Abstract
Primary cilia are chemosensors that play a dual role to either activate or repress Hedgehog signaling, depending on presence or absence of ligand respectively. While inhibition of ciliogenesis has been shown to be characteristic of breast cancers, the functional consequence is unknown. Here, for the first time, inhibition of ciliogenesis led to earlier tumor formation, faster tumor growth rate, higher-grade tumor formation, and increased metastasis in the polyoma middle T (PyMT) mouse model of breast cancer. In in vitro model systems, inhibition of ciliogenesis resulted in increased expression of Hedgehog-target genes through a mechanism involving loss of the repressor form of the GLI transcription factor (GLIR) and activation of Hedgehog-target gene expression through cross-talk with transforming growth factor receptor alpha (TGFΑ) signaling. Bioinformatics analysis revealed that increased Hedgehog signaling is frequently associated with increased TGFΑ signaling in patients with triple negative breast cancers (TNBC), a particularly aggressive breast cancer subtype. These results identify a previously unrecognized role for inhibition of ciliogenesis in breast cancer progression. This study identifies inhibition of ciliogenesis as an important event for activation of Hedgehog signaling and progression of breast cancer to a more aggressive, metastatic disease.
Keywords: Primary cilia, Non-Canonical Hedgehog Signaling, Breast Cancer, Metastasis, Triple Negative Breast Cancer (TNBC)
INTRODUCTION
Breast cancer is the leading cause of cancer-related deaths in women worldwide (1). Analysis of human samples demonstrates that Hedgehog signaling is increased in 20–30% of breast cancers. Experimental inhibition of Hedgehog activity in breast cancer models reduces tumor growth and metastasis in vivo suggesting that targeting this pathway is a promising therapeutic strategy. Inhibition of the Hedgehog pathway (SMO inhibitors) has been shown to be effective in the treatment of basal cell carcinoma; however clinical trials using SMO inhibitors were not successful in the treatment of many other Hedgehog-positive cancers including breast cancer. While canonical Hedgehog signaling is dependent on SMO, it is now clear that cancer cells can activate Hedgehog signaling via alternative mechanisms independent of SMO activity and therefore render them resistant to SMO inhibitors. A better understanding of how Hedgehog signaling is activated in breast cancers is needed in order to design novel therapeutic strategies to target this important oncogenic pathway.
Primary cilia are specialized immotile microtubule-based organelles that protrude from the plasma membrane of many vertebrate cell types including mammary epithelial cells (2). The functional significance of primary cilia has been made clear by the discovery that defects in ciliogenesis or their function plays a causal role in a range of severe diseases and developmental disorders. Primary cilia are essential for both activation and repression of Hedgehog signaling, depending on the presence or absence of Hedgehog ligand respectively. Key proteins that regulate Hedgehog signaling, including PTCH, SMO and GLI transcription factors, localize to the primary cilium (3). Canonical activation of the Hedgehog pathway occurs through the binding of Hedgehog ligands to the PTCH receptor, thereby relieving inhibition of SMO (4). Activated SMO then translocates to the cilium resulting in post-translational processing of GLI into its activated form (GLIA). GLIA then translocates to the nucleus for transcriptional activation of Hedgehog-target genes. In the absence of Hedgehog ligand, GLI is post-translationally modified for transcriptional repression (GLIR) and then translocates from the cilium to the nucleus where it represses expression of Hedgehog-target genes. We, along with others, have demonstrated that ciliogenesis is inhibited in breast cancer cells (5–7); however, its biological consequence is not understood. The findings in this report demonstrate for the first time that inhibition of ciliogenesis promotes breast cancer progression to a more aggressive and metastatic disease. Furthermore, our findings facilitate a better understanding of non-canonical activation of Hedgehog signaling in breast cancer by demonstrating for the first time the importance of inhibition of ciliogenesis for Hedgehog-target gene expression.
MATERIALS AND METHODS
Mice.
Mice were bred and maintained in our colony at the University of Arizona Animal Facility. The mice used were Ift88fl/fl mice (Dr. Bradley Yoder, University of Alabama, Birmingham (8)), Kif3afl/fl mice (Dr. Jeremy Reiter, University of California, San Francisco, (9)), and MMTV-PyMT and Ift20fl/fl mice (Dr. Gregory Pazour, University of Massachusetts Medical Center and Jackson labs). We generated Ift88fl/fl/MMTV-PyMT and Ift20fl/fl/MMTV-PyMT mice by crossing Ift88fl/fl or Ift20fl/fl to MMTV-PyMT mice. Recipient mice for mammary fat pad transplant surgeries were SCID/Beige mice (Charles River). All procedures were performed in accordance with our Institutional Animal Care and Use Committee-approved protocol.
Mammary Epithelial Cell Preparation.
Cell preparations were performed with fresh mammary glands, or alternatively, with glands that had been frozen in 90% FBS and 10% DMSO, and were thawed upon epithelial cell harvest. Mammary epithelial cells were isolated based on the method described by Prater et. al. (10). In short, harvested glands from a single mouse were digested for 14–16 hours at 37°C in collagenase/hyaluronidase (StemCell Technologies, Cat. # 07919) in DMEM/F12/H (Gibco, Cat. # 11330–032) supplemented by 50µg/ml gentamicin (Gibco, Cat. # 15750–060). Digested tissue was resuspended in cold HF [HBSS (Gibco, Cat. # 14025–092), with 10mM HEPES (Gibco, Cat. # 15630), and 2% heat-inactivated Fetal Bovine Serum (FBS) (Gibco, Cat. # 16140–071)] and NH4Cl (StemCell Technologies, Cat. # 07800) to lyse red blood cells. After centrifugation to remove the supernatant, trypsin (StemCell Technologies, Cat. # 07400) was added for 1.5 minutes. To dilute the trypsin, 10 ml of cold HF was added, and the mixture was spun to remove the supernatant. Further protease digestion was performed with trypsin (StemCell Technologies, Cat. # 07400) followed by treatment with dispase (StemCell Technologies, Cat. # 07913) and 1mg/ml DNAse (Sigma, Cat. #DN25). The cell prep was then filtered through a 40 µm cell strainer (BD Falcon, Cat. # 352340). The Mouse Epithelial Enrichment Kit (StemCell Technologies, Cat. # 19758) was used for enrichment of epithelial cells according to manufacturer’s instructions. Isolated epithelial cells were directly used for adenovirus treatment for subsequent mammary fat pad transplantations (Ift88fl/fl/PyMT+ and Ift20fl/fl/PyMT+ studies). Alternatively, epithelial cells were plated for a week according to Liu et al. (11) to expand cell number (Kif3afl/fl and Ift88fl/fl studies) and then trypsinized for adenovirus treatment and mammary fat pad transplantation.
Adenovirus Treatment.
Adenovirus transduction was performed at 20MOI using Ad5-CMVeGFP (Ad-GFP, 2×1010 TU/ml) or Ad5-CMV-Cre-eGFP (Ad-Cre-GFP, 5×1010 TU/ml) (University of Iowa Gene Transfer Vector Core). Viraductin Adenovirus kit (Cell Bio Labs Inc., Cat. # Ad-201) was used to infect 1×105 epithelial cells in HBSS containing 10mM HEPES.
Mammary Fat Pad Transplantation.
SCID/Beige 3-week-old pups, between 8 and 11g, were used as recipients for transplant surgeries. Both inguinal fat pads per mouse were cleared surgically prior to epithelial cell transplantation. For tumor growth studies, Ad-GFP-treated epithelial cells were injected into one fat pad, and Ad-Cre-GFP treated cells were injected into the contralateral fat pad of the same mouse. For metastasis studies, Ad-GFP-treated or Ad-Cre-GFP-treated epithelial cells were injected into both fat pads of the same mouse. Adenovirus-treated epithelial cells were resuspended in 65% HBSS, 25% Growth factor reduced Matrigel (BD, Cat. # 356231), and 10% Trypan Blue (Thermo Scientific Hyclone, Cat. # SV3008401) and injected into the cleared inguinal fat pad using a Hamilton microliter syringe. For Ift88fl/fl and Kif3afl/fl surgeries, 5×104 cells were injected per transplant. For Ift88fl/f/MMTV-PyMT and Ift20fl/fl/MMTV-PYMT surgeries, 3×104 cells were injected per transplant.
Tumor Growth.
Inguinal mammary glands were palpated twice per week post-surgery, and measurements were collected using a digital caliper upon detection of a mass that did not regress upon successive palpation. The two longest diameters were measured, and tumor volume (mm3) was calculated with a modified ellipsoid equation: (width2)*(height/2). Mice were euthanized upon reaching maximum tumor burden (10% of body weight). Alternatively, mice were euthanized 23 weeks post-surgery (Ift88fl/fl/MMTV-PyMT mice) or 25 weeks post-surgery (Ift20fl/fl/MMTV-PyMT mice). For premalignant studies, mammary glands were harvested at 5 weeks post-surgery. All mammary tumors were cut into thirds for separate analyses: immersed in RNA-later RNA Stabilization Reagent (Cat. # 76106, Qiagen) and frozen at −80°C for real-time PCR, flash frozen for protein isolation, and fixed in 4% paraformaldehyde (Electron Microscopy Sciences, Cat. #15714-S) at 4°C overnight and paraffin embedded for IHC and immunofluorescence. For metastasis studies, the lungs were harvested after 20 weeks for whole mounting and paraffin embedding.
Kaplan Myer curves were generated, and log rank tests were performed on tumor-free survival data using SPSS 21 (Statistical Package for the Social Sciences; IBM Corporation). Differences in tumor growth rates were analyzed with STATA 12. Tumor volumes for tumor growth rate statistics were first transformed to induce normality. A mixed model with random effects to account for repeated measures per mouse within treatment groups was then used. Mammary gland whole mounts were performed as previously described (2).
Immunocytochemistry and Immunohistochemistry.
Cell and tissue staining were performed as previously described (2, 7, 12). Adenovirus-treated MECs were plated on coverslips (Fisherbrand 12-545-81, 12CIR-1.5) and grown to confluency and then serum starved in 0.1% serum-containing media for 24 hours. BT-549 cells were serum starved in 0.5% serum-containing media for 24 hours.
For all immunofluorescent staining, the primary antibodies used included: ARL13b (1:400, mouse monoclonal IgG2a, UC, Davis/NIH NeuroMab Facility, clone N295B/66), acetylated tubulin (1:1000, mouse monoclonal IgG2B, Sigma, Cat. # T7451, clone 6–11B-1), γ-tubulin (mouse monoclonal IgG1, Sigma, Cat. # T5326, clone GTU-88), CK5 (1:400, rabbit polyclonal, Abcam, Cat. # ab53121). Secondary antibodies used included: Alexa Fluor 555-labeled goat anti-mouse-IgG2B (Invitrogen, Cat. # A21147), Alexa Fluor 633-labeled goat anti-mouse-IgG1 (Invitrogen, Cat. # A21126), Alexa Fluor 546-labeled goat anti-mouse-IgG2a (Invitrogen, Cat#A21133) and Alexa Fluor 488-labeled goat anti-rabbit-IgG (Invitrogen, Cat. # A11034). Hoechst 33342 (Cat. # H3570, Invitrogen) was used as a counterstain. Slides were mounted with 1.5 coverslips (0.16–0.19 mm thickness, Fisher Scientific, Cat# 12–544B) using Prolong Gold Antifade mounting media (Cat. # P36934, Invitrogen).
For immunohistochemistry using the primary antibody recognizing SLUG (SNAI2) (1:400, C19G7, Cell Signaling, rabbit monoclonal, Cat. #9585), the primary antibody was detected with a universal anti-rabbit, anti-mouse secondary antibody conjugated to peroxidase (ImmPress Universal Reagent, Vector Laboratories, Cat. # MP-7500). 3,3’-diaminobenzidine (DAB, Dako, Cat. # K3467) was used as the peroxidase substrate. The tissues were counterstained with Hematoxylin 1 (Thermo Scientific, Cat# 7221). Tissue slides with secondary antibody only were used as a negative control. Slides were mounted with Faramount Aqueous Mounting Media (Dako, Cat# S3025) using 1.5 coverslips (0.16–0.19 mm thickness, Fisher Scientific, Cat# 12–544B).
Immunofluorescence Imaging and Analysis.
Immunohistochemistry was performed as previously described (2, 7, 12). At least three images were acquired per tissue to quantitate cilia, with at least 150 total nuclei per sample. The presence of cilia in adjacent stroma, as well as centrosomes in the tumor tissue (γ-tubulin), was used as internal staining quality control. Primary cilia were quantified manually from maximum projections. The percentage of ciliated cells was quantified for basal cells (CK5+) and luminal epithelial cells (CK5-) separately.
RNA Isolation and Real Time PCR.
Tumor samples that were used for PCR analysis were selected as pairs where Cilia+/PyMT+ and Cilia-/PyMT+ tumors from the same mouse had an equal percentage of cancer cells (ratio of 1.0 +/−0.2). This selection criteria served as a control for variations in gene expression due to disproportionate ratios of cancer cells to the stroma. The RNeasy Lipid Tissue Mini kit (Qiagen, Cat. # 74804) was used for RNA isolation according to manufacturer’s instructions. Mammary glands frozen in RNAlater RNA Stabilization reagent were thawed and homogenized in Qiazol Lysis Reagent (Qiagen, Cat. # 79306) using a Powergen Model 1000 Homogenizer (Fisher Scientific). For RNA isolation from cell lines, RNA was harvested from cells in 24 wells using the RNeasy mini kit (Qiagen, Cat. # 74104) according to manufacturer’s instructions. RNA was on-column Dnase-treated using an RNase-Free Dnase set (Qiagen, Cat. # 79254). All RNA samples were checked for concentration and A260/280 and A260/230 ratios using a NanoDrop 1000 (Thermo Scientific). RNA samples were run on a polyacrylamide gel to check for ribosomal bands and RNA degradation. Only samples of high quality (260/280 ratios of 1.8–2.1 and 260/230 ratios of above 1.5, and minimal degradation) were used for further analysis. First strand cDNA was generated with 1 µg of RNA using the Maxima First Strand cDNA Synthesis Kit or the Ambion Retroscript Kit for RT-qPCR (Thermo Scientific, Cat. # K1641 and #AM1710). Real-time PCR was performed using the EXPRESS SYBR GreenER qPCR Supermix Universal kit (Invitrogen, Cat. # A10314). A total of 20 ng of cDNA was used per reaction, and reactions were performed in duplicates. A final primer concentration of 300nM was used.
The primer sequences for mouse used were:
mGli1-F: 5’-GCGGAAGGAATTCGTGTGCC-3’,
mGli1-R: 5’-CGACCGAAGGTGCGTCTTGA-3’,
mGli2-F: 5’-TTTGCCGATTGACATGAGACA-3’,
mGli2-R: 5’-GGTGGGAGGCCCGTGTAC-3’,
mPtch1-F: 5’-CTCTGGAGCAGATTTCCAAGG-3’,
mPtch1-R: 5’-TGCCGCAGTTCTTTTGAATG-3’,
mSnai2-F: 5’-GGCTGCTTCAAGGACACATT-3’,
mSnai2-R: 5’-TTGGAGCAGTTTTTGCACTG-3’,
mβ-Actin-F: 5’-CACAGCTTCTTTGCAGCTCCTT-3’,
mβ-Actin-R: 5’-CGTCATCCATGGCGAACTG-3’.
The following primer sequences for human were used:
hGLI1-F: 5’- GAAGTCATACTCACGCCTCGAA-3’,
hGLI1-R: 5’- CAGCCAGGGAGCTTACATACAT-3’,
hβ-ACTIN-F: 5’ AGAGCTACGAGCTGCCTGAC-3’,
hβ-ACTIN-R: 5’- AGCACTGTGTTGGCGTACAG3’.
Real time PCR was run on a standard ABI 7500 or a Roche Light Cycles 480 II. Relative mRNA fold change was calculated using the 2^(−(∆∆Ct)) method, using β-actin as a loading control. Fold changes were calculated between paired tumors from a single mouse treated with Ad-Cre-GFP and Ad-GFP (grown in separate mammary glands in the same mouse). Average fold changes and standard error were calculated per gene across mice. Two-sided paired t-tests were performed on paired fold changes.
Protein Isolation and Western Blotting.
Flash frozen tissue was submerged in RIPA buffer (150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris, pH8.0) with protease inhibitors (Roche #04693159001) and phosphatase inhibitors (0.37 mg/ml sodium orthovanadate, 0.42 mg/ml sodium fluoride, 0.012 mg/ml ammonium molybdate) and homogenized using an electric homogenizer (Kinematica Polytron). Homogenized lysates were briefly sonicated, spun at 4,000 RPM for 20 minutes at 4°C, and the supernatant was aliquoted to a new tube. Cell lysates from cell lines were harvested by incubating RIPA buffer on plated cells. Lysates were then briefly sonicated, spun at 12,000xg, and the supernatant was aliquoted to a new tube. Protein concentrations were measured using a Pierce BCA assay (Thermo Scientific). Protein lysates were run on an SDS-PAGE gel and blotted to an Immobilon-FL membrane (Millipore #IPFL00010). The membrane was blocked with Odyssey blocking buffer (Li-Cor # 927–4000) and then probed with antibodies overnight at 4°C diluted in Odyssey blocking buffer. Antibodies used were against: phospho-Akt (p-Akt, Ser 473, clone 587F11,mouse monoclonal, Cell Signaling, Cat. # 4051; 1:1000), phospho-ERK1/2 (p-ERK1/2, Thr202/Tyr204, Clone D13.14.48, rabbit monoclonal, Cell Signaling, Cat. #4370; 1:2000), GLI3 (R&D Systems, Cat. # AF3690, 1:500), and β-Actin (clone AC-15, mouse monoclonal, Sigma, Cat. # A1978; 1:10,000). Membranes were then incubated with LI-COR fluorescent secondary antibodies (1:10,000), and membranes were imaged using an LI-COR Odyssey Imaging System.
Maintenance of Cell Lines.
BT-549 cells were a kind gift from the laboratory of Ghassan Mouneimne. NIH 3T3 cells were obtained from the ATCC. BT-549 cells were maintained in RPMI 1640 with L-glutamine (CellGro, Cat. # 10–040-CV), sodium pyruvate (Gibco, Cat. # 11360–070), 1μg/ml bovine insulin (Sigma, Cat. # I5500), 10% FBS, 10mM HEPES (Gibco, Cat. # 15630–080), and antibiotic. NIH 3T3 cells were maintained in DMEM with 4.5g/L glucose, L-glutamine and sodium pyruvate (CellGro, Cat. # 10–013-CV), 10% FBS (HyClone, Lot # AUE34991, Cat. # SH30541.03), and antibiotic (Gibco, Cat. # 15240–062). This cell line was authenticated by the ATCC testing service, and mycoplasma testing was performed immediately prior to use of this cell line for experiments described in this study.
Ligand Treatments.
BT-549 cells were seeded in a 24-well plate as to be at sub-confluency at the time of lysate harvest (4×104 for NIH 3T3 cells, 2×104 for BT549 cells). Twenty-four hours post-seeding, complete media was replaced with serum-reduced media (0.5% FBS). After twenty-four hours of serum-starvation, cells were treated with 200nM SAG or 100ng/ml TGFΑ (R&D Systems, Cat. # 239-A-100μg) in serum-reduced media for 24 hours. Cell lysates were then collected for RNA harvesting.
Drug Treatments.
For Forskolin treatments, BT-549 cells were seeded in a 6-well plate so as to be at confluency at time of lysate harvest (1.5×105). Forty-eight hours post-seeding, complete media was replaced with serum-reduced media (0.5% FBS), or with complete media. After twenty-four hours of serum-starvation, cells were treated with 100μM Forskolin (Sigma, Cat. # F6886) in serum-reduced or complete media for 24 hours. Cell lysates were then collected for RNA and protein harvesting.
Statistical Analysis.
The remaining statistics performed were with paired two-sided T-tests using Microsoft Excel. A P-value less than 0.05 was considered significant.
RESULTS
The goal of the current study is to investigate if inhibition of primary cilia contributes to tumor formation. We first assessed whether inhibiting ciliogenesis in the mammary gland is sufficient to cause tumor formation. Loss of expression of the Kif3A or Ift88 genes, which are required for intraflagellar transport (IFT), have been shown to inhibit ciliogenesis (13, 14). Primary mammary epithelial cells (MECs) from mice harboring floxed Kif3A or Ift88 genes were isolated and treated with an adenovirus expressing GFP-tagged Cre-recombinase (Ad-Cre-GFP) to delete the Kif3A (Kif3A-) or Ift88 (Ift88-) alleles, or cells were treated with an adenovirus expressing GFP alone (Ad-GFP) as a control (Kif3A+ or Ift88+). Two models were utilized to validate that any phenotypes observed were due to inhibition of ciliogenesis and not due to non-cilia related functions of KIF3A or IFT88. We confirmed that ciliogenesis was inhibited in epithelial cells of Kif3a- and Ift88- glands (Fig.S1 A, B). Next, mice were followed for 16–17 weeks post-transplant and glands were found to be morphologically normal, with no indication of tumor growth (Fig. S1C). Glands lacked histological abnormalities such as hyperplasia, dysplasia, or carcinoma in situ (Fig.S1D). Others have also reported that loss of Ift88 in the mammary gland in the mouse does not result in tumor formation, even up to 18 months of age (15). Together, this data indicate that loss of primary cilia alone is not sufficient to cause tumor formation.
Given that cancer is a multi-hit disease and that inhibition of ciliogenesis is a common/early event in human breast cancer, we hypothesized that inhibition of ciliogenesis synergizes with other oncogenic events to promote tumor progression. We tested this hypothesis in the polyoma middle T antigen (PyMT) breast cancer model (16). To inhibit ciliogenesis in vivo, we crossed MMTV-PyMT mice with two different mouse models that allow for Cre-specific removal of genes required for ciliogenesis (mice carrying floxed alleles of either Ift88 or Ift20 treated with Ad-Cre-GFP or Ad-GFP). We validated that ciliogenesis was inhibited in epithelial cells of Ift88-/PyMT+ and Ift20-/PyMT+ glands (Fig.1 A, B; Fig. S2).
Fig. 1. Inhibition of Ciliogenesis Increases Tumorigenesis, Rate of Tumor Growth and Tumor Grade in The PyMT Breast Cancer Model.
A, ICC staining of Ift88fl/fl/PyMT+ mammary tissues sections for ARL13b (cilia marker; red), γ-tubulin (centrosome marker; green), CK5 (myoepithelial cell marker; white) and Hoechst (nuclei). The lumen (lum) and stromal (str) regions of the mammary gland are separated by a dashed line. Arrowheads point to primary cilia in Ad-GFP-treated glands, and to centrosomes without cilia in Ad-Cre-GFP-treated glands. B, The bar graph represents quantification of the percent ciliated epithelial cells (basal (bas), luminal (lum), total epithelial cells (all)) from Ift88fl/fl/PyMT+ (Ad-GFP (blue), n=3; Ad-Cre-GFP (red), n=3) and Ift20fl/fl/PyMT+ (Ad-GFP, n=4; Ad-Cre-GFP, n=4) mammary glands. Error bars indicate SE; P values were determined by the Student t-test (paired, two-tailed); *P<0.05. C, Kaplan-Meier curves for tumor-free survival and D, average tumor volume versus time graphed for transplants of Ift88fl/fl/PyMT+ (n=9) and Ift20fl/fl/PyMT+ (n=6) mammary epithelial cells treated with Ad-Cre-GFP (red) and Ad-GFP (blue). Error bars indicate SE; log-rank test and mixed model P-value displayed. Representative images of E, whole mammary tumors, and F, H&E staining of mammary tumor sections from end stage mammary tumors. H&E staining depicts a Grade 1 tumor from an Ad-GFP treated transplant and a Grade 3 tumor from an Ad-Cre-GFP treated transplant. G, The bar graph represents the percent of tumors that were graded as in situ, grade 1, 2 or 3. Ift88fl/fl/PyMT+ (Ad-GFP, n=9; Ad-Cre-GFP, n=9) and Ift20fl/fl/PyMT+ (Ad-GFP, n=6; Ad-Cre-GFP, n=6).
We investigated if tumor onset is enhanced by inhibition of ciliogenesis in the context of PyMT expression. IFT88/PyMT+ and IFT20/PyMT+ transplants treated with Ad-Cre-GFP or Ad-GFP were analyzed for tumor growth. We observed significantly earlier tumor formation in cilia-negative glands (Cilia-/PyMT+) compared to cilia-positive glands (Cilia+/PyMT+), upon deletion of Ift88 and Ift20 (Fig. 1C). Palpable tumors formed 4–6 weeks earlier in Cilia-/PyMT+ glands versus Cilia+/PyMT+ glands (Table S1, Table S2). These data support the conclusion that inhibition of ciliogenesis decreases the time to tumor formation in the PyMT model.
To determine if inhibiting ciliogenesis leads to earlier tumor formation by increasing the frequency of premalignant lesions, we harvested mammary glands 5 weeks post-transplantation and examined premalignant lesions in Ift88/PyMT+ and Ift20/PyMT+ transplants treated with Ad-Cre-GFP or Ad-GFP. Tissue sections from the glands were stained with H&E to quantify the percent of epithelium as normal, hyperplasia, and carcinoma in situ (CIS). Inhibition of ciliogenesis did not increase the frequency of hyperplasia or CIS in Ift88-/PyMT+ and Ift20-/PyMT+ transplants (Fig. S3). These findings suggest that inhibition of ciliogenesis in the PyMT model enhances the growth of malignant lesions, rather than increasing progression from normal to premalignant lesions.
We also analyzed tumor growth rate in Ift88/PyMT+ and Ift20/PyMT+ transplants treated with Ad-Cre-GFP or Ad-GFP. The tumor growth rate was found to be significantly increased in Ift88-/PyMT tumors (Fig. 1D, E). The tumor growth rate was also increased in Ift20-/PyMT tumors, but this was not statistically significant, likely due to a smaller sample size. These data indicate that inhibition of ciliogenesis increases the rate of tumor growth by cooperating with the PyMT oncogene.
To determine if inhibition of ciliogenesis altered the histological features of the tumors, we harvested end-stage tumors and analyzed tissue sections with H&E staining. Tumors were blindly graded by a certified pathologist based on increased disorganization of tissue architecture and increased nuclear pleomorphism (pre-malignant lesion-in situ and invasive cancer grade 1, 2 and 3) (Fig. 1F). We termed grade 1 as low grade, while grades 2 and 3 were grouped as high grade. While we observed in situ only Cilia+/PyMT+ tumors, we did not observe any in situ only Cilia-/PyMT+ tumors (Fig. 1G). A similar frequency of grade 1 tumors was observed in Cilia-/PyMT+ and the respective Cilia+/PyMT tumors (Fig. 1G). However, Cilia-/PyMT+ tumors had a higher frequency of high-grade tumors compared to Cilia+/PyMT+ tumors (Fig. 1G). Taken together, these data suggest that inhibition of ciliogenesis is an important step in the progression to more aggressive, invasive disease.
The PyMT oncoprotein is known to constitutively activate the RAS/MAPK and PI3K/AKT pathways (17). We validated activation of these pathways in Cilia+/PyMT+, and Cilia-/PyMT+ derived tumors by Western blotting for p-ERK1/2 and p-AKT using tumor lysates (Fig. 2A, Fig. S5A). We observed that inhibition of ciliogenesis did not increase the amount of p-ERK1/2 or p-AKT in the presence of PyMT. As mentioned, PyMT results in constitutive activation of these pathways; therefore, it is not surprising to find that inhibition of ciliogenesis does not increase signaling of PI3K/AKT or RAS/MAPK in this model.
Fig. 2. Inhibition of Ciliogenesis Decreases Production of GLIR and Increases Hedgehog Signaling in PyMT-positive Tumors.
Western blot of lysates isolated from Ift88fl/fl/PyMT+ or Ift20fl/fl/PyMT+ mammary tissue treated with Ad-Cre-GFP or Ad-GFP for A, phospho-ERK1/2 (p-ERK1/2) or phospho-AKT (p-AKT) (representative blot from three mice) and B, cell lysates from primary mammary epithelial cells from Ift88fl/fl/PyMT+ or Ift20fl/fl/PyMT+ glands treated with Ad-Cre-GFP or Ad-GFP blotted for GLI3R protein levels normalized to β-Actin levels. C, Bar graph represents the combined analysis of quantitative real time-PCR of Hedgehog-target genes (Gli1, Ptch1, Gli2, Slug) of RNA isolated from Ift88fl/fl/PyMT+ and Ift20fl/fl/PyMT+ mammary tumors. Number of tumors analyzed for each gene are: Gli1 (Ift88fl/fl/PyMT, n=7; Ift20fl/fl/PyMT+, n=5); Ptch1 (Ift88fl/fl/PyMT, n=7; Ift20fl/fl/PyMT+, n=3); Gli2 (Ift88fl/fl/PyMT, n=7; Ift20fl/fl/PyMT+, n=3); Slug (Ift88fl/fl/PyMT, n=5; Ift20fl/fl/PyMT+, n=5).. Average fold change compared to Ad-GFP is plotted. P values were determined by the Student t-test (paired, two-tailed); *P<0.05. Error bars represent standard error. D, Representative images of IHC staining of tumor tissues for SLUG. Arrows point to SLUG-positive nuclei.
The known role of primary cilia in Hedgehog signaling prompted us to investigate Hedgehog signaling in our Cilia+/PyMT+ and Cilia-/PyMT+ model. Primary cilia inhibit Hedgehog pathway activity via processing of GLI transcription factors into their repressor forms (GLIR) (4, 18, 19). Phosphorylation of GLI3 by protein kinase A (PKA) and the partial proteolytic cleavage via the βTrCP/Cul1 complex is required for endogenous GLI3 processing into GLI3R (4). We found the protein level of the proteolytically cleaved GLI3R was decreased when ciliogenesis was inhibited in PYMT+ mammary epithelial cells (MECs) Fig. 2B, Fig. S4, Fig. S5B). These data demonstrate that inhibition of ciliogenesis decreases processing of GLI3 into its repressor form in the PyMT model of breast cancer.
Next, we tested the hypothesis that inhibition of ciliogenesis, and decreased expression of GLI3R, results in increased expression of Hedgehog-target genes in the PyMT breast cancer model. Gene expression of downstream Hedgehog-target genes was analyzed by qPCR from RNA isolated from whole-tumors (in vivo) from Cilia+/PyMT+, and Cilia-/PyMT+ derived tumors (analysis of both IFT88/PyMT+ and IFT20/PyMT+ tumors). Specifically, relative gene expression of Gli1, Gli2, Ptch1 and the pro-migratory gene Slug were analyzed. We observed that inhibition of ciliogenesis resulted in an increase in expression of Gli1, Ptch1, Gli2, and Slug (Fig. 2C). While the increased transcription of Gli1 and Ptch1 was not statistically significant, there was a trend towards an increase in expression when ciliogenesis was inhibited (p=0.08 and p=0.23). Immunostaining of tumor tissues for expression of SLUG protein confirmed that expression of SLUG was increased when ciliogenesis was inhibited (Fig. 2D). We have not found antibodies that work for immunohistochemistry with specificity for murine GLI1, GLI2, and PTCH1 protein.
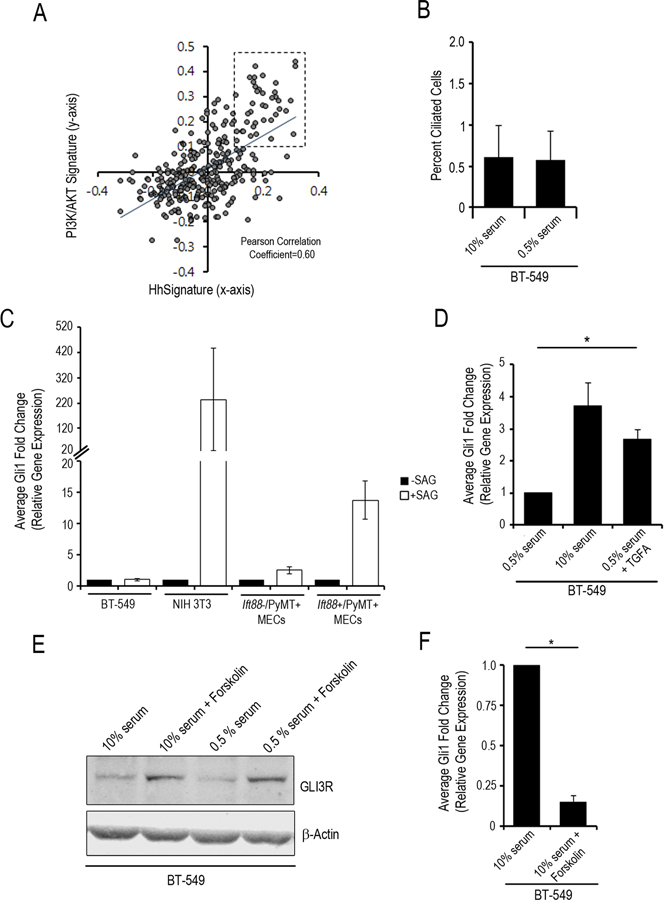
Given that breast cancer cells do not have the primary cilia needed for activation of the Hedgehog pathway via Hedgehog ligands or SMO activation, we hypothesized that loss of GLIR leaves Hedgehog-target genes vulnerable to activation by another pathway (non-canonical activation). To determine which pathway may be responsible for activation of Hedgehog-target genes in human breast cancer we utilized a bioinformatic approach. PyMT is a membrane scaffold protein that is a valuable model of human breast cancer because it functions like constitutively activated receptor tyrosine kinases (RTKs) (e.g. EGFR and ERBB2) to activate the PI3K/AKT and RAS/MAPK pathways. Overactivation of RTKs, including EGFR and ERBB2, are observed frequently in human breast cancer (20). To determine if the PI3K/AKT and RAS/MAPK pathways are associated with Hedgehog-target gene expression in human breast cancers we analyzed these signaling pathways in a published breast cancer microarray dataset (21). First, we identified patients with high levels of expression of Hedgehog-target genes as well as patients with high levels of expression of genes associated with PI3K/AKT and/or RAS/MAPK signaling (Fig. 3A). To accomplish this, we utilized gene expression signatures that represent activation of signaling of the Hedgehog, PI3K/AKT and RAS/MAPK pathways (Table S3). The gene signatures are based on comprehensive transcriptional outputs that have been validated by others and provide a tool to identify patients with increased signaling in the respective pathways (22–24). Analysis showed a positive correlation between patients with activated Hedgehog signaling and patients with activated PI3K/AKT (Fig. 3A). Using this bioinformatics approach, we determined that 39/295 breast cancer patients (13%) have both high Hedgehog and PI3K signaling. Of the 39 patients with high Hedgehog and PI3K/AKT signatures, 16 are also positive for the RAS/MAPK signature. To determine what RTK may be responsible for PI3K/AKT and RAS/MAPK signaling, we analyzed the 39 patients with high Hedgehog and PI3K/AKT signaling (Hedgehog+/PI3K+) to determine which of the known RTKs and their ligands are co-expressed. We observed that expression of EGFR, as well as the EGFR ligand transforming growth factor alpha (TGFΑ), was the most frequently expressed in this subset of patients (80% of Hedgehog+/PI3K+ patients) (Fig. 3A). Further analysis of the 39 patients with high Hedgehog and PI3K/AKT signaling revealed that these patients are negative for expression of ER and ERBB2 indicating that these patients are among the triple negative breast cancer subtype (TNBC). This bioinformatics data indicates that Hedgehog signaling is active in a subset of breast cancers and further supports published findings by others that Hedgehog signaling is increased in patients with TNBC (25, 26). This bioinformatics analysis also supports the use of the PyMT model to provide insight into the interaction/cross-talk between the PI3K/AKT and RAS/MAPK signaling pathways with that of the Hedgehog signaling pathway.
Fig. 3. Inhibition of Ciliogenesis Resulted in Decreased Processing of GLI3R and Induction by TGFA for Expression of Hedgehog-Target Genes.
A, Scatter plot represents the analysis of gene expression signature for PI3K/AKT and Hedgehog pathways. Each circle represents a single patient of the 295 breast cancer patients analyzed. 13% of breast cancers have transcriptional programs consistent with high PI3K/AKT and high Hedgehog signaling (box, upper right quadrant; Pearson Correlation analysis). Dashed box represents the 39 patients positive for both the Hedgehog and PI3K/AKT signatures. B, Bar graph represents quantitation of percent ciliated BT-549 cells treated with 0.5% serum or 10% FBS for 24 hours. Bar graphs represent average fold change (as compared to untreated cells) in expression of Gli1 based on quantitative real time-PCR analysis of BT-549, NIH-3T3, and cultured mammary epithelial cells treated with C, SAG (fold change from –SAG; black bars –SAG, white bars +SAG;), D, and BT-549, NIH-3T3 treated with 0.5% serum, 10% serum or 0.5% serum + TGFA (fold change from BT-549 0.5% serum) and E, 10% FBS or 10% serum + forskolin (fold change from 10% serum alone). F, Western blot for GLI3R protein levels normalized to β-Actin levels in BT-549 cells treated with 10% serum, 10% serum + forskolin, 0.5% serum or 0.5% serum + forskolin. G, Bar graph of average fold change for analysis of Gli1 expression based on quantitative real time-PCR analysis of BT-549 cells. Error bars indicate SE; P values were determined by the Student t-test (paired, two-tailed); *P<0.05. All experiments performed in duplicate (B-G).
To investigate if Hedgehog-target genes can be induced by TGFΑ in the absence of cilia we utilized the human TNBC cell line, BT-549. We analyzed expression of primary cilia in BT-549 cells and found that expression of primary cilia on these cells is rare (Fig. 3B) indicating that ciliogenesis is inhibited in BT-549 cells. To investigate if Hedgehog signaling can be induced in these cells via the canonical pathway (Hedgehog ligand), we treated BT-549 cells with a potent Hedgehog receptor agonist, Smo agonist (SAG) and performed qPCR to examine the transcriptional expression of Gli1 as a reporter of activated Hedgehog signaling. As a positive control, we also treated normal, Cilia+ cells (NIH-3T3) with SAG. As expected, SAG induces expression of Gli1 in NIH-3T3 cells (fold change = 234.5) (Fig. 3C). In contrast, our results demonstrated that treatment of BT-549 cells with SAG did not increase expression of Gli1 (fold change=1) (Fig. 3C). In addition, Ift88-/PyMT+ MECs did not respond to SAG as compared to Ift88+/PyMT+ MECs (fold change=13.7) (Fig. 3C). These results indicate that Hedgehog signaling is not activated in cilia minus BT-549 and MEC cells via SMO activation.
Next, we treated BT-549 cells with 10% serum or with TGFA and observed a statistically significant increase in Gli1 expression (Fig. 3D). In contrast, TGFA did not induce Gli1 expression in Cilia+ NIH-3T3 cells (data not shown). It was previously reported that TNF-α induces Gli1 expression through the mTOR/S6K1 pathway in esophageal adenocarcinoma cells (27). However, TNF-α did not induce expression of Gli1 in the BT-549 cells (data not shown). We also did not find high gene expression of TNF-α in the Hedgehog+/PI3K+ breast cancer patients in our bioinformatics analysis, suggesting that TNF-α is irrelevant to Hedgehog signaling in TNBC. The observation that TGFΑ induces Gli1 expression in BT-549 cells is consistent with our hypothesis and in vivo data suggesting that activation of PI3K and/or RAS signaling induces expression of Hedgehog-target genes when ciliogenesis is inhibited.
As discussed, phosphorylation of GLI3 by PKA and its subsequent proteolytic processing is enhanced by localization of the involved proteins to the primary cilium. Consistent with our findings in the Cilia-/PyMT+ mouse model (Fig. 2B), GLI3R levels were also found to be low in the Cilia- BT-549 cells (Fig. 3E; Fig. S6). Forskolin is known to activate PKA by increasing cAMP levels. We treated BT-549 cells with Forskolin to determine if this can induce processing of GLI3 into GLI3R and inhibit expression of Hedgehog-target genes. Treatment of BT-549 cells with Forskolin resulted in increased levels of GLI3R protein (Fig.3E). Treatment of BT-549 cells with Forskolin also inhibited induction of Gli1 expression (Fig.3F). These data support the hypothesis that loss of GLIR leaves Hedgehog-target genes vulnerable to non-canonical activation.
The increased expression of the Hedgehog-target gene Slug, which has been demonstrated to be a pro-migratory gene (28), prompted us to further investigate if inhibiting ciliogenesis results in increased metastasis. Ift88-/PyMT+ and Ift88+/PyMT+ MECs were injected into cleared fat pads of recipient mice. We then followed mice for 20 weeks and harvested the lungs from tumor-bearing mice to measure lung metastases. The number of metastatic lesions was quantitated and found to be more frequent in lungs from mice with Cilia-/PyMT+ tumors (Fig. 4A, B). These data indicate that inhibiting ciliogenesis in the PyMT model results in a more metastatic phenotype.
Fig. 4. Inhibition of Ciliogenesis Increases Metastasis.
A, Representative images of whole mount and H&E staining of lungs from Ift88fl/fl/PyMT+ (Ad-GFP, n=3; Ad-Cre-GFP, n=3) tumor-bearing mice. Arrowhead points to examples of tumor nodules; dashed outline highlights the whole mount tumor nodules. B, Bar graph represents quantitation of tumor nodules that metastasized from the mammary gland to the lung. Error bars indicate SE; P values were determined by the Student t-test (paired, two-tailed); *P<0.05. C, Model representing the synergy by which inhibition of ciliogenesis cooperates with RTK signaling to activate expression of Hedgehog-target genes.
DISCUSSION
We and others have documented that ciliogenesis is inhibited in breast cancer; however, its biological consequence is not understood. The findings in this report demonstrate for the first time that inhibition of ciliogenesis promotes breast cancer progression to a more aggressive and metastatic disease. In support of the functional importance of inhibition of ciliogenesis in human breast cancer, we found that inhibition of ciliogenesis resulted in increased Hedgehog-target gene expression.
While it is well established that Hedgehog signaling plays a role in the progression of breast cancers to a more aggressive, metastatic disease, and is associated with TNBC and decreased survival; it is not fully understood how breast cancers activate Hedgehog-target gene expression in the absence of cilia. Indeed, consistent with findings in pancreatic, prostate and esophageal cancer (29–31), we showed that breast cancer cells do not activate Hedgehog-target gene expression through the canonical pathway, demonstrating that Hedgehog-target gene expression was not dependent on SMO activity. Using a genetically engineered mouse model of breast cancer, a human breast cancer cell line, pharmacologic treatment and interrogation of patient datasets, we provide data that suggests a possible mechanism for how breast cancers activate Hedgehog-target gene expression (Model, Fig. 4C). We have identified that when ciliogenesis was inhibited, breast cancer cells lose the ability to process the repressor form of the GLI transcription factor (GLIR). Our data also demonstrated that pharmacologic rescue of GLI3R expression resulted in decreased expression of Hedgehog-target genes in breast cancer cells. Our results are consistent with results published from mouse models of basal cell carcinoma and medulloblastoma (32, 33). In these studies, the authors demonstrated that exogenous expression of a constitutively activated form of GLIA resulted in increased tumorigenesis when ciliogenesis was inhibited. They further demonstrated that GLIR expression was decreased when ciliogenesis was inhibited and concluded that the offset balance between GLIA and GLIR was responsible for increased expression of Hedgehog-target genes and the observed increase in tumorigenesis. Further studies are needed to determine if GLIA is required for activation of Hedgehog-target genes in breast cancer.
In this study, we further investigated the mechanism of activation of non-canonical Hedgehog signaling in breast cancer. Unlike the cilia/Hedgehog studies modeled in basal cell carcinoma and medulloblastoma, expression of a mutant, constitutively active form of GLI has not been reported in human breast cancer and is therefore unlikely to account for increased Hedgehog signaling in breast cancer. Our bioinformatic analysis revealed that the PI3K and RAS signaling pathways, as well as their upstream activators EGFR and TGFA, are co-expressed in Hedgehog-positive human breast cancers. Based on our genetic data and its relevance in human disease we hypothesized that PI3K and RAS signaling provide the necessary cross-talk for increased expression of Hedgehog-target genes. We utilized cilia-negative human breast cancer cells and demonstrated that TGFΑ induced Hedgehog-target gene expression. Importantly, TGFΑ could not induce Hedgehog-target gene expression in cilia-positive cells. This finding further supports the biological importance of inhibition of ciliogenesis in breast cancer. What is not yet clear is if the TGFΑ-induced expression of Hedgehog-target genes is dependent on activation of GLI1 or GLI2. However, our results do indicate that Gli2 expression was increased in the Cilia-/PyMT+ tumors. It is widely accepted in the literature that GLI2 require posttranslational modification to become activated and that in normal cells cilia are essential for this posttranslational modification to occur (18, 19). It is possible that TGFΑ activates the PI3K and/or RAS pathway and this, in turn, leads to post-translational modification of GLI into its activated form. Indeed, treatment of breast cancer cell lines with specific chemical and genetic inhibitors of the PI3K/AKT pathway resulted in a significant decrease in Hedgehog signaling (34). A second paper demonstrated that in Esophageal adenocarcinoma, the PI3K/AKT pathway promoted the posttranslational processing (phosphorylation) of GLI transcription factors into their activated form and that this was dependent on mTOR/S6K1 activation (27). Our findings facilitate a better understanding of non-canonical activation of Hedgehog signaling in breast cancer by demonstrating for the first time the importance of inhibition of ciliogenesis for Hedgehog-target gene expression. A deeper exploration of the mechanism by which TGFΑ induces Hedgehog-target gene expression in cilia-negative cells is ongoing.
As discussed, we provide evidence that inhibition of ciliogenesis resulted in non-canonical activation of Hedgehog signaling in breast cancer. The observed increased expression of Hedgehog-target genes in breast cancer cells was not dependent on the classical expression of Hedgehog ligands or the activation of SMO. This is an important finding because the majority of Hedgehog pathway inhibitors that have been designed and that are being used clinically target canonical Hedgehog signaling through inhibition of Hedgehog ligands and SMO (3). As discussed, recent literature supports the conclusion that targeting canonical Hedgehog signaling is ineffective in the treatment of Hedgehog-positive breast cancer cells. Understanding the mechanisms by which inhibition of ciliogenesis cooperates with PI3K and/or RAS pathways to activate non-canonical Hedgehog signaling will provide novel treatment strategies. Pharmacologically re-expressing primary cilia may be a novel approach to inhibition of the Hedgehog pathway in breast cancers. Indeed, in vitro studies have shown that pharmacological inhibition of the Aurora A Kinase/HDAC6 cascade can result in re-expression of cilia in cholangiocarcinoma and ovarian cancer cell lines (35, 36). Others have also demonstrated that ciliogenesis can be turned on in breast cancer cell lines by knockdown of Nek2 or Kif24 (37). Our results also invite speculation regarding Hedgehog and PI3K/RAS combination therapy for the treatment of TNBC and for avoiding drug resistance frequently seen with single-agent targeting.
Supplementary Material
IMPLICATIONS.
These findings change the way we understand how cancer cells turn on a critical signaling pathways and a provide rationale for developing novel therapeutic approaches to target non-canonical Hedgehog signaling for the treatment of breast cancer.
ACKNOWLEDGEMENTS
The authors would like to acknowledge the University of Arizona Cancer Center core facilities including the Tissue Acquisition and Cellular/Molecular Analysis Service for tissue processing, and H&E staining, the Genomics Shared Service for assistance with real-time RT-PCR and the Experimental Mouse Shared Service for help with in vivo studies.
FINANCIAL SUPPORT. National Cancer Institute of the National Institutes of Health award numbers P30 CA023074 (KMM) and T32 CA009213 (NBH), National Institute of Child Health and Human Development of the National Institutes of Health award number R00 HD056965 (KMM), AACR Institutional Research Grant number 74-001-34-IRG from the American Cancer Society (KMM), and the Better Than Ever Foundation (KMM).
list of abbreviations used:
- Smo
Smoothened
- Ptch
Patched
- IFT
Intraflagellar transport
- PyMT
Polyoma Middle T
- H&E
Hematoxylin and Eosin
- CIS
carcinoma in situ
- INV
invasive cancer
- Ac-Tub
acetylated tubulin
- γ-tub
γ-tubulin
- TNBC
triple negative breast cancer
- RTKs
receptor tyrosine kinases
- TGFΑ
transforming growth factor receptor alpha
- SAG
Smoothened agonist
- PKA
protein kinase A
- RT
room temperature
- IHC
immunohistochemistry
- ICC
Immunocytochemistry
Footnotes
Conflict of Interest. The authors declare that they have no competing interests.
REFERENCES
- 1.ACS. Global Cancer Facts and Figures 2nd Edition. In: Society AC, editor. Atlanta, GA: American Cancer Society; 2011. [Google Scholar]
- 2.McDermott KM, Liu BY, Tisty TD, Pazour GJ. Primary Cilia Regulate Branching Morphogenesis during Mammary Gland Development. Current Biology. 2010;20(8):731–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hassounah NB, Bunch TA, McDermott KM. Molecular pathways: the role of primary cilia in cancer progression and therapeutics with a focus on Hedgehog signaling. Clin Cancer Res. 2012;18(9):2429–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Briscoe J, Therond PP. The mechanisms of Hedgehog signaling and its roles in development and disease. Nat Rev Mol Cell Biol. 2013. 14(7):416–29. [DOI] [PubMed] [Google Scholar]
- 5.Nobutani K, Shimono Y, Yoshida M, Mizutani K, Minami A, Kono S, et al. Absence of primary cilia in cell cycle-arrested human breast cancer cells. Genes to Cells. 2014;19(2):141–52. [DOI] [PubMed] [Google Scholar]
- 6.Yuan K, Frolova N, Xie Y, Wang DZ, Cook L, Kwon YJ, et al. Primary Cilia Are Decreased in Breast Cancer: Analysis of a Collection of Human Breast Cancer Cell Lines and Tissues. Journal of Histochemistry & Cytochemistry. 2010;58(10):857–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menzl I, LeBeau L, Pandey R, Hassounah NB, W Li F, Nagle R, et al. Loss of primary cilia occurs early in breast cancer development. Cilia. 2014;3(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haycraft CJ, Zhang Q, Song B, Jackson WS, Detloff PJ, Serra R, et al. Intraflagellar transport is essential for endochondral bone formation. Development. 2007;134(2):307–16. [DOI] [PubMed] [Google Scholar]
- 9.Lin FM, Hiesberger T, Cordes K, Sinclair AM, Goldstein LSB, Somlo S, et al. Kidney-specific inactivation of the KIF3A subunit of kinesin-II inhibits renal ciliogenesis and produces polycystic kidney disease. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(9):5286–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prater M, Shehata M, Watson CJ, Stingl J. Enzymatic dissociation, flow cytometric analysis, and culture of normal mouse mammary tissue. Methods in molecular biology (Clifton, NJ). 2013;946:395–409. [DOI] [PubMed] [Google Scholar]
- 11.Liu X, Ory V, Chapman S, Yuan H, Albanese C, Kallakury B, et al. ROCK Inhibitor and Feeder Cells Induce the Conditional Reprogramming of Epithelial Cells. American Journal of Pathology. 2012;180(2):599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hassounah NB, Nagle R, Saboda K, Roe DJ, Dalkin BL, McDermott KM. Primary Cilia Are Lost in Preinvasive and Invasive Prostate Cancer. PLoS One. 2013;8(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marszalek JR, Ruiz-Lozano P, Roberts E, Chien KR, Goldstein LSB. Situs inversus and embryonic ciliary morphogenesis defects in mouse mutants lacking the KIF3A subunit of kinesin-II. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(9):5043–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cano DA, Murcia NS, Pazour GJ, Hebrok M. orpk mouse model of polycystic kidney disease reveals essential role of primary cilia in pancreatic tissue organization. Development. 2004;131(14):3457–67. [DOI] [PubMed] [Google Scholar]
- 15.Mitchell EH, Serra R. Normal mammary development and function in mice with Ift88 deleted in MMTV- and K14-Cre expressing cells. Cilia. 2014;3(1):4-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin EY, Jones JG, Li P, Zhu UY, Whitney KD, Muller WJ, et al. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. American Journal of Pathology. 2003;163(5):2113–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fluck MM, Schaffhausen BS. Lessons in signaling and tumorigenesis from polyomavirus middle T antigen. Microbiol Mol Biol Rev. 2009;73(3):542–63, Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu A, Wang B, Niswander LA. Mouse intraflagellar transport proteins regulate both the activator and repressor functions of Gli transcription factors. Development. 2005;132(13):3103–11. [DOI] [PubMed] [Google Scholar]
- 19.May SR, Ashique AM, Karlen M, Wang B, Shen Y, Zarbalis K, et al. Loss of the retrograde motor for IFT disrupts localization of Smo to cilia and prevents the expression of both activator and repressor functions of Gli. Dev Biol. 2005;287(2):378–89. [DOI] [PubMed] [Google Scholar]
- 20.Templeton AJ, Diez-Gonzalez L, Ace O, Vera-Badillo F, Seruga B, Jordan J, et al. Prognostic relevance of receptor tyrosine kinase expression in breast cancer: A meta-analysis. Cancer Treat Rev. 2014. [DOI] [PubMed]
- 21.Rodriguez-Viciana P, Collins C, Fried M. Polyoma and SV40 proteins differentially regulate PP2A to activate distinct cellular signaling pathways involved in growth control. Proc Natl Acad Sci U S A. 2006;103(51):19290–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loboda A, Nebozhyn M, Klinghoffer R, Frazier J, Chastain M, Arthur W, et al. A gene expression signature of RAS pathway dependence predicts response to PI3K and RAS pathway inhibitors and expands the population of RAS pathway activated tumors. BMC Med Genomics. 2010;3:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loboda A, Nebozhyn M, Cheng C, Vessey R, Huang P, Dai H, et al. Biomarker discovery: identification of a growth factor gene signature. Clin Pharmacol Ther. 2009;86(1):92–6. [DOI] [PubMed] [Google Scholar]
- 24.Shi T, Mazumdar T, Devecchio J, Duan ZH, Agyeman A, Aziz M, et al. cDNA microarray gene expression profiling of hedgehog signaling pathway inhibition in human colon cancer cells. PLoS ONE. 2010;5(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tao Y, Mao J, Zhang Q, Li L. Overexpression of Hedgehog signaling molecules and its involvement in triple-negative breast cancer. Oncol Lett. 2012;2(5):995–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Im S, Choi HJ, Yoo C, Jung JH, Jeon YW, Suh YJ, et al. Hedgehog related protein expression in breast cancer: gli-2 is associated with poor overall survival. Korean J Pathol. 2013;47(2):116–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Ding Q, Yen CJ, Xia W, Izzo JG, Lang JY, et al. The crosstalk of mTOR/S6K1 and Hedgehog pathways. Cancer Cell. 2012;21(3):374–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen H, Zhu G, Li Y, Padia RN, Dong Z, Pan ZK, et al. Extracellular Signal-Regulated Kinase Signaling Pathway Regulates Breast Cancer Cell Migration by Maintaining slug Expression. Cancer Research. 2009;69(24):9228–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nolan-Stevaux O, Lau J, Truitt ML, Chu GC, Hebrok M, Fernandez-Zapico ME, et al. GLI1 is regulated through Smoothened-independent mechanisms in neoplastic pancreatic ducts and mediates PDAC cell survival and transformation. Genes & Development. 2009;23(1):24–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang JX, Lipinski R, Shaw A, Gipp J, Bushman W. Lack of demonstrable autocrine hedgehog signaling in human prostate cancer cell lines. Journal of Urology. 2007;177(3):1179–85. [DOI] [PubMed] [Google Scholar]
- 31.Zhou J, Zhu G, Huang J, Li L, Du Y, Gao Y, et al. Non-canonical GLI1/2 activation by PI3K/AKT signaling in renal cell carcinoma: A novel potential therapeutic target. Cancer Lett. 2016;370(2):313–23. [DOI] [PubMed] [Google Scholar]
- 32.Wong SY, Seol AD, So PL, Ermilov AN, Bichakjian CK, Epstein EH Jr., et al. Primary cilia can both mediate and suppress Hedgehog pathway-dependent tumorigenesis. Nat Med. 2009;15(9):1055–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han YG, Kim HJ, Dlugosz AA, Ellison DW, Gilbertson RJ, Alvarez-Buylla A. Dual and opposing roles of primary cilia in medulloblastoma development. Nat Med. 2009;15(9):1062–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramaswamy B, Lu Y, Teng KY, Nuovo G, Li X, Shapiro CL, et al. Hedgehog signaling is a novel therapeutic target in tamoxifen-resistant breast cancer aberrantly activated by PI3K/AKT pathway. Cancer Res. 2012;72(19):5048–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Egeberg D, Lethan M, Manguso R, Schneider L, Awan A, Jorgensen T, et al. Primary cilia and aberrant cell signaling in epithelial ovarian cancer. Cilia. 2012;1(15). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gradilone S, Radtke B, Bogert P, Huang B, Gajdos G, Larusso N. HDAC6 inhibition restores ciliary expression and decreases tumor growth. Cancer Research. 2013. [DOI] [PMC free article] [PubMed]
- 37.Kim S, Lee K, Choi JH, Ringstad N, Dynlacht BD. Nek2 activation of Kif24 ensures cilium disassembly during the cell cycle. Nat Commun. 2015;6:8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.