Abstract
The ongoing crisis due to the global pandemic caused by a highly contagious coronavirus (Coronavirus disease – 2019; COVID-19) and the lack of either proven effective therapy or a vaccine has made diagnostic a valuable tool in disease tracking and prevention. The complex nature of this newly emerging virus calls for scientists’ attention to find the most reliable, highly sensitive, and selective detection techniques for better control or spread of the disease. Reverse transcriptase-polymerase chain reaction (RT-PCR) and serology-based tests are currently being used. However, the speed and accuracy of these tests may not meet the current demand; thus, alternative technology platforms are being developed. Nano biosensor technology platforms have been established as a promising diagnostic tool for rapid and accurate detection of viruses as well as other life-threatening diseases even in resource-limited settings. This review aims to provide a short overview of recent advancements in molecular and biosensor-based diagnosis of viruses, including the human coronaviruses, and highlight the challenges and future perspectives of these detection technologies.
Keywords: Biosensors, RT-PCR, Coronavirus, SARS-CoV, MERS-CoV, COVID-19
Abbreviations: S, spike protein; Hel, helicase; E, envelope protein; N, nucleocapsid protein; RdRp, RNA dependent RNA polymerase; NA, not available
1. Introduction
Coronaviruses are a group of single-stranded RNA viruses of size ranging from 26.4 to 31.7 kb [1], which are one of the largest RNA viruses belong to Coronaviridae family and Orthocoronavirinae subfamily isolated originally from domestic chickens in 1930 in North Dakota, USA [2]. The sub-family of Orthocoronavirinae comprises four genera; alpha (α), beta (β), gamma (γ) and delta (δ) coronavirus. Coronaviruses infect birds (γ and δ) and several mammalian species (mainly α and β), including humans. Human coronaviruses (HCoVs) were first discovered in the 1960s and till date, six coronaviruses have been identified, including the HCoVs-NL63 and HCoVs-229E (α-Coronaviruses) and HCoVs-OC43, HCoVs-HKU1 (β-Coronaviruses), severe acute respiratory syndrome-CoV (SARS-CoV), and Middle East respiratory syndrome-CoV (MERS-CoV) [1,3].
In December 2019, a deadly infectious respiratory disease emerged in Wuhan, Hubei Province, China, which was caused by a newly evolved coronavirus, that has infected over 29.3 million people across 230 countries around the world with nearly 929,000 deaths (Johns Hopkins University and Medicine database; https://coronavirus.jhu.edu/map.html). The disease is now known as SARS-coronavirus 2 (SARS-CoV-2) or COVID-19, which is closely related to SARS-CoV. Coronavirus family including SARS-CoV-2 virus is an enveloped RNA (30 kb) virus decorated with spike protein (S), hemagglutinin-esterase dimer (HE), a membrane glycoprotein (M), an envelope protein (E), and a nucleocapsid protein (N) [4]. RNA dependent RNA polymerase (RdRp) is a key catalytic subunit for RNA synthesis and viral replication in a host [5]. The receptor-binding domain of spike protein (S) binds to human angiotensin-converting enzyme 2 (ACE2), which is identified as the receptor in the human host similar to SARS-CoV [6]. The spike protein comprises two subunits (S1 and S2) found on the surface of the viral particles and has been reported to be highly immunogenic and promotes entry into cells and is the main target of circulating antibodies [4].
The COVID-19 is an acute respiratory disease and the patients with underlying conditions are most susceptible including the elderly (>60 years age), obese and immunocompromised individuals, diabetic patients, and individuals suffering from cancer and heart disease. The infected patients suffer from pneumonia and sepsis, often with grave prognosis (2–3% mortality).
During infection, the S protein binds to the host receptor, ACE2 on human epithelial cells in the respiratory tract and the virus enters the cells [6]. The virus propagates in the alveolar epithelial cells, causes cell damage, and induces a severe inflammatory response. The respiratory symptoms of the COVID-19 disease are accompanied by fever (~100.4 °F), cough, and myalgia or fatigue and patients may recover after an illness lasting for 10–14 days. The disease may progress to pneumonia and patients with underlying conditions may suffer from fatal acute respiratory distress syndrome [7]. In March 2020, the World Health Organization declared the SARS-CoV-2 as pandemic and implemented a public health emergency of international concern [3].
In response to coronavirus pandemic, the discovery of rapid and highly sensitive and specific methods including molecular and biosensor-based methods is crucial in various clinical settings. Human coronaviruses are highly contagious respiratory tract viruses that became the prioritized public health threat throughout the globe [1]. Like other pathogenic viruses, these viruses evolved with continuous mutations in animals and adapted the environment then jumped to humans very quickly. Due to the lack of effective therapeutic drugs as well as preventive vaccines, quick and accurate diagnosis and management are critical for tackling the outbreak particularly in developing countries where resources are limited [8]. Therefore, there is an urgent need for a diagnostic platform to detect, track, and contain this life-threatening highly infectious virus readily.
2. Present coronavirus detection approaches
2.1. RT-PCR-based diagnostics
At present, the reverse transcriptase (RT)-PCR-based molecular test is used for the SARS-CoV-2 diagnosis [9]. As SARS-CoV-2 is an RNA virus, reverse transcriptase enzyme is used to synthesize cDNA which is then amplified by using DNA-polymerase and a set of specific primers such as those targeting genes encoding ORF1b, RdRp, E, S, N or a combination of several target genes (Table 1, Table 2 ) [[10], [11], [12], [13]]. The accuracy of the RT-PCR-based diagnosis depends on specimen collection, disease progression, the specificity of the primer sets, and strain involvement.
Table 1.
Comparison between the currently used molecular and serological methods for SARS-CoV-2 detection.
| Diagnostic method | Target gene or protein | Specificity/Sensitivity | Time needed | References |
|---|---|---|---|---|
| RT-qPCR | RdRp, E, S, N | 71% (sensitivity) | 4 h | [13] |
| rRT-PCR | N, E | 95% | Several hours | [33] |
| RT-PCR | RdRp/Hel, S, and N | 95% | Several hours | [34] |
| 29,870-bp, excluding the poly (A) tail | NA | NA | [35] | |
| E, RdRp, S, N, ORF1ab | NA | NA | [36] | |
| RdRP, N, E, and S | NA | NA | [37] | |
| E, RdRp | Low sensitivity | NA | [38] | |
| RT-digital PCR | Molecular | 100%/90% | 4 h | [39] |
| LAMP-based RT-qPCR |
Molecular | 97.6% (42/43) (sensitivity) | 30 min | [40] |
| ELISA (IgM + IgG) | Serological | 100%/87.3% | 24 h | [41] |
| Lateral flow immunochromatographic strip (LFICS) | Point-of-care device | 90.63%/88.66% | 15 min | [42] |
| Chest CT scan | Physical imaging | 25%/97% | 15–30 min | [43] |
Table 2.
Rapid methods including biosensors used for the detection of coronaviruses.
| Virus target | Biosensor type | Viral target molecule | Detection limit | Assay time | References |
|---|---|---|---|---|---|
| SARS-CoV | Localized surface plasmon coupled fluorescence (LSPCF) fiber-optic biosensor | SARS-CoV N (GST-N) protein | ~1 pg/mL | The very early stage of infection | [64] |
| Nanowire Biosensors | Nucleocapsid (N) protein | 0.6 nM | 45 min | [65] | |
| Imaging ellipsometry | scFv, b1 and h12 | 2.2 μg/mL (b1) and 34 μg/ml (h12) | 40 min | [66] | |
| Indirect immunofluorescence | SARS-CoV-IgG | NA | Several hours | [67] | |
| MERS-CoV | RT-LAMP-VF | N gene of MERS-CoV | 1 × 101 copies/μl of MERS-CoV RNA | 35 min | [68] |
| One-pot RT-LAMP | N gene of MERS-CoV | 4 × 103 to 4 × 10−1 RNA copies | 60 min | [69] | |
| Multiplex Paper-based Colorimetric Sensor | Viral RNA | 1.53 nM | 7–10 min | [70] | |
| RT-LAMP (Loop-Mediated Isothermal Amplification) | Viral RNA | 3.4 synthetic RNA molecules | 30–45 min | [71] | |
| Luciferase-based Biosensors | Papain-like protease (PLpro) and the 3-chymotrypsin-like protease (3CLpro) | ~12.5 μM | Several hours | [72] | |
| SARS-Cov-2 | Localized surface plasmon resonance | Nucleic Acid | 0.22 pM | [73] | |
| Lanthanide-Doped Nanoparticles-based Lateral Flow Immunoassay | Nucleocapsid phosphoprotein | NA | 10 min | [74] | |
| CRISPER/Cas-12 a based detection with the naked eye | Nucleic Acid | 10 copies | 45 min | [20] | |
| DNA Endonuclease-Targeted CRISPR Trans Reporter (DETECTR); (Lateral Flow detection) | Nucleic Acid | 10 copies/μl input | 45 min (with manual RNA extraction) | [21] | |
| Reverse transcription recombinase-aided amplification (RT-RAA) | Nucleic Acid | 2 copies | 15 min (with manual RNA extraction) | [18] | |
| Field-Effect Transistor-based Biosensor | Spike protein | 2.42 × 102 copies/mL in a clinical sample | Few minutes | [75] | |
| RT-LAMP + lateral flow | ORF 1a/b | 12 copies | 1 h | [76] | |
| RT-LAMP | Nucleic Acid | 80 copies of viral RNA/mL | 30 min | [11] | |
| RT-LAMP | Nucleic Acid | NA | 30–45 min | [77] |
The virus primarily affects the lower respiratory tract. Even though in some cases viral RNA can be detected from saliva, nasal and pharyngeal swabs, and bronchoalveolar lavage. However, the collection of the lower respiratory samples requires both a suction device and a skilled operator. An earlier study showed that nasal swabs and throat swabs collected ~8 days post-disease onset had a low positive rate compared to bronchoalveolar lavage samples, especially in mild to asymptotic cases. The viral colonization of the lower respiratory tract and the collection of respiratory specimens cause a high false-negative rate in real-time RT-PCR tests [14].
In some samples of hospitalized patients, RT-PCR tests showed false-negative and fluctuating trends, which may be caused by insufficient virus particles in the sample [15]. False-negative results also may occur by mutations in the target regions of the SARS-CoV-2 genome. Although most of the RT-PCR assays are based on the conserved regions of the viral genome, variability causing mismatches between the primers and probes and the target sequences can lead to a decrease in assay performance and possible false-negative results. In this context, multiple genes can be targeted at a time for amplification to avoid false results [16]. Recently, SARS-CoV-2 detection assays using seven different primer-probe sets in one complete testing kit was applied to clinical samples, where all assays evaluated were highly specific, and those using the N2 and the E -gene sets, were found to be more sensitive than others [12]. Besides, the use of a 5′ nuclease probe in the real-time quantitative assay can minimize the false positive rate due to an increase in signal specificity [17].
RT-PCR assay is generally time-consuming, labor-intensive, and requires certified laboratory and skilled technicians. These limitations make RT-PCR an inconvenient tool for quick patient screening and therefore, the search for a precise, rapid and user-friendly high throughput screening test to identify SARS-CoV-2 infected patients on a large scale is urgently needed for timely therapeutic intervention and to prevent virus transmission.
To overcome these drawbacks, the RT-PCR method has been modified with other amplification process or point-of-care assays. A reverse transcription recombinase-aided amplification (RT-RAA) assay for SARS-CoV-2 using a portable instrument was developed in China [18]. The system requires the pre-extraction of RNA from samples before analysis. The RAA uses a recombinase that binds tightly to the primer to form a complex. A specific fluorescent probe is included in the amplification system to obtain results within 15 min [18].
Another reverse transcription loop-mediated isothermal amplification (RT-LAMP) was developed to detect the ORF1ab and S genes of SARS-CoV-2 in 18–20 min. In this point-of-care testing platform, positive results could be judged by the naked eye with a color change from orange to green [19]. DNA Endonuclease-Targeted CRISPR Trans Reporter (DETECTR) is a comparatively new method of SARS-Cov-2 detection [20]. This assay involves simultaneous reverse transcription and isothermal amplification using loop-mediated amplification (RT–LAMP) for RNA, followed by Cas12 detection of predefined coronavirus sequences, after which cleavage of a reporter molecule confirms detection of the E and N genes of SARS-CoV-2. Cas12 fluorescent signals can be visualized by lateral flow assay within 5 min [21].
2.2. Serological diagnostics
Serological tests can play a critical role in the fight against COVID-19 by helping healthcare professionals to identify individuals who have overcome infection in the past and have developed an immune response. COVID-19 disease is reported to induce acute antibody (IgM, IgG) response in all patients and the levels of anti-viral antibodies are plateaued within 6-days after seroconversion [22]. Therefore, a “serology” test can be performed to measure anti-viral antibodies in the blood and can potentially be used in patients with negative RT-PCR or asymptomatic patients [23]. Serological/antibody-based assays are rapid and efficient for the screening of many pathogenic infections, as pathogen-specific IgM and IgG antibodies can be detected with various immunoassays [24], which has relatively high throughput capacity and less stringent specimen requirements (uniformity in serum collection) than viral RNA-based assays [14]. Detecting IgM and IgG against the SERS-CoV-2 offers an alternate means to diagnose a patient. Rapid lateral flow-based assays for anti- COVID-19 antibodies (IgM and IgG) are under development which will play an important role in the epidemiological investigation of the disease [9]. This method can be used to quickly screen all febrile patients effectively, as large-scale confirmation or exclusion of patients is essential for preventing the spread of disease and for an epidemiological survey. However, there is a concern if the serology test could be useful for the diagnosis of asymptomatic or symptomatic patients who may not mount a detectable immune response. Interestingly, anti-SARS-CoV-2 antibodies (seroconversion requires at least 7 days of infection in a patient) have the potentials to neutralize the virus and to protect people from developing future infections [25]. Therefore, the convalescent plasma has been used as therapy for the treatment of critically ill COVID-19 patients [26,27].
A comparison between the molecular test (i.e., RT-PCR) and serological test (i.e., immunoassay) showed that the molecular test has better sensitivity and specificity. Hence, enhancements to the current molecular test are required for improving the diagnosis. But studies have shown that high levels of PCR inhibitors in samples hinder the PCR sensitivity [28]. Furthermore, to avoid potential cross-reaction with other endemic coronaviruses as well as the possible genetic drift of SARS-CoV-2, at least two molecular targets should be included in the assay. Equally unappreciated is the need for a broad screening tool to determine the actual mortality rate as well as other epidemiological markers. Finally, the importance of rapid development of integrated, random access, point-of-care molecular devices for the accurate diagnosis of SARS-CoV-2 infections cannot be overemphasized. In the future, this may potentially be used to help determine, together with other clinical data, that such individuals are no longer susceptible to infection and can return to work. Also, these test results can aid in determining who may donate a part of their convalescent plasma for possible treatment of seriously ill patients.
2.3. Chest CT scan
At present, computed tomography (CT) scans have been employed for the diagnosis of COVID-19. CT scan is an X-ray-based diagnosis tool where the cross-sectional view of the patient's lung is examined from a computer system connected to the apparatus [[29], [30], [31]]. However, a CT scan requires skilled human resources and is expensive. It is limited to only central hospitals and healthcare facilities. This diagnostic technique is not pathogen-specific and does not work for mild pre-symptomatic and asymptomatic infections without pneumonia. The X-ray image needs to be interpreted by more than one radiologist to have confidence in the diagnosis. Lately, a 3D deep scan framework for the diagnosis of COVID-19 has been developed [31,32]. The results indicated that the reported model achieved a specificity of 96% and a sensitivity of 90% when assessing the pathology of lungs in COVID patients.
3. Biosensors for detection of coronaviruses
In a biosensor, biorecognition elements bind the target analyte, a transducer propagates the amplified signal and a computer digitally interrogates the data [44,45]. Based on the types of transducers used, biosensors are categorized into optical, electrical, electrochemical, and mass (piezoelectric) sensors. Improved sensitivity, automated data acquisition, portability, and high throughput capabilities make the biosensor devices highly attractive [46]. The first successful and widely popular biosensor was built for the detection of blood glucose. Since then, research in the area of biosensor development has exploded in the last two decades for the detection of bacteria, viruses, toxins, enzymes, proteins, allergens, heavy metals, pollutants, pesticides, and drugs residues. Biosensors are increasingly being developed for pathogen detection and disease diagnosis and the success of the biosensor depends on the specificity and affinity of biorecognition molecules towards the target analytes [[47], [48], [49]]. Various biorecognition elements are now used including a nucleic acid or gene sequence, antibody, aptamer, antimicrobial peptides, enzymes, bacteriophages, molecularly imprinted polymers, peptide nucleic acid (PNA), and host cell receptor molecules used by pathogens [50,51].
In modern biosensor platforms, gold nanoparticles (GNPs) have been applied to enhance the detection sensitivity. GNPs have distinct optical features such as high biocompatibility, stability, special electronic and catalytic activity, and high electron transfer rate [52,53].
Among the microbial pathogens, reports of biosensors against bacterial pathogens are numerous while scanty for human viral pathogens [54] (Table 2). This may be due to the lack of laboratory facilities to grow and maintain viruses since they require a living host (animals or cell lines). Sometimes, the lack of a cell culture model (example, norovirus) significantly impedes virus research. Moreover, a majority of viruses are highly infectious and contagious, thus requiring a class II, III, or class IV biological containment facility which is expensive to set up. Nevertheless, a few biosensors are developed for viral pathogens [54]. A magnetic bead mediation surface plasmon resonance biosensor platform was reported for HIV-1 viral protease [55] or viral load in the blood [56]. A bead-based electrochemical biosensor was reported for the detection of hemagglutinin protein in the influenza virus [57]. Biosensors have been developed for Zika virus [58], avian influenza (H7N9) [59], swine influenza (H1N1) [60], dengue virus [61], Ebola virus [62], and coronavirus disease-19 [63].
3.1. Field-effect transistor based biosensor (BioFRT)
A field-effect transistor (FET) based biosensing device consists of a three-electrode structure containing the drain, source, and gate and it has been developed for the viral disease diagnosis and detection of small molecules. FET is an electric biosensor and monitors changes in the surface potential after the target molecule binds to the biorecognition element immobilized on the highly conductive chip surface [78]. FET-based biosensors utilize semiconductor materials such as graphene, molybdenum disulfide, titanium oxide, zinc oxide, and gallium nitride. It is highly sensitive allowing prompt detection of a small amount of analyte to the probes immobilized on the conducting channel [79]. Graphene-based FET biosensors can sense the changes in the surrounding area on their surface and offer the best sensing milieu for low-noise and ultrasensitive detection due to their high carrier mobility, high electronic conductivity and experimental flexibility [80]; therefore, the graphene-based FET platform is well suited for sensitive immunological diagnosis [81]. FET-based biosensor using graphene oxide has been developed for the detection of Ebola virus antigen at nanogram quantities in real-time [82].
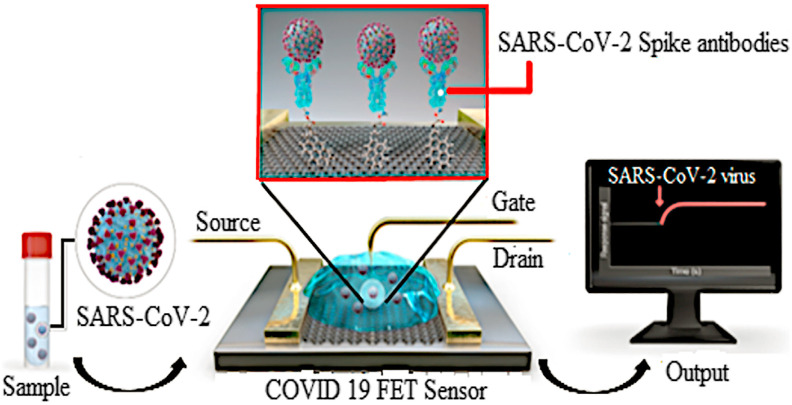
Seo et al. [75] have developed a FET-based biosensor for the detection of COVID-19. The biosensor was developed by using a spike protein of SARS-CoV-2 immobilized onto the FET graphene sheet (a two-dimensional sheet of hexagonal oriented carbon atom) with 1-pyrene butyric acid N-hydroxy succinimide ester (PBASE) (Fig. 1 ). The team used the spike protein of the virus due to its sequence diversity among the coronaviruses and they have evaluated its performance by enzyme-linked immunosorbent assay (ELISA) before immobilizing onto the FET surface. The sensor was designed to detect the SARS-CoV-2 virus in clinical samples with a 1 fg/mL detection limit. Most importantly, the device showed no significant cross-reactivity with the MERS-CoV antigen. Hence, the sensor developed by this research group provides fast, highly sensitive, and specific detection of the SARS-CoV-2 virus in clinical samples.
Fig. 1.
Schematic illustration of the operating procedure of graphene-based FET for detection of COVID-19 (Source: Seo et al., 2020 [75]).
3.2. Localized surface plasmon coupled fluorescence (LSPCF) fiber-optic biosensor
The LSPCF fiber-optic biosensor is excited by localized surface plasmon where it comprises a complex of biomolecules arranged in a sandwich configuration immobilized on the surface of an optical fiber. This biosensor has enormous advantages; simple, easy to operate, and early diagnosis of disease [83]. LSPCF sensor has been developed for the detection of the swine flu virus (H1N1) [60] and SARS CoV [64]. Huang et al. [64] integrated the LSP technique and the sandwich immunoassay to detect the SARS-CoV nucleocapsid protein (N protein) in the patient serum even one day after infection. The detection sensitivity was recorded to be 1 pg/mL for recombinant SARS-CoV N (GST-N) protein in blood/serum. Such improved sensitivity would allow the diagnosis of the SARS patient at the early onset of the disease. Notably, the linear correlation between the fluorescence signal and the amount of GST-N protein was attained from 0.1 pg/mL - 1 ng/mL. Using the same monoclonal antibodies, a 104-fold increase in detection limit has been recorded using LSPCF fiber-optic biosensor in contrast with commercialized antigen capture ELISA. Moreover, the LSPCF system is a highly effective chip-based assay for both quantitative and qualitative measurements of viral proteins in the serum. The ultra-low detection limit of this biosensor is attributed to several properties. First and foremost, the LSPCF stimulates and boosts high-efficiency fluorescence near the gold nanoparticle (GNP) surface. Second, the intensity of fluorescence is amplified as a result of a high number of fluorophores found on each fluorescence – labeled probe and excited at once. Third, protein A on the fluorescence labeled probe is impregnated on the surface of GNP, acts as a spacer as well as a linker by networking to the secondary antibody labeled fluorophores. Last, in the conventional setup, the fluorescence signal is situated at the distal proximity of the optical fiber.
3.3. Surface plasmon resonance sensor
Surface plasmon resonance (SPR) is generated during optical illumination of a metal surface and is widely used to monitor binding events between two molecules. Traditional SPR-based diagnostic platforms have several advantages; high-throughput, sensitive, label-free, economical, easy-to-use, and real-time monitoring capabilities [49,84]. Recently SPR sensor was successfully developed for the detection of several viral pathogens including Dengue, Ebola, and Zika [85] and the detection limit for Dengue was 0.08 pM of Dengue Protein (DENV-2 E-proteins) suggesting SPR's utility as a sensitive detection platform for viruses.
3.4. Electrochemical immunosensor
The electrochemical transducer-based biosensors are the most popularly used biosensors for the detection of viruses including the influenza virus (H1N1, H5N1, H7N9) [86,87], Hepatitis B virus [88], Zika virus [89], Dengue virus [90] and MERS-CoV [91]. The immunosensor for MERS-CoV detection was developed on carbon electrodes array (DEP) for multiplexed detection of different coronaviruses simultaneously [91]. This competitive immunosensor allows one step, selective and sensitive detection of MERS-CoV that can be effectively applied in spiked nasal samples with significant recovery efficiency. The detection centered on the actual competition between immobilized MERS-CoV spike protein S1 and the free virus in the sample for a specified concentration of antibody added to the sample. The output of the sensor was detected by visualizing the changes in square wave voltammetry (SWV) signal current peak upon placing MERS-CoV antigen at various concentrations. The electrochemical measurements using ferrocyanide/ferricyanide as a probe has been used for the generation of the voltammetry signal. A mixture containing 5 mM solution of ferrocyanide/ferricyanide ([Fe (CN)6]3−/4-) (1:1 ratio) in 0 0.1 M PBS, pH 7.4 has been used to measure the electrochemical signal. The immunosensor response was calculated as (Io-I)/Io%, where (Io) is the SWV current measured at the antigen-modified electrodes after blocking with 1% BSA and (I) is the current measured after the immunosensor was incubated with a mixture of anti-HCoV antibody or MERS-CoV antibody with different concentrations of free target analytes (MERS-CoV or HCoV) [91]. This immunosensor showed a very low detection limit (1 pg/ml) and low cross-reactivity [91]. This sensor was also able to detect HCoV at 0.4 pg/ml in 20 min [91].
3.5. Dual-functional plasmonic photothermal biosensors
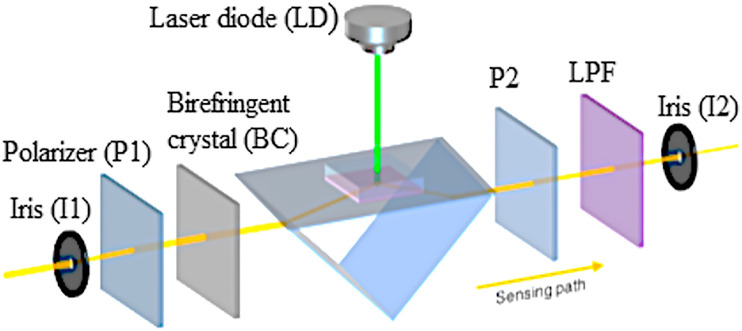
The dual-functional plasmonic biosensing concept combines the plasmonic photothermal (PPT) effect and the localized surface plasmon resonance (LSPR) sensing transduction on a single gold nanoisland (AuNI) chip (Fig. 2 ) [73]. This type of biosensor is ideal for providing reliable clinical diagnosis and continuous monitoring of coronaviruses [73]. In this sensor, researchers combined plasmonic resonances of PPT and LSPR transducer at two different angles of incidence and excited at two different wavelengths, which significantly enhanced the stability, sensitivity, and reliability of the sensor. With this arrangement, the LSPR sensing unit attained a real-time and label-free detection of viral sequences including RNA-dependent RNA polymerase (RdRp), open reading frame 1 ab (ORF1ab), and E genes from SARS-CoV-2. More importantly, the in situ PPT enhancement on the AuNI chips intensely upgraded the kinetics of hybridization as well as the specificity of genomic detection. Similar sequences such as RdRp genes from SARS-CoV and SARS-CoV-2 can be accurately differentiated with the in situ PPT improvement. Due to the urgent need of biosensor to detect the COVID-19 within a short time, the dual-functional LSPR biosensor could attain a reliable and relatively simple platform to enhance the accuracy in clinical settings [73].
Fig. 2.
A schematic illustration of the arrangement and sensing pathway of the dual-functional plasmonic biosensing system (Source: Qiu et al., 2020 [73]).
3.6. Multiplex paper-based colorimetric (MPBC) sensor
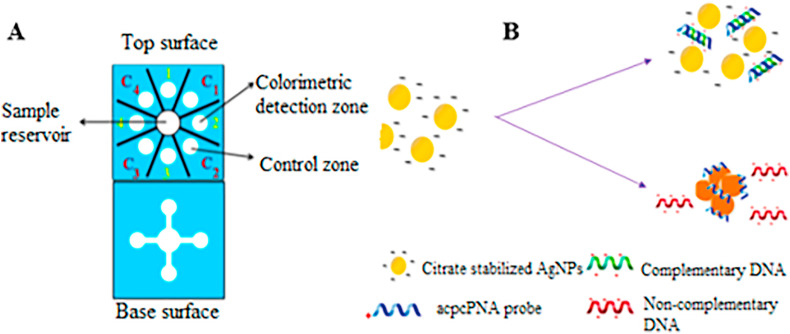
The paper-based analytical devices (PADs) are point-of-care devices that are currently receiving increasing interest because they are portable, cheap, simple, and disposable [92]. Today, PADs have been widely utilized for various settings from clinical diagnosis to environmental studies [93,94]. Colorimetric assays integrated with PADs are advantageous due to their simplicity, without sophisticated external components and the capacity to generate semiquantitative results [95,96]. Furthermore, incorporation of gold (AuNPs) and silver (AgNPs) nanoparticles enhances optical signals of the sensor platforms for pathogen detection. Teengam et al. [70] developed a multiplex colorimetric PAD by adding silver nanoparticles (Fig. 3 ) for real-time detection of viral and bacterial DNA, including MERS-CoV, human papillomavirus (HPV) and Mycobacterium tuberculosis (Mtb). The team utilized AgNPs and colorimetric reagent, for the detection of bacterial/viral nucleic acid (DNA) based on peptide nucleic acid probe (PNA)-induced nanoparticle aggregation. These DNA sensors revealed high selectivity against noncomplementary as well as the single or dual-base mismatch of target DNA. The viral DNA detection limit was 1.53, 1.03, and 1.27 nM, for MERS-CoV, HPV, and Mtb, respectively. Hence, the developed multiplex colorimetric PAD might be a disposable alternative tool for rapid screening and detection of infectious diseases at a low cost [70].
Fig. 3.
A schematic illustration of MPBC DNA sensor (A) Multiplex colorimetric PAD (B) The process of AgNP aggregation in the presence of complementary and non-complementary DNA (Source: Teengam et al., 2017 [70]).
PNA probes are considered to be advantageous over RNA or DNA probes with the fact that they are easy to synthesize, relatively stable, and hybridize easily to the complementary nucleic acid strands. The positive charge of the C-terminus end of the lysine amino acid of the pyrrolidinyl peptide nucleic acid (acpcPNA) probe causes sideway accumulation of citrate anion-stabilized AgNPs in the absence of complementary DNA. Whereas in the presence of target DNA, detectable color changes were produced following the formation of the anionic DNA-acpcPNA duplex as a consequence of the dispersion of the AgNPs produced by electrostatic repulsion (Fig. 3). The sensitivity and selectivity of this assay were dependent on ionic strength, DNA strand mismatches, and AgNP and PNA concentration [70].
4. Challenges and future perspectives
The complex interaction of infectious pathogens and factors such as the radical increase in worldwide animal and human movement, bushmeat harvesting, rapid demographic, and ecological and climate changes have led to the emergence of new highly pathogenic diseases around the globe [[97], [98], [99], [100], [101]]. Disease surveillance and diagnosis are crucial and the most influential elements of all public health services [102]. Accurate and fast disease detection strategy is a critical step to minimize or eradicate the spreading of infectious diseases [103,104]. The accuracy, as well as sensitivity and/or specificity of diagnostic techniques, have their impact on the success of the prevention, control, or overall eradication of the disease. Disease investigation and detection strategies can be organized in different platforms starting from simple handheld devices to sophisticated laboratory-based high throughput instruments [45,46,105,106].
Disease caused by coronaviruses became the major public health threat worldwide [1]. It is highly contagious thus there is a great demand for diagnostic/detection tool that is rapid, accurate, user-friendly to support the healthcare industry. RT-PCR has been the key diagnostic tool to combat the COVD-19 pandemic and is considered a major diagnostic tool for several other viral diseases. However, this method is labor-intensive and often requires several days to obtain results, which is counter-productive for tracing/tracking since the SARS-CoV-2 spreads rapidly. The application of nano biosensors (using nanoparticles and nanomaterials) has the potential to deliver results quickly with high accuracy. However, biosensors may encounter many challenges in the clinical settings, such as biofouling which restricts the widespread utilization of biosensors [107]. It has been investigated that the durability or functionality of immobilized sensors is diminished by particle buildup i.e., biofouling can considerably impede the influx of analyte to the detector. Another challenge is that some of the immunosensors are not fully reversible, hence only one immunoassay can be performed. Nevertheless, research has been done toward the development of renewable antibody surfaces [108].
Some of the biosensor technologies are limited to laboratory use only and are not suitable for field deployment and require a highly qualified operator. Besides, factors such as; reduced reproducibility, surface preparation/immobilization condition, incubation time and temperature, sample preparation, types of biological fluid used, and sample loading may restrict the use of this technology for coronavirus detection within a short period. Hence, more comprehensive research is needed to develop a universal and high-quality multiplexed biosensor device for point-of-care deployment is essential to fight the pandemics.
Given the urgent need for fast and accurate detection of COVID-19, the biomarkers-based sensor also can play a key role as it will minimize the virus detection time and reduce the risk of virus transmission at the time of sample collection and diagnosis. This can be further achieved by combining the biosensor with the microfluidics system wherein a small amount of sample can be tested during diagnosis avoiding the transmission risk at the point of diagnosis. Therefore, further proteomic analysis is needed to identify the specific biomarkers from the COVID-19 patients for the development of a robust diagnostic tool.
It is anticipated that the collective efforts of the researchers around the globe would yield highly sensitive diagnostic tools but point-of-care diagnosis in real-time (instantaneous) remain elusive because of the unpredictable virus load in a sample which largely depends on the type of patient (asymptomatic vs symptomatic) being tested and the sample preparation efficiency. The most promising approach would be to develop a biosensor-based high throughput screening test that can quickly rule out the negative samples and only presumptive positive samples can be retested with a molecular method for confirmation [45]. Such strategies would accelerate diagnosis and aid tracking and prevention of COVID-19.
Declaration of competing interest
The authors declare no conflict of interests.
Acknowledgments
Biosensor related research in the author's (MS and AKB) laboratory is supported by the Center for Food Safety Engineering at Purdue University.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mcp.2020.101662.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Shereen M.A., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henry R. Etymologia: coronavirus. Emerg. Infect. Dis. 2020;26(5):1027. [Google Scholar]
- 3.Wu D., Wu T., Liu Q., Yang Z. The SARS-CoV-2 outbreak: what we know. Int. J. Infect. Dis. 2020;94:44–48. doi: 10.1016/j.ijid.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;180:1–12. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., Zhu C., Hu T., Hua T., Zhang B., Yang X., Li J., Yang H., Liu Z., Xu W., Guddat L.W., Wang Q., Lou Z., Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368(6492):779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ye Z.-W., Yuan S., Yuen K.-S., Fung S.-Y., Chan C.-P., Jin D.-Y. Zoonotic origins of human coronaviruses. Int. J. Biol. Sci. 2020;16(10):1686–1697. doi: 10.7150/ijbs.45472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Azhar E.I., El-Kafrawy S.A., Farraj S.A., Hassan A.M., Al-Saeed M.S., Hashem A.M., Madani T.A. Evidence for camel-to-human transmission of MERS coronavirus. N. Engl. J. Med. 2014;370(26):2499–2505. doi: 10.1056/NEJMoa1401505. [DOI] [PubMed] [Google Scholar]
- 9.Tang Y.-W., Schmitz J.E., Persing D.H., Stratton C.W. The laboratory diagnosis of COVID-19 infection: current issues and challenges. J. Clin. Microbiol. 2020;58:e00512–20. doi: 10.1128/JCM.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu D.K.W., Pan Y., Cheng S.M.S., Hui K.P.Y., Krishnan P., Liu Y., Ng D.Y.M., Wan C.K.C., Yang P., Wang Q., Peiris M., Poon L.L.M. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 2020;66(4):549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu F., Yan L., Wang N., Yang S., Wang L., Tang Y., Gao G., Wang S., Ma C., Xie R. Quantitative detection and viral load analysis of SARS-CoV-2 in infected patients. Clin. Infect. Dis. 2020;71(15):793–798. doi: 10.1093/cid/ciaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nalla A.K., Casto A.M., Huang M.-L.W., Perchetti G.A., Sampoleo R., Shrestha L., Wei Y., Zhu H., Jerome K.R., Greninger A.L. Comparative performance of SARS-CoV-2 detection assays using seven different primer-probe sets and one assay kit. J. Clin. Microbiol. 2020;58(6) doi: 10.1128/JCM.00557-20. e00557-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Won J., Lee S., Park M., Kim T.Y., Park M.G., Choi B.Y., Kim D., Chang H., Kim V.N., Lee C.J. Development of a laboratory-safe and low-cost detection protocol for SARS-CoV-2 of the coronavirus disease 2019 (COVID-19) Exp. Neurobiol. 2020;29(2):107–119. doi: 10.5607/en20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu W., Liu L., Kou G., Zheng Y., Ding Y., Ni W., Wang Q., Tan L., Wu W., Tang S., Zhou X., Zheng S. Evaluation of nucleocapsid and spike protein-based enzyme linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00461-20. e00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y., Yao L., Li J., Chen L., Song Y., Cai Z., Yang C. Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19. J. Med. Virol. 2020;92(7):903–908. doi: 10.1002/jmv.25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tahamtan A., Ardebili A. Real-time RT-PCR in COVID-19 detection: issues affecting the results. Expert Rev. Mol. Diagn. 2020;20(5):453–454. doi: 10.1080/14737159.2020.1757437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poon L.L., Chan K.H., Wong O.K., Yam W.C., Yuen K.Y., Guan Y., Lo Y.D., Peiris J.S. Early diagnosis of SARS coronavirus infection by real time RT-PCR. J. Clin. Virol. 2003;28(3):233–238. doi: 10.1016/j.jcv.2003.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J., Cai K., He X., Shen X., Wang J., Liu J., Xu J., Qiu F., Lei W., Cui L., Ge Y., Wu T., Zhang Y., Yan H., Chen Y., Yu J., Ma X., Shi H., Zhang R., Li X., Gao Y., Niu P., Tan W., Wu G., Jiang Y., Xu W., Ma X. Multiple-centre clinical evaluation of an ultrafast single-tube assay for SARS-CoV-2 RNA. Clin. Microbiol. Infect. 2020;26(8):1076–1081. doi: 10.1016/j.cmi.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan C., Cui J., Huang L., Du B., Chen L., Xue G., Li S., Zhang W., Zhao L., Sun Y. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin. Microbiol. Infect. 2020;26(6):773–779. doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X., Zhong M., Liu Y., Ma P., Dang L., Meng Q., Wan W., Ma X., Liu J., Yang G. Rapid and sensitive detection of COVID-19 using CRISPR/Cas12a-based detection with naked eye readout, CRISPR/Cas12a-NER. Sci. Bull. 2020;65:1436–1439. doi: 10.1016/j.scib.2020.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38(7):870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Long Q.-X., Liu B.-Z., Deng H.-J., Wu G.-C., Deng K., Chen Y.-K., Liao P., Qiu J.-F., Lin Y., Cai X.-F., Wang D.-Q., Hu Y., Ren J.-H., Tang N., Xu Y.-Y., Yu L.-H., Mo Z., Gong F., Zhang X.-L., Tian W.-G., Hu L., Zhang X.-X., Xiang J.-L., Du H.-X., Liu H.-W., Lang C.-H., Luo X.-H., Wu S.-B., Cui X.-P., Zhou Z., Zhu M.-M., Wang J., Xue C.-J., Li X.-F., Wang L., Li Z.-J., Wang K., Niu C.-C., Yang Q.-J., Tang X.-J., Zhang Y., Liu X.-M., Li J.-J., Zhang D.-C., Zhang F., Liu P., Yuan J., Li Q., Hu J.-L., Chen J., Huang A.-L. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26(6):845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 23.Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J., Bermudez-Gonzalez M., Kleiner G., Aydillo T., Miorin L., Fierer D.S., Lugo L.A., Kojic E.M., Stoever J., Liu S.T.H., Cunningham-Rundles C., Felgner P.L., Moran T., García-Sastre A., Caplivski D., Cheng A.C., Kedzierska K., Vapalahti O., Hepojoki J.M., Simon V., Krammer F. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11(1):1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poh C.M., Carissimo G., Bei W., Amrun S.N., Lee C.Y.-P., Chee R.S.-L., Yeo N.K.-W., Lee W.-H., Leo Y.-S., Mark I. Two linear epitopes on the SARS-CoV-2 spike protein that elicit neutralising antibodies in COVID-19 patients. Nat. Commun. 2020;11(1):2806. doi: 10.1038/s41467-020-16638-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen C., Wang Z., Zhao F., Yang Y., Li J., Yuan J., Wang F., Li D., Yang M., Xing L. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. J. Am. Med. Assoc. 2020;323(16):1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ni L., Ye F., Cheng M.-L., Feng Y., Deng Y.-Q., Zhao H., Wei P., Ge J., Gou M., Li X., Sun L., Cao T., Wang P., Zhou C., Zhang R., Liang P., Guo H., Wang X., Qin C.-F., Chen F., Dong C. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity. 2020;52(6):971–977. doi: 10.1016/j.immuni.2020.04.023. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pang J., Wang M.X., Ang I.Y.H., Tan S.H.X., Lewis R.F., Chen J.I.P., Gutierrez R.A., Gwee S.X.W., Chua P.E.Y., Yang Q. Potential rapid diagnostics, vaccine and therapeutics for 2019 novel coronavirus (2019-nCoV): a systematic review. J. Clin. Med. 2020;9(3):623. doi: 10.3390/jcm9030623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bai H.X., Hsieh B., Xiong Z., Halsey K., Choi J.W., Tran T.M.L., Pan I., Shi L.-B., Wang D.-C., Mei J. Performance of Radiologists in Differentiating COVID-19 from Viral Pneumonia on Chest CT. Radiology. 2020 doi: 10.1148/radiol.2020200823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou S., Wang Y., Zhu T., Xia L. CT features of coronavirus disease 2019 (COVID-19) pneumonia in 62 patients in Wuhan, China. Am. J. Roentgenol. 2020;214(6):1287–1294. doi: 10.2214/AJR.20.22975. [DOI] [PubMed] [Google Scholar]
- 31.Li Y., Xia L. Coronavirus disease 2019 (COVID-19): role of chest CT in diagnosis and management. Am. J. Roentgenol. 2020;214(6):1280–1286. doi: 10.2214/AJR.20.22954. [DOI] [PubMed] [Google Scholar]
- 32.Fang Y., Zhang H., Xie J., Lin M., Ying L., Pang P., Ji W. 2020. Sensitivity of Chest CT for COVID-19: Comparison to RT-PCR, Radiology; p. 200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishige T., Murata S., Taniguchi T., Miyabe A., Kitamura K., Kawasaki K., Nishimura M., Igari H., Matsushita K. Highly sensitive detection of SARS-CoV-2 RNA by multiplex rRT-PCR for molecular diagnosis of COVID-19 by clinical laboratories. Clin. Chim. Acta. 2020;507:139–142. doi: 10.1016/j.cca.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan J.F., Yip C.C., To K.K., Tang T.H., Wong S.C., Leung K.H., Fung A.Y., Ng A.C., Zou Z., Tsoi H.W., Choi G.K., Tam A.R., Cheng V.C., Chan K.H., Tsang O.T., Yuen K.Y. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/hel real-time reverse transcription-PCR assay validated in vitro and with clinical specimens. J. Clin. Microbiol. 2020;58(5) doi: 10.1128/JCM.00310-20. e00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lv D.F., Ying Q.M., Weng Y.S., Shen C.B., Chu J.G., Kong J.P., Sun D.H., Gao X., Weng X.B., Chen X.Q. Dynamic change process of target genes by RT-PCR testing of SARS-Cov-2 during the course of a Coronavirus Disease 2019 patient. Clin. Chim. Acta. 2020;506:172–175. doi: 10.1016/j.cca.2020.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Kasteren P.B., van der Veer B., van den Brink S., Wijsman L., de Jonge J., van den Brandt A., Molenkamp R., Reusken C., Meijer A. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. J. Clin. Virol. 2020;128:104412. doi: 10.1016/j.jcv.2020.104412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park M., Won J., Choi B.Y., Lee C.J. Optimization of primer sets and detection protocols for SARS-CoV-2 of coronavirus disease 2019 (COVID-19) using PCR and real-time PCR. Exp. Mol. Med. 2020;52(6):963–977. doi: 10.1038/s12276-020-0452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tagarro A., Epalza C., Santos M., Sanz-Santaeufemia F.J., Otheo E., Moraleda C., Calvo C. JAMA Pediatr; 2020. Screening and severity of coronavirus disease 2019 (COVID-19) in children in madrid, Spain. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao Z., Cui H., Song W., Ru X., Zhou W., Yu X. BioRxiv; 2020. A Simple Magnetic Nanoparticles-Based Viral RNA Extraction Method for Efficient Detection of SARS-CoV-2. Submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu L., Wu S., Hao X., Li X., Liu X., Ye S., Han H., Dong X., Li X., Li J. Rapid detection of COVID-19 coronavirus using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform. Clin. Chem. 2020;66:975–977. doi: 10.1093/clinchem/hvaa102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiang J., Yan M., Li H., Liu T., Lin C., Huang S., Shen C. MedRxiv; 2020. Evaluation of Enzyme-Linked Immunoassay and Colloidal Gold-Immunochromatographic Assay Kit for Detection of Novel Coronavirus (SARS-Cov-2) Causing an Outbreak of Pneumonia (COVID-19), MedRxiv. Submitted for publication. [DOI] [Google Scholar]
- 42.Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S., Sun R., Wang Y., Hu B., Chen W. Development and clinical application of a rapid IgM‐IgG combined antibody test for SARS‐CoV‐2 infection diagnosis. J. Med. Virol. 2020;2020:1–7. doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of Chest CT and RT-PCR Testing in Coronavirus Disease 2019 (COVID-19) in China: a Report of 1014 Cases. Radiology. 2020 doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhunia A.K. Biosensors and bio-based methods for the separation and detection of foodborne pathogens. Adv. Food Nutr. Res. 2008;54:1–44. doi: 10.1016/S1043-4526(07)00001-0. [DOI] [PubMed] [Google Scholar]
- 45.Bhunia A.K. One day to one hour: how quickly can foodborne pathogens be detected? Future Microbiol. 2014;9(8):935–946. doi: 10.2217/fmb.14.61. [DOI] [PubMed] [Google Scholar]
- 46.A.K. Bhunia, C.R. Taitt, M.S. Kim, High throughput screening strategies and technology platforms for detection of pathogens: an introduction, in: A.K. Bhunia, M.S. Kim, C.R. Taitt (Eds.), High Throughput Screening for Food Safety Assessment: Biosensor Technologies, Hyperspectral Imaging and Practical Applications2015, pp. 1-9.
- 47.Holford T.R.J., Davis F., Higson S.P.J. Recent trends in antibody based sensors. Biosens. Bioelectron. 2012;34(1):12–24. doi: 10.1016/j.bios.2011.10.023. [DOI] [PubMed] [Google Scholar]
- 48.Esfandyarpour R., Esfandyarpour H., Harris J.S., Davis R.W. Simulation and fabrication of a new novel 3D injectable biosensor for high throughput genomics and proteomics in a lab-on-a-chip device. Nanotechnology. 2013;24(46):465301. doi: 10.1088/0957-4484/24/46/465301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bhunia A.K., Nanduri V., Bae E., Hirleman E.D. In: Encyclopedia of Industrial Biotechnology. Flickinger M.C., editor. John Wiley & Sons, Inc.; Hoboken: 2010. Biosensors, foodborne pathogen detection. [Google Scholar]
- 50.B. Byrne, N. Gilmartin, R.S. Lakshmanan, R. O'Kennedy, Antibodies, Enzymes, and Nucleic Acid Sensors for High Throughput Screening of Microbes and Toxins in Food, High Throughput Screening for Food Safety Assessment, Elsevier2015, pp. 25-80.
- 51.Bazin I., Tria S.A., Hayat A., Marty J.-L. New biorecognition molecules in biosensors for the detection of toxins. Biosens. Biolectron. 2017;87:285–298. doi: 10.1016/j.bios.2016.06.083. [DOI] [PubMed] [Google Scholar]
- 52.Zhang D., Huarng M.C., Alocilja E.C. A multiplex nanoparticle-based bio-barcoded DNA sensor for the simultaneous detection of multiple pathogens. Biosens. Bioelectron. 2010;26(4):1736–1742. doi: 10.1016/j.bios.2010.08.012. [DOI] [PubMed] [Google Scholar]
- 53.Yu C., Irudayaraj J. Multiplex biosensor using gold nanorods. Anal. Chem. 2007;79(2):572–579. doi: 10.1021/ac061730d. [DOI] [PubMed] [Google Scholar]
- 54.Caygill R.L., Blair G.E., Millner P.A. A review on viral biosensors to detect human pathogens. Anal. Chim. Acta. 2010;681(1):8–15. doi: 10.1016/j.aca.2010.09.038. [DOI] [PubMed] [Google Scholar]
- 55.Esseghaier C., Ng A., Zourob M. A novel and rapid assay for HIV-1 protease detection using magnetic bead mediation. Biosens. Bioelectron. 2013;41:335–341. doi: 10.1016/j.bios.2012.08.049. [DOI] [PubMed] [Google Scholar]
- 56.Shafiee H., Lidstone E.A., Jahangir M., Inci F., Hanhauser E., Henrich T.J., Kuritzkes D.R., Cunningham B.T., Demirci U. Nanostructured optical photonic crystal biosensor for HIV viral load measurement. Sci. Rep. 2014;4:4116. doi: 10.1038/srep04116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krejcova L., Nejdl L., Hynek D., Krizkova S., Kopel P., Adam V., Kizek R. Beads-based electrochemical assay for the detection of influenza hemagglutinin labeled with CdTe quantum dots. Molecules. 2013;18(12):15573–15586. doi: 10.3390/molecules181215573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Afsahi S., Lerner M.B., Goldstein J.M., Lee J., Tang X., Bagarozzi D.A., Jr., Pan D., Locascio L., Walker A., Barron F. Novel graphene-based biosensor for early detection of Zika virus infection. Biosens. Bioelectron. 2018;100:85–88. doi: 10.1016/j.bios.2017.08.051. [DOI] [PubMed] [Google Scholar]
- 59.Dong S., Zhao R., Zhu J., Lu X., Li Y., Qiu S., Jia L., Jiao X., Song S., Fan C. Electrochemical DNA biosensor based on a tetrahedral nanostructure probe for the detection of avian influenza A (H7N9) virus. ACS Appl. Mater. Interfaces. 2015;7(16):8834–8842. doi: 10.1021/acsami.5b01438. [DOI] [PubMed] [Google Scholar]
- 60.Chang Y.-F., Wang S.-F., Huang J.C., Su L.-C., Yao L., Li Y.-C., Wu S.-C., Chen Y.-M.A., Hsieh J.-P., Chou C. Detection of swine-origin influenza A (H1N1) viruses using a localized surface plasmon coupled fluorescence fiber-optic biosensor. Biosens. Bioelectron. 2010;26(3):1068–1073. doi: 10.1016/j.bios.2010.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nguyen B.T.T., Peh A.E.K., Chee C.Y.L., Fink K., Chow V.T.K., Ng M.M.L., Toh C.-S. Electrochemical impedance spectroscopy characterization of nanoporous alumina dengue virus biosensor. Bioelectrochemistry. 2012;88:15–21. doi: 10.1016/j.bioelechem.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 62.Sharma P.K., Kumar J.S., Singh V.V., Biswas U., Sarkar S.S., Alam S.I., Dash P.K., Boopathi M., Ganesan K., Jain R. Surface plasmon resonance sensing of Ebola virus: a biological threat. Anal. Bioanal. Chem. 2020;412:4101–4112. doi: 10.1007/s00216-020-02641-5. [DOI] [PubMed] [Google Scholar]
- 63.Cui F., Zhou H.S. Diagnostic methods and potential portable biosensors for coronavirus disease 2019. Biosens. Bioelectron. 2020;165:112349. doi: 10.1016/j.bios.2020.112349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang J.C., Chang Y.-F., Chen K.-H., Su L.-C., Lee C.-W., Chen C.-C., Chen Y.-M.A., Chou C. Detection of severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein in human serum using a localized surface plasmon coupled fluorescence fiber-optic biosensor. Biosens. Bioelectron. 2009;25(2):320–325. doi: 10.1016/j.bios.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ishikawa F.N., Chang H.-K., Curreli M., Liao H.-I., Olson C.A., Chen P.-C., Zhang R., Roberts R.W., Sun R., Cote R.J. Label-free, electrical detection of the SARS virus N-protein with nanowire biosensors utilizing antibody mimics as capture probes. ACS Nano. 2009;3(5):1219–1224. doi: 10.1021/nn900086c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Qi C., Duan J.-Z., Wang Z.-H., Chen Y.-Y., Zhang P.-H., Zhan L., Yan X.-Y., Cao W.-C., Jin G. Investigation of interaction between two neutralizing monoclonal antibodies and SARS virus using biosensor based on imaging ellipsometry. Biomed. Microdevices. 2006;8(3):247–253. doi: 10.1007/s10544-006-8305-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chan P.K., Ng K.-C., Chan R.C., Lam R.K., Chow V.C., Hui M., Wu A., Lee N., Yap F.H., Cheng F.W. Immunofluorescence assay for serologic diagnosis of SARS. Emerg. Infect. Dis. 2004;10(3):530. doi: 10.3201/eid1003.030493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang P., Wang H., Cao Z., Jin H., Chi H., Zhao J., Yu B., Yan F., Hu X., Wu F., Jiao C., Hou P., Xu S., Zhao Y., Feng N., Wang J., Sun W., Wang T., Gao Y., Yang S., Xia X. A rapid and specific assay for the detection of MERS-CoV. Front. Microbiol. 2018;9 doi: 10.3389/fmicb.2018.01101. 1101-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee S.H., Baek Y.H., Kim Y.-H., Choi Y.-K., Song M.-S., Ahn J.-Y. One-pot reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) for detecting MERS-CoV. Front. Microbiol. 2017;7 doi: 10.3389/fmicb.2016.02166. 2166-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Teengam P., Siangproh W., Tuantranont A., Vilaivan T., Chailapakul O., Henry C.S. Multiplex paper-based colorimetric DNA sensor using pyrrolidinyl peptide nucleic acid-induced AgNPs aggregation for detecting MERS-CoV, MTB, and HPV oligonucleotides. Anal. Chem. 2017;89(10):5428–5435. doi: 10.1021/acs.analchem.7b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bhadra S., Jiang Y.S., Kumar M.R., Johnson R.F., Hensley L.E., Ellington A.D. Real-time sequence-validated loop-mediated isothermal amplification assays for detection of Middle East respiratory syndrome coronavirus (MERS-CoV) PloS One. 2015;10(4) doi: 10.1371/journal.pone.0123126. e0123126-e0123126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kilianski A., Mielech A., Baez-Santos Y.M, Mesecar A.D., Baker S.C. MERS-CoV papain-like protease has deISGylating and deubiquitinating activities. Virology. 2014;450-451:64–70. doi: 10.1016/j.virol.2013.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Qiu G., Gai Z., Tao Y., Schmitt J., Kullak-Ublick G.A., Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020;14(5):5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- 74.Chen Z., Zhang Z., Zhai X., Li Y., Lin L., Zhao H., Bian L., Li P., Yu L., Wu Y. Rapid and sensitive detection of anti-SARS-CoV-2 IgG, using lanthanide-doped nanoparticles-based lateral flow immunoassay. Anal. Chem. 2020;92(10):7226–7231. doi: 10.1021/acs.analchem.0c00784. [DOI] [PubMed] [Google Scholar]
- 75.Seo G., Lee G., Kim M.J., Baek S.-H., Choi M., Ku K.B., Lee C.-S., Jun S., Park D., Kim H.G., Kim S.-J., Lee J.-O., Kim B.T., Park E.C., Kim S.I. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14(4):5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 76.Zhu X., Wang X., Han L., Chen T., Wang L., Li H., Li S., He L., Fu X., Chen S., Xing M., Chen H., Wang Y. Vol. 166. Biosens Bioelectron; 2020. p. 112437. (Multiplex reverse transcription loop-mediated isothermal amplification combined with nanoparticle-based lateral flow biosensor for the diagnosis of COVID-19). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lamb L.E., Bartolone S.N., Ward E., Chancellor M.B. Rapid detection of novel coronavirus/Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) by reverse transcription-loop-mediated isothermal amplification. PloS One. 2020;15(6) doi: 10.1371/journal.pone.0234682. e0234682-e0234682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nehra A., Singh K.P. Current trends in nanomaterial embedded field effect transistor-based biosensor. Biosens. Bioelectron. 2015;74:731–743. doi: 10.1016/j.bios.2015.07.030. [DOI] [PubMed] [Google Scholar]
- 79.Janissen R., Sahoo P.K., Santos C.A., da Silva A.M., von Zuben A.A.G., Souto D.E.P., Costa A.D.T., Celedon P., Zanchin N.I.T., Almeida D.B., Oliveira D.S., Kubota L.T., Cesar C.L., Souza A.P.d., Cotta M.A. InP nanowire biosensor with tailored biofunctionalization: ultrasensitive and highly selective disease biomarker detection. Nano Lett. 2017;17(10):5938–5949. doi: 10.1021/acs.nanolett.7b01803. [DOI] [PubMed] [Google Scholar]
- 80.Geim A.K., Novoselov K.S. The rise of graphene. Nat. Mat. 2007;6(3):183–191. doi: 10.1038/nmat1849. [DOI] [PubMed] [Google Scholar]
- 81.Zhou L., Mao H., Wu C., Tang L., Wu Z., Sun H., Zhang H., Zhou H., Jia C., Jin Q., Chen X., Zhao J. Label-free graphene biosensor targeting cancer molecules based on non-covalent modification. Biosens. Bioelectron. 2017;87:701–707. doi: 10.1016/j.bios.2016.09.025. [DOI] [PubMed] [Google Scholar]
- 82.Chen Y., Ren R., Pu H., Guo X., Chang J., Zhou G., Mao S., Kron M., Chen J. Field-effect transistor biosensor for rapid detection of Ebola antigen. Sci. Rep. 2017;7(1):1–8. doi: 10.1038/s41598-017-11387-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hsieh B.-Y., Chang Y.-F., Ng M.-Y., Liu W.-C., Lin C.-H., Wu H.-T., Chou C. Localized surface plasmon coupled fluorescence fiber-optic biosensor with gold nanoparticles. Anal. Chem. 2007;79(9):3487–3493. doi: 10.1021/ac0624389. [DOI] [PubMed] [Google Scholar]
- 84.Omar N.A.S., Fen Y.W., Abdullah J., Kamil Y.M., Daniyal W.M.E.M.M., Sadrolhosseini A.R., Mahdi M.A. Sensitive detection of dengue virus type 2 E-proteins signals using self-assembled monolayers/reduced graphene oxide-PAMAM dendrimer thin film-SPR optical sensor. Sci. Rep. 2020;10(1):2374. doi: 10.1038/s41598-020-59388-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Saylan Y., Erdem Ö., Ünal S., Denizli A. An alternative medical diagnosis method: biosensors for virus detection. Biosensors. 2019;9(2):65. doi: 10.3390/bios9020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Han J.-H., Lee D., Chew C.H.C., Kim T., Pak J.J. A multi-virus detectable microfluidic electrochemical immunosensor for simultaneous detection of H1N1, H5N1, and H7N9 virus using ZnO nanorods for sensitivity enhancement. Sensor. Actuator. B Chem. 2016;228:36–42. [Google Scholar]
- 87.Singh R., Hong S., Jang J. Label-free detection of influenza viruses using a reduced graphene oxide-based electrochemical immunosensor integrated with a microfluidic platform. Sci. Rep. 2017;7:42771. doi: 10.1038/srep42771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Alizadeh N., Hallaj R., Salimi A. A highly sensitive electrochemical immunosensor for hepatitis B virus surface antigen detection based on Hemin/G-quadruplex horseradish peroxidase-mimicking DNAzyme-signal amplification. Biosens. Bioelectron. 2017;94:184–192. doi: 10.1016/j.bios.2017.02.039. [DOI] [PubMed] [Google Scholar]
- 89.Kaushik A., Yndart A., Kumar S., Jayant R.D., Vashist A., Brown A.N., Li C.-Z., Nair M. A sensitive electrochemical immunosensor for label-free detection of Zika-virus protein. Sci. Rep. 2018;8(1):1–5. doi: 10.1038/s41598-018-28035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nawaz M.H., Hayat A., Catanante G., Latif U., Marty J.L. Development of a portable and disposable NS1 based electrochemical immunosensor for early diagnosis of dengue virus. Anal. Chim. Acta. 2018;1026:1–7. doi: 10.1016/j.aca.2018.04.032. [DOI] [PubMed] [Google Scholar]
- 91.Layqah L.A., Eissa S. An electrochemical immunosensor for the corona virus associated with the Middle East respiratory syndrome using an array of gold nanoparticle-modified carbon electrodes. Microchimica Acta. 2019;186(4):224. doi: 10.1007/s00604-019-3345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yetisen A.K., Akram M.S., Lowe C.R. Paper-based microfluidic point-of-care diagnostic devices. Lab Chip. 2013;13(12):2210–2251. doi: 10.1039/c3lc50169h. [DOI] [PubMed] [Google Scholar]
- 93.Cate D.M., Adkins J.A., Mettakoonpitak J., Henry C.S. Recent developments in paper-based microfluidic devices. Anal. Chem. 2015;87(1):19–41. doi: 10.1021/ac503968p. [DOI] [PubMed] [Google Scholar]
- 94.Mettakoonpitak J., Boehle K., Nantaphol S., Teengam P., Adkins J.A., Srisa‐Art M., Henry C.S. Electrochemistry on paper‐based analytical devices: a review. Electroanalysis. 2016;28(7):1420–1436. [Google Scholar]
- 95.Nery E.W., Kubota L.T. Sensing approaches on paper-based devices: a review. Anal. Bioanal. Chem. 2013;405(24):7573–7595. doi: 10.1007/s00216-013-6911-4. [DOI] [PubMed] [Google Scholar]
- 96.Liana D.D., Raguse B., Gooding J.J., Chow E. Recent advances in paper-based sensors. Sensors. 2012;12(9):11505–11526. doi: 10.3390/s120911505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Morens D.M., Folkers G.K., Fauci A.S. The challenge of emerging and re-emerging infectious diseases. Nature. 2004;430(6996):242–249. doi: 10.1038/nature02759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Morens D.M., Fauci A.S. Emerging infectious diseases: threats to human health and global stability. PLoS Pathog. 2013;9(7) doi: 10.1371/journal.ppat.1003467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nichol S.T., Arikawa J., Kawaoka Y. Emerging viral diseases. Proc. Natl. Acad. Sci. U. S. A. 2000;97(23):12411–12412. doi: 10.1073/pnas.210382297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.L.A. Kurpiers, B. Schulte-Herbrüggen, I. Ejotre, D.M. Reeder, Bushmeat and Emerging Infectious Diseases: Lessons from Africa, Problematic Wildlife, Springer2016, pp. 507-551.
- 101.Bhunia A.K. In: Foodborne Microbial Pathogens: Mechanisms and Pathogenesis. Bhunia A.K., editor. Springer New York; New York, NY: 2018. Introduction to foodborne pathogens; pp. 1–23. [Google Scholar]
- 102.Cunningham A.A., Daszak P., Wood J.L.N. One Health, emerging infectious diseases and wildlife: two decades of progress? Phil. Trans. R. Soc. B. 2017;372(1725):20160167. doi: 10.1098/rstb.2016.0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bissonnette L., Bergeron M.G. Infectious disease management through point-of-care personalized medicine molecular diagnostic technologies. J. Personalized Med. 2012;2(2):50–70. doi: 10.3390/jpm2020050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ince J., McNally A. Development of rapid, automated diagnostics for infectious disease: advances and challenges. Expet Rev. Med. Dev. 2009;6(6):641–651. doi: 10.1586/erd.09.46. [DOI] [PubMed] [Google Scholar]
- 105.Bates M., Zumla A. Rapid infectious diseases diagnostics using Smartphones. Ann. Transl. Med. 2015;3(15) doi: 10.3978/j.issn.2305-5839.2015.07.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Binder S., Levitt A.M., Sacks J.J., Hughes J.M. Emerging infectious diseases: public health issues for the 21st century. Science. 1999;284(5418):1311–1313. doi: 10.1126/science.284.5418.1311. [DOI] [PubMed] [Google Scholar]
- 107.Wisniewski N., Reichert M. Methods for reducing biosensor membrane biofouling. Colloids Surf. B Biointerfaces. 2000;18(3):197–219. doi: 10.1016/s0927-7765(99)00148-4. [DOI] [PubMed] [Google Scholar]
- 108.Canziani G.A., Klakamp S., Myszka D.G. Kinetic screening of antibodies from crude hybridoma samples using Biacore. Anal. Biochem. 2004;325(2):301–307. doi: 10.1016/j.ab.2003.11.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.