Abstract
Background
Processing and quantitative analysis of magnetic resonance spectroscopy (MRS) data are far from standardized and require interfacing with third-party software. Here, we present Osprey, a fully integrated open-source data analysis pipeline for MRS data, with seamless integration of pre-processing, linear-combination modelling, quantification, and data visualization.
New Method
Osprey loads multiple common MRS data formats, performs phased-array coil combination, frequency-and phase-correction of individual transients, signal averaging and Fourier transformation. Linear combination modelling of the processed spectrum is carried out using simulated basis sets and a spline baseline. The MRS voxel is coregistered to an anatomical image, which is segmented for tissue correction and quantification is performed based upon modelling parameters and tissue segmentation. The results of each analysis step are visualized in the Osprey GUI. The analysis pipeline is demonstrated in 12 PRESS, 11 MEGA-PRESS, and 8 HERMES datasets acquired in healthy subjects.
Results
Osprey successfully loads, processes, models, and quantifies MRS data acquired with a variety of conventional and spectral editing techniques.
Comparison with Existing Method(s)
Osprey is the first MRS software to combine uniform pre-processing, linear-combination modelling, tissue correction and quantification into a coherent ecosystem. Compared to existing compiled, often closed-source modelling software, Osprey’s open-source code philosophy allows researchers to integrate state-of-the-art data processing and modelling routines, and potentially converge towards standardization of analysis.
Conclusions
Osprey combines robust, peer-reviewed data processing methods into a modular workflow that is easily augmented by community developers, allowing the rapid implementation of new methods.
Keywords: Magnetic resonance spectroscopy, pre-processing, linear-combination modelling, quantification, tissue correction
1. Introduction
Magnetic resonance spectroscopy (MRS) is the only methodology that can determine the levels of neurochemicals in living tissue non-invasively, providing a unique window on the neurobiology of the human brain in health and pathology. Over the course of several decades, the field has developed a wide range of data acquisition, processing, and quantitative analysis methods (Harris et al., 2017; Landheer et al., 2019; Öz et al., 2020; Wilson et al., 2019).
In general, state-of-the-art analysis of MRS data can be divided into three fundamental steps:
-
Pre-processing of raw data that has been exported directly from the scanner or obtained from an institutional archiving system (PACS). Currently, no convention for a standardized MRS data format exists. Instead, each major vendor has developed proprietary file formats that store data in varying degrees of ‘rawness’ and limited information on acquisition parameters. Some of the most widely used file formats (DICOM MRS, Philips SDAT/SPAR, Siemens RDA, GE P-file) contain data that have been pre-processed to varying extents in the vendor-native online reconstruction environment. On-scanner processing relies on proprietary vendor-specific reconstruction code and is therefore neither standardized nor publicly documented.
Data processing can include basic low-level operations on the raw time-domain data (down-sampling, zero-filling, filtering, truncating, Fourier transformation), higher-level operations to improve critical signal properties like linewidth and signal-to-noise ratio (weighted receiver-coil combination, alignment of individual averages), and operations to address acquisition-related artefacts (removal of residual water signal, eddy-current correction).
-
Modelling of the processed spectral data is performed to derive quantitative parameters that allow conclusions to be drawn about the levels of individual metabolites. The complexity of this process ranges from simple peak integration, through single-resonance modelling, to linear-combination algorithms to decompose spectra into their constituent signals. While simple models are easy to implement, the full information content of an MRS spectrum can only be unlocked by modelling it fully. The most widely used linear-combination algorithms are exclusively implemented in third-party compiled software, either open-source (e.g. Tarquin (Wilson et al., 2011), Vespa (Soher et al., 2011)), or closed-source academic (e.g. the AQSES (Poullet et al., 2007) algorithm in jMRUI (Stefan et al., 2009)) or commercial (LCModel (Provencher, 1993)).
Despite their widespread use, all common fitting software packages are developed and maintained by small groups of researchers (or even a single individual), who often critically rely on third-party funding to keep the project alive.
Quantification is here used to describe the process of converting quantitative modelling parameters into biologically meaningful estimates of metabolite levels. Depending on the complexity of the quantification method, this may entail simply taking ratios of peak areas, or include more sophisticated calculations such as correcting for the fraction of cerebrospinal fluid (CSF), tissue-specific relaxation correction (Gasparovic et al., 2006), or accounting for assumed differences of metabolite abundance between tissue classes (Harris et al., 2015).
The core task of modelling is usually performed by third-party software, which typically has limited capability of pre-processing and quantification. This forces researchers to create their own local pipeline, starting from a rich diversity of scanner-specific export formats, choosing an appropriate set of processing steps, and finally exporting the processed data into a format that is accepted by the modelling software. Many modelling software solutions include the calculation of signal amplitude ratios to a reference (creatine, N-acetyl aspartate, water), but at the time of writing, none of them allow direct incorporation of tissue-specific segmentation or metabolite-specific relaxation information. Therefore, researchers must, again, develop local custom code to import the modelling results, apply corrections, and calculate final quantitative measures. In contrast to popular imaging modalities like functional MRI, which have established publicly available analysis frameworks and software environments open to community contributions, a widely used standardized pipeline is currently not available for MRS data analysis.
We have identified several issues with these practices. In short, the current best practices are not only inefficient, but severely hinder the more widespread use of MRS as a research tool, and curb its potential as a clinical one:
a. Waste of resources
If every lab resorts to writing custom code to carry out the same task, a lot of time, energy, and funding is wasted into duplicate efforts.
b. Methodological heterogeneity
The lack of a single analysis pipeline to include pre-processing, modelling and quantification forces all labs to rely on local custom code. Additionally, many labs conducting advanced methodological MRS research rely on their own, long-established pipelines (including either local or third-party modelling) and keep methodological developments local. This is detrimental to standardization, comparability, and transparency of data analysis.
c. High-entry threshold
Developing custom analysis code represents an often insurmountable effort for new research groups for whom MRS is a potential tool, but not a primary focus, and who might not have the background knowledge, resources, or technical expertise to create a processing pipeline from scratch.
d. Inertia and slow evolution
New methodological developments take longer than necessary to gain acceptance and become widely adopted, because newcomers struggle to implement them if the code is not made publicly available right away. Integrating new developments into compiled third-party software requires larger programming efforts, with low incentives for the developer to devote resources to this task.
e. Dependence and vulnerability
To evolve and be maintained, third-party software is critically dependent on its developers. They may transition to different positions (or leave academia altogether), run out of funding, lack time or staff support, or simply have no incentive to actively develop the software. This is particularly true for closed-source or commercial software, or tools that are maintained by single individuals.
Here we describe a new open-source MATLAB-based toolbox “Osprey”. Osprey is an all-in-one software package that combines all steps of state-of-the-art pre-processing, linear-combination modelling, quantification, and visualization of MRS data. The Osprey framework is designed as a modular, fully open-source environment to flexibly adopt future methodological developments, accelerate their adaptation, and foster standardization. The entire source code of Osprey is available in the public domain, inviting improvements, bug fixes, and addition of state-of-the-art technical developments from the community.
2. Methods
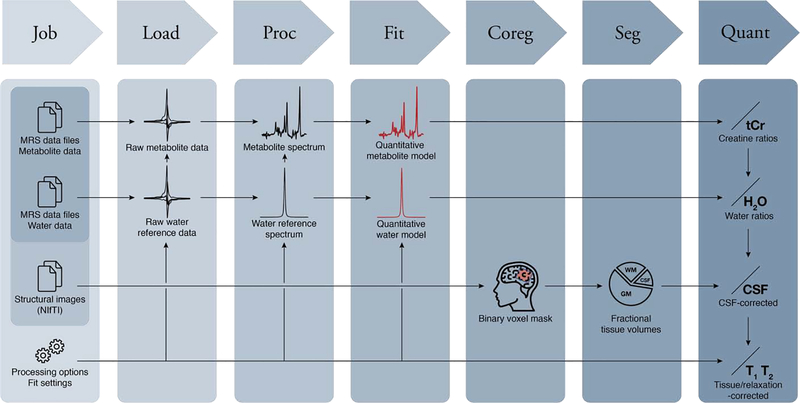
The Osprey workflow is summarized in Fig. 1. Osprey consists of seven separate modules – Job, Load, Proc, Fit, Coreg, Seg, and Quant, all of which are sequentially called by simple MATLAB commands. Alternatively, users can conduct the entire workflow in a graphical user interface designed to minimize the amount of user input. The Osprey code builds upon functions and the organizational structure of FID-A (Simpson et al., 2017), an open-source collection of MATLAB scripts for simulating, loading, and processing MRS data. At the time of writing, the various signal processing and modeling functions require the Optimization Toolbox (to model the spectra) and the Statistics and Machine Learning Toolbox (for statistical analysis). To use the Osprey GUI, the GUI Layout Toolbox (Sampson, 2019) and Widgets Toolbox (Jackey, 2019) need to be installed from the MATLAB File Exchange. Osprey has been tested using MATLAB versions 2017a and newer.
Figure 1:
The Osprey workflow with the seven modules Job, Load, Proc(ess), Fit, Coreg(ister), Seg(ment), and Quant(ify).
Additional details and MATLAB commands are summarized in section S1 of the Supplementary Material.
2.1. Job
The only user interaction that is required to specify and conduct an Osprey data analysis is to define a Job in a job file. The job system allows easy batch processing of multiple datasets. The job file is a text file containing the paths to the MRS data files and structural image files to be processed, and parameters to control data processing and linear-combination modelling. In addition, the user must specify an output folder that Osprey will use to save exported data, coregistration and segmentation images, and quantitative result tables.
Osprey distinguishes between three classes of MRS data files:
Metabolite (water-suppressed) data. These are a mandatory input.
Lineshape reference data. These are an optional input, acquired with the same sequence as the metabolite data, but without water suppression, and used to perform eddy-current correction (Klose, 1990) of the metabolite data. If only lineshape reference data are provided, this signal is also used to calculate water-scaled concentration estimates.
Short-TE reference data. These are another optional input. If the user provides short-TE reference data, they will be used to derive water-scaled concentration estimates (and lineshape data are only used for eddy-current correction). Using short-TE water as the concentration reference standard reduces T2-weighting of the water reference signal (and associated correction errors) compared to long-TE water data.
In addition to the paths to the raw data files, the job file must specify the type of sequence (with or without spectral editing, such as MEGA (Mescher et al., 1998; Rothman et al., 1993), HERMES (Chan et al., 2016; Saleh et al., 2016), or HERCULES (Oeltzschner et al., 2019a)), the target molecules of spectral editing experiments, and options for the fitting process, which are explained in Section 2.4.
2.2. Load
Osprey supports most common raw and processed file formats from major MRI vendors. This includes Philips (SDAT/SPAR, DATA/LIST), Siemens (RDA, TWIX) and GE (P-file) data. Additionally, single-file or multi-file DICOM datasets can be loaded. At the time of writing, single-voxel conventional and various J-difference-edited data from many sequence implementations are supported.
Upon calling the Load command, Osprey determines the file format, loads the raw spectroscopic data (FIDs) as well as all header information necessary for subsequent modules. The Load module also performs receiver-coil combination weighted with the ratio of the signal to the square of the noise. If lineshape reference or short-TE water data are provided, they are used instead of the metabolite data to determine the phasing and weighting parameters.
2.3. Proc
The Proc (Process) module performs all necessary steps to translate the raw, un-aligned, un-averaged data into spectra that are ready to be modeled.
The pre-processing pipeline includes the following steps: 1) alignment of individual averages using spectral registration in the time domain (Near et al., 2015); 2) averaging; 3) Fourier transformation; 4) automatic determination of the correct polarity of the spectrum since some water-suppression schemes can result in negative residual water peaks; 5) residual water removal by singular-value decomposition of the signal (Barkhuijsen et al., 1987) and subtracting components between 4.6 and 4.8 ppm; 6) linear baseline correction (based on the mean of 100 data points at the far edges of the frequency domain spectrum; 7) correct frequency referencing based on a single-Lorentzian fit to the 2.01 ppm NAA singlet. If lineshape reference data are available, the Klose method (Klose, 1990) is applied to correct the metabolite spectra for eddy-currents.
For J-difference-edited experiments (MEGA, HERMES, HERCULES), sub-spectra are aligned by minimizing the frequency-domain difference signal within a particular frequency range containing identical signal in pairs of sub-spectra. As an example, the edit-ON and edit-OFF spectra in GABA-edited MEGA data usually share a considerable residual water signal that is used for alignment. In contrast, the residual water signal is suppressed in the edit-ON spectra of GSH-edited MEGA data due to the editing pulse being applied at 4.56 ppm. In this case, the 2.01 ppm NAA signal is used for alignment. After sub-spectrum alignment, difference and sum spectra are calculated and stored.
In addition to performing the automated processing, Osprey determines several quality-control parameters. Linewidth is determined as the full-width half-maximum of a single-Lorentzian fit to the NAA peak (between 1.8 and 2.2 ppm). SNR is determined as the ratio between the amplitude of the NAA peak and the standard deviation of the detrended noise between −2 and 0 ppm. The frequency drift over the course of the experiment is determined based on the creatine signal in every single average (creatine and choline signals that nominally appear at 3.02 and 3.20 ppm are modeled by two Lorentzians).
The Proc module can optionally export the fully processed metabolite and water reference spectra to output subfolders, in formats readable by external third-party fitting software (LCModel, Tarquin, jMRUI, Vespa). This feature allows users to perform traditional data modelling, with the benefit of improved SNR and linewidth resulting from optimized coil-combination and alignment of individual averages – features that the established software solutions currently do not offer.
2.4. Fit
The Fit module models the processed spectra passed from the Proc module with a linear combination of basis functions. The default Osprey model is designed to mimic several key features of the algorithms implemented in LCModel and Tarquin. Several fit options are specified in the job file, e.g. the frequency range over which the spectrum is to be modelled, and the baseline flexibility (as controlled by the minimum ppm-spacing between neighboring knots of the cubic baseline spline).
The Fit module requires a sequence-specific basis set, which is automatically selected by Osprey based on the vendor, sequence type, and sequence parameters. Several sequence-specific basis sets for commonly used implementations of PRESS, MEGA-PRESS, HERMES, and HERCULES are included, derived from fast spatially resolved density-matrix simulations (Zhang et al., 2017) using ideal excitation and shaped refocusing pulses. Basis functions for macromolecule and lipid functions are added by generating Gauss-shaped signals with properties summarized in Tables S1 and S2 in section S3 of the Supplementary Material.
The Fit module interpolates the basis set to match the resolution (data points per ppm) of the processed spectra. All spectra of a single dataset (water-suppressed and water-unsuppressed) are then scaled to the basis set to facilitate convergence of the subsequent optimization.
Prior to the full analysis, two preparation steps are carried out. First, for optimal frequency referencing, the spectrum is cross-correlated with a sum of unit delta functions at 2.01 ppm, 3.03 ppm, and 3.21 ppm, representing the major landmark singlets from NAA, Cr, and Cho, respectively, and the frequency shift corresponding to the offset of the cross-correlation function is applied to the spectrum. Second, to obtain good starting values for the phase and linebroadening parameters, a preliminary fit is performed with a reduced basis set only including the basis functions of NAA, Cr, PCh, Glu, and ml, and a more flexible baseline with a knot spacing of 0.15 ppm. The final phase and linebroadening estimates from this preliminary fit are used as starting values for the full fit. Together with the initial referencing step, selecting a reasonable starting point for the non-linear parameters helps stabilize the optimization problem.
Osprey fits the real part of the frequency-domain spectrum Y(v) using a model similar to the one used by the LCModel algorithm. The NM simulated time-domain metabolite basis functions mm(t) in the basis set receive the same Gaussian linebroadening γ, and individual Lorentzian linebroadenings αm and frequency shifts ωm (m = 1,..., NM) before they are Fourier-transformed into the frequency domain:
| Equation 1 |
To account for deviations from a perfect Voigtian lineshape as determined by the linebroadening parameters, the frequency-domain basis spectra are then convolved with an arbitrary, unregularized lineshape model. This supplementary lineshape model S has a length equal to a spectral width of 2.5 times the coarse estimate of the FWHM of the spectrum that was estimated during the initial referencing step. S is normalized, so that this convolution does not impact the integral of signals, and is initialized as a unit delta function at the central point.
The smooth baseline is constructed as a linear combination of NB normalized, equally spaced cubic B-spline basis functions Bj(v) with coefficients βj (j = 1, …, NB), with knots 2 and (NB — 1) located on the edges of the fit range, and two additional knots outside the modeled range. NB is determined from the fit range based upon the minimum knot spacing condition that the user specifies in the job file. In contrast to the LCModel algorithm, the default Osprey model does not currently include baseline regularization. To prevent an unreasonably flexible baseline without using a regularizer, the default Osprey spline knot spacing is increased to 0.4 ppm, compared to the default LCModel ‘DKNTMN’ setting of 0.15 ppm.
The model spectrum is constructed from these components as follows:
| Equation 2 |
Here, φ0 represents the global zero-order phase correction; φ1(v) the global first-order (linear) phase correction; and Am the amplitude of each metabolite/MM/lipid basis function.
To minimize the sum of squares of the difference between the data Y(v) and the model , the Fit module uses an implementation of the popular Levenberg-Marquardt (Levenberg, 1944; Marquardt, 1963) non-linear least-squares optimization algorithm that allows hard constraints to be imposed on the parameters (Dentler, 2015). The amplitude parameters applied to the metabolite and baseline spline basis functions occur linearly in the model, and are determined at each iteration of the non-linear algorithm with a limited-memory algorithm for bound constrained optimization (L-BFGS-B) (Becker, 2015; Byrd et al., 1995; Zhu et al., 1997), constraining the metabolite amplitudes to be non-negative (Ai ≥ 0). Default hard constraints on non-linear parameters and weak soft constraints on macromolecule and lipid amplitudes are imposed to stabilize the solution, and are defined as they are in LCModel and Tarquin (Table S3 in section S4 of the Supplementary Material).
Water-unsuppressed data are modelled from a simulated water resonance with the same constrained non-linear least-squares algorithm using a simplified model, only including the following six hard-constrained modelling parameters: zero-order phase (−2π ≤ φ0 ≤ 2π), first order phase , Gaussian linebroadening , Lorentzian linebroadening (0 Hz ≤ α ≤ 50 Hz), frequency shift (−15 Hz < ω < 15 Hz), and amplitude A. No baseline is included in the water model.
2.5. Coreg
The Coreg module uses information about size, position and orientation of the MRS voxel in scanner-space coordinates to create a binary voxel mask, i.e., a 3D image in which the values 1 and 0 represent locations inside or outside the MRS voxel, respectively. The binary voxel mask is then transformed to the same coordinate system as the structural image (in NlfTI format) that the user provides in the job file. This step ensures that the voxel mask is coregistered to the structural image, and reproduces the original voxel placement.
2.6. Seg
The Seg module invokes the SPM12 segmentation function to segment the structural image into gray matter (GM), white matter (WM), and cerebrospinal fluid (CSF). The coregistered voxel mask that was created by the Coreg module is then overlaid with the GM, WM, and CSF tissue probability maps. The Seg module then calculates fractional tissue volumes fvol for GM, WM and CSF according to:
| Equation 3 |
with the tissue probabilities pk (n) for the tissue class k ∈ GM, WM, CSF and the n-th image voxel (n ∈ 1, 2, …, N), where N represents the number of image voxels within the MRS voxel.
2.7. Quant
The Quant (Quantify) module calculates various quantitative outputs, depending on the available modelling parameters that have been determined during the Fit process:
- Ratios of the metabolite signal amplitudes Smet to the total creatine amplitude StCr = SCr + SPCr are always determined, regardless of whether water data have been provided, according to
These tCr ratios are reported as raw ratios, i.e. no relaxation correction or accounting for tissue composition is applied. In difference-edited experiments, the Cr resonances are usually subtracted out in the difference spectra. In these cases, the tCr reference is determined from the edit-OFF spectrum if the fit option ‘Separate’ has been selected, and from the sum spectrum if the fit option ‘Concatenated’ has been selected.Equation 4 - When an unsuppressed water signal is provided, Osprey can report water-scaled metabolite estimates. If both lineshape reference data (i.e. data with the same TE as the water-suppressed data) and additional short-TE water data are available, the latter will be used as the water-scaling reference signal. Osprey reports water-scaled metabolite estimates according to Equation 5, analogous to the LCModel water scaling procedure, which does not account for tissue composition and assumes pure white matter by default:
Here, [H20] is the molal concentration of pure MR-visible water, i.e. 55.5 mol/kg of MR-visible water (Gasparovic et al., 2006; Knight-Scott et al., 2003); is the relative water density of white matter (0.65); TRW, TRmet, TEW, TEmet are the repetition and echo times of the water-unsuppressed and water-suppressed acquisitions; T1w and T2w are averaged relaxation times for tissue water (for brain data at 3T, T1w = 1100 ms and T2w= 95 ms (Wansapura et al., 1999)); T1met and T2met are the averaged relaxation times of all metabolites and generated from a lookup table that can be modified by the user. No tissue correction is applied. These water-scaled estimates are calculated even if no tissue composition information (from the Seg module) is available.Equation 5 - When unsuppressed water data and tissue segmentation are available, Osprey calculates water-scaled metabolite estimates corrected for the volume fraction of CSF in the voxel according to
Here, is the water-scaled metabolite estimate obtained in the previous step, and fvol,CSF is the fractional tissue volume of CSF as determined by the Seg module.Equation 6 - Finally, Osprey derives fully tissue-and-relaxation-corrected molal concentration estimates according to the Gasparovic method (Gasparovic et al., 2006), according to
Here, fGM, fWM and fCSF are the molal water fractions for GM, WM and CSF, which are derived from the volume fractions according toEquation 7
with the relative water densities , , and . Tissue-specific relaxation corrections are calculated according to and . Relaxation times for metabolites and water at 3T field strength were adapted from several widely used references (Edden et al., 2012; Mlynarik et al., 2001; Puts et al., 2013; Wansapura et al., 1999; Wyss et al., 2018).Equation 8
For GABA-edited spectra, Osprey calculates an additional ‘alpha correction’ metric (Harris et al., 2015) that normalizes for the fact that GABA levels are higher in gray matter than in white matter (Jensen et al., 2005).
Osprey saves all available quantitative results for the entire job as comma-separated value (CSV) tables to a subfolder “QuantifyResults” in the output folder. This easily accessible format provides a direct interface to external third-party software for subsequent statistical analysis and visualization (e.g. R, SPSS).
2.8. GUI
While all Osprey analysis steps can be carried out with simple MATLAB console commands, the Osprey GUI serves as the central hub for data visualization and quality assessment. Once a job file is loaded, all analysis steps can be triggered with dedicated GUI buttons, and the results from each step can be viewed on separate tabs, for each sub-spectrum from each dataset in the job. The ‘Load’ tab displays the pre-aligned, pre-averaged data, while the ‘Process’ tab shows the aligned individual transients, the final averaged spectra that are passed on to the Fit module, as well as information about the frequency history of the experiment pre- and post-alignment, and the basic quality assessment metrics (linewidth and SNR). The ‘Fit’ tab shows the spectrum, the complete fit, residual, baseline, and individual basis function contributions. The ‘Coreg/Seg’ tabs visualizes the results of voxel coregistration and segmentation, and the ‘Quantify’ tab features tables with all available quantitative outcome measures.
The additional ‘Overview’ tab provides useful summary visualizations of batched jobs with many datasets. It provides visualization of mean spectra with overlaid ribbon plots of the standard deviation; mean fit, residual, and baseline; raincloud plots (Allen et al., 2019; Whitaker et al., 2019) of quantitative results for quick assessment of the population distributions of metabolite estimates; and interactive display of correlation plots between metabolite estimates. If the job file was specified in CSV format and included assignment of each dataset to a group (e.g. patients or control subjects), the data in each raincloud and correlation plot is separated by the group variable.
2.9. Demonstration
To demonstrate the versatility of the processing and modelling capabilities of Osprey for various acquisition techniques, several single-voxel MRS datasets from the Big GABA dataset (Mikkelsen et al., 2019, 2017) were loaded, processed, and modelled (see section S5 of the Supplementary Materials for details on how to obtain the data from the Big GABA online repository).
Twelve PRESS datasets acquired on a 3.0T GE scanner (GE Healthcare, Milwaukee, United States) were selected. Parameters included: TR/TE = 2000/35 ms; 32 averages; 30 × 30 × 30 mm3 voxel in midline parietal cortex; 5 kHz bandwidth with 4096 data points. For comparison and validation, the PRESS datasets were also modelled with LCModel and Tarquin, using default settings, and the same basis functions as for the Osprey analysis. Estimates of total NAA (tNAA = NAA + NAAG), total choline (tCho = GPC + PCh), myo-inositol (mI) and Glx (glutamate + glutamine) were calculated with respect to total creatine (tCr = Cr + PCr). Agreement of metabolite estimates across software tools was determined based on group comparison and correlation analysis. Metabolite estimates were tested for normal distribution using Shapiro-Wilk testing, and for normal distribution of variances using Fligner-Killeen’s test, with a post-hoc pairwise Fligner-Killeen’s test and Bonferroni correction for the number of pairwise comparisons. Group means were then compared using ANOVA, Kruskal-Wallis rank testing, or Welch’s ANOVA, depending on whether variances were normal and different. Finally, post-hoc paired t-tests with equal or non-equal variances were performed accordingly. Additionally, Pearson correlation coefficients were determined separately for each metabolite and each pair of software tools.
Further, eleven GABA-edited MEGA-PRESS datasets from the Big GABA repository, acquired on a 3.0T Philips scanner (Philips Healthcare, Best, The Netherlands), were loaded, processed, and modeled. Parameters that differed from the PRESS parameters included: TE = 68 ms; 320 averages; 15-ms editing pulses applied at 1.9 ppm (edit-ON) and 7.5 ppm (edit-OFF).
Finally, eight GABA/GSH-edited HERMES datasets, acquired on a 3.0T Philips scanner (Philips Healthcare, Best, The Netherlands), were loaded, processed, and modelled. Parameters that differed from the MEGA-PRESS parameters included: TE = 80 ms; 20-ms editing pulses applied in the GABA/GSH HERMES scheme (Saleh et al., 2016).
3. Results
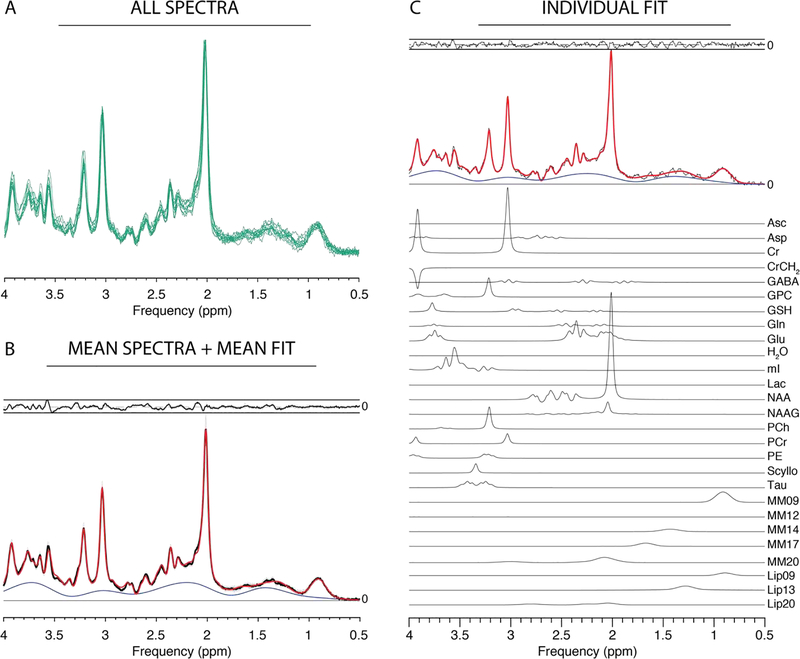
All twelve PRESS datasets were successfully loaded and processed in Osprey, and are plotted in Figure 2A. The mean of these processed spectra (and ± one standard deviation range) is shown in Figure 2B, along with the mean of the model spectra and the mean modelling residual. NAA SNR was 150 ± 24 and NAA linewidth was 7.8 ± 1.2 Hz. The results of quantification of these spectra are summarized in the following. tCr ratios of tNAA, tCho, mI, and Glx were 1.46 ± 0.07, 0.18 ± 0.01, 0.75 ± 0.07, and 1.38 ± 0.07, respectively. Water-scaled estimates of tNAA, tCho, mI and Glx, for example, were 19.40 ± 1.27 i.u., 2.42 ± 0.22 i.u., 8.88 ± 0.96 i.u., and 18.42 ± 1.34 i.u., respectively.
Figure 2:
Results from the Osprey processing and modelling of PRESS data. (A) Individual spectra; (B) mean spectra (black) +/− SD (gray ribbons); mean fit (red), mean residual (above). (C) Example fit with contributions from individual metabolites (Asc ascorbate; Asp aspartate; Cr creatine; GABA y-aminobutyric acid; GPC glycerophosphocholine; GSH glutathione; Gln glutamine; Glu glutamate; mI myo-inositol; Lac lactate; NAA N-acetylaspartate; NAAG N-acetylaspartylglutamate; PCh phosphocholine; PCr phosphocreatine; PE phosphoethanolamine; Scyllo scyllo-inositol; Tau taurine).
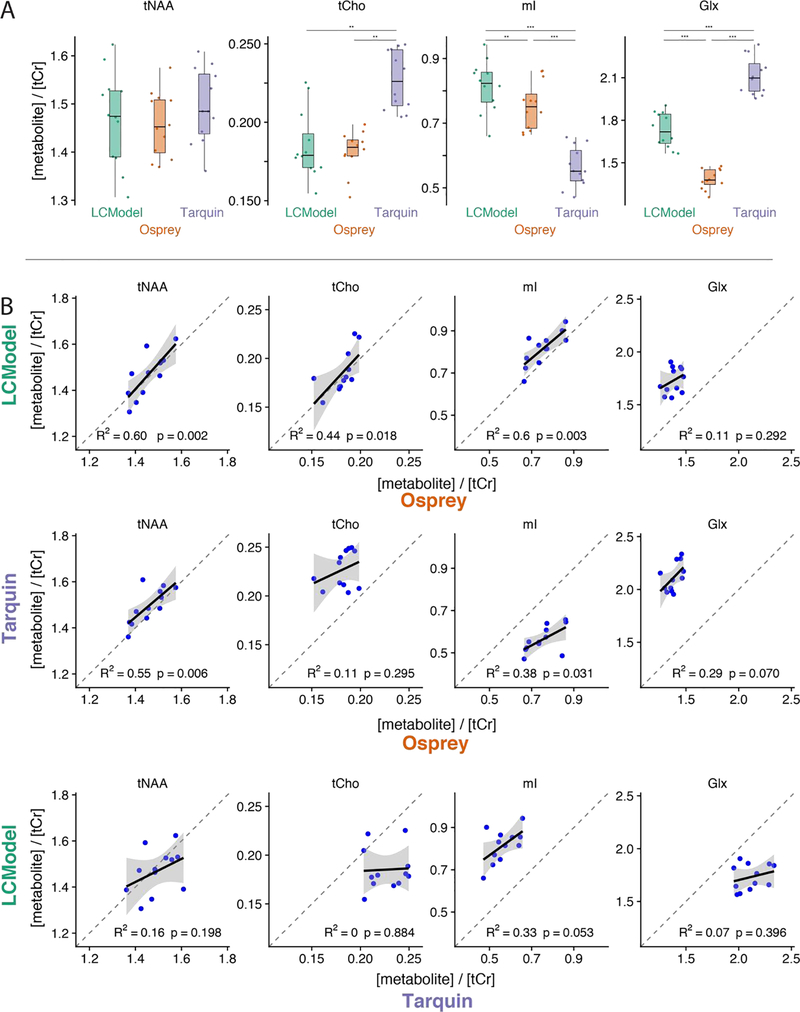
At group level, tNAA/tCr estimates agree well between the three tools, while Tarquin estimates higher tCho/tCr and Glx/tCr, and lower mI/tCr values than LCModel and Osprey (Figure 3A). Figure 3B shows cross-tool correlation results across the twelve datasets for the four metabolite-to-creatine ratios. Osprey estimates of tNAA/tCr, tCho/tCr and mI/tCr show significant positive correlations of moderate strength with LCModel estimates. Likewise, tNAA/tCr and mI/tCr estimates from Osprey and Tarquin correlate positively and moderately strong. In contrast, LCModel and Tarquin correlations do not reach significance.
Figure 3:
Comparison of metabolite estimates (relative to tCr) for 12 PRESS datasets. (A) Distributions for each metabolite and software; (B) scatterplots for each metabolite and pair of software (upper row: LCModelvs. Osprey, middle row: Tarquin vs. Osprey, bottom row: LCModel vs. Tarquin). One asterisk indicates p < 0.05; two asterisks indicate p < 0.01; three asterisks indicate p < 0.001.
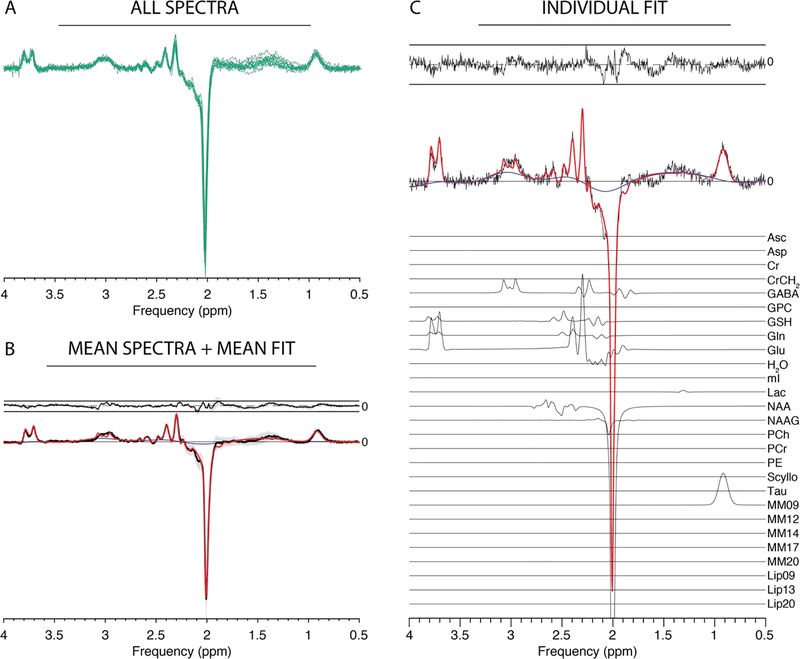
GABA-edited MEGA-PRESS datasets were successfully loaded, processed and modelled in ‘Separate’ mode, as summarized in Figure 4A and B. NAA SNR was 214 ± 32, while the NAA linewidth was 5.1 ± 0.6 Hz. GABA levels were quantified as 0.27 ± 0.07 (tCr ratio) and 1.67 ± 0.31 i.u. (water-scaled), respectively.
Figure 4:
Results from the Osprey processing and modelling of GABA-edited MEGA-PRESS data. (A) Individual spectra; (B) mean spectra (black) +/− SD (gray ribbons); mean fit (red), mean residual (above). (C) Example fit with contributions from individual metabolites.
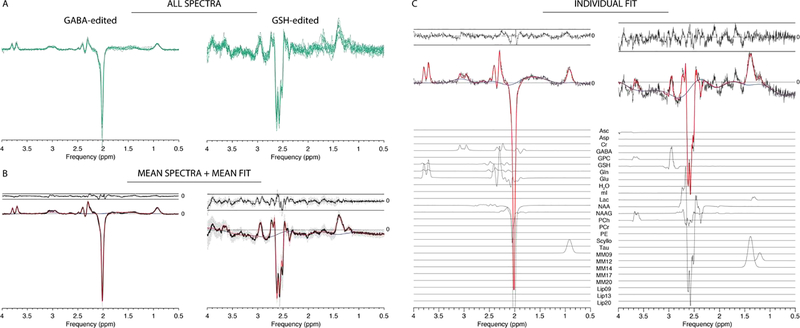
Similarly, the HERMES data are summarized in Figure 5. NAA SNR was 215 ± 24 with a NAA linewidth of 5.3 ± 0.5 Hz. GABA and GSH levels were estimated as 0.19 ± 0.04 and 0.19 ± 0.03 (tCr ratios), and 2.03 ± 0.28 i.u. and 1.94 ± 0.35 i.u. (water-scaled), respectively.
Figure 5:
Results from the Osprey processing and separate-mode modelling of GABA/GSH-edited HERMES data. (A) Individual spectra; (B) mean spectra (black) +/− SD (gray ribbons); mean fit (red), mean residual (above). (C) Example fit with contributions from individual metabolites.
The figures show individual spectra overlaid (green lines) in panel A, demonstrating consistent high-quality data resulting from the Osprey processing pipeline. Panels B display mean spectra (black solid lines), the standard deviation of the spectra (gray ribbon plots), mean model fits (red) and mean residuals (above the spectra) across all datasets. Panels C show representative linear-combination modelling. The fits approximate the data well with a relatively smooth baseline.
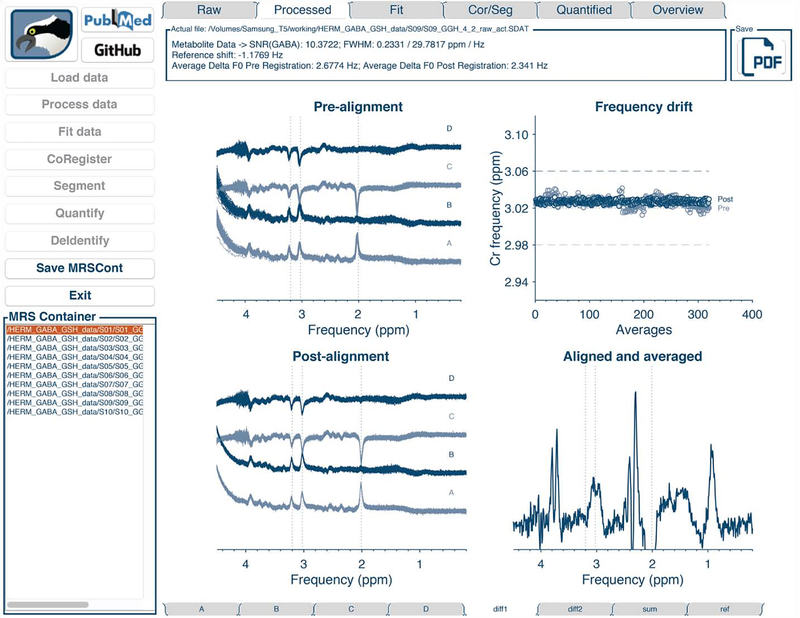
The structure of the GUI is shown in Figure 6. The workflow buttons corresponding to the different Osprey modules are in the left column, as well as a list of loaded datasets from which the user selects the dataset to be displayed. The tabs above the data display panels are used to switch between the analysis stages, and the tabs below correspond to different sub-spectra (here A, B, C, D, sum and difference spectra for HERMES data, along with water reference data).
Figure 6:
The Osprey GUI with workflow buttons (left), list of datasets (bottom left), analysis stage selection tabs (top row), sub-spectrum selection tabs (bottom row), and data display panel. The figure shows a GABA-GSH-editedHERMES dataset, with sub-experiments A, B, C, and D.
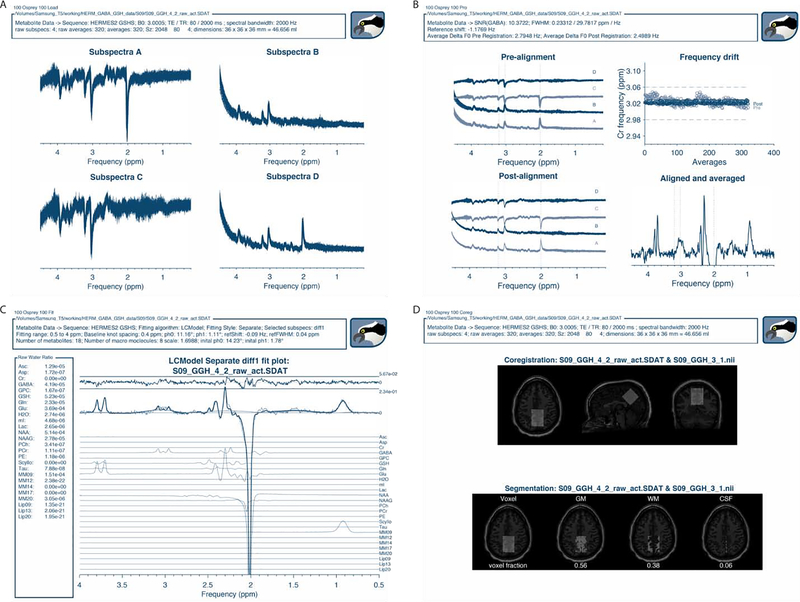
Figure 7 shows the GUI data display panels corresponding to the various stages of analysis of a single HERMES dataset, exemplifying the visualization of spectral-editing sub-experiments.
Figure 7:
Osprey visualization of the analysis of a HERMES dataset. (A) ‘Load’ visualization; (B) ‘Proc ‘ visualization with individual transients and frequency drift plot before and after spectral alignment; (C) ‘Fit’ visualization of the GABA-edited HERMES difference spectrum; (D) ‘Coreg’ and ‘Seg’ visualization of voxel coregistration and segmentation, including display of tissue fractions.
4. Discussion
The magnetic resonance spectrum of the human brain is rich with biochemical information, but extracting that information is a challenging task due to the overlapped nature of the metabolite spectra and the broad in-vivo linewidth. Quantitative MRS measurement outcomes are known to vary considerably depending on field strength, scanner vendor, localization technique, acquisition parameters, and choice of data processing and quantification practices. Recently, the MRS community has led efforts to converge towards standardized data acquisition (Deelchand et al., 2019a; Öz et al., 2020; Saleh et al., 2019; Wilson et al., 2019). In contrast, consensus on the processing and analysis of data has been slower to emerge.
The most commonly used strategy for quantitative analysis of MR spectra is linear-combination modelling. As a result of the methodological diversity and lack of standardized processing pipelines, most researchers have developed their own code to prepare their data for third-party quantification software. This practice is problematic for a number of reasons: a) methodological heterogeneity and opacity diminish comparability and reproducibility of quantitative MRS studies; b) benchmarking and subsequent adaptation of methodological progress is considerably slowed down; c) researchers new to the field experience a high-level entry threshold; d) strong dependency on engagement, support, and funding situation of third-party software developers leaves the community vulnerable.
Our new toolkit ‘Osprey’ seeks to address these issues by providing the community with a freely available open-source environment that unifies all steps of modern MRS data analysis – processing, modelling, quantification – into a common framework. Osprey is designed to: a) reduce methodological heterogeneity with its built-in standardized processing pipeline, modelling and quantification routines. These can serve as a substrate and starting point for the development of advanced data analysis methods, such as coil-combination or spectral alignment routines; b) accelerate the benchmarking, critical evaluation and finally integration of improved methods into the modular workflow; c) be immediately deployed by novice MRS users who seek to obtain quantitative results from MRS data using a single software solution, and who lack the resources to develop analysis code of their own; d) reduce the dependency of the MRS community on continued development of third-party linear-combination modelling software. Instead, the model code is directly accessible, modifiable, and exchangeable, allowing the research community to study the factors influencing results of linear-combination modelling, and compare or improve modelling algorithms.
Across the demonstration sample of PRESS datasets, Osprey estimates of the major metabolites are broadly comparable to estimates from LCModel and Tarquin, while the agreement between LCModel and Tarquin was weaker than expected. Between-software agreement generally appears to be best for tNAA, while ml and Glx estimates are less consistent. In spite of the small sample size of this demonstration, the results do appear to be representative. A broader analysis of the full “Big-GABA PRESS” dataset indicates that correlations of metabolite estimates between tools were moderate at best. While tNAA and tCho appear to be more consistently estimated by LCModel, Tarquin and Osprey, mI and Glx estimates agree less well, most likely due to different baseline estimation strategies (Zöllner et al., 2020). The influence of critical experimental parameters and analysis settings that critically affect performance and results of different modeling algorithms also remain under-studied, for example the influential ‘DKNTMN’ parameter that determines the degree of spline baseline flexibility in LCModel (Bhogal et al., 2017; Marjanska and Terpstra, 2019), or interactions of metabolite estimates with linewidth or SNR (Near, 2014). For difference-editing experiments, there is currently no consensus on optimal linear-combination strategies. In particular, co-edited macromolecular signals are difficult to simulate appropriately, since the spin systems of the contributing molecules are poorly characterized. Currently, co-edited signals representing the MM09, MM12 and MM14 resonances are therefore empirically parametrized in Osprey as Gaussian-shaped. Using Osprey, advanced modeling strategies for edited experiments can be explored, most notably for GABA editing experiments with unresolved macromolecular contamination issues, e.g. whether to include macromolecular and homocarnosine basis functions (Deelchand et al., 2019b), whether to impose soft constraints on co-edited macromolecules (Murdoch and Dydak, 2011), whether to increase baseline stiffness, whether to constrain the model by incorporating fit information from the sum spectrum (Oeltzschner et al., 2019a, 2019b), how to perform appropriate GABA-specific tissue correction strategies (Harris et al., 2015) etc. Osprey facilitates methodological investigations like these through its job system that allows many datasets to be batch-processed by modifying a single text file. While large-scale repositories of MRS data are still rare, projects like Big GABA (https://www.nitrc.org/proiects/biggaba/) already provide publicly available datasets that can be easily deployed to benchmark the performance of analysis methods with high statistical power.
Many other MRI modalities suffer from susceptibility to ‘processing bias’, and their communities have developed and adopted de-facto-standardized data processing and analysis toolboxes. Notable examples are, among others, SPM (Friston, 2007) and FSL (Jenkinson et al., 2012) for fMRI analysis, BART (Uecker et al., 2014) for parallel imaging reconstruction, and the Spinal Cord Toolbox (De Leener et al., 2017). In contrast, no such common framework currently exists for MRS, but rather a diverse field of tools mostly dedicated to modelling or visualization. LCModel continues to be the most widely used spectral analysis tool, despite its current cost of 13,300 USD, restrictive licensing, limited ongoing development by a single software engineer, and lack of built-in pre-processing functions. jMRUI offers basic functions to manipulate spectra and a variety of time-domain modelling algorithms (AMARES, QUEST, AQSES), but requires a high degree of user interaction and expertise, making it less suitable for novice researchers and reproducible processing of large datasets. Tarquin supports automated processing, but, like jMRUI, requires pre-processed spectra to model, and leaves a lot of freedom in choosing modelling options to the user. Gannet is an open-source toolbox with a similar all-inclusive workflow as Osprey, but is limited to simple peak integration of spectral-edited data. The MATLAB toolbox OXSA (Purvis et al., 2017) is conceptually similar to Osprey, but currently limited to processing DICOM data, and implements the AMARES algorithm (Vanhamme et al., 1997), which fits spectra in the time-domain using series of singlets with user-imposed prior knowledge, an approach notably different from linear-combination modelling of simulated metabolite basis functions. SIVIC (Crane et al., 2013) does not offer data pre-processing or linear-combination modelling at all, and is primarily dedicated to visualizing MRSI data and interfacing with radiological PAC systems. INSPECTOR (Juchem, 2018) offers automated analysis, but has not been peer-reviewed and is only distributed as closed-source obfuscated MATLAB executable code, as is the more interactive MRspa (Center for Magnetic Resonance Research, University of Minnesota, 2018).
All these existing software solutions either run as compiled executables or closed-source MATLAB applications, and while the source code is publicly available for some (Tarquin, SIVIC, Vespa), community-sourced modifications are either impossible, or may require substantial modifications downstream and local recompiling. In contrast, the entire Osprey source code (written in MATLAB) is publicly available in a single repository at https://github.com/schorschinho/osprey. At the time of writing, Osprey functions are primarily tailored to processing, modelling, and quantifying data from widely used single-voxel 1H MRS sequences designed to detect common metabolites in the human brain in-vivo. The modularity of the pipeline allows developers and users to implement analogous workflows for data acquired with different localization techniques, at different field strength, from other nuclei and body parts, animals, or phantoms, simply by branching out the workflow using ‘if statements and flags. Our group has previously disseminated the open-source processing toolkit Gannet (Edden et al., 2014), which has since been continuously developed in close interaction with its user base. We are confident that this pool of collaborators (and potential code contributors) will provide a solid foundation for continued development support for Osprey in the future.
5. Conclusions
Osprey is a new, open-source software environment for the pre-processing, linear-combination modelling, quantification and visualization of magnetic resonance spectroscopy data. It is hoped that the availability of such a tool will improve the standardization and accessibility of MRS data processing, while enabling further investigation and rapid adoption of new methodology.
Supplementary Material
Highlights.
Osprey is an open-source analysis toolbox for magnetic resonance spectroscopy data.
Includes pre-processing, linear-combination modelling, and quantification.
Includes a graphical user interface for visualization of each analysis step.
Acknowledgements
This work was supported by NIH grants K99AG062230, R21AG060245, R01EB016089, R01EB023963, and P41EB015909. None of the authors have a competing interest to declare.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allen M, Poggiali D, Whitaker K, Marshall TR, Kievit RA, 2019. Raincloud plots: a multi-platform tool for robust data visualization. Wellcome Open Res. 4, 63 10.12688/wellcomeopenres.15191.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkhuijsen H, De Beer R, Van Ormondt D, 1987. Improved Algorithm for Noniterative Time-Domain Model Fitting to Exponentially Damped Magnetic Resonance Signals. J. Magn. Reson 73, 553–557. [Google Scholar]
- Becker S, 2015. LBFGSB (L-BFGS-B) mex wrapper - File Exchange - MATLAB Central [WWW Document]. URL https://www.mathworks.com/matlabcentral/fileexchange/35104 (accessed 4.15.20).
- Bhogal AA, Schür RR, Houtepen LC, van de Bank B, Boer VO, Marsman A, Barker PB, Scheenen TWJ, Wijnen JP, Vinkers CH, Klomp DWJ, 2017. 1H-MRS processing parameters affect metabolite quantification: The urgent need for uniform and transparent standardization. NMR Biomed. 30, e3804 10.1002/nbm.3804 [DOI] [PubMed] [Google Scholar]
- Byrd RH, Lu Peihuang., Nocedal Jorge., Zhu Ciyou., 1995. A Limited Memory Algorithm for Bound Constrained Optimization. SIAM J. Sci. Comput 16, 1190–1208. 10.1137/0916069 [DOI] [Google Scholar]
- Center for Magnetic Resonance Research, University of Minnesota, 2018. MRspa: Magnetic Resonance signal processing and analysis [WWW Document]. URL https://www.cmrr.umn.edu/downloads/mrspa/ (accessed 4.15.20).
- Chan KL, Puts NAJ, Schär M, Barker PB, Edden RAE, 2016. HERMES: Hadamard encoding and reconstruction of MEGA-edited spectroscopy. Magn. Reson. Med 76, 11–19. 10.1002/mrm.26233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane JC, Olson MP, Nelson SJ, 2013. SIVIC: Open-Source, Standards-Based Software for DICOM MR Spectroscopy Workflows. Int. J. Biomed. Imaging 2013, 12 10.1155/2013/169526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leener B, Lévy S, Dupont SM, Fonov VS, Stikov N, Louis Collins D, Callot V, Cohen-Adad J, 2017. SCT: Spinal Cord Toolbox, an open-source software for processing spinal cord MRI data. NeuroImage 145, 24–43. 10.1016/j.neuroimage.2016.10.009 [DOI] [PubMed] [Google Scholar]
- Deelchand DK, Berrington A, Noeske R, Joers JM, Arani A, Gillen J, Schär M, Nielsen J-F, Peltier S, Seraji-Bozorgzad N, Landheer K, Juchem C, Soher BJ, Noll DC, Kantarci K, Ratai EM, Mareci TH, Barker PB, Öz G, 2019a. Across-vendor standardization of semi-LASER for single-voxel MRS at 3T. NMR Biomed. n/a, e4218 10.1002/nbm.4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deelchand DK, Marjańska M, Henry P-G, Terpstra M, 2019b. MEGA-PRESS of GABA+: Influences of acquisition parameters. NMR Biomed. n/a, e4199 10.1002/nbm.4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dentler A, 2015. Levenberg-Marquardt toolbox - File Exchange - MATLAB Central [WWW Document]. URL https://www.mathworks.com/matlabcentral/fileexchange/53449 (accessed 4.15.20).
- Edden RA, Intrapiromkul J, Zhu H, Cheng Y, Barker PB, 2012. Measuring T2 in vivo with J-difference editing: application to GABA at 3 Tesla. J Magn Reson Imaging 35, 229–234. 10.1002/jmri.22865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edden RA, Puts NA, Harris AD, Barker PB, Evans CJ, 2014. Gannet: A batch-processing tool for the quantitative analysis of gamma-aminobutyric acid-edited MR spectroscopy spectra. J Magn Reson Imaging 40, 1445–1452. 10.1002/jmri.24478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston KJ, 2007. Statistical parametric mapping the analysis of funtional brain images. Elsevier/Academic Press, Amsterdam; Boston. [Google Scholar]
- Gasparovic C, Song T, Devier D, Bockholt HJ, Caprihan A, Mullins PG, Posse S, Jung RE, Morrison LA, 2006. Use of tissue water as a concentration reference for proton spectroscopic imaging. Magn. Reson. Med 55, 1219–1226. 10.1002/mrm.20901 [DOI] [PubMed] [Google Scholar]
- Harris AD, Puts NAJ, Edden RAE, 2015. Tissue correction for GABA-edited MRS: Considerations of voxel composition, tissue segmentation, and tissue relaxations. J. Magn. Reson. Imaging 42 10.1002/jmri.24903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AD, Saleh MG, Edden RAE, 2017. Edited 1H magnetic resonance spectroscopy in vivo: Methods and metabolites. Magn. Reson. Med 77, 1377–1389. 10.1002/mrm.26619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackey R, 2019. Widgets Toolbox [WWW Document]. URL https://www.mathworks.com/matlabcentral/fileexchange/66235-widgets-toolbox (accessed 4.22.20).
- Jenkinson M, Beckmann CF, Behrens TEJ, Woolrich MW, Smith SM, 2012. FSL. NeuroImage, 20 YEARS OF fMRI 62, 782–790. 10.1016/j.neuroimage.2011.09.015 [DOI] [PubMed] [Google Scholar]
- Jensen JE, deB. Frederick B, Renshaw PF, 2005. Grey and white matter GABA level differences in the human brain using two-dimensional,J-resolved spectroscopic imaging. NMR Biomed. 18, 570–576. 10.1002/nbm.994 [DOI] [PubMed] [Google Scholar]
- Juchem C, 2018. INSPECTOR - A Tool for Teaching Magnetic Resonance Spectroscopy, in: Proc Int Soc Magn Reson Med. p. 5666. [Google Scholar]
- Klose U, 1990. In vivo proton spectroscopy in presence of eddy currents. Magn. Reson. Med 14, 26–30. 10.1002/mrm.1910140104 [DOI] [PubMed] [Google Scholar]
- Knight-Scott J, Haley AP, Rossmiller SR, Farace E, Mai VM, Christopher JM, Manning CA, Simnad VI, Siragy HM, 2003. Molality as a unit of measure for expressing 1H MRS brain metabolite concentrations in vivo. Magn. Reson. Imaging 21, 787–797. 10.1016/S0730-725X(03)00179-6 [DOI] [PubMed] [Google Scholar]
- Landheer K, Schulte RF, Treacy MS, Swanberg KM, Juchem C, 2019. Theoretical description of modern (1) H in Vivo magnetic resonance spectroscopic pulse sequences. J Magn Reson Imaging. 10.1002/jmri.26846 [DOI] [PubMed] [Google Scholar]
- Levenberg K, 1944. A method for the solution of certain non-linear problems in least squares. Q. Appl. Math 2, 164–168. 10.1090/qam/10666 [DOI] [Google Scholar]
- Marjanska M, Terpstra M, 2019. Influence of fitting approaches in LCModel on MRS quantification focusing on age-specific macromolecules and the spline baseline. NMR Biomed e4197 10.1002/nbm.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt DW, 1963. An Algorithm for Least-Squares Estimation of Nonlinear Parameters. J. Soc. Ind. Appl. Math 11, 431–441. 10.1137/0111030 [DOI] [Google Scholar]
- Mescher M, Merkle H, Kirsch J, Garwood M, Gruetter R, 1998. Simultaneous in vivo spectral editing and water suppression. NMR Biomed. 11, 266–272. [DOI] [PubMed] [Google Scholar]
- Mikkelsen M, Barker PB, Bhattacharyya PK, Brix MK, Buur PF, Cecil KM, Chan KL, Chen DY-T, Craven AR, Cuypers K, Dacko M, Duncan NW, Dydak U, Edmondson DA, Ende G, Ersland L, Gao F, Greenhouse I, Harris AD, He N, Heba S, Hoggard N, Hsu T-W, Jansen JFA, Kangarlu A, Lange T, Lebel RM, Li Y, Lin C-YE, Liou J-K, Lirng J-F, Liu F, Ma R, Maes C, Moreno-Ortega M, Murray SO, Noah S, Noeske R, Noseworthy MD, Oeltzschner G, Prisciandaro JJ, Puts NAJ, Roberts TPL, Sack M, Sailasuta N, Saleh MG, Schallmo M-P, Simard N, Swinnen SP, Tegenthoff M, Truong P, Wang G, Wilkinson ID, Wittsack H-J, Xu H, Yan F, Zhang C, Zipunnikov V, Zöllner HJ, Edden RAE, 2017. Big GABA: Edited MR spectroscopy at 24 research sites. NeuroImage 159, 32–45. 10.1016/j.neuroimage.2017.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen M, Rimbault DL, Barker PB, Bhattacharyya PK, Brix MK, Buur PF, Cecil KM, Chan KL, Chen DY, Craven AR, Cuypers K, Dacko M, Duncan NW, Dydak U, Edmondson DA, Ende G, Ersland L, Forbes MA, Gao F, Greenhouse I, Harris AD, He N, Heba S, Hoggard N, Hsu TW, Jansen JFA, Kangarlu A, Lange T, Lebel RM, Li Y, Lin CE, Liou JK, Lirng JF, Liu F, Long JR, Ma R, Maes C, Moreno-Ortega M, Murray SO, Noah S, Noeske R, Noseworthy MD, Oeltzschner G, Porges EC, Prisciandaro JJ, Puts NAJ, Roberts TPL, Sack M, Sailasuta N, Saleh MG, Schallmo MP, Simard N, Stoffers D, Swinnen SP, Tegenthoff M, Truong P, Wang G, Wilkinson ID, Wittsack HJ, Woods AJ, Xu H, Yan F, Zhang C, Zipunnikov V, Zollner HJ, Edden RAE, 2019. Big GABA II: Water-referenced edited MR spectroscopy at 25 research sites. Neuroimage 191, 537–548. 10.1016/j.neuroimage.2019.02.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlynárik V, Gruber S, Moser E, 2001. Proton T1 and T2 relaxation times of human brain metabolites at 3 Tesla. NMR Biomed. 14, 325–331. 10.1002/nbm.713 [DOI] [PubMed] [Google Scholar]
- Murdoch JB, Dydak U, 2011. Modeling MEGA-PRESS macromolecules for a better grasp of GABA, in: Proc. Intl. Soc. Magn. Reson. Med Presented at the Intl. Soc. Magn. Reson. Med., Montreal, p. 1394. [Google Scholar]
- Near J, 2014. Investigating the effect of spectral linewidth on metabolite measurement bias in short-TE MRS, in: Proc Int Soc Magn Reson Med. p. 2915. [Google Scholar]
- Near J, Edden R, Evans CJ, Paquin R, Harris A, Jezzard P, 2015. Frequency and phase drift correction of magnetic resonance spectroscopy data by spectral registration in the time domain. Magn. Reson. Med 73, 44–50. 10.1002/mrm.25094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeltzschner G, Saleh MG, Rimbault D, Mikkelsen M, Chan KL, Puts NAJ, Edden RAE, 2019a. Advanced Hadamard-encoded editing of seven low-concentration brain metabolites: Principles of HERCULES. NeuroImage 185, 181–190. 10.1016/J.NEUROIMAGE.2018.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeltzschner G, Zupan G, Edden RAE, 2019b. Simultaneous linear-combination modeling of MEGA-PRESS sum and difference spectra without soft constraints, in: Proc. Intl. Soc. Magn. Reson. Med Presented at the Intl. Soc. Magn. Reson. Med., Montreal, p. 510. [Google Scholar]
- Öz G, Deelchand DK, Wijnen JP, Mlynárik V, Xin L, Mekle R, Noeske R, Scheenen TWJ, Tkáč I, 2020. Advanced single voxel 1H magnetic resonance spectroscopy techniques in humans: Experts’ consensus recommendations. NMR Biomed. n/a, e4236 10.1002/nbm.4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poullet J-B, Sima DM, Simonetti AW, Neuter BD, Vanhamme L, Lemmerling P, Huffel SV, 2007. An automated quantitation of short echo time MRS spectra in an open source software environment: AQSES. NMR Biomed. 20, 493–504. 10.1002/nbm.1112 [DOI] [PubMed] [Google Scholar]
- Provencher SW, 1993. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn. Reson. Med 30, 672–9. [DOI] [PubMed] [Google Scholar]
- Purvis LAB, Clarke WT, Biasiolli L, Valkovič L, Robson MD, Rodgers CT, 2017. OXSA: An open-source magnetic resonance spectroscopy analysis toolbox in MATLAB. PLOS ONE 12, e0185356 10.1371/journal.pone.0185356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puts NAJ, Barker PB, Edden RAE, 2013. Measuring the longitudinal relaxation time of GABA in vivo at 3 tesla. J. Magn. Reson. Imaging 37, 999–1003. 10.1002/jmri.23817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman DL, Petroff OA, Behar KL, Mattson RH, 1993. Localized 1H NMR measurements of gamma-aminobutyric acid in human brain in vivo. Proc. Natl. Acad. Sci. U. S. A 90, 5662–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh MG, Oeltzschner G, Chan KL, Puts NAJ, Mikkelsen M, Sch??r M, Harris AD, Edden RAE, 2016. Simultaneous edited MRS of GABA and glutathione. Neuroimage 142, 576–582. https://doi.org/10.1016Zj.neuroimage.2016.07.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh MG, Rimbault D, Mikkelsen M, Oeltzschner G, Wang AM, Jiang D, Alhamud A, Near J, Schär M, Noeske R, Murdoch JB, Ersland L, Craven AR, Dwyer GE, Grüner ER, Pan L, Ahn S, Edden RAE, 2019. Multi-vendor standardized sequence for edited magnetic resonance spectroscopy. Neuroimage 189, 425–431. 10.1016/j.neuroimage.2019.01.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson D, 2019. GUI Layout Toolbox [WWW Document]. URL https://www.mathworks.com/matlabcentral/fileexchange/47982-gui-layout-toolbox (accessed 4.22.20).
- Simpson R, Devenyi GA, Jezzard P, Hennessy TJ, Near J, 2017. Advanced processing and simulation of MRS data using the FID appliance (FID-A)-An open source, MATLAB-based toolkit. Magn. Reson. Med 77, 23–33. 10.1002/mrm.26091 [DOI] [PubMed] [Google Scholar]
- Soher BJ, Semanchuk P, Todd D, Steinberg J, Young K, 2011. VeSPA: integrated applications for RF pulse design, spectral simulation and MRS data analysis., in: Proc Int Soc Magn Reson Med p. 1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan D, Cesare FD, Andrasescu A, Popa E, Lazariev A, Vescovo E, Strbak O, Williams S, Starcuk Z, Cabanas M, Ormondt D. van, Graveron-Demilly D, 2009. Quantitation of magnetic resonance spectroscopy signals: the jMRUI software package. Meas. Sci. Technol 20, 104035 10.1088/0957-0233/20/10/104035 [DOI] [Google Scholar]
- Uecker M, Lai P, Murphy MJ, Virtue P, Elad M, Pauly JM, Vasanawala SS, Lustig M, 2014. ESPIRiT—an eigenvalue approach to autocalibrating parallel MRI: Where SENSE meets GRAPPA. Magn. Reson. Med 71, 990–1001. 10.1002/mrm.24751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhamme L, van den Boogaart A, Van Huffel S, 1997. Improved Method for Accurate and Efficient Quantification of MRS Data with Use of Prior Knowledge. J. Magn. Reson 129, 35–43. 10.1006/jmre.1997.1244 [DOI] [PubMed] [Google Scholar]
- Wansapura JP, Holland SK, Dunn RS, Ball WS, 1999. NMR relaxation times in the human brain at 3.0 tesla. J. Magn. Reson. Imaging 9, 531–538. [DOI] [PubMed] [Google Scholar]
- Whitaker K, Rhys-Marshall T, van Mourik T, Martinez P, Poggiali D, Ye H, Klug M, 2019. RainCloudPlots/RainCloudPlots: WellcomeOpenResearch. Zenodo. 10.5281/zenodo.3368186 [DOI] [Google Scholar]
- Wilson M, Andronesi O, Barker PB, Bartha R, Bizzi A, Bolan PJ, Brindle KM, Choi I, Cudalbu C, Dydak U, Emir UE, Gonzalez RG, Gruber S, Gruetter R, Gupta RK, Heerschap A, Henning A, Hetherington HP, Huppi PS, Hurd RE, Kantarci K, Kauppinen RA, Klomp DWJ, Kreis R, Kruiskamp MJ, Leach MO, Lin AP, Luijten PR, Marjanska M, Maudsley AA, Meyerhoff DJ, Mountford CE, Mullins PG, Murdoch JB, Nelson SJ, Noeske R, Öz G, Pan JW, Peet AC, Poptani H, Posse S, Ratai E, Salibi N, Scheenen TWJ, Smith ICP, Soher BJ, Tkac I, Vigneron DB, Howe FA, 2019. Methodological consensus on clinical proton MRS of the brain: Review and recommendations. Magn. Reson. Med 82, 527–550. 10.1002/mrm.27742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M, Reynolds G, Kauppinen RA, Arvanitis TN, Peet AC, 2011. A constrained least-squares approach to the automated quantitation of in vivo 1H magnetic resonance spectroscopy data. Magn. Reson. Med 65, 1–12. 10.1002/mrm.22579 [DOI] [PubMed] [Google Scholar]
- Wyss PO, Bianchini C, Scheidegger M, Giapitzakis IA, Hock A, Fuchs A, Henning A, 2018. In vivo estimation of transverse relaxation time constant (T 2 ) of 17 human brain metabolites at 3T. Magn. Reson. Med 10.1002/mrm.27067 [DOI] [PubMed] [Google Scholar]
- Zhang Y, An L, Shen J, 2017. Fast computation of full density matrix of multispin systems for spatially localized in vivo magnetic resonance spectroscopy. Med. Phys 44, 4169–4178. 10.1002/mp.12375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Byrd RH, Lu P, Nocedal J, 1997. Algorithm 778: L-BFGS-B: Fortran subroutines for large-scale bound-constrained optimization. ACM Trans. Math. Softw. TOMS 23, 550–560. 10.1145/279232.279236 [DOI] [Google Scholar]
- Zöllner HJ, Povazan M, Hui SCN, Tapper S, Edden RAE, Oeltzschner G, 2020. Comparison of algorithms for linear-combination modelling of short-echo-time magnetic resonance spectra. bioRxiv 2020.06.05.136796. https://doi.org/10.110½020.06.05.136796 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.