with 10,000 bootstrap replicates with a custom R script.37
Error bars represent the SEM. Asterisks indicate statistical significance (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
We then tested activity, seizure susceptibility, and gross neurological and locomotor function in adult flies upon pan-neuronal knockdown. Evaluation of locomotor activity with a monitor system (Trikinetics) and a 12 h day-night rhythm for 4 days did not reveal any alterations in spontaneous activity or sleeping behavior (Figure S6). After inducing a mechanical shock by vortexing the flies for 10 s in the bang sensitivity assay,38,39 we did not observe a seizure phenotype (Figure S7). However, when tapping flies down in a vial and assessing their innate behavior to walk up immediately in the climbing assay,39,40 locomotor ability was severely impaired for both RNAi lines (Figure 4C).
Furthermore, we tested whether knockdown of CG4266 also impairs complex learning and memory behavior by utilizing the courtship conditioning paradigm. This assay is based on the reduction of male courtship behavior in response to sexual rejection of a non-receptive pre-mated female,41 as described in more detail previously42 and in Supplemental Material and Methods. By adjusting the time between training and testing periods, both learning (test immediately after training) and short-term memory (test 1 h after training) can be assessed. Knockdown of CG4266 in the mushroom body, the fly center for learning and memory,43 resulted in a significantly impaired learning performance for both RNAi lines and a significantly impaired short-term memory upon knockdown with RNAi line 1. RNAi line 2 showed the same tendency but without reaching significance (Figure 4D). Also pan-neuronal knockdown of CG4266 resulted in significant but less consistent learning and memory impairment for the two different RNAi lines (Figure S8).
Taken together, these results suggest that CG4266 plays a crucial role in neurological functioning and complex learning and memory processes. This is consistent with the neurodevelopmental phenotypes and cognitive impairments seen in humans with SCAF4 loss-of-function variants.
In conclusion, our results suggest that heterozygous truncating/likely gene-disrupting variants in SCAF4 cause a variable neurodevelopmental disorder and that impaired SCAF4 function results in altered mRNA processing and splicing.
Data and Code Availability
The raw datasets supporting the current study have not been deposited in a public repository because of data safety and privacy regulations.
Consortia
UCLA Clinical Genomics Center: Stanley F. Nelson, Wayne W. Grody, Hane Lee, Joshua L. Deignan, Sung-Hae Kang, Valerie A. Arboleda, T. Niroshi Senaratne, Naghmeh Dorrani, Marina S. Dutra-Clarke, Jessica Kianmahd, Franceska L. Hinkamp, Ahna M. Neustadt, Julian A. Martinez-Agosto, Brent L. Fogel, and Fabiola Quintero-Rivera.
Acknowledgment
We thank all individuals and families for participating in this study. We especially thank Laila Distel, Christine Suchy, and Petra Rothe for excellent technical assistance. We thank Christian T. Thiel for the in-house NGS tool and André Reis from the NGS facility at the institute of Human Genetics, Erlangen. Fly stocks were obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) and the Vienna Drosophila Resource Center. Antibodies from the Developmental Studies Hybridoma Bank (created by the NICHD of the NIH) were used in this study. C.Z. is supported by grants from the German Research Foundation (DFG) (ZW184/3-1, ZW184/6-1, and 270949263/GRK2162) and by the IZKF Erlangen (E31). G.M.M. is supported by the National Institute of Neurological Disorders and Stroke (NINDS) under award number K08NS092898, Jordan’s Guardian Angels, and the Brotman Baty Institute.
Declaration of Interests
The Department of Molecular and Human Genetics at Baylor College of Medicine receives revenue from clinical genetic testing completed at Baylor Genetics Laboratories. J.J. and K.Mc. are employees of GeneDx, Inc. The remaining authors declare no competing interests.
Published: July 29, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.06.019.
Contributor Information
Christiane Zweier, Email: christiane.zweier@uk-erlangen.de.
UCLA Clinical Genomics Center:
Stanley F. Nelson, Wayne W. Grody, Hane Lee, Joshua L. Deignan, Sung-Hae Kang, Valerie A. Arboleda, T. Niroshi Senaratne, Naghmeh Dorrani, Marina S. Dutra-Clarke, Jessica Kianmahd, Franceska L. Hinkamp, Ahna M. Neustadt, Julian A. Martinez-Agosto, Brent L. Fogel, and Fabiola Quintero-Rivera
Web Resources
eulerr, https://cran.r-project.org/web/packages/eulerr/index.html
Gene Ontology, http://geneontology.org/
gnomAD Browser, https://gnomad.broadinstitute.org/
MutationTaster, http://www.mutationtaster.org/
OMIM, https://www.omim.org/
pheatmaps, https://cran.r-project.org/package=pheatmap
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
UCSC Genome Browser, https://genome.ucsc.edu
Vienna Drosophila Research Center, https://stockcenter.vdrc.at/control/main
Supplemental Data
References
- 1.Cramer P. RNA polymerase II structure: from core to functional complexes. Curr. Opin. Genet. Dev. 2004;14:218–226. doi: 10.1016/j.gde.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Orphanides G., Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- 3.Kornberg R.D. Eukaryotic transcriptional control. Trends Cell Biol. 1999;9:M46–M49. [PubMed] [Google Scholar]
- 4.Lee T.I., Young R.A. Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 2000;34:77–137. doi: 10.1146/annurev.genet.34.1.77. [DOI] [PubMed] [Google Scholar]
- 5.Haijes H.A., Koster M.J.E., Rehmann H., Li D., Hakonarson H., Cappuccio G., Hancarova M., Lehalle D., Reardon W., Schaefer G.B. De Novo Heterozygous POLR2A Variants Cause a Neurodevelopmental Syndrome with Profound Infantile-Onset Hypotonia. Am. J. Hum. Genet. 2019;105:283–301. doi: 10.1016/j.ajhg.2019.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cormack B.P., Struhl K. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell. 1992;69:685–696. doi: 10.1016/0092-8674(92)90232-2. [DOI] [PubMed] [Google Scholar]
- 7.Mizzen C.A., Yang X.-J., Kokubo T., Brownell J.E., Bannister A.J., Owen-Hughes T., Workman J., Wang L., Berger S.L., Kouzarides T. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 8.O’Rawe J.A., Wu Y., Dörfel M.J., Rope A.F., Au P.Y., Parboosingh J.S., Moon S., Kousi M., Kosma K., Smith C.S. TAF1 Variants Are Associated with Dysmorphic Features, Intellectual Disability, and Neurological Manifestations. Am. J. Hum. Genet. 2015;97:922–932. doi: 10.1016/j.ajhg.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tawamie H., Martianov I., Wohlfahrt N., Buchert R., Mengus G., Uebe S., Janiri L., Hirsch F.W., Schumacher J., Ferrazzi F. Hypomorphic Pathogenic Variants in TAF13 Are Associated with Autosomal-Recessive Intellectual Disability and Microcephaly. Am. J. Hum. Genet. 2017;100:555–561. doi: 10.1016/j.ajhg.2017.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oegema R., Baillat D., Schot R., van Unen L.M., Brooks A., Kia S.K., Hoogeboom A.J.M., Xia Z., Li W., Cesaroni M. Human mutations in integrator complex subunits link transcriptome integrity to brain development. PLoS Genet. 2017;13:e1006809. doi: 10.1371/journal.pgen.1006809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hauer N.N., Popp B., Schoeller E., Schuhmann S., Heath K.E., Hisado-Oliva A., Klinger P., Kraus C., Trautmann U., Zenker M. Clinical relevance of systematic phenotyping and exome sequencing in patients with short stature. Genet. Med. 2018;20:630–638. doi: 10.1038/gim.2017.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cunningham F., Achuthan P., Akanni W., Allen J., Amode M.R., Armean I.M., Bennett R., Bhai J., Billis K., Boddu S. Ensembl 2019. Nucleic Acids Res. 2019;47(D1):D745–D751. doi: 10.1093/nar/gky1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuryev A., Patturajan M., Litingtung Y., Joshi R.V., Gentile C., Gebara M., Corden J.L. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc. Natl. Acad. Sci. USA. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gregersen L.H., Mitter R., Ugalde A.P., Nojima T., Proudfoot N.J., Agami R., Stewart A., Svejstrup J.Q. SCAF4 and SCAF8, mRNA Anti-Terminator Proteins. Cell. 2019;177:1797–1813.e18. doi: 10.1016/j.cell.2019.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv. 2019 doi: 10.1101/531210. [DOI] [Google Scholar]
- 16.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Firth H.V., Richards S.M., Bevan A.P., Clayton S., Corpas M., Rajan D., Van Vooren S., Moreau Y., Pettett R.M., Carter N.P. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am. J. Hum. Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deciphering Developmental Disorders Study Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwarz J.M., Rödelsperger C., Schuelke M., Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 20.Adzhubei I., Jordan D.M., Sunyaev S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protocols Human Genet. 2013;Chapter 7 doi: 10.1002/0471142905.hg0720s76. Unit7.20-Unit27.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sim N.-L., Kumar P., Hu J., Henikoff S., Schneider G., Ng P.C. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40:W452-7. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jagadeesh K.A., Wenger A.M., Berger M.J., Guturu H., Stenson P.D., Cooper D.N., Bernstein J.A., Bejerano G. M-CAP eliminates a majority of variants of uncertain significance in clinical exomes at high sensitivity. Nat. Genet. 2016;48:1581–1586. doi: 10.1038/ng.3703. [DOI] [PubMed] [Google Scholar]
- 23.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal. 2011;17:10–12. [Google Scholar]
- 25.Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fabregat A., Sidiropoulos K., Viteri G., Forner O., Marin-Garcia P., Arnau V., D’Eustachio P., Stein L., Hermjakob H. Reactome pathway analysis: a high-performance in-memory approach. BMC Bioinformatics. 2017;18 doi: 10.1186/s12859-017-1559-2. 142–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mi H., Muruganujan A., Ebert D., Huang X., Thomas P.D. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019;47(D1):D419–D426. doi: 10.1093/nar/gky1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., The Gene Ontology Consortium Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The Gene Ontology Consortium Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 2017;45(D1):D331–D338. doi: 10.1093/nar/gkw1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soneson C., Love M., Robinson M. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 2015;4:1521. doi: 10.12688/f1000research.7563.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43 doi: 10.1093/nar/gkv007. e47–e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vitting-Seerup K., Sandelin A. The Landscape of Isoform Switches in Human Cancers. Mol. Cancer Res. 2017;15:1206–1220. doi: 10.1158/1541-7786.MCR-16-0459. [DOI] [PubMed] [Google Scholar]
- 33.Perkins L.A., Holderbaum L., Tao R., Hu Y., Sopko R., McCall K., Yang-Zhou D., Flockhart I., Binari R., Shim H.S. The Transgenic RNAi Project at Harvard Medical School: Resources and Validation. Genetics. 2015;201:843–852. doi: 10.1534/genetics.115.180208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brand A.H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 35.Jan L.Y., Jan Y.N. Properties of the larval neuromuscular junction in Drosophila melanogaster. J. Physiol. 1976;262:189–214. doi: 10.1113/jphysiol.1976.sp011592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ehmann N., Owald D., Kittel R.J. Drosophila active zones: From molecules to behaviour. Neurosci. Res. 2018;127:14–24. doi: 10.1016/j.neures.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 37.Gregor A., Kramer J.M., van der Voet M., Schanze I., Uebe S., Donders R., Reis A., Schenck A., Zweier C. Altered GPM6A/M6 dosage impairs cognition and causes phenotypes responsive to cholesterol in human and Drosophila. Hum. Mutat. 2014;35:1495–1505. doi: 10.1002/humu.22697. [DOI] [PubMed] [Google Scholar]
- 38.Kuebler D., Tanouye M.A. Modifications of seizure susceptibility in Drosophila. J. Neurophysiol. 2000;83:998–1009. doi: 10.1152/jn.2000.83.2.998. [DOI] [PubMed] [Google Scholar]
- 39.Straub J., Konrad E.D.H., Grüner J., Toutain A., Bok L.A., Cho M.T., Crawford H.P., Dubbs H., Douglas G., Jobling R., Deciphering Developmental Disorders Study Missense Variants in RHOBTB2 Cause a Developmental and Epileptic Encephalopathy in Humans, and Altered Levels Cause Neurological Defects in Drosophila. Am. J. Hum. Genet. 2018;102:44–57. doi: 10.1016/j.ajhg.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palladino M.J., Hadley T.J., Ganetzky B. Temperature-sensitive paralytic mutants are enriched for those causing neurodegeneration in Drosophila. Genetics. 2002;161:1197–1208. doi: 10.1093/genetics/161.3.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siegel R.W., Hall J.C. Conditioned responses in courtship behavior of normal and mutant Drosophila. Proc. Natl. Acad. Sci. USA. 1979;76:3430–3434. doi: 10.1073/pnas.76.7.3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Konrad E.D.H., Nardini N., Caliebe A., Nagel I., Young D., Horvath G., Santoro S.L., Shuss C., Ziegler A., Bonneau D., DDD Study CTCF variants in 39 individuals with a variable neurodevelopmental disorder broaden the mutational and clinical spectrum. Genet. Med. 2019;21:2723–2733. doi: 10.1038/s41436-019-0585-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zars T. Behavioral functions of the insect mushroom bodies. Curr. Opin. Neurobiol. 2000;10:790–795. doi: 10.1016/s0959-4388(00)00147-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw datasets supporting the current study have not been deposited in a public repository because of data safety and privacy regulations.