 A parthenolide-derivative with favourable pharmacokinetic properties and good activity against drug-resistant chronic lymphocytic leukaemia is reported.
A parthenolide-derivative with favourable pharmacokinetic properties and good activity against drug-resistant chronic lymphocytic leukaemia is reported.
Abstract
Parthenolide is a natural product that exhibits anti-leukaemic activity, however, its clinical use is limited by its poor bioavailability. It may be extracted from feverfew and protocols for growing, extracting and derivatising it are reported. A novel parthenolide derivative with good bioavailability and pharmacological properties was identified through a screening cascade based on in vitro anti-leukaemic activity and calculated “drug-likeness” properties, in vitro and in vivo pharmacokinetics studies and hERG liability testing. In vitro studies showed the most promising derivative to have comparable anti-leukaemic activity to DMAPT, a previously described parthenolide derivative. The newly identified compound was shown to have pro-oxidant activity and in silico molecular docking studies indicate a prodrug mode of action. A synthesis scheme is presented for the production of amine 7 used in the generation of 5f.
Introduction
Chronic lymphocytic leukaemia (CLL) is the most common adult leukaemia with >3000 new cases in the UK annually.1 Its variable clinical course ranges from stable indolent disease that does not require treatment, to rapidly progressive disease that necessitates immediate therapeutic intervention.2 Clinical response rates to current therapies are strongly influenced by genetic changes including disruption of DNA damage response (DDR) genes ATM and p53.3 Tumours with a DDR defect are refractory to chemotherapeutics because they are unable to initiate apoptosis in response to therapy-induced DNA damage. This means tumours with or that develop this DDR defect are not treatable with typical DNA-damaging chemotherapies. Furthermore, since most CLL patients are over the age of 60 (median age >70) many have comorbidities that preclude the use of aggressive chemotherapeutic regimens. This highlights a need to develop alternative, less aggressive, therapies for the treatment of CLL.4
An attractive potential therapeutic is parthenolide (PTL, 1), Fig. 1, upper. PTL (1) is a natural product isolated from feverfew (Tanacetum parthenium) and varietals thereof (Fig. 1, lower),5feverfew being so-named due to its use in traditional remedies.6 PTL (1) demonstrates effective and selective anti-CLL activity in vitro,7 and through its pro-oxidant activity, can target CLL cells independently of their p53 status.8 Furthermore, co-authors of this report9 are among those who have shown that ATM-deficiency disrupts redox homeostasis, increasing further the sensitivity of this DDR-defective subtype of CLL to PTL.10 In addition to inducing oxidative stress, PTL has been shown to target tumour cells by suppressing pro-survival and proliferation signalling through the NF-κB pathway,11 by inhibiting JAK–STAT kinase activity and thereby preventing STAT-mediated transcription of anti-apoptotic genes and by inhibiting DNMT1 and HDACs leading to activation of epigenetically silenced tumour suppressor genes.12 Furthermore, through inhibition of NF-κB, PTL is able to reverse resistance to a number of chemotherapeutics including paclitaxel, oxaliplatin, doxorubicin and metoxantrone.13 Despite numerous beneficial anti-tumour activities demonstrated in vitro, the clinical utility of PTL is limited by its poor bioavailability and pharmacokinetics.14
Fig. 1. Upper: Structure of parthenolide (PTL, 1) and dimethyl amino-PTL (DMAPT) (2a) respectively; lower flowers of feverfew and a button-like (Tanacetum parthenium ‘Flore Pleno’) feverfew varietal.
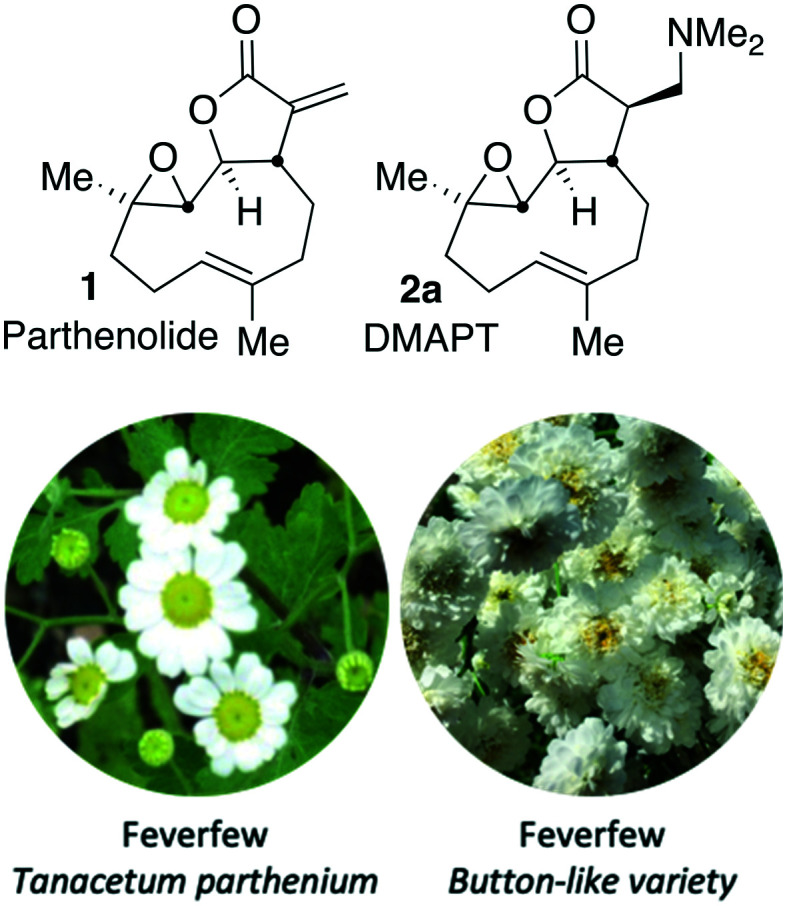
Crooks and co-workers pioneered the derivatisation of PTL and identified a promising candidate for the treatment of acute myeloid leukaemia (AML),15 a dimethyl amino-PTL (DMAPT) derivative, compound 2a depicted in Fig. 1.14,16 This derivative of PTL (1), DMAPT (2a) and related compounds, have found utility across different diseases with activities against a range of cancer-types reported.16u PTL has been shown to induce oxidative stress in cancer cells,17 and recently DMAPT (2a) was confirmed to induce oxidative stress in CLL cells.9 There are a number of reports on the use and derivatisation of PTL, with a view to addressing various cancer types,16z,18 but the only PTL derivative to be applied to CLL remains compound 2a.19 Previous studies have used molecular docking to investigate 1 and 2a which have suggested that interaction with IKKβ is a plausible mechanism for their action against haematological and solid tumours.18g
It is reasoned that retro-Michael-type chemistry, that reveals PTL at the site of interest, may be at the origin of the activity of amino-PTL derivatives, meaning amino-PTL derivatives (such as 2a) serve as excellent prodrugs for PTL delivery.16ac,ad Our interest in drug-like nitrogen-containing motifs20 prompted us to investigate analogues of DMAPT (2a) in the search for increased activity, favourable pharmacokinetic properties and minimal toxicological burdens, with the ambition of addressing drug-resistant CLL in elderly patients who may suffer more intensely from adverse effects of other therapies. Herein a synthesis cascade to explore the SAR of PTL derivatives is described and findings pertaining to these aims presented.
Results and discussion
Source of Parthenolide
Since 2013 feverfew and varietals have been cultivated at Winterbourne House and Garden,21 a UK visitor attraction, museum and heritage centre adjunct to the University of Birmingham campus (UK), located 52.4527° N. Annual crops of plant material have been obtained (Fig. 2), and PTL extracted. Plants in the late stage of flowering were found to contain the highest extractable content of PTL.
Fig. 2. Feverfew cultivation effort, Winterbourne Botanic Garden, prior to harvesting feverfew; (inset) recrystallised PTL 1.
An extraction procedure previously reported by co-authors of this report was used (detailed in ESI†)9 to access recrystalised, analytically pure, PTL (1), (see ESI†).
Derivatisation of parthenolide
The addition of nucleophilic primary and secondary amines to PTL has been reported to proceed with high diastereoselectivity via a conjugate addition to the exocyclic Michael acceptor double bond part of the lactone unit.14,16a,18f,22 The high selectivity for reaction of nucleophilic amines at the α,β-unsaturated lactone provides evidence of the compatibility of the epoxide motif contained within PTL and its derivatives thus obtained within a drug discovery programme. That is, epoxides display unwanted reactivity towards nucleophiles, and when this reactivity is unleashed in vivo off-target effects can render them undesirable motifs in medicinal chemistry.23 In related research, Long et al. reported the semi-synthesis of PTL and its cyclopropyl (in place of the epoxide part) analogue,24 the epoxide-free analogue was more stable to acid-mediated degradation, showing there may be scope to improve the drug-like properties of PTL analogues by further scaffold manipulation (although that was not probed herein).
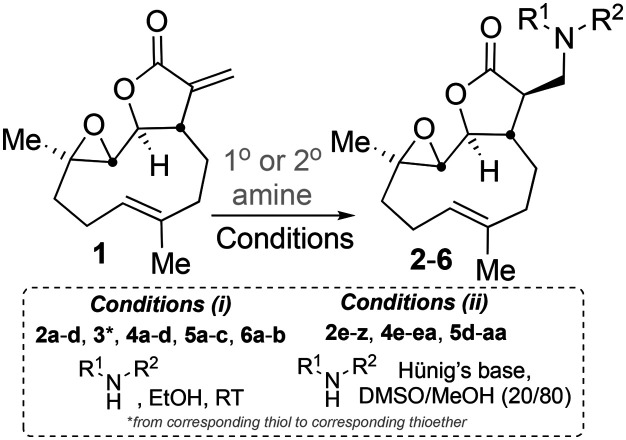
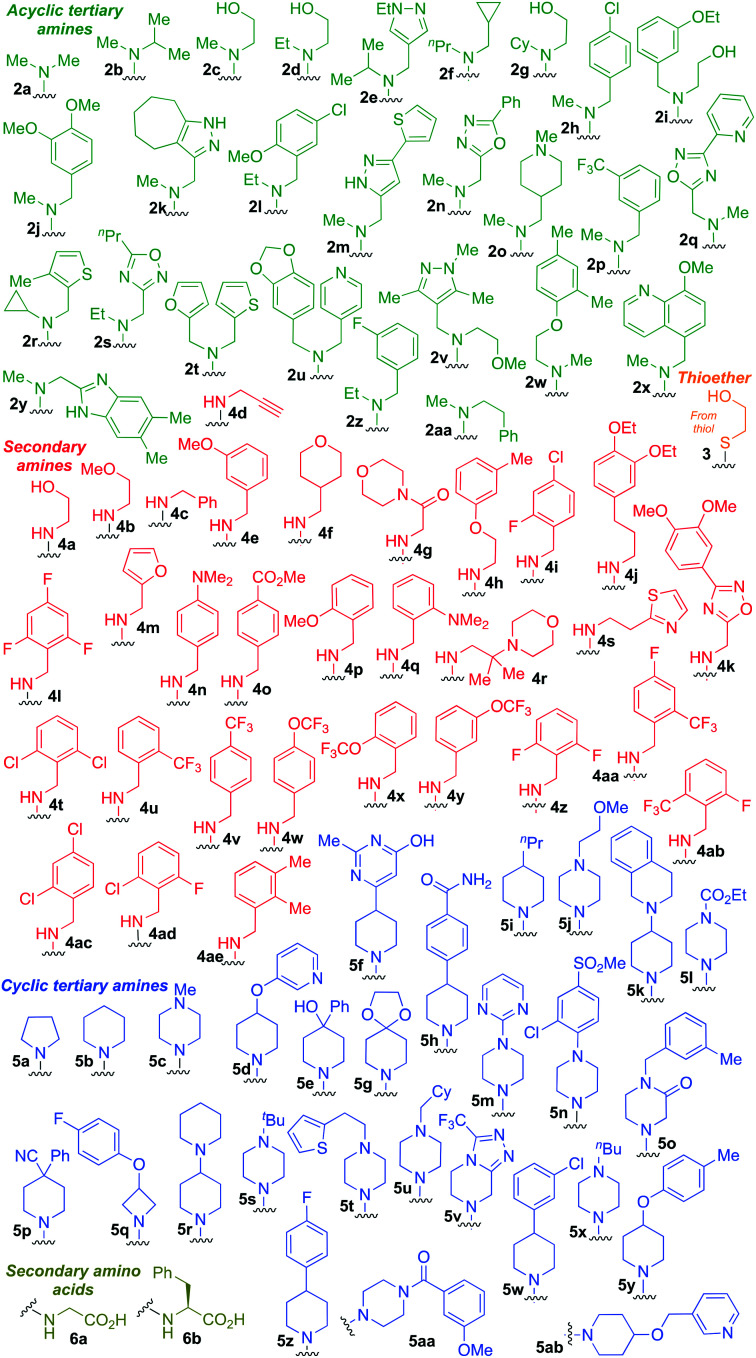
Taking advantage of the ease of derivatisation of the lactone part of parthenolide a small initial library of amino (and related) PTL derivatives was generated using an established synthetic protocol (Scheme 1 – conditions i and Fig. 3), resulting in the isolation of compounds 2a–d, 3, 4a–d, 5a–c and 6a & b. Following the successful synthesis of this small collection of PTL derivatives (Scheme 1 - conditions i), and in order to rapidly expand the library of amino-PTL derivatives available, a more high-throughput approach that allows for a library of products to be created with a range of functionalities and desirable drug-like properties was sought.25
Scheme 1. Synthesis of PTL derivatives 2 (tertiary amines), 3 (a thioether), 4 (acyclic secondary amines), 5 (cyclic tertiary amines) and 6 (amino acid derivatives). Two protocols: Conditions (i) – Addition of nucleophile in ethanol at room temperature; conditions (ii) – addition of nucleophile in dimethylsulphoxide/methanol (20/80) with Hünig's base at room temperature. All product R1 and R2 groups shown in Fig. 3.
Fig. 3. Summary of PTL derivatives' amine (and one thioether) parts, prepared according to Scheme 1.
Under slightly modified conditions from those previously employed,9,26 PTL was reacted with 120 primary or secondary amines that were selected based on calculated properties of the potential products. Used as supplied or obtained (whether neutral or as salts thereof) the amines reacted with PTL 1 in ethanol (Scheme 1 – conditions i) or in DMSO (20%) /methanol (80%) in the presence of Hünig's base (Scheme 1 – conditions ii).
The reaction mixtures were filtered and purified by reverse-phase HPLC of the 120 attempted synthesise, 76 successfully delivered products of sufficient purity (ideally >85% purity; 65 of which were obtained in >95% purity), as judged by HPLC or proton NMR spectroscopy). The materials obtained (shown in Fig. 3) were thus deemed suitable to progress to in vitro activity screening for anti CLL activity. Of the 76 compounds successfully synthesised and progressed to in vitro screening eleven have been previously reported elsewhere in the peer reviewed or patent literature, namely: 2a,16af2b,142c,14,16a 3,16a,w,27 4a,144c,16a4d,285a,14,16w,27–29 5b,14,27–29 5c27b,29b and 6a.16y
Anti-chronic lymphocytic leukaemia activity
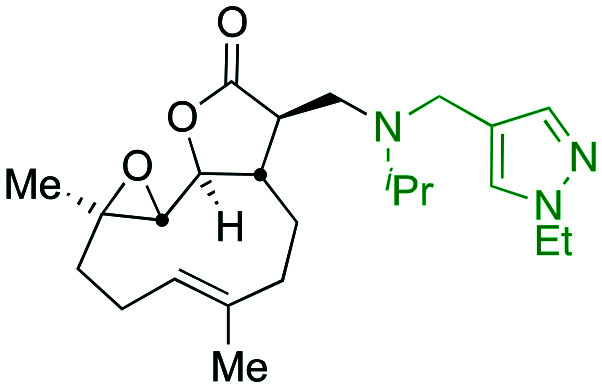
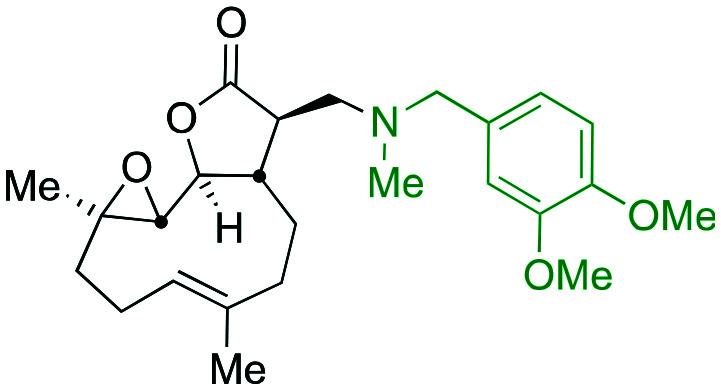
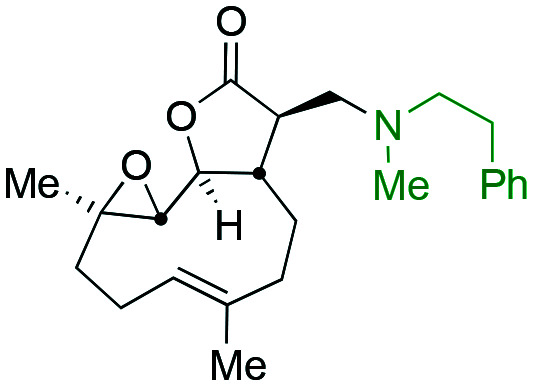
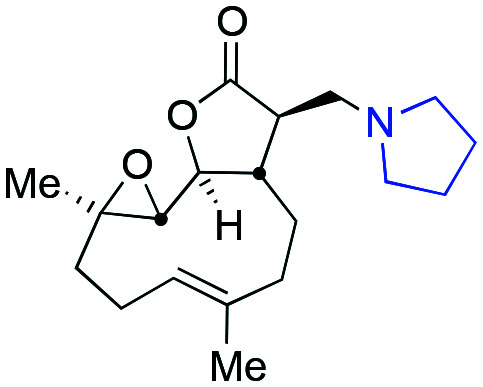
Defective p53-signalling increases genomic instability and drives tumour progression. A high frequency of p53 inactivation is found in relapsed, treatment-resistant, high risk CLL.30 The MEC1 cell line (obtained from the American type culture collection (Manassas, VA 20110 USA) expresses a mutated form of p53 and as previously shown, is resistant to clinically relevant DNA-damaging agents such as cyclophosphamide and is therefore representative of treatment-refractory, progressive CLL for which there is an urgent unmet clinical need.31 The in vitro antileukaemic activity of the PTL-derivatives against the MEC1 CLL cell line was determined using the alamarBlue® (ThermoFischer Scientific) assay and the results are summarised in Table S2.† 32 Among the active compounds 2c, 2d, 2e, 2f, 2g, 2j, 2aa, 5a, 5b, 5d, 5e, 5f, 5h, 5ab and 6b (Table S2,† entries 4 to 8, 11, 12, 29, 62, 63, 65 to 67, 69, 89 and 91 respectively) gave EC50 values <15 μM. Owing to resource availability a subset of up to ten of these compounds was prioritised for further investigation. Compounds 2d, 2e, 2j, 2aa, 5a, 5d, 5e, 5f, 5ab and 6b were selected on the basis of reasons including ease of synthetic access, diversity across the chemical space probed and novelty. Those prepared under conditions (i)Scheme 1 (2d, 5a and 6b) were available on a scale and in a purity to directly permit follow-up studies, whereas compounds prepared under conditions (ii)Scheme 1 were prepared on a small scale so were resynthesised. These seven compounds (2e, 2j, 2aa, 5d, 5e, 5f and 5ab) were isolated on >30 mg scale in high purity and progressed to more following studies.
In vitro DMPK
Compounds 2d, 2e, 2f, 2g, 2j, 2aa, 5a, 5d, 5e, 5f, 5ab and 6b were subjected to in vitro DMPK testing. Seven of the ten PTL derivatives (2e, 5a, 5b, 5d, 5f, 5ab and 6b) exhibited greater than two hours stability in aqueous solution and were retained for further investigation. The other three (2d, 2j and 2aa) were not retained for further investigation due to poor stability (T1/2 at pH 7.4: 13, 107 and 76 min, Table 1, entries 1, 3 and 4 respectively). One obvious aspect contributing to observed stability is the propensity for PTL derivatives of this type to undergo a retro-amination (reverse-Michael-type mechanism), indeed this reverse reaction is often claimed as a pro-drug mode of action.29a,33 Whether poor stability in this test is as a result of retro-amination or any other process was not determined, either way the compound of interest does not survive long enough in solution to be retained for further study. It is a reasonable assertion that activity observed for these unstable compounds could well be ascribed to free PTL generated rapidly in the assay.
Table 1. Screening cascade for favourable drug-like properties of the ten most promising compounds ([—] indicates not tested).
| Entry | Compound | EC50 ave./μM | LipE pEC50–clog P | Stability pH 7.4 T1/2/min | Microsomal stability (mouse) |
Hepatocyte stability (mouse) |
Caco-2 pH 7.4 mean Papp (10–6 cm s–1) |
||||
| A to B | A to B | CLint/μL min–1 per million cells | T 1/2/min | A to B | B to A | Efflux ratio | |||||
| 1 |
|
4.7 | 3.31 | 13 | — | — | — | — | — | — | — |
| 2 |
|
5.6 | 1.61 | >120 | 828 | 2 | — | — | — | — | — |
| 3 |
|
5.6 | 1.48 | 107 | — | — | — | — | — | — | — |
| 4 |
|
5.9 | 2.52 | 76 | — | — | — | — | — | — | — |
| 5 |
|
7.7 | 2.35 | >120 | 19 | 75 | 22.0 | 65.0 | 15.9 | 16.2 | 1.0 |
| 6 |
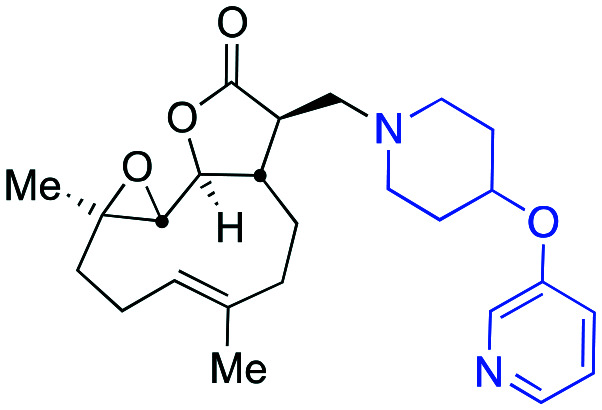
|
5.4 | 2.48 | >120 | 251 | 6 | — | — | — | — | — |
| 7 |
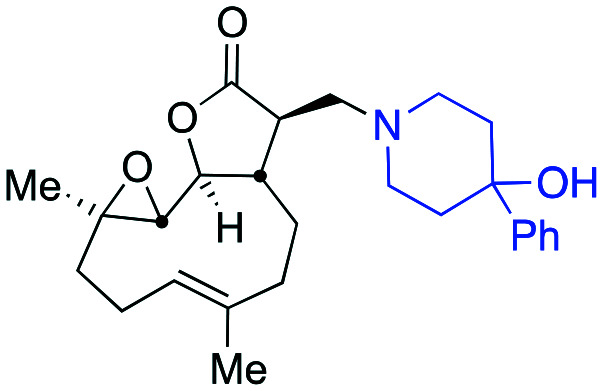
|
6.0 | 1.90 | >120 | 391 | 4 | — | — | — | — | — |
| 8 |
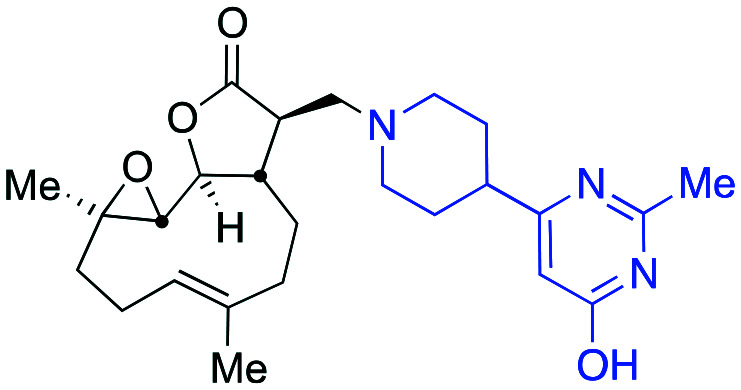
|
7.7 | 3.69 | >120 | 18 | 77 | 22.0 | 64.0 | 2.7 | 2.8 | 1.1 |
| 9 |
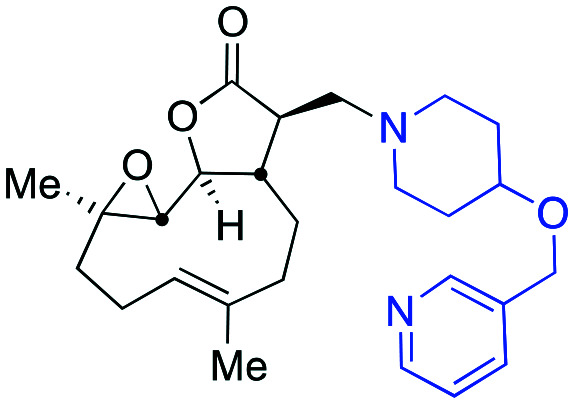
|
7.6 | 2.29 | >120 | 128 | 11 | — | — | 10.9 | 9.1 | 0.8 |
| 10 |
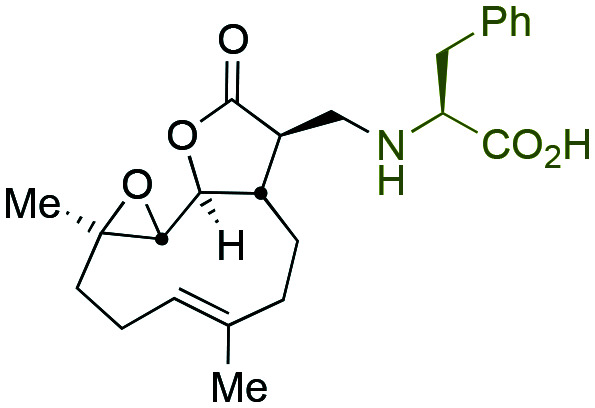
|
13.5 | 3.66 | >120 | 9 | 160 | 16.0 | 88.0 | 1.5 | 4.2 | 2.7 |
Microsomal and hepatocyte stability assays are in vitro ADME assays used to determine metabolic stability of compounds through measuring intrinsic clearance (CLint) by liver microsomes or by liver hepatocytes. Compounds 2e, 5d, 5e and 5ab (Table 1, entries 2, 6, 7 and 9) exhibited CLint of >100 μL min–1 mg–1 in the microsomal stability assay indicating rapid clearance and as such, were removed from further investigation. Compounds 5a, 5f and 6b displayed the lowest microsomal intrinsic clearances, 19 μL, 18 μL and 9 μL min–1 mg–1, respectively (Table 1, entries 5, 8 and 10) and were therefore retained for further study.34 As a secondary screen for metabolic stability, compounds 5a, 5f and 6b were subjected to the hepatocyte stability assay and this confirmed their suitability for progression to the Caco-2 permeability assay (Table 1, entries 5, 8 and 10). In order to determine the likelihood of the retained compounds being suitable for oral dosing a Caco-2 permeability assay, which predicts intestinal permeability and drug efflux, was conducted.35 Compounds 5a and 5f (Table 1, entries 5 and 8) gave efflux ratios close to unity (1.0 and 1.1 respectively), whereas compound 6b (Table 1, entry 10) gave an efflux ratio of 2.7. Thus, compounds 5a and 5f were retained and selected for in vivo pharmacokinetic studies.
The parameter lipophilic ligand efficiency (LipE) allows activity of a compound in a given assay to be tensioned against the lipophilicity of the compound. The LipE parameter has become increasingly useful and important in medicinal chemistry drug discovery decision making.36 LipE allows the quality of hits to be compared, i.e. deconvolution of activity arising due to better chemistry away from increased activity due to enhancement in lipophilicity alone. It has been claimed that consideration of LipE alongside other chemoinformatic parameters in a drug discovery programme can result in the identification of compounds with superior in vivo properties.37 As such the calculated log P values (see Table S2†) were in determination of the LipE for the active compounds detailed in Table 1. Notably, the two compounds identified as superior through the conducted screening cascade of this manuscript (5a and 5f) have the same 7.7 μM EC50 (pEC50 5.11) values against the MEC1 cell line, whereas the calculated log P values differ by more than one log unit 2.76 versus 1.42 (respectively). Thus, the corresponding LipE values of 2.35 versus 3.69 for 5a and 5f respectively in the probed MEC1 cell line assay (Table 1, entries 5 and 8) reveal that whilst compound 5f has an equivalent activity to 5a, its activity may arise from advantageous chemical features and not from lipophilicity alone.38 Coupled with the data presented in Table S2,† consideration of the LipE indicates compound 5f to offer a potential advantage in a drug discovery programme.
In vivo pharmacokinetics
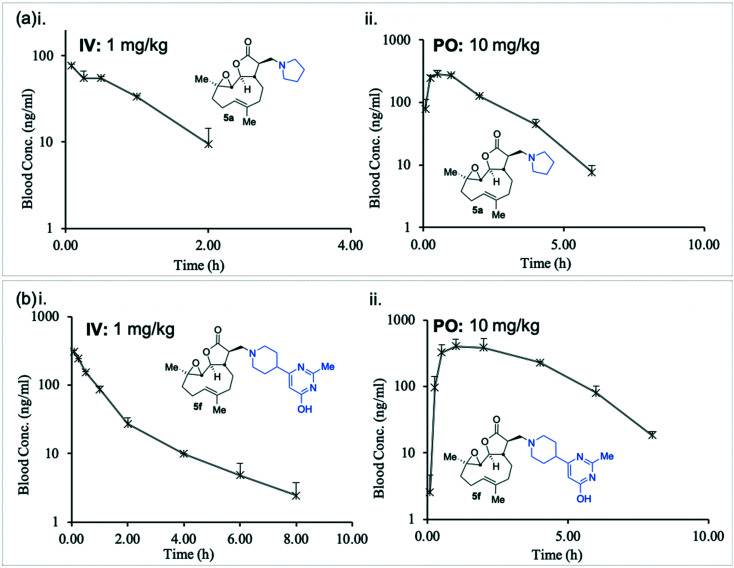
In order to determine 5a and 5f are suitable for use as pharmaceutical agents their pharmacokinetic (PK) parameters were next probed. A murine intravenous PK study of compounds 5a and 5f at 1.0 mg kg–1 was conducted and their concentrations in blood were measured with time (Fig. 4(a)i and (b)i and Table 2).
Fig. 4. Graphical representation of murine in vivo pharmacokinetic study, concentrations of 5a (a) and 5f (b) i. IV (1 mg kg–1); ii. PO (10 mg kg–1). Data summarised in Table 2.
Table 2. Data obtained from murine in vivo pharmacokinetic study, concentrations of 5a and 5f (IV (1 mg kg–1) and PO (10 mg kg–1)). Data represented graphically in Fig. 4 .
| Entry | Compound | EC50 ave./μM | Mouse PK (IV) |
Mouse PK (PO) |
||||||
| Cl/mL min–1 kg–1 | T 1/2/h | VSS/L kg–1 | UC (0-inf)/ng h mL–1 | T max/h | C max/ng mL–1 | F% | UC (0-inf)/ng h mL–1 | |||
| 1 |
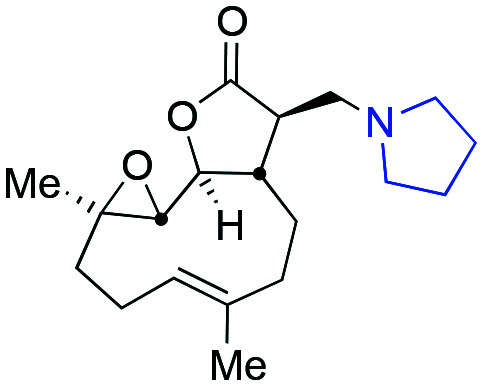
|
7.7 | 207 | 0.66 | 11.6 | 80.5 | 0.5 | 283 | 95.8 | 711 |
| 2 |
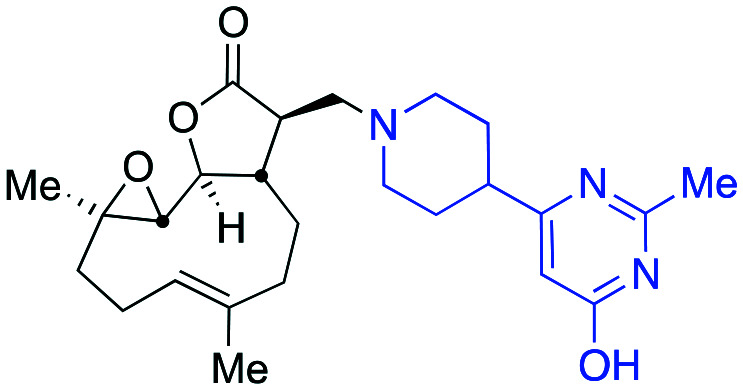
|
7.7 | 58 | 1.14 | 4.62 | 289 | 1 | 404 | 58.5 | 1696 |
Unlike the similarity between in vitro intrinsic clearance between 5a and 5f (Table 1), the in vivo clearance of 5a was nearly four times that of 5f (207 versus 58 mL min–1 kg–1, Table 2). The plasma half-life (T1/2) of 5f was approximately twice that of 5a (1.14 versus 0.66 hours, Table 2), and the total body exposure to 5f was more than three times greater than 5a (289 versus 80.5 ng h mL–1, Table 2).
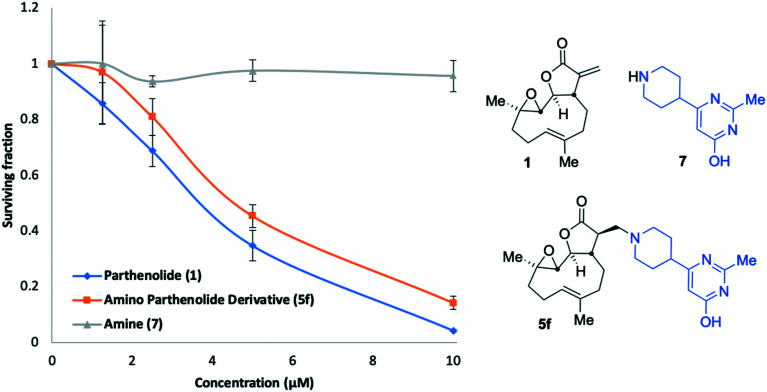
The in vivo oral bioavailability of compounds 5a and 5f was compared in a murine PK study at 10 mg kg–1 (Fig. 4(a)ii and (b)ii and Table 2). In keeping with the intravenous findings, the murine PO half-life of 5f was twice as long as 5a (1.0 versus 0.5 hours, Table 2) and the Cmax attained by 5f was approximately 40% higher than that achieved by 5a (404 versus 283 ng mL–1). The total body exposure was over two times greater for 5f than 5a (1696 versus 711 ng h mL–1, Table 2), indicating superior bioavailability. Compound 5f was therefore identified as a promising potential new agent with desirable pharmacological properties towards a therapy for drug-resistant CLL.
Comparison of the activity of 5f with 1 (PTL) and 2a (DMAPT)
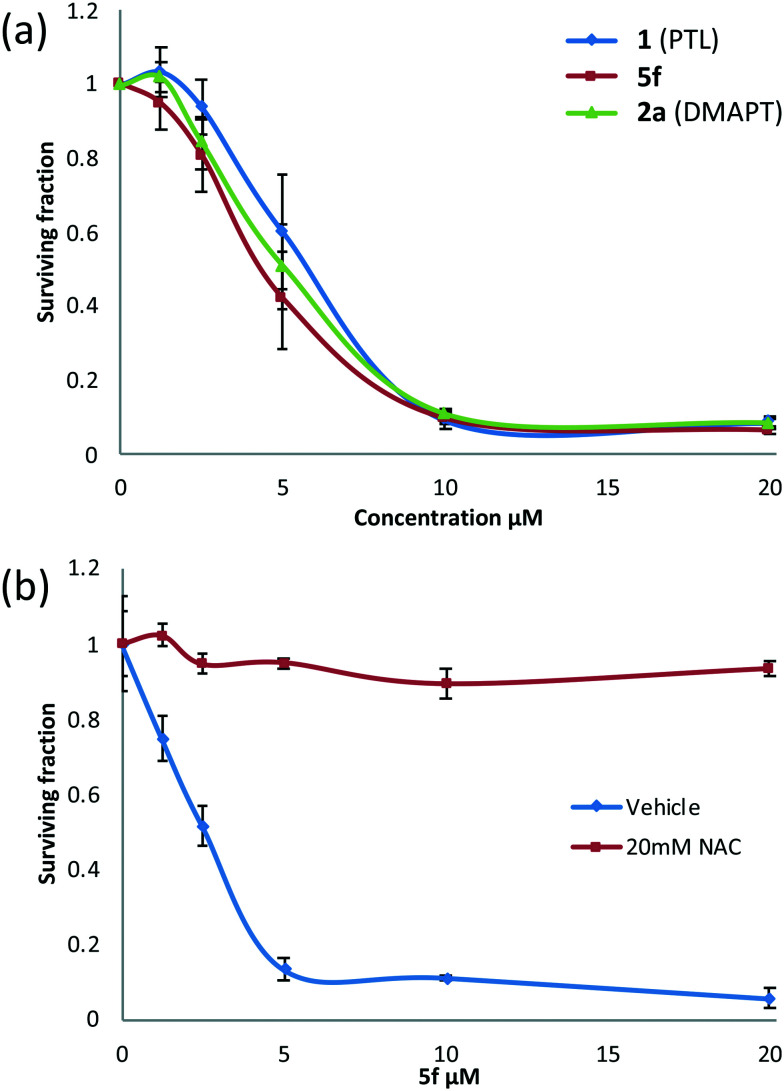
Following the identification of 5f as the most promising drug-like derivative of PTL from the aforementioned parallel/high-throughput screening, the anti-leukaemic activity of 5f was tested once more and compared directly with that of PTL (1) and DMAPT (2a), which serve as comparators and positive controls. Fig. 5(a) shows that the activity of 5f (n = 5) (EC50 = 4.5 μM) was not significantly different to the activity of PTL (n = 5) or DMAPT (n = 5) with EC50 values of 6.2 μM and 5.6 μM, respectively (Table S2,† entries 1 and 2) indicating that it has comparable activity with the parent and literature compounds (1 and 2a respectively). Pre-treatment with N-acetylcysteine (NAC) abrogated activity demonstrating that similar to PTL and other derivatives, 5f is a pro-oxidant and induces cell death through induction of oxidative stress, Fig. 5(b). Furthermore, in silico modelling suggests that 5f is likely to function as a pro-drug and deliver PTL to also inhibit IKKβ and NF-κB, Fig. S3 and S4.†
Fig. 5. Concentration versus activity plots (alamarBlue® assay) showing (a) anti-leukaemic activity of 5fversus PTL and DMAPT in MEC1 cells and (b) the effect of NAC pretreatment on the activity of 5f.
In vitro hERG liability testing
Early toxicological screening is an important step in derisking drug discovery programmes and is increasingly accessible.39 Since an unacceptable human ether a go-go related gene (hERG) liability is an all too common reason for failure at the candidate selection stage, a hERG assay was conducted to ensure the compounds being developed did not carry this liability.40
Three PTL derivatives (2a, 5a and 5f) were tested for inhibition of the hERG K+ channel (against cisapride as a positive control) using IonWorks patch clamp electrophysiology. In assessment of any hERG liability, compound 5a was compared to compound 5f which had already been identified as a superior pre-candidate compound; whilst 2a (DMAPT) is specifically not the focus of this manuscript it was deemed appropriate to contrast any hERG liability of this the literature-reported anti-AML compound.16af Eight-point concentration-response curves were generated using three-fold serial dilutions from a maximum final test concentration of 100 μM, results are summarised in Fig. S6.†
The three PTL derivatives compared (2a, 5a and 5f) in this hERG liability assay all performed well displaying IC50 values >100 μM (i.e. inhibition of less than 50% at the top 100 μM test concentration). At said top (100 μM) test concentration the novel compound 5f elicited only a 24% inhibition, whereas compounds 2a and 5a performed slightly less well with inhibitions of 40 and 45% (Fig. S6 and Table S4,† entries 1 and 2) respectively. Thus, hERG liability study, confirms PTL derivatives tested to represent a minimal risk according to results of the in vitro tests conducted. Among the compounds tested 5f displayed the lowest hERG liability and was flagged as inactive (Table S4,† entry 3) confirming compound 5f as the most promising lead from the studies conducted herein.
Synthetic access to compound 5f
Through identification of compound 5f as the resulting pre-candidate from the described screening cascade in earlier sections the requirement to provide more material for this and onwards studies presented a new and unexpected problem. Whilst it was possible to synthesise compound 5f in 83% isolated yield in high purity via a hybrid of the earlier mentioned protocols (Scheme 2a), the availability of amine 7 caused some issues, namely during the course of this work resupply of amine 7 (or salts thereof) became challenging. This uncertain supply chain led us to probe the synthesis and set about synthesising our own material. Despite the compound being listed in the catalogues of numerous suppliers, a literature search (SciFinder) revealed very few instances of the motif occurring in the peer reviewed41 or patent literature.42 The synthesis of amine 7 had not, to the best of our knowledge at the time of writing this report, been reported in the peer reviewed literature. As such a synthetic protocol was proposed (Scheme 2b).
Scheme 2. (a) The optimised synthesis of 5f; (b) the designed and executed plan for the synthesis of the required amine 7.
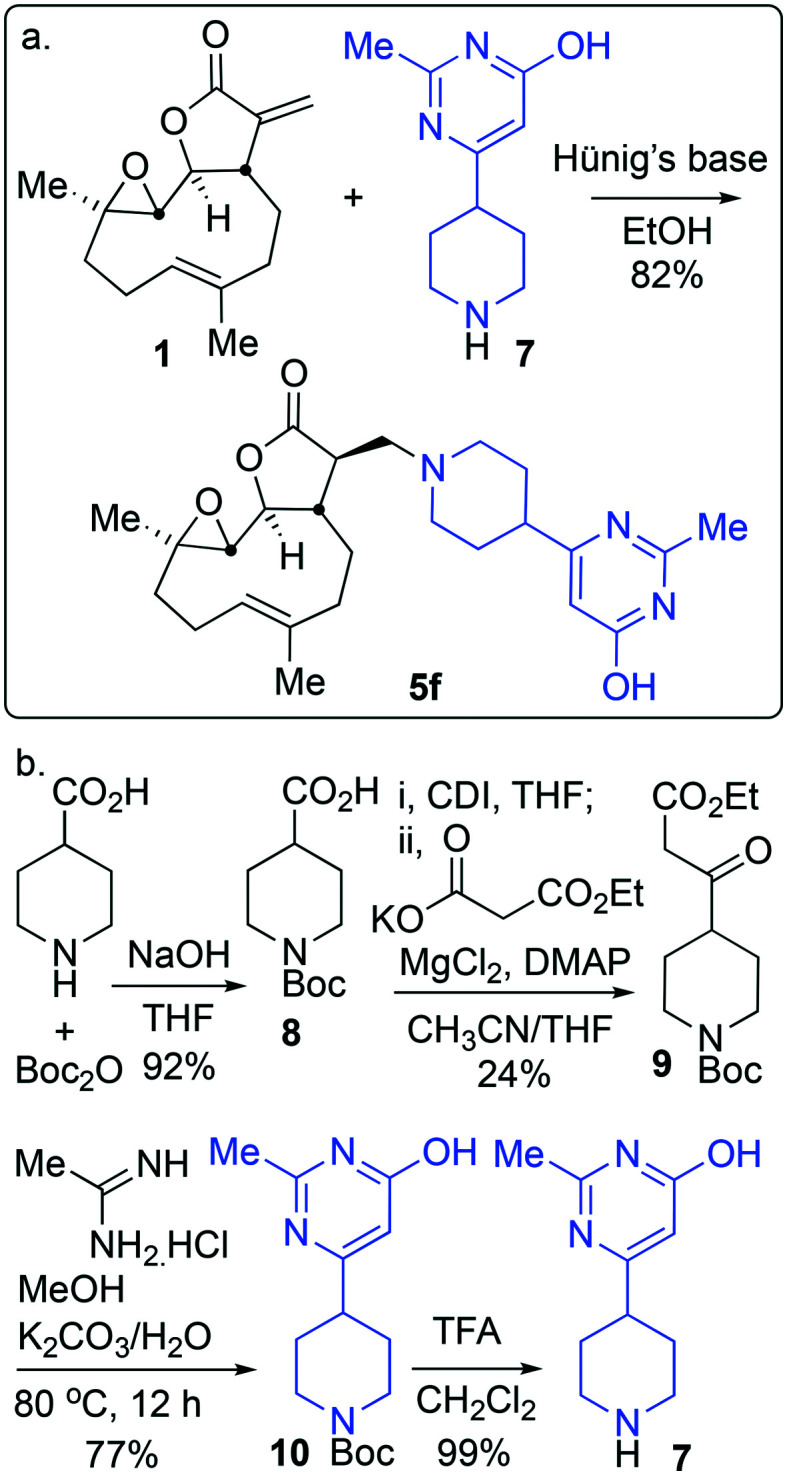
Amine 7 was successfully prepared as follows: 4-Piperidinecarboxylic acid was N-Boc protected to deliver 8 in 92% yield.43 In order to confidently assign the NMR spectra of compound 8 J-MOD, HSQC and 1H–1H COSY were helpful, particularly in establishing that a broad resonance in the proton-decoupled 13C NMR spectrum, centred on 42.8 ppm, corresponds to the ring-carbons bonded to nitrogen. The carboxylic acid of 8 was converted to the corresponding ethyl-3-keto-propanoate 9 in 24% yield. Conversion of 9 to the N-Boc protected congener 10, was achieved by treatment with acetamidine hydrochloride and potassium carbonate, furnishing 10 in 77% yield. Treatment with trifluoroacetic acid delivered desired compound 7 in 99% yield for the Boc-deprotection step (scale 200 mg, 17% over four linear steps). Thus, evidencing accessibility of amine 7 and with it in hand, it was timely to probe any activity arising from the amine alone. MEC1 sensitivity to 1, 5f and 7 was compared in parallel using the alamarBlue® assay (Fig. 6). EC50 values were determined (CalcuSyn Version 2.11, BIOSOFT) for 1 (3.6 μM ± 0.46) and 5f (4.3 μM ± 0.60). In contrast, an EC50 could not be determined for amine 7 since MEC1 cells were unaffected by equivalent concentrations of it indicating that 7 is not a source of cytotoxicity observed in this assay. A pre-peer-reviewed preprint of this article was deposited and may be viewed elsewhere.44
Fig. 6. Concentration versus activity plots (alamarBlue® assay) showing anti-leukaemic activity of 1 (PTL), 5f and amine 7 in MEC1 cells.
Conclusions
The cultivation of feverfew, from which PTL 1 was extracted, was described. The isolation of PTL (1) permitted the synthesis of a library of derivatives, through smooth 1,4-addition to PTL's exocyclic Michael-acceptor C C double bond. The library thus synthesised displayed good coverage of medicinally relevant chemical space. The compounds synthesised were tested for activity against the MEC1 CLL cell line. These compounds showed no particular trend in activity across the window of already acceptable medicinal chemistry parameters, such as pKa, cLog D, cLog P, TPSA, Fsp3 and molecular weight. The DMPK screening cascade described identified compound 5f as the most promising from activity, safety profile and ADME property stand-points. Direct in vitro comparison showed 5f exhibited comparable activity to DMAPT against the MEC1 cell line and pre-treatment with NAC suggested a pro-oxidant mechanism of action. Molecular docking studies with components of the NF-κB pathway also support a pro-drug mode of action of the compounds involving release of 1 and covalent interaction with one or more proteins involved in that pathway. This is partially supported by the lack of activity of the thioether adduct, compound 3, that is far less prone to undergo a reverse reaction to reveal 1, although lack of activity cannot be ascribed to this phenomenon alone. Taken together, these findings confirm 5f as a promising motif for further study and elaboration towards treatments for drug resistant CLL.
Notes
Ruth Roberts and Michael J. Morton are co-founders and co-directors of Apconix Ltd, an integrated toxicology and ion channel company that provides expert advice on nonclinical aspects of drug discovery and drug development to academia, industry and non-for-profit organisations. The remaining authors declare no competing interests.
Author Contributions
All authors contributed critically to devising and executing this research. A detailed description of the specific contribution of each author is given in the ESI.†
Conflicts of interest
There is no conflict of interest to declare.
Supplementary Material
Acknowledgments
AA and JSF are grateful for the support of internally allocated MRC Confidence in Concepts (14/15) and EPSRC (ISF 2012) pump-priming grants. The University of Birmingham are thanked for support, not only of the scientists and research facilities but also via Winterbourne House and Garden. Their crucial, critical and extremely valuable support includes the staff who have dedicated time, resources and effort to the cultivation of feverfew and varieties thereof. The CASE network has facilitated cooperation, training and networking opportunities.46 Professor Jonathan Frampton (University of Birmingham) is thanked for help throughout and providing support that was crucial at the project initiation stage. The authors thank the ERDF AWM II for support. DTP thanks The School of Chemistry for a studentship. Huy Van Nguyen (University of Birmingham) is thanked for assistance with aspects of experiments conducted in the latter stages. JSF is extremely grateful to the Royal Society Industry Fellowship scheme (2012/R1) and the EPSRC for funding (EP/J003220/1). JSF also acknowledges the importance of the training received through a residential Royal Society of Chemistry Drug Discovery training workshop. Dr. Chi Tsang and Dr. Peter Ashton are thanked for helpful discussions about mass spectrometry. Dr. Cécile S. Le Duff and Dr. Neil Spencer gave advice on aspects of NMR spectroscopy. The laboratory of TS is grateful for the support of Bloodwise (14031). AGL thanks Liverpool John Moores University for support. Dr. Sam Butterworth (University of Manchester, formerly of University of Birmingham), Dr. David Fox (Vulpine Science and Learning) and Dr. John Liddle (GSK) are thanked for advice, mentorship and training through agreements with the University of Birmingham and internal advisory panels. Past and present members of the research group headed by JSF are thanked where appropriate for efforts in harvesting feverfew and extraction of PTL.
Footnotes
†Electronic supplementary information (ESI) available: General experimental details, aspects relating to cultivation of feverfew, the PTL extraction protocol, synthesis, biological assays computational information and spectrums are detailed in the supporting information. Methods described and references cited therein,45 should be referred to in regard of the docking, crystallographic, synthetic and toxicological studies detailed therein. See DOI: 10.1039/c9md00297a
References
- (a) Pfeil A. M., Imfeld P., Pettengell R., Jick S. S., Szucs T. D., Meier C. R., Schwenkglenks M. Ann. Hematol. 2015;94:421–429. doi: 10.1007/s00277-014-2217-7. [DOI] [PubMed] [Google Scholar]; (b) Montillo M. Leuk. Suppl. 2012;1:S31. doi: 10.1038/leusup.2012.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Parikh S. A. Blood Cancer J. 2018;8:93. doi: 10.1038/s41408-018-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Dighiero G., Hamblin T. J. Lancet. 2008;371:1017–1029. doi: 10.1016/S0140-6736(08)60456-0. [DOI] [PubMed] [Google Scholar]
- (a) Baliakas P., Iskas M., Gardiner A., Davis Z., Plevova K., Nguyen-Khac F., Malcikova J., Anagnostopoulos A., Glide S., Mould S., Stepanovska K., Brejcha M., Belessi C., Davi F., Pospisilova S., Athanasiadou A., Stamatopoulos K., Oscier D. Am. J. Hematol. 2013;89:249–255. doi: 10.1002/ajh.23618. [DOI] [PubMed] [Google Scholar]; (b) Döhner H., Stilgenbauer S., Benner A., Leupolt E., Kröber A., Bullinger L., Döhner K., Bentz M., Lichter P. N. Engl. J. Med. 2000;343:1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]; (c) Pepper C., Lin T. T., Pratt G., Hewamana S., Brennan P., Hiller L., Hills R., Ward R., Starczynski J., Austen B., Hooper L., Stankovic T., Fegan C. Blood. 2008;112:3807–3817. doi: 10.1182/blood-2008-05-157131. [DOI] [PubMed] [Google Scholar]
- (a) Stout E. P., Choi M. Y., Castro J. E., Molinski T. F. J. Med. Chem. 2014;57:5085–5093. doi: 10.1021/jm4016922. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Brunhofer-Bolzer G., Le T., Dyckmanns N., Knaus H. A., Pausz C., Freund P., Jäger U., Erker T., Vanura K. J. Med. Chem. 2015;58:1244–1253. doi: 10.1021/jm501848m. [DOI] [PubMed] [Google Scholar]; (c) Chen H., Song P., Diao Y., Hao Y., Dou D., Wang W., Fang X., Wang Y., Zhao Z., Ding J., Li H., Xie H., Xu Y. MedChemComm. 2018;9:697–704. doi: 10.1039/c8md00019k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Brown A. M. G., Lowe K. C., Davey M. R., Power J. B., Knight D. W., Heptinstall S. Phytochem. Anal. 1996;7:86–91. [Google Scholar]; (b) Zhou J. Z., Kou X., Stevenson D. J. Agric. Food Chem. 1999;47:1018–1022. doi: 10.1021/jf980798u. [DOI] [PubMed] [Google Scholar]; (c) Pasquali M. C., Alizai S., Rostron C., Berry M. I. J. Pharm. Pharmacol. 2005;57:S72. [Google Scholar]; (d) Seca A. M. L., Silva A. M. S. and Pinto D. C. G. A., in Stud. Nat. Prod. Chem., ed. A. U. Rahman, Elsevier, 2017, vol. 52, pp. 337–372. [Google Scholar]; (e) babaei G., Aliarab A., Abroon S., Rasmi Y., Aziz S. G.-G. Biomed. Pharmacother. 2018;106:239–246. doi: 10.1016/j.biopha.2018.06.131. [DOI] [PubMed] [Google Scholar]
- (a) Biggs M. J., Johnson E. S., Persaud N. P., Ratcliffe D. M. Lancet. 1982;2:776. doi: 10.1016/s0140-6736(82)90965-5. [DOI] [PubMed] [Google Scholar]; (b) Hylands D. M., Hylands P. J., Johnson E. S., Kadam N. P., Macrae K. D. Br. Med. J. 1985;291:1128. doi: 10.1136/bmj.291.6495.569. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Johnson E. S., Kadam N. P., Hylands D. M., Hylands P. J. Br. Med. J. 1985;291:569–573. doi: 10.1136/bmj.291.6495.569. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Johnson E. S., Kadam N. P., Anderson D., Jenkinson P. C., Dewdney R. S., Blowers S. D. Hum. Toxicol. 1987;6:533–534. doi: 10.1177/096032718700600616. [DOI] [PubMed] [Google Scholar]; (e) Vogler B. K., Pittler M. H., Ernst E. Cephalalgia. 1998;18:704–708. doi: 10.1046/j.1468-2982.1998.1810704.x. [DOI] [PubMed] [Google Scholar]; (f) Ernst E., Pittler M. H. Public Health Nutr. 2000;3:509–514. doi: 10.1017/s1368980000000598. [DOI] [PubMed] [Google Scholar]; (g) Pittler M. H., Vogler B. K. and Ernst E., Cochrane Database Syst. Rev., 2000, 10.1002/14651858.CD002286, CD002286. [DOI] [PubMed]; (h) Pittler M. H. and Ernst E., Cochrane Database Syst. Rev., 2004, 10.1002/14651858.CD002286.pub2, CD002286. [DOI] [PubMed]; (i) Pareek A., Rathore G., Suthar M., Bansal V. Pharmacogn Rev. 2011:103. doi: 10.4103/0973-7847.79105. [DOI] [PMC free article] [PubMed] [Google Scholar]; (j) Chavez M. L., Chavez P. I. Hosp. Pharm. 1999;34:436–461. [Google Scholar]
- Wu R. P., Hayashi T., Cottam H. B., Jin G., Yao S., Wu C. C. N., Rosenbach M. D., Corr M., Schwab R. B., Carson D. A. Proc. Natl. Acad. Sci. U. S. A. 2010;107:7479. doi: 10.1073/pnas.1002890107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele A. J., Jones D. T., Ganeshaguru K., Duke V. M., Yogashangary B. C., North J. M., Lowdell M. W., Kottaridis P. D., Mehta A. B., Prentice A. G., Hoffbrand A. V., Wickremasinghe R. G. Leukemia. 2006;20:1073–1079. doi: 10.1038/sj.leu.2404230. [DOI] [PubMed] [Google Scholar]
- Agathanggelou A., Weston V. J., Perry T., Davies N. J., Skowronska A., Payne D. T., Fossey J. S., Oldreive C. E., Wei W., Pratt G., Parry H., Oscier D., Coles S. J., Hole P. S., Darley R. L., McMahon M., Hayes J. D., Moss P., Stewart G. S., Taylor A. M. R., Stankovic T. Haematologica. 2015;100:1076–1085. doi: 10.3324/haematol.2014.115170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosentino C., Grieco D., Costanzo V. EMBO J. 2011;30:546–555. doi: 10.1038/emboj.2010.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele A. J., Jones D. T., Ganeshaguru K., Duke V. M., Yogashangary B. C., North J. M., Lowdell M. W., Kottaridis P. D., Mehta A. B., Prentice A. G., Hoffbrand A. V., Wickremasinghe R. G. Leukemia. 2006;20:1073. doi: 10.1038/sj.leu.2404230. [DOI] [PubMed] [Google Scholar]
- (a) Wen J., You K.-R., Lee S.-Y., Song C.-H., Kim D.-G. J. Biol. Chem. 2002;277:38954–38964. doi: 10.1074/jbc.M203842200. [DOI] [PubMed] [Google Scholar]; (b) Liu Z., Liu S., Xie Z., Pavlovicz R. E., Wu J., Chen P., Aimiuwu J., Pang J., Bhasin D., Neviani P., Fuchs J. R., Plass C., Li P.-K., Li C., Huang T. H. M., Wu L.-C., Rush L., Wang H., Perrotti D., Marcucci G., Chan K. K. J. Pharmacol. Exp. Ther. 2009;329:505. doi: 10.1124/jpet.108.147934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Patel N. M., Nozaki S., Shortle N. H., Bhat-Nakshatri P., Newton T. R., Rice S., Gelfanov V., Boswell S. H., Goulet R. J., Sledge G. W., Nakshatri H. Oncogene. 2000;19:4159. doi: 10.1038/sj.onc.1203768. [DOI] [PubMed] [Google Scholar]; (b) Fang L. J., Shao X. T., Wang S., Lu G. H., Xu T., Zhou J. Y. Planta Med. 2010;76:258–264. doi: 10.1055/s-0029-1186083. [DOI] [PubMed] [Google Scholar]; (c) Carlisi D., De Blasio A., Drago-Ferrante R., Di Fiore R., Buttitta G., Morreale M., Scerri C., Vento R., Tesoriere G. Cell Death Discovery. 2017;3:17078. doi: 10.1038/cddiscovery.2017.78. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Knight D. W. Nat. Prod. Rep. 1995;12:271–276. doi: 10.1039/np9951200271. [DOI] [PubMed] [Google Scholar]; (e) Mathema V. B., Koh Y.-S., Thakuri B. C., Sillanpää M. Inflammation. 2012;35:560–565. doi: 10.1007/s10753-011-9346-0. [DOI] [PubMed] [Google Scholar]
- Neelakantan S., Nasim S., Guzman M. L., Jordan C. T., Crooks P. A. Bioorg. Med. Chem. Lett. 2009;19:4346–4349. doi: 10.1016/j.bmcl.2009.05.092. [DOI] [PubMed] [Google Scholar]
- Guzman M. L., Rossi R. M., Li X., Corbett C., Hassane D. C., Bushnell T., Carroll M., Sullivan E., Neelakantan S., Crooks P. A., Jordan C. T. Blood. 2006;108:237. [Google Scholar]
- (a) Nasim S., Crooks P. A. Bioorg. Med. Chem. Lett. 2008;18:3870–3873. doi: 10.1016/j.bmcl.2008.06.050. [DOI] [PubMed] [Google Scholar]; (b) Shanmugam R., Kusumanchi P., Appaiah H., Cheng L., Crooks P., Neelakantan S., Peat T., Klaunig J., Matthews W., Nakshatri H., Sweeney C. J. Int. J. Cancer. 2011;128:2481–2494. doi: 10.1002/ijc.25587. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Lamture G., Crooks P. A., Borrelli M. J. Drug Dev. Res. 2018;79:287–294. doi: 10.1002/ddr.21441. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Kennedy M. A., US2018/0099014A1, 2018.; (e) Wang R., Bhat-Nakshatri P., Padua M. B., Prasad M. S., Anjanappa M., Jacobson M., Finnearty C., Sefcsik V., McElyea K., Redmond R., Sandusky G., Penthala N., Crooks P. A., Liu J., Zimmers T., Nakshatri H. Mol. Cancer Ther. 2017;16:2747–2758. doi: 10.1158/1535-7163.MCT-17-0717. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Morel K. L., Ormsby R. J., Bezak E., Sweeney C. J., Sykes P. J. Radiat. Res. 2017;187:501–512. doi: 10.1667/RR14710.1. [DOI] [PubMed] [Google Scholar]; (g) Mendonca M. S., Turchan W. T., Alpuche M. E., Watson C. N., Estabrook N. C., Chin-Sinex H., Shapiro J. B., Imasuen-Williams I. E., Rangel G., Gilley D. P., Huda N., Crooks P. A., Shapiro R. H. Free Radical Biol. Med. 2017;112:318–326. doi: 10.1016/j.freeradbiomed.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Janganati V., Crooks P., Ponder J. and Jordan C., WO2017/132528A1, 2017.; (i) Carlisi D., Buttitta G., Di Fiore R., Scerri C., Drago-Ferrante R., Vento R., Tesoriere G. Cell Death Dis. 2016;7:e2194. doi: 10.1038/cddis.2016.94. [DOI] [PMC free article] [PubMed] [Google Scholar]; (j) Nakshatri H., Appaiah H. N., Anjanappa M., Gilley D., Tanaka H., Badve S., Crooks P. A., Mathews W., Sweeney C., Bhat-Nakshatri P. Cell Death Dis. 2015;6:e1608. doi: 10.1038/cddis.2014.569. [DOI] [PMC free article] [PubMed] [Google Scholar]; (k) Kempema A. M., Widen J. C., Hexum J. K., Andrews T. E., Wang D., Rathe S. K., Meece F. A., Noble K. E., Sachs Z., Largaespada D. A., Harki D. A. Bioorg. Med. Chem. 2015;23:4737–4745. doi: 10.1016/j.bmc.2015.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]; (l) Karmakar A., Xu Y., Mustafa T., Kannarpady G., Bratton S. M., Radominska-Pandya A., Crooks P. A., Biris A. S. RSC Adv. 2015;5:2411–2420. doi: 10.1039/c4ra10871j. [DOI] [PMC free article] [PubMed] [Google Scholar]; (m) Janganati V., Penthala N. R., Crooks P. A. and Jordan C. T., US2015/0133444A1, 2015.; (n) Bommagani S., Crooks P., Penthala N. R., Janganati V., Jordan C. T. and Ponder J., US2015/0203508A1, 2015.; (o) Viennois E., Xiao B., Ayyadurai S., Wang L., Wang P. G., Zhang Q., Chen Y., Merlin D. Lab. Invest. 2014;94:950–965. doi: 10.1038/labinvest.2014.89. [DOI] [PubMed] [Google Scholar]; (p) Song J. M., Qian X., Upadhyaya P., Hong K. H., Kassie F. Curr. Cancer Drug Targets. 2014;14:59–69. doi: 10.2174/15680096113136660108. [DOI] [PubMed] [Google Scholar]; (q) Janganati V., Penthala N. R., Madadi N. R., Chen Z., Crooks P. A. Bioorg. Med. Chem. Lett. 2014;24:3499–3502. doi: 10.1016/j.bmcl.2014.05.059. [DOI] [PMC free article] [PubMed] [Google Scholar]; (r) Yip-Schneider M. T., Wu H., Stantz K., Agaram N., Crooks P. A., Schmidt C. M. BMC Cancer. 2013;13:194. doi: 10.1186/1471-2407-13-194. [DOI] [PMC free article] [PubMed] [Google Scholar]; (s) Yip-Schneider M. T., Wu H., Hruban R. H., Lowy A. M., Crooks P. A., Schmidt C. M. Pancreas. 2013;42:160–167. doi: 10.1097/MPA.0b013e318254f455. [DOI] [PubMed] [Google Scholar]; (t) Ramachandran P. V., Nicponski D. R., Nair H. N. G., Helppi M. A., Gagare P. D., Schmidt C. M., Yip-Schneider M. T. Bioorg. Med. Chem. Lett. 2013;23:6911–6914. doi: 10.1016/j.bmcl.2013.09.065. [DOI] [PubMed] [Google Scholar]; (u) Ghantous A., Sinjab A., Herceg Z., Darwiche N. Drug Discovery Today. 2013;18:894–905. doi: 10.1016/j.drudis.2013.05.005. [DOI] [PubMed] [Google Scholar]; (v) D'Anneo A., Carlisi D., Lauricella M., Puleio R., Martinez R., Di Bella S., Di Marco P., Emanuele S., Di Fiore R., Guercio A., Vento R., Tesoriere G. Cell Death Dis. 2013;4:e891. doi: 10.1038/cddis.2013.415. [DOI] [PMC free article] [PubMed] [Google Scholar]; (w) Michelle R., Marcus Tullius S., Mauro Vicentini C., Harold Hilarion F., Luciana S., Francisco Jaime Bezerra Mendonca J., Marcelo Sobral da S., Vicente de Paulo E. Lett. Drug Des. Discovery. 2012;9:881–890. [Google Scholar]; (x) Harki D. A. and Wang D., WO2012/145678A1, 2012.; (y) Zhang S. and Xiong G., CN101270102A, 2008.; (z) Flores-Lopez G., Moreno-Lorenzana D., Ayala-Sanchez M., Aviles-Vazquez S., Torres-Martinez H., Crooks P. A., Guzman M. L., Mayani H., Chávez-González A. J. Cell. Mol. Med. 2018;22:4899–4912. doi: 10.1111/jcmm.13755. [DOI] [PMC free article] [PubMed] [Google Scholar]; (aa) Shi C., Wang Y., Guo Y., Chen Y., Liu N. Oncotarget. 2017;8:35009–35018. doi: 10.18632/oncotarget.16557. [DOI] [PMC free article] [PubMed] [Google Scholar]; (ab) Penthala N. R., Janganati V., Alpe T. L., Apana S. M., Berridge M. S., Crooks P. A., Borrelli M. J. Bioorg. Med. Chem. Lett. 2016;26:5883–5886. doi: 10.1016/j.bmcl.2016.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; (ac) Hexum J. K., Becker C. M., Kempema A. M., Ohlfest J. R., Largaespada D. A., Harki D. A. Bioorg. Med. Chem. Lett. 2015;25:2493–2495. doi: 10.1016/j.bmcl.2015.04.058. [DOI] [PubMed] [Google Scholar]; (ad) Shanmugam K., Kusumanchi P., Cheng L., Crooks P., Neelakantan S., Matthews W., Nakshatri H., Sweeny C. J. Drug Discovery Today. 2010;15:322. doi: 10.1002/pros.21141. [DOI] [PubMed] [Google Scholar]; (ae) Peese R. Drug Discovery Today. 2010;15:322. [Google Scholar]; (af) Guzman M. L., Rossi R. M., Neelakantan S., Li X., Corbett C. A., Hassane D. C., Becker M. W., Bennett J. M., Sullivan E., Lachowicz J. L., Vaughan A., Sweeney C. J., Matthews W., Carroll M., Liesveld J. L., Crooks P. A., Jordan C. T. Blood. 2007;110:4427–4435. doi: 10.1182/blood-2007-05-090621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Wen J., You K. R., Lee S. Y., Song C. H., Kim D. G. J. Biol. Chem. 2002;277:38954–38964. doi: 10.1074/jbc.M203842200. [DOI] [PubMed] [Google Scholar]; (b) Pei S., Minhajuddin M., Callahan K. P., Balys M., Ashton J. M., Neering S. J., Lagadinou E. D., Corbett C., Ye H., Liesveld J. L., O'Dwyer K. M., Li Z., Shi L., Greninger P., Settleman J., Benes C., Hagen F. K., Munger J., Crooks P. A., Becker M. W., Jordan C. T. J. Biol. Chem. 2013;288:33542–33558. doi: 10.1074/jbc.M113.511170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Shin M., McGowan A., DiNatale G. J., Chiramanewong T., Cai T., Connor R. E. ACS Omega. 2017;2:7267–7274. doi: 10.1021/acsomega.7b00954. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Liu Z. F., Liu S. J., Xie Z. L., Pavlovicz R. E., Wu J., Chen P., Aimiuwu J., Pang J. X., Bhasin D., Neviani P., Fuchs J. R., Plass C., Li P. K., Li C., Huang T. H. M., Wu L. C., Rush L., Wang H. Y., Perrotti D., Marcucci G., Chan K. K. J. Pharmacol. Exp. Ther. 2009;329:505–514. doi: 10.1124/jpet.108.147934. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Moumou M., El Bouakher A., Allouchi H., El Hakmaoui A., Benharref A., Mathieu V., Guillaumet G., Akssira M. Bioorg. Med. Chem. Lett. 2014;24:4014–4018. doi: 10.1016/j.bmcl.2014.06.019. [DOI] [PubMed] [Google Scholar]; (d) Yang Z., Kuang B., Kang N., Ding Y., Ge W., Lian L., Gao Y., Wei Y., Chen Y., Zhang Q. Eur. J. Med. Chem. 2017;127:296–304. doi: 10.1016/j.ejmech.2016.12.044. [DOI] [PubMed] [Google Scholar]; (e) Penthala N. R., Bommagani S., Janganati V., Macnicol K. B., Cragle C. E., Madadi N. R., Hardy L. L., Macnicol A. M., Crooks P. A. Eur. J. Med. Chem. 2014;85:517–525. doi: 10.1016/j.ejmech.2014.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Taleghani A., Nasseri M. A., Iranshahi M. Bioorg. Chem. 2017;71:128–134. doi: 10.1016/j.bioorg.2017.01.020. [DOI] [PubMed] [Google Scholar]; (g) Janganati V., Ponder J., Balasubramaniam M., Bhat-Nakshatri P., Bar E. E., Nakshatri H., Jordan C. T., Crooks P. A. Eur. J. Med. Chem. 2018;157:562–581. doi: 10.1016/j.ejmech.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Kempema A. M., Widen J. C., Hexum J. K., Andrews T. E., Wang D., Rathe S. K., Meece F. A., Noble K. E., Sachs Z., Largaespada D. A., Harki D. A. Bioorg. Med. Chem. 2015;23:4737–4745. doi: 10.1016/j.bmc.2015.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Long J., Ding Y.-H., Wang P.-P., Zhang Q., Chen Y. Tetrahedron Lett. 2016;57:874–877. [Google Scholar]; (j) Zimmermann S., Fouché G., De Mieri M., Yoshimoto Y., Usuki T., Nthambeleni R., Parkinson J. C., van der Westhuyzen C., Kaiser M., Hamburger M., Adams M. Molecules. 2014;19:3523–3538. doi: 10.3390/molecules19033523. [DOI] [PMC free article] [PubMed] [Google Scholar]; (k) Ren Y., Yu J., Douglas Kinghorn A. Curr. Med. Chem. 2016;23:2397–2420. doi: 10.2174/0929867323666160510123255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewamana S., Alghazal S., Lin T. T., Clement M., Jenkins C., Guzman M. L., Jordan C. T., Neelakantan S., Crooks P. A., Burnett A. K., Pratt G., Fegan C., Rowntree C., Brennan P., Pepper C. Blood. 2008;111:4681. doi: 10.1182/blood-2007-11-125278. [DOI] [PubMed] [Google Scholar]
- (a) Feula A., Male L., Fossey J. S. Org. Lett. 2010;12:5044–5047. doi: 10.1021/ol102215e. [DOI] [PubMed] [Google Scholar]; (b) Archer R. M., Hutchby M., Winn C. L., Fossey J. S., Bull S. D. Tetrahedron. 2015;71:8838–8847. [Google Scholar]; (c) Brittain W. D. G., Buckley B. R., Fossey J. S. Chem. Commun. 2015;51:17217–17220. doi: 10.1039/c5cc04886a. [DOI] [PubMed] [Google Scholar]; (d) Feula A., Dhillon S. S., Byravan R., Sangha M., Ebanks R., Hama Salih M. A., Spencer N., Male L., Magyary I., Deng W.-P., Müller F., Fossey J. S. Org. Biomol. Chem. 2013;11:5083–5093. doi: 10.1039/c3ob41007b. [DOI] [PubMed] [Google Scholar]; (e) Hama Salih M. A., Male L., Spencer N., Fossey J. S. Org. Chem. Front. 2015;2:1445–1449. [Google Scholar]; (f) Zhao Y., van Nguyen H., Male L., Craven P., Buckley B. R., Fossey J. S. Organometallics. 2018;37:4224–4241. doi: 10.1021/acs.organomet.8b00539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterbourne House and Garden, www.winterbourne.org.uk, (accessed 9th December 2018).
- Zoubir M., Zeroual A., El Idrissi M., El Haib A., Moumou M., Hammal R., Mazoir N., Benharref A., El Hajbi A. J. Mater. Environ. Sci. 2017;8:990–996. [Google Scholar]
- Baell J. B., Nissink J. W. M. ACS Chem. Biol. 2018;13:36–44. doi: 10.1021/acschembio.7b00903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J., Ding Y.-H., Wang P.-P., Zhang Q., Chen Y. J. Org. Chem. 2013;78:10512–10518. doi: 10.1021/jo401606q. [DOI] [PubMed] [Google Scholar]
- (a) Pajouhesh H., Lenz G. R. NeuroRx. 2005;2:541–553. doi: 10.1602/neurorx.2.4.541. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Hughes J. P., Rees S., Kalindjian S. B., Philpott K. L. Br. J. Pharmacol. 2010;162:1239–1249. doi: 10.1111/j.1476-5381.2010.01127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Lovering F., Bikker J., Humblet C. J. Med. Chem. 2009;52:6752–6756. doi: 10.1021/jm901241e. [DOI] [PubMed] [Google Scholar]
- See supplementary material for model amination reactions using tulipane as substrate
- (a) Hwang D.-R., Wu Y.-S., Chang C.-W., Lien T.-W., Chen W.-C., Tan U.-K., Hsu J. T. A., Hsieh H.-P. Bioorg. Med. Chem. 2006;14:83–91. doi: 10.1016/j.bmc.2005.07.055. [DOI] [PubMed] [Google Scholar]; (b) Hsieh H.-P., Hsu T.-A. and Hwang D.-R., US2004/0229936A1, 2004.
- Crooks P. A., Jordan C. T. and Wei X., US2005/0272716A1, 2005.
- (a) Woods J. R., Mo H., Bieberich A. A., Alavanja T., Colby D. A. J. Med. Chem. 2011;54:7934–7941. doi: 10.1021/jm201114t. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Crooks P. A., Jordan C. T. and Wei X., WO2005/007103A2, 2005.
- Lukas S. Curr. Cancer Drug Targets. 2016;16:701–709. doi: 10.2174/1568009616666160408150032. [DOI] [PubMed] [Google Scholar]
- (a) Stacchini A., Aragno M., Vallario A., Alfarano A., Circosta P., Gottardi D., Faldella A., Rege-Cambrin G., Thunberg U., Nilsson K., Caligaris-Cappio F. Leuk. Res. 1999;23:127–136. doi: 10.1016/s0145-2126(98)00154-4. [DOI] [PubMed] [Google Scholar]; (b) Kandaswamy R., Sava G. P., Speedy H. E., Beà S., Martín-Subero J. I., Studd J. B., Migliorini G., Law Philip J., Puente X. S., Martín-García D., Salaverria I., Gutiérrez-Abril J., López-Otín C., Catovsky D., Allan J. M., Campo E., Houlston R. S. Cell Rep. 2016;16:2061–2067. doi: 10.1016/j.celrep.2016.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Agathanggelou A., Smith E., Davies N. J., Kwok M., Zlatanou A., Oldreive C. E., Mao J., Da Costa D., Yadollahi S., Perry T., Kearns P., Skowronska A., Yates E., Parry H., Hillmen P., Reverdy C., Delansorne R., Paneesha S., Pratt G., Moss P., Taylor A. M. R., Stewart G. S., Stankovic T. Blood. 2017;130:156. doi: 10.1182/blood-2016-12-758219. [DOI] [PubMed] [Google Scholar]
- (a) Nociari M. M., Shalev A., Benias P., Russo C. J. Immunol. Methods. 1998;213:157–167. doi: 10.1016/s0022-1759(98)00028-3. [DOI] [PubMed] [Google Scholar]; (b) Nakayama G. R., Caton M. C., Nova M. P., Parandoosh Z. J. Immunol. Methods. 1997;204:205–208. doi: 10.1016/s0022-1759(97)00043-4. [DOI] [PubMed] [Google Scholar]
- Woods J. R., Mo H., Bieberich A. A., Alavanja T., Colby D. A. MedChemComm. 2013;4:27–33. [Google Scholar]
- Obach R. S. Drug Metab. Dispos. 1999;27:1350. [PubMed] [Google Scholar]
- (a) Hidalgo I. J., Raub T. J., Borchardt R. T. Gastroenterology. 1989;96:736–749. [PubMed] [Google Scholar]; (b) Wang Z., Hop C. E. C. A., Leung K. H., Pang J. J. Mass Spectrom. 2000;35:71–76. doi: 10.1002/(SICI)1096-9888(200001)35:1<71::AID-JMS915>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]; (c) Luo S., Pal D., Shah S. J., Kwatra D., Paturi K. D., Mitra A. K. Mol. Pharmaceutics. 2010;7:412–420. doi: 10.1021/mp900193e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman-Cook K. D., Hoffman R. L., Johnson T. W. Future Med. Chem. 2013;5:113–115. doi: 10.4155/fmc.12.208. [DOI] [PubMed] [Google Scholar]
- Johnson T. W., Gallego R. A., Edwards M. P. J. Med. Chem. 2018;61:6401–6420. doi: 10.1021/acs.jmedchem.8b00077. [DOI] [PubMed] [Google Scholar]
- Compound 2a (DMAPT) has a LipE in the alamarBlue assay of activity against the MEC1 cell line of 3.19
- Szymanski P., Markowicz M., Mikiciuk-Olasik E. Int. J. Mol. Sci. 2012;13:427–452. doi: 10.3390/ijms13010427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Pollard C. E., Abi Gerges N., Bridgland-Taylor M. H., Easter A., Hammond T. G., Valentin J. P. Br. J. Pharmacol. 2010;159:12–21. doi: 10.1111/j.1476-5381.2009.00207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Rampe D., Brown A. M. J. Pharmacol. Toxicol. Methods. 2013;68:13–22. doi: 10.1016/j.vascn.2013.03.005. [DOI] [PubMed] [Google Scholar]; (c) Ekins S., Crumb W. J., Sarazan R. D., Wikel J. H., Wrighton S. A. J. Pharmacol. Exp. Ther. 2002;301:427. doi: 10.1124/jpet.301.2.427. [DOI] [PubMed] [Google Scholar]
- Zheng Z., Huang X.-P., Mangano T. J., Zou R., Chen X., Zaidi S. A., Roth B. L., Stevens R. C., Katritch V. J. Med. Chem. 2017;60:3070–3081. doi: 10.1021/acs.jmedchem.7b00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., O'Connor S. J., Gunn D., Newcom J., Chen J., Yi L., Zhang H.-J., Hunyadi L. M. and Natero R., WO2004/058727A1, 2004.
- (a) Nimbarte V. D., Murtuza H., Phaniraj S., Shrivastava S., Naidu V. G. M., Satheesh Kumar N., Atcha K. R. Med. Chem. Res. 2014;23:2178–2197. [Google Scholar]; (b) Qu C., Ding M., Zhu Y., Lu Y., Du J., Miller M., Tian J., Zhu J., Xu J., Wen M., Er-Bu A. G. A., Wang J., Xiao Y., Wu M., McManus O. B., Li M., Wu J., Luo H.-R., Cao Z., Shen B., Wang H., Zhu M. X., Hong X. J. Med. Chem. 2017;60:4680–4692. doi: 10.1021/acs.jmedchem.7b00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Payne D. T., Ampolu B., Bland N., Brown J. T., Dutton M. J., Fitton C. A., Gulliver A., Hale L., Hamza D., Jones G., Lane R., Leach A. G., Male L., Merisor E. G., Morton M. J., Quy A. S., Roberts R., Scarll R., Schulz-Utermoehl T., Stankovic T., Stevenson B., Fossey J. S. and Agathanggelou A., ChemRxiv, 2019, 10.26434/chemrxiv.7629680. [DOI] [PMC free article] [PubMed]
- (a) Vidal M., García-Arriagada M., Rezende M. C., Domínguez M. Synthesis. 2016;48:4246–4252. [Google Scholar]; (b) Jamal W., Bari A., Mothana R. A., Basudan O., Mohammed M. S., Ng S. W. Asian J. Chem. 2014;26:5183–5185. [Google Scholar]; (c) Bryant G., Plant Propagation A to Z: Growing Plants for Free, David & Charles Ltd, 2004. [Google Scholar]; (d) Royal Horticultural Society: Tanacetum parthenium, feverfew, https://www.rhs.org.uk/Plants/17986/i-Tanacetum-parthenium-i/Details, (accessed 19th December 2018).; (e) The Plant List, http://www.theplantlist.org/tpl1.1/record/gcc-36373, (accessed 11th December 2018).; (f) Zherebker A. Y., Airapetyan D., Konstantinov A. I., Kostyukevich Y. I., Kononikhin A. S., Popov I. A., Zaitsev K. V., Nikolaev E. N., Perminova I. V. Analyst. 2015;140:4708–4719. doi: 10.1039/c5an00602c. [DOI] [PubMed] [Google Scholar]; (g) Bolton G. L., e-EROS Encyclopedia of Reagents for Organic Synthesis, 2001. [Google Scholar]; (h) Huck L., Berton M., de la Hoz A., Díaz-Ortiz A., Alcázar J. Green Chem. 2017;19:1420–1424. [Google Scholar]; (i) Wang F., Li J., Sinn A. L., Knabe W. E., Khanna M., Jo I., Silver J. M., Oh K., Li L., Sandusky G. E., Sledge G. W., Nakshatri H., Jones D. R., Pollok K. E., Meroueh S. O. J. Med. Chem. 2011;54:7193–7205. doi: 10.1021/jm200782y. [DOI] [PMC free article] [PubMed] [Google Scholar]; (j) Bashford K. E., Burton M. B., Cameron S., Cooper A. L., Hogg R. D., Kane P. D., MacManus D. A., Matrunola C. A., Moody C. J., Robertson A. A. B., Warne M. R. Tetrahedron Lett. 2003;44:1627–1629. [Google Scholar]; (k) Kazulkin D. N. and Kochubey V. S., EP2719696A1, 2011.; (l) Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., Olson A. J. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]; (m) Trott O., Olson A. J. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]; (n) Xu G., Lo Y.-C., Li Q., Napolitano G., Wu X., Jiang X., Dreano M., Karin M., Wu H. Nature. 2011;472:325. doi: 10.1038/nature09853. [DOI] [PMC free article] [PubMed] [Google Scholar]; (o) Garcia-Pineres A. J., Castro V., Mora G., Schmidt T. J., Strunck E., Pahl H. L., Merfort I. J. Biol. Chem. 2001;276:39713–39720. doi: 10.1074/jbc.M101985200. [DOI] [PubMed] [Google Scholar]; (p) Goldberg F. W., Kettle J. G., Kogej T., Perry M. W. D., Tomkinson N. P. Drug Discovery Today. 2015;20:11–17. doi: 10.1016/j.drudis.2014.09.023. [DOI] [PubMed] [Google Scholar]; (q) Dennington R., Keith T. A. and Millam J. M., GaussView 6, 2016.; (r) Chen Y.-Q., Ghosh S., Ghosh G. Nat. Struct. Biol. 1998;5:67. doi: 10.1038/nsb0198-67. [DOI] [PubMed] [Google Scholar]; (s) Meyers J., Carter M., Mok N. Y., Brown N. Future Med. Chem. 2016;8:1753–1767. doi: 10.4155/fmc-2016-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]; (t) O'Boyle N. M., Banck M., James C. A., Morley C., Vandermeersch T., Hutchison G. R. Aust. J. Chem. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]; (u) Firth N. C., Brown N., Blagg J. J. Chem. Inf. Model. 2012;52:2516–2525. doi: 10.1021/ci300293f. [DOI] [PMC free article] [PubMed] [Google Scholar]; (v) Leeson P. D., Davis A. M. J. Med. Chem. 2004;47:6338–6348. doi: 10.1021/jm049717d. [DOI] [PubMed] [Google Scholar]
- (a) Fossey J. S., Brittain W. D. G. Org. Chem. Front. 2015;2:101–105. [Google Scholar]; (b) Payne D. T., Fossey J. S., Elmes R. B. P. Supramol. Chem. 2016;28:921–931. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.