Abstract
Autozygosity is associated with an increased risk of genetic rare disease, thus being a relevant factor for clinical genetic studies. More than 2400 exome sequencing data sets were analyzed and screened for autozygosity on the basis of detection of >1 Mbp runs of homozygosity (ROHs). A model was built to predict if an individual is likely to be a consanguineous offspring (accuracy, 98%), and probability of consanguinity ranges were established according to the total ROH size. Application of the model resulted in the reclassification of the consanguinity status of 12% of the patients. The analysis of a subset of 79 consanguineous cases with the Rare Disease (RD)–Connect Genome-Phenome Analysis Platform, combining variant filtering and homozygosity mapping, enabled a 50% reduction in the number of candidate variants and the identification of homozygous pathogenic variants in 41 patients, with an overall diagnostic yield of 52%. The newly defined consanguinity ranges provide, for the first time, specific ROH thresholds to estimate inbreeding within a pedigree on disparate exome sequencing data, enabling confirmation or (re)classification of consanguineous status, hence increasing the efficiency of molecular diagnosis and reporting on secondary consanguinity findings, as recommended by American College of Medical Genetics and Genomics guidelines.
It is estimated that 350 million individuals worldwide experience one of approximately 7000 existing rare diseases (RDs).1 The low prevalence of each disease and the high heterogeneity and variability of clinical symptoms make diagnosis and accessibility to appropriate treatment a real challenge. As emphasized by the International Rare Disease Research Consortium, progress in this field requires identification of RDs and their causes to develop appropriate treatments,2 and as 80% of RDs are thought to have a genetic origin, particular emphasis has been placed on the rapidly expanding development of genomic technologies. The next-generation sequencing era has enabled cost-effective sequencing of RD patients' exomes or genomes, bringing these approaches into diagnostics.3 However, the interpretation of the genome is still a real challenge for molecular geneticists, and innovative bioinformatics solutions combining genomic and clinical data are crucial for reaching a diagnosis.4,5 Data sharing and analysis platforms, such as the RD-Connect Genome-Phenome Analysis Platform (GPAP; https://platform.rd-connect.eu, registration required, last accessed May 9, 2020),5,6 have emerged to provide methods and standardized analyses of phenotypic and (gen)omic data to facilitate the mutation detection processes.
Autozygosity, as a result of consanguineous mating, has long been known to be a risk factor for RDs of genetic origin through a variety of effects, such as reduction in genetic variation, increased frequency of homozygous genotypes for deleterious alleles, and lower population viability.7 The deleterious consequences in populations with higher prevalence of consanguinity, due to physical or cultural isolation, have been widely reported (reviewed in Fareed and Afzal8), and many rare recessive disease genes have been identified by homozygosity mapping in which large regions flanking the disease-causing variant are expected to be identical by descent in affected individuals whose parents are related.9,10 In addition, about one-third of autosomal recessive rare disorders occurring in families with no known consanguinity are caused by homozygous variants located in regions likely identical by descent.11
Next-generation sequencing technologies allow precise detection of genomic regions where a reduction in heterozygosity is evident and offer the opportunity to estimate autozygosity at the exome and genome level.12 Different software, such as HomozygosityMapper,13 PLINK,14 HomSI,15 and H3M2,16 has been developed for the detection of runs of homozygosity (ROHs) from exome and genome sequencing data.12,17 The identification of autozygous regions through the detection of contiguous lengths of homozygous segments of the genome where the two haplotypes inherited are identical has been applied in multiple population genomic studies (reviewed in Ceballos et al18). Different homozygosity mapping software presents specific advantages and limitations, as reviewed in Howrigan et al19 and Oliveira et al20. Part of the limitations encompass the use of exome sequencing (ES), which by definition fragments genomic data, thus interfering with the identification of homozygous regions. Recently, optimized protocols for homozygosity mapping based on ES and using PLINK software have been published, with promising results.10,21,22
Herein, we report on the integration of genomic analysis and autozygosity assessment on the basis of the detection of long [>1 megabase (Mb)] ROHs in >2400 ES data sets from the RD-Connect GPAP. A subset of these measurements was used to generate a model to determine the likelihood of an individual being the offspring of consanguineous parents. To assess this approach, individuals were classified according to these consanguinity ranges, and a subset of consanguineous offspring was subsequently analyzed in the RD-Connect GPAP, applying ROH-specific region filtering to identify the disease-causing variant(s). To our knowledge, this is the first study providing thresholds based on total ROH length to estimate consanguinity from ES data regardless of sequencing center and protocol used and the largest study attempting to combine ES and ROH detection approaches to identify genetic defects of different types of RDs, reaching a diagnostic yield of 52% in consanguineous probands.
The consanguinity ranges defined herein for ES data will facilitate inbreeding estimation in clinical laboratories and enable confirmation, or (re)classification, of consanguineous cases, hence increasing the efficiency of molecular diagnosis and reporting on secondary consanguinity findings, as recommended by American College of Medical Genetics and Genomics (ACMG) guidelines.23
Materials and Methods
Subjects
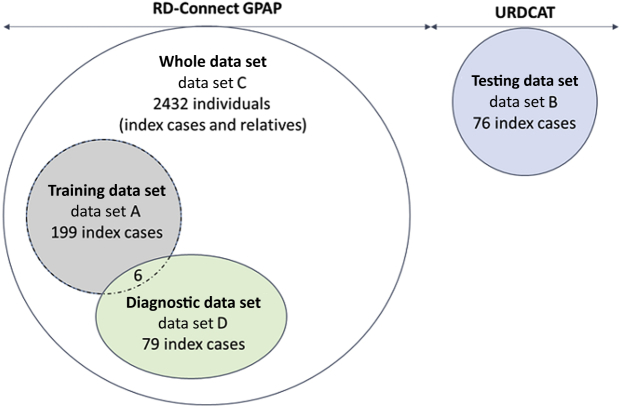
This study includes clinical and genomic data from 2432 individuals collated within the RD-Connect GPAP (data set C) (Figure 1) and 76 individuals from an independent project, the Undiagnosed Rare Disease Program of Catalonia (URDCAT; https://www.urdcat.cat/home, last accessed May 9, 2020) (data set B) (Figure 1). Clinical information concerning reported consanguinity and ethnicity classification, according to the Ontology of Precision Medicine and Investigation (OPMI; http://www.ontobee.org/ontology/OPMI, last accessed December 1, 2019) database, were obtained for each individual, where available. As required by the RD-Connect and URDCAT adherence agreements, patient consent allowing the sharing of pseudonymized clinical information with international collaborators and researchers was obtained for all individuals included in this study. This study adheres to the principles set out in the Declaration of Helsinki.
Figure 1.
Description of the different data sets used in this study. Data sets used to train, test, and evaluate the model described in this study. Data set A, referred to as training data set, includes 199 index cases for which presence or absence of consanguinity was determined by kinship analysis and was used to define the logistic regression model. Data set B, referred to as testing data set, includes 76 index cases from the Undiagnosed Rare Disease Program of Catalonia (URDCAT) for which the presence or absence of consanguinity status was determined by kinship analysis and was used to test our model. Data set C, referred to as whole data set, includes 2432 individuals (index cases and relatives) from the Rare Disease (RD)–Connect Genome-Phenome Analysis Platform (GPAP) to which our model was applied. Data set D, referred to as diagnostic data set, includes 79 index cases in which genomic data were combined with run of homozygosity results to identify the pathogenic variants responsible for different types of rare disease.
Data Sets Used to Establish the Consanguinity Model
Different data sets and subsets were used to train, test, and apply the model described in this study (Figure 1). Data set A, referred to as training data set, includes 199 index cases for which presence/absence of consanguinity was determined by kinship analysis and was used to define the logistic regression model. Data set B, referred to as testing data set, includes 76 index cases from URDCAT for which presence/absence of consanguinity status was determined by kinship analysis and was used to test our model. Data set C, referred to as whole data set, includes 2432 individuals (index cases and relatives) from the RD-Connect GPAP to which our model was applied. Finally, data set D, referred to as diagnostic data set, includes 79 index cases from data set C in which genomic data were combined with ROH results to identify the pathogenic variants responsible for different types of RDs.
Genomic Data Processing
In total, ES data derived from 2432 individuals from RD-Connect GPAP, sequenced using six different exome capture kit protocols [Nextera Rapid Exome (Illumina, San Diego, CA), Nimblegen SeqCap EZ MedExome (Roche, Basel, Switzerland), SureSelect version 5 (Agilent, Santa Clara, CA), Broad Custom Exome (Broad Institute, Cambridge, MA), Nextera Expanded Exome (Illumina), and Illumina TruSeq Expanded Exome], with target capture sizes ranging from 37 Mb to 62 Mb, and 76 individuals from URDCAT, sequenced using five different exome capture kit protocols [Nimblegen SeqCapEZ Exome (Roche) and Agilent SureSelect version 3, version 4, version 5, and version 6 (Agilent)], with target capture sizes ranging from 50 to 64 Mb, were included in the study. In all cases, sequencing reads were processed using the RD-Connect GPAP standardized analysis pipeline based on GATK3.6 best practices, as described in Laurie et al,24 and the resultant variant calls were used for ROH detection and made available for analysis through the RD-Connect GPAP.
Detection of Homozygous Regions
Quality filtering of the processed data (VCF files) was performed to minimize the impact of low-quality variant calls resulting from sequencing artifacts, misalignment, or low coverage. Insertions/deletions were discounted, and only single-nucleotide variants covered by a minimum read depth of 10 reads and a genotype quality of at least 90 were included in the analysis. For each individual, ROHs were identified using PLINK version 1.9014-homozyg option, applying the optimal parameters defined by Kancheva et al,21. This method is designed for whole exome sequencing data and assumes intronic and intergenic regions to be homozygous when surrounded by two detected homozygous coding regions.21 PLINK was run for each sample to identify ROH size with a minimum length of 1 Mb to exclude common shorter ROHs. Plots were generated using RStudio version 1.0.143 (RStudio, Boston, MA).
Establishing Consanguinity Ranges for ES Data
For this study, consanguineous individuals were defined as being the offspring of third degree (equivalent to being first cousins) or more closely related parents (ie, having a kinship coefficient >0.045).25 To build a logistic regression model to predict if an individual is likely to be a consanguineous offspring according to the total ROH size, we first identified a subset of samples for which presence or absence of consanguinity had been clinically reported and subsequently experimentally confirmed by trio kinship analysis using -relatedness2 from vcftools. In total, 98 index cases were confirmed as consanguineous offspring (kinship coefficient > 0.045) and 101 were confirmed as nonconsanguineous (kinship coefficient < 0.045). These cases (199 in total) were included in data set A (Figure 1), referred as training data set. Two thirds of data set A, 62 consanguineous cases and 66 nonconsanguineous cases, was used to train a logistic model, and the remaining 71 samples were used to define four consanguinity ranges: nonconsanguineous (consanguinity probability < 0.05), uncertain (0.05 < consanguinity probability < 0.5), probably consanguineous (0.5 < consanguinity probability < 0.95), and consanguineous (consanguinity probability > 0.95). The accuracy of the model is 98%, with a P value of 0.05.
Precision of the Consanguinity Model
The specificity, sensitivity, and accuracy of the model for predicting consanguinity status was evaluated using data set B, referred to as testing data set (Figure 1), which includes 76 index cases from URDCAT for which presence or absence of consanguinity status was determined by kinship analysis.
Genomic Data Analysis and Interpretation
Genomic data were analyzed using the RD-Connect GPAP, which enables the combination of variant filtering and homozygosity mapping. Identification of putative disease-causing variants in consanguineous or probably consanguineous individuals was achieved by applying the following filters: homozygous variant, minimum depth of coverage of 10, variants classified as having a high (disruptive) or moderate (amino acid change) impact on the protein, according to SnpEff, and observed population allele frequency <0.02, according to gnomAD,26 ExAC,27 and 1000 Genomes Project28 databases. When no interesting variants were identified, other inheritances and genotypes were assessed (eg, autosomal recessive inheritance associated with compound heterozygous variants and X-linked inheritance). Candidate variants were classified following the ACMG standards and guidelines for interpretation29 and proposed to the corresponding submitter for confirmation of molecular diagnosis.
Statistical Analysis
A paired samples Wilcoxon test was performed for the comparison of the number of rare homozygous variant with or without applying a 1-Mb ROH filter. Statistical significance was determined as P < 0.05.
Results
Consanguinity Model and Ranges
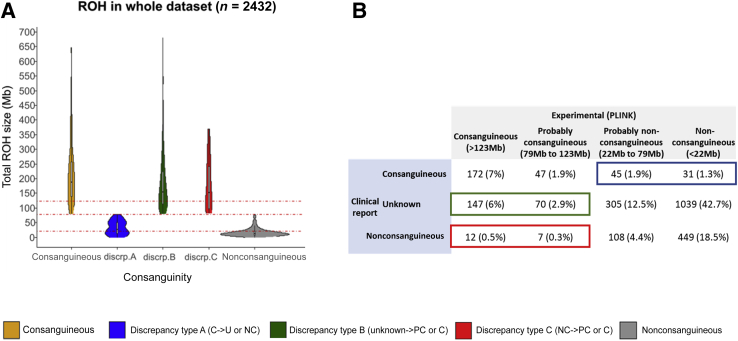
To predict if an individual is likely to be a consanguineous offspring, according to the total ROH size identified from ES data, we analyzed ROH results from data set A, the training data set (Figure 1 and Figure 2A). Consanguinity was defined as unions contracted between individuals biologically related as first cousins (equivalent to third-degree relationship) or closer. We built a logistic regression model to define the probability of consanguinity according to the total ROH size identified by ROH analysis (Figure 2B). We used this model and probabilities of being a consanguineous offspring of 5%, 50%, and 95% to define four consanguinity ranges: nonconsanguineous (total ROH size < 22 Mb), uncertain (22 Mb < total ROH size < 79 Mb), probably consanguineous (79 Mb < total ROH size < 123 Mb), and consanguineous (total ROH size > 123 Mb) (Table 1). If we consider the percentage of the genome that is homozygous (Froh) assuming a total autosomal genomic length of 2691 Mb for GRCh37/hg19 (https://www.ncbi.nlm.nih.gov/assembly, accession number GRCh38.p13), consanguinity ranges can be extrapolated as follows: nonconsanguineous (Froh < 0.8% of the genome), uncertain (0.8% < Froh < 2.9%), probably consanguineous (2.9% < Froh < 4.6%), and consanguineous (total ROH size > 4.6%) (Table 1). The robustness of this approach was tested using an independent data set B, defined as testing data set (Figure 1). The sensitivity (true-positive rate), specificity (true-negative rate), and accuracy (degree of closeness to a true value) of the test were 80%, 99%, and 97%, respectively (Figure 2B and Supplemental Table S1). According to the established thresholds, 194 of the 199 cases included in the training data set were correctly classified, and five nonconsanguineous cases were incorrectly classified as (probably) consanguineous (Supplemental Table S2). Two of the five cases (C4 and C5) having a total ROH size of 82.4 and 80 Mb, respectively, were close to the defined threshold of 79 Mb. All cases presented an ES median coverage between 57 and 94.
Figure 2.
Consanguinity model and ranges defined using the training data set. A: Distribution of the total run of homozygosity (ROH) size across samples from the training data set (199 individuals) for which presence or absence of consanguinity was determined by kinship analysis. B: Linear regression model used to define the probability of consanguinity on the basis of total ROH size per sample of the training data set. Sensitivity, specificity, and accuracy of the model have been calculated by analyzing the Undiagnosed Rare Disease Program of Catalonia testing data set (Supplemental Table S1). C: Total ROH size distribution of the training data set, categorized by the different ethnicities reported. D: Total ROH size distribution of the training data set, categorized by exome capture kits. Colors indicate the presence (orange) or absence (gray) of consanguinity for an individual, as determined by kinship analysis of the parents (third-degree or closer relation classified as consanguineous); and dotted red lines, the defined consanguinity thresholds [22, 79, and 123 megabases (Mb)]. Points C1 to C5 indicate the five nonconsanguineous cases incorrectly classified as (probably) consanguineous (Supplemental Table S2). n = 98 consanguineous samples (A); n = 101 nonconsanguineous samples (A).
Table 1.
Consanguinity Classification, According to the Model Described in This Study
| ROH interval, Mb | Froh, (%)∗ | Experimental offspring consanguinity classification |
|---|---|---|
| >123 | >4.6 | Consanguineous |
| 79–123 | 2.9–4-6 | Probably consanguineous |
| 22–79 | 0.8–2.9 | Uncertain |
| <22 | <0.8 | Nonconsanguineous |
Mb, megabase; ROH, run of homozygosity.
Froh is defined as the percentage of the genome that is homozygous compared with the total autosomal genomic length (approximately 2691 Mb for GRCh37/hg19).
Ethnicity Effect on Total ROH Size Detection
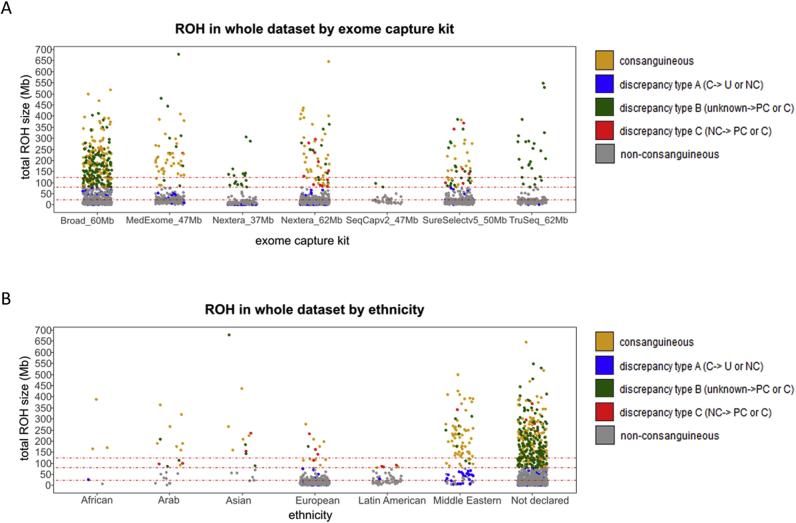
In some populations in North Africa, West Asia, or South India, consanguineous marriages are culturally and socially favored. This fact, together with existing consanguinity in isolated populations, results in almost 10% of the world population either being married to a biological relative or being a consanguineous offspring.8 To know to which extent population origin may affect the consanguinity ranges defined above, total ROH size per individual was assessed across the different ethnicities reported in the training data set (Figure 2C). When analyzing the nonconsanguineous cohort across different ethnicities, total ROH size medians were, as expected, within the nonconsanguineous range for European, Latin American, and Middle Eastern individuals (Figure 2C). The median total ROH size was above the nonconsanguineous range in two different ethnicities: Arabs and Asians. Indeed, two of the incorrectly classified cases mentioned above (C2 and C3) (Supplemental Table S2) are of Asian origin. However, because of the scarce number of individuals in each of these nonconsanguineous data sets (Arabs = 2, and Asians = 3) (Supplemental Table S3), results were not confirmed statistically. Similar tendencies were observed when analyzing the mean length of the homozygous segments by ethnicity (Supplemental Table S3).
Effect of Exome Capture Kit on Detected Total ROH Size
The algorithm used herein to identify ROH regions uses a sliding window that scans along single-nucleotide variant data to detect nonheterozygous stretches. ES experiments target the evaluation of specific regions of the genome that differ between exome capture kits. It was hypothesized that the regions captured in each exome capture kit might affect the total ROH size, and thus interfere with the determination of consanguinity status using the ranges identified above. Therefore, we analyzed the total ROH size across the exome capture kits from the training data set. Six different exome capture kits with target capture sizes ranging from 37 to 62 Mb were assessed (Figure 2D). When analyzing the nonconsanguineous cohort across the different exome capture kits, all total ROH size medians were, as expected, within the nonconsanguineous range for all of the kits tested. Similar results were observed when analyzing the mean length of the segments by exome capture kit (Supplemental Table S3).
Consanguinity Model Assessment
The consanguinity ranges defined in the first part of this study were applied to the whole data set (Figure 1), 2432 individuals (index cases and relatives) from the RD-Connect GPAP. Total ROH size was computed for each individual and classified according to the comparison of the consanguinity status experimentally conferred and its corresponding clinical record. Individuals were classified as consanguineous or nonconsanguineous when no discrepancies were found between clinical records and experimental conferred consanguinity status. Otherwise, three types of discrepancies were defined: discrepancy type A, when the individual was reported as consanguineous but experimentally classified as uncertain or nonconsanguineous; discrepancy type B, when the individual was reported as of unknown consanguinity but experimentally classified as probably consanguineous or consanguineous; and discrepancy type C, when the individual was reported as nonconsanguineous but experimentally classified as probably consanguineous or consanguineous (Figure 3A). According to our model, consanguinity was confirmed in 219 of 295 reported cases (74%) and enabled the reclassification of 95 individuals: 76 individuals (3.1%) from consanguineous to nonconsanguineous status (discrepancy type A) and 19 individuals (0.8%) from nonconsanguineous to consanguineous status (discrepancy type C). Moreover, the model was able to detect and classify 217 potential consanguineous cases (8.9% of the data, discrepancy type B) (Figure 3B). Individuals were also clustered by exome capture kit and ethnicity (Supplemental Figure S1). No significant differences were found among the nonconsanguineous cohort between the different exome capture kits (Supplemental Figure S1A). Nonconsanguineous Arabs, Asians, and Latin Americans showed a tendency of increased total ROH size compared with European populations (Supplemental Figure S1B).
Figure 3.
Consanguinity model assessment through the analysis of 2432 ES data sets. A: Distribution of the total ROH size across 2432 exome sequencing data sets, classified according to the comparison of the consanguinity status experimentally conferred and that of the corresponding clinical record. Individuals are classified as consanguineous (C; orange) when stated in the clinical record and conferred experimentally; discrepancy type A (blue) when stated as consanguineous in the clinical record and experimentally conferred as uncertain (U) or nonconsanguineous (NC); discrepancy type B (green) when stated as of unknown consanguinity in the clinical record (Unknown) and experimentally conferred as (probably) consanguineous (PC); and discrepancy type C (red) when stated as nonconsanguineous in the clinical record and experimentally conferred as (probably) consanguineous. B: Number of individuals within each experimentally conferred consanguinity range as a function of the consanguinity status reported in the clinical record. Red dotted lines indicate the defined consanguinity thresholds (22, 79, and 123 megabases). discrp., discrepancy type; ROH, run of homozygosity.
Uniparental Disomy Assessment
To identify complete uniparental disomy, samples with at least one ROH >30 Mb were further analyzed. A total of 22 samples from 2432 met that criterion (0.9%) (Supplemental Table S4). In eight of these samples, the corresponding runs represented >30% of the total ROH size detected in that sample. All eight individuals were classified as consanguineous or likely consanguineous, according to our model, and four were index cases affected by a rare disorder.
Application to Molecular Diagnostics Workflow
To assess the impact of proper identification of consanguinity and the usage of ROH for diagnosis, we analyzed a subset of 79 undiagnosed index cases from the whole data set, which were consanguineous or had a discrepant consanguineous status (diagnostic data set) (Figure 1).
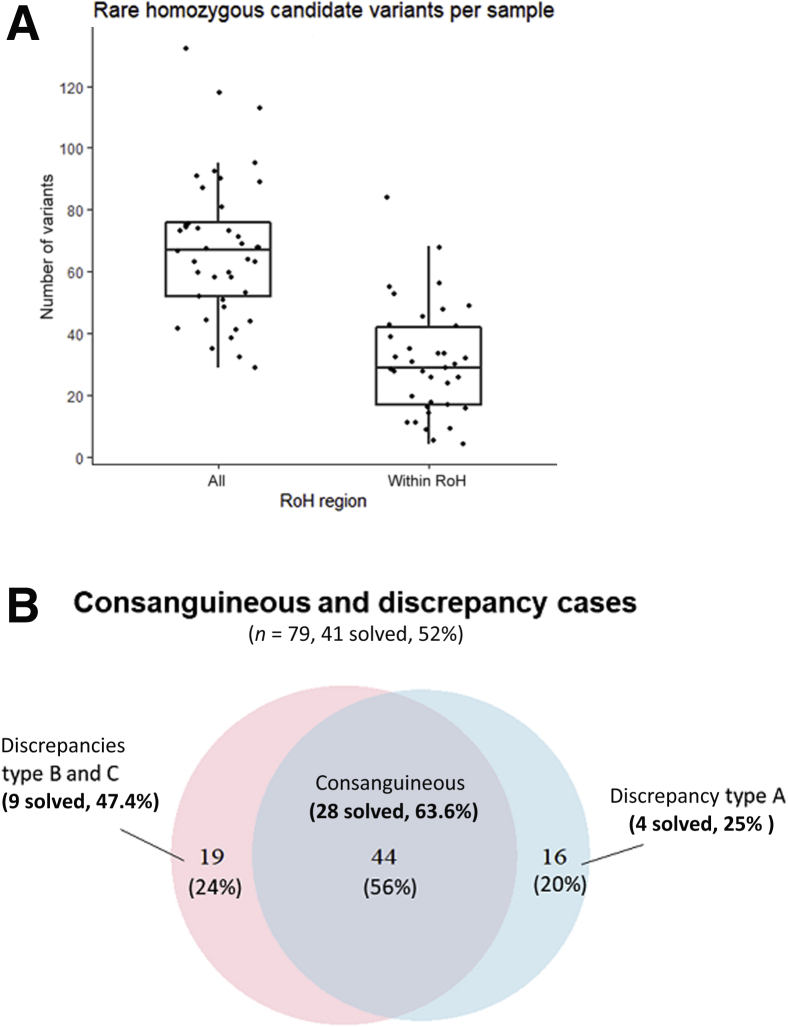
When applying our model classification to this diagnostic data set, consanguinity was confirmed in 44 of 60 reported cases (73%), and 35 individuals were reclassified: 16 individuals (46% of reclassified) from consanguineous to nonconsanguineous status (discrepancy type A), 5 individuals (14% of reclassified) from nonconsanguineous to consanguineous status (discrepancy type C), and 14 individuals (40% of reclassified) from unknown to consanguineous status (discrepancy type B). On the basis of these results, we used the RD-Connect GPAP to conduct variant filtering with or without homozygosity mapping (ROH region > 1 Mb) in all 79 cases from the diagnostic data set. The number of resulting candidate variants was counted for each individual, and results showed an overall 50% decrease of the number of variants to be further evaluated when filtering to variants within an ROH region (Figure 4A). The candidate variants for each of these 79 cases were classified according to ACMG criteria,29 and those in known disease-causing genes were reported to the clinicians having submitted the cases to the RD-Connect GPAP. The clinicians confirmed one of the variants was the cause of the disorder in 41 cases (diagnostic rate = 52%) (Figure 4B); in all cases, it was a likely pathogenic or pathogenic variant, according to ACMG criteria. In total, 28 of 44 cases (63.6%) clinically reported and classified as consanguineous by our model were solved; 9 of 19 (47.7%) only classified as consanguineous by our model (discrepancies type B and C) were solved, and 4 of 16 (25%) reported as consanguineous and classified as uncertain or nonconsanguineous by our model (discrepancy type A) were solved. Results from the 13 discrepant solved cases (discrepancy types A, B, and C) are shown (Table 2. All causative variants from which patients were experimentally classified as consanguineous or probably consanguineous (cases 1, 2, 4, 5, 8, 9, 10, 11, and 12) were found within ROH regions (>1 Mb). Patients from cases 3, 6, 7, and 13 were reported as consanguineous but not experimentally confirmed. In two of these patients (cases 7 and 13), the causative variant was not found in an ROH (Table 2).
Figure 4.
Combination of variant filtering and homozygosity mapping to identify pathogenic variants. A: Comparison of the number of rare homozygous variants identified before and after combining variant filtering with homozygosity mapping [ie, filtering to variants in run of homozygosity (ROH) regions >1 megabase in length; P = 1.179 × 10−7]. B: Distribution of the 79 cases reported as consanguineous or classified as consanguineous by our model. In blue, cases reported as consanguineous; and in red, cases experimentally classified as probably consanguineous or consanguineous. The intersection of both sections corresponds to 44 cases both reported and experimentally classified as consanguineous. Discrepancy type A, individual stated as consanguineous in the clinical record but experimentally conferred as uncertain or nonconsanguineous; discrepancy type B, when status of consanguinity is unknown in the clinical record and experimentally (probably) consanguineous; and discrepancy type C, when stated as nonconsanguineous in the clinical record but experimentally (probably) consanguineous. n = 60 (B, cases reported as consanguineous); n = 63 (B, cases experimentally classified as probably consanguineous or consanguineous); n = 44 (B, cases both reported and experimentally classified as consanguineous).
Table 2.
List of Cases from the Diagnostic Data Set in Which Reported Consanguinity Status and Experimentally Inferred Status Are Discordant
| Case | ROH_length, Mb | Exome capture kit | Ethnicity | Consanguinity status |
Gene | Disease | Variant in a 1-Mb ROH | |
|---|---|---|---|---|---|---|---|---|
| Reported | Experimental | |||||||
| 1 | 194.6 | Nextera_62 Mb | Unknown | Nonconsanguineous | Consanguineous | MLTK | Congenital fiber-type disproportion myopathy (ORPHA: 2020) | Yes |
| 2 | 124.3 | Nextera_62 Mb | Unknown | Unknown | Consanguineous | ADCK3 | Autosomal recessive ataxia due to ubiquinone deficiency (ORPHA: 139485) | Yes |
| 3 | 21.7 | SureSelectv5_50 Mb | Unknown | Consanguineous | Nonconsanguineous | MUSK | Postsynaptic congenital myasthenic syndromes (ORPHA: 98913) | Yes |
| 4 | 369.4 | SureSelectv5_50 Mb | Unknown | Nonconsanguineous | Consanguineous | MUSK | Postsynaptic congenital myasthenic syndromes (ORPHA: 98913) | Yes |
| 5 | 262.1 | MedExome_47 Mb | Unknown | Unknown | Consanguineous | POMK | Autosomal recessive limb-girdle muscular dystrophy (ORPHA: 102015) | Yes |
| 6 | 31.2 | MedExome_47 Mb | Unknown | Consanguineous | Uncertain | PTPN23 | Epileptic encephalopathy with hypomyelination and brain atrophy (PMID: 29899372) | Yes |
| 7 | 34.4 | MedExome_47 Mb | White | Consanguineous | Uncertain | ARV1 | Early-onset epileptic encephalopathy (ORPHA: 442835) | No |
| 8 | 88.9 | SureSelectv5_50 Mb | Middle Eastern | Unknown | Probably consanguineous | HACE1 | Spastic paraplegia–severe developmental delay–epilepsy syndrome (ORPHA: 464282) | Yes |
| 9 | 301.7 | MedExome_47 Mb | Middle Eastern | Unknown | Consanguineous | SYNJ1 | Early-onset epileptic encephalopathy (ORPHA: 442835) | Yes |
| 10 | 231.2 | MedExome_47 Mb | White | Nonconsanguineous | Consanguineous | PLEKHG5 | Autosomal recessive intermediate Charcot-Marie-Tooth disease type C (ORPHA: 369867) | Yes |
| 11 | 444.4 | MedExome_47 Mb | Unknown | Unknown | Consanguineous | COLQ | Synaptic congenital myasthenic syndromes (ORPHA: 98915) | Yes |
| 12 | 310.2 | MedExome_47 Mb | Middle Eastern | Unknown | Consanguineous | CRTAP | Rare disorder with pigmented sclera (ORPHA: 519296) | Yes |
| 13 | 8.6 | MedExome_47 Mb | Middle Eastern | Consanguineous | Nonconsanguineous | UPF3B | X-linked nonsyndromic intellectual disability (ORPHA: 777) | No |
Mb, megabase; ORPHA, Orphanet Ontology Code; PMID, Pubmed ID; ROH, run of homozygosity.
Discussion
Next-generation sequencing technologies, and more specifically ES, are increasingly used for diagnostics and require methods and approaches to decrease the turnaround time of mutation detection. Autozygosity is known to be associated with an increased risk of genetic RD and is thus a relevant factor to take into consideration when undertaking clinical genetic studies. In this study, we present an approach to detect consanguineous offspring from ES data and increase the efficiency of molecular diagnosis by combining variant filtering and homozygosity mapping. To demonstrate the usefulness of this approach, we have analyzed ES data from the RD-Connect GPAP.
We have established a model to determine if an individual is likely to be a consanguineous offspring, according to the total ROH size called by the PLINK software in the training data set. Our model has enabled the classification of four ranges: nonconsanguineous (total ROH size < 22 Mb or Froh < 0.8%), uncertain (22 Mb < total ROH size < 79 Mb or 0.8% < Froh < 2.9%), probably consanguineous (79 Mb < total ROH size < 123 Mb or 2.9% < Froh < 4.6%), and consanguineous (total ROH size > 123 Mb or Froh > 4.6%). When tested with an independent data set, the model showed high accuracy (97%), sensitivity (80%), and specificity (99%). The consanguineous threshold we define (Froh = 4.6%) falls into the lower end of the CI (Froh = 4.6%–8.3%) reported for a third-degree consanguineous offspring using single-nucleotide polymorphism arrays.30 This may be explained by the fact that ES has more data points (single-nucleotide polymorphisms) than single-nucleotide polymorphism arrays, thus enabling the usage of 1-Mb length as minimum ROH segment for ES21 instead of the 2 to 5 Mb recommended for single-nucleotide polymorphism arrays.30 These differences emphasize the relevance of defining specific ROH (and Froh) thresholds for each different genomic technique and the importance of having specific ranges for ES data, as defined herein. Because we did not identify any significant correlation with the number of ROHs per individual, we did not include this parameter in our model. However, size of the longest ROH and its corresponding percentage from the total ROH size per individual were checked to assess for any possible complete uniparental disomy.31 In our cohort, 8 cases (0.3%) presented an ROH >30 Mb and >30% of the total ROH size for the sample. Although those individuals were classified as (likely) consanguineous, we cannot discard the prediction could be masked by the presence of uniparental disomy or a large deletion.
As demonstrated in our study, the proposed model is independent from the sequencing facility, size, and type of the exome capture kit used to perform ES as no variations between total ROH size from nonconsanguineous individuals were observed between kits. Therefore, the thresholds established herein for identification and reporting of possible offspring consanguinity can be applied in any clinical laboratory, independent of the sequencing facility and the library approach used for ES. However, the model should be adapted if the data set to be tested is from a population in which consanguinity may be elevated because of culturally and socially favored consanguineous marriages and/or geographic isolation.8 Our data set is strongly biased toward European individuals, and similar studies still need to be performed on larger ES data sets to set specific consanguinity ranges by ethnicity as our results showed a tendency toward increased total ROH size in Arabs and Asians. Indeed, two of the cases incorrectly classified by our model as consanguineous were of Asian origin. This result emphasizes the importance of recording ethnic and/or continental origin when submitting cases for molecular testing.
The analysis of the total ROH size distribution across 2432 ES data sets from the RD-Connect GPAP enabled us to reclassify the consanguinity status of 12.8% of the patients, either through the detection of possible unstated consanguinity or by challenging the consanguinity reported in the corresponding clinical record. To highlight the utility of this approach in molecular diagnostics, we have analyzed and interpreted a subset of 79 index cases, looking for rare homozygous variants within ROH regions (>1 Mb). The analysis was done using the RD-Connect GPAP, which enables the routine combination of variant filtering and homozygosity mapping. Although this analysis focused only on variants in known disease-causing genes, this workflow may also be useful to identify new causative genes. In experimentally classified consanguineous cases, the number of candidate variants to be assessed was reduced by 50% when filtering by ROH regions. This drastic reduction of candidate variants facilitates and accelerates the identification of causative variants by clinical geneticists. After genomic analysis through the RD-Connect GPAP, causative variants were identified and confirmed in 41 cases (diagnostic rate = 52%). All the causative variants from the 37 cases (92.5%) classified as consanguineous by our model were found in an ROH region, supporting the fact that filtering data from experimentally classified consanguineous cases by ROH regions is a robust approach. Indeed, nine of these solved cases (22.5%) were not declared as consanguineous in their clinical record, which might have had misled geneticists as to which filtering approach to follow. Four additional solved cases that were stated to be consanguineous in the clinical record were not classified as consanguineous by our model, with two being uncertain. In two of the cases, the causative variants were found in an ROH region; and in the other two cases, the causative variants were not found in an ROH region. The former could be indicative of more distant identical by descent inheritance. We aimed to provide robust thresholds with available tools to assess relatedness. Consequently, to ensure proper sensitivity, consanguinity was defined for an individual as being the offspring of up to third-degree (first cousin) related parents. We are aware that more distant identical by descent might also be detectable with ROH sizes falling in the gray area of the uncertain range. Thus, when a more distant consanguinity is suspected, we also encourage genomic analyses to start by performing a combined ROH-genomic variants filtering approach. A patient's clinical diagnosis should also be taken into account before undertaking this approach, as, for example, individuals with severe developmental disorders are known to be enriched for damaging de novo variants regardless of their level of consanguinity.32
As the analytical approach described herein may lead to the discovery of a consanguineous mating between the proband's parents, laboratories are encouraged to develop a reporting policy to effectively and accurately communicate these findings, as recommended by the ACMG guidelines.23 Therefore, the inclusion in laboratory reports of the total ROH size and the corresponding experimental consanguinity classification described herein would help clinicians to correlate laboratory results with family history and cultural traditions and to investigate any concern of abuse, as recommended by the ACMG.23
In summary, we have defined a method to identify and easily report inbreeding within a pedigree from ES data, enabling confirmation or (re)classification of consanguineous status. Furthermore, we have implemented and demonstrated the usefulness of combining variant filtering and homozygosity mapping routinely in the RD-Connect GPAP to filter to variants within ROH and thus facilitate data filtering and interpretation. This method can be easily implemented systematically in a clinical diagnostic setting for data analysis and reporting, according to ACMG guidelines regarding consanguinity as a secondary finding of genomic testing.23 Altogether, the described approach increases diagnostic yield and improves turnaround time, hence reducing costs and contributing to the International Rare Disease Research Consortium vision “to enable all people living with a rare disease to receive an accurate diagnosis, care and available therapy within one year of coming to medical attention.”2,pp.21
Acknowledgments
We thank Rare Disease (RD)–Connect Genome-Phenome Analysis Platform and the Undiagnosed Rare Disease Programme of Catalonia (URD-Cat) data contributors, who have contributed the data used to perform the analysis described in this study: Alessandra Renieri, Ali Dursun, Antoni Matilla-Duenas, Bru Cormand, Carlo Rivolta, Carmen Ayuso, Carmen Espinós, Christian Scerri, Dilek Yalnizoglu, Doriette Soler, Eva Morava, Fabrizio Barbetti, Francesca Forzano, Francesca Mari, Francesco Muntoni, Frederic Tort, Henry James Houlden, Maria-Isabel Tejada, Jan Senderek, Javier Benitez, Javier Corral De La Calle, Jordi Serra, José Ma Millán, Jose Segovia, Juan Ramon Gimeno Blanes, Judith Armstrong, Koksal Ozgul, Laura Vilarinho, Lluis Montoliu, Manuel Posada, Maria Antonietta Mencarelli, Marina Mora, Paola Bianchi, Pavel Seeman, Perry M. Elliott, Alessandra Ferlini, Alexis Brice, Brunhilde Wirth, Francesco Muntoni, Mike Hanna, Sarah Tabrizi, Thomas Klockgether, Vincent Timmerman, Volker Straub, Semra Hiz Kurul, Yavuz Oktay, Serdal Gungor, Ahmet Yaramis, Uluc Yis, Alfons Macaya, Antonia Ribes, Aurora Pujol, Conxi Lázaro, Daniel Grinberg, Eduardo Tizzano, Francesc Cardellach, Francesc Palau, Montse Milà, Pia Gallano, Rafael Artuch, Ramon MartíSeves, Gonzalo Villanueva, Silvia Vidal, Gloria Garrabou, Susanna Balcells, Roser Urreizti, Estrella López, Ivon Cuscó, Irene Valenzuela, and Maria Sabater.
Footnotes
Supported by European Union projects RD-Connect, Solve-RD, and European Joint Programme of Rare Diseases (EJP-RD) grants FP7 305444, H2020 779257, and H2020 825575; Instituto de Salud Carlos III grants PT13/0001/0044 and PT17/0009/0019; Instituto Nacional de Bioinformática; ELIXIR Implementation Studies; European Union projects BBMRI-LPC EU FP7 313010, NeurOmics EU FP7 305121, and Undiagnosed Rare Disease Program of Catalonia (Departament de Salut, Generalitat de CatalunyaSLT002/16/00174); Canadian Institutes of Health Research Foundation grant FDN-167281 (H.L.); the European Research Council309548 (R.H.); the Wellcome Investigator Award 109915/Z/15/Z (R.H.); the Medical Research Council (United Kingdom) MR/N025431/1 (R.H.); the Wellcome Trust Pathfinder Scheme 201064/Z/16/Z (R.H. and H.L.); the Newton Fund (United Kingdom/Turkey) MR/N027302/1 (R.H. and H.L.); the Spanish Ministry of Economy, Industry and Competitiveness to the European Molecular Biology Laboratory (EMBL) partnership; the Centro de Excelencia Severo Ochoa; the Centres de Recerca de Catalunya (CERCA) Program/Generalitat de Catalunya; the Generalitat de Catalunya through the Department of Health and Department of Business and Knowledge; the Spanish Ministry of Economy, Industry and Competitiveness with funds from the European Regional Development Fund corresponding to the 2014 to 2020 Smart Growth Operating Program.
Disclosures: None declared.
Supplemental material for this article can be found at http://doi.org/10.1016/j.jmoldx.2020.06.008.
Contributor Information
Sergi Beltran, Email: sergi.beltran@cnag.crg.eu.
RD–Connect Genome-Phenome Analysis Platform and URD-Cat Data Contributors:
Alessandra Renieri, Ali Dursun, Antoni Matilla-Duenas, Bru Cormand, Carlo Rivolta, Carmen Ayuso, Carmen Espinós, Christian Scerri, Dilek Yalnizoglu, Doriette Soler, Eva Morava, Fabrizio Barbetti, Francesca Forzano, Francesca Mari, Francesco Muntoni, Frederic Tort, Henry James Houlden, Maria-Isabel Tejada, Jan Senderek, Javier Benitez, Javier Corral De La Calle, Jordi Serra, José Ma Millán, Jose Segovia, Juan Ramon Gimeno Blanes, Judith Armstrong, Koksal Ozgul, Laura Vilarinho, Lluis Montoliu, Manuel Posada, Maria Antonietta Mencarelli, Marina Mora, Paola Bianchi, Pavel Seeman, Perry M. Elliott, Alessandra Ferlini, Alexis Brice, Brunhilde Wirth, Francesco Muntoni, Mike Hanna, Sarah Tabrizi, Thomas Klockgether, Vincent Timmerman, Volker Straub, Semra Hiz Kurul, Yavuz Oktay, Serdal Gungor, Ahmet Yaramis, Uluc Yis, Alfons Macaya, Antonia Ribes, Aurora Pujol, Conxi Lázaro, Daniel Grinberg, Eduardo Tizzano, Francesc Cardellach, Francesc Palau, Montse Milà, Pia Gallano, Rafael Artuch, Ramon MartiSeves, Gonzalo Villanueva, Silvia Vidal, Gloria Garrabou, Susanna Balcells, Roser Urreizti, Estrella López, Ivon Cuscó, Irene Valenzuela, and Maria Sabater
Authors' Contributions
L.M., S.L., and A.P. analyzed data (integration of SNV-indel and ROH analysis); D.P. developed the Rare Disease (RD)–Connect platform to integrate the analysis of ROH data; E.M. designed the logistic regression model; G.B. processed and analyzed Undiagnosed Rare Disease Program of Catalonia samples; R.T., R.H., L.P.J., O.R., I.G., G.-J.v.O., and H.L. recruited patient/data and coordinated molecular diagnosis feedback from each of the projects involved in this study; S.B., S.L., and L.M. designed the study and wrote the manuscript; all authors revised the article and read and approved the final manuscript. L.M. and S. B. are the guarantors of this work and, as such, had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Supplemental Data
Supplemental Figure S1.
Ethnicity and exome capture kit assessment through the analysis of 2432 exome sequencing (ES) data sets. A: Total run of homozygosity (ROH) size distribution of the whole data set, classified by ES capture kits. B: Total ROH size distribution of the whole data set, according to different ethnicities reported. Dotted red lines indicate the defined consanguinity thresholds (22, 79, and 123 megabases). C, consanguineous; NC, nonconsanguineous; PC, probably consanguineous; U, uncertain; unknown, no consanguinity stated in the clinical record.
References
- 1.Nguengang Wakap S., Lambert D.M., Olry A., Rodwell C., Gueydan C., Lanneau V., Murphy D., Le Cam Y., Rath A. Estimating cumulative point prevalence of rare diseases: analysis of the Orphanet database. Eur J Hum Genet. 2020;28:165–173. doi: 10.1038/s41431-019-0508-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Austin C.P., Cutillo C.M., Lau L.P.L., Jonker A.H., Rath A., Julkowska D., Thomson D., Terry S.F., de Montleau B., Ardigò D., Hivert V., Boycott K.M., Baynam G., Kaufmann P., Taruscio D., Lochmüller H., Suematsu M., Incerti C., Draghia-Akli R., Norstedt I., Wang L., Dawkins H.J.S., International Rare Diseases Research Consortium (IRDiRC) Future of rare diseases research 2017-2027: an IRDiRC perspective. Clin Transl Sci. 2018;11:21–27. doi: 10.1111/cts.12500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boycott K.M., Hartley T., Biesecker L.G., Gibbs R.A., Innes A.M., Riess O., Belmont J., Dunwoodie S.L., Jojic N., Lassmann T., Mackay D., Temple I.K., Visel A., Baynam G. A: Diagnosis for all rare genetic diseases: the horizon and the next frontiers. Cell. 2019;177:32–37. doi: 10.1016/j.cell.2019.02.040. [DOI] [PubMed] [Google Scholar]
- 4.Boycott K.M., Hartley T., Biesecker L.G., Gibbs R.A., Innes A.M., Riess O., Belmont J., Dunwoodie S.L., Jojic N., Lassmann T., Mackay D., Temple I.K., Visel A., Baynam G. International cooperation to enable the diagnosis of all rare genetic diseases. Am J Hum Genet. 2017;100:695–705. doi: 10.1016/j.ajhg.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lochmüller H., Badowska D.M., Thompson R., Knoers N.V., Aartsma-Rus A., Gut I., Wood L., Harmuth T., Durudas A., Graessner H., Schaefer F., Riess O. RD-connect, NeurOmics and EURenOmics: collaborative European initiative for rare diseases. Eur J Hum Genet. 2018;26:778–785. doi: 10.1038/s41431-018-0115-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thompson R., Johnston L., Taruscio D., Monaco L., Béroud C., Gut I.G., Hansson M.G., 't Hoen P.B., Patrinos G.P., Dawkins H., Ensini M., Zatloukal K., Koubi D., Heslop E., Paschall J.E., Posada M., Robinson P.N., Bushby K., Lochmüller H. RD-connect: an integrated platform connecting databases, registries, biobanks and clinical bioinformatics for rare disease research. J Gen Intern Med. 2014;29 Suppl 3:S780–S787. doi: 10.1007/s11606-014-2908-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ouborg N.J., Pertoldi C., Loeschcke V., Bijlsma R.K., Hedrick P.W. Conservation genetics in transition to conservation genomics. Trends Genet. 2010;26:177–187. doi: 10.1016/j.tig.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Fareed M.,, Afzal M. Genetics of consanguinity and inbreeding in health and disease. Ann Hum Biol. 2017;44:99–107. doi: 10.1080/03014460.2016.1265148. [DOI] [PubMed] [Google Scholar]
- 9.Matthijs G., Rymen D., Millón M.B., Souche E., Race V. Approaches to homozygosity mapping and exome sequencing for the identification of novel types of CDG. Glycoconj J. 2013;30:67–76. doi: 10.1007/s10719-012-9445-7. [DOI] [PubMed] [Google Scholar]
- 10.Vahidnezhad H., Youssefian L., Jazayeri A., Uitto J. Research techniques made simple: genome-wide homozygosity/autozygosity mapping is a powerful tool for identifying candidate genes in autosomal recessive genetic diseases. J Invest Dermatol. 2018;138:1893–1900. doi: 10.1016/j.jid.2018.06.170. [DOI] [PubMed] [Google Scholar]
- 11.Posey J.E., O'Donnell-Luria A.H., Chong J.X., Harel T., Jhangiani S.N., Coban Akdemir Z.H. Insights into genetics, human biology and disease gleaned from family based genomic studies. Genet Med. 2019;21:798–812. doi: 10.1038/s41436-018-0408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pippucci T., Magi A., Gialluisi A., Romeo G. Detection of runs of homozygosity from whole exome sequencing data: state of the art and perspectives for clinical, population and epidemiological studies. Hum Hered. 2014;77:63–72. doi: 10.1159/000362412. [DOI] [PubMed] [Google Scholar]
- 13.Seelow D., Schuelke M., Hildebrandt F., Nürnberg P. HomozygosityMapper--an interactive approach to homozygosity mapping. Nucleic Acids Res. 2009;37:W593–W599. doi: 10.1093/nar/gkp369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Görmez Z., Bakir-Gungor B., Sagiroglu M.S. HomSI: a homozygous stretch identifier from next-generation sequencing data. Bioinformatics. 2014;30:445–447. doi: 10.1093/bioinformatics/btt686. [DOI] [PubMed] [Google Scholar]
- 16.Magi A., Tattini L., Palombo F., Benelli M., Gialluisi A., Giusti B., Abbate R., Seri M., Gensini G.F., Romeo G., Pippucci T. H3M2: detection of runs of homozygosity from whole-exome sequencing data. Bioinformatics. 2014;30:2852–2859. doi: 10.1093/bioinformatics/btu401. [DOI] [PubMed] [Google Scholar]
- 17.Gibson J., Morton N.E., Collins Extended tracts of homozygosity in outbred human populations. Hum Mol Genet. 2006;15:789–795. doi: 10.1093/hmg/ddi493. [DOI] [PubMed] [Google Scholar]
- 18.Ceballos F.C., Joshi P.K., Clark D.W., Ramsay M., Wilson J.F. Runs of homozygosity: windows into population history and trait architecture. Nat Rev Genet. 2018;19:220–234. doi: 10.1038/nrg.2017.109. [DOI] [PubMed] [Google Scholar]
- 19.Howrigan D.P., Simonson M.A., Keller M.C. Detecting autozygosity through runs of homozygosity: a comparison of three autozygosity detection algorithms. BMC Genomics. 2011;12:460. doi: 10.1186/1471-2164-12-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oliveira J., Pereira R., Santos R., Sousa M. Homozygosity mapping using whole-exome sequencing: a valuable approach for pathogenic variant identification in genetic diseases. Bioinformatics (Biostec) 2017;3:210–216. [Google Scholar]
- 21.Kancheva D., Atkinson D., De Rijk P., Zimon M., Chamova T., Mitev V., Yaramis A., Maria Fabrizi G., Topaloglu H., Tournev I., Parman Y., Parma Y., Battaloglu E., Estrada-Cuzcano A., Jordanova A. Novel mutations in genes causing hereditary spastic paraplegia and Charcot-Marie-Tooth neuropathy identified by an optimized protocol for homozygosity mapping based on whole-exome sequencing. Genet Med. 2016;18:600–607. doi: 10.1038/gim.2015.139. [DOI] [PubMed] [Google Scholar]
- 22.Masingue M., Perrot J., Carlier R.Y., Piguet-Lacroix G., Latour P., Stojkovic T. WES homozygosity mapping in a recessive form of Charcot-Marie-Tooth neuropathy reveals intronic GDAP1 variant leading to a premature stop codon. Neurogenetics. 2018;19:67–76. doi: 10.1007/s10048-018-0539-7. [DOI] [PubMed] [Google Scholar]
- 23.Rehder C.W., David K.L., Hirsch B., Toriello H.V., Wilson C.M., Kearney H.M. American College of Medical Genetics and Genomics: standards and guidelines for documenting suspected consanguinity as an incidental finding of genomic testing. Genet Med. 2013;15:150–152. doi: 10.1038/gim.2012.169. [DOI] [PubMed] [Google Scholar]
- 24.Laurie S., Fernandez-Callejo M., Marco-Sola S., Trotta J.R., Camps J., Chacón A., Espinosa A., Gut M., Gut I., Heath S., Beltran S. From wet-lab to variations: concordance and speed of bioinformatics pipelines for whole genome and whole exome sequencing. Hum Mutat. 2016;37:1263–1271. doi: 10.1002/humu.23114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manichaikul A., Mychaleckyj J.C., Rich S.S., Daly K., Sale M., Chen W.M. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.1000 Genomes Project Consortium. Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., Voelkerding K., Rehm H.L., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sund K.L., Rehder C.W. Detection and reporting of homozygosity associated with consanguinity in the clinical laboratory. Hum Hered. 2014;77:217–224. doi: 10.1159/000362448. [DOI] [PubMed] [Google Scholar]
- 31.Bis D.M., Schüle R., Reichbauer J., Synofzik M., Rattay T.W., Soehn A., de Jonghe P., Schöls L., Züchner S. Uniparental disomy determined by whole-exome sequencing in a spectrum of rare motoneuron diseases and ataxias. Mol Genet Genomic Med. 2017;5:280–286. doi: 10.1002/mgg3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deciphering Developmental Disorders Study Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.