Figure 2. CD160 Is a Monomer in Solution and Adopts a Unique Ig Structure.
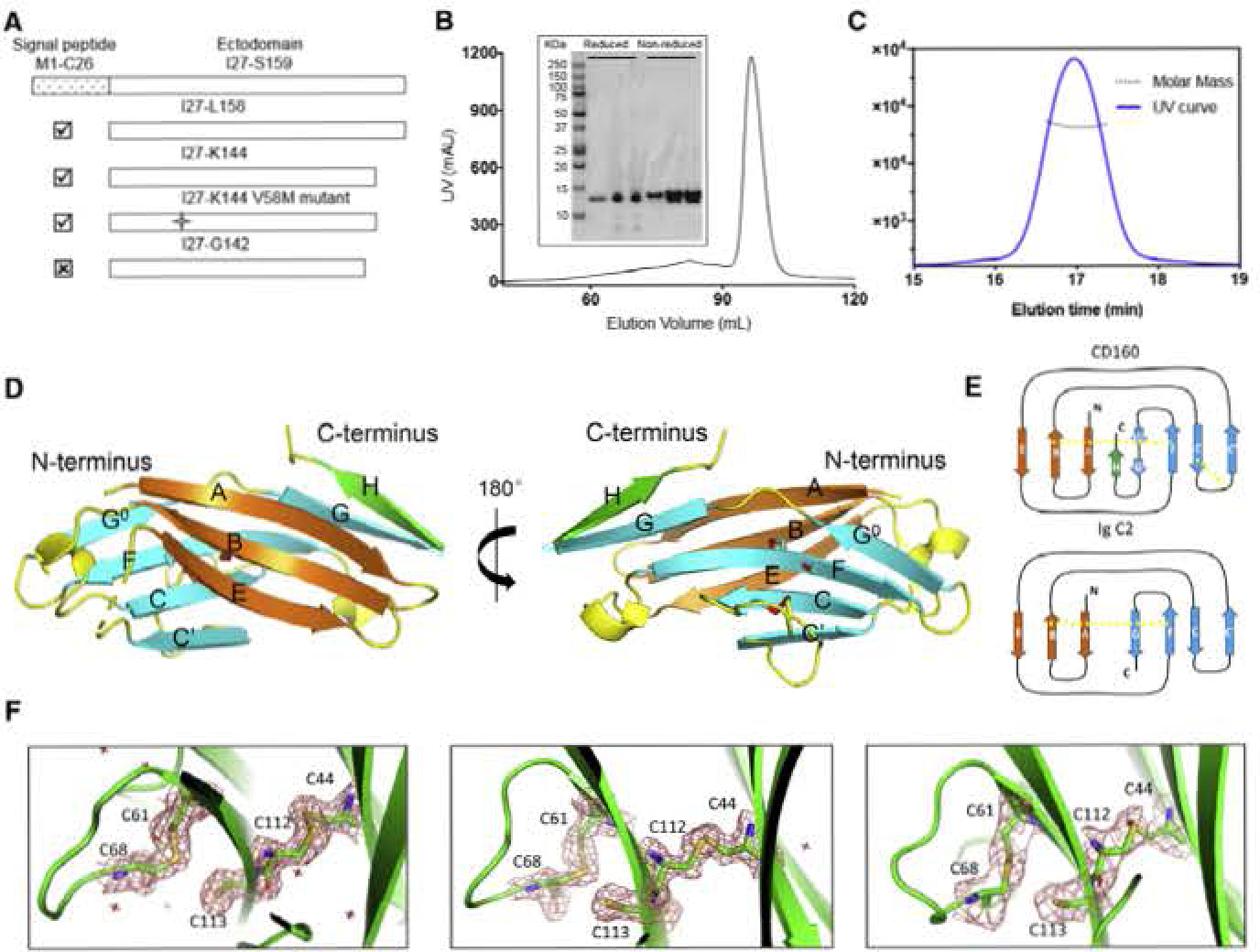
(A) Different construct designs of human CD160. Top panel shows the organization of the human GD160 ectodomain. Cross mark (☒) indicates that this construct was not successfully refolded, whereas a checkmark (☑) indicates successful refolding of the construct.
(B) Size-exclusion chromatography (SEC) demonstrates that refolded CD160 ((27-K144) runs as a single monodisperse peak. Proteins from different SEC fractions were subjected to reducing or non-reducing SDS-RAGE analysis.
(C) SEC-MALS analysis of the refolded CDI60 demonstrates that the molecular weight of CDI60 is 13.2 ± 0.6 kDa. corresponding to the calculated molecular weight of monomeric CD160. The mean value and standard deviation are from three independent experiments
(D) The back β-strand sheet is colored orange and the front β-strand sheet is colored cyan; the unique H strand is colored green; the connecting loops are colored yellow. Two disulfide bonds formed between the B and F strands, and the C and C′ strands are shown as slicks. The cysteine sulfur atoms are colored red. The unpaired C113 residue on strand F is also shown as a stick. Dashed lines indicate loops missing from the structure.
(E) Schematic presentation of the differences between CD160 and the canonical Ig C2 fold. The intramolecular disulfide bonds are indicated by yellow dashed lines.
(F) Clear electron densities for C113 and the adjacent disultide bond formed by the C44-C112 and C61-C68 pairs in the structures of CD160 (left), SeMet V58M GD160 (middle), and sc-CD16Q:HVEM (right). 2Fo-FC electron density maps were contoured at 1.5σ and shown as red mesh.
See also Figure S1.
