Abstract
Benzo[a]pyrene (BaP) is a widespread environmental carcinogen activated by cytochrome P450 (P450) enzymes. In Hepatic P450 Reductase Null (HRN) and Reductase Conditional Null (RCN) mice, P450 oxidoreductase (Por) is deleted specifically in hepatocytes, resulting in the loss of essentially all hepatic P450 function. Treatment of HRN mice with a single i.p. or oral dose of BaP (12.5 or 125mg/kg body weight) resulted in higher DNA adduct levels in liver (up to 10-fold) than in wild-type (WT) mice, indicating that hepatic P450s appear to be more important for BaP detoxification in vivo. Similar results were obtained in RCN mice. We tested whether differences between hepatocytes and non-hepatocytes in P450 activity may underlie the increased liver BaP-DNA binding in HRN mice. Cellular localisation by immunohistochemistry of BaP-DNA adducts showed that HRN mice have ample capacity for formation of BaP-DNA adducts in liver, indicating that the metabolic process does not result in the generation of a reactive species different from that formed in WT mice. However, increased protein expression of cytochrome b5 in hepatic microsomes of HRN relative to WT mice suggests that cytochrome b5 may modulate the P450-mediated bioactivation of BaP in HRN mice, partially substituting the function of Por.
Keywords: Benzo[a]pyrene, Cytochrome P450, Cytochrome P450 oxidoreductase, DNA adducts, Immunohistochemistry, Polycyclic aromatic hydrocarbon
1. Introduction
Environmental factors and individual genetic susceptibility play an important role in many human cancers (Wild, 2009). Polycyclic aromatic hydrocarbons(PAHs), of which benzo[a]pyrene (BaP) is the most commonly studied and measured, are formed by the incomplete combustion of organic matter (Baird et al., 2005). Human exposure to PAHs is unavoidable and environmental sources include tobacco smoking, ambient air pollution and diet (Phillips, 1999, 2002). Numerous epidemiological studies have implicated BaP and other PAHs are implicated as causative agents in human cancer, particularly lung and colon cancer (Gunter et al., 2007; IARC, 2010; Kucab et al., 2010).
BaP requires metabolic activation prior to reaction with DNA, and DNA adduct formation is an essential step by which it and other carcinogenic PAHs exert their biological effects (Lemieux et al., 2011; Luch and Baird, 2005). The initial oxidation of BaP is catalysed by cytochrome P450 (P450)-dependent monooxy-geneases, of which CYP1A1 and CYP1B1 are two of the most important enzymes in this process (Hamouchene et al., 2011). The resulting epoxide is then converted to a dihydrodiol by microsomal epoxide hydrolase (mEH), which leads, via further bioactivation by CYP1A1 or CYP1B1, to the formation of the ultimately reactive species, BaP-7,8-dihydrodiol-9,10-epoxide (BPDE). The expression of P450s, such as CYP1A1, is known to be upregulated by the aryl hydrocarbon receptor (AHR) (Hockley et al., 2007). BaP can bind to and activate the AHR thereby enhancing its own metabolic activation. Ultimately, BPDE reacts with DNA, forming adducts preferentially at guanine residues; the most abundant DNA adduct is 10-(deoxyguanosin-N2-yl)-7,8,9-trihydroxy-7,8,9,10-tetrahydro-BaP (dG-N2-BPDE) (Phillips, 2005). The level of BaP-DNA adducts in cells is most probably the result of a balance between their formation and their loss through DNA repair processes, cell turnover, and/or apoptosis. Collectively, BaP genotoxicity depends on various factors: (i) metabolism of BaP by phase I enzymes (activation) to reactive DNA-binding species; (ii) detoxification of reactive BaP metabolites by both phase I and phase II enzymes (conjugation); (iii) rate of repair of BaP-DNA adducts; and (iv) BaP-induced expression of genes such as those encoding enzymes involved in activation/detoxification or in DNA damage response (Uno et al., 2004).
In contrast to in vitro studies showing the role of Cyp1a1 in metabolic activation of BaP, several studies indicate that in vivo Cyp1a1 is more important in the detoxification than in the metabolic activation of BaP (Arlt et al., 2008; Uno et al., 2001, 2004). It has been found that Cyp1a1(−/−) mice administered repeated oral doses of BaP (125 mg/kg body weight [bw]/day) die within approximately 28 days due to immunosuppression, whereas wildtype (WT) mice thus treated remain healthy for at least 1 year on this regimen (Uno et al., 2004). Using the Hepatic P450 Reductase Null (HRN) mouse model we also showed that hepatic P450 enzymes appear to be more important for detoxification of BaP in vivo (Arlt et al., 2008). In HRN mice P450 oxidoreductase (Por), the electron donor to P450s, is deleted specifically in hepatocytes, resulting in the loss of essentially all hepatic P450 function (Henderson et al., 2003); we found that the levels of dG-N2-BPDE in livers of these mice treated intraperitoneally with BaP were higher than in WT mice (Arlt et al., 2008).
The metabolic process (i.e. the specific enzyme(s) involved) by which DNA-binding species are generated in liver from BaP in HRN mice is not known, but it is clear that the process does not result in the generation of a different reactive species from that which is formed in WT mice. One hypothesis is that cell-specific differences in the activity of Cyp enzymes responsible for the metabolic activation of BaP in the liver (e.g. hepatocytes versus non-hepatocytes) may underlie the increased DNA binding by BaP in the livers of HRN mice. However, using our previous analysis of DNA adduct formation,32P-postlabelling or mass spectrometry, it is difficult to answer this question because DNA is extracted from whole tissue. Thus, in the present study we have used immunohistochemistry (IHC) staining with anti-BPDE-DNA antiserum (John et al., 2009; Pratt et al., 2007, 2011; van Gijssel et al., 2002) to explore the localisation of BaP-derived DNA adducts within the liver of HRN mice. In addition, we have studied BaP-DNA adduct formation by 32P-postlabelling in a second mouse model, the P450 Reductase Conditional Null (RCN) mouse (Finn et al., 2007), to confirm results previously obtained in the HRN mouse model.
2. Methods
2.1. Chemicals
BaP (>96%) was purchased from Sigma–Aldrich (St. Louis, MO). All other chemicals were of analytical purity or better.
2.2. Animal treatment
All animal experiments were carried out under license in accordance with the law, and with local ethical approval.
HRN (Porlox/lox/CreALB) mice on a C57BL/6 background used in this study were derived as described previously (Henderson et al., 2003). Mice homozygous for loxP sites at the Por locus (Porlox/lox) were used as wild-type (WT). BaP was dissolved in corn oil at a concentration of 1.25 or 12.5mg/mL. Groups of female HRN and WT mice (3 months old, 25–30 g) were treated i.p. or orally with 12.5 or 125 mg/kg bw BaP (n = 3) for 1 day. Control mice (n = 3) received corn oil only. Animals were killed 24 h after the single dose. Several organs (liver, lung, forestomach, glandular stomach, kidney, spleen and colon) were removed, snap frozen and stored at −80 °C until analysis. For IHC organ sections of the liver were fixed in PBS containing 4% paraformaldehyde, and subsequently subjected to paraffin embedding and sectioning.
RCN (Porlox/lox/CreCYP1A1) mice (Finn et al., 2007) on a C57BL/6 background were bred in-house at the Medical Research Institute (Dundee, UK). In brief, Por floxed mice (Porlox/lox) (Henderson et al., 2003) were crossed with a transgenic line expressing Cre recombinase under the control of the rat CYP1A1 promoter (Ireland et al., 2004) to generate the mouse line (Porlox/lox/CreCYP1A1). Pretreatment of RCN mice with 3-methylcholanthrene (3-MC; 40 mg/kg bw i.p. in corn oil) 2 weeks before BaP treatment resulted in hepatic POR loss (Arlt et al., 2011). BaP was dissolved in corn oil at a concentration of 12.5mg/mL. Groups of female RCN mice (3 months old, 25–30g) were treated i.p. with 125 mg/kg bw (n = 3) of BaP for 1 day. Control mice (n = 3) received corn oil only. Animals were sacrificed 24h after the single dose. Organs for 32P-postlabelling were collected as described above.
2.3. BaP-DNA adduct detection by 32P-postlabelling analysis
Genomic DNA from whole tissue was isolated by a standard phenol-chloroform extraction method and DNA adducts were measured for each DNA sample using the nuclease P1 enrichment version of the 32P-postlabelling method as described previously (Arlt et al., 2008; Phillips and Arlt, 2007).
2.4. BaP-DNA adduct detection by immunohistochemistry
Rabbit polyclonal antibodies, elicited against BPDE-modified DNA (rabbit#30 bleed 6/30/78) (Poirier et al., 1980; Pratt et al., 2011; Weston et al., 1989), were employed for detection of dG-N2-BPDE. For IHC, the BPDE-DNA antiserum was pre-absorbed with calf-thymus DNA to reduce non-specific background staining. In addition, some of the specific BPDE-DNA antiserum was absorbed with the immunogen BPDE-modified DNA (1.3% modified, 13.9 nmol dG-N2-BPDE adducts) so as to eliminate the specific BPDE-DNA immunoglobulins. This was used as a control for unknown samples.
Three serial 5 μm sections (a, b, c) of paraffin-embedded liver tissue were mounted on positively charged glass slides, which were incubated at 60 °C for 1h, deparaffinised by a series of xylene/ethanol washes, and subjected to antigen retrieval by microwaving in the presence of Antigen Retrieval Citra solution (Biogenex, San Ramon, CA). Subsequently, slide “a” was stained with specific BPDE-DNA antiserum, absorbed only with calf thymus DNA; slide “b” was stained with haematoxylin, for visualisation of the nuclei; and slide “c” was stained with BPDE-DNA antiserum absorbed with immunogen BPDE-DNA and unmodified DNA. Rabbit antisera were diluted 1:20,000 in antibody diluent (Ventana Medical Systems Inc., Tucson, AZ) before use.
Slides were stained using the Nexes IHC (Ventana Medical Systems Inc.) automated slide staining system as previously described (van Gijssel et al., 2004). The antiserum incubation was for 20min (for slides “a” and “c”) with no counterstaining. Slide “b” was incubated with normal rabbit serum and counterstained with modified haematoxylin-OS, a modified form of Mayer’s haematoxylin (Vector Labs, Burlingame, CA, USA) for 1min. Stained slides were subsequently rinsed for 1min in water containing Dawn dishwashing liquid soap (Proctor and Gamble, Cincinnati, OH), and rinsed 3–4 times in deionised water to remove the soap. The stained slides were subsequently air dried, and mounted with cover slips using Permount mounting media (Fisher Scientific) and subsequently scanned at 20× resolution using the ScanScope AT digital scanner (Aperio, Vista, CA, USA). The ‘digital’ scans of the slides were subsequently maintained on the ‘Spectrum’ database (Aperio) and visualised using ImageScope software (Aperio).
2.5. Expression of xenobiotic-metabolising enzymes by Western blotting
HRN and WT mice were treated i.p. daily for 5 days with 125 mg/kg bw BaP (n = 3) as described previously (Arlt et al., 2008). Control mice (n = 3) received corn oil only. Hepatic microsomes from HRN and WT mice were isolated as reported. Pooled microsomal fractions were used for further analysis. Western blot analysis of cytochrome b5 (Cyt b5) and microsomal epoxide hydrolase (mEH) was performed using sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE); 75 μg microsomal protein was subjected to 15% SDS-PAGE. After migration, proteins were transferred onto polyvinylidene difluoride (PVDF) membranes. Cytochrome b5 protein was probed with rabbit polyclonal anti-cytochrome b5 antibody (1:750; ab69801; Abcam, MA, USA) and mEH with rabbit polyclonal anti-EH antibody (1:1000; ab76226; Abcam) overnight at 4 °C. Glyceraldehyde phosphate dehydrogenase (GAPDH; 1:750; Millipore, MA, USA) was used as loading control. The antigen–antibody complex was visualised with an alkaline phosphatise-conjugated goat anti-rabbit IgG antibody and 5-bromo-4-chloro-3-indolylphosphate/nitrobluetetrazolium as chromogenic substrate (Kotrbova et al., 2011; Stiborova et al., 2002).
3. Results
3.1. DNA adduct formation in mice
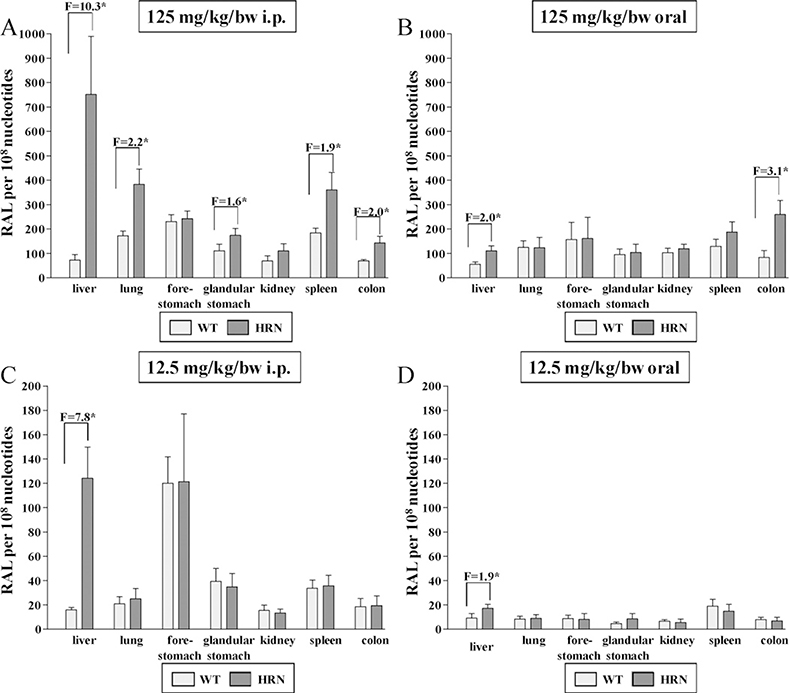
The DNA adduct pattern in organs (liver, lung, forestomach, glandular stomach, kidney, spleen and colon) of HRN and WT mice treated i.p. or orally with a single dose of BaP(12.5 or 125 mg/kg bw) consisted of a single spot analysed by TLC-32P-postlabelling, previously identified as dG-N2-BPDE (Arlt et al., 2008). No DNA adducts were detected in control animals (data not shown). DNA adduct formation was dose-dependent both after i.p. and oral treatment (Fig. 1). After i.p. administration the results at the higher dose were in concordance with our previous study (Arlt et al., 2008) showing a ~10-fold higher DNA binding by BaP in the livers and elevated levels in several extra-hepatic tissues of HRN mice (i.e. lung, glandular stomach, spleen and colon) compared with WT mice (Fig. 1A). At the lower dose BaP-DNA adduct levels were substantially lower in all tissues, but as with the higher dose they were higher (~8-fold) in the livers of HRN mice than in WT mice (Fig. 1C). However, differences between HRN and WT mice in DNA adduct formation were not observed in extra-hepatic tissues with the lower dose of BaP. After oral administration of 12.5 or 125 mg/kg bw BaP, DNA adduct formation by BaP was overall lower compared to i.p. administration (compare Fig. 1B and D). Again, DNA binding by BaP in the livers of HRN mice was higher relative to WT mice, but this effect was less pronounced (only ~2-fold) than after i.p. administration. Overall, no difference in DNA binding by BaP was observed in extra-hepatic tissues, independent of the dose, except for the colon in the higher dose group which showed 3-fold elevated DNA adducts in HRN mice relative to WT mice (Fig. 1B).
Fig. 1.
Quantitative TLC 32P-postlabelling analysis of dG-N2-BPDE adducts in organs of HRN and WT mice treated i.p. (A and C) or orally (B and D) with 12.5 (C and D) or 125 mg/kg bw BaP (A and B) for 24h. F = fold increase in DNA binding in HRN mice compared to WT mice. Values are given as means ± SD (n = 3); each DNA sample was determined by two postlabelled analyses. Comparison was performed by t-test analysis: *P <0.01 different from WT. RAL, relative adduct labelling.
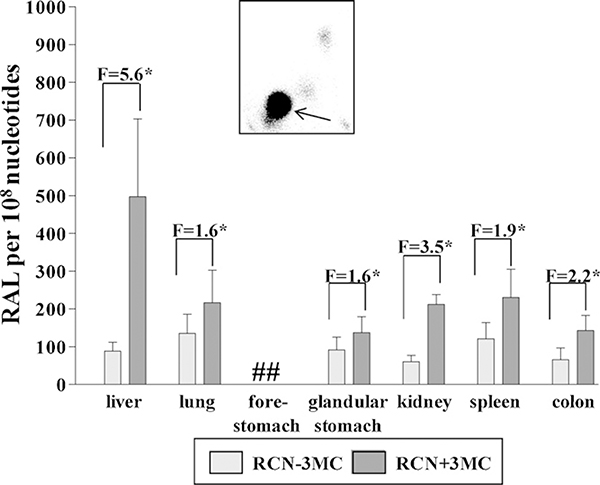
After treatment of RCN mice with a single i.p. dose of 125 mg/kg body weight BaP, the DNA adduct pattern on TLC again consisted of a single spot (i.e. dG-N2-BPDE) with all organs from BaP-treated animals (Fig. 2, insert), whereas no DNA adducts were detected in control animals (data not shown). Quantitative analysis revealed that BaP-induced DNA adduct levels were significantly higher (~6-fold) in livers of RCN mice that lack hepatic Por activity compared to RCN mice with active Por; elevated adduct levels (~2-fold) were also found in all extra-hepatic tissues (Fig. 2). Interestingly, elevated levels of DNA adducts were found in kidney of RCN mice that lack hepatic Por activity, however, no difference in DNA adduct levels in the kidney were observed in the HRN model (compare Fig. 1).
Fig. 2.
Quantitative TLC 32P-postlabelling analysis of dG-N2-BPDE adducts in organs of RCN mice treated i.p. with 125 mg/kg bw BaP for 24h without and with 3-MC pretreatment (single dose of 40 mg/kg bw 3-MC 14 days before BaP treatment. F = fold increase in DNA binding in RCN mice treated with BaP compared with RCN mice treated with BaP and inducer (i.e. 3-MC). Values are given as means ± SD (n = 3); each DNA sample was determined by two postlabelled analyses. Comparison was performed by t-test analysis: *P <0.01 different from RCN mice treated with BaP but without inducer. RAL, relative adduct labelling. Inset: Typical autoradiographic profile of BaP-derived DNA adducts obtained by 32P-postlabelling; solvent conditions for the resolution of 32P-labelled adducts on polyethyleneimine-cellulose thin-layer chromatography were: D1, 1.0M sodium phosphate, pH 6; D3, 4.0M lithium formate, 7.0M urea, pH 3.5; D4, 0.8M LiCl, 0.5M Tris, 8.5M urea, pH 8. The arrow indicates the position of the 5’-32P-labelled biphosphate dG-N2-BPDE adduct. ##, not determined (DNA sample lost).
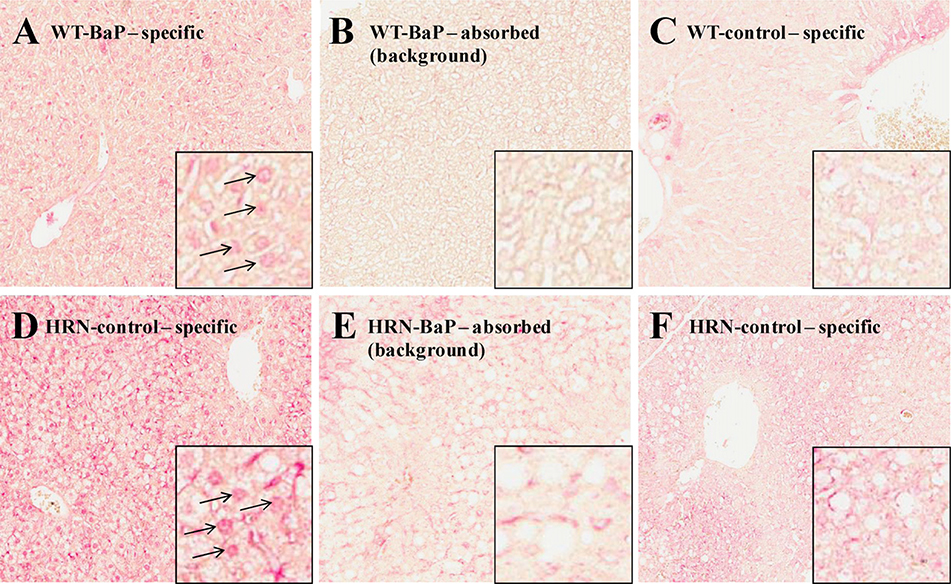
IHC has the capacity to show localisation of adducts, and to show magnitude of adduct formation by differences in colour intensity. Fig. 3 shows liver sections from BaP-treated (at 125 mg/kg body weight, i.p.) WT (Fig. 3A) and HRN (Fig. 3D) mice incubated with the specific anti-BPDE-DNA antiserum, and showing nuclear pink colour intensity indicating BaP-derived DNA adducts. In Fig. 3B and E, adjacent sections from WT and HRN mouse livers, respectively, were stained with the immunogen (BPDE-DNA)-absorbed anti-BPDE-DNA antiserum, and the pink nuclear staining disappeared, indicating that the specific staining was due to BaP-derived DNA adducts. Further evidence that the nuclear staining observed in Fig. 3A–D is specific is shown in Fig. 3C and F, where unexposed WT and HRN mice, respectively, showed no evidence of BaP-derived DNA adducts. Therefore the staining shown in Fig. 3A and D shows nuclear localisation of BaP-derived DNA adducts; the fact that the adducts were nuclear was confirmed by blue staining of the nuclei with haematoxylin (data not shown). The adduct signal in the HRN (Fig. 3D) mice was much stronger compared to the WT (Fig. 3A) mice but experimental conditions did not allow any further semi-quantitation; the effect was so striking that the difference were strongly evident without talking any additional steps. These observations correspond well with the DNA adduct data obtained by 32P-postlabelling. While Fig. 3 shows a representative mouse from each group, all 3 mice from each group showed similar patterns of staining (data not shown). As there were relatively few non-hepatocytes, we were not able to distinguish a difference in nuclear BaP-derived DNA adduct staining between hepatocytes and non-hepatocytes in livers from either WT or HRN mice.
Fig.3.
Immunostaining (20×) for BPDE-DNA adducts in hepatic tissue sections of HRN and WT mice treated i.p. with 125 mg/kg bw BaP for 24h. Staining of BaP-treated mice (A and D) with specific BPDE-DNA antiserum; the arrows indicate stained nuclei detecting dG-N2-BPDE adducts. Staining of BaP-treated mice (B and E) with BPDE-DNA-absorbed BPDE-DNA antiserum (background). No staining was observed in control (untreated) animals (C and F).
3.2. The effect of BaP on expression of hepatic biotransforming enzymes
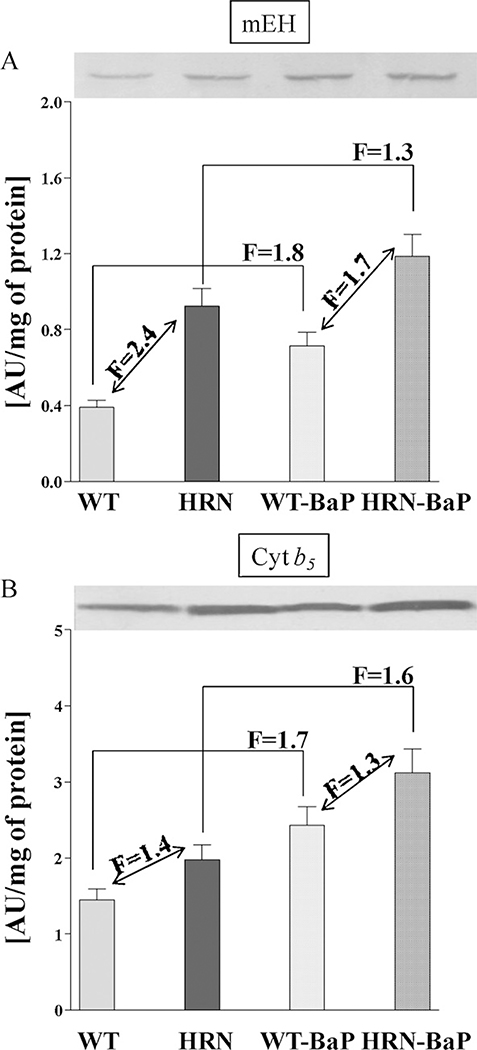
Using Western blot analysis we previously examined the protein expression levels of the BaP-activating enzymes Cyp1a1 and prostaglandin H synthase (Ptgs) as well as Por in hepatic microsomes isolated from HRN and WT mice treated with 125 mg/kg bw BaP for 5 days (Arlt et al., 2008). To investigate further the role of BaP-metabolising enzymes we determined the expression of mEH and a component of a Cyp-monooxygenase system, cytochrome b5 (Cyt b5) (Yamazaki et al.,2002), in the same samples (Fig.4). The levels of mEH were ~2.4-fold higher in hepatic microsomes isolated from HRN relative to WT mice; expression of mEH was induced (~1.3-fold in HRN mice) by BaP treatment. Levels of cytochrome b5 were higher (~1.4 fold) in hepatic microsomes from HRN mice relative to WT mice. In addition, treatment with BaP increased the expression by ~1.6-fold in both HRN and WT mice.
Fig. 4.
Expression of mEH and cytochrome b5 (Cyt b5) in livers of HRN and WT mice, control (untreated) mice or mice treated i.p. daily with 125 mg/kg bw BaP for 5 days. Pooled hepatic microsomal samples were used for analyses as described in Section 2. Values are given as means ± SD of 3 determinations.
4. Discussion
We have previously used the HRN model to investigate hepatic versus extra-hepatic P450-mediated carcinogen metabolism (Arlt et al., 2005, 2006, 2008; Levova et al., 2011; Stiborova et al., 2008). We found that hepatic P450s seem to be more important for detoxification of BaP in vivo despite being important for its bioactivation in vitro (Arlt et al., 2008). Compared to WT mice, BaP-DNA adduct levels were up to 13-fold higher in liver, and elevated in several extra-hepatic tissues, of HRN mice relative to WT mice, after a single i.p. dose of 125 mg/kg sbw BaP.
In HRN mice, the deletion of the Por gene occurs neonatally and although HRN mice develop normally, they exhibit a number of phenotypic changes associated with the loss of P450 function, including hepatic lipid accumulation, reduced bile acid production, increased constitutive P450 expression, and decreased plasma cholesterol and triglyceride levels (Henderson et al., 2003), which may have an impact on the pharmacokinetics/metabolism of xenobiotics studied. In RCN mice, hepatic Por can be deleted conditionally using a rat CYP1A1 promoter to drive Cre recombinase expression (Finn et al., 2007). Thus the use of this promoter provides a tightly regulated method for controlling expression of the transgene in vivo shortly before the animal experiment by the administration of an inducer (i.e. 3-MC) that acts through the aryl hydrocarbon receptor. In addition, RCN mice can be used as their own control. As shown previously, administration of 40 mg/kg bw (i.p.) 3-MC led to a complete and specific deletion of the hepatic Por gene within 14 days; no expression of Por was observed by Western blotting in hepatic microsomes isolated from RCN mice pretreated with 3-MC (Arlt et al., 2011). Further, it is noteworthy that 14 days after a single i.p. dose of 40 mg/kg bw 3-MC, there is no hepatic Cyp1a protein evident (Finn et al., 2007), although some other hepatic Cyps are induced, consistent with the elevated hepatic P450 expression seen in the HRN model and driven by lipid accumulation (Finn et al., 2009). Collectively our results indicate that the previously observed phenomenon, whereby hepatic P450 enzymes appear to more important for BaP detoxification in vivo, is also seen at considerably lower BaP doses in the HRN model. Further, results obtained in the RCN model fully confirm the data obtained in the HRN mouse model.
In general, i.p. administration results in the uptake of BaP by mesenteric veins and lymphatic system that go directly to the liver, bypassing the gastrointestinal tract. It has been pointed out that important pharmacokinetic differences depend on the route of administration (Uno et al., 2004). After oral administration, BaP uptake should be via the gastrointestinal tract and hence to the liver. Indeed, DNA adduct formation after oral administration of BaP was overall lower compared with i.p. administration. Again, DNA binding by BaP in the livers of HRN mice was higher than in WT mice, but this effect was less pronounced than after i.p. administration, indicating that after oral administration the first-pass metabolism of BaP occurs in the gastrointestinal tract.
The metabolic process (i.e. the specific enzyme(s) involved) by which DNA-binding species are generated by BaP in the liver of HRN mice (or RCN mice lacking hepatic POR) in the present study is not known, but it is clear that the process did not result in the generation of a reactive species different from that formed in WT mice. One hypothesis was that cell-specific differences in the activity of P450 enzymes responsible for the metabolic activation of BaP may underlie the increased DNA binding by BaP in the livers of HRN mice. We used IHC to investigate the localisation of BaP-derived DNA adducts within the liver (i.e. hepatocytes versus non-hepatocytes). This may be of importance as in the HRN mouse model hepatic Por is only deleted in hepatocytes which account for the majority of cells in the liver. However, it is clear from the 32P-postlabelling analysis, and confirmed by IHC, that HRN mice have ample capacity for formation of BaP-derived DNA adducts in their liver.
Since hepatic P450 enzyme activity had been essentially obliterated by the conditional deletion of Por in hepatocytes, the level of BaP activation to DNA adducts in hepatocytes in HRN mice is difficult to rationalise. Our studies and those of others (Kondraganti et al., 2003; Sagredo et al., 2009) may suggest that metabolic activation of BaP in vivo is mediated by P450-independent and/or Ahr-independent pathways, but our own studies have shown that the end result, i.e. the nature of the DNA adduct (i.e. dG-N2-BPDE) formed, is the same. BaP is a good substrate not only for CYP1A1 but also for PTGS. However, we previously found no protein expression of Ptgs1 or Ptgs2 in hepatic microsomes of HRN and WT mice (Arlt et al., 2008). Microsomal epoxide hydrolase (mEH) is important for the hydrolysis of the BaP-7,8-epoxide formed during P450-mediated oxidation generating BaP-7,8-dihydrodiol which subsequently leads to the formation of BPDE, the ultimate reactive metabolite binding covalently to DNA. The levels of mEH were ~2.4-fold higher in hepatic microsomes isolated from HRN relative to WT mice (Fig. 4); expression of mEH was induced (~1.3-fold in HRN mice) by BaP treatment. These findings indicate that the different expression of mEH in HRN and WT mice might be one reason contributing to the increased DNA adduct formation by BaP in HRN mice.
For many years microsomal cytochrome b5 (Cyt b5) has been proposed to modulate the activity of P450 enzymes (Schenkman and Jansson, 2003). The modulation of P450 activity by cytochrome b5 is reported to be both substrate- and P450-specific, with evidence of both stimulation and inhibition of substrate metabolism (Yamazaki et al., 2002). Two mechanisms of cytochrome b5-mediated modulation of P450 catalysis have been suggested: it can affect the P450 catalytic activities by donating the second electron to P450 in a P450 catalytic cycle and/or act as an allosteric modifier of the oxygenase. We previously found that hepatic Cyp1a protein induction was higher in HRN mice compared to WT mice, while, EROD activity, a measure for Cyp1a-mediated enzyme activity, was ~3.4-fold lower in microsomes isolated from livers of BaP-treated HRN mice relative to BaP-treated WT mice (Arlt et al., 2008). In the present study we found that the protein levels of cytochrome b5 were higher (~1.4 fold) in hepatic microsomes from HRN mice relative to WT mice (Fig. 4). In addition, treatment with BaP increased the expression by ~1.6-fold in both HRN and WT mice. Thus, in HRN mice cytochrome b5 may not only modulate the P450-mediated (i.e. Cyp1a1) bioactivation of BaP in vivo, basically partially substituting Por in its function, but it may also stimulate P450-mediated metabolic activation of BaP because of its increased expression induced by BaP. This hypothesis is in concordance with recent findings that cytochrome b5 causes a shift in the oxidation by CYP1A1 and CYP1A2 of the anticancer drug ellipticine from detoxification to activation, leading to increased DNA adduct formation by ellipticine in vitro (Kotrbova et al., 2011). Therefore, it would be interesting to examine the potential role of cytochrome b5 in the metabolic activation of BaP in vivo. Recently, a mouse model has been developed in which cytochrome b5 has been deleted in all tissues [cytochrome b5 complete null (BCN)] (McLaughlin et al., 2010), allowing investigation of the general function of cytochrome b5 in BaP metabolism in vivo. To dissect the role of hepatic cytochrome b5 versus hepatic Por in Cyp-mediated BaP bioactivation a liver-specific cytochrome b5 conditional knockout mouse (Cytb5lox/lox/CreALB; HBN mouse) has been generated (Finn et al., 2008) with or without the expression of hepatic Por (Henderson and Wolf, unpublished data). In the future we will test BaP in these mouse models, which potentially provide improved models to investigate the balance between P450-mediated activation and detoxification of BaP in vivo.
HIGHLIGHTS.
We studied the P450-mediated metabolism of BaP in HRN mouse that lack hepatic Por.
Hepatic P450 appear to be more important for BaP detoxification in vivo.
HRN mice have ample capacity for the formation of BaP-DNA adducts in the liver compared to WT mice.
Cytochrome b5 may modulate P450-mediated bioactivation of BaP in HRN mice.
Acknowledgements
This study was supported by Cancer Research UK and ECNIS2 (Environmental Cancer Risk, Nutrition and Individual Susceptibility) European Union Network of Excellence. Work at Charles University is supported by the Grant Agency of the Czech Republic (grant P301/10/0356) and by the University grant UNCE#42. We would like to acknowledge the Comparative Molecular Pathology Unit at the National Cancer Institute, National Institutes of Health for their assistance with the slide scanning.
Abbreviations
- BaP
benzo[a]pyrene
- BPDE
benzo[a]pyrene-7,8-dihydrodiol-9,10-epoxide
- Cyt b5
cytochrome b5
- P450
cytochrome P450
- HRN
Hepatic P450 Reductase Null
- IHC
immunohistochemistry
- mEH
microsomal epoxide hydrolase
- PAH
polycyclic aromatic hydrocarbon
- POR
cytochrome P450 oxidoreductase
- PTGS
prostaglandin H synthase
- RCN
P450 Reductase Conditional Null
- TLC
thin-layer chromatography
Footnotes
Conflict of interest
None declared.
References
- Arlt VM, Henderson CJ, Wolf CR, Schmeiser HH, Phillips DH, Stiborova M, 2006. Bioactivation of 3-aminobenzanthrone, a human metabolite of the environmental pollutant 3-nitrobenzanthrone: evidence for DNA adduct formation mediated by cytochrome P450 enzymes and peroxidases. Cancer Letters 234, 220–231. [DOI] [PubMed] [Google Scholar]
- Arlt VM, Singh R, Stiborova M, Gamboa da Costa G, Frei E, Evans JD, Farmer PB, Wolf CR, Henderson CJ, Phillips DH, 2011. Effect of hepatic cytochrome P450 (P450) oxidoreductase deficiency on 2-amino-1-methyl6-phenylimidazo[4,5-b]pyridine-DNA adduct formation in P450 reductase conditional null mice. Drug Metabolism and Disposition 39, 2169–2173. [DOI] [PubMed] [Google Scholar]
- Arlt VM, Stiborova M, Henderson CJ, Osborne MR, Bieler CA, Frei E, Martinek V, Sopko B, Wolf CR, Schmeiser HH, Phillips DH, 2005. Environmental pollutant and potent mutagen 3-nitrobenzanthrone forms DNA adducts after reduction by NAD(P)H:quinone oxidoreductase and conjugation by acetyltransferases and sulfotransferases in human hepatic cytosols. Cancer Research 65, 2644–2652. [DOI] [PubMed] [Google Scholar]
- Arlt VM, Stiborova M, Henderson CJ, Thiemann M, Frei E, Aimova D, Singh R, Gamboa da Costa G, Schmitz OJ, Farmer PB, Wolf CR, Phillips DH, 2008. Metabolic activation of benzo[a]pyrene invitro by hepatic cytochrome P450 contrasts with detoxification in vivo: experiments with hepatic cytochrome P450 reductase null mice. Carcinogenesis 29, 656–665. [DOI] [PubMed] [Google Scholar]
- Baird WM, Hooven LA, Mahadevan B, 2005. Carcinogenic polycyclic aromatic hydrocarbon-DNA adducts and mechanism of action.Environmental and Molecular Mutagenesis 45, 106–114. [DOI] [PubMed] [Google Scholar]
- Finn RD, Henderson CJ, Scott CL, Wolf CR, 2009. Unsaturated fatty acid regulation of cytochrome P450 expression via a CAR-dependent pathway. Biochemical Journal 417, 43–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, McLaren AW, Carrie D, Henderson CJ, Wolf CR, 2007. Conditional deletion of cytochrome p450 oxidoreductase in the liver and gastrointestinal tract: a new model for studying the functions of the p450 system. Journal of Pharmacology and Experimental Therapeutics 322, 40–47. [DOI] [PubMed] [Google Scholar]
- Finn RD, McLaughlin LA, Ronseaux S, Rosewell I, Houston JB, Henderson CJ, Wolf CR, 2008. Defining the in vivo role for cytochrome b5 in cytochrome P450 function through the conditional hepatic deletion of microsomal cytochrome b5. The Journal of Biological Chemistry 283, 31385–31393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunter MJ, Divi RL, Kulldorff M, Vermeulen R, Haverkos KJ, Kuo MM, Strickland P, Poirier MC, Rothman N, Sinha R, 2007. Leukocyte polycyclic aromatic hydrocarbon-DNA adduct formation and colorectal adenoma. Carcinogenesis 28, 1426–1429. [DOI] [PubMed] [Google Scholar]
- Hamouchene H, Arlt VM, Giddings I, Phillips DH, 2011. Influence of cell cycle on responses of MCF-7 cells to benzo[a]pyrene. BMC Genomics 12, 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson CJ, Otto DM, Carrie D, Magnuson MA, McLaren AW, Rosewell I, Wolf CR, 2003. Inactivation of the hepatic cytochrome P450 system by conditional deletion of hepatic cytochrome P450 reductase. Journal of Biological Chemistry 278, 13480–13486. [DOI] [PubMed] [Google Scholar]
- Hockley SL, Arlt VM, Brewer D, Te Poele R, Workman P, Giddings I, Phillips DH, 2007. AHR- and DNA-damage-mediated gene expression responses induced by benzo(a)pyrene in human cell lines.Chemical Research in Toxicology 20, 1797–1810. [DOI] [PubMed] [Google Scholar]
- IARC, 2010. Some non-heterocyclic polycyclic aromatic hydrocarbons and some related exposures. In: IARC Monograph on the Evaluation of Carcinogenic Risks to Humans 92. [PMC free article] [PubMed] [Google Scholar]
- Ireland H, Kemp R, Houghton C, Howard L, Clarke AR, Sansom OJ, Winton DJ, 2004. Inducible Cre-mediated control of gene expression in the murine gastrointestinal tract: effect of loss of beta-catenin. Gastroenterology 126, 1236–1246. [DOI] [PubMed] [Google Scholar]
- John K, Ragavan N, Pratt MM, Singh PB, Al-Buheissi S, Matanhelia SS, Phillips DH, Poirier MC, Martin FL, 2009. Quantification of phase I/II metabolizing enzyme gene expression and polycyclic aromatic hydrocarbon-DNA adduct levels in human prostate. Prostate 69, 505–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondraganti SR, Fernandez-Salguero P, Gonzalez FJ, Ramos KS, Jiang W, Moorthy B, 2003. Polycyclic aromatic hydrocarbon-inducible DNA adducts: evidence by 32P-postlabeling and use of knockout mice for Ah receptor-independent mechanisms of metabolic activation in vivo. International Journal of Cancer 103, 5–11. [DOI] [PubMed] [Google Scholar]
- Kotrbova V, Mrazova B, Moserova M, Martinek V, Hodek P, Hudecek J, Frei E, Stiborova M, 2011. Cytochrome b(5) shifts oxidation of the anticancer drug ellipticine by cytochromes P450 1A1 and 1A2 from its detoxication to activation, thereby modulating its pharmacological efficacy. Biochemical Pharmacology 82, 669–680. [DOI] [PubMed] [Google Scholar]
- Kucab JE, Phillips DH, Arlt VM, 2010. Linking environmental carcinogen exposure to TP53 mutations in human tumours using the human TP53 knock-in (Hupki) mouse model. FEBS Journal 277, 2567–2583. [DOI] [PubMed] [Google Scholar]
- Lemieux CL, Douglas GR, Gingerich J, Phonethepswath S, Torous DK, Dertinger SD, Phillips DH, Arlt VM, White PA, 2011. Simultaneous measurement of benzo[a]pyrene-induced Pig-a and lacZ mutations, micronuclei and dna adducts in muta(TM) mouse. Environmental and Molecular Mutagenesis 52, 756–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levova K, Moserova M, Kotrbova V, Sulc M, Henderson CJ, Wolf CR, Phillips DH, Frei E, Schmeiser HH, Mares J, Arlt VM, Stiborova M, 2011. Role of cytochromes P450 1A1/2 in detoxication and activation of carcinogenic aristolochic acid I: studies with the hepatic NADPH:Cytochrome P450 Reductase Null (HRN) mouse model. Toxicological Sciences 121, 43–56. [DOI] [PubMed] [Google Scholar]
- Luch A, Baird WM, 2005. Metabolic activation and detoxification of polycyclic aromatic hydrocarbons Imperial College Press, London, pp. 19–96. [Google Scholar]
- McLaughlin LA,Ronseaux S,Finn RD,Henderson CJ,RolandWolf C,2010Deletion of microsomal cytochrome b5 profoundly affects hepatic and extrahepatic drug metabolism. Molecular Pharmacology 78, 269–278. [DOI] [PubMed] [Google Scholar]
- Phillips DH, 1999. Polycyclic aromatic hydrocarbons in the diet. Mutation Research 443, 139–147. [DOI] [PubMed] [Google Scholar]
- Phillips DH, 2002. Smoking-related DNA and protein adducts in human tissues. Carcinogenesis 23, 1979–2004. [DOI] [PubMed] [Google Scholar]
- Phillips DH, 2005. Macromolecular adducts as biomarkers of human exposure to polycyclic aromatic hydrocarbons Imperial College Press, London, pp. 137–169. [Google Scholar]
- Phillips DH, Arlt VM, 2007. The 32P-postlabeling assay for DNA adducts. Nature Protocols 2, 2772–2781. [DOI] [PubMed] [Google Scholar]
- Poirier MC, Santella R, Weinstein IB, Grunberger D, Yuspa SH, 1980. Quantitation of benzo(a)pyrene-deoxyguanosine adducts by radioimmunoassay. Cancer Research 40, 412–416. [PubMed] [Google Scholar]
- Pratt MM, John K, MacLean AB, Afework S, Phillips DH, Poirier MC, 2011. Polycyclic aromatic hydrocarbon (PAH) exposure and DNA adduct semiquantitation in archived human tissues. International Journal of Environmental Research and Public Health 8, 2675–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt MM, Sirajuddin P, Poirier MC, Schiffman M, Glass AG, Scott DR, Rush BB, Olivero OA, Castle PE, 2007. Polycyclic aromatic hydrocarbon-DNA adducts in cervix of women infected with carcinogenic human papillomavirus types: an immunohistochemistry study. Mutation Research 624, 114–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagredo C, Mollerup S, Cole KJ, Phillips DH, Uppstad H, Ovrebo S, 2009. Biotransformation of benzo[a]pyrene in Ahr knockout mice is dependent on time and route of exposure. Chemical Research in Toxicology 22, 584–591. [DOI] [PubMed] [Google Scholar]
- Schenkman JB, Jansson I, 2003. The many roles of cytochrome b5. Pharmacology & Therapeutics 97, 139–152. [DOI] [PubMed] [Google Scholar]
- Stiborova M, Arlt VM, Henderson CJ, Wolf CR, Kotrbova V, Moserova M, Hudecek J, Phillips DH, Frei E, 2008. Role of hepatic cytochromes P450 in bioactivation of the anticancer drug ellipticine: studies with the hepatic NADPH:Cytochrome P450 reductase null mouse. Toxicology and Applied Pharmacology 226, 318–327. [DOI] [PubMed] [Google Scholar]
- Stiborova M, Martinek V, Rydlova H, Hodek P, Frei E, 2002. Sudan I is a potential carcinogen for humans: evidence for its metabolic activation and detoxication by human recombinant cytochrome P450 1A1 and liver microsomes. Cancer Research 62, 5678–5684. [PubMed] [Google Scholar]
- Uno S, Dalton TP, Derkenne S, Curran CP, Miller ML, Shertzer HG, Nebert DW, 2004. Oral exposure to benzo[a]pyrene in the mouse: detoxication by inducible cytochrome P450 is more important than metabolic activation. Molecular Pharmacology 65, 1225–1237. [DOI] [PubMed] [Google Scholar]
- Uno S, Dalton TP, Shertzer HG, Genter MB, Warshawsky D, Talaska G, Nebert DW, 2001. Benzo[a]pyrene-induced toxicity: paradoxical protection in Cyp1a1(−/−) knockout mice having increased hepatic BaP-DNA adduct levels. Biochemical and Biophysical Research Communications 289, 1049–1056. [DOI] [PubMed] [Google Scholar]
- van Gijssel HE, Divi RL, Olivero OA, Roth MJ, Wang GQ, Dawsey SM, Albert PS, Qiao YL, Taylor PR, Dong ZW, Schrager JA, Kleiner DE, Poirier MC, 2002. Semiquantitation of polycyclic aromatic hydrocarbon-DNA adducts in human esophagus by immunohistochemistry and the automated cellular imaging system. Cancer Epidemiology: Biomarkers and Prevention 11, 1622–1629. [PubMed] [Google Scholar]
- van Gijssel HE, Schild LJ, Watt DL, Roth MJ, Wang GQ, Dawsey SM, Albert PS, Qiao YL, Taylor PR, Dong ZW, Poirier MC, 2004. Polycyclic aromatic hydrocarbon-DNA adducts determined by semiquantitative immunohistochemistry in human esophageal biopsies taken in 1985. Mutation Research 547, 55–62. [DOI] [PubMed] [Google Scholar]
- Weston A, Manchester DK, Poirier MC, Choi JS, Trivers GE, Mann DL, Harris CC, 1989. Derivative fluorescence spectral analysis of polycyclic aromatic hydrocarbon-DNA adducts in human placenta. Chemical Research in Toxicology 2, 104–108. [DOI] [PubMed] [Google Scholar]
- Wild CP, 2009. Environmental exposure measurement in cancer epidemiology. Mutagenesis 24, 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H, Nakamura M, Komatsu T, Ohyama K, Hatanaka N, Asahi S, Shimada N, Guengerich FP, Shimada T, Nakajima M, Yokoi T, 2002. Roles of NADPH-P450 reductase and apo- and holo-cytochrome b5 on xenobiotic oxidations catalyzed by 12 recombinant human cytochrome P450s expressed in membranes of Escherichia coli. Protein Expression and Purification 24, 329–337. [DOI] [PubMed] [Google Scholar]