Abstract
The FilmArray® Pneumonia plus Panel (FAPP) is a new multiplex molecular test for hospital-acquired pneumonia (HAP), which can rapidly detect 18 bacteria, 9 viruses, and 7 resistance genes. We aimed to compare the diagnosis performance of FAPP with conventional testing in 100 intensive care unit (ICU) patients who required mechanical ventilation, with clinically suspected HAP. A total of 237 samples [76 bronchoalveolar lavages (BALDS) and 82 endotracheal aspirates (ETADS) obtained at HAP diagnosis, and 79 ETA obtained during follow-up (ETATT)], were analyzed independently by routine microbiology testing and FAPP. 58 patients had paired BALDS and ETADS. The positivity thresholds of semi-quantified bacteria were 103–104 CFUs/mL or 104 copies/mL for BAL, and 105 CFUs/mL or copies/mL for ETA. Respiratory commensals (H. influenzae, S. aureus, E. coli, S. pneumoniae) were the most common pathogens. Discordant results for bacterial identification were observed in 33/76 (43.4%) BALDS and 36/82 (43.9%) ETADS, and in most cases, FAPP identified one supplemental bacteria (23/33 BALDS and 21/36 ETADS). An absence of growth, or polybacterial cultures, explained almost equally the majority of the non-detections in culture. No linear relationship was observed between bin and CFUs/mL variables. Concordant results between paired BALDS and ETADS were obtained in 46/58 (79.3%) patients with FAPP. One of the 17 resistance genes detected with FAPP (mecA/C and MREJ) was not confirmed by conventional testing. Overall, FAPP enhanced the positivity rate of diagnostic testing, with increased recognition of coinfections. Implementing this strategy may allow clinicians to make more timely and informed decisions.
Keywords: multiplex syndromic testing, hospital-acquired pneumonia, rapid diagnosis, coinfection, antibiotic resistance
Introduction
Hospital-acquired pneumonia (HAP) is the most frequent cause of nosocomial infection in intensive care unit (ICU) patients, with dramatic effects on patients’ outcomes. International experts have developed guidelines to prevent and improve the management of HAP (Kalil et al., 2016; Torres et al., 2017). Among strategies proposed, optimization of empiric antimicrobial therapy is of major importance. This entails administrating early appropriate antimicrobial therapy, while limiting overuse of broad-spectrum antibiotics. Hence, European guidelines suggest using narrow-spectrum empiric therapy (amoxicillin-clavulanate, cefotaxime, ceftriaxone, and fluoroquinolones) in patients without risk factors for multidrug-resistant (MDR) pathogens in case of early-onset HAP (first 4 days of hospitalization). However, making such choice is not so obvious in ICU patients, and adherence to guidelines is associated with a high rate of unnecessary broad-spectrum antibiotics (Roquilly et al., 2016; Ekren et al., 2018).
Microbiological confirmation of HAP is a crucial step for tailoring antibiotic therapy. Nevertheless, current culture methods take 48–72 h to obtain antimicrobial susceptibility results. Moreover, traditional techniques fail to recover pathogens in up to 30% of clinically-diagnosed HAP (Roquilly et al., 2019). Recently, syndromic multiplex molecular tests have emerged as powerful tools for rapid diagnostics (meningitis/encephalitis, gastroenteritis, bacteraemia, pneumonia) (Couturier and Bard, 2019; Poole and Clark, 2020). Initially based on qualitative DNA detection, those approaches were not suitable for diagnosing pneumonia caused by common colonizers of the upper airways (e.g., Streptococcus pneumoniae, Haemophilus influenzae). The FilmArray® Pneumonia plus Panel (FAPP) is a new panel for HAP, which offers potential advantage to detect and quantify in a single test, 27 respiratory pathogens (18 bacteria, 9 viruses) and 7 antibiotic resistance genes.
The aim of this study was to assess the performances of this new molecular test on bronchoscopy specimens [bronchoalveolar lavages (BAL) and/or endotracheal aspirates (ETA)] from 100 ICU patients with HAP requiring mechanical ventilation.
Materials and Methods
Ethic and Study Design
The study protocol was approved by our local Ethical Committee (GNEDS, Nantes, France). Patients and relatives were informed of the trial. Consent was waived according to French law.
Population and Specimen Collection
The study was conducted at the Nantes University Hospital (France), in 3 ICUs located on two sites spaced 10 km apart. We recruited 100 critically ill adult patients receiving mechanical ventilation with clinically suspected HAP, between October 2018 and January 2020 (Table 1). Pneumonia was suspected based on European guidelines, if there were the following criteria: a new or persistent radiological pulmonary infiltrate without another obvious cause combined with two clinical signs among fever, purulent endotracheal secretions, hyperleukocytosis or leukopenia, and increasing oxygen requirements (Torres et al., 2017). Patients underwent a bronchoscopy with BAL and/or ETA at the time of suspicion of HAP (BALDS (for DIAGNOSIS) and ETADS, respectively). In addition, if an ETA was collected 2–3 days later, as part of a routine clinical care, the specimen [ETATT(for TREATMENT)] was also sent to the laboratory for microbiological analysis. A total of 237 respiratory specimens were analyzed (76 BALDS, 82 ETADS, and 79 ETATT). Both BALDS and ETADS were collected in 58 patients.
TABLE 1.
Characteristics of patients at onset of pneumonia.
| Patient characteristics | n = 100 |
| Age (years), Median (range) | 57 (19–85) |
| Male sex, n (%) | 81 (81%) |
| Median length of hospital stay before pneumonia, days | 6 |
| Median length of ICU stay before pneumonia, days | 5 |
| Early-onset pneumonia, n (%) | 33 (33%) |
| Late-onset pneumonia, n (%) | 67 (67%) |
| Ventilator-associated pneumonia, n (%) | 87 (87%) |
| Most pejorative PaO2/FiO2 at day 1, Median (range) | 135 (56–309) |
| Risk factors for MRSAa, n (%) | 6 (6%) |
| Previous isolation of ESBL-Enterobacteria, n (%) | 2 (2%) |
| Antibiotics use during the previous 90 days, n (%) | 40 (40%) |
| Antibiotics use before sampling at the time of HAP diagnosis | 25 (25%) |
aRecent colonization by MRSA, chronic skin lesions, chronic renal replacement therapy.
Microbiological Testing
The respiratory specimens were analyzed in parallel by routine microbiology testing and FAPP, as soon as they arrived at the microbiology laboratory. The turnaround times from samples to validated results were recorded. Results of routine microbiology testing were analyzed independently of FAPP.
Routine Microbiology Testing
Gram staining and bacteriological cultures were performed for all respiratory specimens according to the French REMIC recommendations (Société Française de Microbiologie [SFM], 2018). Briefly, 10 μL of the samples were seeded directly (for BAL), or after dilution (1:100 after fluidification for ETA), onto Columbia horse blood (Oxoid), Chocolate agar (BD), and chromogenic UriSelect4 agar (Biorad) plates, and if necessary (cases of chronic obstructive pulmonary disease) on Chapman (bioMérieux) and Cetrimide (Biorad) selective agar plates, then incubated at 37°C in 5% CO2 for 24–48 h, as necessary. Plates were examined daily for bacterial growth. Streptococcus pneumoniae, Haemophilus influenzae, Staphylococcus aureus, Enterobacteriales, Pseudomonas aeruginosa, Stenotrophomonas maltophilia, and any other largely predominant pathogen were searched on the plates. In accordance with current guidelines, the positivity thresholds were 105 CFUs/mL for ETA and 104 CFUs/mL for BAL, but in BAL, potential pathogens that were present in pure culture at 103 CFUs/mL and associated with many leukocytes at Gram staining were reported as positive. Culture results were considered as negative if there was no significant growth or a normal non-pathogenic flora. Bacterial isolates were identified by mass spectrometry (BioMérieux). Antimicrobial susceptibility testing (AST) was performed according to the CA-SFM/EUCAST guidelines (Société Française de Microbiologie [SFM], 2019), using Vitek2 AST cards. Based on phenotypic susceptibility results, additional tests were performed if required, for ESBLs (Mastdiscs™ D68C), carbapenemases (CORIS BioConcept RESIST-3 O.K.N. immuno-chromatographic test and/or in-house real-time PCRs for the blaKPC, blaOXA–48, blaVIM, blaIMP, and blaNDM genes), and methicillin-resistance detection (Alere™ PBP2A culture colony test and BDMAX™ StaphSR). Furthermore, when requested by the Clinicians, the presence in respiratory samples of Mycoplasma pneumoniae and/or respiratory viruses was investigated by real-time PCR (Fast Track Diagnostics® Respiratory Pathogens 21 qPCR assay for viruses and/or in-house real-time PCR for M. pneumoniae).
FilmArray Pneumonia Plus Panel Assay
The BioFire® FilmArray® Pneumonia plus Panel (bioMérieux) was performed according to the manufacturer’s instructions, with a handling time of ∼5 min. Briefly, the respiratory sample collected with a flocked swab (∼ 200 μL) and then mixed with a sample buffer, was injected along with an hydration solution in the reagent pouch “Pneumonia plus Panel,” which was then inserted into the FilmArray® instrument. The test consisted of automated nucleic acid extraction, purification, amplification, detection, and analysis with each target reported as “detected” or “not detected.” A semi-quantitative measurement reported into bins (i.e., 104, 105, 106, and ≥ 107 bacterial DNA copies/mL) was provided for 15 bacteria, if detected. The panel included 15 bacteria, 3 atypical bacteria, 9 viruses, and 7 antimicrobial resistance genes (Table 2). Each resistance marker was reported only if the potential microorganism harboring the gene was concomitantly detected in the sample. Clinicians were left blinded to the FAPP results.
TABLE 2.
FilmArray® pneumonia plus panel targets.
| Variables |
| 15 Bacteria reported into bins (104, 105, 106, and ≥ 107 DNA copies/mL) |
| Acinetobacter calcoaceticus baumannii complex |
| Enterobacter cloacae complex |
| Escherichia coli |
| Haemophilus influenza |
| Klebsiella aerogenes |
| Klebsiella oxytoca |
| Klebsiella pneumoniae group |
| Moraxella catarrhalis |
| Proteus spp. |
| Pseudomonas aeruginosa |
| Serratia marcescens |
| Staphylococcus aureus |
| Streptococcus agalactiae |
| Streptococcus pneumoniae |
| Streptococcus pyogenes |
| 3 Atypical bacteria |
| Chlamydophilia pneumoniae |
| Legionella pneumophila |
| Mycoplasma pneumoniae |
| 9 Viruses |
| Adenovirus |
| Coronavirus (229E, OC43, HKU1, NL63) |
| human Metapneumovirus |
| Influenza A |
| Influenza B |
| MERS CoV |
| Parainfluenza viruses |
| Rhinovirus/Enterovirus |
| RSV |
| 7 Antimicrobial resistance genes |
| MRSA genes (mecA/C and MREJ) |
| Carbapenemases (blaKPC, blaNDM, blaOXA–48–like, blaVIM, blaIMP) |
| ESBL (blaCTX–M) |
Data Analysis
BAL were considered as positive with FAPP when at least one microbial target was detected (at ≥104 copies/mL for semi-quantified bacteria). For ETA, in order to match the culture threshold that differentiate commensalism from pathogenicity (≥105 CFUs/mL), we set up a bin threshold of ≥105 copies/mL to consider the 15 semi-quantitative bacterial targets as positive. The agreement between FAPP and culture was measured for each bacterial pathogen in the form of negative percent agreement (NPA), positive percent agreement (PPA) and overall percent agreement (OPA), and their two-sided 95 percent confidence intervals. In order to explain discrepant results, cultures were reread after routine final reports in light of results obtained with FAPP. Concordance was calculated based on the original culture reading.
Results
Summary of FAPP Findings
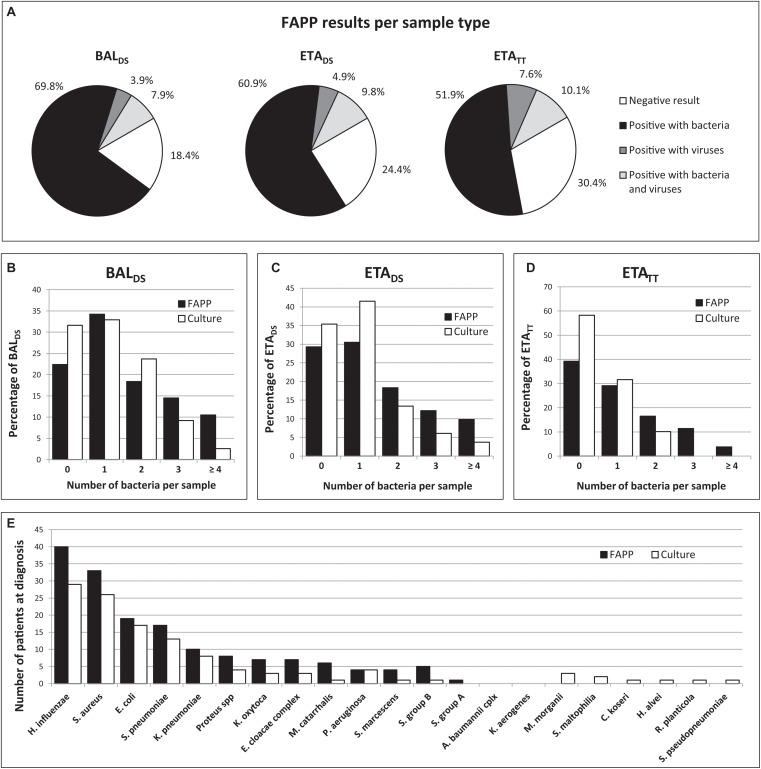
At the time of HAP diagnosis, FAPP yielded positive results with significant levels (i.e., ≥ 104 bin in BAL and ≥ 105 bin in ETA for semi-quantified bacteria) in 82/100 patients. Thus, as shown in Figure 1A, 81.6% (62/76) BALDS, and 75.6% (62/82) ETADS were positive for at least one target. Of these, more than half were positive for at least two pathogens (36/62 (58.1%) for BALDS, and 36/62 (58.1%) for ETADS), leading to the diagnosis of coinfection in 49/100 patients (Figure 1). Multiple detections per positive specimen were not higher in ETADS than in BALDS, since bacteria with bin results of 104 were considered as negative in ETA (it represented 23 bacteria in 21 ETADS). Of note, if the 104 cutoff had been used for ETA, 84.1% (69/82) ETADS would have been positive, and multiple targets would have been detected in 60.9% (42/69) of these specimens (Figure 2). A maximum of 7 pathogens (6 bacteria and one human rhinovirus/enterovirus) was detected in one patient (BALDS and ETADS). The most common pathogens detected at diagnosis were H. influenzae, S. aureus, E. coli, S. pneumoniae, and K. pneumoniae, which were found in 40 (40%), 33 (33%), 19 (19%), 17 (17%), and 10 (10%) patients, respectively (Figure 1). The panel identified 6 viruses at diagnosis [human rhinovirus/enterovirus (5 patients), coronavirus (4 patients), influenza A (3 patients), adenovirus (2 patients), parainfluenza viruses (2 patients), and RSV (1 patient)] in 16/100 patients (11.8% (9/76) BALDS, and 14.6% (12/82) ETADS). In most cases, it corresponded to viral-bacterial co-infections (12 patients, including one with multiple viruses (adenovirus and influenza A) and S. pneumoniae) (Supplementary Table S1). An atypical bacteria (M. pneumoniae) was detected with other bacteria in one patient. The positivity rate of ETATT obtained during follow-up was 69.6% (55/79), and 38 bacteria were below the 105 cutoff in 29 ETATT (Figures 1A, 2). Four types of resistance genes were detected in 8 patients: mecA/C and MREJ (one patient), and the CTX-M ESBL (7 patients), either alone (5 patients) or combined with a carbapenemase (blaNDM in one patient, and blaOXA–48–like in one another). The median turnaround time (from sample collection to results) was 4 h 15 min (BALDS or ETADS).
FIGURE 1.
Summary of FAPP and culture results. FAPP results distribution per pathogen category and sample type (A). Number of bacteria per sample with FAPP compared to culture in BALDS (B), ETADS (C), and ETATT (D). FAPP results compared to culture results for bacterial detection (E).
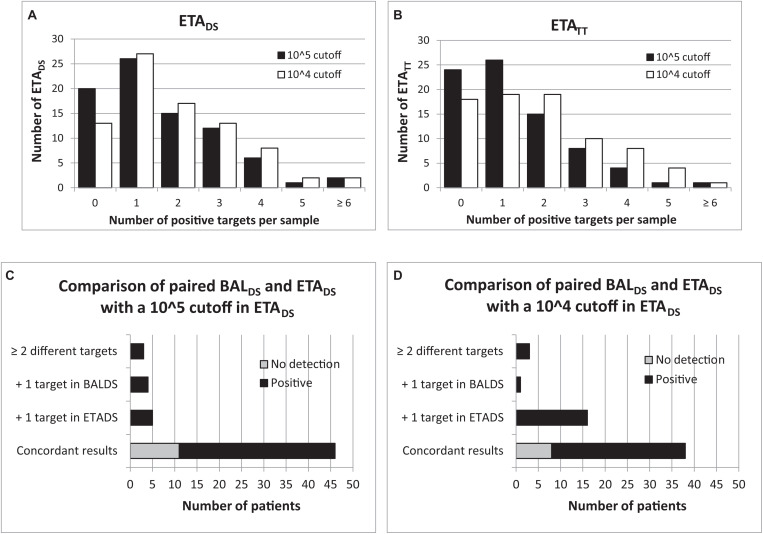
FIGURE 2.
Comparison of 104 and 105 cutoffs with FAPP for ETA. Number of positive detections per sample for each threshold, in ETA collected at diagnosis (A) and 2–3 days later (B). Comparison of paired BALDS and ETADS for each threshold (C,D).
Summary of Routine Microbiology Testing
At HAP diagnosis, culture identified one or more bacteria in 73/100 patients (52/76 (68.4%) BALDS and 53/82 (64.6%) ETADS), and respiratory viruses were detected in 8/35 (22.9%) patients who benefited from a Fast Track multiplex PCR routinely ordered by clinicians, yielding an overall positive detection in 78/100 patients. Two or more bacterial pathogens were identified and reported in 32/100 patients, in a higher proportion of BALDS (27/52, 51.9%) than ETADS (19/53, 35.8%), certainly because BAL are more distal than ETA and are normally not contaminated. Indeed, this property might have encouraged microbiologists to identify and report any bacteria found in these distal specimens rather than concluding to “polymicrobial flora.” Thus, only 25.0% (6/24) of culture-negative BALDS had results reported as “mixed bacterial flora” vs. 41.4% (12/29) of culture-negative ETADS (Supplementary Table S1). The most frequent bacteria detected by culture were H. influenzae, S. aureus, E. coli, S. pneumoniae, and K. pneumoniae in 29 (29%), 26 (26%), 17 (17%), 13 (13%), and 8 (8%) patients, respectively (Figure 1). Culture showed a lower positivity rate of 41.8% (33/79) for ETATT collected during follow-up, with a high proportion of culture-negative results reported as “no growth” (35/46, 76.1%) (Supplementary Table S1). Regarding AST, Enterobacteriaceae resistant to third-generation cephalosporins were found on average 2 days after specimens collection, in 11/100 patients. In 7 cases, it was ESBL-producing strains (K. pneumoniae or E. coli), while in the 4 others cases, high-level cephalosporinases were confirmed with additional tests (Mastdiscs™ D68C), in strains of E. cloacae complex (2 patients), S. marcescens (1 patient), and E. coli (1 patient). Two ESBL-producing K. pneumoniae that were resistant to ertapenem ± imipenem, were also confirmed to be carbapenemase (NDM or OXA-48 like) producers, by means of an immuno-chromatographic test (CORIS BioConcept RESIST-3 O.K.N.) performed 2 days after specimen collection. All strains of P. aeruginosa detected in 4 patients were susceptible to ceftazidime. The mean turnaround time from sample collection to results validation was 70 h for BALDS, and 64 h for ETADS.
Comparison of FAPP and Routine Microbiology Testing
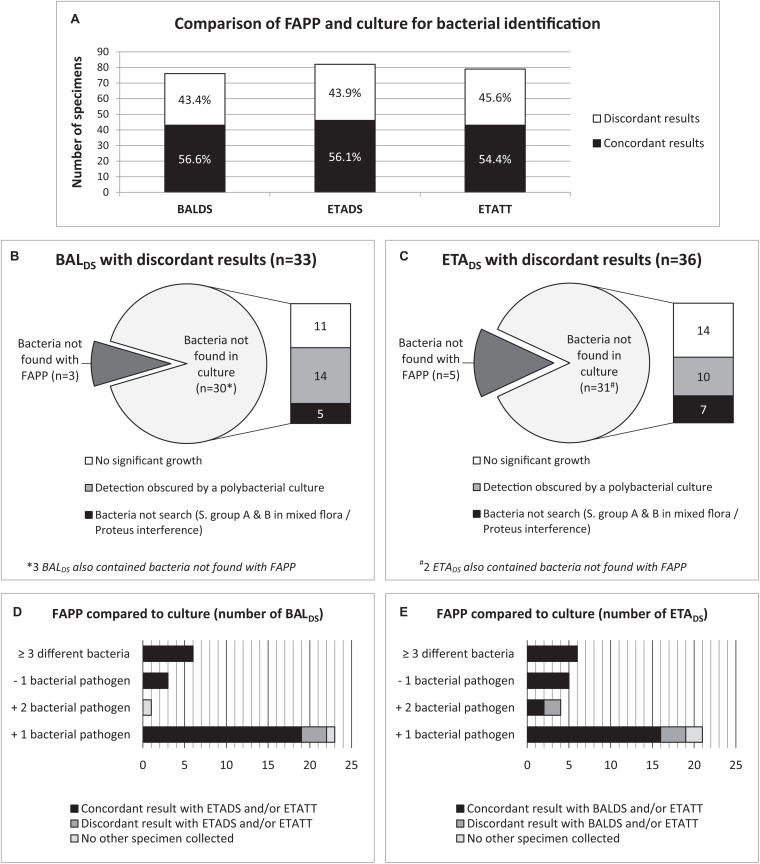
In total, at HAP diagnosis, just over half of the specimens were concordant for the bacterial identification (43/76 (56.6%) BALDS and 46/82 (56.1%) ETADS) (Figure 3 and Table 3). In most of the discordant specimens (23/33 (69.7%) BALDS and 21/36 (58.3%) ETADS), FAPP identified one supplemental bacterial pathogen, which was most often confirmed by FAPP in the paired respiratory sample and/or in the ETATT collected 2–3 days later (Figure 3). By rereading the plates in light of FAPP results after final report, we showed that an absence of significant growth, or polybacterial cultures impeding the accurate visualization of non-predominant pathogens, explained almost equally the majority of the non-detections in culture (Figure 3). In the rest of the cases, the corresponding bacteria had not been searched on the plates (S. pyogenes or S. agalactiae in mixed flora, or because of an impossibility due to Proteus invasion) (Figure 3). Furthermore, in 8 patients [5/76 (6.6%) BALDS and 6/82 (7.3%) ETADS], culture yielded bacteria that were not targeted by FAPP (Citrobacter koseri, Hafnia alvei, Morganella morganii, Raoultella planticola, Stenotrophomonas maltophilia, and Streptococcus pseudopneumoniae), and two FAPP false-negative results were observed: K. oxytoca (one BALDS with a pure culture at 103 CFUs/mL), and H. influenzae (one polymicrobial ETADS with H. influenzae at > 105 CFUs/mL) (Figure 1E and Table 3). The atypical bacteria M. pneumoniae found in one patient with FAPP, had not been searched with conventional methods at the time of HAP diagnosis, but was subsequently confirmed with an in-house real-time PCR. The performance data for each FAPP bacterial target are provided in Table 3. The overall percent agreement (OPA) between FAPP and culture results ranged from 88 to 100% in BALDS, and 82 to 100% in ETADS. Only three bacterial species of the panel had an OPA bellow 95%: H. influenzae and S. aureus in BALDS and ETADS, and K. oxytoca in BALDS.
FIGURE 3.
Analysis of discordant results between FAPP and culture at diagnosis, by rereading cultures in light of FAPP results, and by investigating the concordance with FAPP results from the paired ETADS/BALDS and/or ETATT if collected. Concordance and discordance between FAPP and culture identifications (A). Causes of discordant results in BALDS (B) and ETADS (C). Discrepancy investigations in BALDS (D) and ETADS (E).
TABLE 3.
Comparison of FAPP and culture for each FAPP bacterial target, at the time of HAP diagnosis.
| FAPP Bacterial targets | BALDS (number of results for FAPP/culture) | ETADS(number of results for FAPP/culture) | ||||||||||||
| (+ / +) | (+ /−) | (−/ +) | (−/−) | PPA (95% CI) | NPA (95% CI) | OPA (95% CI) | (+ / +) | (+ /−) | (−/ +) | (−/−) | PPA (95% CI) | NPA (95% CI) | OPA (95% CI) | |
| A. baumannii | 0 | 0 | 0 | 76 | NA | 100 [100; 100] | 100 [100; 100] | 0 | 0 | 0 | 82 | NA | 100 [100; 100] | 100 [100; 100] |
| E. cloacae | 2 | 3 | 0 | 71 | 100 [100; 100] | 96 [92; 100] | 96 [92; 100] | 2 | 3 | 0 | 77 | 100 [100; 100] | 96 [92; 100] | 96 [92; 100] |
| E. coli | 15 | 2 | 0 | 59 | 100 [100; 100] | 97 [94; 100] | 97 [94; 100] | 9 | 0 | 0 | 73 | 100 [100; 100] | 100 [100; 100] | 100 [100; 100] |
| H. influenzae | 23 | 9 | 0 | 44 | 100 [100; 100] | 83 [73; 93] | 88 [81; 95] | 20 | 14 | 1 | 47 | 95 [86; 100] | 77 [66; 88] | 82 [73; 90] |
| K. aerogenes | 0 | 0 | 0 | 76 | NA | 100 [100; 100] | 100 [100; 100] | 0 | 0 | 0 | 82 | NA | 100 [100; 100] | 100 [100; 100] |
| K. oxytoca | 2 | 4 | 1 | 69 | 67 [13; 100] | 95 [89;100] | 93 [88; 99] | 2 | 4 | 0 | 76 | 100 [100; 100] | 95 [90; 100] | 95 [90; 100] |
| K. pneumoniae | 7 | 2 | 0 | 67 | 100 [100; 100] | 97 [94; 100] | 97 [94; 100] | 6 | 1 | 0 | 75 | 100 [100; 100] | 99 [96; 100] | 99 [96; 100] |
| M. catarrhalis | 0 | 2 | 0 | 74 | NA | 97 [94; 100] | 97 [94; 100] | 1 | 4 | 0 | 77 | 100 [100; 100] | 95 [90; 100] | 95 [90; 100] |
| Proteus spp. | 1 | 4 | 0 | 71 | 100 [100; 100] | 95 [90; 100] | 95 [90; 100] | 4 | 4 | 0 | 74 | 100 [100; 100] | 95 [90; 100] | 95 [90; 100] |
| P. aeruginosa | 3 | 0 | 0 | 73 | 100 [100; 100] | 100 [100; 100] | 100 [100; 100] | 3 | 0 | 0 | 79 | 100 [100; 100] | 100 [100; 100] | 100 [100; 100] |
| S. marcescens | 1 | 1 | 0 | 74 | 100 [100; 100] | 99 [96; 100] | 99 [96; 100] | 0 | 3 | 0 | 79 | NA | 96 [92; 100] | 96 [92; 100] |
| S. aureus | 17 | 6 | 0 | 53 | 100 [100; 100] | 90 [82; 98] | 92 [86; 98] | 19 | 5 | 0 | 58 | 100 [100; 100] | 92 [85; 99] | 94 [89; 99] |
| S. agalactiae | 1 | 3 | 0 | 72 | 100 [100; 100] | 96 [92; 100] | 96 [92; 100] | 0 | 4 | 0 | 78 | NA | 95 [90; 100] | 95 [90; 100] |
| S. pneumoniae | 11 | 4 | 0 | 61 | 100 [100; 100] | 94 [88; 100] | 95 [90; 100] | 9 | 4 | 0 | 69 | 100 [100; 100] | 95 [90; 100] | 95 [90; 100] |
| S. pyogenes | 0 | 1 | 0 | 75 | NA | 99 [96; 100] | 99 [96; 100] | 0 | 0 | 0 | 82 | NA | 100 [100; 100] | 100 [100; 100] |
| Total | 83 | 41 | 1 | 1015 | 99 [96; 100] | 96 [95; 97] | 96 [95; 97] | 75 | 46 | 1 | 1108 | 99 [96; 100] | 96 [95; 97] | 96 [95; 97] |
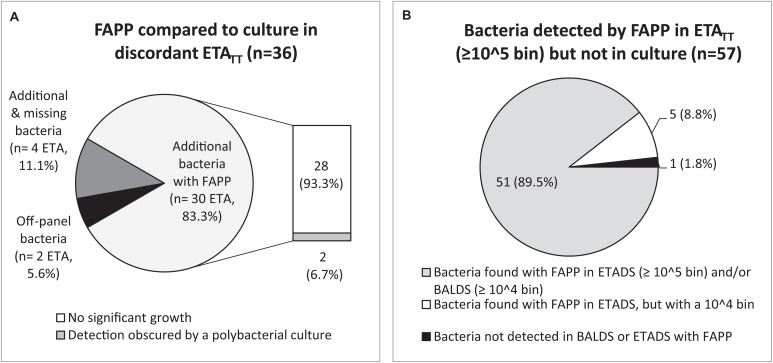
Regarding the 79 ETATT obtained under treatment, 45.6% (36/79) had discordant results between both methods (Figures 3A, 4 and Supplementary Table S1). Not surprisingly, most discrepancies (28/36, 77.8%) were explained by no growth of bacteria identified with FAPP (Figure 4A). The vast majority of the 57 FAPP-positive bacterial targets that were not reported by routine culture, had already been detected by FAPP at diagnosis, either above positive threshold values (51/57, 89.5%), or not (bin result of 104 in ETADS) in a few cases (5/57, 8.8%) (Figure 4B).
FIGURE 4.
Analysis of discordant results between FAPP and culture in ETATT. Causes of discordant results in ETATT (A). Discrepancy investigations for each additional bacteria detected by FAPP (B).
Regarding FAPP semi-quantitative results, most bacteria with bin results of 104 in ETA (i.e., below our positivity threshold) were not reported in culture (23/23 (100%) in ETADS, and 36/38 (94.7%) in ETATT). On the other hand, for patients with ETA at diagnosis and 2–3 days later, 38.9% (7/18) of the detections with a bin value of 104 in ETADS were positive again in ETATT with a higher bin value (≥ 105 copies/mL). No linear relationship was observed between the bin and CFUs/mL variables (Supplementary Table S1). However, semi-quantitative culture results were not stratified into log10 ranges above positive thresholds (105 CFUs/mL for ETA and 104 CFUs/mL for BAL).
Eighteen resistance markers were detected with FAPP in 15 samples (2 mecA/C and MREJ, 13 blaCTX–M, 2 blaNDM, and 1 blaOXA–48–like) (Supplementary Table S1). All ESBL and carbapenemases were confirmed by standard laboratory protocols (AST and additional tests performed in routine). Among both methicillin-resistant S. aureus (MRSA) detected with FAPP, one found at 104 bin in ETATT did not grow in culture. The other corresponded to a false-positive mecA/C and MREJ result since a methicillin-susceptible S. aureus (MSSA) was found in culture. This result was repeatable after retesting with FAPP, but none of the comparator methods (BDMAX™ StaphSR performed on the same BALDS, or Alere™ PBP2A testing and cefoxitin susceptibility testing performed on several colonies) found a MRSA. No additional cases of methicillin-resistance, ESBL, or carbapenemase production were found with routine microbiology testing.
Lastly, based on FAPP results, an initial antibiotic therapy by amoxicillin-clavulanate could have been proposed in 83/100 patients, whose results ruled out pathogens with chromosomally-encoded cephalosporinase (i.e., P. aeruginosa, A. baumannii, E. cloacae complex, K. aerogenes, and S. marcescens) and/or resistance markers of the panel. However, this antibiotic would have not been optimal in 7/83 (8.4%) patients. In fact, in those cases, culture brought to light bacterial strains with acquired resistance to amoxicillin-clavulanate (2 H. influenzae and 2 E. coli, in 4 patients), or pathogens not targeted by FAPP and naturally resistant to amoxicillin-clavulanate (1 H. alvei, 1 M. morganii, and 1 S. maltophilia, in 3 patients). A medico-economic evaluation is ongoing to determine what impacts FAPP results would have had on care and antibiotics prescribing (Guillotin et al., in preparation).
Comparison of Paired BALDS and ETADS Specimens
Among the 58 patients with paired BALDS and ETADS, 46 (79.3%) had the same pathogen(s) (or no pathogen) identified in both samples with FAPP. Of the 12 discrepancies observed, 5 were due to detection of one more pathogen in ETADS (2 viruses, and 3 bacteria at 105 bin), 4 to detection of one additional bacteria in BALDS (3 of which were also detected in ETADS, but considered as negative since at 104 bin in ETADS). In the 3 latter cases, the difference relied on two pathogens. If bacteria with a 104 bin had been considered as positive in ETADS, the agreement between both types of specimens would have been less satisfactory, with 38/58 (65.5%) concordant results (Figures 2C,D). Regarding culture, concordant results were obtained in 48/58 (82.8%) paired specimens. In most of the discordant cases (7/10), there was at least one additional pathogen detected in BALDS. At last, only two of all discordant pairs (n = 20 with FAPP and/or culture) were confirmed with both methods (similar results between FAPP and culture) (Supplementary Table S1).
Discussion
At first developed for the detection of widely circulating respiratory viruses and selected atypical bacteria, syndromic molecular tests for respiratory tract infections continuously expand their breadth of coverage to improve diagnostic accuracy. FAPP and the Curetis® Unyvero Hospitalized Pneumonia Panel, are the first two, FDA approved and CE marked, commercially available platforms which target a large number of lower respiratory tract pathogens and resistance genes from aspirates or BAL fluids (Collins et al., 2020; Murphy et al., 2020). There are no published prospective studies comparing the performances of both plateforms, but regarding their technical characteristics, FAPP offers a shorter turnaround time (75 min vs. 4–5 h), a smaller footprint, and the possibility to detect viral pathogens and to semi-quantify bacteria (Poole and Clark, 2020). In this study, this test was compared to routine microbiological methods using 237 prospectively collected BAL and ETA specimens obtained from 100 ICU patients at the time of suspected HAP and, if possible, at a later timepoint during follow-up.
As expected, implementation of FAPP shortened the delay in getting results (4 h 15 min on average, one ICU setting being located 10 km away from the laboratory vs. 64–70 h with culture). In accordance with recent evaluations (Lee et al., 2019; Buchan et al., 2020; Murphy et al., 2020; Yoo et al., 2020), FAPP increased the positivity rate of diagnostic testing (81.6% for BALDS, and 75.6% for ETADS), enabling identification of additional bacteria in 39.5% BALDS and 37.8% ETADS. The most common pathogens detected were consistently the same across both methods (i.e., in order of prevalence, H. influenzae, S. aureus, E. coli, S. pneumoniae, and K. pneumoniae). This pathogen distribution, which mostly corresponded to bacterial species that are part of the normal throat flora, was not really different from that described in community-acquired pneumonia. According to the latest European surveillance report on healthcare-associated infections acquired in ICU in 2017, P. aeruginosa was the most common microorganism associated with pneumonia (19.9%), followed by S. aureus (18.5%), Klebsiella spp. (15.2%), and E. coli (13.5%). In the majority of cases, pneumonia was associated with intubation, and HAP episodes occurred after an average length of ICU stay of 7.3–12.1 days, depending on the country (European Centre for Disease Prevention and Control [ECDC], 2019). In our study, whatever the method used, P. aeruginosa was identified in only 4/100 patients, including three who did not present classic risk factors for MDR pathogens (no previous antimicrobial therapy or hospitalization in the preceding 90 days, and length of ICU stay of 4–6 days) (Torres et al., 2017; European Centre for Disease Prevention and Control [ECDC], 2019). The most common pathogen of our study, H. influenzae, was detected with FAPP in 40/100 patients at diagnosis, after a median length of ICU stay of 4 days, but was less frequently found in culture (29/100 patients). In line with our data, the majority of discrepancies previously reported between FAPP and culture, concerned the same fastidious bacteria, and were explained by the higher sensitivity of the molecular test and/or antibiotics consumption before sampling (Lee et al., 2019; Yoo et al., 2020). Here, in just over half of the discrepant cases, H. influenzae grew on the enriched medium used for culture, but was overgrown by other pathogens or commensal bacteria, and was therefore not detected and/or not reported. Thus, whether detection of H. influenzae represents true infection or colonization will be an important area for future research. It is less a question for S. aureus, which is a member of the normal nasal flora in about 30% of the population, but can also be regarded as an aggressive and life-threatening bacterial pathogen (Laux et al., 2019). However, in the same manner as for H. influenzae, discrepant results obtained for S. aureus in 7 patients (FAPP-positive but culture-negative), were not always explained by no bacterial growth. As noted previously, these findings pointed the limits of bacterial cultures, which are subject to interpretation and based on selection of dominant species assigned to play a pathogenic role, the minority species being not considered (Buchan et al., 2020; Murphy et al., 2020). These results confirmed the need to inoculate selective agars for enhancing detection of specific bacteria in lower airways (Chapin and Doern, 1983; Doern and Brogden-Torres, 1992). Moreover, a significant part of discrepancies was linked to a lack of growth in culture [11/33 (33.3%) for BALDS, 14/36 (38.9%) for ETADS, and 28/36 (77.8%) for ETATT]. A quater (25/100) of the patients enrolled in the study had received antibiotics before sampling at the time of HAP diagnosis, while ETATT were collected under antibiotic treatment. Thus, in our view, these culture-negative detections most likely corresponded to pathogens present at low abundances (i.e., below the limit of detection in culture) or to remnant DNA from non-viable bacteria, notably in supplemental ETATT, rather than non-specific amplifications. In fact, FAPP results from ETATT and/or paired BALDS or ETADS allowed to verify a lot of FAPP-positive results for bacteria that had been undetected by culture. As a result, FAPP may prove useful to guide treatment in situations of diagnostic uncertainty where patients have received antibiotics before sampling, and/or have unfavorable treatment outcomes after obtaining culture, because the higher sensitivity of this method decreases the likelihood to miss out on pathogens of the panel.
An important finding of this study, was that the implementation of FAPP increased the number of coinfections detected compared to conventional methods. Thus, the multiplex panel identified mixed infections in 49/100 patients (58.1% of positive BALDS or ETADS), compared to 32/100 patients (51.9% and 34.6% of positive BALDS and ETADS, respectively) by culture. These data corroborate other published results, and outline that the true incidence of polymicrobial HAP is probably underestimated with conventional techniques (Lee et al., 2019; Buchan et al., 2020; Murphy et al., 2020; Yoo et al., 2020). It remains to be evaluated whether detection of more pathogens will increase cure rates, and not adversely result in unnecessary consumption of broad-spectrum antimicrobials. New research avenues have emerged in recent years about the pathophysiology of HAP, because their rate of clinical cure does not commonly exceed 70% (Roquilly et al., 2019). It has been demonstrated that healthy lungs harbor a diverse and dynamic microbiota, which is profoundly altered in critically ill patients, and would play a role in the development of pneumonia. Future progress in this field should help understand how to appreciate lower abundance taxa of the microbiome, over other most numerous species (Panzer et al., 2018; Roquilly et al., 2019).
In our study, viruses were identified in 16/100 patients with FAPP, but in half of them no viral testing had been ordered, including one with an influenza A. As this virus can be responsible for severe pneumonia, and can represent a potential source of intra-hospital transmission, FAPP demonstrated a concrete benefit in that case (Loubet et al., 2017; Van Someren Gréve et al., 2018). Conventional testing for respiratory viruses other than influenza, has not been universally embraced as a standard of care, especially because viral carriage is not uncommon in patients with HAP (Loubet et al., 2017; Torres et al., 2017; Papazian et al., 2020). Furthermore, while the interaction between influenza and S. pneumoniae or S. aureus is a major contributor to influenza mortality in community-acquired pneumonia, the consequences of viral-bacterial coinfection on the prognosis of HAP is still unclear (Loubet et al., 2017; Van Someren Gréve et al., 2018). In our study, the majority (75%) of the 16 patients with identified viruses, were coinfected with bacteria, and 4 patients were infected with a single virus (influenza A, RSV, coronavirus, or human rhinovirus/enterovirus). Furthermore, in our opinion, additional viral targets (herpes simplex virus and cytomegalovirus) might be relevant if added to the panel, because reactivation of these viruses are indeed quite frequent in ICU patients, causing nosocomial viral pneumonia that can evolve into acute respiratory distress syndrome (ARDS) (Papazian et al., 2020).
One special feature of FAPP, is its ability to provide semiquantitative assessment of bacterial DNA targets to help in interpretation. Here, we showed that in BAL, 104 copies/mL corresponded to bacterial counts of ∼103–104 CFU/mL. In ETA, bacteria with bin results of 104 copies/mL were not found in culture in 96.7% of the cases (59/61). However, a small proportion (38.9%) of targets quantified as 104 copies/mL in ETADS, were recovered later with higher bin values in ETATT. Thus, we show that in those potentially contaminated samples, targets quantified as 104 copies/mL by FAPP, can be reported as negative to provide results concordant with those routinely reported by culture, in accordance with current guidelines (≥ 105 CFU/mL) (Buchan et al., 2020). Nonetheless, this raises the important question of whether low concentration culture-negative detections with FAPP are adding value in the care of ICU patients. This issue is discussed in the medico-economic evaluation coupled with this study (Guillotin et al., in preparation). We found no correlation between bin ≥ 105 and culture concentrations in both types of specimens. However, the plating method used in the present study did not allow accurate determination of relative quantities beyond 104 CFU/mL for BAL, and 105 CFU/mL for ETA.
An originality of this work lies on the inclusion of 58 patients from whom both BALDS and ETADS were collected, and could be compared. Latest European and American guidelines for the management of HAP provide divergent recommendations on sampling techniques to prioritize for diagnosis of HAP. While scientific societies from North America place a high value on non-invasive sampling with semiquantitative cultures (i.e., ETA), European guidelines suggest obtaining distal quantitative samples with invasive techniques to improve the accuracy of results, and reduce overutilization of antibiotics (Kalil et al., 2016; Torres et al., 2017). In fact, endotracheal aspirates may overestimate the presence of bacteria, but they can be performed more quickly and simply, with fewer complications and resources. In our study, provided that a 105 copies/mL threshold was applied for ETA, those specimens appeared equally accurate as BAL for the diagnosis of HAP (concordance obtained in 79.3% of patients with FAPP vs. 82.8% patients for conventional culture).
Finally, this study examined if when compared to culture, informations supplied by FAPP would have had positive impacts on antibiotics prescribing. Regarding the adequacy of bacteria targeted by FAPP, five Gram-negative species including three resistant to amoxicillin-clavulanate (H. alvei, M. morganii, and S. maltophilia) in 3/100 patients, were missed by the panel. On the other hand, the molecular test led to an increased identification of respiratory pathogens, and to the rapid detection of some genotypic markers of resistance in 8 patients. Thus, in total, for covering FAPP findings, the narrow-spectrum antibiotic amoxicillin-clavulanate could have been a therapeutic option in the majority of patients (83%). Nonetheless, natural or acquired resistances to amoxicillin-clavulanate would have gone unnoticed in 8.4% of them. All carbapenemase and/or ESBL-producing strains were correctly detected with the multiplex panel (AST agreed with FAPP). However, it should be noted that the overall prevalence of antimicrobial resistance was low in our study, and it should also be kept in mind that a lack of detection of resistance genes does not necessarily means susceptibility to antibiotics as there are resistance mechanisms that are not detected by FAPP (i.e., derepressed or plasmidic cephalosporinases, or non-CTX-M ESBL). Regarding methicillin resistance, consistent with previous observations, we noticed the false-positive detection of mecA/C and MREJ genes in one specimen containing a MSSA in culture (Yoo et al., 2020). Since this respiratory sample was polymicrobial, we hypothesized that it could have contained both a methicillin-resistant coagulase-negative Staphylococcus carrying mecA/C, and a MSSA with an empty SCCmec cassette (thus positive for MRJE) (Murphy et al., 2020).
Conclusion
In conclusion, our study demonstrates that FAPP provides results at a speed and sensitivity never possible before, and may allow clinicians to make more informed decisions about antibiotics use and isolation of patients. There is still room for improvements in terms of breadth (amoxicillin-clavulanate naturally resistant Gram-negative bacilli), resistance (MRSA), and cost, but this culture-independent technique may achieve more reliable identification of causative agents than culture. There will be a learning curve for physicians to establish how best to use FAPP results in the management of ICU patients with HAP. To achieve maximum benefit from this new molecular test, nuances in results interpretation might be applied on the basis of clinical presentation, timing of initial antimicrobial therapy (fresh vs. post-treatment samples), sampling type (BAL vs. ETA), and local bacterial ecology and resistance patterns. We are currently assessing the impact of this platform on antibiotic use and patients outcome in our hospital, and are evaluating if an algorithm-based treatment plan guided by FAPP would be of great benefit.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by the Groupe Nantais d’Ethique dans le Domaine de la Santé (GNEDS). Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author Contributions
LC, AR, and SG were involved in all the aspects of the study and were the guarantors for the data. BG, MB, KL, BR, and AR performed the clinical procedures. LC, TD, EP, and SG performed the laboratory procedures. LC, TD, FG, CP, M-AV, and SG analyzed the data. LC and AR wrote the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Clinical Research and Innovation Department of the Nantes University Hospital for their helpful contribution to this study.
Footnotes
Funding. This work has been supported by the bioMérieux BioFire Medical, France. The authors maintained control over all the aspects of the study and over the content of the publication.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.02080/full#supplementary-material
FAPP and original culture reading results.
References
- Buchan B. W., Windham S., Balada-Llasat J. M., Leber A., Harrington A., Relich R., et al. (2020). Practical comparison of the BioFire® FilmArray® Pneumonia Panel to routine diagnostic methods and potential impact on antimicrobial stewardship in adult hospitalized patients with lower respiratory tract infections. J. Clin. Microbiol. 29:JCM.00135-20. 10.1128/JCM.00135-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapin K. C., Doern G. V. (1983). Selective media for recovery of Haemophilus influenzae from specimens contaminated with upper respiratory tract microbial flora. J. Clin. Microbiol. 17 1163–1165. 10.1128/jcm.17.6.1163-1165.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins M. E., Popowitch E. B., Miller M. B. (2020). Evaluation of a novel multiplex PCR panel compared to quantitative bacterial culture for diagnosis of lower respiratory tract infections. J. Clin. Microbiol. 58:e02013-19. 10.1128/JCM.02013-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier M. R., Bard J. D. (2019). Direct-from-specimen pathogen identification: evolution of syndromic panels. Clin. Lab. Med. 39 433–451. 10.1016/j.cll.2019.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doern G. V., Brogden-Torres B. (1992). Optimum use of selective plated media in primary processing of respiratory tract specimens from patients with cystic fibrosis. J. Clin. Microbiol. 30 2740–2742. 10.1128/jcm.30.10.2740-2742.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekren P. K., Ranzani O. T., Ceccato A., Li Bassi G., Muñoz Conejero E., Ferrer M., et al. (2018). Evaluation of the 2016 infectious diseases society of America/American thoracic society guideline criteria for risk of multidrug-resistant pathogens in patients with hospital-acquired and ventilator-associated pneumonia in the ICU. Am. J. Respir. Crit. Care Med. 197 826–830. 10.1164/rccm.201708-1717LE [DOI] [PubMed] [Google Scholar]
- European Centre for Disease Prevention and Control [ECDC] (2019). Healthcare-Associated Infections Acquired in Intensive Care Units, in ECDC. Annual epidemiological report for 2017 Stockholm: ECDC. [Google Scholar]
- Kalil A. C., Metersky M. L., Klompas M., Muscedere J., Sweeney D. A., Palmer L. B., et al. (2016). Management of adults with hospital-acquired and Ventilator-associated pneumonia: 2016 clinical practice guidelines by the infectious diseases society of America and the American thoracic society. Clin. Infect. Dis. 63:e0061-11. 10.1093/cid/ciw353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laux C., Peschel A., Krismer B. (2019). Staphylococcus aureus colonization of the human nose and interaction with other microbiome members. Microbiol. Spectr. 7:GPP3-0029-2018. 10.1128/microbiolspec.GPP3-0029-2018 [DOI] [PubMed] [Google Scholar]
- Lee S. H., Ruan S. Y., Pan S. C., Lee T. F., Chien J. Y., Hsueh P. R. (2019). Performance of a multiplex PCR pneumonia panel for the identification of respiratory pathogens and the main determinants of resistance from the lower respiratory tract specimens of adult patients in intensive care units. J. Microbiol. Immunol. Infect. 52 920–928. 10.1016/j.jmii.2019.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loubet P., Voiriot G., Houhou-Fidouh N., Neuville M., Bouadma L., Lescure F. X., et al. (2017). Impact of respiratory viruses in hospital-acquired pneumonia in the intensive care unit: a single-center retrospective study. J. Clin. Virol. 91 52–57. 10.1016/j.jcv.2017.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy C. N., Fowler R., Balada-Llasat J. M., Carroll A., Stone H., Akerele O., et al. (2020). Multicenter evaluation of the BioFire® FilmArray® Pneumonia/Pneumonia plus panel for the detection and quantification of agents of lower respiratory tract infection. J. Clin. Microbiol. 58:e0128-20. 10.1128/JCM.00128-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzer A. R., Lynch S. V., Langelier C., Christie J. D., McCauley K., Nelson M., et al. (2018). Lung microbiota is related to smoking status and to development of acute respiratory distress syndrome in critically ill trauma patients. Am. J. Respir. Crit. Care Med. 197 621–631. 10.1164/rccm.201702-0441OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papazian L., Klompas M., Luyt C. E. (2020). Ventilator-associated pneumonia in adults: a narrative review. Intensive Care Med. 46 888–906. 10.1007/s00134-020-05980-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole S., Clark T. W. (2020). Rapid syndromic molecular testing in pneumonia: the current landscape and future potential. J. Infect. 80 1–7. 10.1016/j.jinf.2019.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roquilly A., Feuillet F., Seguin P., Lasocki S., Cinotti R., Launey Y., et al. (2016). Empiric antimicrobial therapy for ventilator-associated pneumonia after brain injury. Eur. Respir. J. 47 1219–1228. 10.1183/13993003.01314-2015 [DOI] [PubMed] [Google Scholar]
- Roquilly A., Torres A., Villadangos J. A., Netea M. G., Dickson R., Becher B., et al. (2019). Pathophysiological role of respiratory dysbiosis in hospital-acquired pneumonia. Lancet Respir. Med. 7 710–720. 10.1016/S2213-2600(19)30140-7 [DOI] [PubMed] [Google Scholar]
- Société Française de Microbiologie [SFM] (2018). Rémic – Référentiel en Microbiologie Médicale, 6th Edn, Paris: SFM. [Google Scholar]
- Société Française de Microbiologie [SFM] (2019). European Committee on Antimicrobial Susceptibility Testing. Comité de l’antibiogramme de la Société Française de Microbiologie – Recommandations 2019. Paris: SFM. [Google Scholar]
- Torres A., Niederman M. S., Chastre J., Ewig S., Fernandez-Vandellos P., Hanberger H., et al. (2017). International ERS/ESICM/ESCMID/ALAT guidelines for the management of hospital-acquired pneumonia and ventilator-associated pneumonia: guidelines for the management of hospital-acquired pneumonia (HAP)/ventilator-associated pneumonia (VAP) of the European respiratory society (ERS), European society of intensive care medicine (ESICM), European society of clinical microbiology and infectious diseases (ESCMID) and Asociación Latino americana del Tórax (ALAT). Eur. Respir. J. 50:1700582. 10.1183/13993003.00582-2017 [DOI] [PubMed] [Google Scholar]
- Van Someren Gréve F., Juffermans N. P., Bos L. D. J., Binnekade J. M., Braber A., Cremer O. L., et al. (2018). Respiratory viruses in invasively ventilated critically ill patients-A prospective multicenter observational study. Crit. Care Med. 46 29–36. 10.1097/CCM.0000000000002752 [DOI] [PubMed] [Google Scholar]
- Yoo I. Y., Huh K., Shim H. J., Yun S. A., Chung Y. N., Kang O. K., et al. (2020). Evaluation of the BioFire® FilmArray® Pneumonia panel for rapid detection of respiratory bacterial pathogens and antibiotic resistance genes in sputum and endotracheal aspirate specimens. Int. J. Infect. Dis. 95 326–331. 10.1016/j.ijid.2020.03.024 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FAPP and original culture reading results.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.