Dear Editor,
The COVID-19 pandemic worldwide is caused by a novel coronavirus SARS-CoV-2 (the severe acute respiratory syndrome coronavirus 2).1 After viral invasion into the host cells, the ~30 kb viral genome RNA injected is translated into structural and non-structural proteins to replicate viral genome and assemble more viral particles. Many copies of nucleocapsid (N) protein can bind to viral genome RNA and pack it into ~100 nm particles, assisting membrane (M) and envelope (E) proteins to efficiently assemble the viral envelope.2 The exact molecular mechanism by which N protein packs up the viral genome still remains elusive.
An N protein of SARS-CoV-2 consists of an N-terminal RNA-binding domain (NTD) and a C-terminal dimerization domain (CTD) and shares ~90% sequence identity with N protein of SARS-CoV (Supplementary information, Fig. S1a). The regions located between the N-terminus and NTD, between NTD and CTD, and between CTD and the C-terminus of the N protein of SARS-CoV-2 (thereafter referred to as N protein) are predicted to be intrinsically disordered (Supplementary information, Fig. S1b, c). At neutral pH, the N protein is positively charged (+24 e), consistent with its strong binding affinity with negatively charged RNA, and this has also been validated by the nucleotide contaminant in N protein purification (Supplementary information, Fig. S2a, b). The gel filtration and dynamic light scattering results further suggested the oligmerization of N protein (Supplementary information, Fig. S2c–f). Altogether, the sequence and structure features of N protein are similar to those of other proteins that have been reported to undergo liquid–liquid phase separation (LLPS) with nucleic acids.3 Thus, we hypothesized that N protein may also undergo LLPS with viral genome RNA and potentially facilitate viral assembly.
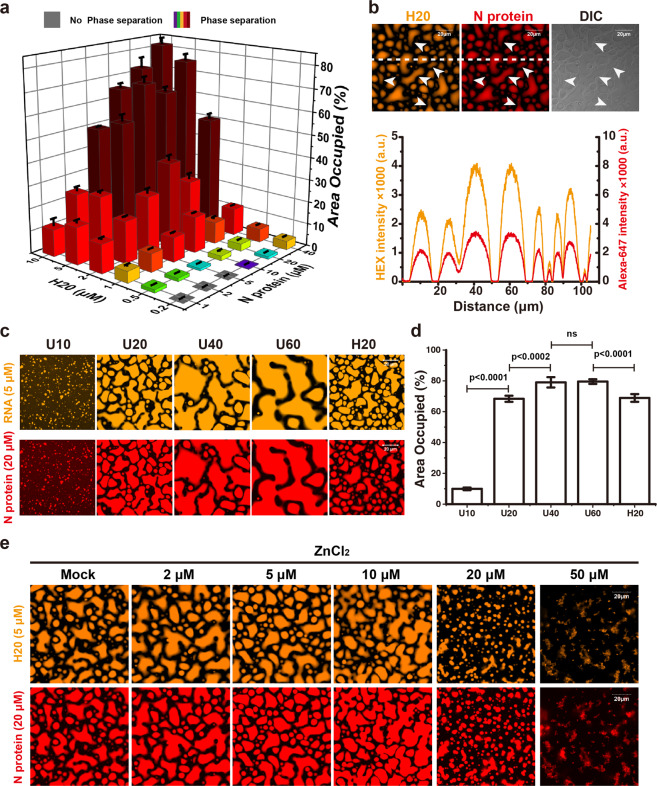
To test this hypothesis, we directly mixed the Alexa-488-labeled recombinant full-length N protein (488-N) with 20-nt single strand RNA (ssRNA; A20) (Supplementary information, Table S1). We observed formation of liquid-like droplets when N protein or RNA concentration was higher than certain thresholds (Supplementary information, Fig. S3a). The shapes of LLPS droplets were determined by the ratio of N protein and ssRNA concentrations. At fixed N protein concentration, typical liquid-like droplets were formed at higher RNA concentrations, whereas denser, solid-like sediments were observed at lower RNA concentrations. To further quantify the properties of N protein/RNA LLPS, we established the phase diagram of Alexa-647-labeled N protein (647-N) with another 20-nt ssRNA labeled with HEX (H20) (Fig. 1a; Supplementary information, Fig. S3b and Table S1). We found that H20 and 647-N co-localized well within the phase-separated droplets (Fig. 1b), further confirming that the observed phase-separated droplets were indeed formed by N protein and ssRNA. Moreover, the observed LLPS is the intrinsic biochemical property of N protein and ssRNA, as it was still present even with no or low percentage of crowding polymer PEG8000 (Supplementary information, Fig. S3c). Upon mixing, 647-N and H20 formed micrometer-sized droplets rapidly within minutes, and the small droplets further fused into larger ones (Supplementary information, Fig. S4a–c). Even after 0.5–1 h, we still could observe ongoing droplet fusion (Fig. 1b).
Fig. 1. N protein of SARS-CoV-2 undergoes LLPS with ssRNA in vitro.
a Phase diagram of a 20-nt RNA H20 separating with N protein of SARS-CoV-2. The histogram shows the percentage of area occupied by N protein/H20 droplets. b Upper panel, representative fluorescence and DIC images of LLPS droplets formed by H20 RNA (orange) and N protein (red) after 1-h mixing. White arrows indicate ongoing fusion events. Bottom panel, fluorescence intensity profiles of two fluorescence channels (H20 RNA, orange; N protein, red) along the white dashed line in the upper panels. c The length of ssRNA modulates the N protein/RNA LLPS. Representative fluorescence images of HEX-labeled ssRNA (orange, 5 μM) of different lengths forming phase-separated liquid droplets with Alexa-647-labeled N protein (red, 20 μM). d Comparison of the percentage of area occupied by droplets in each field of view in c. e Zn2+ promotes N protein/RNA LLPS. Representative fluorescence images of Alexa-647-labeled N protein (20 μM) mixed with H20 RNA (5 μM) to form phase-separated liquid droplets in the presence of the indicated concentrations of Zn2+. Scale bars, 20 μm. Error bars refer to SD of five independent experiments.
When 488-N was injected into the phase-separated solution formed by 647-N and H20, we found that the fluorescence intensity of Alexa-488 in droplets increased gradually along with the decrease of Alexa-647 fluorescence intensity (Supplementary information, Fig. S4d). When H20 was injected into the phase-separated solution that had already been formed by 488-N and A20, HEX fluorescence also appeared gradually inside the original droplets. Both the intensity of HEX fluorescence in each droplet and the number of droplets with HEX fluorescence increased over time (Supplementary information, Fig. S4e). All these results suggest that both N protein and ssRNA are exchangeable between the dense and solute phases.
The sequences of H20 and A20 are totally different, and both can phase separate with N protein and exhibit similar phase diagram (Fig. 1a; Supplementary information, Fig. S3a, b), thus the ability of N protein to undergo LLPS with short RNA is independent of short RNA sequence. This is consistent with the previously reported finding that the binding of SARS-CoV’s N protein with RNA was not dependent on RNA sequence4 and also agrees with the fact that N protein could bind various sites on genome RNAs.5 It should be noted that these results do not rule out the possibility that N protein may undergo stronger LLPS with longer RNAs containing specific sequences or secondary structures.
When mixing N protein with 5 μM HEX-labeled poly-U RNA oligos of lengths ranging from 10 nt to 60 nt (U10, U20, U40 and U60) to induce LLPS in vitro, we found that N protein could form larger droplets with longer ssRNAs (Fig. 1c), indicating the enhancing effect of RNA length on LLPS. The percentage of area occupied by the dense phase relative to the solute phase6 increased rapidly from ~10% for 10-nt RNA (U10) to ~70% for 20-nt RNA (U20) and reached ~80% for 40-nt and 60-nt RNAs (U40 and U60) (Fig. 1d). When inducing N protein/RNA LLPS by using these poly-U oligos with the same mass and the same concentrations of N protein, we found that the longest ssRNA, U60, could phase separate with N protein to form much larger droplets compared with other shorter oligoes (Supplementary information, Fig. S5a), confirming that the LLPS is indeed dependent on ssRNA length.
The genome RNA of SARS-CoV-2 is about 30 kb, much longer than RNA oligoes that we used above. We next tested long RNA in the LLPS. Two longer RNAs, 85-nt (A85) RNA and 1541-nt (N1541, the positive strand RNA of N protein), were tested. Both A85 and N1541 RNAs could form droplets with N protein, and the droplets turned to be solid-like structure instead of droplets as the length of RNA increased (Supplementary information, Fig. S5b, c). We thereby hypothesized that the viral packaging of SARS-CoV-2 might be driven by N protein/RNA LLPS. However, the exact role of N protein/RNA LLPS in viral packaging remains to be further validated.
To test whether divalent cation could modulate N protein/RNA LLPS, we performed LLPS experiments in the presence of various cations. We found that Zn2+ had a significant effect on enhancing the LLPS (Fig. 1e; Supplementary information, Fig. S6), while Mg2+, Mn2+ and Ca2+ did not (Supplementary information, Fig. S6). Moreover, Ni2+ and Cu2+ could also modulate the LLPS at a relative lower concentration (10 μM) compared with Zn2+ (20 μM) (Supplementary information, Fig. S6).
Zn2+ physiologically regulates cell functions,7 and its regulation on LLPS has been reported previously.8,9 We further investigated how the concentration of Zn2+ affected N protein/RNA LLPS. We found that 20 μM or higher concentration of Zn2+ could remarkably enhance the LLPS and turn the LLPS droplets to solid-like condensates, whereas lower concentrations of Zn2+ (< 10 μM) could not (Fig. 1e). When 0.5 mM Zn2+ was added into freshly made N protein/RNA droplets, liquid-like droplets turned to be denser, solid-like structures, which could recover to liquid-like droplets after addition of 50 mM EDTA (Supplementary information, Fig. S7a, b). In addition, negative staining EM (Supplementary information, Fig. S7c) or cryo-EM (Supplementary information, Fig. S7d) imaging of N protein/RNA LLPS in the presence and absence of Zn2+ revealed similar loose filament-like structures as those observed in the RNP particles of another β-coronavirus MHV,10 supporting the potential role of N protein/RNA LLPS in viral assembly.
N protein alone formed small condensates in the presence of 20 μM Zn2+ and became more aggregative when Zn2+ concentration was increased (Supplementary information, Fig. S7e). In contrast, H20 alone did not show significant change upon supplementation with 20 μM Zn2+ (Supplementary information, Fig. S7d). The result indicates that Zn2+ promotes N protein/RNA LLPS via inducing N protein oligomerization.
Because the free cytoplasmic Zn2+ concentration is very low, the physiological relevance of Zn2+-promoted N protein/RNA LLPS is still not clear. Nevertheless, A recent study suggests that Chloroquine is a zinc ionophore.11 Chloroquine has been shown to effectively inhibit the infection of SARS-CoV-2,12 but the mechanism is still not known. We speculate that Chloroquine may inhibit N protein/RNA LLPS through chelating the free Zn2+ in cytosol.
To identify the essential regions of N protein to facilitate LLPS with RNAs, we designed a panel of N protein mutants (Supplementary information, Fig. S8a). Interestingly, the deletion of CTD (dCTD) dramatically attenuated the droplet formation, whereas the deletions of NTD (dNTD), a basic amino acid-rich sequence (dBRS) or both (ddRBD) only mildly attenuated N protein/RNA LLPS (Supplementary information, Fig. S8b). These results are in agreement with previous findings that N protein harbors extra RNA-binding sites other than NTD (NTD, CTD and intrinsic disorder regions are all involved in RNA binding), and that the oligomerization of N protein mediated mainly by the CTD plays an essential role in N protein/RNA LLPS. We further tested the ability of N protein single domain only (NTD or CTD) to undergo phase separation with H20. We found that NTD alone could not induce LLPS, whereas CTD alone had weak ability to induce phase separation with H20 (Supplementary information, Fig. S8b). This result indicates that CTD could also bind RNA without affecting the oligomerization states.
Interestingly, droplet formation could be enhanced by the addition of 20 μM Zn2+ for all these N protein mutants (Supplementary information, Fig. S8c), even the NTD domain only mutant, which failed to form phase-separated droplets with H20 in the absence of Zn2+ (Supplementary information, Fig. S8b). This suggests that Zn2+ promotes new interactions between NTDs and RNA, or it triggers N protein dimerization or multimerization via NTD. Zn2+ also significantly modulated the phase separation of CTD/H20 to form smaller droplets (Supplementary information, Fig. S8c), suggesting that the CTD also contributes to Zn2+-enhanced N protein/RNA LLPS.
In summary, we revealed that N protein and RNA underwent LLPS. The LLPS is dependent on the length and concentration of ssRNA. N protein forms typical sphere-like droplets with short ssRNAs, but solid-like structures with long ssRNAs. We further identified that the LLPS could be enhanced by Zn2+. Our findings suggest that N protein/RNA LLPS may be essential for SARS-CoV-2 viral assembly, which may shed light on developing intervention strategies to prevent COVID-19 pandemic by disrupting the LLPS and viral assembly.
Supplementary information
Acknowledgements
The authors would like to thank Prof. Tao Xu and Prof. Hong Zhang for helpful discussions and Prof. Fei Sun, Drs. Gang Ji, Xiaojun Huang and Qingshan Luo for helping with EM data acquisition. The EM experiments were performed at Center for Biological Imaging (CBI; http://cbi.ibp.ac.cn), Institute of Biophysics, Chinese Academy of Sciences. This work is supported by grants from the Ministry of Science and Technology of China (2019YFA0707001 to J.L., 2017ZX10203205 to W.C.), the National Natural Science Foundation of China (11672317 to J.L., 31971237 to W.C.), Zhejiang University special scientific research fund for COVID-19 prevention and control (2020XGZX077 to W.C.), and the COVID-19 emergency tackling research project of Shandong University (2020XGB03 to P.-H.W.).
Author contributions
H.C., W.C., and J.L. conceived and designed the experiments. H.C. performed the experiments and analyzed the data; Y.C., X.H., W.H., M.S., Y.Z., P.-H.W., and G.S. participated in experiments; H.C., W.C. and J.L. wrote the manuscript.
Competing interests
The authors declare no competing interests.
Contributor Information
Wei Chen, Email: jackweichen@zju.edu.cn.
Jizhong Lou, Email: jlou@ibp.ac.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41422-020-00408-2.
References
- 1.Wu F, et al. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Masters PS. Virology. 2019;537:198–207. doi: 10.1016/j.virol.2019.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanders DW, et al. Cell. 2020;181:306–324. doi: 10.1016/j.cell.2020.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsieh PK, et al. J. Virol. 2005;79:13848–13855. doi: 10.1128/JVI.79.22.13848-13855.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang CK, et al. J. Virol. 2009;83:2255–2264. doi: 10.1128/JVI.02001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L, et al. Cell Res. 2020;30:393–407. doi: 10.1038/s41422-020-0288-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maret W. Int. J. Mol. Sci. 2017;18:2285. doi: 10.3390/ijms18112285. [DOI] [Google Scholar]
- 8.Du M, Chen ZJ. Science. 2018;361:704–709. doi: 10.1126/science.aat1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rayman JB, Karl KA, Kandel ER. Cell Rep. 2018;22:59–71. doi: 10.1016/j.celrep.2017.12.036. [DOI] [PubMed] [Google Scholar]
- 10.Gui M, et al. Protein Cell. 2017;8:219–224. doi: 10.1007/s13238-016-0352-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xue J, et al. PLoS One. 2014;9:e109180. doi: 10.1371/journal.pone.0109180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang M, et al. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.