Abstract
The tumor suppressor gene deleted in liver cancer-1 (DLC1), which encodes a protein with strong RhoGAP (GTPase activating protein) activity and weak Cdc42GAP activity, is inactivated in various human malignancies. Following Dlc1 inactivation, mouse embryo fibroblasts (MEFs) with a conditional Dlc1 knockout allele reproducibly underwent neoplastic transformation. In addition to inactivation of Dlc1 and increased activity of Rho and Cdc42, transformation depended on the subsequent decreased expression of the Cdk4/6 inhibitors p15Ink4b and p16Ink4a together with increased expression and activation of Cdk4/6. The level of expression of these cell cycle regulatory genes was relevant to human tumors with low DLC1 expression. Analysis of publicly available annotated datasets of lung and colon cancer with gene expression microarray profiles indicated that, in pairwise comparisons, low DLC1 expression occurred frequently together (p<0.01) with downregulation of p15Ink4b or p16Ink4a or upregulation of CDK4 or CDK6. In addition, an unfavorable prognosis (p<0.05) was associated with low DLC1 and low p15Ink4b in lung cancer and colon cancer, low DLC1 and low p16Ink4a in lung cancer, low DLC1 and high CDK4 in lung cancer, and low DLC1 and high CDK6 in colon cancer. Thus, several genes and biochemical activities collaborate with the inactivation of DLC1 to give rise to cell transformation in MEFs, and the identified genes are relevant to human tumors with low DLC1 expression.
INTRODUCTION
Carcinogenesis is a multistep process that includes the activation of genes that promote cancer together with inactivation of those that restrict its development (1, 2). DLC1 is a tumor suppressor gene that is inactivated, via genetic and epigenetic mechanisms, in a variety of human malignancies, including cancers of the lung, breast, colorectum, and prostate (2–4). DLC1 is the prototypic member of a multigene family that includes two closely related genes, DLC2 and DLC3, which have been studied less extensively, but are reported to be frequently down-regulated in human cancer and have growth suppressive effects.
The principal protein encoded by human DLC1 is a RhoGAP (GTPase activating protein) whose mouse Dlc1 counterpart is highly homologous (Fig. 1A)(5). In addition to the RhoGAP domain, located in the C-terminal half of the protein, the human DLC1 and mouse Dlc1 proteins have a SAM (sterile alpha motif) domain at their N-terminus and a START (StAR-related lipid transfer) domain at the C-terminus. In vitro studies of Rho family GTPases indicate that DLC1 can efficiently catalyze the inactivation of RhoGTP to RhoGDP, is less active in converting Cdc42GTP to Cdc42GDP, and lacks detectable activity against RacGTP (6). DLC1 has been shown to negatively regulate RhoGTP in cells, but an in vivo role in the regulation of Cdc42 has not been determined.
Figure 1. Disruption of Dlc1 gene expression in Mouse embryo fibroblasts (MEFs) derived from conditional Dlc1 knock-out mouse.
(A) Domain structure of mouse Dlc1 protein. (B) Genomic structure of conditional Dlc1 gene (Dlc1fl/fl), which expresses wild type DLC1. Two LoxP sites flank exon 4. Infection with Adenovirus encoding Cre-recombinase (Adeno-Cre) induces deletion of sequences between the LoxP sites, disrupting Dlc1 (Dlc1Δ4/ Δ 4), while the control virus vector (Adeno-Vt) does not. (C) Verification of Dlc1 gene disruption by Cre-recombinase (top). Characterization of Dlc1 transcript in MEF-Vt and MEF-Cre cells by RT-PCR exon 4 specific primers. The smaller sized PCR product from MEF-Cre cells reflects the deletion of exon 4 in mouse genome. validation of Dlc1 knock-out by protein expression (bottom). MEF-Vt or MEF-Cre cells, derived from two independent mouse litters, analyzed by immunoprecipitation followed by immunoblotting with anti-DLC1 (428) antibody. Endogenous DLC1 from human melanoma cell line 553B serves as a positive control. (D) Elevated RhoGTP level in early passage MEF-Cre cells. MEF-Vt and MEF-Cre cells derived from mouse MEF(A) were analyzed by Rhotekin or Pak-1 pull-down assays for RhoGTP and Cdc42GTP, respective. In vivo RhoGTP and Cdc42GTP levels, as well as the total RhoA or Cdc42 proteins, are shown. (E) In early passage cells, v-Ras oncogene expression induces greater transformation of MEF-Cre than MEF-Vt. Cells were analyzed by growth in soft agar for 4 weeks. Quantization of colony number (top) and representative images (bottom) are shown. (F) Ras protein levels in MEFs with and without v-Ras. Validation of MEF-Vt and MEF-Cre cells with v-Ras infection. Cells were analyzed by Raf-RBD pull-down assay to measure RasGTP. In vivo RasGTP and total Ras levels are shown.
Most experimental studies of DLC1 have been conducted with cancer-derived cell lines in which, because their endogenous DLC1 expression has been down-regulated, the analyses have focused on phenotypic changes resulting from the expression of exogenous wild type and mutant DLC1. These analyses have determined that constitutive expression of exogenous DLC1 can revert biological parameters associated with oncogenic transformation, with the RhoGAP activity of DLC1 being necessary, but not sufficient, for these biological phenotypes (7–12). Conversely, in vivo infusion of hepatoblasts, from p53−/− mice that had been transduced with a Myc oncogene and an inhibitory RNA against Dlc1 led to hepatocellular carcinoma (13). Although this latter model was informative, human cancer is thought to result from the combination of several genetic and epigenetic changes that occur over time, rather than from the simultaneous inactivation of a tumor suppressor gene and activation of an oncogene.
It has not been determined experimentally whether inactivation of Dlc1 by itself can lead to neoplasia or what genes might collaborate with loss of Dlc1 to produce oncogenic transformation. To this end, we here describe the development of mouse embryo fibroblasts (MEFs) whose two endogenous Dlc1 alleles can be conditionally disrupted by the Cre recombinase because a Dlc1 exon has been engineered to have flanking loxP sites. Using this system, we have evaluated the short-term and long-term phenotypic consequences of disrupting both Dlc1 alleles in the cultured MEFs. The results indicate that while disruption of Dlc1 does not immediately induce oncogenic transformation, longer-term culturing of MEFs with disrupted Dlc1 reproducibly leads to neoplastic transformation. This process is associated with several transformation-dependent changes, including loss of the Cdk4/6 inhibitors p15Ink4b (p15) and p16Ink4a (p16) (14) together with increased RhoGTP and Cdc42GTP (15). The genetic findings in the MEFs are relevant to human malignancies, as DLC1 is frequently downregulated together with the CDK inhibitors or with upregulated CDK4 or CDK6, and their level of expression may be associated with prognosis.
Materials and Methods
Generation of a conditional knockout allele of the mouse Dlc1 gene
The strategy for generating a conditional knockout allele of the mouse Dlc1 gene involved using homologous recombination in embryonic stem (ES) cells to insert loxP sites on either side of exon 4. The methods used for gene targeting and for genotyping ES cells and mice will be described in detail elsewhere (Durkin et al, in preparation). Briefly, a targeting vector was constructed in which a single LoxP site was inserted 0.97 kb upstream of exon 4, and a LoxP- and frt-flanked neomycin-resistance cassette (neo) was inserted 0.61 kb downstream of exon 4. The vector was introduced into ES cells, and correctly targeted clones were identified and injected into blastocysts to generate chimeric mice that transmitted the conditional knockout allele (Dlc1neo) to their progeny. The neo cassette was removed by breeding Dlc1neo/wt mice with transgenic mice that constitutively express the Flpe recombinase. The “floxed” allele (Dlc1fl) present in the offspring retains a LoxP site in intron 4 after excision of frt-flanked segment (Fig. 1B). The Dlc1fl/wt mice were mated to obtain Dlc1fl/fl homozygotes, which were viable and apparently normal as expected. The genetic background of the mice used in this study was approximately 93% C57BL/6 and 7% 129/Sv.
Isolation of Mouse Embryo Fibroblasts, Adenovirus infection, and RT-PCR analysis
Embryos from timed matings of Dlc1fl/fl males and females were dissected out on day 12–13 of gestation. After decapitation and removal of the liver and other viscera, the embryos were minced, incubated in trypsin/EDTA solution at 37oC, and triturated several times. The dissociated cells were seeded in tissue culture flasks and cultured in DMEM with 10% FBS.
The high titer adenoviruses (6.8–9.5 X107pfu/μl), with or without Cre recombinase (designated Adeno-Cre and Adeno-Vector [Adeno-Vt], respectively), were produced and purified by the Adneovirus production Group, Gene Expression Laboratory, SAIC, Frederick. The adenoviruses, at multiplicity of infection of 5 in 2 ml of serum-free DMEM containing 4 μg/ml of Polybrene, were added to cultures of the Dlc1fl/fl MEFs (3.5 X 106 cells/100 mm dish) for 30 min. at 37oC with rocking every 5 min. The dishes were then supplemented with complete medium (DMEM with 10% FBS) and incubated for 48 hours.
After 2 passages, the adenovirus-infected MEFs were analyzed by RT-PCR to verify deletion of exon 4 in the Dlc1 mRNA. Briefly, total RNA was isolated from MEF-Cre and MEF-Vt cells using TRIzol reagent as recommended by the manufacturer (Invitrogen, Carlsbad, CA). First strand cDNA was synthesized from 1 μg of the total, using a SuperScript ®III kit (Invitrogen). PCR was performed with a forward primer in exon 3 (5’- gct ggt caa gag aga aca tg) and a reverse primer in exon 5 (5’- gtc tgt cag cat gct ttc gtg), which yield products of 307 bp and 243 bp from transcripts with and without exon 4, respectively.
Retroviral v-RasH infection of MEF-Vt and MEF-Cre cells, cell proliferation assay, soft agar colony growth assay, and isolation of single colonies from agar
The replication-defective v-RasH (1423) expressing retroviral genome pseodotyped by the Moloney murine leukemia virus (Mo-MuLV) and control virus Mo-MuLV have been described previously (16, 17). The infected MEFs were grown for several days so that the vast majority of the cells in the 1423 v-RasH infected cells had become morphologically transformed.
For the monolayer proliferation assay, MEF-Vt and MEF-Cre cells (1 X 105/well) were plated in a 12-well dish, and grown for three days in DMEM containing high (10%) or low (1%) FBS. Cell numbers were counted daily in triplicate, using a Cellmetor Auto T4 counter (Nexcelom Bioscience). The cells were also analyzed for growth in soft agar for 3–4 weeks, and colonies were photographed microscopically and quantified with a colony counter as described (10). In some experiments, the ROCK inhibitor Y27362 (5 nM), JNK inhibitor VIII (10 nM), or CDK4/6 inhibitor IV (10 nM) (all from EMD Millipore) was added to the agar plates every 5–6 days, to test the contribution of the respective kinase to cell growth in agar.
To isolate clonally-derived cells with disrupted Dlc1, several single colonies, from spontaneously transformed longer-term passaged MEF-Cre cells (without v-RasH infection), were isolated from soft agar plates and dispersed for monolayer growth after trypsinization. They were named MEF-Cre AIG (anchorage-independent growth) clones, and their homozygosity for Dlc1fl/fl disruption to Dlc1 Δ4/Δ4 verified by RT-PCR.
Mouse tumorigenesis studies
The mouse studies were approved by the NCI Animal Care and Use Committee and conducted in compliance with the approved protocols. For tumor xenografts, longer-passaged MEF-Vt and MEF-Cre cells were washed with cold PBS, diluted to 108/ml with serum-free medium/matrigel basement membrane matrix (Becton Dickinson Labware) at a ratio of 3:1, and injected subcutaneously into NOD/SCID mice (107 cells per injection). The animals were monitored for tumor growth, and five weeks post-injection tumors were excised and weighed.
Transfection and transwell migration assay
The isolated homozygous Dlc1 Δ4/Δ4 knockout clone, AIG-5, was used in stable transfections to express GFP and GFP-tagged human DLC1 (wt or R718A, a RhoGAP-deficient mutant) (10), as well as PEGFP-C1 (clontech), pcDNA3-p16Ink4a (from Beverly Mock, NCI) and pcDNA3-p15Ink4b (from Linda Wolff, NCI). CTCCells were observed for cell proliferation and morphological changes. The transwell cell migration assay was previously described (18).
MEF-Cre cells were transfected with HA-tagged rat ROKa (ROCK2, from Thomas Leung, Singapore) followed by treatment with or without inhibitor Y27362 in the culture. MEF-Cre cells treated with JNK inhibitor VIII or Cdk 4/6 inhibitor IV (EMD Chemical) were verified by immunoblotting to detect phosphorylation of c-jun (S68), phosho-Cyclin D1 (T286) and phospho-RB (S780) (Cell Signaling Technology), respectively.
Immunofluorescent staining and confocal microscopy
MEF-Vt and MEF-Cre AIG-5 cells expressing GFP or GFP-tagged human DLC1 were seeded on glass coverslips and incubated for 24 h. Cells were fixed with 4% paraformaldehyde, permeabilized with 0.25% Triton X-100 in PBS, and then blocked with 5% goat serum in PBS. The cells were incubated with a 1:100 dilution (in PBS) of DLC1 (428) or Ras (Millipore) primary antibodies at 4°C overnight. After thorough washing in PBS, cells were incubated 1:250 with the appropriate Alexa-conjugated secondary antibodies for 1 hour. To visualize actin or nuclei, cells were incubated with phalloidin (1:50; Invitrogen) for 1 h. After staining, cells were thoroughly washed with PBS and mounted with gel mounting solution (Biomeda Corporation, Foster City, CA). Fluorescent-labeled cells were viewed in a Zeiss 510 UV confocal microscope, using a 63X objective.
Immunoprecipitation, immunoblotting, Raf-RBD, Rhotekin-RBD, and Pak-1-RBD pull-down assays
Cells were lysed with Golden Lysis Buffer (12). Equal amounts of protein from cell extracts were used for immunoprecipitation by anti DLC1 (428) (12) or commercial antibody indicated in the figures. RasGTP, RhoGTP, and Cdc42GTP were assayed, respectively, by Raf-RBD, Rhotekin-RBD, and Pak-1-RBD (Millipore), followed, respectively, by anti-Ras (Millipore), anti-RhoA (Cytoskeleton), and anti-Cdc42 (Santa Cruz) blotting, based on the manufacturers’ instructions. For each blot, horseradish peroxidase-conjugated anti- rabbit or anti-mouse immunoglobulin G (GE Heathcare) was used for the second reaction at 1:10,000 dilution. Immunocomplexes were visualized by enhanced chemiluminescence (ECL), using an ECL kit (GE Healthcare)
siRNA Transfection
Control siRNA and validated siRNAs for mouse or human p15 and p16 and mouse Cdk4 and Cdk6 were all from Qiagen. Target sequences are listed in Supplementary Table 1. MEF cells or human NSCLC A549 cells (from Curt Harris, NCI) were transfected with siRNAs for 18–24 hours using lipofectamine 2000, followed by a change of media. After 3–4 days, cell extracts were prepared for various protein assays. For biologic assays, MEFs were seeded for colony growth in soft agar the day after siRNA transfection, or for cell migration 3–4 days after transfection.
Bioinformatic analysis
The Gene expression microarray analyses in this study use data from Arrayexpress of the European Institute of Bioinformatics (EBI), which is publicly available at http://www.ebi.ac.uk/arrayexpress. The analyses include 3 independent experiments with NSCLC and 3 independent experiments with colon cancer for Affymetrix U133 plus 2 chips. The CEL files with raw data from each experiment were directly downloaded from the EBI website and normalized with CEL file quality control evaluation using 3’ Expression Arrays Robust Multi-array Analysis (RMA) from the Affymetrix software Expression Console (http://affymetix.com). The normalized expression values are the probe set intensity on a log-2 scale. Since data from E-GEOD-3629 has no raw data available, processed data from the original author was used for the analysis. The specific probe sets for DLC1, CDKN2A (p16), CDKN2B (p15), CDK4, and CDK6 are 224822_at, 209644_x_at, 236313_at, 202246_s_at, and 243000_at respectively. Probe 209644_x_at is used to detect p16INK4A. Since this gene generates several transcript variants which differ in their first exon, expression levels measured using 209644_x_at cannot exclude the detection of p14ARF.
Statistical analysis
The gene regulation was analyzed to compare different expression values between cancers and controls. For individual genes from each experiment, the median expression level (50% quartile) is considered as a cut-off. Values higher than the cut-off were categorized as “high”; other values were categorized as “low”. The Kaplan-Meier survival and Chi Square analyses were performed using statistical computing and graphics software R (www.r-project.org). P < 0.05 was considered statistically significant. For survival analysis, the values higher or lower than the median in each gene group were placed in “high”, “low”, or two different combinations for the analysis. All survival times were adjusted to months.
RESULTS
Conditional Dlc1 disruption induces increased RhoGTP and increased susceptibility to ras transformation
Constitutive disruption of mouse Dlc1 leads to embryonic lethality (5, 19). To develop an endogenous Dlc1 allele that can be conditionally disrupted, we used gene targeting to introduce LoxP sites on both sides of exon 4 of the Dlc1 gene (Fig. 1B). Mouse embryo fibroblasts (MEFs) were then derived from 12–13 day old embryos that had been bred to homozygosity for this allele (Dlc1fl/fl). Exposure of the MEFs to Cre recombinase should induce deletion of the 1.65 kb LoxP-flanked genomic DNA segment. This deletion results in loss of the 64-nt exon 4 sequence in the Dlc1 transcript (Fig. 1B), which produces a reading frame shift after codon 64, leading to premature translation termination and loss of full-length Dlc1 protein expression.
MEFs homozygous for the conditional allele were infected with a Cre-encoding adenovirus (Adeno-Cre), which induced disruption of the Dlc1fl/fl alleles and produced inactive Dlc1Δ4/Δ4 alleles, as determined by RT-PCR and loss of detectable full-length Dlc1 protein for two independently derived MEF lines, MEF-A, which was selected for further analysis, and MEF-B (Fig. 1C). These changes were not induced by the control Adenovirus vector (Adeno-Vt).
Consistent with the strong RhoGAP activity of DLC1, disruption of Dlc1 was associated with increased RhoGTP, as determined by a Rhotekin pull-down assay carried out following Adeno-Cre infection (Fig. 1D, upper panels). However, there was little difference between the level of Cdc42GTP in MEF-Cre and control MEF-Vt (Fig. 1D, lower panels), perhaps because the Cdc42GAP activity of DLC1 is less efficient than its RhoGAP activity. MEFs with constitutively disrupted p190B RhoGAP are also reported to have increased RhoGTP (20), while there does not appear to be a change for MEFs with constitutively disrupted p190A RhoGAP (21). Despite their increased RhoGTP, the MEF-Cre cells, studied within two passages of Adeno-Cre infection, did not grow in soft agar, indicating that loss of Dlc1 does not lead directly to transformation (Fig. 1E).
To examine whether the early passaged MEF-Cre cells might be more susceptible to transformation by an oncogene, they were transduced with mutant Ras (v-RasH [v-Ras]), which induced a greater degree of transformation in MEF-Cre than in MEF-Vt transductants, as determined by the anchorage-independent growth of large colonies in agar (Fig. 1E). The MEF-Vt and MEF-Cre cells displayed a similar increase in RasGTP (Fig. 1F), indicating that the stronger transformation of the MEF-Cre cells was not attributable to differences in RasGTP levels. After submission of this manuscript, a study reported that the incidence of metastatic thymic tumors was increased in mice heterozygous for a gene-trapped Dlc1 allele after Cre-mediated induction of oncogenic K-ras, compared to mice with two wild type Dlc1 alleles (22). These results are analogous to our finding that reduced Dlc1 activity enhances the transforming ability of v-Ras.
Longer-term cultured MEFs with disrupted Dlc1 become spontaneously transformed in association with p15Ink4b and p16Ink4a inactivation
Although early passaged MEF-Cre cells did not display a transformed phenotype, they did undergo neoplastic transformation after longer-term cultivation of 15–20 passages. Transformation depended on the loss of Dlc1 in the MEF-Cre cells, as identically cultured MEF-Vt cells did not become transformed. Unlike the MEF-Vt cells, the MEF-Cre cells grew to high density in monolayer culture in high serum (10%, Fig. 2A, upper panel) or low serum (1%; Fig. 2A, lower panel), underwent anchorage-independent growth in soft agar (Fig. 2B), and formed tumor xenografts in immunocompromised mice (Fig. 2C). The spontaneously transformed MEF-Cre cells were less transformed than the MEF-Cre v-Ras transductants at the same passage level, and were similar to MEF-Vt v-Ras transductants, as determined by monolayer growth and soft agar growth (Fig. 2A and 2B).
Figure 2. Later passage MEF-Cre cells, but not MEF-Vt cells, become spontaneously transformed.
(A) Later passage MEF-Cre cells proliferate faster than MEF-Vt cells under high or low serum conditions. Equal numbers of MEF cells were cultured with 10% FBS (top) or 1% FBS (bottom) for three days. The mean number of cells from each day is plotted. (B) Cell growth in soft agar. Later passage MEF cells with or without v-Ras were grown in soft agar for 5 weeks. Quantitation of colony number (top) and representative images (bottom) are shown. (C) Later passage MEF-Cre cells are tumorigenic. Equal numbers of cells were injected into nude mice and grown for 5 weeks. The mean tumor weight is plotted. (D) Biochemical analysis of later passage MEFs. In vivo RhoGTP (top) and RasGTP (bottom) are shown.
The cells were analyzed for RasGTP, RhoGTP, and Cdc42GTP. The levels of RasGTP remained low in the later passaged MEF-Vt and MEF-Cre, and remained elevated to a similar degree in both v-Ras lines (Fig. 2D, upper panels). RhoGTP and Cdc42GTP were higher in the later passaged spontaneously transformed MEF-Cre cells than in the later passaged MEF-Vt cells (Fig. 2D, middle and bottom panels). The RhoGTP and Cdc42GTP levels were even higher, and similar to each other, in the Ras-transformed MEF-Vt and MEF-Cre cells, as Ras-transformation of MEFs depends on the activity of both Rho and Cdc42 (23, 24). Consistent with their transformed phenotype and elevated RhoGTP levels, the later passaged MEF-Cre cells had more stress fibers than MEF-Vt, and the v-Ras expressing MEF-Vt and MEF-Cre also had abundant stress fibers that appeared to be thicker than in MEF-Cre (Supplementary Fig. 1).
The Cdk4/6 inhibitors p15 and p16 have been reported to become down-regulated during passage of other MEFs (25, 26). We therefore examined their level of expression in early and later passaged cells. p15 and p16 were expressed in early passaged MEF-Vt and MEF-Cre cells, but were no longer detectable in later passaged cells of either line, and loss of their expression was associated in both lines with a concomitant increase in Cdk4/6 expression and phosphoryation of Cyclin D1 (Fig. 3A). We also verified that the phenomenon seen with the MEF-Cre cells was reproducible, in that adeno-Cre infection of independently derived MEFs resulted in loss of Dlc1 expression and increased RhoGTP after short-term passage and cell transformation after longer term passage (Supplementary Fig. 2).
Figure 3. Transformation of MEF-Cre cells is affected by the status of p15 and p16 expression.
(A) p15 and p16 expression become undetectable in later passage MEFs, leading to up-regulation of Cdk4/6 and phospho-cyclin D1. Cell extracts from MEF-Vt and MEF-Cre cells at early (~6o) or later (~25 o) passages were analyzed by immunobloting with anti-p15, p16, Cdk4, Cdk6, phospho-Cyclin D1 (T286), and anti-β-actin antibodies. (B and C) siRNA knock-down of p15 or p16 in early passage of MEF-Cre cells (A) and A549 cells (C, right) results in colony growth in soft agar. Cell extracts from siRNA transfected cells were analyzed to verify reduced expression of p15 and p16. Cells were grown in soft agar for 4 weeks. Unlike NSCLC H1703 cells, which contain detectable DLC1 protein, DLC1 expression is undetectable in NSCLC A549 cells (left). (D) Genotyping of isolated colonies from MEF-Cre mass culture grown in soft agar. Individual colonies isolated from agar colonies, designated anchorage-independent growth (AIG) clones, were analyzed by RT-PCR using Dlc1 exon 4-specific primers (top) or GAPDH primers (bottom). (E) Inverse relationship between p16 expression and CdK6 level in MEFs. The later passage MEF-Vt, MEF-Cre and derived AIG clones were analyzed for expression of p16 and Cdk6. β-actin blot serves as a loading control. (F) The presence of p16 in AIG-2 line grew poorly in soft agar. The indicted MEFs were grown in soft agar, and the quantization of colony number (top) and representative images (bottom) are shown.
It is well known that endogenous p15 and p16 negatively regulate the growth of human tumor cells, but it has not been determined whether such regulation is relevant to tumor cells in which DLC1 has been down-regulated (14, 27). Therefore, we identified a p15- and p16-expressing human tumor line, the A549 non-small cell lung cancer (NSCLC) line, in which endogenous DLC1 had been down-regulated (Fig. 3C). The siRNA-induced reduction of p15 or p16 increased the ability of the A549 cells to grow in agar, indicating that p15 and p16 negatively regulate growth of human tumor cells that do not express DLC1. Furthermore, in earlier passaged non-transformed MEF-Vt cells that expressed p15 and p16, decreased p15 or p16 expression increased their growth potential, since siRNA-mediated reduction of p15 and p16 led to some anchorage-independent growth in soft agar (Fig. 3B).
Spontaneous transformation of MEFs with disrupted Dlc1 depends on loss of Dlc1, p15, and p16 and activation of Cdk4/6, ROCK, and Jnk
Before determining whether the identified changes in gene expression and activity were mechanistically involved in transformation of the late passaged MEF-Cre cells, we first developed clonally derived lines, to ensure the Dlc1 alleles were disrupted in all cells, and characterized the clones. To obtain clonal lines, individual colonies were isolated after growth in soft agar, and their clonality for Dlc1 disruption was analyzed by RT-PCR (Fig. 3D). While the Dlc1 alleles were uniformly disrupted in some colonies (AIG-2 and AIG-5), one colony (AIG-7) contained a mixture of intact and disrupted Dlc1 alleles. An interesting correlation was that a later passaged MEF-Cre clone whose Dlc1 alleles had been disrupted but grew poorly in soft agar (AIG-2) was found to continue to express p16, but not p15, while both p15 and p16 were down-regulated in the the AIG-5 clone, which grew efficiently in soft agar (Fig. 3E and 3F; data shown only for p16).
As expected from the RT-PCR results, AIG-5 cells were negative for endogenous Dlc1 protein by microscopy, while the longer-term passaged MEF-Vt cells were positive (Figure 4A, top left panels). The high RhoGTP level in the AIG-5 cells was associated with increased stress fiber formation, a widely studied phenotype that depends on the Rho-associated protein kinases (ROCKs: ROCK1 and ROCK2)(28, 29), which are activated by Rho (Figure 4A, bottom left panels). By contrast, the low RhoGTP level in the longer-term passaged MEF-Vt cells was associated with less stress fiber formation.
Figure 4. Spontaneous transformation of MEF-Cre cells can be reversed by re-expression of human wild-type DLC1.
(A) Dlc1 expression in MEF-Cre cells affects stress fiber formation. Dlc1 expression of in MEF-Vt cells, its absence in MEF-Cre (AIG-5), and the derived stable clones expressing wild type DLC1 (GFPDLC1) and GAP-dead mutant (GFPR718A) were validated by anti-DLC1 (428) antibody staining (in red). The RhoGTP activity-related stress fiber formation of each cell type is shown by green costaining with phalloidin. The scale bar = 20 μm. (B) RhoGTP and Cdc42GTP levels vary inversely with the expression of DLC1. MEF-Cre clone AIG-5, as well as human NSCLC line H1703, stably expressing GFP, GFPDLC1, or GFPR718A, were analyzed by Rhotekin (top) and Pak-1 (middle) pull-down assays for Cdc42GTP and RhoGTP, respectively, and GFP-DLC1 (bottom). (C-D) Spontaneous transformation of MEF-Cre (AIG-5) cells can be reversed by re-expression of human wild-type DLC1 but not GAP-dead mutant R718A. Representative images of monolayer growth, colony growth in soft agar, and transwell migration (C) and quantitation of agar colonies and migrated cells (D).
We assessed whether the inactivation of Dlc1, p15, and 16, as well as increased expression of Cdk4 and Cdk6 and increased activity of Rho, Cdc42, and Cdk4/6, which were the identified changes associated with MEF-Cre transformation, contributed mechanistically to the transformed phenotype. We used genetic approaches to evaluate the role of the Dcl1, p15, p16, Cdk4, and Cdk6 genes in MEF-Cre transformation. Pharmacological inhibitors were used to examine whether the activities of several protein kinase were relevant to the transformation process: these included Cdk4/6, the RhoGTP –dependent kinase ROCK, and JNK, a downstream target of Cdc42GTP (30).
Stable transfection of wild type DLC1 in the AIG-5 clone induced morphological reversion that was associated with reductions in RhoGTP, Cdc42GTP, stress fiber formation, growth in soft agar, and transwell cell migration (Fig. 4A–D). All of these phenotypes, including morphological reversion, depended on the GAP activity of DLC1, as a DLC1 GAP-dead mutant (R718A) was deficient for inducing these changes, and were not accompanied by re-expression of p15 or p16 (data not shown). These findings confirm that reversion depended on the GAP activity of DLC1 and indicate, in addition, that DLC1 possesses in vivo Cdc42GAP activity in addition to its RhoGAP activity. In vivo Cdc42GAP activity of ectopically expressed DLC1 was also detected in the H1703 human NSCLC line (Fig. 4B, right panels).
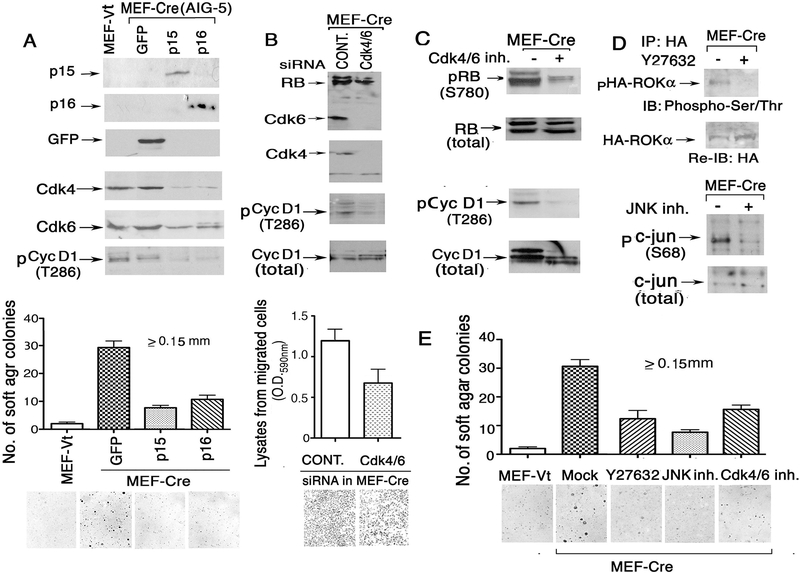
Stable transfection of p15 and p16 reverted the transformed phenotype, as determined by reduced anchorage-independent growth in soft agar (Fig. 5A). Conversely, siRNA-mediated reduction of Cdk4 and Cdk6 inhibited cell migration (Fig. 5B). Pharmacologic studies indicated that transformation also depended on the activities of Cdk4/6, ROCK, and Jnk, as the Cdk4/6 inhibitor, the ROCK inhibitor Y27632, and the Jnk inhibitor VIII reduced their respective enzymatic activity and suppressed growth in soft agar (Fig. 5C–E).
Figure 5. Transformation of later passage MEF-Cre cells can be partially reverted by p15 or p16 re-expression, siRNA knock-down of Cdk4/6, or pathway-specific inhibitors.
(A) Re-expression of p15 or p16 in MEF-Cre cells reduces RhoGTP, Cdc42GTP, Cdk4/6, pCyclin D1, and growth in soft agar. MEF-Cre AIG-5 cells stably expressing GFP, p15, or p16 were analyzed for various biochemical properties (top) and agar growth (bottom). (B) siRNA knock-down of Cdk4/6 can inhibit phosphorylation of Cyc D1 and cell migration. Reduced Cdk4 and Cdk6 expression was verified by immunoblotting. (C) Inhibition of Cdk4/6 activity in MEF-Cre cells reduces phosphorylation of Cdk4/6 substrates. pCyclin D1 (T286) and pRB (S780) levels are validated by immunoblots. (D) Validation of ROCK and Jnk pathway-specific inhibitors in signaling. Top panels: The inhibitory effect of Y27632 on transfected rat ROKα-HA activity in MEF-Cre cells was assayed by anti-HA immunoprecipitation followed by Phospho-Ser/Thr immunoblotting. The membrane was re-probed by anti-HA to show the loading control (bottom). Bottom panels: MEF-Cre cells treated with JNK inhibitor VIII were assayed by phospho-c-jun (S78) and c-jun antibody blots. (E) Partial reversion of soft agar growth of MEF-Cre cells treated with the pathway-specific inhibitors in (C) and (D). Cells were grown in agar for 5 weeks. Quantitation of colony numbers (top) and representative images of agar colonies (bottom) are shown.
In human cancer, down-regulation of DLC1 together with low p15 or p16 or with high CDK4 or CDK6 may have prognostic significance
To examine whether the genes implicated in MEF-Cre transformation might have relevance to human cancer, we searched publicly available cancer datasets with mRNA gene expression profiling, focusing on DLC1, p15, p16, CDK4, and CDK6, as those were the genes whose changes in expression were associated with MEF transformation. Down-regulation of DLC1 occurred together with down-regulation of p15 and/or p16 in NSCLC cancer and colon cancer (Table 1a), and together with upregulation of CDK4 and CDK6 (Table 1b). In addition, we analyzed two annotated NSCLC cancer cohorts (62 annotated cases in cohort I and 81 annotated cases in cohort II) and one annotated colon cancer cohort (37 annotated cases) for whether the level of DLC1 expression together with that of any one of these genes might have prognostic implications. An unfavorable prognosis was found to be associated with low DLC1 and low p15 in one of the NSCLC cohorts (cohort II) and the colon cancer cohort, with low DLC1 and low p16 in both NSCLC cohorts, with low DLC1 and high CDK4 in one of the NSCLC cohorts (cohort I), and low DLC1 and high CDK6 in the colon cancer cohort (Fig. 6 and Supplementary Fig. 2).
Table 1a.
Down-regulation of DLC1 and p15/p16 gene expression in human cancers
| DLC1↓ | DLC1↓ p15↓ | DLC1↓ p16↓ | DLC1↓p15↓p16↓ | |||||
|---|---|---|---|---|---|---|---|---|
| Lung Cancer (E-GEOD-18842) | ||||||||
| Lung Control (n=45) | n=2 | p < 0.001 | n=2 | p < 0.001 | n=0 | p < 0.001 | n=0 | p < 0.001 |
| Tumor (n=46) | n=44 | n=36 | n=18 | n=18 | ||||
| Lung Cancer (E-GEOD-19188) | ||||||||
| Normal (n=50) | n=1 | p < 0.001 | n=1 | p < 0.001 | n=0 | p < 0.01 | n=0 | p < 0.01 |
| Tumor (n=68) | n=58 | n=43 | n=11 | n=11 | ||||
| Colon Cancer (E-GEOD-3629) | ||||||||
| Non-Cancer (n=53) | n=7 | p < 0.001 | n=4 | p < 0.001 | n=1 | p < 0.001 | n=1 | p < 0.001 |
| Colorectal Cancer (n=68) | n=53 | n=39 | n=52 | n=39 | ||||
| Colon Cancer (E-GEOD-23878) | ||||||||
| Control (n=24) | n=8 | p = 0.044 | n=0 | p < 0.001 | n=3 | p = 0.216 | n=0 | p = 0.032 |
| Colorectal Cancer (n=35) | n=21 | n=15 | n=9 | n=6 | ||||
Table 1b.
The reverse expression pattern of DLC1 and CDK4/6 in human cancers
| DLC1↓ | DLC1↓ CDK4↑ | DLC1↓ CDK6↑ | DLC1↓ CDK4/6↑ | |||||
|---|---|---|---|---|---|---|---|---|
| Lung Cancer (E-GEOD-18842) | ||||||||
| Lung Control (n=45) | n=2 | p < 0.001 | n=1 | p < 0.001 | n=0 |
p < 0.001 |
n=0 | p < 0.001 |
| Tumor (n=46) | n=44 | n=41 | n=36 | n=34 | ||||
| Lung Cancer (E-GEOD-19188) | ||||||||
| Normal (n=50) | n=1 | p < 0.001 | n=1 | p < 0.001 | n=0 | p< 0.01 | n=0 | p < 0.001 |
| Tumor (n=68) | n=58 | n=48 | n=44 | n=37 | ||||
| Colon Cancer (E-GEOD-23878) | ||||||||
| Control (n=24) | n=8 | p=0.044 | n=0 | p <0.001 | n=0 | p<0.001 | n=0 | p <0.001 |
| Colorectal Cancer (n=35) | n=21 | n=19 | n=11 | n=11 | ||||
The data from Arrayexpress of European Bioinformatics Institute are reanalyzed to study the co-down-regulation of DLC1 and CDK4/6 gene expression from selected cancers. The contents in the parenthesis are the ID number of array experiments. Chi Square analysis is performed to study whether the down regulation is statistically significant.
Figure 6. Prognostic significance in NSCLC and colon cancer of co-down regulation of DLC1 and p15 or p16 or down-regulation of DLC1 with up-regulation of CDK4 or CDK6.
(A-C) Kaplan-Meier analysis of DLC1, p15, p16, CDK4, and CDK6 gene expression in NSCLC (A and B) and colon cancer (C). Low vs. high expression of each gene by itself does not have prognostic value in these cohorts (p>0.05), but there is prognostic significance when low DLC1 is combined with low p15 (B and C), with low p16 (A and B), with high CDK4 (A, “DL4H”), and with high CDK6 (C, “DL6H”). The p value for each comparison is shown.
DISCUSSION
In this report, we found that conditional disruption of both Dlc1 alleles in MEFs reproducibly led to their spontaneous tumorigenic transformation after several passages in culture, a phenotype that was not seen in MEFs with intact Dlc1 that had an identical passage history. This system may mimic some aspects of human tumors, in that the oncogenic transformation occurs over time (1), rather than resulting from the simultaneously forced inactivation of tumor suppressor genes and/or activation of oncogenes. Compared with MEFs that carry a constitutively disrupted growth regulatory gene, conditional disruption means the gene does not need to be inactivated until after the cells have been placed in culture, which makes it easier to rigorously examine the short-term effects of disrupting the gene and compare these effects with those resulting from longer-term disruption. This system also has theoretical advantages over the more typical MEFs which have constitutively disrupted genes, as developmentally related compensatory changes might lead to inappropriate interpretation when these MEFs are compared with wild type MEFs.
MEFs carrying the conditionally disrupted Dlc1 alleles are well suited to identifying cell-autonomous candidate genes and biochemical activities that may have cooperated over time with Dlc1 inactivation to produce the observed oncogenic phenotype. When such modifications are identified in other systems, the findings often remain as correlations (31). With the MEF system, however, the putative contribution of identified candidate genes can readily be evaluated by genetic manipulation, while that of their biochemical activities can be assessed pharmacologically. Using this approach, we identified several genes whose expression and/or activities are altered in conjunction with the transformed phenotype, and confirmed their mechanistic role in maintaining the phenotype. Despite this being an in vitro mouse fibroblast system, we were able to verify that our genetic findings have relevance to human epithelial tumors.
In the MEFs, one set of results identified reduced expression of the CDK 4/6 inhibitors p15 and p16 (14, 27) as passage-dependent changes, whether Dlc1 was disrupted or intact (Zindy et al., 1997). The lack of dependence on Dlc1 disruption increased the theoretical possibility that loss of p15 and p16 expression might not have contributed to the transformed state in the MEFs. However, re-expression of p16 or p15 induced reversion of transformation, verifying their role in its maintenance, while siRNA-mediated reduction of either gene in non-transformed early passaged MEF-Cre cells increased their anchorage-independent growth. This role was also reinforced by identification and analysis of an unusual MEF clone that still expressed p16 and grew poorly in agar, although it did not express Dlc1 or p15; siRNA-mediated reduction of its p16 expression enhanced its ability to grow in soft agar.
The reduced p15 and p16 in the transformed MEFs was associated with an increase in Cdk4/6 expression and activity, which contributed to their transformation. There were also transformation-associated biochemical changes not accompanied by alterations in steady-state protein levels, including increases in RhoGTP and Cdc42GTP and their respective targets, ROCK and Jnk (28–30). Using pharmacologic inhibitors, we established a role in transformation for the enzymatic activities of ROCK and Jnk.
Given the apparently sequential nature of the observed changes in gene expression and activity, it was theoretically possible that later changes rendered the fully transformed phenotype independent of some earlier changes. However, that was found not to be the case, which suggests that the process of oncogenic transformation in this system occurs primarily by a series of cooperative changes that continue to be mechanistically important even after development of the neoplastic state. The later passaged MEF-Vt cells, which have high Cdk4/6 activity but are not morphologically transformed because they have intact Dlc1 alleles, may have phenotypic similarities to the HBEC human lung epithelial cells, which constitutively express CDK4 and telomerase (hTERT) but are not transformed (32).
Our findings also verify that DLC1 can regulate Cdc42 in cells. Until now, a role for DLC1 in Cdc42 regulation has been confined to the weak in vitro Cdc42GAP activity mediated by the RhoGAP domain of DLC1, which contrasts with its strong, well documented in vitro and in vivo RhoGAP activity (6). Here, the transformed MEFs were found to contain high levels of RhoGTP and Cdc42GTP, which could be reduced by re-expression of DLC1. This reduction of active Rho and Cdc42 depends on the GAP activity of DLC1, since the GAP-dead DLC1 mutant did not alter the level of RhoGTP or Cdc42GTP.
Taken together, our observations suggest a multifactorial model for the genes and functions identified here as contributing to neoplastic transformation of the MEFs initiated by inactivation of Dlc1. Key features of the model include the inactivation of Dlc1, p15, and p16, and their consequences, which include activation of Cdk4/6, Rho, and ROCK, and Cdc42 and Jnk. We recognize that additional changes, not identified here, probably also contribute to this process.
It is noteworthy that the five genes whose expression in the MEFs was altered in a transformation-dependent manner – Dlc1, p16, p15, Cdk4, and Cdk6 – are altered in human cancer. Indeed, we found in publicly available datasets with mRNA gene expression profiling that down-regulation of DLC1 expression occurs frequently in conjunction with reductions in expression of p15 and/or p16 or upregulation of CDK4 and/or CDK6 in NSCLC and colon cancer. Furthermore, downregulation of DLC1 and p15 or p16 was associated with an unfavorable prognosis in annotated NSCLC and colon cancer datasets, despite a relatively small number of annotated tumors in each dataset. Analogous findings were seen in the same datasets for low DLC1 expression together with high CDK4 or CDK6. Such associations are not limited to NSCLC and colorectal cancer. In lymphocytic leukemia, promoter methylation of DLC1, p15, and p16 is reported to be associated with a more advanced stage (33). Their association with a poorer prognosis or a more advanced tumor stage implies that these changes make important contributions to disease outcome.
These results validate the clinical relevance of the genes identified in the transformed MEFs. The clinical findings contribute to the growing list of tumors in which reduced DLC1 expression, either by itself or as part of a multigene signature, is associated with poor prognosis and/or advanced stage (34–36).
Supplementary Material
ACKNOWLEDGMENTS
We thank Zhizhong Fei for technical assistance, Juraj Bies, Rita Humeniuk, and Affymetrix for technical advice, Thomas Leung, Silvio Gutkind, Beverly Mock, and Linda Wolff for reagents, Curt Harris for NSCLC lines H1703 and A549, and Ed Harlow for helpful discussions. This research was supported by the Intramural Research Program, National Institutes of Health, National Cancer Institute, Center for Cancer Research.
References
- 1.Balmain A, Gray J, Ponder B. The genetics and genomics of cancer. Nat Genet. 2003;33 Suppl:238–44. [DOI] [PubMed] [Google Scholar]
- 2.Kim TY, Vigil D, Der CJ, Juliano RL. Role of DLC-1, a tumor suppressor protein with RhoGAP activity, in regulation of the cytoskeleton and cell motility. Cancer Metastasis Rev. 2009;28:77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Durkin ME, Yuan BZ, Zhou X, Zimonjic DB, Lowy DR, Thorgeirsson SS, et al. DLC-1:a Rho GTPase-activating protein and tumour suppressor. J Cell Mol Med. 2007;11:1185–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seng TJ, Low JS, Li H, Cui Y, Goh HK, Wong ML, et al. The major 8p22 tumor suppressor DLC1 is frequently silenced by methylation in both endemic and sporadic nasopharyngeal, esophageal, and cervical carcinomas, and inhibits tumor cell colony formation. Oncogene. 2007;26:934–44. [DOI] [PubMed] [Google Scholar]
- 5.Durkin ME, Avner MR, Huh CG, Yuan BZ, Thorgeirsson SS, Popescu NC. DLC-1, a Rho GTPase-activating protein with tumor suppressor function, is essential for embryonic development. FEBS Lett. 2005;579:1191–6. [DOI] [PubMed] [Google Scholar]
- 6.Healy KD, Hodgson L, Kim TY, Shutes A, Maddileti S, Juliano RL, et al. DLC-1 suppresses non-small cell lung cancer growth and invasion by RhoGAP-dependent and independent mechanisms. Mol Carcinog. 2008;47:326–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong CM, Yam JW, Ching YP, Yau TO, Leung TH, Jin DY, et al. Rho GTPase-activating protein deleted in liver cancer suppresses cell proliferation and invasion in hepatocellular carcinoma. Cancer Res. 2005;65:8861–8. [DOI] [PubMed] [Google Scholar]
- 8.Yam JW, Ko FC, Chan CY, Jin DY, Ng IO. Interaction of deleted in liver cancer 1 with tensin2 in caveolae and implications in tumor suppression. Cancer Res. 2006;66:8367–72. [DOI] [PubMed] [Google Scholar]
- 9.Liao YC, Si L, deVere White RW, Lo SH. The phosphotyrosine-independent interaction of DLC-1 and the SH2 domain of cten regulates focal adhesion localization and growth suppression activity of DLC-1. J Cell Biol. 2007;176:43–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qian X, Li G, Asmussen HK, Asnaghi L, Vass WC, Braverman R, et al. Oncogenic inhibition by a deleted in liver cancer gene requires cooperation between tensin binding and Rho-specific GTPase-activating protein activities. Proc Natl Acad Sci U S A. 2007;104:9012–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang X, Popescu NC, Zimonjic DB. DLC1 interaction with S100A10 mediates inhibition of in vitro cell invasion and tumorigenicity of lung cancer cells through a RhoGAP-independent mechanism. Cancer Res. 2011;71:2916–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li G, Du X, Vass WC, Papageorge AG, Lowy DR, Qian X. Full activity of the deleted in liver cancer 1 (DLC1) tumor suppressor depends on an LD-like motif that binds talin and focal adhesion kinase (FAK). Proc Natl Acad Sci U S A. 2011;108:17129–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xue W, Krasnitz A, Lucito R, Sordella R, Vanaelst L, Cordon-Cardo C, et al. DLC1 is a chromosome 8p tumor suppressor whose loss promotes hepatocellular carcinoma. Genes Dev. 2008;22:1439–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Canepa ET, Scassa ME, Ceruti JM, Marazita MC, Carcagno AL, Sirkin PF, et al. INK4 proteins, a family of mammalian CDK inhibitors with novel biological functions. IUBMB Life. 2007;59:419–26. [DOI] [PubMed] [Google Scholar]
- 15.Vigil D, Cherfils J, Rossman KL, Der CJ. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat Rev Cancer. 2010;10:842–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willumsen BM, Vass WC, Velu TJ, Papageorge AG, Schiller JT, Lowy DR. The bovine papillomavirus E5 oncogene can cooperate with ras: identification of p21 amino acids critical for transformation by c-rasH but not v-rasH. Mol Cell Biol. 1991;11:6026–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lowy DR, Rands E, Scolnick EM. Helper-independent transformation by unintegrated Harvey sarcoma virus DNA. J Virol. 1978;26:291–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qian X, Li G, Vass WC, Papageorge A, Walker RC, Asnaghi L, et al. The Tensin-3 protein, including its SH2 domain, is phosphorylated by Src and contributes to tumorigenesis and metastasis. Cancer Cell. 2009;16:246–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sabbir MG, Wigle N, Loewen S, Gu Y, Buse C, Hicks GG, et al. Identification and characterization of Dlc1 isoforms in the mouse and study of the biological function of a single gene trapped isoform. BMC Biol. 2010;8:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sordella R, Classon M, Hu KQ, Matheson SF, Brouns MR, Fine B, et al. Modulation of CREB activity by the Rho GTPase regulates cell and organism size during mouse embryonic development. Dev Cell. 2002;2:553–65. [DOI] [PubMed] [Google Scholar]
- 21.Brouns MR, Matheson SF, Hu KQ, Delalle I, Caviness VS, Silver J, et al. The adhesion signaling molecule p190 RhoGAP is required for morphogenetic processes in neural development. Development. 2000;127:4891–903. [DOI] [PubMed] [Google Scholar]
- 22.Sabbir MG, Prieditis H, Ravinsky E, Mowat MR. The Role of Dlc1 Isoform 2 in K-Ras2(G12D) Induced Thymic Cancer. PLoS One. 2012;7:e40302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qiu RG, Abo A, McCormick F, Symons M. Cdc42 regulates anchorage-independent growth and is necessary for Ras transformation. Mol Cell Biol. 1997;17:3449–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiu RG, Chen J, McCormick F, Symons M. A role for Rho in Ras transformation. Proc Natl Acad Sci U S A. 1995;92:11781–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quelle DE, Zindy F, Ashmun RA, Sherr CJ. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83:993–1000. [DOI] [PubMed] [Google Scholar]
- 26.Zindy F, Quelle DE, Roussel MF, Sherr CJ. Expression of the p16INK4a tumor suppressor versus other INK4 family members during mouse development and aging. Oncogene. 1997;15:203–11. [DOI] [PubMed] [Google Scholar]
- 27.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9:153–66. [DOI] [PubMed] [Google Scholar]
- 28.Narumiya S, Tanji M, Ishizaki T. Rho signaling, ROCK and mDia1, in transformation, metastasis and invasion. Cancer Metastasis Rev. 2009;28:65–76. [DOI] [PubMed] [Google Scholar]
- 29.Street CA, Bryan BA. Rho kinase proteins--pleiotropic modulators of cell survival and apoptosis. Anticancer Res. 2011;31:3645–57. [PMC free article] [PubMed] [Google Scholar]
- 30.Warner SJ, Yashiro H, Longmore GD. The Cdc42/Par6/aPKC polarity complex regulates apoptosis-induced compensatory proliferation in epithelia. Curr Biol. 2010;20:677–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheung AF, Carter AM, Kostova KK, Woodruff JF, Crowley D, Bronson RT, et al. Complete deletion of Apc results in severe polyposis in mice. Oncogene. 2010;29:1857–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramirez RD, Sheridan S, Girard L, Sato M, Kim Y, Pollack J, et al. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res. 2004;64:9027–34. [DOI] [PubMed] [Google Scholar]
- 33.Forsterova K, Votavova H, Schwarz J, Karban J, Stuka C, Trneny M. Advanced rai stage in patients with chronic lymphocytic leukaemia correlates with simultaneous hypermethylation of plural tumour suppressor genes. Folia Biol (Praha). 2010;56:158–64. [DOI] [PubMed] [Google Scholar]
- 34.Roessler S, Long EL, Budhu A, Chen Y, Zhao X, Ji J, et al. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology. 2012;142:957–66 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tripathi SC, Kaur J, Matta A, Gao X, Sun B, Chauhan SS, et al. Loss of DLC1 is an independent prognostic factor in patients with oral squamous cell carcinoma. Mod Pathol. 2012;25:14–25. [DOI] [PubMed] [Google Scholar]
- 36.Zhang T, Zheng J, Liu C, Lu Y. Expression of DLC-1 in clear cell renal cell carcinoma: prognostic significance for progression and metastasis. Urol Int. 2009;82:380–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.