Abstract
The “isomorphic subtype of diffuse astrocytoma” was identified histologically in 2004 as a supratentorial, highly differentiated glioma with low cellularity, low proliferation and focal diffuse brain infiltration. Patients typically had seizures since childhood and all were operated on as adults. To define the position of these lesions among brain tumours, we histologically, molecularly and clinically analysed 26 histologically prototypical isomorphic diffuse gliomas. Immunohistochemically, they were GFAP-positive, MAP2-, OLIG2- and CD34-negative, nuclear ATRX-expression was retained and proliferation was low. All 24 cases sequenced were IDH-wildtype. In cluster analyses of DNA-methylation data, isomorphic diffuse gliomas formed a group clearly distinct from other glial/glio-neuronal brain tumours and normal hemispheric tissue, most closely related to paediatric MYB/MYBL1-altered diffuse astrocytomas and angiocentric gliomas. Half of the isomorphic diffuse gliomas had copy number alterations of MYBL1 or MYB (13/25, 52%). Gene fusions of MYBL1 or MYB with various gene partners were identified in 11/22 (50%) and were associated with an increased RNA-expression of the respective MYB-family gene. Integrating copy number alterations and available RNA sequencing data, 20/26 (77%) of isomorphic diffuse gliomas demonstrated MYBL1 (54%) or MYB (23%) alterations. Clinically, 89% of patients were seizure-free after surgery and all had a good outcome. In summary, we here define a distinct benign tumour class belonging to the family of MYB/MYBL1-altered gliomas. Isomorphic diffuse glioma occurs both in children and adults, has a concise morphology, frequent MYBL1- and MYB-alterations and a specific DNA-methylation profile. As an exclusively histological diagnosis may be very challenging and as paediatric MYB/MYBL1-altered diffuse astrocytomas may have the same gene fusions, we consider DNA-methylation profiling very helpful for their identification.
Keywords: glioma, isomorphic diffuse glioma, epilepsy, MYB, MYBL1, gene fusion
Introduction
Genome-wide DNA-methylation profiling and identification of recurrent genomic alterations have become important tools for both refining the molecular classification of known brain tumour entities and the detection of novel entities and subclasses of brain tumours [e.g. 21,50,51,35,28,59,40,7,43,15,23]. One enigmatic glioma entity that has not yet been molecularly characterized in detail is the “isomorphic subtype of diffuse astrocytoma”. It was first proposed as a separate entity in 2004 as a supratentorial, epilepsy-associated, highly differentiated, microscopically diffuse glial tumour with low cellularity and low proliferation [3,46]. While all patients in the original description were adults at operation (median age 32 years), most had a history of epileptic seizures since childhood. On cMRI, the tumours displayed a mass effect, a homogenous signal increase on FLAIR and T2-weighted images and a signal decrease on T1-weighted images. Contrast enhancement was not observed [3,2,55]. Lacking additional evidence of a truly distinct entity, these isomorphic diffuse gliomas have not yet been incorporated into the WHO classification of brain tumours [32]. Using the histological criteria described in 2004, they are rare even among epilepsy associated tumours, accounting for approximately 0.8% to 1.9% in such a setting [4,2]. In a more recent analysis of the same tumours, then termed “isomorphic neuroepithelial tumours” (INET), all isomorphic diffuse gliomas were IDH1 R132H-negative, suggesting that they differ from diffuse astrocytoma, IDH-mutant [2]. In accordance with this, isomorphic diffuse gliomas had a generally benign clinical course without malignant progression [3,46].
In particular when occurring in adults, isomorphic diffuse gliomas may be very problematic to classify. According to the current WHO classification, they fall into the provisional category of diffuse astrocytoma, IDH-wildtype. This category mostly consists of misclassified high-grade gliomas, in particular IDH-wildtype glioblastomas, and a few IDH-wildtype low-grade gliomas [41,5]. Thus, a further characterization and definition of isomorphic diffuse gliomas is required to allow a reliable identification.
Some recurrent genomic alterations in glioma subgroups and glio-neuronal tumours are well established such as IDH-mutations [18,58] and BRAF mutations and fusions [45,26]. In addition, especially for paediatric low-grade gliomas novel genomic alterations have been detected in recent years. These include gene fusions of FGFR1–3, NTRK2, PRKCA and MYB/MYBL1 with different fusion partners, or different rearrangements and mutations of these genes [59,37,39,24,54,25,20]. To analyse whether the “isomorphic subtype of diffuse astrocytoma” is a separate tumour entity with distinct molecular alterations, we performed genome-wide DNA-methylation profiling and copy number analyses from FFPE-samples of 26 histologically typical isomorphic diffuse gliomas and, from a subset, IDH1/2 sequencing and RNA sequencing. Moreover, clinical data of these tumours were evaluated and compared to those of other low-grade glioma entities.
Materials and Methods
Tissue samples, MRI and clinical data
Most study cases were diagnosed histologically by members of the International League against Epilepsy (ILAE) brain tumour study group and associated partners since the entity proposal in 2004. Others were obtained from further collaborating institutions (Institute of Pathology, University of Bern, Switzerland; Department of Neuropathology, Sainte-Anne Hospital, Paris, France; Department of Neuropathology, Tübingen, Germany). Formalin-fixed and paraffin-embedded (FFPE) tissue and clinical data were collected at the Department of Neuropathology of the University Hospital Heidelberg (Heidelberg, Germany). Representative MRI-images of 11 tumours were evaluated by an experienced neuroradiologist (D. Schwarz). Tissue collection and processing and data collection were in accordance with local ethical approvals.
Histology and immunohistochemistry
For all cases, in addition to the histological review at the local centres, a review of an H&E-staining was done by two experienced neuropathologists (DC, AW). For all cases with sufficient material, immunohistochemical staining was performed on a Ventana BenchMark Ultra Immunostainer using either the OptiView DAB IHC Detection Kit or the ultraView Universal DAB Detection Kit (Ventana Medical Systems, Tucson, Arizona, USA). Antibodies were directed against: Alpha Thalassemia/Mental Retardation Syndrome X-Linked- (ATRX) protein (BSB3296, 1:2000, pretreatment using CC1-buffer, OptiView; BioSB, Santa Barbara, CA, USA), CD34 (Ventana Kit 790–2927, pretreatment CC1, OptiView; Ventana Medical Systems), oligodendrocyte transcription factor (OLIG2; Abcam ab109186, 1:100, pretreatment CC1, ultraView; Abcam, Cambridge, UK), glial fibrillary acid protein (GFAP; Agilent Dako Z0334, 1:1000, no pretreatment, ultraView; Agilent Technologies, Inc., Santa Clara, CA, USA), microtubule-associated protein 2 (MAP2; Sigma-Aldrich M4403, 1:15000, pretreatment CC1, ultraView; Merck KGaA, Darmstadt, Germany), Ki67 (Agilent Dako M7240, 1:100, pretreatment CC1, OptiView; Agilent), p53 (Novocastra NCL-p53-DO1, 1:50, pretreatment CC1, ultraView; Leica Biosystems, Nussloch, Germany), and IDH1 R132H (internal clone H14 [9], 1:2, pretreatment CC2, ultraView; exemplary stainings only). Stained slides were scanned on a NanoZoomer Digital Slide Scanner (Hamamatsu, Japan) or an Aperio AT2 Scanner (Aperio Technologies, Vista, California, USA) and photographed using Aperio ImageScope software v12.3.2.8013.
DNA and RNA extraction
DNA and, if sufficient tissue was available, RNA were extracted from FFPE tissue of representative tumour areas with the highest tumour cell content. An automated extraction was done with a Maxwell system (Promega, Fitchburg, WI, USA) and the Maxwell 16 FFPE Plus LEV DNA Purification Kit or the Maxwell 16 LEV RNA FFPE Kit according to the manufacturer’s instructions. The DNA concentrations were determined with the Invitrogen Qubit dsDNA BR Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA) and a FLUOstar Omega Microplate Reader (BMG Labtech GmbH, Ortenberg, Germany). RNA concentrations and quality were assessed using the Agilent RNA 6000 Nano Assay and an Agilent Bioanalyser 2100 (Agilent Technologies) following the protocols provided by the manufacturer.
IDH sequencing
Targeted sanger sequencing of IDH1 and IDH2 was done with 20 ng of DNA as previously described [18] using the following primers: IDH1 forward 5′-TGATGAGAAGAGGGTTGAGGA-3′, reverse 5′-GCAAAATCACATTATTGCCAAC-3′; IDH2 forward 5′-GCTGCAGTGGGACCACTATT-3′, reverse 5′-CTCCACCCTGGCCTACCT-3’. Sequences were determined using an ABI 3500 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) and the Sequence Pilot version 3.1 software (JSI Medical systems GmbH, Ettenheim, Germany).
DNA-methylation profiling
DNA-methylation profiling of all samples was performed with 200–500 ng of DNA using the Infinium HumanMethylation450 BeadChip array (450k) or the Infinium MethylationEPIC BeadChip array (850k; Illumina, Inc., San Diego, CA, USA) at the Genomics and Proteomics Core Facility of the German Cancer Research Center (DKFZ) according to the protocols provided by the manufacturer. Filtering was performed as previously described [7], and genome-wide copy number analyses were done using the conumee package in R [22]. Unprocessed IDAT files can be downloaded from the NCBI Gene Expression Omnibus (GEO), accession number GSE136361.
Copy number alterations of genomic segments were inferred from the methylation array data based on the R-package conumee [22] after additional baseline correction (https://github.com/dstichel/conumee) with a cut-off of 0.1 for gains and −0.1 for losses on log2-scale. Summary copy number profiles were created by summarizing these data from the samples. The status of both the MYBL1- and MYB-loci and neighbouring regions within 2 Mb from MYBL1 and MYB was determined by evaluating the respective probes according to the IlluminaHumanMethylation450kmanifest [16] or IlluminaHumanMethylationEPICmanifest [17], respectively. Results were verified manually using the Integrative Genomics Viewer [42] to exclude a misinterpretation in single cases with high background noise or very focal alterations.
Reference cases for an unsupervised hierarchical cluster analysis and a t-SNE-analysis included well-characterized cases belonging to the following methylation classes of the DNA methylation-based brain tumour classifier [7 and www.molecularneuropathology.org/mnp/classifier/2]: hemispheric cortex (n = 9); white matter (n = 9); low grade glioma, dysembryoplastic neuroepithelial tumour (n = 13); low grade glioma, ganglioglioma (n = 15); low grade glioma, subclass hemispheric pilocytic astrocytoma and ganglioglioma (n = 10); low grade glioma, subclass midline pilocytic astrocytoma (n = 15); low grade glioma, subclass posterior fossa pilocytic astrocytoma (n = 11); (anaplastic) pleomorphic xanthoastrocytoma (n = 15); anaplastic astrocytoma with piloid features [40] (n = 15); diffuse midline glioma H3 K27M mutant (n = 14); glioblastoma, IDH wildtype, H3.3 G34 mutant (n = 10); glioblastoma, IDH wildtype, subclass mesenchymal (n = 15); glioblastoma, IDH wildtype, subclass midline (n = 10); glioblastoma, IDH wildtype, subclass RTK I (n = 15); glioblastoma, IDH wildtype, subclass RTK II (n = 8). Moreover, as isomorphic diffuse gliomas have in the past often been diagnosed as diffuse astrocytoma, WHO grade II, cases with the integrated diagnosis of diffuse astrocytoma, IDH-mutant, WHO grade II (n = 8) were selected for comparison. In addition, tumours with the integrated diagnosis of angiocentric glioma, i.e. tumours with a histology compatible with this diagnosis, a methylation profile typical of angiocentric glioma and confirmed MYB-QKI fusions in all cases with sufficient RNA for RNA sequencing, were included in the analyses (n = 15). This was done because we noticed that both angiocentric gliomas and isomorphic diffuse gliomas are classified as methylation class low-grade glioma, MYB/MYBL1 with the recently published DNA methylation-based brain tumour classifier [7]. The test cases were further compared to published DNA-methylation data from seven paediatric MYB/MYBL1-altered diffuse astrocytomas [37].
RNA sequencing and RT-PCR
RNA sequencing of samples for which RNA of sufficient quality and quantity was available (n = 22) was performed on a NextSeq 500 (Illumina) as described previously [44,49]. Fusion detection was done using Arriba (https://github.com/suhrig/arriba/). Fusions of the MYBL1- and MYB-genes were verified by RT-PCR. cDNA was generated using TruSeq RNA Access (Illumina, Inc., San Diego, CA, USA). Primer sequences are available upon request. For one case in which a MAML2-MYBL1 fusion was detected by RNA sequencing from FFPE-tissue, the inverse MYBL1-MAML2 fusion was verified by RT-PCR from fresh frozen tissue of the same tumour (#12).
Expected counts of MYBL1 and MYB expression were calculated using rsem [29]. RNA sequencing data from pilocytic astrocytomas (n = 5) were used for comparison. MYBL1 expression of individual MYBL1-altered isomorphic diffuse gliomas was regarded as increased if it exceeded the expression of that of all pilocytic astrocytomas and that of all MYB-altered isomorphic diffuse gliomas. MYB expression of individual MYB-altered isomorphic diffuse gliomas was regarded as increased if it exceeded the expression of that of all pilocytic astrocytomas and that of all MYBL1-altered isomorphic diffuse gliomas.
Statistics
For an unsupervised hierarchical cluster analysis of isomorphic diffuse gliomas and reference classes, we selected the 10,000 most variably methylated CpG sites across the dataset according to median absolute deviation. Clustering was done using the Euclidean distance and Ward linkage after adjustment for FFPE versus frozen material. For unsupervised 2D representation of pairwise sample correlations, dimensionality reduction by t-distributed stochastic neighbor embedding (t-SNE) was done with the R-package Rtsne (https://github.com/jkrijthe/Rtsne) using the 10,000 most variably methylated CpG sites according to the standard deviation, 1000 iterations and a perplexity value of 10.
RNA-expression data were compared with a Kruskal-Wallis test followed by post hoc Dunn’s multiple comparisons test using Prism 8.02 (GraphPad, La Jolla, CA, USA). p-values of less than 0.05 were considered significant.
Results
Isomorphic diffuse gliomas are monomorphic IDH-wildtype gliomas with moderately increased cell density
As a first step, we histologically identified tumours in accordance with the criteria specified for the “isomorphic subtype of diffuse astrocytomas” or “isomorphic neuroepithelial tumour” [3,2]. These cases were mainly diagnosed by members of the ILAE brain tumour study group and associated partners since the proposal of the entity in 2004. One case of the original series [3] was available for further analyses in this study (our case #19). The other 25 cases represent newly identified specimens. Histologically, the tumours shared a low to moderate increase in cellular density in an often slightly pronounced neurofibrillary matrix (Fig. 1a–e). The tumour cells typically had monomorphic, round nuclei with finely dispersed chromatin and inconspicuous nucleoli (insets in Fig. 1a–d). Occasionally, the nuclei were very small and chromatin-dense, which may be a result of fixation conditions (Fig. 1e). In many cases, the neurofibrillary matrix showed some degree of microcystic changes (Fig. 1 c, d, e). In these areas, the tumour cell bodies could be distinguished and frequently had many fine glial processes (inset Fig. 1d). One tumour focally showed myxoid changes of the tumour matrix with interspersed neurons, resembling the glio-neuronal element of dysembryoplastic neuroepithelial tumours (DNT; Fig. 1f), while other areas showed a typical isomorphic morphology.
Fig. 1.
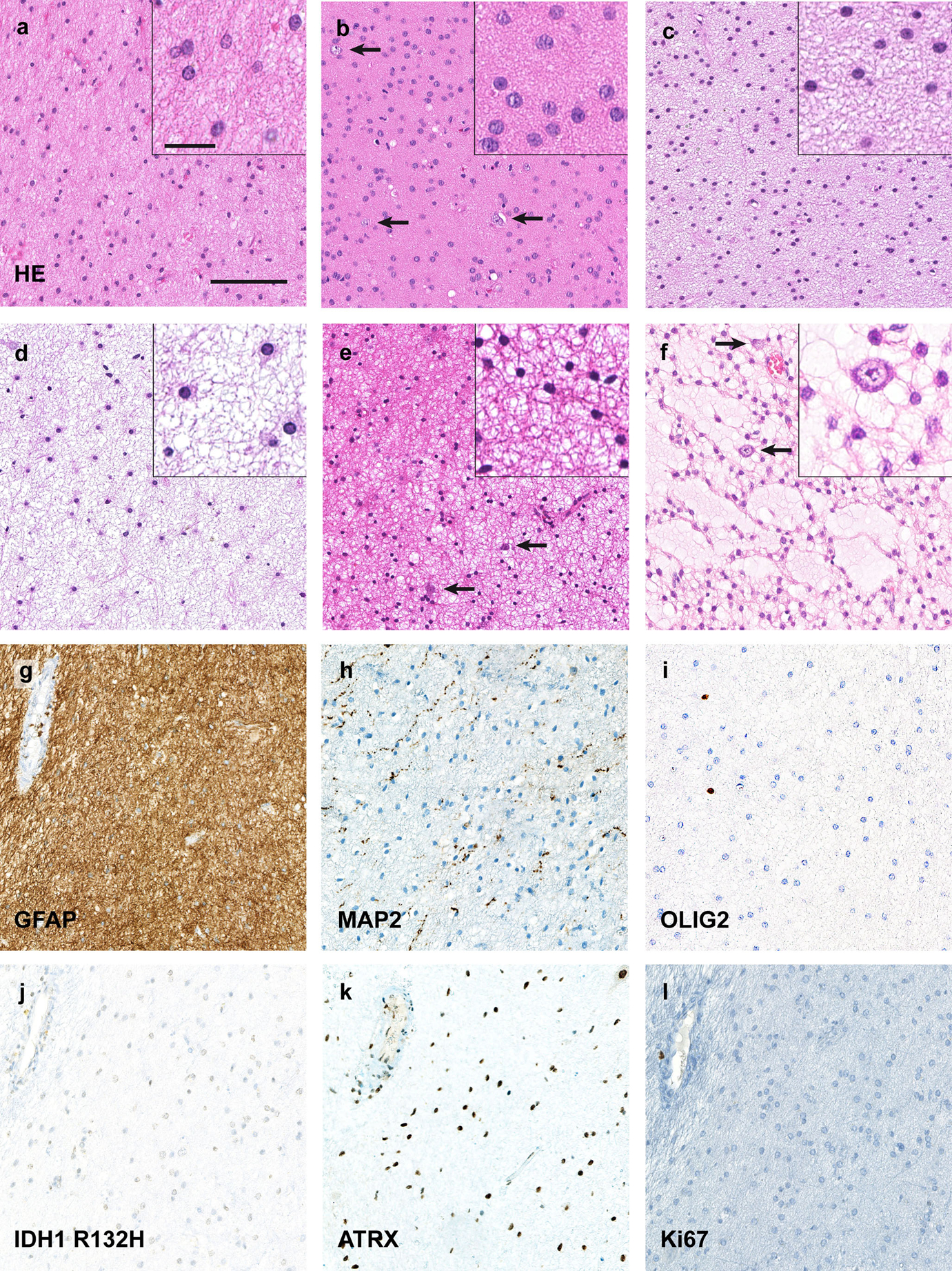
Isomorphic diffuse gliomas are monomorphic, IDH-wildtype tumours with low proliferation. a–d Histologically typical isomorphic diffuse gliomas showing round, only minimally pleomorphic nuclei, often with speckled chromatin. The cell density is slightly (a, d) to moderately (b, c) increased. The fibrillary matrix between the tumour cells can have slight (c) to extensive (d) microcystic changes. In some tumours scattered pre-existing neurons (arrows in b, e, f) are observed. e Occasionally, isomorphic diffuse gliomas may have smaller, more condensed round nuclei resembling those of oligodendrocytes. f One case focally showed myxoid changes of the matrix with scattered residual neurons, resembling the glio-neuronal element of dysembryoplastic neuroepithelial tumours. In other areas, the tumour also showed a clearly isomorphic growth. g–l Immunohistochemically, isomorphic diffuse gliomas are GFAP-positive (g; same tumour as in a) whereas MAP2 only labels residual neurons and neuronal processes (h). The tumours are OLIG2-negative (i) and IDH1 R132H-negative (j) with a retained nuclear expression of ATRX (k). The Ki67 proliferation index is below 1% (l). Scale bars 200 μm in a for a–l, 25 μm in the inset in a for all insets in a–f
In some of the cases, it was challenging to differentiate between tumour and normal white matter. In 23/26 tumours, the tumour cells microscopically diffusely infiltrated the adjacent brain structures such as the neocortex. Thus, scattered residual neurons were visible in most of the isomorphic diffuse gliomas (arrows in Fig. 1 b, e, f).
Immunohistochemistry of different epitopes was done on 19 cases while OLIG2 could only be stained on 14 cases. The tumour cells of all cases analysed were embedded in a glial fibrillary acid protein (GFAP) positive matrix (Fig. 1g) and expressed this protein. They were negative for microtubule-associated protein 2 (MAP2), whereas residual neurons or neuronal processes between the tumour cells were frequently positive (Fig. 1h). The tumour cells neither expressed OLIG2 (Fig. 1i) nor CD34, with only blood vessels staining positive for CD34 (data not shown). IDH1/2-wildtype status was verified by Sanger sequencing of 24 cases with sufficient DNA. An exemplary IDH1 R132H staining is shown in Figure 1j. The Alpha Thalassemia/Mental Retardation Syndrome X-Linked- (ATRX) protein was retained in the tumour cell nuclei in all cases (Fig. 1k). Proliferation was below 1% in all cases analysed (Fig. 1l) and mitoses were not identified. A prominent accumulation of the p53-protein was never observed (data not shown). The vasculature was inconspicuous. Necrosis was not observed.
Isomorphic diffuse gliomas present as mass lesions on MRI
Next we wanted to evaluate whether MRI-images of isomorphic diffuse glioma are indicative of intraaxial mass lesions (n = 11). Due to the retrospective nature of the imaging data, not all imaging modalities were available for all eleven cases. In agreement with published MRI data of single isomorphic diffuse gliomas [3,2,55], isomorphic diffuse gliomas were hyperintense both in FLAIR and T2-weighted images (n = 9/9 each; Fig. 2a, b). On the contrary, they were hypointense in T1-weighted images (n = 8/8; Fig. 2c) and contrast enhancement was never observed (n = 8/8; Fig. 2c). The majority of the lesions had a diameter larger than 1 cm (n = 9/11, 82%), one a diameter of about one centimetre and only one lesion was smaller. Most lesions were well confined (n = 9/10, 90%), mainly with a very sharp margin (n = 6/10, 60%, Fig. 2). Only one lesion showed a slightly more extensive growth in white matter extending into the corpus callosum. The signal of the lesions was suggestive of a microcystic structure (n = 9/9, 100%). Two lesions contained large cysts (n = 2/9, 22%). Overall, the MRI imaging was highly indicative of a tumour diagnosis and was most compatible with a low-grade glioma or glioneuronal tumour like dysembryoplastic neuroepithelial tumour.
Fig. 2.
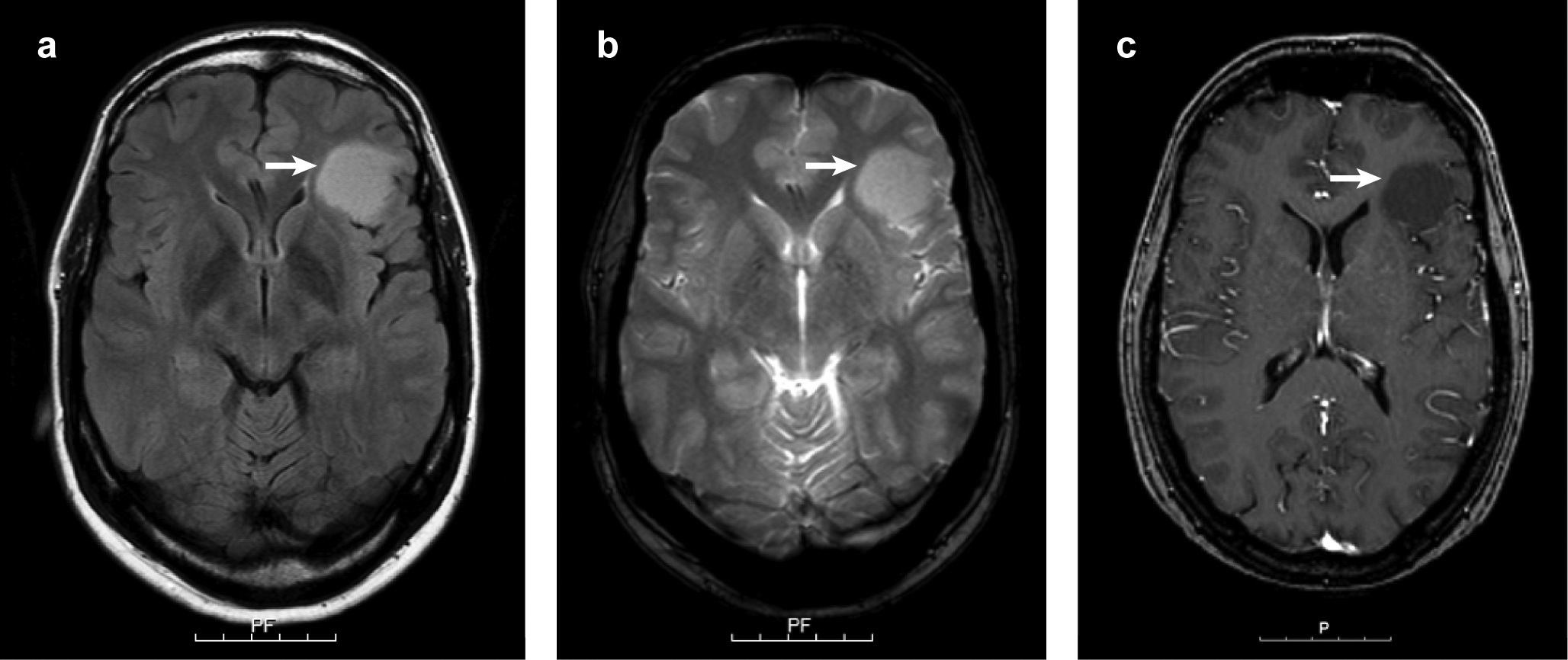
Representative MRI images of an isomorphic diffuse glioma. The tumour is hyperintense in FLAIR (arrow in a) and T2-weighted images (arrow in b) while hypointense in the T1-sequence (arrow in c). The tumour did not show contrast enhancement (c). Scale bars 5 cm with 1 cm intervals.
Isomorphic diffuse glioma is a distinct molecular tumour entity defined by a specific DNA-methylation profile
We have previously demonstrated that DNA-methylation profiling is a powerful tool to molecularly define clinico-pathological brain tumour entities [6,40,48,12,50,35,7]. Thus, we performed a genome wide DNA-methylation analysis of all 26 isomorphic diffuse gliomas. Both in an unsupervised hierarchical cluster analysis and in a t-distributed stochastic neighbor embedding (t-SNE) analysis together with 207 cases from 17 reference methylation classes of glial/glio-neuronal tumours plus normal white matter and cortex, isomorphic diffuse gliomas consistently formed a distinct cluster (Fig. 3a, b). The cluster was clearly separate from all other reference cases including diffuse astrocytoma, IDH-mutant, low-grade IDH-wildtype glio-neuronal tumours such as DNT and ganglioglioma, but also white matter and cortex. Also, it did not show any resemblance to IDH-wildtype high-grade glioma methylation classes. The closest relation was found to the cluster of angiocentric gliomas, which however appeared rather distinct in t-SNE analyses. These data indicate that isomorphic diffuse glioma is a distinct molecular tumour entity with a DNA-methylation profile closely related to angiocentric glioma.
Fig. 3.
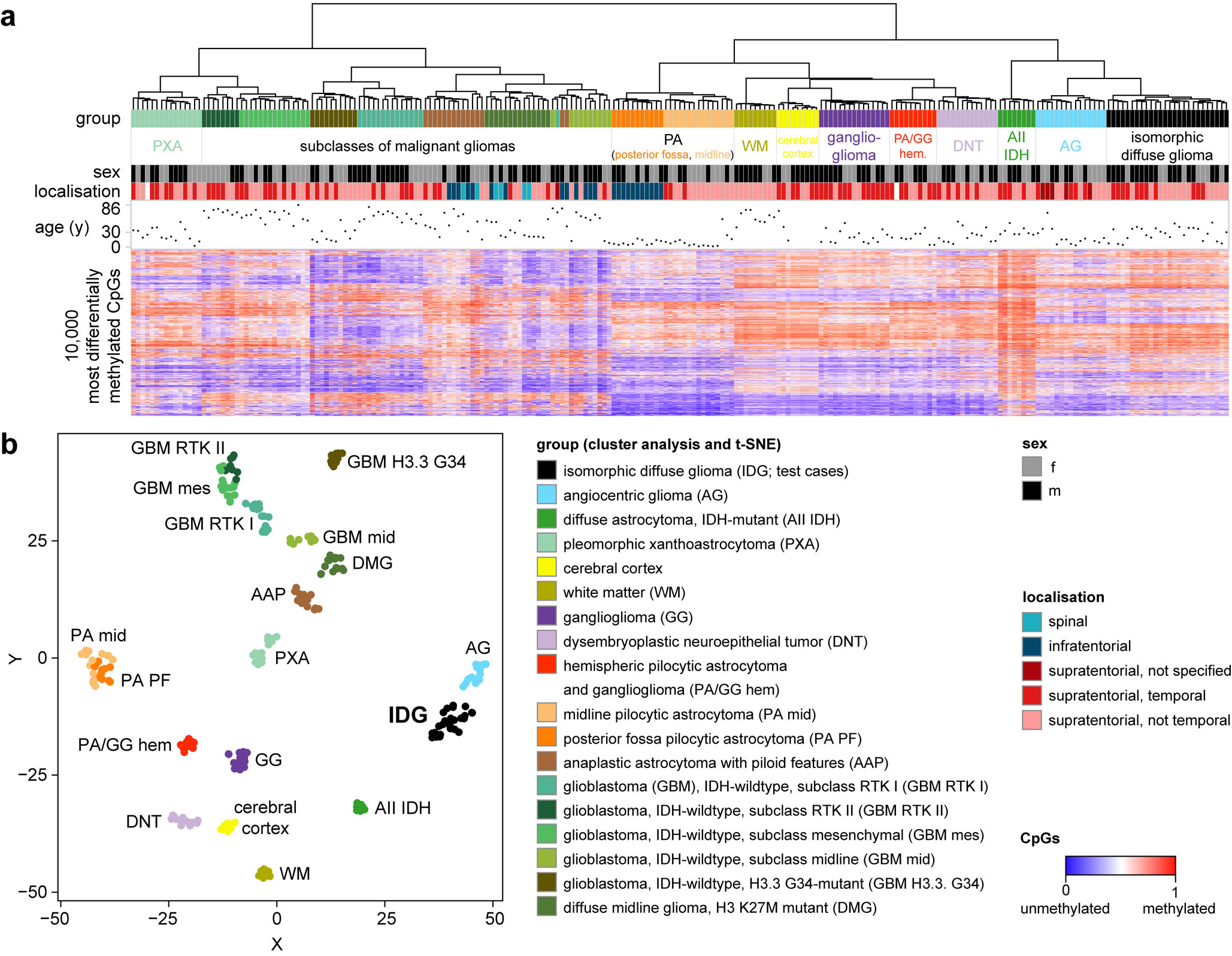
Isomorphic diffuse glioma forms a distinct methylation cluster. a Unsupervised hierarchical cluster analysis of 26 isomorphic diffuse gliomas with 246 cases from 17 reference classes. b t-distributed stochastic neighbor embedding (t-SNE) analysis of the same cases. Isomorphic diffuse glioma forms a cluster distinct from other glial/glio-neuronal tumour entities and normal cortex and white matter. The closest relation was found to the cluster of angiocentric glioma.
Isomorphic diffuse gliomas have frequent copy number alterations of MYBL1 and MYB
We next analysed copy number profiles (CNP) calculated from the methylation array data (n = 25 evaluable; in case #14 the background noise was too high to allow an analysis). The CNPs analysed showed only few copy number alterations. However, the analysis revealed that 10/25 tumours (40%) had focal copy number alterations affecting the MYBL1-locus (8q13.1; Fig. 4a, b, Supplementary Fig. 1) and 3/25 (12%) showed copy number alterations of the MYB-locus (6q23.3; Fig. 4c, d, Supplementary Fig. 1). For MYBL1, the alterations included focal gains (n = 5, Fig. 4a), losses (n = 4, Fig. 4b) and complex rearrangements of chromosome 8 (n = 1). Of the tumours with rearrangements of the MYB-locus, one tumour had a MYB-gain (Fig. 4c), one had a MYB-loss (Fig. 4d) and one tumour showed losses in distances of approximately 750 kb 5’ and 460 kb 3’ from the MYB-locus, likely indicating a more complex rearrangement of the MYB-locus. One additional tumour showed both a loss of the MYB-locus on 6q and a gain of the MYBL1-locus on 8q. As it seems unlikely that the tumour has alterations both of MYBL1 and MYB, possibly one of these alterations represents an artefact.
Fig. 4.
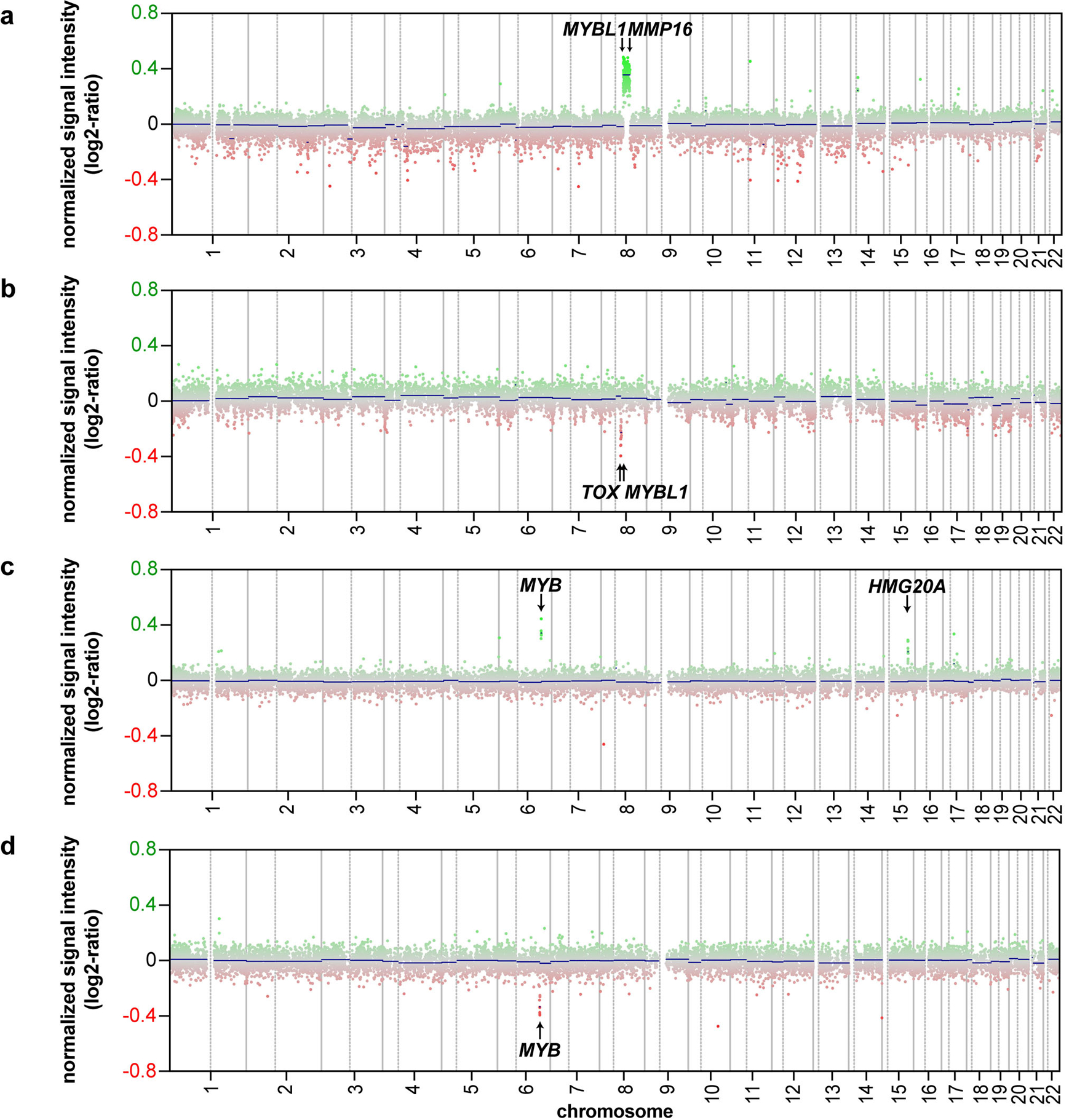
A subset of isomorphic diffuse gliomas has copy number alterations of the MYBL1- and MYB-genes. a–d Exemplary copy number profiles (CNP) of isomorphic diffuse gliomas. Gains are shown in green, losses in red. (a) Gain from MYBL1 to MMP16 (# 2; similar CNP in # 1). (b) Loss from MYBL1 to TOX (# 5). (c) Gain of MYB plus gain of HMG20A on chromosome 15 (# 15). (d) Loss of MYB and the region 3’ of MYB (# 17).
Of all cases with CNP alterations of MYBL1 or MYB, eleven cases (85%) had broader gains/losses of MYBL1 or MYB and adjacent loci which were evident from a low resolution CNP. Two tumours (15%) had very small gains/losses identifiable only when the CNPs were investigated in detail using the integrated genomic viewer [42]. The same holds true for the case with alterations of both MYBL1 and MYB.
Of the remaining CNPs without MYBL1/MYB copy number alterations, one showed a gain at the 3’ end of 6p. Three tumours had changes which were neither on chromosome 6 nor 8, while the CNPs of seven cases were balanced. Altogether, we detected alterations of the MYBL1- or MYB-loci in 13/25 (52%) of the isomorphic diffuse gliomas analysed (Supplementary Fig. 1; Table 1; Supplementary Table 1).
Table 1.
Characteristics of the 26 study cases
| # | Sex | Age at surgery (years) | Epilepsy | Localisation | MYBL1- or MYB-alterations copy number profile | Fusion transcripts | Increased MYB- or MYBL1-expression | Summary alterations chr. 6 + 8 (all methods) |
|---|---|---|---|---|---|---|---|---|
| 1 | m | 50 | yes | frontal | MYBL1 | MYBL1-MMP16 | MYBL1 | MYBL1 |
| 2 | m | 39 | yes | temporal pole | MYBL1 | MYBL1-MMP16 | MYBL1 | MYBL1 |
| 3 | f | 37 | yes | temporal | MYBL1 | 5’ of MYBL1-MMP16 | MYBL1 | MYBL1 |
| 4 | f | 8 | yes | centro-parietal | MYBL1 | MYBL1-RP11–586K2.1 (intergenic) | MYBL1 | MYBL1 |
| 5 | m | 6 | yes | temporal | MYBL1 | MYBL1-TOX | no | MYBL1 |
| 6 | m | 38 | yes | temporal | MYBL1 | MYBL1-ZFHX4 | no | MYBL1 |
| 7 | m | 30 | yes | occipital | MYBL1 | no relevant fusion detected | MYBL1 | MYBL1 |
| 8 | m | 24 | yes | temporal pole | MYBL1 | no relevant fusion detected | MYBL1 | MYBL1 |
| 9 | m | 37 | yes | temporal | MYBL1 | no relevant fusion detected | no | MYBL1 |
| 10 | f | 24 | yes | opercular/insular | MYBL1 | no relevant fusion detected | no | MYBL1 |
| 11 | f | 19 | no | temporal | - | MYBL1-RAD51B | MYBL1 | MYBL1 |
| 12 | m | 19 | yes | occipital | - | MYBL1-MAML2 | MYBL1 | MYBL1 |
| 13 | m | 36 | yes | temporal | - | MYBL1-RP11–492M23.2 (intergenic) | MYBL1 | MYBL1 |
| 14 | m | 39 | yes | frontal | not evaluable | no relevant fusion detected | MYBL1 | MYBL1 |
| 15 | m | 4 | yes | temporo-parietal | MYB | MYB-HMG20A | MYB | MYB |
| 16 | m | 7 | yes | frontal | MYB | no relevant fusion detected | no | MYB |
| 17 | m | 48 | no | parieto-occipital | MYB | N/A | N/A | MYB |
| 18 | m | 42 | yes | temporal | - | MYB-PCDHGA1 | MYB | MYB |
| 19 | f | 28 | yes | frontal | - | MYB-PCDHGA1 | MYB | MYB |
| 20 | m | 11 | yes | frontal | - | no relevant fusion detected | MYB | MYB |
| 21 | f | 31 | yes | parietal | - | no relevant fusion detected | no | none |
| 22 | m | 37 | yes | frontal | - | no relevant fusion detected | no | none |
| 23 | m | 20 | yes | parietal | - | N/A | N/A | none |
| 24 | m | 41 | yes | occipital | - | N/A | N/A | chr. 6, different alteration |
| 25 | f | 12 | yes | temporal | - | N/A | N/A | none |
| 26 | f | 27 | yes | fronto-parietal | MYBL1, MYB | no relevant fusion detected | no | (MYB and MYBL1) |
MYBL1 or MYB gene fusions and corresponding RNA overexpression are frequent in isomorphic diffuse gliomas
The focal structural alterations described above may indicate the presence of gene fusions. To confirm such fusions, we performed RNA sequencing of all cases with sufficient material (n = 22). MYBL1 fusions were detected in 8/22 tumours (36%; Fig. 5a). The only recurrent fusion partner of MYBL1 was MMP16 in 2/22 tumours (9%; Fig. 5a). Further fusion partners of MYBL1, validated by RT-PCR, were RAD51B, MAML2, ZFHX4 and TOX as well as intergenic sites on chromosomes 8 and 10 (n = 1 each). In all detected fusion genes, MYBL1 was truncated after exon 8, 9 or 10, resulting in a loss of the negative regulatory C-myb domain [53,14] (Fig. 5a).
Fig. 5.
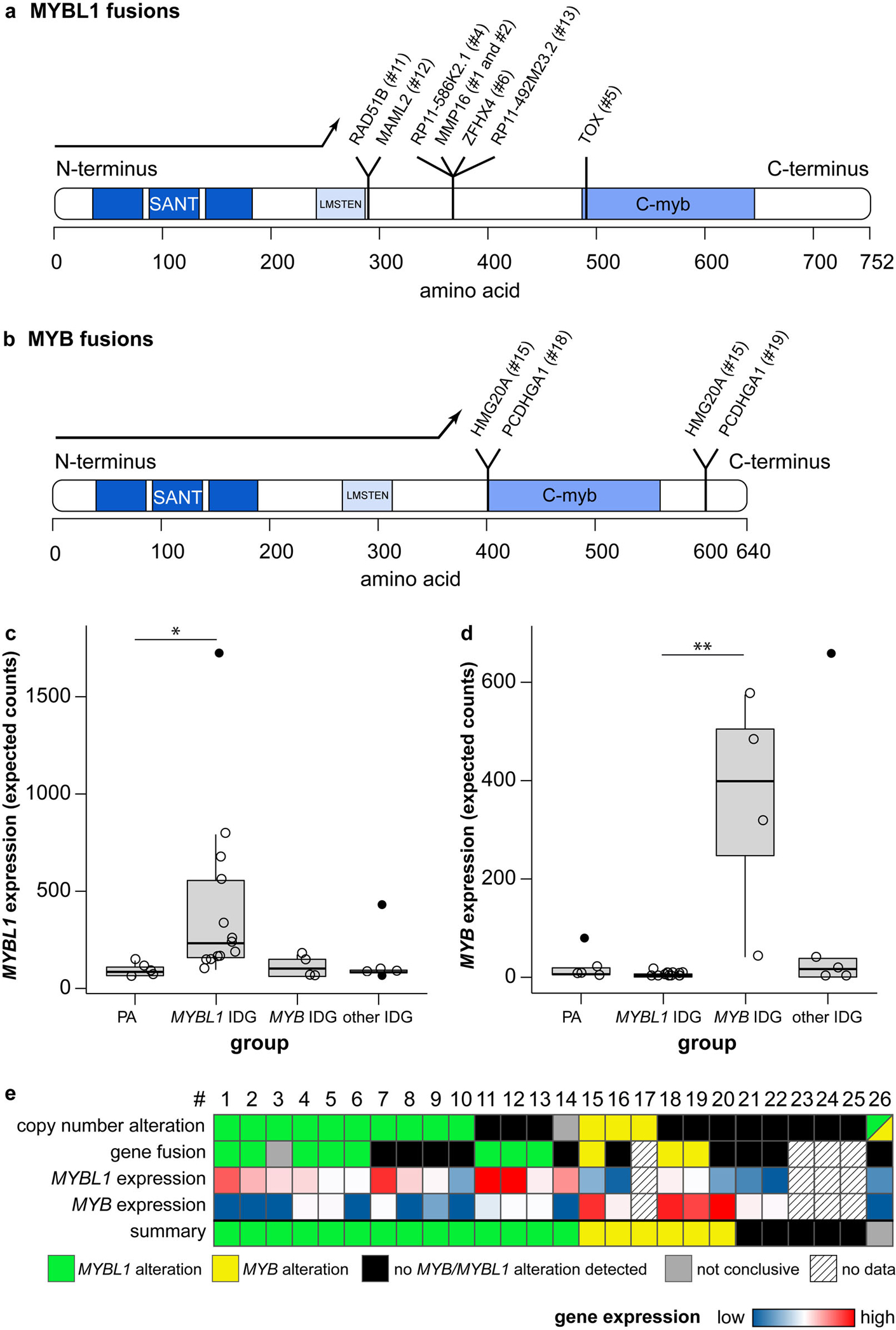
Isomorphic diffuse gliomas have fusions of the MYBL1- and MYB-genes and show a corresponding mRNA-overexpression. a, b Canonical MYBL1- (a) and MYB-protein (b) with loci of fusions to different partners. Protein domains in blue: “SANT” DNA-binding domain, “LMSTEN” transactivating domain, “C-myb” C-terminal negative regulatory domain. Fusions result in a deletion of the C-terminal negative regulatory C-myb domain of MYBL1 (a) or MYB (b), or the 3’ miRNA-binding sites of MYB (not included in depiction as no part of the MYB protein). (c, d) Many isomorphic diffuse gliomas show an overexpression of MYBL1 (c) or MYB (d). Compared were pilocytic astrocytomas (“PA”), isomorphic diffuse gliomas with MYBL1-alterations (“MYBL1 IDG”) or MYB-alterations (“MYB IDG”) in the CNP and/or RNA sequencing, and isomorphic diffuse gliomas without evidence of MYBL1/MYB-alterations (“other IDG”). Outliers are depicted with filled circles. p = 0.022 for PA versus MYBL1 IDG in c, p = 0.009 for MYBL1 versus MYB IDG in d (p-values adjusted for multiple comparisons). (e) Summary of alterations of MYBL1 and MYB in the 26 isomorphic diffuse gliomas of this study detected with different methods. For better differentiation, the range of the colour scale for the MYBL1 expression does not account for the outlier. #: case number
In an additional case, RNA sequencing indicated that the region 5’ of MYBL1 was fused to MMP16 (not shown). Together with a MYBL1-gain in the CNP, this may indicate a complex rearrangement involving the MYBL1-locus that was not fully resolvable with the techniques used, e.g. resulting in an enhancer hijacking or alternative transcription start site.
Some fusions were associated with corresponding changes of the CNP: The MYBL1-MMP16 fused cases showed corresponding duplications in the CNP (Fig. 4a) while the MYBL1-TOX fusion was associated with a focal loss of 8q between MYBL1 and TOX (Fig. 4b). However, there was only a partial overlap of the cases with MYBL1 fusions in RNA sequencing and those with copy number alterations of MYBL1 (Fig. 5e), which may be related to missed fusions in RNA sequencing due to degradation of RNA in FFPE tissue.
MYB fusions were detected in 3/22 (14%) cases (Fig. 5b). Two cases with balanced CNP had MYB-PCDHGA1 fusions. In a case with a MYB-gain (Fig. 4c), we detected two MYB-HMG20A fusions with two different breakpoints in MYB, likely representing splice variants. In addition to the gain of MYB, also a gain of HMG20A on 15q was visible in the CNP of this case (Fig. 4c) While some of the fusions resulted in a truncation of MYB before the negative regulatory C-myb domain reminiscent of the MYBL1 fusions, others resulted in a truncation downstream of the C-myb domain leading to a loss of the 3’ miRNA-binding sites of MYB. This has also been shown to lead to an activation of MYB [10,57,60].
In sum, we detected MYBL1 or MYB fusions in 11/22 (50%) of the cases analysed by RNA sequencing (Fig. 5e; Table 1; Supplementary Table 1). In an additional case, the breakpoint detected was 5’ of MYBL1 while the CNP suggests that MYBL1 may also be involved.
Deletion of the C-terminal regions of MYBL1 or MYB has been shown to result in an upregulation of the corresponding gene [1,36]. We therefore quantified the MYBL1 and MYB expression from the RNA sequencing data. Quantification of the mRNA-expression of MYBL1 confirmed an increased MYBL1 expression in tumours with corresponding alterations in the CNP and/or RNA sequencing (“MYBL1 IDG”; n = 13) as compared to pilocytic astrocytomas (“PA”; n = 5; adjusted p = 0.022; Fig. 5c). There was a trend of an increased MYBL1 expression in MYBL1-altered isomorphic diffuse gliomas compared to isomorphic diffuse gliomas without detected MYBL1/MYB-alteration (“other IDG”; n = 5; p = 0.06). Expression in MYBL1 IDG did not significantly differ from that in MYB-altered isomorphic diffuse gliomas (“MYB IDG”; n = 4), possibly due to the small sample size of the latter (p = 0.32; PA vs. MYB IDG; PA vs. other IDG and MYB IDG vs. other IDG p > 0.99). The isomorphic diffuse glioma with a rearrangement with breakpoint 5’ of MYBL1 (#3) also had an increased MYBL1 expression. Of the isomorphic diffuse gliomas without detected MYBL1/MYB-alteration, one case showed a clearly increased MYBL1 expression, possibly indicating an undetected rearrangement of MYBL1 or some other process resulting in MYBL1 induction.
Isomorphic diffuse gliomas with a MYB-alteration in the CNP and/or RNA sequencing (n = 4) showed a higher expression of MYB than MYBL1 IDG (adjusted p = 0.009; Fig. 5d). Expression between the other groups did not differ (PA vs. MYBL1 IDG p = 0.51; MYB IDG vs. other IDG p = 0.47; PA vs. MYBL1 IDG, PA vs. other IDG and MYBL1 IDG vs. other IDG p > 0.99). One case in the group of isomorphic diffuse gliomas without detected MYBL1/MYB-alteration showed an increased MYB expression, even higher than that of the other MYB-altered tumours. This possibly indicates an undetected rearrangement of MYB or some other process resulting in MYB induction.
Altogether, 14/22 of isomorphic diffuse gliomas (64%) showed an increased expression of MYBL1 or MYB.
In summary of all findings from the copy number analyses and RNA sequencing, our data indicates that at least 77% of isomorphic diffuse gliomas have alterations of MYBL1 or MYB (n = 20/26, 77%; Fig. 5e; Table 1; Supplementary Table 1). Of note, for 3/5 (60%) cases for which we did not detect copy number alterations of MYBL1 or MYB, RNA sequencing data are not available. It is possible that these cases have fusions of a MYB-family gene that are not evident from the CNP. Moreover, in two cases with RNA-seq data but without clear MYBL1-or MYB-alteration, including the case with indication of both a MYB- and MYBL1-alteration, the read counts were comparatively low which may be due to a low quality of the FFPE RNA in these cases. Also in other cases, we may have missed gene fusions due to RNA degradation. Thus, the percentage of MYBL1/MYB-altered isomorphic diffuse gliomas may be even higher than detected by our testing. Alternatively, other molecular alterations with a similar functional consequence not detectable by our testing modalities may also be present.
Isomorphic diffuse gliomas are related to paediatric MYB/MYBL1-altered diffuse astrocytomas
To see how the isomorphic diffuse gliomas in our cohort relate to paediatric MYB/MYBL1-altered diffuse astrocytomas, we did an additional unsupervised hierarchical cluster analysis of the cases contained in Fig. 3 with published methylation data from seven paediatric MYB/MYBL1-altered diffuse astrocytomas [37]. Of note, while these paediatric cases were related to the isomorphic diffuse gliomas, they still formed a cluster clearly distinct from all but two paediatric isomorphic diffuse gliomas (Supplementary Fig. 2a). This does not seem to be an effect solely related to the age of the patients at operation, as 4/6 (67%) paediatric isomorphic diffuse gliomas clearly mixed with their adult counterparts in the separate cluster exclusively containing isomorphic diffuse gliomas (Supplementary Fig. 2a). Also, in a t-SNE analysis of the same cases (Supplementary Fig. 2b), most paediatric MYB/MYBL1-altered diffuse astrocytomas separated from isomorphic diffuse glioma with the identical two isomorphic diffuse gliomas falling in close proximity. In addition, two paediatric MYB/MYBL1-altered diffuse astrocytomas intermingled with the neighbouring isomorphic diffuse gliomas. Thus, while the majority of isomorphic diffuse gliomas clearly separated from the paediatric MYB/MYBL1-altered diffuse astrocytomas with these analyses, the data indicate a close relation of these two groups, probably with partial overlap. An analysis of a larger number of cases will be needed to define the whole spectrum of MYB/MYBL1-altered gliomas.
Isomorphic diffuse gliomas are epilepsy-associated tumours with a good prognosis
Analysis of clinical data demonstrated that in our series of 26 cases, eight patients were female and 18 male (approaching a significant difference; p=0.08, binomial test). All tumours were supratentorial and almost half of the tumours had a temporal localisation (n = 11/26; 42%). Twenty-four of 26 patients (92%) had epileptic seizures. Epilepsy started during childhood in 17/21 patients (81%) with a median age at onset of epilepsy of 10 ± 6 years (range 1–35 years; n = 19). However, only 6/26 patients (23%) were operated on as children (median age at surgery for these cases 8 ± 3 years, range 4–12 years; all with epilepsy). Thus, the overall median age at surgery was 29 ± 14 years (range 4–50 years, n = 26) and the median time to operation after onset of epilepsy was as long as 15 ± 12 years (range 1–41 years, n = 18). Tumours were resected completely in 16/22 patients (73%), incompletely in 5/22 (23%) and in one patient only a stereotactic biopsy was taken.
None of the patients were reported to receive additional radio- or chemotherapy and none of the patients died during follow up. The median available follow-up time was 2.5 ± 3.0 years after surgery (range 0.3–9.6 years, n = 18; Fig. 6a) as most patients were lost to follow up relatively shortly after resection. Only one patient showed further growth of residual tumour and was re-operated after three years (Fig. 6b). At that time, the tumour was removed completely, and no recurrence was observed afterwards (6.4 years of further follow-up).
Fig. 6.
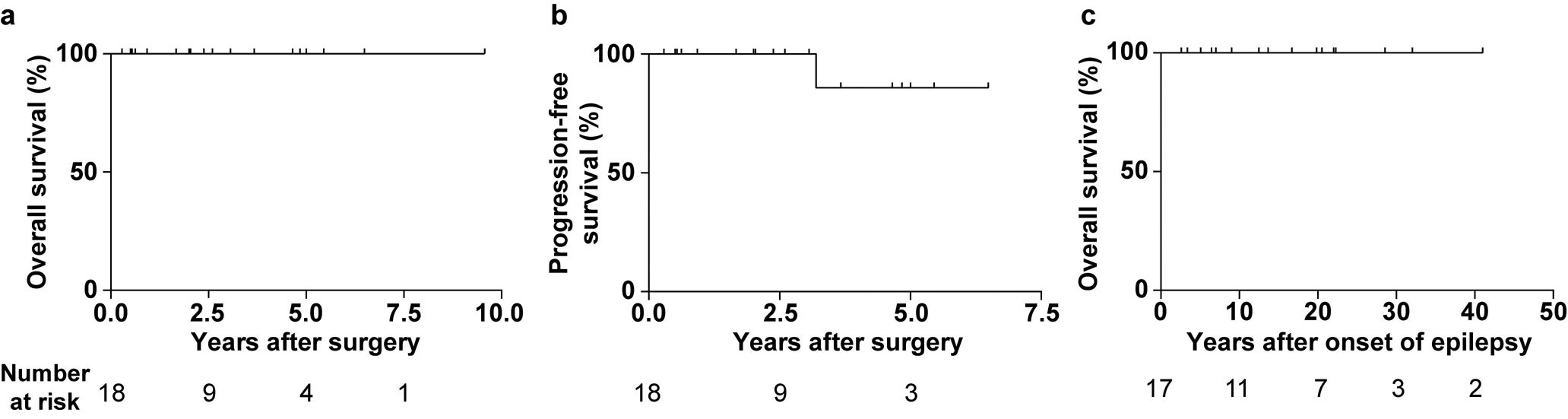
Patients with isomorphic diffuse gliomas have a good prognosis. (a) Overall survival after surgery of 18 patients with isomorphic diffuse glioma. (b) Progression-free survival after surgery of the same patients. One patient showed a growth of residual tumour and underwent a second surgery three years after the first operation with complete removal of the tumour (recurrence-free since). (c) Overall survival after onset of epilepsy of 17 patients with isomorphic diffuse glioma. Note that no patient with isomorphic diffuse glioma died during follow-up.
We next analysed the outcome of seizures after surgery. Of 18 patients with epileptic seizures for whom follow-up data was available, 16 (89%) were permanently seizure-free after surgery and the other two had a reduction in seizure frequency. Thus, it is very likely that the seizures were tumour related. Many patients had had seizures for decades before surgery. We therefore calculated the survival after onset of epilepsy. The median time after onset of epilepsy until patients were lost to follow-up after surgery was 17 ± 12 years (range 3–41 years, n = 17; Fig. 6c). This indicates a benign course of these tumours.
Discussion
In this study, we demonstrate that isomorphic diffuse glioma is an IDH-wildtype tumour class that is molecularly distinct from other established glial/glio-neuronal tumour entities, belongs to the family of MYB/MYBL1-altered gliomas and should be included in the differential diagnoses of low-grade epilepsy-associated tumours in both children and adults. Most tumours histologically showed the typical isomorphic growth pattern. While the nuclei of the tumour cells were occasionally more condensed and smaller than in typical isomorphic diffuse glioma, reminiscent of oligodendrocytes or oligodendrocyte-like nuclei of DNT, only one single tumour focally contained a small area resembling the glio-neuronal element of DNT (Fig. 1f). However, as opposed to DNT, in the remaining areas of this tumour the cells showed an isomorphic growth pattern, tumour cells were OLIG2-negative and the methylation profiles of both morphologically different areas indicated isomorphic diffuse glioma. Isomorphic diffuse glioma histologically also separates from ganglioglioma as it does not contain clearly dysplastic binucleated ganglion cells, typically has a highly isomorphic phenotype and is CD34-negative. Further, the morphology is more isomorphic than what is usually encountered in infiltration zones of IDH-wildtype glioblastomas and the proliferation was exceedingly low. Due to the low cell density, some isomorphic diffuse gliomas may even be difficult to distinguish from normal white matter, or rare histological patterns such as “Mild Malformation of Cortical Development with Oligodendroglial Hyperplasia in Frontal Lobe Epilepsy” [47] though both of these would express OLIG2.
When investigated as a series, isomorphic diffuse glioma clearly has a prototypical morphology and typical immunoprofile. However, because of the low incidence, we believe that recognition in clinical routine by histology alone will likely not be possible and additional molecular testing such as methylation profiling may be required. We think that the combination of histological analyses with DNA-methylation profiling [7] is highly suitable to confirm the diagnosis of isomorphic diffuse glioma. In the current version of the DNA methylation-based brain tumour classifier (V11b4) [7], only one methylation class of “low-grade glioma, MYB/MYBL1-altered” has been defined, containing both isomorphic diffuse gliomas and angiocentric gliomas. However, within this methylation class we can now demonstrate the presence of distinct methylation subgroups as shown by clustering and t-SNE analyses, in which isomorphic diffuse glioma and angiocentric glioma form two separate groups (Fig. 3). It is possible that inclusion of additional cases will lead to even more subgroups among MYB/MYBL1-altered gliomas.
MYB or MYBL1 fusions result in an activation and overexpression of the respective gene [36,27] through loss of the C-terminal negative regulatory domain (C-myb) [56,34]. MYB fusions also lead to a loss of the 3’ UTR binding sites for several microRNAs that also negatively regulate MYB mRNA stability and translation [10,30,11]. MYB/MYBL1 fusions have been detected in a variety of tumour entities such as adenoid cystic carcinoma [36,19], haematological neoplasias [31,38,52] and, in the CNS, angiocentric glioma [1] and paediatric diffuse gliomas/astrocytomas [59,39,37,54,13,1]. We never observed the highly prototypical MYB-QKI fusions of angiocentric gliomas in isomorphic diffuse gliomas while some fusions were the same as in “paediatric diffuse astrocytomas”, i.e. paediatric MYB/MYBL1-altered gliomas other than angiocentric glioma with an astrocytic morphology ([59,39,37,54,1]; sequencing data for n = 12 cases available). However, in isomorphic diffuse gliomas, MYBL1 fusions were the most abundant, while fusions in previously-published paediatric diffuse astrocytomas primarily affected MYB.
Importantly, our study clarifies that MYB/MYBL1-altered gliomas other than angiocentric gliomas also exist in adults, as to date only paediatric cases have been described [59,39,54,37,1]. While there is a close relation between isomorphic diffuse glioma and the published paediatric MYB/MYBL1-altered diffuse gliomas or possibly even a partial overlap, cluster analyses indicate that nonetheless, as angiocentric glioma, isomorphic diffuse glioma represents a distinct tumour class in the family of MYB/MYBL1-altered gliomas (Supplementary Fig. 2).
In five cases, no MYB/MYBL1-alterations were identified by our analyses. For three of these cases, RNA-sequencing data were not available. We expect that for the other two cases and also further cases with MYB/MYBL1-copy number alterations but without detected MYB or MYBL1 fusion, technical reasons due to low RNA quality extracted from the FFPE material may have hindered the identification of alterations. Thus, the rate of MYB/MYBL1-alterations in this series may be even higher. Alternatively, different alterations, outside the scope of our testing, with equivalent biological consequences may also explain these cases. We cannot rule out the possibility that non-tumorous pathological processes like reactive changes may acquire a methylation profile reminiscent of isomorphic diffuse glioma. For our series, it is unlikely that we included non-tumorous processes as all cases were operated as a space occupying lesions, but for diagnostic processes caution should be taken when only the methylation profile suggests the diagnosis of isomorphic diffuse glioma.
How should isomorphic diffuse gliomas be classified?
Isomorphic diffuse gliomas microscopically showed a focal diffuse growth into pre-existing brain. This contrasts with the MRI images of isomorphic diffuse gliomas in which they were well circumscribed (Fig. 2). There is an ongoing debate about the usage of the term “diffuse” when describing a brain tumour. While one interpretation focusses on the extensive spread of tumour cells throughout the brain such as in diffuse glioma, IDH-mutant [8], another usage refers to the local diffuse growth into adjacent brain structures as seen in isomorphic diffuse gliomas. The second interpretation was implemented in the recent cIMPACT-NOW update 4 on “paediatric-type” diffuse gliomas, that includes MYB- and MYBL1-altered diffuse gliomas [13]. Using the term “isomorphic diffuse glioma”, we chose to follow this latter proposal.
The 2014 ISN-Haarlem Consensus Guidelines for Nervous System Tumour Classification and Grading [33] proposed a four-layered diagnosis in which the histological classification, the WHO grade and the molecular information available should be incorporated into an integrated diagnosis. Table 2 shows a proposal of a layered diagnosis of isomorphic diffuse glioma based on the 2014 ISN-Haarlem guidelines. We highly recommend complementing the histological classification by DNA methylation analyses, if available, as a diagnosis based on histology alone may be very challenging in some cases. Other techniques such as copy number analyses of the MYB- and MYBL1-loci and detection of fusions of these genes may prove helpful as well. However, isomorphic diffuse glioma seems to share certain gene fusions with paediatric MYB/MYBL1-altered gliomas so that a distinction between these two subclasses may not be possible with these methods. For certain entities such as isomorphic diffuse glioma, the future integrated diagnosis might depend on methylation profiling as the defining layer, once this has been validated by independent institutions. In such a system, tumours may be grouped in methylation class families as is already implemented in the current DNA-methylation-based brain tumour classifier version [7]. This could be implemented in a format such as “Family - Type - Subtypes”. For isomorphic diffuse glioma, the classification might be “Family: Low-grade glial or mixed glio-neuronal tumour, Type: MYB/MYBL1-altered glioma, Subtype: isomorphic diffuse glioma”. Such steps will obviously require further validation of DNA methylation-based tumour classifications and an increased availability of this technique.
Table 2.
Report format for the diagnosis of isomorphic diffuse glioma. Molecular data in brackets are optional.
| Layer | Report format for integrated diagnosis – 2014 ISN-Haarlem guidelines [33] | Report format for isomorphic diffuse glioma | Example |
|---|---|---|---|
| 1 | Integrated diagnosis (incorporating all tissue-based information) | Isomorphic diffuse glioma, [DNA methylation group isomorphic diffuse glioma MYBL1/MYB,*] [MYBL1/MYB- rearrangement,] [MYBL1/MYB fusion,] WHO grade not assigned (WHO grade I proposed) | Isomorphic diffuse glioma, DNA methylation group isomorphic diffuse glioma MYBL1/MYB, MYBL1-rearrangement, MYBL1-MMP16 fusion, WHO grade not assigned (WHO grade I proposed) |
| 2 | Histological classification | Diffuse glioma with low proliferation | Diffuse glioma with low proliferation |
| 3 | WHO grade | not assigned (WHO grade I proposed) | not assigned (WHO grade I proposed) |
| 4 | Molecular information | [DNA methylation group isomorphic diffuse glioma MYBL1/MYB, *] [MYBL1/MYB-rearrangement,] [MYBL1/MYB fusion] | DNA methylation group isomorphic diffuse glioma MYBL1/MYB, MYBL1-rearrangement, MYBL1-MMP16 fusion |
highly recommended.
In summary, isomorphic diffuse glioma is a morphologically recognizable and molecularly distinct IDH-wildtype glioma with alterations of the MYBL1- and, less frequently, the MYB-gene that exists both in paediatric and in adult patients. Isomorphic diffuse glioma has a specific DNA-methylation profile, related to but distinct from that of angiocentric glioma and paediatric MYB/MYBL1-altered diffuse astrocytomas. Patients have a good prognosis.
Supplementary Material
Acknowledgements
We thank K. Böhmer, A. Habel, S. Kocher, U. Lass, K. Lindenberg, R. Quan, S. Sprengart, U. Vogel, M. Werner and V. Zeller for excellent technical assistance. We thank the Genomics and Proteomics Core Facility of the German Cancer Research Center (DKFZ) for the performance of DNA-methylation analyses. A.K. Wefers is a fellow of the Physician Scientist-Program of the Medical Faculty of Heidelberg. This study was supported by an International League against Epilepsy (ILAE) grant to D. Capper. I. Blumcke was supported by the European Union (FP6 DESIRE Grant Agreement #602531). M. Mittelbronn would like to thank the Luxembourg National Research Fund (FNR) for the support (FNR PEARL P16/BM/11192868 grant). K. Ligon is funded by the Paediatric Low Grade Astrocytoma Foundation and NCI R01CA215489. T. Jacques is funded by The Brain Tumour Charity, Children with Cancer UK, Great Ormond Street Hospital Children’s Charity, Cancer Research UK, the Olivia Hodson Cancer Fund and the National Institute of Health Research. T. Jacques’s work is partly funded by the NIHR GOSH BRC. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health. We thank the biomedical scientists in the Division of Neuropathology, the National Hospital for Neurology and Neurosurgery (NHNN) for excellent technical assistance. We also thank clinicians and neuropathologists for referring cases for molecular analysis. Part of the study was funded by the National Institute for Health Research to UCLH Biomedical research centre (BRC399/NS/RB/101410). S. Brandner and Z. Jaunmuktane are also supported by the Department of Health’s NIHR Biomedical Research Centre’s funding scheme.
Additional funding was provided by the Brain Tumour Charity (UK) for the Everest Centre for Paediatric Low-Grade Brain Tumour Research.
Footnotes
Conflict of interest
D. Capper, D. T. W. Jones, A. von Deimling and S. M. Pfister declare that under the No. EP3067432A1 a patent was applied for a “DNA-methylation based method for classifying tumor species”. D. Capper and A. von Deimling are patent holders of “Methods for the diagnosis and the prognosis of a brain tumor”, the IDH1 R132H-specific antibody used in this manuscript (US 8367347 B2). The patent is under the administrative supervision of the DKFZ technology transfer office.
References
- 1.Bandopadhayay P, Ramkissoon LA, Jain P, Bergthold G, Wala J, Zeid R et al. (2016) MYB-QKI rearrangements in angiocentric glioma drive tumorigenicity through a tripartite mechanism. Nat Genet 48:273–282. doi: 10.1038/ng.3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blumcke I, Aronica E, Urbach H, Alexopoulos A, Gonzalez-Martinez JA (2014) A neuropathology-based approach to epilepsy surgery in brain tumors and proposal for a new terminology use for long-term epilepsy-associated brain tumors. Acta Neuropathol 128:39–54. doi: 10.1007/s00401-014-1288-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blumcke I, Luyken C, Urbach H, Schramm J, Wiestler O (2004) An isomorphic subtype of long-term epilepsy-associated astrocytomas associated with benign prognosis. Acta Neuropathol 107:381–388. doi: 10.1007/s00401-004-0833-3 [DOI] [PubMed] [Google Scholar]
- 4.Blumcke I, Spreafico R, Haaker G, Coras R, Kobow K, Bien CG et al. (2017) Histopathological Findings in Brain Tissue Obtained during Epilepsy Surgery. New Engl J Med 377:1648–1656. doi: 10.1056/NEJMoa1703784 [DOI] [PubMed] [Google Scholar]
- 5.Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, Cooper LA et al. (2015) Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N Engl J Med 372:2481–2498. doi: 10.1056/NEJMoa1402121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Capper D, Engel NW, Stichel D, Lechner M, Glöss S, Schmid S et al. (2018) DNA methylation-based reclassification of olfactory neuroblastoma. Acta Neuropathol 136:255–271. doi: 10.1007/s00401-018-1854-7 [DOI] [PubMed] [Google Scholar]
- 7.Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D et al. (2018) DNA methylation-based classification of central nervous system tumours. Nature 555:469–474. doi: 10.1038/nature26000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Capper D, Weißert S, Balss J, Habel A, Meyer J, Jäger D et al. (2010) Characterization of R132H Mutation-specific IDH1 Antibody Binding in Brain Tumors. Brain Pathol 20:245–254. doi: 10.1111/j.1750-3639.2009.00352.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Capper D, Zentgraf H, Balss J, Hartmann C, von Deimling A (2009) Monoclonal antibody specific for IDH1 R132H mutation. Acta Neuropathol 118:599–601. doi: 10.1007/s00401-009-0595-z [DOI] [PubMed] [Google Scholar]
- 10.Chung EY, Dews M, Cozma D, Yu D, Wentzel EA, Chang T-C et al. (2008) c-Myb oncoprotein is an essential target of the dleu2 tumor suppressor microRNA cluster. Cancer Biol Ther 7:1758–1764. doi: 10.4161/cbt.7.11.6722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ciceri G, Dehorter N, Sols I, Huang ZJ, Maravall M, Marin O (2013) Lineage-specific laminar organization of cortical GABAergic interneurons. Nat Neurosci 16:1199–1210. doi: 10.1038/nn.3485 [DOI] [PubMed] [Google Scholar]
- 12.Deng MY, Sill M, Chiang J, Schittenhelm J, Ebinger M, Schuhmann MU et al. (2018) Molecularly defined diffuse leptomeningeal glioneuronal tumor (DLGNT) comprises two subgroups with distinct clinical and genetic features. Acta Neuropathol 136:239–253. doi: 10.1007/s00401-018-1865-4 [DOI] [PubMed] [Google Scholar]
- 13.Ellison DW, Hawkins C, Jones DTW, Onar-Thomas A, Pfister SM, Reifenberger G et al. (2019) cIMPACT-NOW update 4: diffuse gliomas characterized by MYB, MYBL1, or FGFR1 alterations or BRAFV600 E mutation. Acta Neuropathol:Online first doi: 10.1007/s00401-019-01987-0 [DOI] [PubMed] [Google Scholar]
- 14.Gonda TJ, Buckmaster C, Ramsay RG (1989) Activation of c-myb by carboxy-terminal truncation: relationship to transformation of murine haemopoietic cells in vitro. The EMBO journal 8:1777–1783. doi: 10.1002/j.1460-2075.1989.tb03571.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gröbner SN, Worst BC, Weischenfeldt J, Buchhalter I, Kleinheinz K, Rudneva VA et al. (2018) The landscape of genomic alterations across childhood cancers. Nature 555:321–327. doi: 10.1038/nature25480 [DOI] [PubMed] [Google Scholar]
- 16.Hansen KD, Aryee M (2012) IlluminaHumanMethylation450kmanifest: Annotation for Illumina’s 450k methylation arrays. R package version 0.4.0 doi: 10.18129/B9.bioc.IlluminaHumanMethylation450kmanifest [DOI] [Google Scholar]
- 17.Hansen KD, Aryee M (2016) IlluminaHumanMethylationEPICmanifest: Manifest for Illumina’s EPIC methylation arrays. R package version 0.3.0 doi: 10.18129/B9.bioc.IlluminaHumanMethylationEPICmanifest [DOI] [Google Scholar]
- 18.Hartmann C, Meyer J, Balss J, Capper D, Mueller W, Christians A et al. (2009) Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol 118:469–474. doi: 10.1007/s00401-009-0561-9 [DOI] [PubMed] [Google Scholar]
- 19.Ho AS, Kannan K, Roy DM, Morris LGT, Ganly I, Katabi N et al. (2013) The mutational landscape of adenoid cystic carcinoma. Nat Genet 45:791–798. doi: 10.1038/ng.2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hou Y, Pinheiro J, Sahm F, Reuss DE, Schrimpf D, Stichel D et al. (2019) Papillary glioneuronal tumor (PGNT) exhibits a characteristic methylation profile and fusions involving PRKCA. Acta Neuropathol. doi: 10.1007/s00401-019-01969-2 [DOI] [PubMed] [Google Scholar]
- 21.Hovestadt V, Remke M, Kool M, Pietsch T, Northcott P, Fischer R et al. (2013) Robust molecular subgrouping and copy-number profiling of medulloblastoma from small amounts of archival tumour material using high-density DNA methylation arrays. Acta Neuropathol 125:913–916. doi: 10.1007/s00401-013-1126-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hovestadt V, Zapatka M conumee: Enhanced copy-number variation analysis using Illumina DNA methylation arrays. R package version 1.9.0 doi: 10.18129/B9.bioc.conumee [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johann PD, Bens S, Oyen F, Wagener R, Giannini C, Perry A et al. (2018) Sellar Region Atypical Teratoid/Rhabdoid Tumors (ATRT) in Adults Display DNA Methylation Profiles of the ATRT-MYC Subgroup. The American Journal of Surgical Pathology 42:506–511. doi: 10.1097/PAS.0000000000001023 [DOI] [PubMed] [Google Scholar]
- 24.Jones DTW, Hutter B, Jager N, Korshunov A, Kool M, Warnatz H-J et al. (2013) Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet 45:927–932. doi: 10.1038/ng.2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones DTW, Kieran MW, Bouffet E, Alexandrescu S, Bandopadhayay P, Bornhorst M et al. (2017) Pediatric low-grade gliomas: next biologically driven steps. Neuro Oncol 22:160–173. doi: 10.1093/neuonc/nox141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones DTW, Kocialkowski S, Liu L, Pearson DM, Bäcklund LM, Ichimura K et al. (2008) Tandem Duplication Producing a Novel Oncogenic BRAF Fusion Gene Defines the Majority of Pilocytic Astrocytomas. Cancer Res 68:8673. doi: 10.1158/0008-5472.CAN-08-2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanei-Ishii C, MacMillan EM, Nomura T, Sarai A, Ramsay RG, Aimoto S et al. (1992) Transactivation and transformation by Myb are negatively regulated by a leucine-zipper structure. Proceedings of the National Academy of Sciences 89:3088–3092. doi: 10.1073/pnas.89.7.3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koelsche C, Mynarek M, Schrimpf D, Bertero L, Serrano J, Sahm F et al. (2018) Primary intracranial spindle cell sarcoma with rhabdomyosarcoma-like features share a highly distinct methylation profile and DICER1 mutations. Acta Neuropathol 136:327–337. doi: 10.1007/s00401-018-1871-6 [DOI] [PubMed] [Google Scholar]
- 29.Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin Y-C, Kuo M-W, Yu J, Kuo H-H, Lin R-J, Lo W-L et al. (2008) c-Myb Is an Evolutionary Conserved miR-150 Target and miR-150/c-Myb Interaction Is Important for Embryonic Development. Mol Biol Evol 25:2189–2198. doi: 10.1093/molbev/msn165 [DOI] [PubMed] [Google Scholar]
- 31.Liu Y, Easton J, Shao Y, Maciaszek J, Wang Z, Wilkinson MR et al. (2017) The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet 49:1211–1218. doi: 10.1038/ng.3909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK (2016) WHO Classification of Tumours of the Central Nervous System. Revised 4th edition edn. IARC, Lyon [Google Scholar]
- 33.Louis DN, Perry A, Burger P, Ellison DW, Reifenberger G, von Deimling A et al. (2014) International Society of Neuropathology-Haarlem Consensus Guidelines, for Nervous System Tumor Classification and Grading. Brain Pathol 24:671–672. doi: 10.1111/bpa.12171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oh IH, Reddy EP (1999) The myb gene family in cell growth, differentiation and apoptosis. Oncogene 18:3017–3033. doi: 10.1038/sj.onc.1202839 [DOI] [PubMed] [Google Scholar]
- 35.Pajtler KW, Witt H, Sill M, Jones DTW, Hovestadt V, Kratochwil F et al. (2015) Molecular Classification of Ependymal Tumors across All CNS Compartments, Histopathological Grades, and Age Groups. Cancer Cell 27:728–743. doi: 10.1016/j.ccell.2015.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Persson M, Andrén Y, Mark J, Horlings HM, Persson F, Stenman G (2009) Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proceedings of the National Academy of Sciences 106:18740–18744. doi: 10.1073/pnas.0909114106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qaddoumi I, Orisme W, Wen J, Santiago T, Gupta K, Dalton JD et al. (2016) Genetic alterations in uncommon low-grade neuroepithelial tumors: BRAF, FGFR1, and MYB mutations occur at high frequency and align with morphology. Acta Neuropathol 131:833–845. doi: 10.1007/s00401-016-1539-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quelen C, Lippert E, Struski S, Demur C, Soler G, Prade N et al. (2011) Identification of a transforming MYB-GATA1 fusion gene in acute basophilic leukemia: a new entity in male infants. Blood 117:5719–5722. doi: 10.1182/blood-2011-01-333013 [DOI] [PubMed] [Google Scholar]
- 39.Ramkissoon LA, Horowitz PM, Craig JM, Ramkissoon SH, Rich BE, Schumacher SE et al. (2013) Genomic analysis of diffuse pediatric low-grade gliomas identifies recurrent oncogenic truncating rearrangements in the transcription factor MYBL1. Proceedings of the National Academy of Sciences 110:8188–8193. doi: 10.1073/pnas.1300252110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reinhardt A, Stichel D, Schrimpf D, Sahm F, Korshunov A, Reuss DE et al. (2018) Anaplastic astrocytoma with piloid features, a novel molecular class of IDH wildtype glioma with recurrent MAPK pathway, CDKN2A/B and ATRX alterations. Acta Neuropathol 136:273–291. doi: 10.1007/s00401-018-1837-8 [DOI] [PubMed] [Google Scholar]
- 41.Reuss DE, Kratz A, Sahm F, Capper D, Schrimpf D, Koelsche C et al. (2015) Adult IDH wild type astrocytomas biologically and clinically resolve into other tumor entities. Acta Neuropathol 130:407–417. doi: 10.1007/s00401-015-1454-8 [DOI] [PubMed] [Google Scholar]
- 42.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G et al. (2011) Integrative genomics viewer. Nat Biotechnol 29:24–26. doi: 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sahm F, Reuss D, Koelsche C, Capper D, Schittenhelm J, Heim S et al. (2014) Farewell to oligoastrocytoma: in situ molecular genetics favor classification as either oligodendroglioma or astrocytoma. Acta Neuropathol 128:551–559. doi: 10.1007/s00401-014-1326-7 [DOI] [PubMed] [Google Scholar]
- 44.Sahm F, Schrimpf D, Stichel D, Jones DTW, Hielscher T, Schefzyk S et al. (2017) DNA methylation-based classification and grading system for meningioma: a multicentre, retrospective analysis. The Lancet Oncology 18:682–694. doi: 10.1016/S1470-2045(17)30155-9 [DOI] [PubMed] [Google Scholar]
- 45.Schindler G, Capper D, Meyer J, Janzarik W, Omran H, Herold-Mende C et al. (2011) Analysis of BRAF V600E mutation in 1,320 nervous system tumors reveals high mutation frequencies in pleomorphic xanthoastrocytoma, ganglioglioma and extra-cerebellar pilocytic astrocytoma. Acta Neuropathol 121:397–405. doi: 10.1007/s00401-011-0802-6 [DOI] [PubMed] [Google Scholar]
- 46.Schramm J, Luyken C, Urbach H, Fimmers R, Blumcke I (2004) Evidence for a clinically distinct new subtype of grade II astrocytomas in patients with long-term epilepsy. Neurosurgery 55:340–347. doi: 10.1227/01.NEU.0000129546.38675.1B [DOI] [PubMed] [Google Scholar]
- 47.Schurr J, Coras R, Rössler K, Pieper T, Kudernatsch M, Holthausen H et al. (2017) Mild Malformation of Cortical Development with Oligodendroglial Hyperplasia in Frontal Lobe Epilepsy: A New Clinico-Pathological Entity. Brain Pathol 27:26–35. doi: 10.1111/bpa.12347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sievers P, Stichel D, Schrimpf D, Sahm F, Koelsche C, Reuss DE et al. (2018) FGFR1:TACC1 fusion is a frequent event in molecularly defined extraventricular neurocytoma. Acta Neuropathol 136:293–302. doi: 10.1007/s00401-018-1882-3 [DOI] [PubMed] [Google Scholar]
- 49.Stichel D, Schrimpf D, Casalini B, Meyer J, Wefers AK, Sievers P et al. (2019) Routine RNA sequencing of formalin-fixed paraffin-embedded specimens in neuropathology diagnostics identifies diagnostically and therapeutically relevant gene fusions. Acta Neuropathol First Online: 05 July 2019. doi: 10.1007/s00401-019-02039-3 [DOI] [PubMed] [Google Scholar]
- 50.Sturm D, Orr BA, Toprak UH, Hovestadt V, Jones DTW, Capper D et al. (2016) New Brain Tumor Entities Emerge from Molecular Classification of CNS-PNETs. Cell 164:1060–1072. doi: 10.1016/j.cell.2016.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C et al. (2012) Hotspot Mutations in H3F3A and IDH1 Define Distinct Epigenetic and Biological Subgroups of Glioblastoma. Cancer Cell 22:425–437. doi: 10.1016/j.ccr.2012.08.024 [DOI] [PubMed] [Google Scholar]
- 52.Suzuki K, Suzuki Y, Hama A, Muramatsu H, Nakatochi M, Gunji M et al. (2017) Recurrent MYB rearrangement in blastic plasmacytoid dendritic cell neoplasm. Leukemia 31:1629–1633. doi: 10.1038/leu.2017.101 [DOI] [PubMed] [Google Scholar]
- 53.Takahashi T, Nakagoshi H, Sarai A, Nomura N, Yamamoto T, Ishii S (1995) Human A-myb gene encodes a transcriptional activator containing the negative regulatory domains. FEBS Lett 358:89–96. doi: 10.1016/0014-5793(94)01402-M [DOI] [PubMed] [Google Scholar]
- 54.Tatevossian R, Tang B, Dalton J, Forshew T, Lawson A, Ma J et al. (2010) MYB upregulation and genetic aberrations in a subset of pediatric low-grade gliomas. Acta Neuropathol 120:731–743. doi: 10.1007/s00401-010-0763-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Urbach H, Binder D, von Lehe M, Podlogar M, Bien CG, Becker A et al. (2007) Correlation of MRI and histopathology in epileptogenic parietal and occipital lobe lesions. Seizure 16:608–614. doi: 10.1016/j.seizure.2007.04.009 [DOI] [PubMed] [Google Scholar]
- 56.Weston K (1998) Myb proteins in life, death and differentiation. Curr Opin Genet Dev 8:76–81. doi: 10.1016/S0959-437X(98)80065-8 [DOI] [PubMed] [Google Scholar]
- 57.Xiao C, Calado DP, Galler G, Thai T-H, Patterson HC, Wang J et al. (2007) MiR-150 Controls B Cell Differentiation by Targeting the Transcription Factor c-Myb. Cell 131:146–159. doi: 10.1016/j.cell.2007.07.021 [DOI] [PubMed] [Google Scholar]
- 58.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W et al. (2009) IDH1 and IDH2 Mutations in Gliomas. New Engl J Med 360:765–773. doi: 10.1056/NEJMoa0808710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang J, Wu G, Miller CP, Tatevossian RG, Dalton JD, Tang B et al. (2013) Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet 45:602–612. doi: 10.1038/ng.2611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao H, Kalota A, Jin S, Gewirtz AM (2009) The c-myb proto-oncogene and microRNA-15a comprise an active autoregulatory feedback loop in human hematopoietic cells. Blood 113:505. doi: 10.1182/blood-2008-01-136218 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


