Abstract
Background: Metastasis is the primary cause of death in colorectal cancer (CRC); the underlying mechanisms remain partly unknown. In this study, we aim to investigate the value of HOXA11-AS in survival evaluation and the potential role of HOXA11-AS/miR-149-3p axis in the CRC metastasis.
Methods: The expressions of HOXA11-AS, both in obtained CRC samples and adjacent noncancerous tissues, were analyzed in survival evaluation. Competing endogenous RNAs (CeRNAs) Analysis were employed to reveal the potential relationship between HOXA11-AS and miR-149-3p. It was further confirmed by Quantitative real-time polymerase chain reaction (qRT-PCR) and Dual-luciferase reporter assay. Migration and invasion assay were used to verify the potential role of HOXA11-AS and miR-149-3p in the regulation of CRC metastasis. The potential pathway was explored by Western blot analysis.
Results: The expression of HOXA11-AS in the CRC tissue is significantly higher than the expression in adjacent noncancerous tissue (p<0.0001). High expressions of HOXA11-AS were noticeably correlated with clinicopathologic characteristics including advanced clinical stage (p=0.021), larger tumor size (p<0.001) and frequent tumor recurrence (p=0.001). The overall survival in HOXA11-AS-High group was significantly shorter than the HOXA11-AS-Low group (p<0.001). Advanced clinical stage, tumor size and high expression of HOXA11-AS were showed as independent prognostic prediction factors for the 5-year tumor relapse of CRC patients (p<0.001). HOXA11-AS acts as a potential molecular sponge for miR-149-3p, in the promotion of CRC metastasis. In the miR-149-3p mimic-treated group, the expression of E-cadherin was increased, whereas the expression of N-cadherin, Snail, Slug, TGF-β1, Wnt2b, Twist and C/EBPβ was decreased.
Conclusion: This study demonstrates that high expression of HOXA11-AS is correlated with CRC progression and poor prognosis and may promote metastasis via EMT by modulating miR-149-3p.
Keywords: colorectal cancer (CRC), long non-coding RNA (lncRNA), HOXA11-AS, miR-149-3p, epithelial mesenchymal transition (EMT)
Introduction
Colorectal cancer (CRC) is the third most common cancer in the United States, according to 2019 statistics for both male and female cancers 1, and it was the second most common cancer in China in 2018 2. Although great progress has been made in the past several decades related to the prevention, detection, and treatment of CRC, metastasis is still the primary cause of death 3, and the underlying mechanisms remain partly unknown 4.
Accumulating evidence has demonstrated that long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) play important roles in tumor growth and metastasis and are linked to the activation of oncogenic signaling pathways or inactivation of tumor-suppressive signaling in the nucleus 4, 5. Although most functions of lncRNAs and miRNAs have not been clarified, recent evidence indicates that they are involved in the regulation of CRC proliferation and metastasis 6.
The expression of homeobox A11 antisense RNA (HOXA11-AS), a lncRNA, has been found in diverse human neoplasms, such as glioma, epithelial ovarian cancer, lung adenocarcinoma, gastric cancer, hepatocellular carcinoma, uterine cervix carcinoma, breast cancer, and CRC 7-15. In our previous study, HOXA11-AS was found to function as a competing endogenous RNA (ceRNA) to regulate peptidyl arginine deiminase 2 expression by sponging miR-125a-5p, promoting the metastasis of CRC 15. It was reported that some lncRNAs may be related to several miRNAs in ceRNA regulation of CRC 16. In our previous study, many associated miRNAs including miR-149-3p were found to potentially participate in the function of HOXA11-AS as a ceRNA regulator of CRC metastasis.
In this study, we evaluated whether HOXA11-AS is associated with the prognosis of CRC, as well as the effect and mechanism of the HOXA11-AS/miR-149-3p axis in the regulation of CRC metastasis.
Materials and Methods
Human tissue samples and cell lines
Tissue samples were collected from patients visiting the Department of Colorectal Surgery, First Affiliated Hospital, Zhejiang University (Hangzhou, China). The first 105 CRC samples were collected between June 2014 and September 2014; the second 30 primary CRC samples were obtained from 15 patients with CRC and liver metastasis and 15 patients with CRC without metastasis between April 2016 and September 2016. None of the patients received neoadjuvant therapy. Surgical staging was based on the tumor/node/metastasis (TNM) system. The inclusion criteria include (1) histologically confirmed diagnosis and (2) no previous treatment. The exclusion criteria include (1) serious complications, (2) presence of other malignant diseases, or (3) incomplete follow-up data. For staging, Cancer staging system defined by the United States Joint Commission 8th edition was adopted. The CRC samples were postoperative pathologically confirmed as colorectal adenocarcinoma, and the liver samples were confirmed to be metastatic adenocarcinoma by surgery or biopsy. Samples were collected within 10 min of tumor excision, immersed in RNA later™ Stabilization Solution (Thermo Fisher Scientific, Waltham, MA, USA) immediately, and stored at -80°C until use. Written informed consent was obtained from all patients. This study was approved by the Ethics Committee of First Affiliated Hospital, College of Medicine, Zhejiang University. The human CRC cell lines SW480, SW620, HCT116, HT-29, and RKO were obtained from Tongpai Biological Technology (Shanghai, China). SW480, SW620, HT-29, and HCT116 cells were cultivated in RPMI-1640 medium (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; HyClone, Logan, UT, USA). RKO cells were cultivated in Minimal Essential Media (Gibco) supplemented with 10% FBS. All cells were maintained in a 37°C incubator with a humidified atmosphere containing 5% CO2.
RNA extraction and qRT-PCR
Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. RNA samples were reverse transcribed into cDNA using different primers and the Prime Script Kit (Takara Bio Inc., Otsu, Japan). qRT-PCR was performed in triplicate to amplify lncRNAs and miRNAs using the SYBR Premix Ex Taq™ Kit (Takara Bio Inc.). The expression levels were normalized to that of β-actin, and relative gene expression was calculated as ΔCt = Ctgene - Ctreference. The fold change in gene expression was calculated using the 2-ΔΔCt formula. The primer sequences are shown in Table 1.
Table 1.
The sequence of the primers
| Gene | Forward primer | Reverse primer |
|---|---|---|
| HOXA11-AS | GAGTGTTGGCCTGTCCTCAA | TTGTGCCCAGTTGCCTGTAT |
| miR-149-3p | CGAAAGCACGUAAUCGCCGGUGUAA | |
| miR-149-3p inhibitor | GCUUUCGUGCAUUAGCGGCCACAUU | |
| β-actin | TGAGGATGTCACGGTTCCAG | GTCACCTTCACCGTTCCAGT |
Dual-luciferase reporter assay
Luciferase constructs harboring wild-type (WT) HOXA11-AS or mutant (Mut) HOXA11-AS, which contained a mutation site that abolishes targeting by miR-149-3p, were generated to analyze the interaction between HOXA11-AS and miR-149-3p. 293T cells were seeded onto 24-well plates and co-transfected with a luciferase reporter and miR-149-3p or the negative control (NC). Cells were lysed according to the instructions provided with the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA). Luciferase activity was measured 48 h after transfection using the Panomics Luminometer (Affymetrix, Santa Clara, CA, USA), according to the manufacturer's instructions. Luciferase activity was normalized to that of Renilla luciferase.
Construct generation and transient transfection
A small interfering RNA (siRNA) against HOXA11-AS (HOXA11-AS-siRNA), NC siRNA, miR-149-3p inhibitor, and miR-149-3p mimic were obtained from Gene Pharma (Shanghai, China). The HCT116 human CRC cells (2 × 105 cells) were transfected with these constructs at a final concentration of 25 nmol/L using Lipofectamine 2000 Reagent (Life Technologies, Carlsbad, CA, USA). Cells were transfected with the pcDNA-HOXA11-AS constructs at a final concentration of 1 μg/μL according to the manufacturer's protocol. After transfection for 48 h, total RNA was isolated from the harvested cells using TRIzol reagent (Invitrogen). Empty pEX3 vector and scrambled sequences of the miR-149-3p mimics, miR-149-3p inhibitor, or siRNAs were used as NCs.
Scratch assay
HCT116 cells were seeded at 4.0 × 105/mL. After 48 h, confluent cells were linearly scratched using a 20 μL pipette chip. The scratched region was photographed immediately, 24 h and 48 h after scratching by a microscope equipped with a camera. The photograph was traced to tracing paper, followed by the coloring of cells using image editing software (FireAlpaca, Tokyo, Japan). Subsequently, the area containing cells as a percentage of the total area was determined using the area measurement function of VHX-5000 (Keyence, Osaka, Japan). LIPUS exposure at a frequency of 3 MHz and intensities of 160 and 240 mW/cm2 was performed immediately, 24 h, and 48 h after scratching.
Migration and invasion
Cells were trypsinized and seeded at 50,000/well on both Matrigel-coated (Corning, Tewksbury, MA, USA) and uncoated (BD Biosciences, San Jose, CA, USA) Transwell filters in a 24-well plate. Both groups of cells were allowed to migrate or invade for 12 h toward the lower chambers containing 10% FBS. Three-Step-Stain (Richard-Allan Scientific, San Diego, CA, USA) was applied when the cells migrated or invaded through the uncoated or coated filters. Each filter was counted entirely under the rule of four 10× fields, whereas migration or invasion was quantified as the fold change relative to the control.
Western blot analysis
Cell lysates were homogenized in Cell Lysis Solution (Sigma-Aldrich, St. Louis, MO, USA) and centrifuged at 4ºC for 5 min. Samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis for 3 h and then electrotransferred to polyvinylidene fluoride membranes (Amersham Biosciences, Piscataway, NJ, USA) for an additional 2 h. Antibodies against transforming growth factor beta 1 (TGF-β1), CCAAT-enhancer binding protein beta (C/EBPβ), Snail 1, Wnt2b, Slug, Twist, E-cadherin, and N-cadherin were used for Western blot analysis. The Snail 1 antibody was purchased from Biorbyt Ltd. (Cambridge, UK); the other antibodies were purchased from Abcam (Cambridge, MA, USA). After incubation with a specific antibody against GAPDH at 4ºC overnight, the membranes were washed with 1% Tris-buffered saline containing Tween 20 in triplicate, incubated with the appropriate secondary antibodies for 1 h, and detected by chemiluminescence.
Statistical analysis
All statistical analyses were performed by SPSS 16.0 software (SPSS Inc., Chicago, IL, USA). Significant differences between two groups were estimated by Student's t-test, Pearson's chi-square test or Wilcoxon signed-rank test as appropriate. Pearson's correlation analysis was used to estimate the relationship between the expression of HOXA11-AS and that of miR-149-3p. p<0.05 was considered statistically significant.
Results
HOXA11-AS expression is correlated with CRC progression and poor prognosis
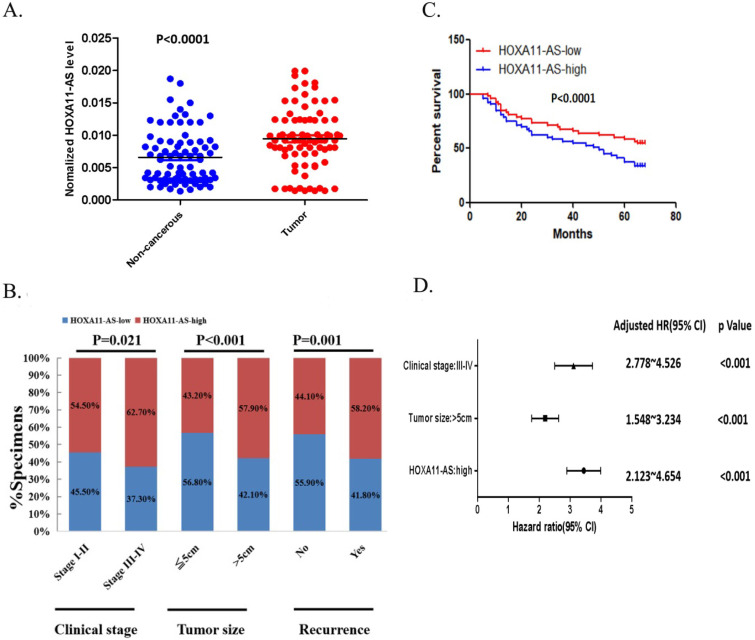
We investigated the clinical significance of HOXA11-AS expression in CRC tissue samples by qRT-PCR. The 105 patients were classified into high and low HOXA11-AS expression groups, according to the median level of relative HOXA11-AS expression in the tumor tissues. As shown in Figure 1A, the expression of HOXA11-AS was significantly upregulated in CRC tissues compared with adjacent noncancerous tissues. Furthermore, high expression of HOXA11-AS was noticeably correlated with clinicopathologic characteristics including advanced clinical stage, large tumor size, and frequent tumor recurrence (Fig. 1B). To evaluate the relationship between HOXA11-AS expression and CRC prognosis, a Kaplan-Meier survival curve was generated. Patients in the high HOXA11-AS group had significantly shorter overall survival compared with those in the low HOXA11-AS group (p<0.001; Fig. 1C). Moreover, advanced clinical stage, large tumor size, and high expression of HOXA11-AS were independent prognostic factors for the 5-year relapse-free survival of CRC patients (Fig. 1D).
Figure 1.
HOXA11-AS expression is correlated with colorectal cancer (CRC) poor prognosis. (A) The expression of HOXA11-AS in the CRC tissue is significantly higher than the expression in adjacent noncancerous tissue (p<0.0001). (B) High expressions of HOXA11-AS were noticeably correlated with clinicopathologic characteristics including advanced clinical stage (p=0.021), larger tumor size (p<0.001) and frequent tumor recurrence (p=0.001). (C) The overall survival in HOXA11-AS-High group was significantly shorter than the HOXA11-AS-Low group (p<0.001) in the Kaplan-Meier survival curves. (D) Advanced clinical stage, tumor size and high expression of HOXA11-AS were showed as independent prognostic prediction factors for the 5-year tumor relapse of CRC patients (p<0.001).
The potential role of the HOXA11-AS/miR-149-3p axis in CRC metastasis
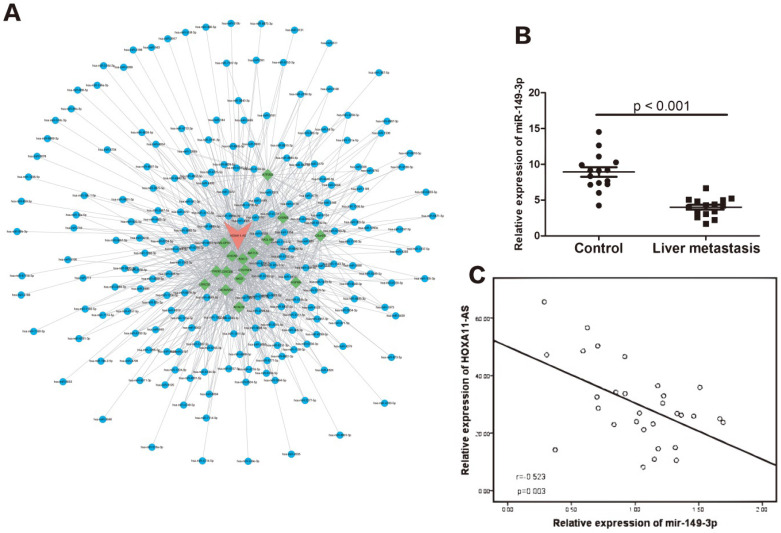
We performed bioinformatics analysis using Starbase 2.0 (http://starbase.sysu.edu.cn) and found that the HOXA11-AS/miR-149-3p axis may function in the regulation of CRC metastasis (Fig. 2A). Compared with the 15 samples of CRC without metastasis, the relative expression of miR-149-3p was significantly lower in the 15 CRC samples with liver metastasis (p<0.001), as shown in Figure 2B. Moreover, a negative correlation was found between the relative expression of miR-149-3p and that of HOXA11-AS (Fig. 2C).
Figure 2.
The potential function of HOXA11-AS/miR-149-3p axis in the regulation of colorectal cancer (CRC) liver metastasis. (A) The competing endogenous RNAs (CeRNAs) analysis showed the potential HOXA11-AS/miR-149-3p pathway. (B) The expression of miR-149-3p in the primary CRC tissue (with liver metastasis) was significantly lower than in the CRC tissue without metastasis (p<0.001). (C) The expression of miR-149-3p was negative correlated with the expression of miR-149-3p in CRC tissues (p=0.003).
HOXA11-AS acts as a molecular sponge for miR-149-3p
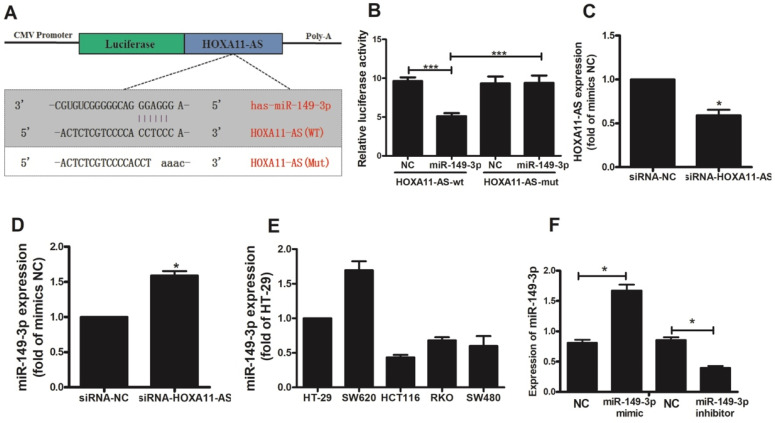
Based on our ceRNA analysis, we found that HOXA11-AS and miR-149-3p contained complementary base pairs (Fig. 3A), indicating that HOXA11-AS may act as a sponge to deregulate miR-149-3p. Thereafter, we generated luciferase reporter constructs harboring WT or Mut HOXA11-AS as the binding sites for investigation. Luciferase assays indicated a significant reduction in luciferase activity after co-transfection of miR-149-3p and a WT HOXA11-AS vector, but not Mut HOXA11-AS (p<0.0001; Fig. 3B). The transfection of HOXA11-AS-siRNA in HCT116 cells was validated by qRT-PCR. HOXA11-AS-siRNA caused a significant decrease (p=0.0226) in HOXA11-AS expression (Fig. 3C). The effect of HOXA11-AS on the endogenous expression of miR-149-3p was further examined. qRT-PCR revealed that knockdown of HOXA11-AS substantially increased the expression of miR-149-3p in HCT116 cells (p=0.011; Fig. 3D). HOXA11-AS expression was detected in all five CRC cell lines (Fig. 3E), and the HCT116 cell line was selected for subsequent analysis because its expression level was lowest. We also constructed a miR-149-3p mimic and miR-149-3p inhibitor and evaluated their effects by qRT-PCR (Fig. 3F).
Figure 3.
HOXA11-AS acts as a molecular sponge for miR-149-3p. (A) The putative miR-149-3p-binding sequence of HOXA11-AS. A mutation was generated in the HOXA11-AS sequence in the complementary site for the seed region of miR-149-3p. (B) Verification of the target relationship between miR-149-3p and HOXA11-AS. (C) HOXA11-AS siRNA transfection into HCT116 cells significantly reduced the expression of HOXA11-AS (p=0.0226). (D) HOXA11-AS siRNA transfection into HCT116 cells significantly increased the expression of miR-149-3p (p=0.011). (E) The relative expression of miR-149-3p in colorectal cell lines HT-29, SW620, HCT116, RKO and SW480. (F) Verification of the effectiveness of miR-149-3p mimic and miR-149-3p inhibitor.
The HOXA11-AS/miR-149-3p axis regulates the metastatic potential of CRC cells
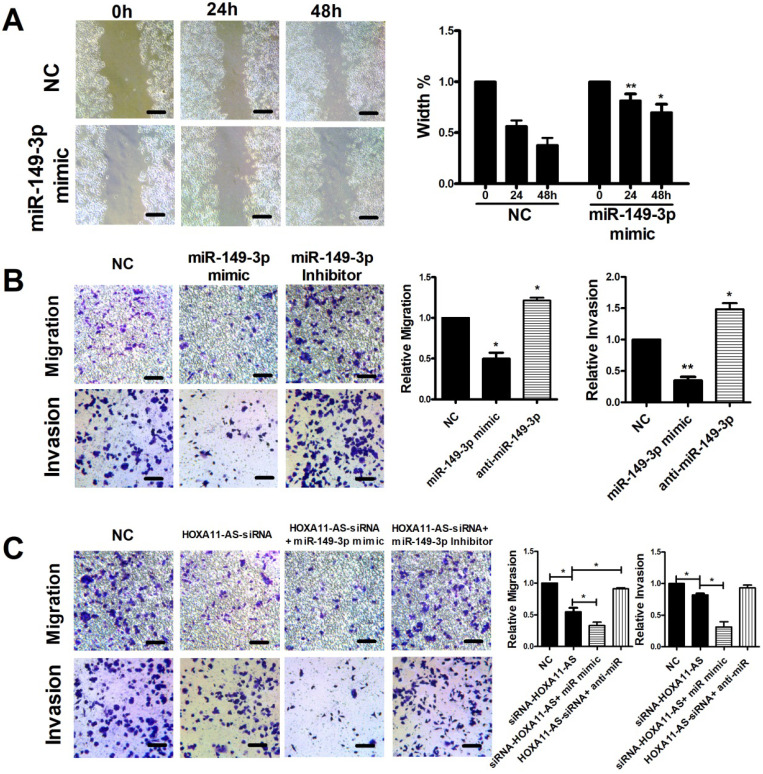
Scratch, migration, and invasion assays were performed to verify the functional roles of miR-149-3p and the HOXA11-AS/miR-149-3p axis in the regulation of CRC metastasis. The scratch assay indicated that overexpression of miR-149-3p markedly reduced the migration of HCT116 cells (Fig. 4A). Moreover, a Transwell assay demonstrated that overexpression of miR-149-3p significantly inhibited cell migration and invasion, whereas miR-149-3p inhibitors promoted migration and invasion of HCT116 cells (Fig. 4B). To further investigate the role of the HOXA11-AS/miR-149-3p axis in the regulation of CRC metastasis, we transfected the NC siRNA, HOXA11-AS-siRNA, HOXA11-AS-siRNA plus the miR-149a-3p mimic, and HOXA11-AS-siRNA plus the miR-149a-3p inhibitor into HCT116 cells. Both the migration and invasion of HCT116 cells were affected by HOXA11-AS-siRNA and further affected by the miR-149a-3p mimic or miR-149a-3p inhibitor (Fig. 4C).
Figure 4.
HOXA11-AS/miR-149-3p axis regulated the metastasis of colon cancer cell. (A) Representative images of the scratch area at 0, 24 and 48 hours post-treatment are shown. Wound closure became significantly lower both 24 and 48h after miR-149-3p treated. (B) miR-149-3p mimic inhibited the migration and invasion of HCT116 cells significantly; while miR-149-3p inhibitor increased the migration and invasion of HCT116 cells significantly. (C) The significant effectiveness of HOXA11-AS/miR-149-3p axis in regulating migration and invasion of HCT116 cells. *: p<0.05, **: p<0.01. Scale bar: 50 µm.
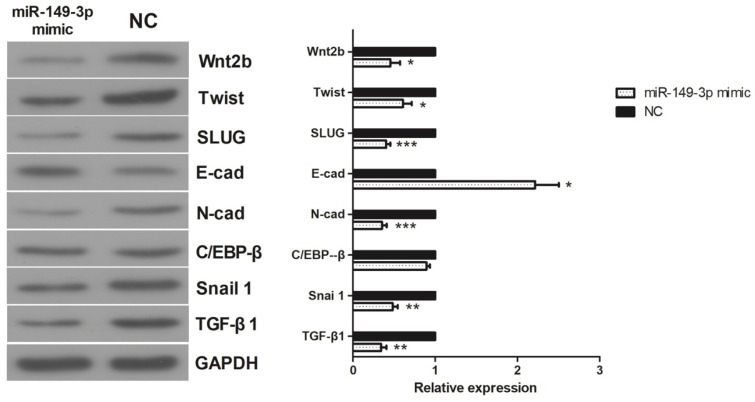
Effects of miR-149-3p on the expression of TGF-β1, C/EBPβ, Snail 1, Wnt2b, Slug, Twist, E-cadherin, and N-cadherin in vitro
The activities of TGF-β1, C/EBPβ, Snail 1, Wnt2b, Slug, Twist, E-cadherin, and N-cadherin were assessed in HCT116 CRC cells. As shown in Figure 5, in the miR-149-3p mimic-treated group, the expression of E-cadherin was increased, whereas the expression of N-cadherin, Snail, Slug, TGF-β1, Wnt2b, Twist and C/EBPβ was decreased. These results suggest that the epithelial-mesenchymal transition (EMT) may play a crucial role in the regulation of miR-149-3p in the metastasis of CRC.
Figure 5.
Western Blot results of TGF-β1, C/EBP β, Snail 1, Wnt 2b, Slug, Twist, E-cadherin and N-cadherin. In miR-149-3p mimics treated group, the expression of E-cadherin was significantly increased; the expression of N-cadherin, Snail, Slug, TGF-β1, Wnt2b and Twist were significantly decreased; the expression of C/EBPβ had no significant difference. *: p<0.05, **: p<0.01, ***: p<0.001.
Discussion
Although extensive studies have elucidated the genetic and epigenetic regulation of CRC oncogenesis, there is still an urgent need to improve the diagnostic and treatment approaches for CRC because of its tremendous heterogeneity and high mortality rates. To this end, establishing effective markers and targets to better understand the underlying mechanisms is of great importance.
In gastric cancer, oral squamous cell carcinoma, osteosarcoma, renal cancer, hepatocellular carcinoma, breast cancer, prostate cancer and thyroid cancer, high expressions of HOXA11-AS have been found; furthermore, the upregulated expression of HOXA11-AS were associated with tumor progression and metastasis 12-14, 17-26. Here, we observed overexpression of HOXA11-AS in CRC tissues, and high expression of HOXA11-AS was positively correlated with more aggressive clinicopathological parameters for the first time. Our results suggested that high HOXA11-AS expression in tumor tissue is an independent risk factor for HCC patients after radical resection. Moreover, a higher protein level of HOXA11-AS was correlated with poorer overall survival in CRC patients. Additionally, univariate analyses showed that higher HOXA11-AS expression, a larger tumor size, more frequent metastasis/recurrence, and an advanced TNM stage were correlated with an increased risk of death in CRC patients. Thus, HOXA11-AS appears to be a potent prognostic marker of survival in CRC.
miR-149-3p potentially plays a key role in the proliferation and metastasis of diverse neoplasms. This miRNA is reportedly upregulated in the plasma of melanoma patients compared with healthy controls 23 and is highly expressed in non-small cell lung cancer tissue 24. Meanwhile, it is downregulated in gastric cancer cells and tissue 20. miR-149-3p was found to enhance prostate cancer cell motility and invasiveness by efficiently downregulating disabled homolog 2-interacting protein, facilitating activation of nuclear factor kappa B signaling, and promoting expression of pro-inflammatory and pro-angiogenic factors 27. However, miR-149-3p was found to be a tumor inhibitor in most previous studies. For example, it reportedly inhibits pancreatic cancer by regulating the Akt1 signaling pathway 28, inhibits gastric cancer via Wnt-1 signaling or the ZEB1-AS1/miR-149-3p axis 29, 30, inhibits the proliferation, migration, and invasion of bladder cancer by targeting S100 calcium-binding protein A4 31, and inhibits renal cell carcinoma by targeting forkhead box protein M1 32. To date, there is no report about the role of miR-149-3p in CRC. In this investigation, we found that the expression of miR-149-3p was significantly decreased (p<0.0001) in the tissue of CRC patients with liver metastasis. Furthermore, the expression of miR-149-3p was negatively correlated with that of HOXA11-AS. Above results inferred that miR-149-3p may inhibits the metastasis of CRC via binding to HOXA11-AS.
In this investigation, we verified that miR-149-3p directly binding to HOXA11-AS in dual-luciferase reporter assay. Moreover, the negative correlation between miR-149-3p and HOXA11-AS was vevified in vitro. The inhibition of miR-149-3p to CRC metastasis was verified in scratch, migration and invasion assays.
EMT is a crucial process in tumor progression that affects the key steps of morphogenesis by converting epithelial cells into cells with mesenchymal attributes 33. EMT is primarily known as a phenotypic transformation process during embryonic development, wound healing, and tissue remodeling 34. Diverse factors, such as Snail, E-cadherin, cyclin D, TGF-β, nodal, N-cadherin, cadherins 6, 7, and 11, epidermal growth factor, TGF-βR, Notch, ErbB, and hepatocyte growth factor/scatter factor, are involved in regulating EMT via the integrin-linked kinase, Wnt, phosphoinositide 3-kinase, and Src signaling pathways 35-44. Moreover, various proteins, such as p38-interacting protein, p38 mitogen-activated protein kinase, and 4.1R, ezrin, radixin, moesin (FERM) protein.
FERM protein, metalloproteases, and extracellular signal-related kinases are involved in the process of EMT 40, 45-47. The significance of EMT in human cancers was revealed by studies that provided morphological and imaging evidence 48, 49. Accumulating evidence has shown that EMT plays a key role in tumor invasion and metastasis 34, 50. The hallmarks of EMT include decreased expression of E-cadherin, as well as increased expression of N-cadherin, Snail, Slug, Twist, TGF-β1, Wnt2b, and C/EBPβ 50-52. In this investigation, overexpression of miR-149-3p led to an increase in E-cadherin expression and decreases in N-cadherin, Snail, Slug, Twist, TGF-β1, Wnt2b, and C/EBPβ expression. Thus, miR-149-3p may inhibit EMT by targeting the abovementioned signaling pathways.
This study demonstrates that high expression of HOXA11-AS is correlated with CRC progression and poor prognosis and may promote metastasis via EMT by modulating miR-149-3p.
Acknowledgments
This work was supported by grants from Natural Science Foundation of China (81401874), Natural Science Foundation of Zhejiang Province (LY20H160033, LQ20H100002), Medical Scientific Research foundation of Zhejiang Province (2017170661), Program of Science and Technology of Zhejiang Province (LGF18H030001) and Ethicon Excellence in Surgery Grant of China Bethune Charitable Foundation (HZB-20181119-1). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- CRC
colorectal cancer
- lncRNA
long non-coding RNA
- miRNAs
micro RNAs
- CeRNAs
competing endogenous RNAs
- HOXA11-AS
HOMEOBOX A11 antisense RNA
- EMT
epithelial mesenchymal transition
- qRT-PCR
Quantitative real-time polymerase chain reaction
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Feng RM, Zong YN, Cao SM. et al. Current cancer situation in China: good or bad news from the 2018 Global Cancer Statistics? Cancer communications. 2019;39(1):22. doi: 10.1186/s40880-019-0368-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nature medicine. 2006;12(8):895–904. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 4.Guo Y, Bao Y, Yang W. Regulatory miRNAs in Colorectal Carcinogenesis and Metastasis. International journal of molecular sciences. 2017. 18(4) [DOI] [PMC free article] [PubMed]
- 5.Xue JY, Huang C, Wang W. et al. HOXA11-AS: a novel regulator in human cancer proliferation and metastasis. OncoTargets and therapy. 2018;11:4387–93. doi: 10.2147/OTT.S166961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ji Q, Zhang L, Liu X. et al. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. British journal of cancer. 2014;111(4):736–48. doi: 10.1038/bjc.2014.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Q, Zhang J, Liu Y. et al. A novel cell cycle-associated lncRNA, HOXA11-AS, is transcribed from the 5-prime end of the HOXA transcript and is a biomarker of progression in glioma. Cancer letters. 2016;373(2):251–9. doi: 10.1016/j.canlet.2016.01.039. [DOI] [PubMed] [Google Scholar]
- 8.Chen J, Fu Z, Ji C. et al. Systematic gene microarray analysis of the lncRNA expression profiles in human uterine cervix carcinoma. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2015;72:83–90. doi: 10.1016/j.biopha.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 9.Richards EJ, Permuth-Wey J, Li Y. et al. A functional variant in HOXA11-AS, a novel long non-coding RNA, inhibits the oncogenic phenotype of epithelial ovarian cancer. Oncotarget. 2015;6(33):34745–57. doi: 10.18632/oncotarget.5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang J, Lv J, Liu Z. Identification of stage-specific biomarkers in lung adenocarcinoma based on RNA-seq data. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36(8):6391–9. doi: 10.1007/s13277-015-3327-0. [DOI] [PubMed] [Google Scholar]
- 11.Li T, Xu C, Cai B. et al. Expression and clinicopathological significance of the lncRNA HOXA11-AS in colorectal cancer. Oncology letters. 2016;12(5):4155–60. doi: 10.3892/ol.2016.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhan M, He K, Xiao J. et al. LncRNA HOXA11-AS promotes hepatocellular carcinoma progression by repressing miR-214-3p. Journal of cellular and molecular medicine. 2018;22(8):3758–3767. doi: 10.1111/jcmm.13633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Z, Chen Z, Fan R. et al. Over-expressed long noncoding RNA HOXA11-AS promotes cell cycle progression and metastasis in gastric cancer. Molecular cancer. 2017;16(1):82. doi: 10.1186/s12943-017-0651-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li W, Jia G, Qu Y. et al. Long Non-Coding RNA (LncRNA) HOXA11-AS Promotes Breast Cancer Invasion and Metastasis by Regulating Epithelial-Mesenchymal Transition. Medical science monitor: international medical journal of experimental and clinical research. 2017;23:3393–3403. doi: 10.12659/MSM.904892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen D, Sun Q, Zhang L. et al. The lncRNA HOXA11-AS functions as a competing endogenous RNA to regulate PADI2 expression by sponging miR-125a-5p in liver metastasis of colorectal cancer. Oncotarget. 2017;8(41):70642–52. doi: 10.18632/oncotarget.19956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Samir N, Matboli M, El-Tayeb H. et al. Competing endogenous RNA network crosstalk reveals novel molecular markers in colorectal cancer. Journal of cellular biochemistry. 2018;119(8):6869–81. doi: 10.1002/jcb.26884. [DOI] [PubMed] [Google Scholar]
- 17.Sun M, Nie F, Wang Y. et al. LncRNA HOXA11-AS Promotes Proliferation and Invasion of Gastric Cancer by Scaffolding the Chromatin Modification Factors PRC2, LSD1, and DNMT1. Cancer research. 2016;76(21):6299–310. doi: 10.1158/0008-5472.CAN-16-0356. [DOI] [PubMed] [Google Scholar]
- 18.Li B, Wang W, Miao S. et al. HOXA11-AS promotes the progression of oral squamous cell carcinoma by targeting the miR-518a-3p/PDK1 axis. Cancer cell international. 2019;19:140. doi: 10.1186/s12935-019-0838-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao K, Fang Y, Wang H. et al. The lncRNA HOXA11-AS regulates Rab3D expression by sponging miR-125a-5p promoting metastasis of osteosarcoma. Cancer management and research. 2019;11:4505–18. doi: 10.2147/CMAR.S196025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang FQ, Zhang JQ, Jin JJ. et al. HOXA11-AS promotes the growth and invasion of renal cancer by sponging miR-146b-5p to upregulate MMP16 expression. Journal of cellular physiology. 2018;233(12):9611–9. doi: 10.1002/jcp.26864. [DOI] [PubMed] [Google Scholar]
- 21.Wang S, Zhang S, He Y. et al. HOXA11-AS regulates JAK-STAT pathway by miR-15a-3p/STAT3 axis to promote the growth and metastasis in liver cancer. Journal of cellular biochemistry. 2019;120(9):15941–51. doi: 10.1002/jcb.28871. [DOI] [PubMed] [Google Scholar]
- 22.Niu X, Yang B, Liu F. et al. LncRNA HOXA11-AS promotes OSCC progression by sponging miR-98-5p to upregulate YBX2 expression. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2020;121:109623. doi: 10.1016/j.biopha.2019.109623. [DOI] [PubMed] [Google Scholar]
- 23.Yu J, Hong JF, Kang J. et al. Promotion of LncRNA HOXA11-AS on the proliferation of hepatocellular carcinoma by regulating the expression of LATS1. European review for medical and pharmacological sciences. 2017;21(15):3402–11. [PubMed] [Google Scholar]
- 24.Xing Z, Li S, Liu Z. et al. CTCF-induced upregulation of HOXA11-AS facilitates cell proliferation and migration by targeting miR-518b/ACTN4 axis in prostate cancer. The Prostate. 2020;80(5):388–98. doi: 10.1002/pros.23953. [DOI] [PubMed] [Google Scholar]
- 25.Yin X, Zhang J, Li C. et al. LncRNA HOXA11-AS accumulation-induced microRNA-761 downregulation regulates cell growth by targeting TRIM29 in papillary thyroid cancer. American journal of translational research. 2019;11(11):6826–37. [PMC free article] [PubMed] [Google Scholar]
- 26.Guo T, Yuan X, Liu DF. et al. LncRNA HOXA11-AS promotes migration and invasion through modulating miR-148a/WNT1/beta-catenin pathway in gastric cancer. Neoplasma. 2020;67(3):492–500. doi: 10.4149/neo_2020_190722N653. [DOI] [PubMed] [Google Scholar]
- 27.Bellazzo A, Di Minin G, Valentino E. et al. Cell-autonomous and cell non-autonomous downregulation of tumor suppressor DAB2IP by microRNA-149-3p promotes aggressiveness of cancer cells. Cell death and differentiation. 2018;25(7):1224–38. doi: 10.1038/s41418-018-0088-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Si L, Xu L, Yin L. et al. Potent effects of dioscin against pancreatic cancer via miR-149-3P-mediated inhibition of the Akt1 signalling pathway. British journal of pharmacology. 2017;174(7):553–68. doi: 10.1111/bph.13718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao D, Jia Z, You L. et al. 18beta-glycyrrhetinic acid suppresses gastric cancer by activation of miR-149-3p-Wnt-1 signaling. Oncotarget. 2016;7(44):71960–73. doi: 10.18632/oncotarget.12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma MH, An JX, Zhang C. et al. ZEB1-AS1 initiates a miRNA-mediated ceRNA network to facilitate gastric cancer progression. Cancer cell international. 2019;19:27. doi: 10.1186/s12935-019-0742-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang D, Du G, Xu A. et al. Expression of miR-149-3p inhibits proliferation, migration, and invasion of bladder cancer by targeting S100A4. American journal of cancer research. 2017;7(11):2209–19. [PMC free article] [PubMed] [Google Scholar]
- 32.Okato A, Arai T, Yamada Y. et al. Dual Strands of Pre-miR-149 Inhibit Cancer Cell Migration and Invasion through Targeting FOXM1 in Renal Cell Carcinoma. International journal of molecular sciences. 2017;18(9):1969. doi: 10.3390/ijms18091969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koehn BH, Schoenberger SP. Tumor immunotherapy: making an immortal army. Nature medicine. 2009;15(7):731–2. doi: 10.1038/nm0709-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thiery JP, Acloque H, Huang RY. et al. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 35.Nakagawa S, Takeichi M. Neural crest cell-cell adhesion controlled by sequential and subpopulation-specific expression of novel cadherins. Development. 1995;121(5):1321–32. doi: 10.1242/dev.121.5.1321. [DOI] [PubMed] [Google Scholar]
- 36.Vallin J, Girault JM, Thiery JP. et al. Xenopus cadherin-11 is expressed in different populations of migrating neural crest cells. Mechanisms of development. 1998;75(1-2):171–4. doi: 10.1016/s0925-4773(98)00099-9. [DOI] [PubMed] [Google Scholar]
- 37.Smolen GA, Schott BJ, Stewart RA. et al. A Rap GTPase interactor, RADIL, mediates migration of neural crest precursors. Genes & development. 2007;21(17):2131–6. doi: 10.1101/gad.1561507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carver EA, Jiang R, Lan Y. et al. The mouse snail gene encodes a key regulator of the epithelial-mesenchymal transition. Molecular and cellular biology. 2001;21(23):8184–8. doi: 10.1128/MCB.21.23.8184-8188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vega S, Morales AV, Ocana OH. et al. Snail blocks the cell cycle and confers resistance to cell death. Genes & development. 2004;18(10):1131–43. doi: 10.1101/gad.294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahmed N, Maines-Bandiera S, Quinn MA. et al. Molecular pathways regulating EGF-induced epithelio-mesenchymal transition in human ovarian surface epithelium. American journal of physiology Cell physiology. 2006;290(6):C1532–42. doi: 10.1152/ajpcell.00478.2005. [DOI] [PubMed] [Google Scholar]
- 41.Mercado-Pimentel ME, Runyan RB. Multiple transforming growth factor-beta isoforms and receptors function during epithelial-mesenchymal cell transformation in the embryonic heart. Cells, tissues, organs. 2007;185(1-3):146–56. doi: 10.1159/000101315. [DOI] [PubMed] [Google Scholar]
- 42.Timmerman LA, Grego-Bessa J, Raya A. et al. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes & development. 2004;18(1):99–115. doi: 10.1101/gad.276304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rivera-Feliciano J, Lee KH, Kong SW. et al. Development of heart valves requires Gata4 expression in endothelial-derived cells. Development. 2006;133(18):3607–18. doi: 10.1242/dev.02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maina F, Pante G, Helmbacher F. et al. Coupling Met to specific pathways results in distinct developmental outcomes. Molecular cell. 2001;7(6):1293–306. doi: 10.1016/s1097-2765(01)00261-1. [DOI] [PubMed] [Google Scholar]
- 45.Zohn IE, Li Y, Skolnik EY. et al. p38 and a p38-interacting protein are critical for downregulation of E-cadherin during mouse gastrulation. Cell. 2006;125(5):957–69. doi: 10.1016/j.cell.2006.03.048. [DOI] [PubMed] [Google Scholar]
- 46.Hirano M, Hashimoto S, Yonemura S. et al. EPB41L5 functions to post-transcriptionally regulate cadherin and integrin during epithelial-mesenchymal transition. The Journal of cell biology. 2008;182(6):1217–30. doi: 10.1083/jcb.200712086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee JD, Silva-Gagliardi NF, Tepass U. et al. The FERM protein Epb4.1l5 is required for organization of the neural plate and for the epithelial-mesenchymal transition at the primitive streak of the mouse embryo. Development. 2007;134(11):2007–16. doi: 10.1242/dev.000885. [DOI] [PubMed] [Google Scholar]
- 48.Prall F. Tumour budding in colorectal carcinoma. Histopathology. 2007;50(1):151–62. doi: 10.1111/j.1365-2559.2006.02551.x. [DOI] [PubMed] [Google Scholar]
- 49.Wyckoff JB, Wang Y, Lin EY. et al. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer research. 2007;67(6):2649–56. doi: 10.1158/0008-5472.CAN-06-1823. [DOI] [PubMed] [Google Scholar]
- 50.Matos ML, Lapyckyj L, Rosso M. et al. Identification of a Novel Human E-Cadherin Splice Variant and Assessment of Its Effects Upon EMT-Related Events. Journal of cellular physiology. 2017;232(6):1368–86. doi: 10.1002/jcp.25622. [DOI] [PubMed] [Google Scholar]
- 51.Zhou P, Wang C, Hu Z. et al. Genistein induces apoptosis of colon cancer cells by reversal of epithelial-to-mesenchymal via a Notch1/NF-kappaB/slug/E-cadherin pathway. BMC cancer. 2017;17(1):813. doi: 10.1186/s12885-017-3829-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Z, Sun J, Li T. et al. Iron chelator-induced up-regulation of Ndrg1 inhibits proliferation and EMT process by targeting Wnt/beta-catenin pathway in colon cancer cells. Biochemical and biophysical research communications. 2018;506(1):114–21. doi: 10.1016/j.bbrc.2018.10.054. [DOI] [PubMed] [Google Scholar]