Transcriptiomic and clinico-pathologocal comparison of mediastinal gray zone lymphoma to related disorders documents intrinsic heterogeneity and refines features relevant to diagnosis and therapy.
Abstract
Mediastinal gray zone lymphoma (MGZL) has immunopathologic features between classical Hodgkin lymphoma (cHL) and primary mediastinal thymic B-cell lymphoma (PMBL), leading to uncertainty regarding its biological relationship to these entities. We performed gene expression profiling from patients with MGZL (20), cHL (18), and PMBL (17) and show MGZL clusters between cHL and PMBL. Expression signatures reveal germinal B-cell and IFN regulatory factor 4 (IRF4) signatures were relatively low in MGZL and cHL compared with PMBL, indicating downregulation of the B-cell program in MGZL, a hallmark of cHL. T-cell and macrophage signatures were higher in MGZL and cHL compared with PMBL, consistent with infiltrating immune cells, which are found in cHL. The NFκB signature was higher in MGZL than PMBL, and like cHL, MGZL and PMBL express NFκB inducing kinase (NIK), indicating noncanonical signaling. These findings indicate that while MGZL has distinctive clustering, it is biologically closer to cHL.
Significance:
We performed comparative gene expression analysis of MGZL, cHL, and PMBL and show most MGZL cases are biologically closer to cHL. MGZL has significantly higher tumor cell density than cHL and greater NFκB activation compared with PMBL, which may explain its greater treatment resistance compared with cHL and PMBL.
This article is highlighted in the In This Issue feature, p. 127
Introduction
Mediastinal gray zone lymphoma (MGZL) is a pathologic entity with morphologic and/or IHC features intermediate between primary mediastinal B-cell lymphoma (PMBL) and mediastinal classical Hodgkin lymphoma (cHL; refs. 1,2,3,4). Historically, these cases were sometimes termed “Hodgkin-like anaplastic large cell lymphoma”, but a connection to what we recognize today as anaplastic large-cell lymphoma was not confirmed (5, 6). As a group, PMBL and cHL are hypothesized to derive from a thymic B cell and share molecular features including gains as well as amplification of the REL/BCL11A locus (2p16.1), JAK2/PDL2 locus (9p24.1), and CIITA locus (16p13.13), which also occur in MGZL (7,8,9,10). PMBL and cHL also have overlapping gene expression profiles, suggesting they lie along a pathobiological continuum with MGZL described as the “missing link” between these entities (2, 8). Methylation profiling of MGZL showed a unique signature, but also shared components with cHL and PMBL, further supporting its association with these entities (9). MGZL also has overlapping clinical features with PMBL and cHL, raising the question of where it lies within the biological and clinical spectrum of mediastinal B-cell lymphomas. To address these questions, we performed gene expression profiling of MGZL, cHL, and PMBL and compared expression signatures of immune cells, NFκB survival pathway, and B-cell differentiation.
Results
Patient Characteristics and Pathobiology
Fifty-five tumor samples comprising 20 MGZL, 18 cHL, and 17 PMBL were analyzed (Table 1). Of these biopsies, 50 were pretreatment, 2 MGZL had unknown treatment status, and 3 cHL were at relapse. The median (range) patient age of the respective groups was 34 (17–77), 28 (19–72) and 33 (17–52) years, and 15 (75%), 8 (44%), and 10 (59%) were male. Patients with cHL and PMBL had features consistent with these diagnoses, whereas patients with MGZL had histologic and/or phenotypic features intermediate between cHL and PMBL according to the World Health Organization Classification (11).
Table 1.
Clinical and pathologic characteristics
| Characteristics | MGZL | cHL | PMBL |
|---|---|---|---|
| Total patients | 20 | 18 | 17 |
| Male gender | 15 (75%) | 8 (44%) | 10 (59%) |
| Median age (years) | 34 | 28 | 33 |
| Age range | 17–77 | 19–72 | 17–52 |
| Mediastinal tumor present | 16/17a (94%) | 18 (100%) | 17 (100%) |
| Pretreatment biopsy | 18/18b (100%) | 15/18 (83%) | 17/17 (100%) |
| MGZL Tumor morphology | |||
| PMBL-Like | 5/20 (25%) | NA | NA |
| cHL-Like | 5/20 (25%) | NA | NA |
| Composite PMBL/cHL-like | 1/20 (5%) | NA | NA |
| Intermediate | 9/20 (45%) | NA | NA |
| Immunophenotype | |||
| CD20 | 19/20 (95%) | 3/16 (19%) | 17 (100%) |
| CD30 | 20/20 (100%) | 18/18 (100%) | 12/14 (85%) |
| CD15 | 16/20 (80%) | 17/18 (95%) | 1/11 (9%) |
| EBER in situ hybridization | 0/11 (0%) | NA | NA |
| Percent tumor cell quartiles | |||
| <25 | 3/17 (18%) | 8/18 (44%) | 3/17 (18%) |
| 25–50 | 4/17 (23.5%) | 7/18 (39%) | 3/17 (18%) |
| 50–75 | 6/17 (35%) | 2/18 (11%) | 2/17 (12%) |
| 75–100 | 4/17 (23.5%) | 1/18 (6%) | 9/17 (53%) |
Abbreviation: NA, not applicable.
aThree cases unknown.
bTwo cases unknown.
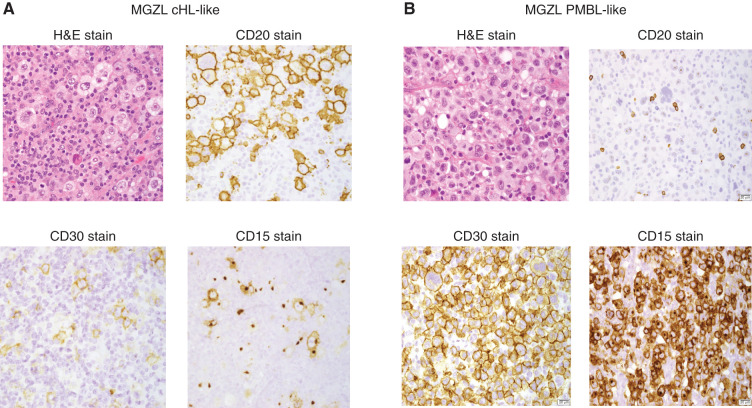
The diagnosis of MGZL is dependent on a discordance between morphology and phenotype, which is inconsistent with classification as either cHL or PMBL. In general, MGZL is relatively rich in tumor cells compared with cHL, and although Hodgkin Reed-Sternberg (HRS) cells may be present, they lack a typical inflammatory background and phenotypically have a stronger B-cell phenotype with expression of CD20 and CD79 compared with cHL. MGZL tumors may have a predominant morphology, which allows further categorization of these tumors as PMBL-like, cHL-like, or rarely composite with two separate components; however, cases with an intermediate morphology are more typically seen. MGZL cHL-like tumors show Reed-Sternberg cells in an inflammatory background with relatively robust CD20 staining (Fig. 1A), whereas cases predominantly composed of large cells as seen in PMBL may lack or only partially express B-cell markers such as CD20, but often strongly express CD30 and CD15 (Fig. 1B). In the current series of 20 MGZL cases, the categories of cHL-like, PMBL-like, and composite were present in 25%, 25%, and 5%, respectively, with 45% of cases showing an intermediate morphology (Table 1). Almost all MGZL cases showed CD20 and CD15 expression and all were positive for CD30 (Table 1). We also assessed EBV encoded RNA in situ (EBER) in 11 MGZL cases and found no positive cases.
Figure 1.
MGZL IHC. A, MGZL cHL-like tumors show Reed-Sternberg cells in an inflammatory background on hematoxylin and eosin (H&E) stain along with robust CD20 and low CD15- and CD30-positive cells. B, MGZL PMBL-like tumors show diffuse large B cells on H&E stain but with robust CD30- and CD15-positive cells and low CD20.
We determined the percent distribution of tumor cells in our MGZL, cHL, and PMBL cohorts as a control for tumor cell density (Table 1). As expected, PMBL had significantly more tumor cells compared with cHL with the median quartile being 75%–100% and 25%–50% cells in each group, respectively (P = 0.003). MGZL also showed significantly higher tumor cells density compared with cHL with a median quartile of 50% to 75% and 25% to 50% cells, respectively (P = 0.015). However, there was no significant difference in tumor cell density between MGZL and PMBL (P = 0.455).
Gene Expression
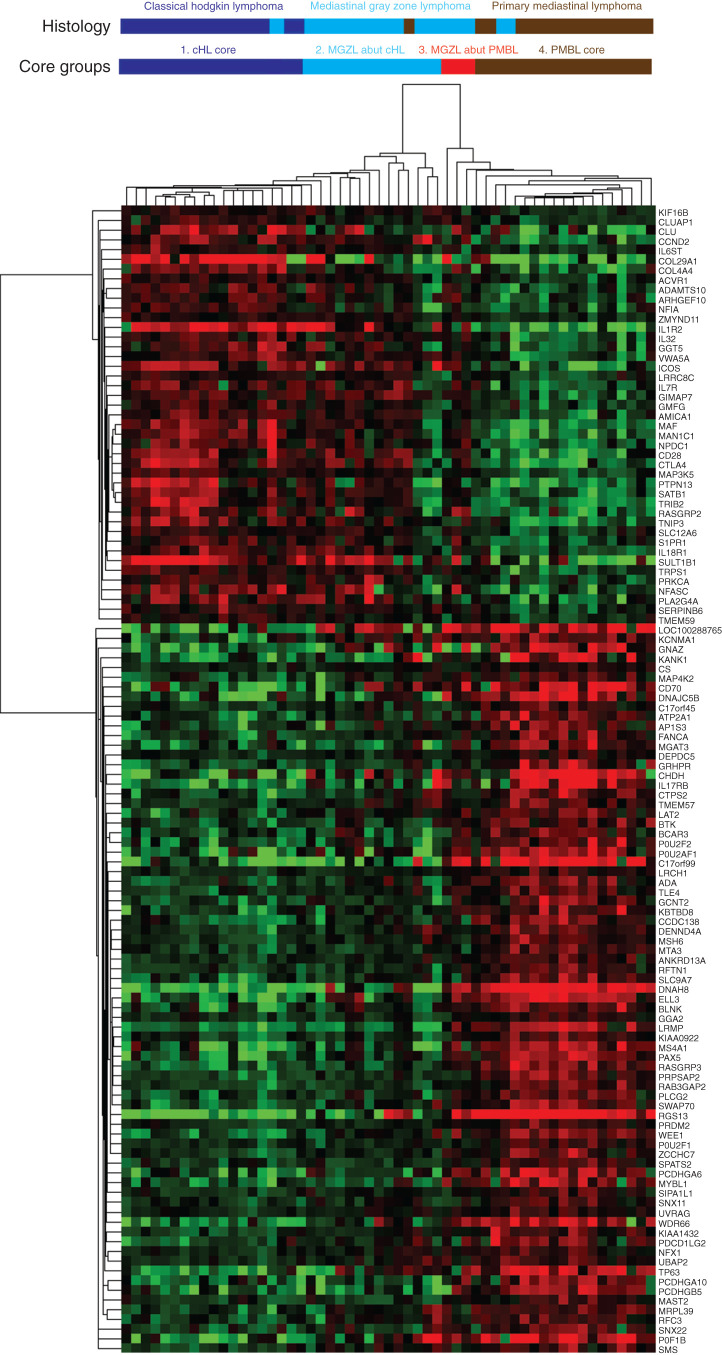
Gene expression profiling was performed on all 55 biopsy samples from patients with MGZL, cHL, and PMBL. On the basis of the hypothesis that MGZL is biologically intermediate between cHL and PMBL, we first analyzed expression differences between cHL and PMBL and identified 103 genes that were significant at P < 10−6 (FDR = 0.0001). The resultant clustering generally placed the MGZL cases between core clusters for cHL and PMBL (Fig. 2). The cHL core, termed cluster group 1, includes all cHL samples and one MGZL, and the PMBL core, termed cluster group 4, includes all but one PMBL and two MGZL cases. The MGZL cases broke down into two cluster groups. One abutted the cHL core, termed cluster group 2, and includes most MGZL samples and one outlier PMBL sample that had an extremely low tumor content, and one abutted the PMBL core, termed cluster group 3, and includes 4 MGZL samples.
Figure 2.
Gene expression clustering of 103 genes in tumor samples. MGZL clustered between cHL and PMBL. Four cluster groups were identified and included a core cHL group (cluster 1), a core PMBL group (cluster 4), and two MGZL groups divided by their abutment to the cHL (cluster 2) and PMBL (cluster 3) core groups. MGZL cluster groups 2 and 3 had gene expression closer to cHL and PMBL, respectively.
This division represents the most stable grouping of samples and recognizes two MGZL clusters distinguished by proximity to cHL or PMBL cases. On the basis of the pathologic recognition of MGZL as cHL-like, PMBL-like, or a composite of the two, we looked at their distribution in the array (Fig. 2). Among 5 cHL-like cases, one was at the edge of cluster group 1 (cHL core), 3 were in cluster group 2 (cHL abutting), and one was in cluster group 3 (PMBL abutting). Of the five cases with PMBL-like features, 3 were in cluster group 3 (PMBL abutting), one was in cluster group 4 (PMBL core), and one in cluster group 2 (cHL abutting). The single case that showed composite features clustered within cluster group 4 (PMBL core), raising the suggestion that the PMBL-like component was overrepresented in the biopsy specimen. The majority of cases with intermediate features were distributed in cluster groups 2 and 3.
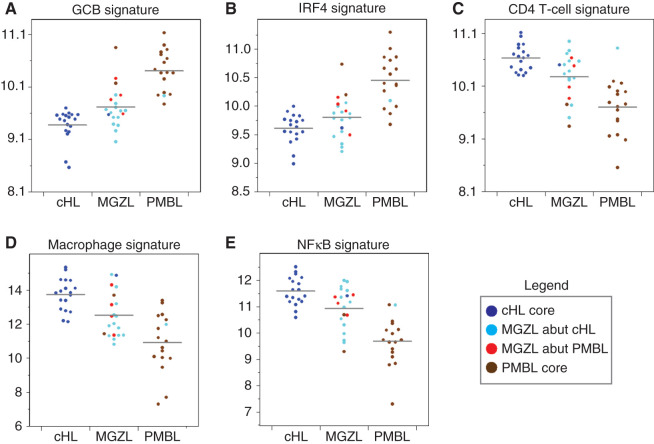
The gene expression signature revealed 43 genes more highly expressed in cHL compared with PMBL (Fig. 2). We explored signatures of cellular differentiation and found that like cHL, MGZL had lower expression of germinal center B-cell genes (GCB) and IFN regulatory factor 4 (IRF4) genes, indicating these tumors down regulate B-cell differentiation relative to PMBL (Fig. 3A and B). Genes associated with infiltrating T cells, including IL6ST, CTLA4, CD28, and ICOS, and immune regulation, including IL1R2, IL32, IL7R, and TNIP3, were of interest because of their association with the inflammatory background found in cHL (11). MGZL tumors showed variable expression of these genes with cluster group 2 abutting cHL showing relatively high levels and cluster group 3 abutting PMBL showing low expression. We also looked at two immune-associated signatures defined as a thymic CD4 T-cell signature and a macrophage cell signature (Fig. 3C and D). In both instances, the median gene expression of MGZL tumors was intermediate between cHL and PMBL, but in many cases closer to cHL. These results show gene expression associated with infiltrating immune cells in MGZL is distributed among cases abutting PMBL and cHL and include cases that are cHL-like and PMBL-like.
Figure 3.
MGZL, cHL, and PMBL gene expression signatures are shown for GCB cells (A), IRF4 (B), CD4 T cells (C), macrophages (D), and NFκB (E). The distribution of the cluster groups within each signature and cell type is color coded as shown in the legend.
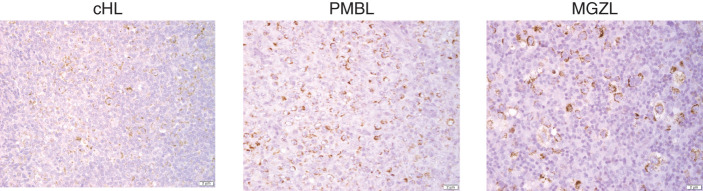
Finally, we explored the nuclear factor κB (NFκB) signaling pathway that regulates cell survival and is expressed in both cHL and PMBL. NFκB expression was highest in cHL with most MGZL cases showing greater expression than PMBL, including those in cluster group 3 that abutted the PMBL core (Fig. 3E). A recent study showed increased copies of the NFκB inducing kinase (NIK) activates the noncanonical NFκB pathway in cHL and accounts for much of the NFκB pathway activation in cHL (12). We also found increased NIK expression by IHC in our MGZL (5/6 positive) and PMBL (5/6 positive) cases, indicating they share a similar mechanism of NFκB activation (Fig. 4).
Figure 4.
Representative IHC of NIK staining in cHL, PMBL, and MGZL. Analysis of NIK staining in MGZL and PMBL showed 5 of 6 positives in both cohorts. Published analysis of NIK staining in cHL showed 30 of 31 positive cases (12).
Discussion
The variable immunopathologic features and distinguishing clinical outcome of MGZL necessitates a greater biological understanding. Our cases show a distribution of tumor morphology that reflects the reported pathologic diversity, with a quarter each showing a PMBL-like and cHL-like morphology and the balance mostly showing an intermediate morphology. The variable morphology of MGZL is highlighted by a recent study of 139 cases where 62% and 38% of cases were identified as cHL-like and large B-cell lymphoma-like, respectively, with only two cases interpreted as having an intermediate morphology (7). This stands in contrast to our own series in which we found an intermediate morphology in 45% of cases, raising the likelihood that the discrepancy between the series reflects the difficulty in such assignments rather than a difference in biology (13). Furthermore, MGZL rarely express EBV, which was reported in 24% of cases in the above series, and not found in any of 11 cases analyzed in our series. Indeed, the presence of EBV raises the likelihood that such cases are EBV+ diffuse large B-cell lymphomas, where Hodgkin-like features are often present, and we believe they should be largely excluded from series of MGZL (11).
The frequency of CD20 and CD15 expression in 95% and 80% of our MGZL cases, respectively, points to a shared biology with PMBL and cHL. Gene expression clustering of our cases showed two stable groupings with most MGZL cases (13) abutting the cHL core and a few cases (4) abutting the PMBL core cases. Although limited in numbers, all but one MGZL case with PMBL-like features either abutted or clustered in the PMBL core, and all but one cHL-like case abutted or clustered in the cHL-core, suggesting an association between gene expression and morphology and phenotype. Of course, most MGZL cases were interpreted as intermediate and abutted the cHL core. Overall, these findings suggest that most MGZL cases have gene expression features more closely aligned with cHL than with PMBL. Notably, this gene expression distribution does not appear to be driven by tumor cell number per se based on our finding that the MGZL and PMBL cohorts have similar tumor cell densities, whereas the cHL cohort is significantly lower.
Analysis of gene expression signatures provides a more granular view of MGZL. Downregulation of the B-cell program as reflected in the GCB signature, which is a feature of cHL, was prominent in most MGZL cases with the cases abutting the PMBL core showing the least down regulation. The IRF4 signature, which is associated with B-cell differentiation and germinal center formation, was also lower in MGZL compared with PMBL and more similar to cHL. Most MGZL cases exhibit somewhat higher expression of NFκB genes compared with PMBL, and like cHL, this appears to be driven by the noncanonical pathway (12). Features of the microenvironment as determined by the T-cell and macrophage signatures were increased in MGZL, indicating another biological similarity to cHL (4).
Because of the rarity of MGZL and its biological diversity, our results are limited by relatively low sample sizes. Nonetheless, the totality of data confirms the diversity of these tumors, but also indicates that most cases are biologically closer to cHL than to PMBL. While this suggests these cases should be treated as cHL, historical data indicates such treatment produces poorer outcomes compared with those observed in cHL (5, 6). The alternative therapeutic strategy to treat these patients with regimens designed for PMBL has also been tested and shows an inferior outcome compared with PMBL, albeit still with a significant rate of cure (4, 14, 15). These data suggest that the poorer outcome of MGZL, with regimens designed for cHL or PMBL, likely reflects the presence of a higher degree of chemotherapy resistance as opposed to an issue with the therapeutic approach per se, possibly related to increased activation of NFκB relative to PMBL and higher tumor cell density relative to cHL. Thus, the totality of data suggests that most MGZL are biologically closer to cHL, but harbor biological features of PMBL and future therapy should integrate active components used in both diseases including anti-CD20 antibodies and immune checkpoint inhibitors.
Methods
Tissue Specimens
Deidentified tissue samples obtained from the Laboratory of Pathology, Center for Cancer Research, National Cancer Institute included MGZL (20 cases), cHL (18 cases), and PMBL (17 cases). Eight MGZL cases in the current series were included in a previously published study ClinicalTrials.gov (NCT00001337). The study samples were from a protocol approved by the Institutional Review Board, NCI or had approved waiver of written informed consent. Histologic review was conducted by the authors (S. Pittaluga, A. Nicolae, and E.S. Jaffe) in accordance with the World Health Organization Classification.
IHC Analysis
IHC was performed on formalin-fixed, paraffin-embedded (FFPE) tissue sections and included CD20, CD3, CD15, CD30, CD79, PAX-5, and OCT-2, and performed as described previously. Positive controls were run with each set of slides and showed appropriate staining patterns. The percentage of tumor cells was assessed by morphology and CD20, PAX5, or CD30 depending on immunoreactivity of the tumor cells. CD30 and CD15 immunoreactivity were not quantitated but scored as positive if there was any staining on the malignant cells. IHC for NIK and EBV-encoded RNA (EBER) in situ hybridization was performed on a subset of cases. All slides were independently reviewed and the scores agreed upon by joint rereview (S. Pittaluga, A. Nicolae, and E.S. Jaffe).
Gene Expression Analysis
The methods used to extract, sequence, and analyze RNA from FFPE samples to generate digital gene expression values were followed as described previously (16). An RNA library employing TruSeq (Illumina Prep Kit v2) was prepared and genes with zero signal on more than 20% of the samples were excluded from analysis.
Statistical Analysis
Two-sided t tests were used to identify genes that were differentially expressed between histologically defined cHL and PMBL samples. Genes were centered such that zero represented the midpoint between the cHL and PMBL averages. Hierarchical clustering was performed using centroid linkage with an uncentered correlation distance metric. Summarized gene expression measure for signatures were generated as follows: For each signature and each sample calculated an initial average of all genes that had 10% or fewer zero values. We then refined this list by excluding those genes that were not correlated (Pearson r > 0.25) with this signature average across all samples. The average of the remaining correlated genes for each sample was then reported as the signature gene expression for that sample. P values for signature gene expression were calculated on the basis of an F-test. An exact Cochran–Armitage test for trend was used to test the difference in the ordered categorical percent tumor cells in three sets of two pathologic groups each; P values are two-tailed.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors' Contributions
S. Pittaluga: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, project administration, writing−review, and editing. A. Nicolae: Resources, validation, investigation, visualization, writing−review, and editing. G.W. Wright: Conceptualization, data curation, software, formal analysis, validation, investigation, methodology, writing−review, and editing. C. Melani: Resources, investigation, writing−review, and editing. M. Rochewski: Resources, investigation, writing−review, and editing. S. Steinberg: Data curation, software, formal analysis, writing−review, and editing. D. Huang: Resources, data curation, software, formal analysis, validation, methodology, writing−review, and editing. L.M. Staudt: Conceptualization, resources, data curation, formal analysis, supervision, validation, investigation, methodology, writing−review, and editing. E.S. Jaffe: Conceptualization, resources, formal analysis, investigation, methodology, writing−review, and editing. W.H. Wilson: Conceptualization, resources, data curation, formal analysis, supervision, validation, investigation, methodology, writing−original draft, project administration, writing−review, and editing.
Acknowledgments
This work was supported by grants from the Center for Cancer Research, NCI.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
#S. Pittaluga and W.H. Wilson contributed equally as the co-senior authors of this article.
Blood Cancer Discov 2020;1:155–61
References
- 1.World Health Organisation. WHO Classification of tumours of haematopoietic and lymphoid tissues (revised 4th edition). Lyon, France: International Agency for Research on Cancer; 2017. [Google Scholar]
- 2.Traverse-Glehen A, Pittaluga S, Gaulard P, Sorbara L, Alonso MA, Raffeld M, et al. Mediastinal gray zone lymphoma: the missing link between classic Hodgkin's lymphoma and mediastinal large B-cell lymphoma. Am J Surg Pathol 2005;29:1411–21. [DOI] [PubMed] [Google Scholar]
- 3.Quintanilla-Martinez L, de Jong D, de Mascarel A, Hsi ED, Kluin P, Natkunam Y, et al. Gray zones around diffuse large B cell lymphoma. Conclusions based on the workshop of the XIV meeting of the European Association for Hematopathology and the Society of Hematopathology in Bordeaux, France. J Hematop 2009;2:211–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson WH, Pittaluga S, Nicolae A, Camphausen K, Shovlin M, Steinberg SM, et al. A prospective study of mediastinal gray-zone lymphoma. Blood 2014;124:1563–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pileri S, Bocchia M, Baroni CD, Martelli M, Falini B, Sabattini E, et al. Anaplastic large cell lymphoma (CD30+/Ki-1+): results of a prospective clinico-pathological study of 69 cases. Br J Haematol 1994;86:513–23. [DOI] [PubMed] [Google Scholar]
- 6.Zinzani PL, Martelli M, Magagnoli M, Zaccaria A, Ronconi F, Cantonetti M, et al. Anaplastic large cell lymphoma Hodgkin's-like: a randomized trial of ABVD versus MACOP-B with and without radiation therapy. Blood 1998;92:790–4. [PubMed] [Google Scholar]
- 7.Sarkozy C, Copie-Bergman C, Damotte D, Ben-Neriah S, Burroni B, Cornillon J, et al. Gray-zone lymphoma between cHL and large B-cell lymphoma: a histopathologic series from the LYSA. Am J Surg Pathol 2019;43:341–51. [DOI] [PubMed] [Google Scholar]
- 8.Rosenwald A, Wright G, Leroy K, Yu X, Gaulard P, Gascoyne RD, et al. Molecular diagnosis of primary mediastinal B cell lymphoma identifies a clinically favorable subgroup of diffuse large B cell lymphoma related to Hodgkin lymphoma. J Exp Med 2003;198:851–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eberle FC, Rodriguez-Canales J, Wei L, Hanson JC, Killian JK, Sun HW, et al. Methylation profiling of mediastinal gray zone lymphoma reveals a distinctive signature with elements shared by classical Hodgkin's lymphoma and primary mediastinal large B-cell lymphoma. Haematologica 2011;96:558–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eberle FC, Salaverria I, Steidl C, Summers TA, Jr, Pittaluga S, Neriah SB, et al. Gray zone lymphoma: chromosomal aberrations with immunophenotypic and clinical correlations. Mod Pathol 2011;24:1586–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016;127:2375–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ranuncolo SM, Pittaluga S, Evbuomwan MO, Jaffe ES, Lewis BA. Hodgkin lymphoma requires stabilized NIK and constitutive RelB expression for survival. Blood 2012;120:3756–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parker K, Venkataraman G. Challenges in the diagnosis of gray zone lymphomas. Surg Pathol Clin 2019;12:709–18. [DOI] [PubMed] [Google Scholar]
- 14.Dunleavy K, Grant C, Eberle FC, Pittaluga S, Jaffe ES, Wilson WH. Gray zone lymphoma: better treated like Hodgkin lymphoma or mediastinal large B-cell lymphoma? Curr Hematol Malig Rep 2012;7:241–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunleavy K, Pittaluga S, Maeda LS, Advani R, Chen CC, Hessler J, et al. Dose-adjusted EPOCH-rituximab therapy in primary mediastinal B-cell lymphoma. N Engl J Med 2013;368:1408–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmitz R, Wright GW, Huang DW, Johnson CA, Phelan JD, Wang JQ, et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N Engl J Med 2018;378:1396–407. [DOI] [PMC free article] [PubMed] [Google Scholar]