Abstract
Recent studies have provided insights into the pathogenesis of coronavirus disease 2019 (COVID-19)1–4. However, the longitudinal immunological correlates of disease outcome remain unclear. Here we serially analysed immune responses in 113 patients with moderate or severe COVID-19. Immune profiling revealed an overall increase in innate cell lineages, with a concomitant reduction in T cell number. An early elevation in cytokine levels was associated with worse disease outcomes. Following an early increase in cytokines, patients with moderate COVID-19 displayed a progressive reduction in type 1 (antiviral) and type 3 (antifungal) responses. By contrast, patients with severe COVID-19 maintained these elevated responses throughout the course of the disease. Moreover, severe COVID-19 was accompanied by an increase in multiple type 2 (anti-helminths) effectors, including interleukin-5 (IL-5), IL-13, immunoglobulin E and eosinophils. Unsupervised clustering analysis identified four immune signatures, representing growth factors (A), type-2/3 cytokines (B), mixed type-1/2/3 cytokines (C), and chemokines (D) that correlated with three distinct disease trajectories. The immune profiles of patients who recovered from moderate COVID-19 were enriched in tissue reparative growth factor signature A, whereas the profiles of those with who developed severe disease had elevated levels of all four signatures. Thus, we have identified a maladapted immune response profile associated with severe COVID-19 and poor clinical outcome, as well as early immune signatures that correlate with divergent disease trajectories.
Introduction
Coronavirus Disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a highly infectious, zoonotic virus that exploits angiotensin-converting enzyme 2 (ACE2)5,6 as a cell entry receptor. Clinical presentation of COVID-19 involves a broad range of symptoms and disease trajectories. Understanding the nature of the immune response that leads to recovery over severe disease is key to developing effective treatment against COVID-19. Coronaviruses, including Severe Acute Respiratory Syndrome (SARS-CoV) and Middle Eastern Respiratory Syndrome (MERS), typically induce strong inflammatory responses and associated lymphopenia7,8. Studies of COVID-19 patients have reported increases in inflammatory monocytes and neutrophils and a sharp decrease in lymphocytes1–4, and an inflammatory milieu containing IL-1β, IL-6, and TNF-α in severe disease1,2,4,9,10. Despite these analyses, immune response dynamics during the course of SARS-CoV-2 infection and its possible correlation with clinical trajectory remain unknown.
Immune responses against pathogens are divided roughly into three types11–13. Type-1 immunity, characterized by T-bet-dependent responses and IFN-γ, is generated against intracellular pathogens including viruses. In type-1 immunity, pathogen clearance is mediated through effector cells including ILC1, NK cells, cytotoxic T lymphocytes, and Th1 cells. Type-2 immunity, which relies on the GATA-3 transcription factor, mediates anti-helminths defense through effector molecules including IL-4, IL-5, IL-13, and IgE designed to expel these pathogens through the concerted action of epithelial cells, mast cells, eosinophils, and basophils. Type-3 immunity, orchestrated by the RORγt-induced cytokines IL-17, IL-22 secreted by ILC3 and Th17 cells, is mounted against fungi and extracellular bacteria to elicit neutrophil-dependent clearance. In this study, we focused on the longitudinal analysis of these three types of immune responses to COVID-19 patients and identified correlations between distinct immune phenotype and disease.
Results
Overview of COVID-19 immunological features
One hundred and thirteen patients with COVID-19 who were admitted to Yale New Haven Hospital (YNHH) between 18 March 2020 and 27 May 2020 were recruited to the Yale IMPACT (Implementing Medical and Public Health Action Against Coronavirus CT) study. We assessed viral RNA load (quantified by quantitative PCR with reverse tran- scription (RT–qPCR) using nasopharyngeal swabs); levels of plasma cytokines and chemokines; and leukocyte profiles (by flow cytometry using freshly isolated peripheral blood mononuclear cells; PBMCs). We performed 253 collections and follow-up measurements on the patient cohort with a range of one to seven longitudinal time-points that occurred 3–51 days after the onset of symptoms. In parallel, we enrolled 108 volunteer healthcare workers (HCWs), whose samples served as healthy controls (SARS-CoV-2-negative by RT–qPCR and serology).
Basic demographic information stratified by disease severity is pro- vided in Extended Data Table 1 and detailed in Supplementary Table 1. Patients who had been admitted to YNHH were stratified into moder- ate and severe disease groups on the basis of supplemental oxygen requirements and admission to the intensive care unit (ICU) (Fig. 1a). Among our cohort, patients who developed moderate or severe dis- ease did not differ significantly with respect to age or sex. Body mass index (BMI) was generally higher among patients with severe disease, and extremes in BMI correlated with an increased relative risk (RR) of mortality (RR BMI ≥ 35: 1.62 (95% confidence interval (CI) 0.81–3.22)) (Extended Data Table 1, Extended Data Fig. 1a, b). Exposure to select therapeutic regimens of interest was assessed in patients with moderate or severe disease (Extended Data Fig. 1c.) Initial presenting symptoms demonstrated a preponderance of headache (54.55%), fever (64.47%), cough (74.03%), and dyspnoea (67.09%) with no significant difference in symptom presentation between patients with moderate disease and those who developed severe disease. Finally, mortality was significantly higher in patients who were admitted to the ICU than in those who were not (27.27% versus 3.75%; P < 0.001) (Extended Data Table 1).
Figure 1. Overview of immunological features in COVID-19 patients.

a, Overview of cohort, including healthy donors (HCWs) and patients with moderate or severe COVID-19. Ordinal scores assigned according to clinical severity scale as described in Methods. D, deceased; ICU, intensive care unit; MV, mechanical ventilation. b, Heat map comparison of the major immune cell populations within PBMCs in patients with moderate (n = 121) or severe (n = 43) COVID-19, or HCSs (n = 43). n values represent a separate time point per subject Subjects are arranged across rows, with each coloured unit indicating the relative distribution of an immune cell population normalized against the same population across all subjects. K-means clustering was used to arrange patients and measurements. Eoso, eosinophil; ncMono, non-classical monocyte; neut, neutrophil; cMono, classical monocyte; intMono, intermediate monocyte; DC2 and DC1, type 2 and 1 dendritic cells, respectively; pDC, plasmacytoid dendritic cell; T-CD8 and T-CD4, CD8+ and CD4+ T cells, respectively; NKT, natural killer T cell; NK, natural killer cell. c, Immune cell subsets plotted as a concentration of millions of cells per millilitre of blood or as a percentage of live single cells. Each dot represents a separate time point per subject (HCW, n = 50; moderate, n = 117; severe, n = 40). d, Correlation matrices across all time points of 71 cytokines from patient blood, comparing patients with moderate and severe disease. Only significant correlations (<0.05) are represented as dots. Pearson’s correlation coefficients from comparisons of cytokine measurements within the same patients are visualized by colour intensity. e-g, Quantification of prominent inflammatory cytokines (e), interferons type I and II (f), and CCL1 and IL-17 (g) presented as log10-transformed concentrations. Each dot represents a separate time point per subject (HCW, n = 50; moderate, n = 117; severe, n = 40). Centre, median; box limits, first and third percentiles; whiskers, 1.5× interquartile range (IQR). Significance determined by two-sided, Wilcoxon rank-sum test.
We analysed PBMC and plasma samples from patients with moderate or severe COVID-19 and healthy HCW donors (Fig. 1a, gating strategy in Extended Data Fig. 9) by flow cytometry and ELISA to quantify leuko- cytes and soluble mediators, respectively. An unsupervised heat map constructed from the main innate and adaptive circulating immune cell types revealed marked changes in patients with COVID-19 compared to uninfected HCW (Fig. 1b). As reported1–4, COVID-19 patients presented with marked reductions in T cell number and frequency in both CD4+ and CD8+ T cells, even after normalization for age as a possible confounder (Extended Data Fig.1d). Granulocytes such as neutrophils and eosinophils are normally excluded from the PBMC fraction following density gradient separation. However, low density granulocytes are present in the PBMC layer from peripheral blood collections in patients with inflammatory diseases14. In patients with COVID-19, increases in monocytes, low-density neutrophils and eosinophils correlated with the severity of disease (Fig. 2c, Extended Data Fig. 2a, b). In addi- tion, patients showed increased activation of T cells and a reduction in expression of the human leukocyte antigen DR isotype (HLA-DR) by circulating monocytes1 (Extended Data Fig. 2c). A complete overview of PBMC cells subsets is presented in Extended Data Fig. 2.
Figure 2: Longitudinal immune profiling of moderate and severe COVID-19 patients.

a, Correlation matrices of 71 cytokines from patient blood comparing cytokine concentrations in patients with moderate or severe disease during the early phase (<10 DfSO) or late phase (>10 DfSO) of disease. Only significant correlations (<0.05) are represented as dots, and Pearson’s correlation coefficient from comparisons of cytokine measurements within each patient is visualized by colour intensity. b, c, Anti-viral interferons (b) and inflammasome-related cytokines (c) plotted as log10 concentrations over time and grouped by disease severity. d–f, Cellular and cytokine measurements representative of type 1 (d), type 2 (e) and type 3 (f) immune responses reported over time in intervals of days (left) and continuously as linear regressions (right). Left, each dot represents a distinct patient and time point arranged in intervals of 5 days until 25 DfSO; dark blue, moderate disease (n = 112), pink, severe disease (n = 40). Dark blue or pink lines pass through the mean at each time interval; error bars denote the s.e.m. Dashed green line, mean from healthy HCWs. Right, regression lines are indicated by the dark blue (moderate) or red (severe) solid lines. Associated Pearson’s correlation coefficients and linear regression significance are coloured accordingly; shading represents 95% CI.
To gain insights into key differences in cytokines, chemokines, and additional immune markers between patients with moderate and severe disease, we correlated the measurements of these soluble pro- teins across all sample collection time-points. (Fig. 1d). We observed a ‘core COVID-19 signature’ that was shared by both moderate severe disease groups and was defined by the following inflammatory cytokines, which correlated positively with each other: IL-1α, IL-1β, IL-17A, IL-12 p70, and IFNα (Fig. 1d). In patients with severe disease, we observed an additional inflammatory cluster defined by thrombopoietin (TPO), IL-33, IL-16, IL-21, IL-23, IFNλ, eotaxin and eotaxin 3 (Fig. 1d). Most of the cytokines linked to cytokine release syndrome (CRS), such as IL-1α, IL-1β, IL-6, IL-10, IL-18 and TNF, showed increased positive associations in patients with severe disease (Fig. 1d–f, Extended Data Fig. 3). These data highlight broad inflammatory changes, involving concomitant release of type 1, type 2 and type 3 cytokines, in patients with severe COVID-19.
Longitudinal immune profiling of COVID-19
Our data presented above, as well as previous single-cell transcriptome and flow cytometry-based studies2,4,15–17, depicted an overt innate and adaptive immune activation in severe COVID-19 patients. Longitu- dinal cytokine correlations, measured in terms of days from symptom onset (DfSO), indicated that major differences in immune phenotypes between moderate and severe disease were apparent after day 10 of infection (Fig. 2a). In the first 10 DfSO, patients with severe or moderate disease displayed similar correlation intensity and markers, including the overall core COVID-19 signature described above (Fig. 2a). After day 10 these markers declined steadily in patients with moderate disease. By contrast, patients with severe COVID-19 maintained elevated levels of these core signature makers. Notably, additional correlations between cytokines emerged in patients with severe disease following day 10 (Fig. 2a). These analyses strongly support the observation (Fig. 1) that TPO and IFNα associate strongly with IFNλ, IL-9, IL-18, IL-21, IL-23, and IL-33 (Fig. 2a). These observations indicate sharp differences in the expression of inflammatory markers along disease progression between patients who exhibit moderate versus severe symptoms of COVID-19.
Temporal analyses of PBMC and soluble proteins in plasma, either by linear regression or grouped intervals, supported distinct courses in disease. IFN-α levels were sustained at higher levels in sever patients while they declined moderate patients (Fig. 2b). Plasma IFN-λ levels increased during the first week of symptom onset in ICU patients and remained elevated in later phases (Fig. 2b). Additionally, inflammasome-induced cytokines, such as IL-1β and IL-18 were also elevated in severe patients compared to patients with moderate disease at most time-points analysed (Fig. 2c). Consistently, IL-1 receptor antagonist (IL-1Ra), induced by IL-1R signalling as a negative feedback regulator18, also showed increased levels in ICU patients from day 10 of disease onset (Extended Data Fig. 4).
With respect to type-1 immunity, an increased number of monocytes was observed at approximately 14 DfSO in severe but not in moderate COVID-19 patients (Fig. 2d). The innate cytokine IL-12, a key inducer of type-1 immunity11,12, displayed a similar pattern to IFN-γ; increasing over time in severe patients but steadily declining in moderate patients (Fig. 2d). By intracellular cytokine staining, CD4+ and CD8+ T cells from patients with moderate disease secreted comparable amounts of IFN-γ to those from severe patients. Together with the severe T cell depletion in severe patients (Fig. 1), our data suggested that secretion of IFN-γ by non-T cells (ILC1, NK), or non-circulating T cells in tissues were the primary contributors to the enhanced levels observed in severe patients (Extended Data Fig. 5).
Type-2 immune markers continued to increase in severe patients over time, as indicated by strong correlations observed in late time points from severe patients (Fig. 2a). Eosinophils and eotaxin-2 increased in severe patients and remained higher than levels measured in moderate patients (Fig. 2e). Type-2 innate immune cytokines, including TSLP and IL-33, did not exhibit significant differences between severe and moderate patients (Fig. 2e). Hallmark type-2 cytokines, including IL-5 (associated with eosinophilia) and IL-13 (Fig. 2e), were enhanced in patients with severe over moderate disease. In contrast, IL-4, was not significantly different. However, IL-4, similar to IL-5 and IL-13, exhibited an upward trend over the course of disease in severe patients (Fig. 2e). A type-2 antibody isotype was also increased; IgE levels were significantly higher in severe patients and continued to increase during the disease course (Fig. 2e).
IL-6, which is linked to CRS, was elevated in patients with severe disease19. Circulating neutrophils did not show a significant increase in our longitudinal analysis (Fig. 2f), although hallmarks of type-3 responses were observed in severe patients, including increased plasma IL-17A and IL-22, as well as IL-17 secretion by circulating CD4 T cells as assessed by intracellular cytokine staining (Fig. 2f, Extended Data Fig. 5). These data identify broad elevation of type-1, type-2 and type-3 signatures in severe cases of COVID-19, with distinct temporal dynamics and quantities between severe and moderate patients.
Viral load correlates with elevated cytokines
We next measured viral load kinetics using serial nasopharyngeal swabs. Although there was no significant difference in viral RNA load between patients with moderate and severe disease at any specific time point ana- lysed, patients with moderate disease showed a steady decline in viral load over the course of disease, whereas those with severe disease did not (Fig. 3a). Regardless of whether patients exhibited moderate or severe disease, viral load correlated significantly with the levels of IFNα, IFNγ, TNF and tumour necrosis factor-related apoptosis-inducing ligand (TRAIL) (Fig. 3b). In addition, several chemokines responsible for monocyte recruit- ment correlated significantly with viral load only in patients with severe disease (Extended Data Fig. 6a, b). These data indicate that nasopharyn- geal viral load correlates with plasma levels of interferons and cytokines.
Figure 3. Early viral and cytokine profiles distinguish moderate and severe outcomes.

a, Viral loads measured by nasopharyngeal swabs are plotted as log10 of genome equivalents against time after symptom onset for patients with moderate disease (n = 112) or severe disease (n = 39). Left, each dot represents a distinct patient and time point arranged in intervals of 5 days until 25 DfSO. Dark blue or pink lines pass through the mean of each measurement; error bars denote s.e.m. Right, longitudinal data plotted over time continuously. Regression lines are shown as dark blue (moderate) or red (severe). Associated linear regression equations, Pearson’s correlation coefficients, and significance are coloured accordingly. Green text is the regression analysis and correlation for all patients. Shading represents 95% CIs. Dashed green line denotes mean threshold for positivity. Dashed grey line indicates mean limit of detection. b, Correlation and linear regression of cytokines plotted, as Log10 of concentration, and viral load by nasopharyngeal swab, plotted as Log10 of genome equivalents (GE), regardless of disease severity (n=151). Each dot represents a unique patient time point; dark blue, moderate disease; red, severe disease. White line indicates the regression line for all patients. The associated linear regression equation, Pearson’s correlation coefficient, and significance are shown in green. Grey shading indicates 95% CIs. Dashed green line denotes mean threshold for positivity. Dashed grey line indicates mean limit of detection. c, Unbiased heat map comparisons of cytokines in PBMCs. Measurements were normalized across all patients. K-means clustering was used to determine clusters 1–3 (cluster 1, n = 46; cluster 2, n = 50; cluster 3, n = 16). d, Distribution of age and length of hospital stay (violin plots; solid lines, median; dotted lines, quartiles.) and frequency of coagulopathy and mortality (bar graphs) within each cluster. e, Top 20 cytokines by mutual information analysis to determine their importance for determining mortality. Significance of comparisons determined by two-sided, Wilcoxon rank-sum test.
Early cytokine profile marks disease outcomes
Next, we investigated whether specific early cytokine responses are asso- ciated with severe COVID-19. To this end, we conducted an unsupervised clustering analysis using baseline measurements collected before 12 DfSO (Fig. 3c). Three main clusters with correlation to distinct disease outcomes emerged. These were characterized by 4 distinct immune signatures: Signature A contained several stromal growth factors including EGF, PDGF, VEGF that are mediators of wound healing and tissue repair20, as well as IL-7, a critical growth factor for lymphocytes. Signature B consisted of eotaxin 3, IL-33, TSLP, along with IL-21, IL-23 and IL-17F, thus representing type-2 & 3 immune effectors. Signature C comprised of mixtures of all immunotypes, including type-1 cytokines (IFN-γ, IL-12 p70, IL-15, IL-2, TNF-α), type-2 (IL-4, IL-5, IL-13), as well as type-3 (IL-1α, IL-1β, IL-17A, IL-17E, IL-22). Finally, signature D contained a number of chemokines involved in leukocyte trafficking including CCL1, 2, 5, 8, 15, 21, 22, 27, CXCL9, 10, 13, and SDF1.
Cluster 1 was comprised primarily of patients with moderate disease who experienced low occurrences of coagulopathy, shorter lengths of hospital stay, and no mortality (Fig. 3c, d). The main characteristics in this cluster were low levels of inflammatory markers and similar or increased levels of parameters in signature “A” containing tissue reparative growth factors (Fig. 3c). Clusters 2 and 3 were characterized by the rise in inflammatory markers, and patients belonging to these clusters had higher incidence of coagulopathy and mortality, which was more pronounced in cluster 3 (Fig. 3c,d). Cluster 2 showed higher levels of markers in signatures “C and D”, which included IFN-α, IL-1Ra and several hallmark type-1, type-2 and type-3 cytokines, than patients in cluster 1, but lower expression of markers in signatures “B, C and D” than in Cluster 3 (Fig. 3c,d). Cluster 3 displayed heightened expression of markers in signatures “B, C and D” than other clusters. Cluster 3 showed particular enrichment in expression of markers in signature “B”, which include several innate cytokines including IFN-λ, TGF-α, TSLP, IL-16, IL-23 and IL-33, and markers linked to coagulopathy, such as TPO (Fig. 3c, d).
We next ranked these parameters obtained at early time points as pre- dictors of severe disease outcomes (Fig. 3e, Extended Data Fig. 6c). In both cases, plasma inflammatory markers were strongly associated with severe disease outcomes. For example, high levels of type I IFN (IFNα) before the first 12 DfSO correlated with longer hospital stays and death (Fig. 3e, Extended Data Fig. 6c). Moreover, patients who ultimately died of COVID-19 exhibited significantly elevated levels of IFNα, IFNλ and IL-1Ra, as well as chemokines associated with monocytes and T cell recruitment and survival such as CCL1, CLL2, macrophage colony stimulating factor (M-CSF), IL-2, IL-16 and CCL21, within the first 12 DfSO (Fig. 3e, Extended Data Fig. 6c). These analyses identify specific immunological markers that appear early in the disease and correlate strongly with poor outcomes and death.
Retrospective analysis of COVID-19 immune correlates
To further evaluate potential drivers of severe COVID-19 outcome in an unbiased manner, we performed unsupervised clustering analysis that included all patients and all time points using cytokines and chemokines (Fig. 4a). Notably, three main clusters of patients emerged and the dis- tribution of patients in early time-point clusters identified in Fig. 3c matched the distribution for the all-time point analysis (Fig. 4a) in 96% of cases. Cluster 1 primarily comprised patients with moderate disease who showed improving clinical signs (Fig. 4a–d, Extended Data Fig. 7). This cluster contained only two deceased patients. Cluster 1 was characterized by low levels of inflammatory markers as well as similar or increased expression of markers in signature A′ (Fig. 4a–d), which mostly matched the signature A markers described in Fig. 3c. Clusters 2 and 3 contained patients with coagulopathy and worsened clinical progression, including most of the deceased patients (Fig. 4a–d, Extended Data Fig. 7).
Figure 4. Immune correlates of COVID-19 outcomes.
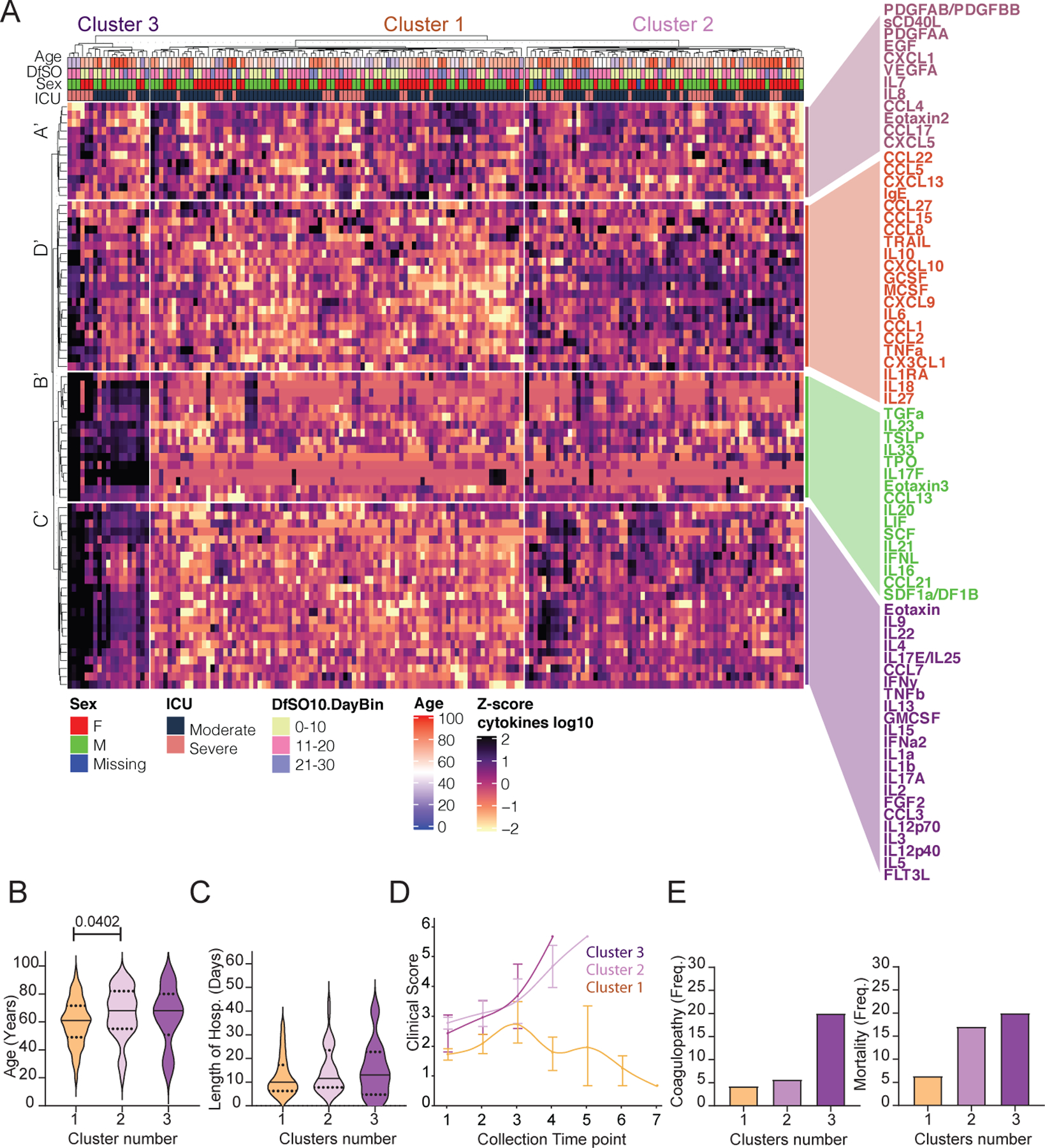
a, Unbiased heat map comparisons of cytokines within peripheral blood mononuclear cells (PBMCs) measured at distinct time points in COVID-19 patients. Measurements were normalized across all patients. K-means clustering was used to determine Clusters 1–3 (Cluster 1, n=84; Cluster 2, n=66; Cluster 3, n=20). b, c, Distribution of age (b) and length of hospital stay (violin plots) (c) of patients within each cluster. For statistical differences, adjusted P values calculated using one-way ANOVA with Tukey’s correction for multiple comparisons are shown (age: F(2, 90) = 3.115; P = 0.0492). Solid lines, median; dotted lines, quartiles. d, Disease progression measured by clinical severity score for patients in each cluster. Data (mean ± s.e.m.) are ordered by the collection time points for each patient, with regular collection intervals of 3–4 days (Extended Data Fig. 7). e, Percentage of patients in each cluster with new-onset coagulopathy or death.
Clusters 2 and 3 were driven by a set of inflammatory markers that fell into signatures B′, C′ and D′ to some extent, which overlapped highly with the ‘core signature’ cytokines and chemokines identified in Fig. 1 as well as with signatures B and C identified in Fig. 3c. These include type 1 immunity markers, including IL-12, chemokines linked to monocyte recruitment and IFNγ; type 2 responses, such as TSLP, chemokines linked to eosinophil recruitment, IL-4, IL-5 and IL-13; and type-3 responses, including IL-23, IL-17A and IL-22. In addition, most CRS- and inflammasome-associated cytokines were enriched in these clusters, including IL-1α, IL-1β, IL-6, IL-18 and TNF (Fig. 4a). These findings were consistent with generalized estimating equations that identified relationships between the risk of death and cytokines or immune cell populations over time (Extended Data Fig. 8). Together, these results identify groups of inflammatory and potentially protective markers that correlated with COVID-19 trajectories. The immune signatures that correlate with recovery (cluster 1) and the immune signatures that correlate with worsening diseases (cluster 2 < cluster 3) were remarkably similar whether we took a prospective (Fig. 3) or retrospective (Fig. 4) approach.
Discussion
Our longitudinal analyses of patients admitted to YNHH with COVID-19 revealed key temporal features of viral load and immune responses that distinguish disease trajectories during hospitalization. Unsupervised clustering revealed three distinct profiles that influenced the evolution and severity of COVID-19. Cluster 1, characterized by low expres- sion of proinflammatory cytokines and enrichment in tissue repair genes, followed a disease trajectory that remained moderate and led to eventual recovery. Clusters 2 and 3 were characterized by highly elevated proinflammatory cytokines (cluster 3 being more intense), worse disease, and death. Thus, in addition to the known CRS-related pro-inflammatory cytokines, we propose these four signatures of immune response profiles that more accurately divide patients into distinct COVID-19 disease courses.
Although nasopharyngeal viral RNA levels were not significantly different between patients with moderate and severe disease at the specific time points, linear regression analyses showed a slower decline of viral loads in patients who were admitted to the ICU. Viral load was highly correlated with IFNα, IFNγ and TNF, suggesting that viral load may drive these cytokines and that interferons may not successfully control the viral replication. Moreover, many interferons, cytokines, and chemokines were elevated early in disease for patients who ulti- mately died of COVID-19. This finding suggests possible pathological roles associated with these host defence factors, as previous reported for SARS-CoV-1 patients21.
Our comprehensive analysis of soluble plasma factors revealed a broad misfiring of immune effectors in COVID-19 patients, with early predictive markers, distinct dynamics between types of immune responses, among moderate and severe disease outcomes. These results suggest that COVID-19 late stage pathology may be driven primarily by host immune responses to SARS-CoV-2 and highlights the need for combination therapy to block other cytokines highly represented by these clusters, including inflammasome-dependent cytokines and type-2 cytokines. We observed a correlation with cytokines linked to the inflammasome pathway, which partially overlap with CRS, including IL-1β and IL-18. Indeed, it is plausible that inflammasome activation, along with a sepsis-like CRS, triggers vascular insults or tissue pathology observed in severe COVID-19 patients22.
Overall, our analyses provide a comprehensive examination of the diverse inflammatory dynamics during COVID-19 and possible contributions by distinct sets of inflammatory mediators towards disease progression. This raises the possibility that early immunological interventions that target inflammatory markers predictive of worse disease outcome are preferred to blocking late-appearing cytokines. Our disease trajectory analyses provide bases for more targeted treatment of COVID-19 patients based on early cytokine markers, as well as therapies targeted to enhancing tissue repair and promoting disease tolerance.
Methods
Ethics statement
This study was approved by Yale Human Research Protection Program Institutional Review Boards (FWA00002571, Protocol ID. 2000027690). Informed consents were obtained from all enrolled patients and healthcare workers.
Patients
135 COVID-19 patients admitted to Yale-New Haven Hospital (YNHH) between March 18th and May 5th, 2020 were included in this study. Nasopharyngeal swabs were collected, as recently described23, approximately every four days for SARS-CoV-2 RT-qPCR analysis where clinically feasible. Paired whole blood for flow cytometry analysis was collected simultaneously in sodium heparin-coated vacutainers and kept on gentle agitation until processing. All blood was processed the same day as collection from patients. Patients were scored for COVID-19 disease severity through review of electronic medical records (EMR) at each longitudinal time point. Scores were assigned by a clinical infectious disease physician according to a custom developed disease severity scale. Moderate disease status (Clinical Score 1, 2 and 3) was defined as: (1) SARS-CoV-2 infection requiring hospitalization without supplemental oxygen, (2) infection requiring non-invasive supplemental oxygen (<3 L / min, sufficient to maintain greater than 92% SpO2), (3) infection requiring non-invasive supplemental oxygen (> 3L supplemental oxygen to maintain SpO2 > 92%, or, required > 2L supplemental oxygen to maintain SpO2 > 92% and had a high sensitivity C-reactive protein (CRP) > 70) and received tocilizumab. Severe disease status (Clinical score 4 and 5) was defined as infection meeting all criteria for clinical score 3 while also requiring admission to the YNHH Intensive Care Unit (ICU) and > 6L supplemental oxygen to maintain SpO2 > 92% (4); or infection requiring invasive mechanical ventilation / extracorporeal membrane oxygenation (ECMO) in addition to glucocorticoid / vasopressor administration (5). Clinical score 6 was assigned for deceased patients. Of note, the use of tocilizumab can increase circulating levels of IL-6 through inhibition of IL-6Rα-mediated degradation. Analysis of our cohort indicate higher plasma levels of IL-6 in both moderate and severe patients that received tocilizumab treatment (Extended data Fig. 1d).
For all patients, days from symptom onset were estimated according to the following scheme: (1) highest priority was given explicit onset dates provided by patients; (2) next highest priority was given to the earliest reported symptom by a patient, and (3) in the absence of direct information regarding symptom onset, we estimated a date through manual assessment of the electronic medical record (EMRs) by an independent clinician. Demographic information was aggregated through a systematic and retrospective review of patient EMRs and was used to construct Extended Table 1. Symptom onset and etiology was recorded through standardized interview with patients or patient surrogates upon enrollment in our study, or alternatively through manual EMR review if no interview was possible due to clinical status. The clinical data was collected using EPIC EHR and REDCap 9.3.6 software.
Viral RNA measurements
RNA concentrations were measured from nasopharyngeal samples by RT-qPCR as previously described23. Briefly, total nucleic acid was extracted from 300 μl of viral transport media (nasopharyngeal swab) using the MagMAX Viral/Pathogen Nucleic Acid Isolation kit (ThermoFisher Scientific) using a modified protocol and eluted into 75 μl of elution buffer.
For SARS-CoV-2 RNA detection, 5 μl of RNA 371 template was tested as previously described24, using the US CDC real-time RT-qPCR primer/probe sets for 2019-nCoV_N1, 2019-nCoV_N2, and the human RNase P (RP) as an extraction control. Virus RNA copies were quantified using a 10-fold dilution standard curve of RNA transcripts that we previously generated24. The lower limit of detection for SARS-CoV-2 genomes assayed by qPCR in nasopharyngeal specimens was established as recently described24. In addition to a technical detection threshold, we also utilized a clinical referral threshold (detection limit) to either: (1) refer asymptomatic HCWs for diagnostic testing at a CLIA-approved laboratory, or (2) cross-validate results from a CLIA-approved laboratory for SARS-CoV-2 qPCR+ individuals upon study enrollment. Individuals above the technical detection threshold, but below the clinical referral threshold, are considered SARS-CoV-2 positive for the purposes of our research study.
Isolation of patient plasma
Plasma samples were collected after whole blood centrifugation at 400 g for 10 minutes at RT without brake. The undiluted serum was then transferred to 15 ml polypropylene conical tubes, and aliquoted and stored at −80 °C for subsequent analysis.
Cytokine and chemokine measurements
Patient serum was isolated as before and aliquots were stored in −80°C. Sera were shipped to Eve Technologies (Calgary, Alberta, Canada) on dry ice, and levels of cytokines and chemokines were measured with Human Cytokine Array/Chemokine Array 71–403 Plex Panel (HD71). All the samples were measured upon the first thaw.
Isolation of PBMCs
Peripheral blood mononuclear cells (PBMCs) were isolated from heparinized whole blood using Histopaque (Sigma-Aldrich, #10771–500ML) density gradient centrifugation in a biosafety level 2+ facility. After isolation of undiluted serum, blood was 1:1 diluted in room temperature PBS and layered over Histopaque in a SepMate tube (Stemcell Technologies; #85460) and centrifuged for 10 minutes at 1200g. The PBMC layer was isolated according to manufacturer’s instructions. Cells were washed twice with PBS prior to counting. Pelleted cells were briefly treated with ACK lysis buffer for 2 minutes and then counted. Percentage viability was estimated using standard Trypan blue staining and an automated cell counter (Thermo-Fisher, #AMQAX1000).
Flow cytometry
Antibody clones and vendors are as follows: BB515 anti-hHLA-DR (G46–6) (1:400) (BD Biosciences), BV785 anti-hCD16 (3G8) (1:100) (BioLegend), PE-Cy7 anti-hCD14 (HCD14) (1:300) (BioLegend), BV605 anti-hCD3 (UCHT1) (1:300) (BioLegend), BV711 anti-hCD19 (SJ25C1) (1:300) (BD Biosciences), AlexaFluor647 anti-hCD1c (L161) (1:150) (BioLegend), Biotin anti-hCD141 (M80) (1:150) (BioLegend), PE-Dazzle594 anti-hCD56 (HCD56) (1:300) (BioLegend), PE anti-hCD304 (12C2) (1:300) (BioLegend), APCFire750 anti-hCD11b (ICRF44) (1:100) (BioLegend), PerCP/Cy5.5 anti-hCD66b (G10F5) (1:200) (BD Biosciences), BV785 anti-hCD4 (SK3) (1:200) (BioLegend), APCFire750 or PE-Cy7 or BV711 anti-hCD8 (SK1) (1:200) (BioLegend), BV421 anti-hCCR7 (G043H7) (1:50) (BioLegend), AlexaFluor 700 anti-hCD45RA (HI100) (1:200) (BD Biosciences), PE anti-hPD1 (EH12.2H7) (1:200) (BioLegend), APC anti-hTIM3 (F38–2E2) (1:50) (BioLegend), BV711 anti-hCD38 (HIT2) (1:200) (BioLegend), BB700 anti-hCXCR5 (RF8B2) (1:50) (BD Biosciences), PE-Cy7 anti-hCD127 (HIL-7R-M21) (1:50) (BioLegend), PE-CF594 anti-hCD25 (BC96) (1:200) (BD Biosciences), BV711 anti-hCD127 (HIL-7R-M21) (1:50) (BD Biosciences), BV421 anti-hIL17a (N49–653) (1:100) (BD Biosciences), AlexaFluor 700 anti-hTNFa (MAb11) (1:100) (BioLegend), PE or APC/Fire750 anti-hIFNy (4S.B3) (1:60) (BioLegend), FITC anti-hGranzymeB (GB11) (1:200) (BioLegend), AlexaFluor 647 anti-hIL-4 (8D4–8) (1:100) (BioLegend), BB700 anti-hCD183/CXCR3 (1C6/CXCR3) (1:100) (BD Biosciences), PE-Cy7 anti-hIL-6 (MQ2–13A5) (1:50) (BioLegend), PE anti-hIL-2 (5344.111) (1:50) (BD Biosciences), BV785 anti-hCD19 (SJ25C1) (1:300) (BioLegend), BV421 anti-hCD138 (MI15) (1:300) (BioLegend), AlexaFluor700 anti-hCD20 (2H7) (1:200) (BioLegend), AlexaFluor 647 anti-hCD27 (M-T271) (1:350) (BioLegend), PE/Dazzle594 anti-hIgD (IA6–2) (1:400) (BioLegend), PE-Cy7 anti-hCD86 (IT2.2) (1:100) (BioLegend), APC/Fire750 anti-hIgM (MHM-88) (1:250) (BioLegend), BV605 anti-hCD24 (ML5) (1:200) (BioLegend), BV421 anti-hCD10 (HI10a) (1:200) (BioLegend), BV421 anti-CDh15 (SSEA-1) (1:200) (BioLegend), AlexaFluor 700 Streptavidin (1:300) (ThermoFisher), BV605 Streptavidin (1:300) (BioLegend). Briefly, freshly isolated PBMCs were plated at 1–2 × 106 cells / well in a 96 well U-bottom plate. Cells were resuspended in Live/Dead Fixable Aqua (ThermoFisher) for 20 minutes at 4°C. Following a wash, cells were then blocked with Human TruStan FcX (BioLegend) for 10 minutes at RT. Cocktails of desired staining antibodies were directly added to this mixture for 30 minutes at RT. For secondary stains, cells were first washed and supernatant aspirated; then to each cell pellet a cocktail of secondary markers was added for 30 minutes at 4°C. Prior to analysis, cells were washed and resuspended in 100 μL of 4% PFA for 30 minutes at 4°C. For intracellular cytokine staining following stimulation, cells were resuspended in 200 μL cRPMI (RPMI-1640 supplemented with 10% FBS, 2 mM L-glutamine, 100 U/ml penicillin, and 100 mg/ml streptomycin, 1mM Sodium Pyruvate, and 50uM 2-Mercaptoethanol) and stored at 4°C overnight. Subsequently, these cells were washed and stimulated with 1X Cell Stimulation Cocktail (eBioscience) in 200 μL cRPMI for 1 hour at 37°C. 50 μL of 5X Stimulation Cocktail (plus protein transport 442 inhibitor) (eBioscience) was added for an additional 4 hours of incubation at 37°C. Following stimulation, cells were washed and resuspended in 100 μL of 4% PFA for 30min at 4°C. To quantify intracellular cytokines, these samples were permeabilized with 1X Permeabilization Buffer from the FOXP3/ Transcription Factor Staining Buffer Set (eBioscience) for 10 minutes at 4°C. All subsequent staining cocktails were made in this buffer. Permeabilized cells were then washed and resuspended in a cocktail containing Human TruStan FcX (BioLegend) for 10 minutes at 4°C. Finally, intracellular staining cocktails were directly added to each sample for 1 hour at 4°C. Following this incubation, cells were washed and prepared for analysis on an Attune NXT (ThermoFisher). Data were analysed using FlowJo software version 10.6 software (Tree Star). The specific et of markers used to identify each subset of cells are summarized in Extended Data Figure 9.
Statistical analysis
Patients and their analyzed features were clustered using the K-means algorithm. The heatmaps were created with the ComplexHeatmap package43. The optimum number of clusters was determined by using the silhouette coefficient analysis, available with the NBClust and factoextra packages44. Before data visualization, each feature was scaled and centered. Multiple group comparisons were analyzed by running both parametric (ANOVA) and non-parametric (Kruskal-Wallis) statistical tests with the Dunn’s post hoc tests. Mutual information analyses were performed using the Caret R package and visualized with ggplot2. Multiple correlation analysis was performed by computing Spearman’s coefficients with the Hmisc package for R and visualized with corrplot by only showing correlations with a pvalue less than 0.05. For generalized linear models (GLM), we calculated the incident risk ratio (IRR) by conducting a Poisson regression with a log link and robust variance estimation; this value approximates the risk ratio estimated by a log-linear model. For generalized estimating equation models (GEE), we calculated the incidence risk ratio (IRR) in the same way as for non-GEE GLM models, assuming an independent correlation structure. All models controlled for participant sex and age.
Extended Data
Extended Data Fig. 1: Age and BMI cohort distributions and Select Medications distributions.

a, b, Aggregated ages (a) and BMIs (b) were collected for patients with moderate, severe, and fatal COVID-19 and relative frequency histograms generated for comparison across disease sub-groups. Gaussian and lognormal distributions were fit through least squares regression and compared for goodness of fit through differential Akaike information criterion (AICc) comparison. All distributions were best described by a Gaussian model except for age in the ‛severe’ disease category, which was best modelled by a lognormal distribution. c, Proportion of patients admitted to YNHH receiving hydroxycholorquine (HCQ), tocilizumab (Toci), methylprednisolone (Solu-medrol), and remdesivir (Rem) are shown, stratified by disease severity. d, Medication and age adjustments for IL-6 and T cell count.
Extended Data Fig. 2: Overview of cellular immune changes in COVID-19 patients.

Immune cell subsets of interest, plotted (a) as a concentration of millions of cells per mL of blood or (b) as a percentage of a parent population. (c) Phenotyping to TCR-activated T cells, cytokine-secreting T cells, and HLA-DR expression within monocytes and neutrophils. Each dot represents a separate time point per subject (HCW, n=49; Moderate, n=114; Severe, n=41). For all boxplots, the centre is drawn through the median of the measurement, while the lower and upper bounds of the box correspond to the first and third percentile. Whiskers beyond these points denote 1.5 x the interquartile range. Significance of comparisons were determined by two-sided, Wilcoxon rank-sum test and indicated as such; p-values accompany their respective comparisons.
Extended Data Fig. 3: Overview cytokine and chemokines profiles of COVID-19 patients.

(a) Quantification of cytokines in the periphery plotted as Log10-transformed concentrations in pg/mL. Each dot represents a separate time point per subject (HCW, n= 47; Moderate, n= 124; Severe, n= 45). For all boxplots, the centre is drawn through the median of the measurement, while the lower and upper bounds of the box correspond to the first and third percentile. Whiskers beyond these points denote 1.5 x the interquartile range. Significance of comparisons were determined by two-sided, Wilcoxon rank-sum test and indicated as such; p-values accompany their respective comparisons.
Extended Data Fig. 4. Longitudinal cytokines and chemokines of COVID-19 patients.

(a) Log10-transformed cytokine concentrations plotted continuously over time according to the days of symptom onset for patients with moderate disease (n= 112) or severe disease (n= 39). The dotted green line represents the mean measurement from uninfected health care workers. Regression lines are indicated by the dark blue (moderate) or red (severe) solid lines. Associated, Pearson’s Correlation Coefficients and linear regression significance are in pink (moderate) or dark blue (severe). 95% confidence intervals for the regression lines are denoted by the pink (moderate) or dark blue (severe) filled areas.
Extended Data Fig. 5: T cell immune profiles in moderate and severe patients.

(a) CD4+ and (b) CD8+ T cell populations of interest, plotted as a percentage of parent populations, over time according to the days following symptom onset for patients with moderate disease (n= 118) or severe disease (n= 41). Each dot represents a distinct patient and time point arranged by intervals of five days until 25 days. Dark blue or pink lines pass through the mean of each measurement at the specified time interval; error bars at this intersection denote the standard error the mean. The dotted green line represents the mean measurement from uninfected health care workers.
Extended Data Fig. 6: Early cytokine profile distinguishes moderate and severe outcomes.

(a) Log10-transformed cytokine concentrations plotted continuously NP viral load (expressed as log10 genomic equivalents (GE)/ml) per within an individual patient and time point. Regression lines are indicated by the dark blue (moderate) or red (severe) solid lines for patients with moderate disease (n= 112) or severe disease (n= 39), respectively. Associated Pearson’s Correlation Coefficients, and linear regression significance are in pink (moderate) or dark blue (severe). 95% confidence intervals for the regression lines are denoted by the pink (moderate) or dark blue (severe) filled areas. (b) Correlation map of highly correlated cytokines with NP viral load in patients with moderate (blue) or severe disease (red). Pearson’s Correlation Coefficients are indicated in grey, connecting the central node, NP viral load, with peripheral nodes; p-values for each correlation are indicated above each peripheral node. (c) Length of hospital stay plotted per patient against an individual’s baseline plasma cytokine measurements (<12 days from symptom onset), which were grouped according to high or low expression (>0.5 Log10 difference): IFNa2 (Hi:12, Lo:13), TNFa (Hi:6, Lo:4), IL4 (Hi:7, Lo:11), IL4 (Hi:8, Lo:6), IL1RA (Hi:8, Lo:7), IL1b (Hi:11, Lo:5), IL6 (Hi:8, Lo:7), IL18 (Hi:5, Lo:5). (d) Baseline plasma cytokine measurements for each patient who was either discharged from the hospital (n=83) or expired during treatment for COVID-19 (n=11). For all boxplots, the centre is drawn through the median of the measurement, while the lower and upper bounds of the box correspond to the first and third percentile. Whiskers beyond these points denote 1.5 x the interquartile range. Significance of comparisons were determined by two-sided, Wilcoxon rank-sum test; p-values accompany their respective comparisons.
Extended Data Fig. 7: Distribution of days from symptom onset stratified by collection time point and select cluster clinical data.

a, Correlation of days from symptom onset and samples collection time points. Violin plots comparing the distributions of days from symptom for each patient ordered by sequential IMPACT study time points (1–8). Study time points 7 and 8 are represented by discrete points for the single patient collected at each. Violin plots display median values (solid line) and associated quartiles (dashed lines). T1–8 (time point 1 to 8). b–h, Aggregated clinical data for patients in clusters 1–3. Displayed are laboratory values at time of admission to YNHH (“admit”); last recorded values from duration of admission (“last”); maximum recorded values from duration of admission (“max”); minimum recorded values from duration of admission (“min”); and average recorded values for duration of admission (“mean”). Scatter plots show cluster means with s.e.m. plotted above and below. Clusters were subsequently compared using ordinary two- way ANVOA and post hoc pairwise comparisons are identified where significant (adjusted P values displayed, Tukey’s method for multiple comparisons).
Extended Data Fig. 8: Risk of ICU admission and death according to biomarkers levels.

Forest plots comparing the risk of death (b) among ill patients. Each effect estimate represents an individual regression estimate with a Poisson family, log link, and robust variance estimation; each model accounts for repeated measures within one individual through the use of generalized estimating equations (GEE). Measurements are divided into three time-periods: 0–11 days after symptom onset, 12–19 days after symptom onset, and ≥20 days after symptom onset. If an individual had more than one measurement of a biomarker during any particular time period, we used the average of all values. Each model controls for participant age and gender.
Extended Data Fig. 9: Gating strategies.

a, Leukocyte gating strategy to identify lymphocytes, granulocytes, monocytes, pDCs, and cDCs in Figs. 1b, c, 2d–f and Extended Data Fig. 2a. b, T cell surface staining gating strategy to identify CD4 and CD8 T cells, TCR-activated T cells, terminally-differentiated T cells, and additional subsets as shown in Extended Data Fig. 2b. c, Intracellular T cell gating strategy to identify CD4 and/or CD8 T cells secreting TNF, IFNγ, IL-6, IL-2, granzyme B, IL-4, and/or IL-17 in Extended Data Figs. 2c, 5a, b.
Table 1:
Unless otherwise noted, relative risks were not statistically significant. Moderate (clinical score 1–3) and severe (clinical score 4–5) disease status were assigned as described in Methods. Percentages of sub-group (moderate or severe) are shown for each category with respective counts in parenthesis. Average age was calculated with accompanying sample standard deviation. Ethnicity and BMI were extracted from most recent electronic medical record (EMR) data. Select COVID-19 risk factors were scored by a clinical infectious disease physician. Presenting symp- toms were recorded through direct interview with patient or surrogate or retrospective EMR review.
| Moderate COVID-19 | Severe COVID-19 | Relative Risk (95% Cl); [*; p-value] | Total | |
|---|---|---|---|---|
| Number | 70.8% (80/113) | 29.2% (33/113) | 113 | |
| Age (years) | 62.66 ± 16.1 | 63.67 ± 19.3 | [n.s.] | 62.96 ± 17.0 |
| Sex | ||||
| Male | 45% (36/80) | 48.48% (16/33) | 1.07 (.70 −1.65) | 46.02% (52/113) |
| Female | 55% (44/80) | 51.52% (17/33) | .94 (.64 −1.38) | 53.98% (61/113) |
| Ethnicity | ||||
| American Indian /Alaskan Native | 0% (0/80) | 0% (0/33) | -- | 0% (0/113) |
| Asian | 1.25% (1/80) | 0% (0/33) | -- | 0.88% (1/113) |
| Black / African American | 27.5% (22 / 80) | 33.33% (11/33) | 1.21 (.67 – 2.21) | 29.2% (33/113) |
| Native Hawaiian / Pacific Islander | 0% (0/80) | 0% (0/33) | 0% (0 /113) | |
| White | 53.75% (43/80) | 54.55% (18/33) | 1.01 (.70 −1.47) | 53.98% (61 /113) |
| Hispanic | 12.5% (10/80) | 12.12% (4/33) | .97 (.33 – 2.87) | 12.39% (14/113) |
| Multiple | 0% (0/80) | 0% (0/33) | -- | 0% (0 /113) |
| Unknown | 5% (4/80) | 0% (0/33) | n.c. | 3.54% (4/113) |
| BMI | ||||
| <18.5 | 0% (0/80) | 6.06% (2/33) | n.c. | 1.77% (2/113) |
| 18.5–24.9 | 21.25% (17/80) | 9.09% (3/33) | .43 (.13–1.36) | 17.7% (20/113) |
| 25.0–29.9 | 32.5% (26/80) | 24.24% (8/33) | .75 (.38 −1.47) | 30.09% (34/113) |
| 30–35 | 27.5% (22/80) | 30.3% (10/33) | 1.10 (.59–2.06) | 28.32% (32 /113) |
| >35 | 18.75% (15/80) | 30.3% (10/33) | 1.62 (.81–3.22) | 22.12% (25/113) |
| COVID Risk Factors | ||||
| None | 27.5% (22/80) | 30.3% (10/33) | 1.10 (.59–2.06) | 28.32% (32/113) |
| Cancer Treatment within 1 year | 7.5% (6/80) | 15.15% (5/33) | 2.02 (.66–6.16) | 9.73% (11/113) |
| Chronic Heart Disease | 27.5% (22/80) | 24.24% (8/33) | .88 (.44 −1.78) | 26.55% (30/113) |
| Hypertension | 53.75% (43/80) | 48.48% (16/33) | .90 (.60–1.35) | 52.21% (59/113) |
| Chronic Lung Disease (asthma, COPD, ILD) | 26.25% (21/80) | 18.18% (6/33) | .69 (.31–1.56) | 23.89% (27/113) |
| Immunosupression | 11.25% (10/80) | 6.06% (2/33) | .52 (.12–2.29) | 9.73% (12/113) |
| Solid Organ Transplant | 6.25% (4 / 80) | 3.03% (1/33) | .60 (.07 – 5.16) | 4.42% (5 /113) |
| HIV+ (with anti-viral treatment; CD4 > 400) | 2.5% (2/80) | 0% (0 / 33) | n.c. | 1.77% (2 /113) |
| Other (Multiple Sclerosis, Rheumatoid Arthritis, Scleroderma, Cirrhosis) | 3.75% (3 / 80) | 3.03% (1/33) | 1.21 (.11–12.91) | 3.54% (4 /113) |
| Presenting Symptoms | ||||
| Headache | 56.9% (33/58) | 47.37% (9/19) | .83 (.49 −1.41) | 54.55% (42/77) |
| Objective Fever (> 100.3 °F / 37.9 °C) | 64.29% (36/56) | 65% (13/20) | 1.01 (.69 −1.47) | 64.47% (49/76) |
| Cough | 77.19% (44/57) | 65% (13/20) | .84 (.59 −1.20) | 74.03% (57/77) |
| Dyspnea | 64.41% (38/59) | 75% (15/20) | 1.16 (.85–1.60) | 67.09% (53/79) |
| Rhinorrhea | 30.36% (17/56) | 35.29% (6/17) | 1.16 (.55 – 2.48) | 31.51% (23/73) |
| Sore Throat | 27.59% (16/58) | 22.22% (4/18) | .81 (.31–2.10) | 26.32% (20/76) |
| Nausea | 48.28% (28/58) | 41.18% (7/17) | .85 (.46 −1.60) | 46.67% (35/75) |
| Vomitting | 31.03% (18/58) | 27.78% (5/18) | .90 (.39 – 2.07) | 30.26% (23/76) |
| Diarrhea | 50% (29/58) | 35.29% (6/17) | .71 (.35–1.41) | 44% (33/75) |
| Abdominal Pain | 31.03% (18/58) | 5.88% (1/17) | .19 (.03–1.32) | 25.33% (19/75) |
| Hypogeusia | 37.04% (20/54) | 33.33% (5/15) | .90 (.41 −1.99) | 36.23% (25/69) |
| Anosmia | 31.37% (16/51) | 33.33% (5/15) | 1.06 (.47 – 2.42) | 31.82% (21/66) |
| All Cause Mortality | 3.75% (3/80) | 27.27% (9/33) | 7.27 *** (2.10 – 25.19) [ p = .0002] | 10.62% (12 / 113) |
Supplementary Material
Acknowledgements
We thank Melissa Linehan for technical and logistical assistance, and thank helpful discussions with Drs. Andrew Wang, Aaron Ring, Craig Wilen and Daniel Mucida. This work was supported by the Women’s Health Research at Yale Pilot Project Program (AI, AR), Fast Grant from Emergent Ventures at the Mercatus Center, Mathers Foundation, and the Ludwig Family Foundation, the Department of Internal Medicine at the Yale School of Medicine, Yale School of Public Health and Beatrice Kleinberg Neuwirth Fund. IMPACT received support from the Yale COVID-19 Research Resource Fund. A.I. is an Investigator of the Howard Hughes Medical Institute. C.L. is a Pew Latin American Fellow. P.Y is supported by Gruber Foundation and the NSF. B.I is supported by NIAID 2T32AI007517–16. CBFV is supported by NOW Rubicon 019.181EN.004.
Yale IMPACT Research Team Authors:
Abeer Obaid11, Alice Lu-Culligan1, Allison Nelson11, Anderson Brito3, Angela Nunez11, Anjelica Martin1, Annie Watkins3, Bertie Geng11, Chaney Kalinich3, Christina Harden3, Codruta Todeasa11, Cole Jensen3, Daniel Kim1, David McDonald11, Denise Shepard11, Edward Courchaine12, Elizabeth B. White3, Eric Song1, Erin Silva11, Eriko Kudo1, Giuseppe DeIuliis9, Harold Rahming11, Hong-Jai Park11, Irene Matos11, Jessica Nouws11, Jordan Valdez11, Joseph Fauver3, Joseph Lim13, Kadi-Ann Rose11, Kelly Anastasio14, Kristina Brower3, Laura Glick11, Lokesh Sharma11, Lorenzo Sewanan11, Lynda Knaggs11, Maksym Minasyan11, Maria Batsu11, Mary Petrone3, Maxine Kuang3, Maura Nakahata11, Melissa Campbell8, Melissa Linehan1, Michael H. Askenase15, Michael Simonov11, Mikhail Smolgovsky11, Nicole Sonnert1, Nida Naushad11, Pavithra Vijayakumar11, Rick Martinello4, Rupak Datta4, Ryan Handoko11, Santos Bermejo11, Sarah Prophet16, Sean Bickerton12, Sofia Velazquez15, Tara Alpert4, Tyler Rice1, William Khoury-Hanold1, Xiaohua Peng11, Yexin Yang1, Yiyun Cao1, Yvette Strong11.
Yale IMPACT Research Team Authors Affiliation:
1 Department of Immunobiology, Yale University School of Medicine, New Haven, CT 06520
3 Department of Epidemiology of Microbial Diseases, Yale School of Public Health, New Haven, CT 06520
4 Department of Medicine, Section of Infectious Diseases, Yale University School of Medicine, New Haven, CT, 06520
9 Department of Medicine, Section of Pulmonary and Critical Care Medicine; Yale University School of Medicine, New Haven, CT 06520
11 Yale School of Medicine, New Haven, CT, 06520.
12 Department of Biochemistry and of Molecular Biology, Yale University School of Medicine, New Haven, CT 06520
13 Yale Viral Hepatitis Program; Yale University School of Medicine, New Haven, CT 06520
14 Yale Center for Clinical Investigation; Yale University School of Medicine, New Haven, CT 06520
15 Department of Neurology, Yale University School of Medicine, New Haven, CT 06520
16 Department of Molecular, Cellular and Developmental Biology, Yale University School of Medicine, New Haven, CT 06520
Footnotes
Competing interests
The authors declare no competing financial interests.
Data Availability Statement
All of the background information of HCWs, clinical information of patients, and raw data used in this study are included in the Supplementary Information Table 1. Additionally, all of the raw fcs files for the flow cytometry analysis are uploaded in ImmPort (https://www.immport.org/shared/home, Study ID: SDY1655).
References
- 1.Giamarellos-Bourboulis EJ et al. Complex Immune Dysregulation in COVID-19 Patients with Severe Respiratory Failure. Cell Host Microbe, doi: 10.1016/j.chom.2020.04.009 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou Z et al. Heightened Innate Immune Responses in the Respiratory Tract of COVID-19 Patients. Cell Host Microbe, doi: 10.1016/j.chom.2020.04.017 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang C et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506, doi: 10.1016/S0140-6736(20)30183-5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mathew D et al. Deep immune profiling of COVID-19 patients reveals patient heterogeneity and distinct immunotypes with implications for therapeutic interventions. bioRxiv, 2020.2005.2020.106401, doi: 10.1101/2020.05.20.106401 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann M et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell, doi: 10.1016/j.cell.2020.02.052 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan R et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367, 1444–1448, doi: 10.1126/science.abb2762 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J & Subbarao K The Immunobiology of SARS. Annual Review of Immunology 25, 443–472, doi: 10.1146/annurev.immunol.25.022106.141706 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Jose RJ & Manuel A COVID-19 cytokine storm: the interplay between inflammation and coagulation. Lancet Respir Med, doi: 10.1016/S2213-2600(20)30216-2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen G et al. Clinical and immunologic features in severe and moderate Coronavirus Disease 2019. J Clin Invest, doi: 10.1172/JCI137244 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen N et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 395, 507–513, doi: 10.1016/S0140-6736(20)30211-7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Annunziato F, Romagnani C & Romagnani S The 3 major types of innate and adaptive cell-mediated effector immunity. J Allergy Clin Immunol 135, 626–635, doi: 10.1016/j.jaci.2014.11.001 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Iwasaki A & Medzhitov R Control of adaptive immunity by the innate immune system. Nat Immunol 16, 343–353, doi: 10.1038/ni.3123 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Shea JJ & Paul WE Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science 327, 1098–1102, doi: 10.1126/science.1178334 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ostendorf L et al. Low-Density Granulocytes Are a Novel Immunopathological Feature in Both Multiple Sclerosis and Neuromyelitis Optica Spectrum Disorder. Front Immunol 10, 2725, doi: 10.3389/fimmu.2019.02725 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalfaoglu B, Almeida-Santos J, Adele Tye C, Satou Y & Ono M T-cell hyperactivation and paralysis in severe COVID-19 infection revealed by single-cell analysis. bioRxiv, 2020.2005.2026.115923, doi: 10.1101/2020.05.26.115923 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blanco-Melo D et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 181, 1036–1045 e1039, doi: 10.1016/j.cell.2020.04.026 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuri-Cervantes L et al. Immunologic perturbations in severe COVID-19/SARS-CoV-2 infection. bioRxiv, 2020.2005.2018.101717, doi: 10.1101/2020.05.18.101717 (2020). [DOI] [Google Scholar]
- 18.Gabay C, Lamacchia C & Palmer G IL-1 pathways in inflammation and human diseases. Nat Rev Rheumatol 6, 232–241, doi: 10.1038/nrrheum.2010.4 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Wang D et al. The regulation of the Treg/Th17 balance by mesenchymal stem cells in human systemic lupus erythematosus. Cell Mol Immunol 14, 423–431, doi: 10.1038/cmi.2015.89 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duffield JS, Lupher M, Thannickal VJ & Wynn TA Host responses in tissue repair and fibrosis. Annu Rev Pathol 8, 241–276, doi: 10.1146/annurev-pathol-020712-163930 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cameron MJ et al. Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome. J Virol 81, 8692–8706, doi: 10.1128/JVI.00527-07 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yap JKY, Moriyama M & Iwasaki A Inflammasomes and Pyroptosis as Therapeutic Targets for COVID-19. J Immunol, doi: 10.4049/jimmunol.2000513 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wyllie AL et al. Saliva is more sensitive for SARS-CoV-2 detection in COVID-19 patients than nasopharyngeal swabs. medRxiv, 2020.2004.2016.20067835, doi: 10.1101/2020.04.16.20067835 (2020). [DOI] [Google Scholar]
- 24.Vogels CBF et al. Analytical sensitivity and efficiency comparisons of SARS-COV-2 qRT-PCR primer-probe sets. medRxiv, 2020.2003.2030.20048108, doi: 10.1101/2020.03.30.20048108 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu Z, Eils R & Schlesner M Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849, doi: 10.1093/bioinformatics/btw313 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Charrad M, Ghazzali N, Boiteau V & Niknafs A NbClust: An R Package for Determining the Relevant Number of Clusters in a Data Set. 2014 61, 36, doi: 10.18637/jss.v061.i06 (2014). [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


