Abstract
Background
COPD is a leading cause of mortality.
Research Question
We hypothesized that applying machine learning to clinical and quantitative CT imaging features would improve mortality prediction in COPD.
Study Design and Methods
We selected 30 clinical, spirometric, and imaging features as inputs for a random survival forest. We used top features in a Cox regression to create a machine learning mortality prediction (MLMP) in COPD model and also assessed the performance of other statistical and machine learning models. We trained the models in subjects with moderate to severe COPD from a subset of subjects in Genetic Epidemiology of COPD (COPDGene) and tested prediction performance in the remainder of individuals with moderate to severe COPD in COPDGene and Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints (ECLIPSE). We compared our model with the BMI, airflow obstruction, dyspnea, exercise capacity (BODE) index; BODE modifications; and the age, dyspnea, and airflow obstruction index.
Results
We included 2,632 participants from COPDGene and 1,268 participants from ECLIPSE. The top predictors of mortality were 6-min walk distance, FEV1 % predicted, and age. The top imaging predictor was pulmonary artery-to-aorta ratio. The MLMP-COPD model resulted in a C index ≥ 0.7 in both COPDGene and ECLIPSE (6.4- and 7.2-year median follow-ups, respectively), significantly better than all tested mortality indexes (P < .05). The MLMP-COPD model had fewer predictors but similar performance to that of other models. The group with the highest BODE scores (7-10) had 64% mortality, whereas the highest mortality group defined by the MLMP-COPD model had 77% mortality (P = .012).
Interpretation
An MLMP-COPD model outperformed four existing models for predicting all-cause mortality across two COPD cohorts. Performance of machine learning was similar to that of traditional statistical methods. The model is available online at: https://cdnm.shinyapps.io/cgmortalityapp/.
Key Words: COPD, machine learning, mortality, prediction, random survival forest
Abbreviations: 6MWD, 6-min walk distance; ADO, age, dyspnea, and airflow obstruction; BODE, BMI, airflow obstruction, dyspnea, and exercise capacity; COPDGene, Genetic Epidemiology of COPD; ECLIPSE, Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints; MLMP, machine learning mortality prediction; PA:A, pulmonary artery to aorta; % LAA < −950 HU, percent emphysema determined by the percent low attenuation area of the lungs < −950 Hounsfield units; Pi10, square root of wall area of a hypothetical airway with internal perimeter of 10 mm; RSF, random survival forest; VIMP, variable importance
FOR EDITORIAL COMMENT, SEE PAGE 846
COPD is one of the leading causes of mortality worldwide.1 Improving performance of mortality prediction models can identify patients with COPD who might benefit from earlier or more specific intervention. One widely used mortality prediction tool is the BMI, airflow obstruction, dyspnea, and exercise capacity (BODE) index.2 Studies that have modified the BODE index by varying the input3, 4, 5 or adding serum biomarkers6 have resulted in similar or slightly improved performance for mortality prediction. Other models include the dyspnea, airflow obstruction, smoking status, exacerbation frequency (DOSE) index7; the age, dyspnea, and airflow obstruction (ADO) index8; and the St. George’s Respiratory Questionnaire score, airflow limitation, and exercise tolerance (SAFE)9 index. A survival analysis in more than 3,500 subjects from 11 COPD cohorts compared predictive accuracy of several of these measures, finding that ADO, BODE, and BODE modifications performed best; after adjusting for age, BODE modifications outperformed ADO.10 A meta-analysis in 16,000 subjects from the COPD Cohorts Collaborative International Assessment (3CIA) initiative found that ADO and updated BODE showed a higher, but not statistically significantly different, area under the curve for predicting 3-year survival vs BODE.11
These prediction models did not include quantitative CT imaging. Several quantitative CT imaging measures of airway,12 emphysematous,13 vascular,14,15 and interstitial16 abnormalities have been independently associated with morbidity and mortality in subjects with COPD. Evaluating many potential predictors is fraught with challenges, which may be addressed by machine learning algorithms that can learn rules from one data set that can be used to make predictions in another data set.17 Machine learning can be broadly defined; some consider traditional statistical methods such as logistic or Cox regression important machine learning tools,18, 19, 20, 21, 22 and in certain cases these perform as well as newer more advanced methods.23 Machine learning algorithms have the potential to improve predictive modeling of health outcomes,24 and digitalization of health records is facilitating incorporation into health care.25 In COPD, machine learning has been used to identify which features are most important for case identification26 and predicting exacerbations.27 Machine learning methods were used to improve prediction of 5-year all-cause mortality in subjects undergoing CT coronary angiography28 and cardiac motion MRI.29
We hypothesized that machine learning methods applied to an expanded set of clinical and quantitative CT imaging features would be useful for identifying the most important predictors of all-cause mortality and improving mortality prediction in moderate to severe COPD compared with BODE, BODE modifications, and ADO. To test this hypothesis, we used data from subjects from the Genetic Epidemiology of COPD (COPDGene) and Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints (ECLIPSE) studies.
Materials and Methods
Study Participants
We included individuals with moderate to severe COPD (postbronchodilator FEV1 < 80% predicted, FEV1/FVC < 0.7; ie, Global Initiative for Chronic Obstructive Lung Disease [GOLD] 2-4 spirometry grades) from the COPDGene and ECLIPSE studies. We selected 30 clinical, spirometric, and imaging features as inputs for a random survival forest. To ensure that the same variables could be examined in both data sets, we selected a set of demographic, clinical, spirometric, and imaging features present in at least 80% of subjects in both data sets (e-Table 1). Notable imaging features included pulmonary artery-to-aorta (PA:A) ratio,30 square root of wall area of a hypothetical airway with internal perimeter of 10 mm (Pi10),31 mean wall area percent,13 percent emphysema determined by the percent low attenuation area of the lungs < −950 Hounsfield units (% LAA < −950 HU),13 and 15th percentile of the lung density histogram on inspiratory scans.32 The primary outcome was time to death from any cause.
Study Design
To develop a mortality prediction model that balanced prediction and interpretability, we applied random survival forests (RSFs) for feature selection followed by Cox regression. We used the randomForestSRC R package33 and obtained variable importance (VIMP) by means of the VIMP method.34 We filtered for multicollinearity of the RSF features, removing collinear variables to ensure all variance inflation factors were < 1035,36 and then applied Cox regression (survival R package).37,38 As a reference for comparison, we used the BODE index,2 exacerbations and BODE,3 updated BODE, and ADO.8 To understand the contribution of RSF to this approach, we also compared a range of feature selection methods and prediction models, including standard statistical methods (see the Methods in e-Appendix 1).
The accuracy of prediction models was assessed by using C indexes (Hmisc R package)36,39 and receiver operating characteristic curves (ROCR R package).40 The C indexes were compared with the compareC R package by using the one-shot method.41. For our main model, we developed a Web-based tool by using the Shiny R package. Additional details are available in the Methods in e-Appendix 1.
Results
Characteristics of Study Participants
Characteristics of subjects with complete data used in analysis are shown in Table 1. Compared with subjects in COPDGene, those in the ECLIPSE study had a longer median follow-up and lower BMI and FEV1 %predicted, and ECLIPSE had a higher proportion of individuals who died. In addition, several quantitative CT imaging measures differed in ECLIPSE (e-Table 1, e-Fig 1). The BODE and the exacerbations and BODE indexes were higher in ECLIPSE, but median scores were the same between samples. The ADO index was higher in subjects in COPDGene.
Table 1.
Demographic Characteristics of Subjects in COPDGene and ECLIPSE Included in Analysis
| Characteristic | COPDGene | ECLIPSE | P Value |
|---|---|---|---|
| No. of subjects | 2,632 | 1,268 | … |
| Sex, female, No. (%) | 1,157 (44.0) | 426 (33.6) | < .001 |
| Age, mean (SD), y | 63.58 (8.92) | 63.51 (7.03) | .8 |
| Race, African American, No. (%) | 485 (18.4) | 0 (0) | < .001 |
| FEV1 % predicted, median (IQR) | 52.00 (36.88-66.60) | 45.95 (35-58.73) | < .001 |
| FVC % predicted, mean (SD) | 77.17 (16.76) | 79.16 (19.82) | .001 |
| GOLD spirometry grade, No. (%) | < .001 | ||
| 2 | 1,407 (53.5) | 518 (40.9) | |
| 3 | 819 (31.1) | 568 (44.8) | |
| 4 | 406 (15.4) | 182 (14.4) | |
| Pack-years cigarette smoking, median (IQR) | 47.50 (36-68) | 45 (32-60) | < .001 |
| Current smoking, No. (%) | 1,041 (39.6) | 431 (34.0) | .001 |
| Dead at 3 y, No. (%) | 221 (8.4) | 121 (9.5) | .3 |
| Dead at 5 y No. (%) | 454 (17.2) | 238 (18.8) | .3 |
| Dead at 8 y No. (%) | 631 (24.0) | 405 (31.9) | < .001 |
| Total dead, No. (%) | 631 (24.0) | 405 (31.9) | < .001 |
| Days followed up, median (IQR) | 2,321 (2,043-2,652) | 2,616 (1,110-2,924) | < .001 |
| 6-Min walk distance, mean (SD), ft | 1,205.97 (392.28) | 1,190.36 (389.77) | .2 |
| BODE, median (IQR) | 3.00 (1-4) | 3.00 (2-5) | < .001 |
| e-BODE, median (IQR) | 3.00 (1-5) | 3.00 (2-5) | < .001 |
| Updated BODE, median (IQR) | 3.00 (1-7) | 3.00 (1-7) | .9 |
| ADO, mean (SD) | 4.60 (1.8) | 3.84 (1.44) | < .001 |
Percentages do not necessarily total 100% because of rounding. ADO = age, dyspnea, and airflow obstruction; BODE = BMI, airflow obstruction, dyspnea, and exercise capacity; e-BODE = exacerbations and BODE; COPDGene = Genetic Epidemiology of COPD; ECLIPSE = Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints; GOLD = Global Initiative for Chronic Obstructive Lung Disease; IQR = interquartile range.
Development of a Mortality Prediction Model
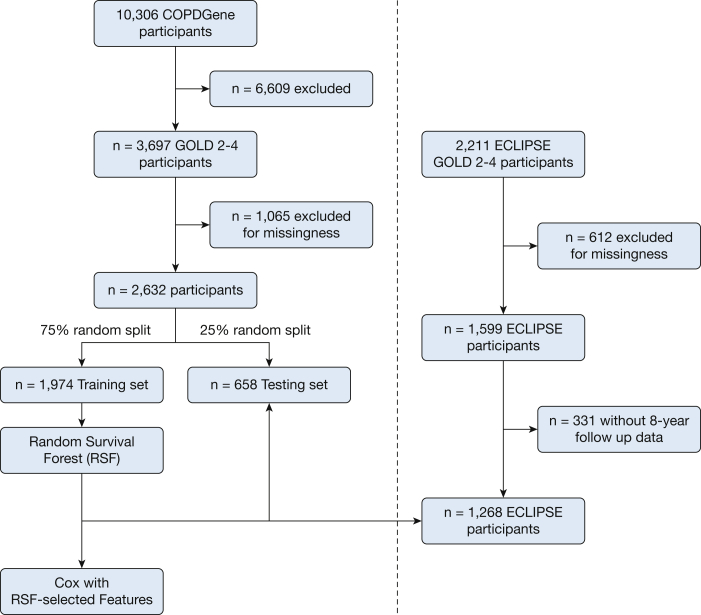
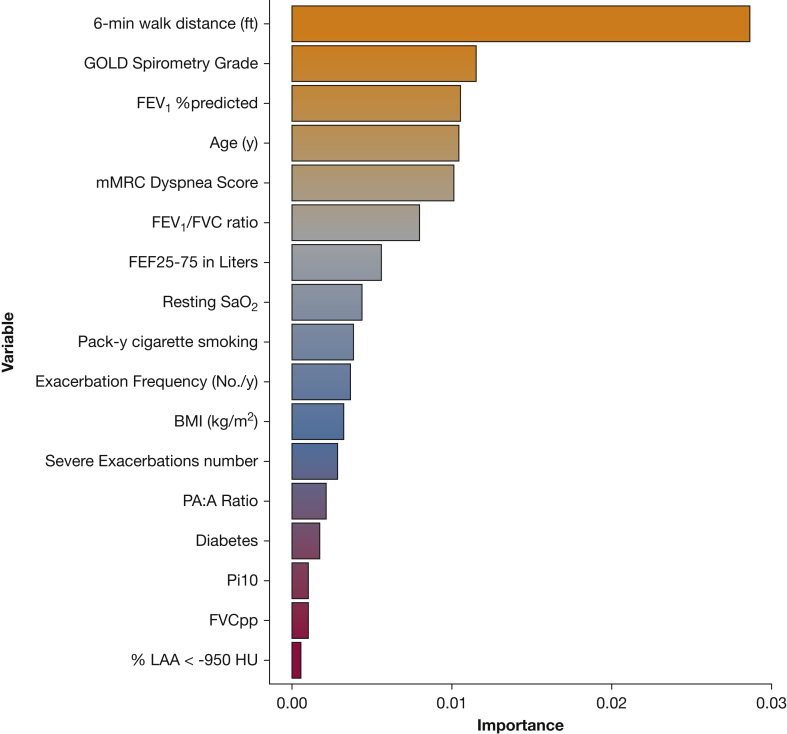
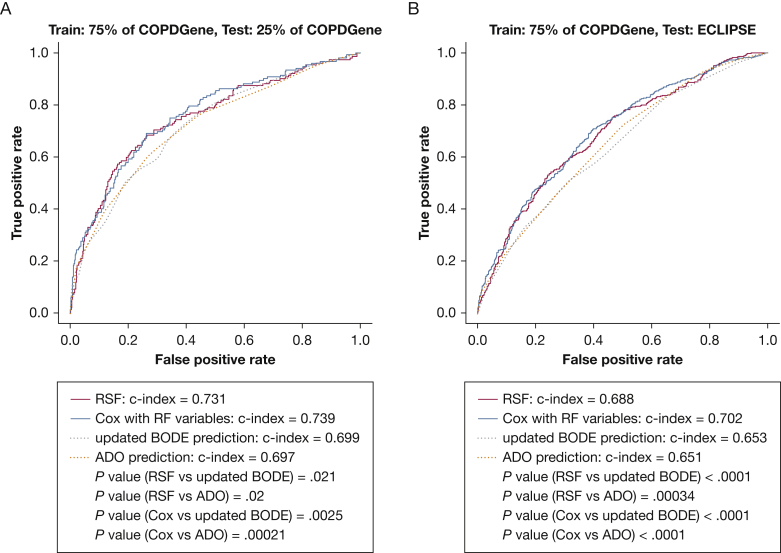
A schematic of the study design is shown in Figure 1. We randomly divided 2,632 participants from COPDGene into training (n = 1,974 [479 deaths]) and testing (n = 658 [152 deaths]) samples (e-Table 1). We chose features present in at least 80% of the cohort (initial feature list in e-Table 2) and used RSFs to select features, identifying the components of the BODE score (BMI, FEV1 % predicted, modified Medical Research Council dyspnea score, and exercise as assessed by the 6-min walk distance [6MWD]), as well as additional clinical (eg, age, diabetes), spirometric (FEV1/FVC ratio, forced expiratory flow 25% to 75%), and imaging (Pi10, mean wall area percent, % LAA < −950 HU, PA:A ratio) features by using the training sample. Features are shown in order of importance in Figure 2. Before Cox regression, features displaying multicollinearity were excluded; the remaining features and associated hazard ratios are shown in Table 2. Using this subset of RSF-selected features, we developed a Cox regression model (further denoted as machine learning mortality prediction [MLMP] in COPD). The MLMP-COPD model had a C index of 0.74 on the testing sample, outperforming BODE, BODE modifications, and ADO (P < .05) (Fig 3A, e-Fig 2A). The regression coefficients for the MLMP-COPD model are shown in e-Table 3, and clinical interpretations of hazard ratios are shown in e-Table 4.
Figure 1.
Schematic of study design. A total of 2,632 participants in COPDGene were randomly split into training (n = 1,974) and testing (n = 658) data sets. A random survival forest algorithm was applied to the training data set, and features chosen by variable importance were used to develop a Cox regression model. Both models were tested in the testing data set of COPDGene and externally in a sample of participants in ECLIPSE (n = 1,268). COPDGene = Genetic Epidemiology of COPD; ECLIPSE = Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints; GOLD = Global Initiative for Chronic Obstructive Lung Disease; RSF = random survival forest.
Figure 2.
Variable importance based on RSF trained in subjects in COPDGene. These features were all used to develop a Cox regression model, and GOLD spirometry grade and FVC % predicted were removed for collinearity. 6MWD = 6-min walk distance; FEF25%-75% = forced expiratory flow 25% to 75%; mMRC = modified Medical Research Council; PA:A = pulmonary artery to aorta; % LAA < −950 HU = percent emphysema determined by the percent low attenuation area of the lungs < −950 Hounsfield units; Pi10 = square root of wall area of a hypothetical airway with internal perimeter of 10 mm; SaO2 = arterial oxygen saturation. See Figure 1 legend for expansion of other abbreviations.
Table 2.
Hazard Ratios for Mortality of Random Survival Forest-Selected Variables Used in MLMP-COPD (Cox Regression) Model Trained on Subjects in COPDGene
| Feature | Unadjusted Hazard Ratio (95% CI) | P Value | Adjusted Hazard Ratio (95% CI) | P Value |
|---|---|---|---|---|
| 6-Min walk distance, per 100 ft | 0.85 (0.83-0.87) | < .0001 | 0.9 (0.87-0.93) | < .0001 |
| FEV1 % predicted | 0.96 (0.957-0.968) | < .0001 | 0.99 (0.974-0.997) | .01 |
| Age | 1.04 (1.03-1.05) | < .0001 | 1.04 (1.03-1.06) | < .0001 |
| mMRC dyspnea score | 1.49 (1.39-1.61) | < .0001 | 1.15 (1.05-1.25) | .002 |
| FEV1/FVC ratioa | 0.009 (0.005-0.02) | < .0001 | 0.1 (0.02-0.45) | .003 |
| FEF25%-75%, L/min | 0.14 (0.1-0.2) | < .0001 | 1.74 (1.02-2.98) | .04 |
| Resting SaO2 | 0.9 (0.88-0.92) | < .0001 | 0.97 (0.948-0.995) | .02 |
| Exacerbations per year | 1.2 (1.14-1.28) | < .0001 | 1.03 (0.96-1.11) | .4 |
| Pack-years cigarette smoking | 1.01 (1.006-1.011) | < .0001 | 1.01 (1.002-1.008) | .0003 |
| BMI, kg/m2 | 0.97 (0.96-0.99) | .00022 | 0.97 (0.95-0.99) | .001 |
| Severe exacerbations | 1.95 (1.6-2.4) | < .0001 | 1.33 (1.05-1.7) | .02 |
| PA:A ratio | 5.8 (3.01-11.3) | < .0001 | 2.7 (1.35-5.55) | .005 |
| Pi10 | 5.5 (3.1-9.7) | < .0001 | 1.6 (0.8-3.3) | .2 |
| Diabetes | 1.4 (1.1-1.8) | .0072 | 1.3 (1.01-1.73) | .04 |
| % LAA < −950 HU | 1.03 (1.027-1.04) | < .0001 | 0.995 (0.98-1.004) | .3 |
Adjusted models include all covariates, and FVC and GOLD spirometry grade were removed for multicollinearity. FEF25%-75% = forced expiratory flow 25% to 75%; MLMP = machine learning mortality prediction; mMRC = modified Medical Research Council; PA:A = pulmonary artery to aorta; % LAA < −950 HU = percent emphysema determined by the percent low attenuation area of the lungs < −950 Hounsfield units; Pi10 = square root of wall area of a hypothetical airway with internal perimeter of 10 mm; SaO2 = arterial oxygen saturation. See Table 1 legend for expansion of other abbreviations.
FEV1/FVC is reported as a ratio from 0 to 1 and not as a percentage.
Figure 3.
Receiver operating characteristic curve comparing the RSF-derived mortality prediction models with updated BODE and ADO. A, Models were trained in 75% of the COPDGene sample (n = 1,974) and tested in the remaining 25% of the COPDGene sample (n = 658). B, Models were trained in 75% of the COPDGene sample (n = 1,974) and tested in the ECLIPSE sample (n = 1,268). ADO = age, dyspnea, and airflow obstruction; BODE = BMI, airflow obstruction, dyspnea, and exercise capacity. See Figure 1 and 2 legends for expansion of other abbreviations.
To test mortality prediction in an external validation sample, we applied the MLMP-COPD model to 1,268 participants in ECLIPSE (Table 1, e-Table 1). As expected, the performance of the models decreased in external validation (C index, 0.7) but retained improved prediction compared with BODE, BODE modifications, and ADO (P < .05 for all) (Fig 3B, e-Fig 2B). We repeated our analyses after normalizing imaging features to the mean values across matched subsets of individuals from the COPDGene and ECLIPSE studies in an attempt to reconcile differences in mean values and distributions of imaging features (see the Methods in e-Appendix 1). Our results were not substantially different (e-Fig 3) from those in the original model. To examine calibration, we plotted expected vs actual survival as shown in e-Figure 4 and tested for evidence of miscalibration by using the Greenwood-Nam-D'Agostino test.42 As shown in e-Table 5, miscalibration evaluated using the Greenwood-Nam-D'Agostino test for the MLMP-COPD model was not statistically significant, which was in contrast to results for the BODE score also shown in e-Table 5.
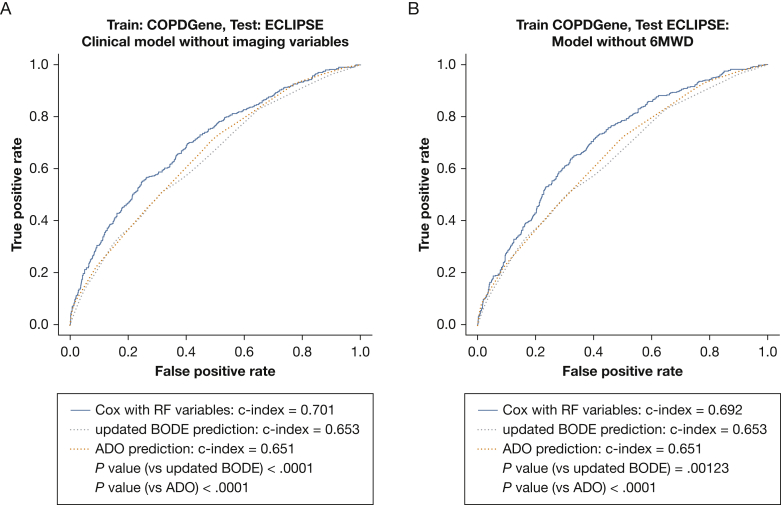
As all of these measured features may have variable availability in clinical practice, we calculated the performance of prediction models excluding select features. A Cox model built after excluding imaging features still improved prediction compared with updated BODE and ADO on the ECLIPSE validation sample (P < .05) (Fig 4A). The regression coefficients for the Cox model without imaging features are shown in e-Table 6. Mortality prediction models of individual quantitative imaging features added to BODE were not superior to updated BODE or ADO (e-Table 7).
Figure 4.
Receiver operating characteristic curve comparing the RSF-derived mortality prediction model with updated BODE and ADO after removing select features. Models were trained in 75% of the COPDGene sample and tested in the ECLIPSE sample. A, Imaging variables (% LAA < −950 HU, PAA, Pi10) were excluded when building the Cox regression model. B, The 6MWD was excluded when building the Cox regression model. See Figure 1, 2, and 3 legends for expansion of abbreviations.
Although theoretically easy to obtain, 6MWD is often not available during initial clinical encounters. When we removed 6MWD and retrained our Cox model, the resulting model (C index of 0.69 on the testing sample) retained improved performance compared with that of updated BODE (C index, 0.65; P < .05) and ADO (C index, 0.65; P < .05) (Fig 4B).
We developed an online Web application (https://cdnm.shinyapps.io/cgmortalityapp/) to facilitate use of the MLMP-COPD model in exploring the relative contributions of risk factors for all-cause mortality in COPD. In addition, we provide a case study of a hypothetical patient to demonstrate the potential usefulness of this application (see the Results in e-Appendix 1, e-Fig 5).
To assess the relative performance of other methods, we evaluated several combinations of feature selection methods and prediction models (see the Methods in e-Appendix 1, e-Table 8). All methods performed similarly, including stepwise regression, although RSFs selected the most parsimonious set of features (n = 15).
Subgroup Analyses
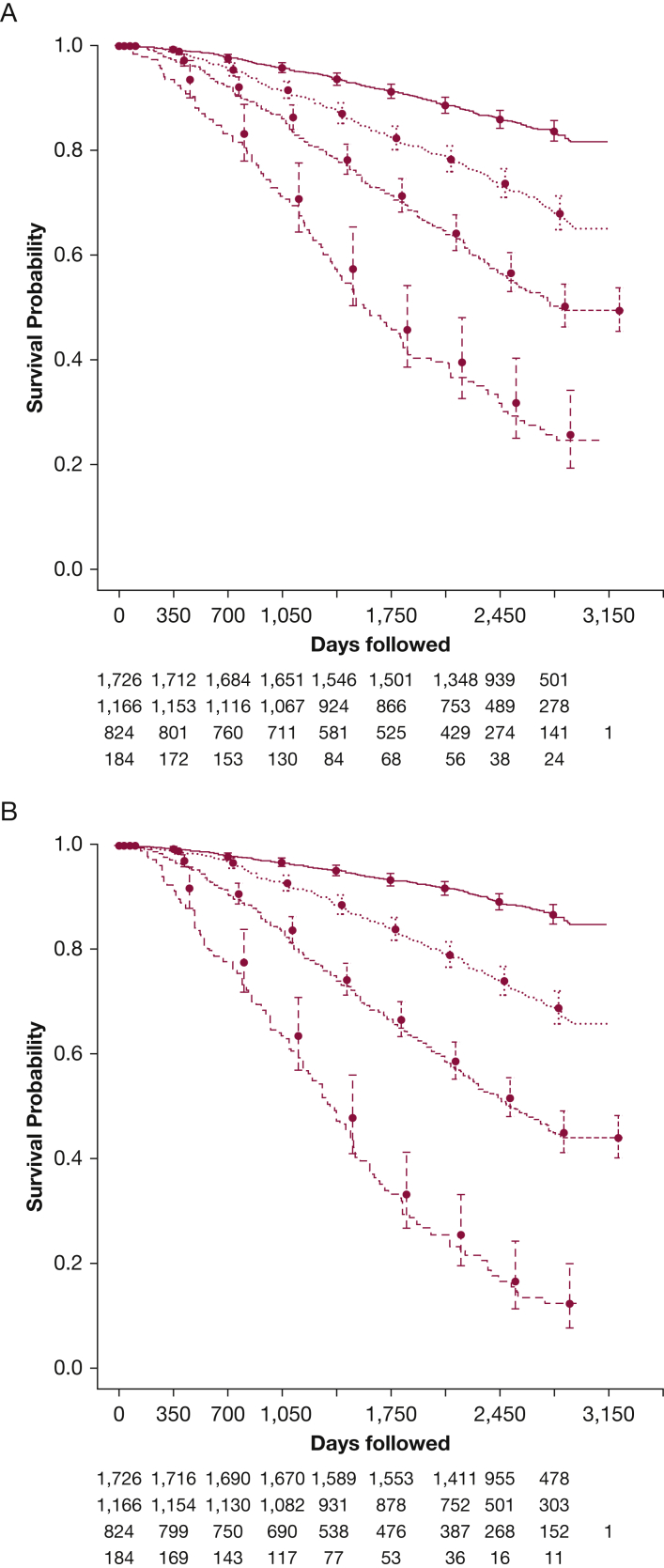
We tested the performance of the MLMP-COPD model in subgroups of ECLIPSE (see the Results in e-Appendix 1, e-Tables 9, 10). We divided individuals into four groups according to BODE score categories (0-2, 3-4, 5-6, 7-10) from Celli et al2 and then used the MLMP-COPD model predictions (log hazard ratio) to stratify participants into four equally sized groups. We compared the strata by using Kaplan-Meier analysis (Fig 5). Characteristics of the sickest group defined by BODE (score 7-10) and our model are shown in Table 3, and the less severe groups in e-Tables 11, 12, and 13. The sickest group had a 19.5% higher relative and 12.5% absolute mortality than did the sickest BODE group (BODE 7-10; 64% vs 77% absolute mortality; P = .012), with shorter median follow-up (1,281 vs 1,193 days; P = .049). The sickest group was also older and had fewer female subjects, higher PA:A ratio, lower % LAA < −950 HU, more comorbid diabetes, more pack-years of cigarette smoking, and lower resting arterial oxygen saturation. However, both groups had similar FEV1 % predicted and 6MWD, but the sickest group had higher BMI with lower modified Medical Research Council and BODE scores (P < .001).
Figure 5.
Kaplan-Meier analysis of subjects in a pooled COPDGene and ECLIPSE sample. A, Subjects were grouped by BODE score (0-2, 3-4, 5-6, 7-10). B, Subjects were stratified into four groups based on the machine learning mortality prediction (MLMP)-COPD model. Participants were ranked according to their calculated risk by the MLMP-COPD model and divided into four groups of mortality risk that were of equal size as those of the BODE groups. Circles indicate the probability of survival at each time point, and the bars are 95% CIs. See Figure 1 and 3 legends for expansion of other abbreviations.
Table 3.
Characteristics of Subjects With the Highest Predicted Mortality
| Characteristic | BODE Group | Model Group | P Value |
|---|---|---|---|
| No. of subjects | 184 | 184 | … |
| Age, mean (SD), y | 63.76 (7.47) | 69.83 (6.44) | < .001 |
| Race, African American, No. (%) | 18 (9.8) | 16 (8.7) | .857 |
| Sex, female, No. (%) | 82 (44.6) | 57 (31.0) | .010 |
| Total dead, No. (%) | 118 (64.1) | 141 (76.6) | .012 |
| Days followed up, median (IQR) | 1,280.50 (940.50-2,243.50) | 1,192.50 (791.75-1,833.75) | .049 |
| BODE, median (IQR) | 7.00 (7.00-8.00) | 6.00 (5.00-7.00) | < .001 |
| 6-Min walk distance, mean (SD), ft | 608.18 (343.20) | 593.19 (263.68) | .639 |
| FEV1 % predicted, median (IQR) | 27.65 (21.60-33.28) | 28.10 (20.88-34.38) | .942 |
| mMRC dyspnea score, No. (%) | .009 | ||
| 1 | 0 (0.0) | 5 (2.7) | |
| 2 | 19 (10.3) | 32 (17.4) | |
| 3 | 58 (31.5) | 64 (34.8) | |
| 4 | 107 (58.2) | 83 (45.1) | |
| FEV1/FVC ratio, median (IQR) | 0.34 (0.28-0.41) | 0.33 (0.27-0.37) | .053 |
| FEF25%-75%, median (IQR), L | 0.24 (0.19-0.33) | 0.23 (0.17-0.30) | .080 |
| Resting SaO2, median (IQR) | 94.00 (91.00-96.00) | 93.00 (88.00-94.25) | < .001 |
| Exacerbation frequency, mean (SD), No./y | 1.26 (1.33) | 1.52 (1.57) | .086 |
| Pack-years cigarette smoking, median (IQR) | 45.00 (34.60-67.62) | 61.40 (40.00-88.92) | < .001 |
| BMI, mean (SD), kg/m2 | 23.81 (6.30) | 25.47 (5.63) | .008 |
| Severe exacerbations, No. (%) | 86 (46.7) | 95 (51.6) | .404 |
| PA:A ratio, mean (SD) | 0.97 (0.16) | 1.02 (0.18) | .006 |
| Pi10, mean (SD) | 4.12 (0.35) | 4.10 (0.37) | .611 |
| Diabetes, No. (%) | 20 (10.9) | 38 (20.7) | .015 |
| % LAA < −950 HU, median (IQR) | 30.80 (18.50-39.40) | 24.48 (15.83-36.96) | .021 |
Discussion
In this study, we applied a machine learning approach to clinical features, including quantitative CT imaging, to develop an all-cause mortality prediction model in moderate to severe COPD. The resulting MLMP-COPD model outperformed BODE, BODE modifications, and ADO across two COPD cohorts. Our model also identified subjects at high risk of death on the basis of on variables not included in the BODE index. We have included an online tool to allow researchers and clinicians to explore the contributions of our model features to predicted COPD survival. Although the MLMP-COPD model is a prediction tool, it also provides valuable insights into how individual risk factors influence mortality in the context of other known predictors of mortality.
We developed the MLMP-COPD model in COPDGene and used ECLIPSE for external validation. External validation in diverse cohorts is essential, as even the BODE index, which reported a C index of 0.74 in the initial study, demonstrates significantly reduced performance in several external studies.5,6,10,43, 44, 45, 46, 47 Compared with prior models of mortality, our model was developed with more subjects with longer follow-ups. The BODE index, which arguably remains the gold standard for mortality prediction, was developed on 207 subjects followed up for 4 years and prospectively validated in 625 subjects.2 By comparison, we developed our model by using 1,974 subjects followed up for a median of 6.4 years from the COPDGene study and validated our model by using data from the ECLIPSE study. Overall, our model was highly consistent with the Galaxy COPD model,48 which found a similar set of important features and was externally validated, although it underestimated mortality in one of the two cohorts.49 However, the current study had five important differences: (1) the use of quantitative imaging features; (2) automated and reproducible feature selection; (3) direct comparison with BODE, BODE modifications, and ADO by using C-statistics; (4) more subjects; and (5) longer follow-up.
Our analysis provides insights into the relative contributions of predictors of mortality. Current smoking was not selected as a predictor, possibly because sicker patients are more likely to quit, confounding the relationship of current smoking and mortality in study populations enriched for subjects with COPD. The 6MWD was ranked substantially higher in VIMP than all other variables. This finding is consistent with those from prior literature, possibly because 6MWD reflects both pulmonary and extrapulmonary (eg, muscle weakness, pulmonary vascular disease) disease manifestations.50, 51, 52 Although the 6MWD, in theory, can be implemented easily, it is not always readily available; thus, we developed a model exclusive of 6MWD that demonstrated improved prediction compared with other mortality prediction indexes. However, we advocate for obtaining 6MWD when possible. Although exacerbation history53 and severe exacerbations requiring hospitalization54,55 are independent predictors of COPD mortality, they have not been evaluated in a single model together. As exacerbation history has been reported to be a predictor of severe exacerbations,56 and mortality increases acutely after a severe exacerbation,55 severe exacerbations may be adequately capturing the mortality risk conferred by exacerbation history. Diabetes was chosen as an important predictor of mortality, which has been previously reported.57,58 The fact that diabetes was chosen as an important predictor of mortality likely reflects the high cardiovascular mortality of COPD but may also support the notion that COPD may be an inflammatory multisystem disease.
Of quantitative imaging features, % LAA < −950 HU, Pi10, and PA:A ratio were chosen by the RSF algorithm, yet only the PA:A ratio was significant in the fully adjusted model. These features likely have a complex relationship with survival, and the PA:A ratio may capture vascular disease less directly captured by other features. Our results are consistent with those of a recent study reporting that PA:A is associated with mortality in patients with COPD after adjusting for BODE variables.59 However, this and other articles reporting the association of individual risk factors with mortality do not directly address whether combinations of these individual features leads to better prediction. In the current study, we observed that adding the PA:A ratio and other CT imaging-based disease features individually to BODE does not significantly improve predictive performance. Instead, a combination of clinical variables and CT imaging features are required to improve mortality prediction over that of existing models.
To our knowledge, our study is the first to evaluate multiple imaging features simultaneously in the context of mortality prediction. Removal of quantitative CT imaging features from the MLMP-COPD model decreased predictive performance only slightly. This finding is consistent with the RSF VIMP measures, which ranked quantitative imaging features as less important than most of the other features included. In contrast, a study using coronary CT angiography found age followed by imaging features as the most important predictors of all-cause mortality.28 This finding is likely consistent with the ability of coronary imaging to measure directly the specific lesions that lead to the most likely cause of death, in contrast to COPD, for which CT imaging features may be more important for describing COPD heterogeneity and play a more limited role in predicting all-cause mortality. Future work may use imaging to define and predict mortality directly in COPD.60,61
When stratifying subjects by using the MLMP-COPD model, the sickest group of the same-sized BODE group (score 7-10) had 19.5% higher mortality. Older age, lower resting arterial oxygen saturation, higher PA:A ratio, lower % LAA < −950 HU, increased pack-years of smoking, and diabetes added prognostic information for these subjects. Thus, it is important to think beyond the BODE variables when trying to identify patients with COPD at the highest risk of death.
RSFs have been used for feature selection in COPD case identification26 and for identifying risk factors for COPD exacerbations.27 The current study differs in that we used a survival implementation of RSFs (ie, time-to-event analysis) to identify predictors of all-cause mortality in people with COPD. Combinations of feature selection methods and traditional prediction models performed similarly to more advanced methods, although the combination of using RSFs for variable selection and Cox yielded the most parsimonious model. This result is perhaps not surprising, as simpler machine learning methods or models have been observed to perform similarly to or better than more advanced methods, depending on the data set.23 In addition, a simpler model that explains the data equally well is more likely to generalize in independent cohorts, presumably because of less overfitting or overparameterization.18 In a survival analysis of 1,371 patients with head and neck cancer, RSF identified the most important predictors of survival, whereas Cox regression performed slightly better than RSF.62 Thus, although the more advanced RSF machine learning method reduced the number of predictors, it provided accuracy similar to that of traditional Cox regression.
Many have attempted, but failed, to develop a mortality prediction model as consistently superior and equally parsimonious as the BODE index.3,5,7, 8, 9,11 Most previously published models were not directly compared with BODE in their initial publication, nor were they externally validated. A 2018 study examined the predictive power of 10 COPD mortality prediction models and reported that none performed significantly better than BODE.11 By contrast, our model demonstrated superior predictive performance for long-term mortality across multiple cohorts in patients with moderate to severe COPD. The MLMP-COPD model, although more accurate than BODE, is more complex; however, even relatively simple measures like BODE, in our experience, are not calculated by hand. The use of online clinical calculators as diagnostic, prognostic, and decision aids has increased in clinical practice.63,64 There is evidence that such point-of-care resources may improve diagnostic accuracy, adherence to guidelines, and accuracy of calculations.65 Therefore, we developed an online Web application that allows users to observe how altering input values affects predicted survival. Although our model should undergo further validation, future endeavors may include automated calculations via the electronic medical record to facilitate clinical implementation.
This study had several limitations. First, the COPDGene and ECLIPSE studies were multicenter case-control studies and not representative of the general population. The MLMP-COPD model should ideally be validated in general population samples. Differences in imaging protocols between the COPDGene and ECLIPSE studies may have accounted for the higher proportion of interstitial lung abnormalities and greater amount of emphysema in ECLIPSE and, thus, diminished the predictive power of quantitative imaging features. However, a simple harmonization (z score transformation) of imaging features did not improve mortality prediction, which could reflect the relatively lower effect of imaging compared with clinical features or the need to both standardize imaging protocols and/or develop more advanced image harmonization techniques. Despite these differences, the MLMP-COPD model performed well across these two heterogeneous populations.
Our study used all-cause mortality, which, although arguably more generalizable, given that many deaths in COPD are not due to respiratory disease, is nevertheless a limitation because of the lack of cause-specific mortality. Age was initially not included in the BODE index to maintain respiratory-specific mortality; however, our model performed better than ADO, which also accounts for age. Age, smoking history, and diabetes may be proxies for cardiovascular disease, which was not included in the model because of differences in assessment and missingness between the cohorts. The BODE index is currently used for selection of lung transplant recipients,66 and it is possible that our score could be used to improve this process, given that the median survival after lung transplant is approximately 5 years67 and BODE predicts out only to 4 years (vs 8 years with the MLMP-COPD model). Additional testing, ideally in a transplant population, is needed before considering its use as a selection tool. Future studies should examine the performance of the MLMP-COPD model in predicting respiratory- and cardiovascular-specific mortality with cause-specific comorbidities. Finally, previous studies have demonstrated the usefulness of adding blood-based biomarkers to predictive models6,47; however, our study did not include biomarkers because of the missingness in these cohorts.
Conclusions
In conclusion, the MLMP-COPD model demonstrated predictive performance superior to that of four prior mortality prediction indexes in subjects with moderate to severe COPD across two large cohorts. Further investigation across diverse populations and investigation of cause-specific mortality will help support the validity of this model.
Acknowledgments
Author contributions: M. M., D. Q., E. A. R., G. M. H., B. J. M., R. T.-S., M. J. M., P. J. C., M. K. H., J. V., E. K. S., B. D. H., and M. H. C. contributed to the study design. M. M., G. M. H., M. J. M. P. J. C., R. S. J. E., G. R. W., J. M. W., E. K. S., B. D. H., and M. H. C. contributed to the acquisition, analysis, or interpretation of the data. All authors contributed to the critical revision of the manuscript for important intellectual content. M. M., D. Q., M. J. M., E. K. S., B. D. H., and M. H. C. contributed to the statistical analysis. E. K. S. and M. H. C. obtained funding.
Financial/nonfinancial disclosures: The authors have reported to CHEST the following: G. M. H. reports personal fees from Boehringer Ingelheim; Genentech, Inc; and the Gerson Lehrman Group. R. T.-S. is an employee of GlaxoSmithKline plc. R. S. J. E. reports personal fees from Boehringer Ingelheim and Toshiba Corporation and is also a founder and co-owner of Quantitative Imaging Solutions LLC. G. R. W. has received grant support from Boehringer Ingelheim and BTG Interventional Medicine plc and other support from Genentech, Inc; GlaxoSmithKline plc; Janssen Pharmaceutica; ModoSpira; Pulmonx Corporation; Quantitative Imaging Solutions LLC; Regeneron Pharmaceuticals, Inc; and Toshiba Corporation. J. M. W. has received research contracts from AstraZeneca plc; Bayer AG; Gilead Sciences, Inc; GlaxoSmithKline plc; and Mereo BioPharma Group plc and advisory and consultancy fees from AstraZeneca plc, Boehringer Ingelheim, GlaxoSmithKline plc, IQVIA, Mereo BioPharma Group plc, and Mylan NV. M. K. H. reports consulting for AstraZeneca plc, Boehringer Ingelheim, and GlaxoSmithKline plc and research support from Novartis and Sunovion Pharmaceuticals Inc. E. K. S. has received grant funding and travel support from GlaxoSmithKline plc and honoraria from Novartis for continuing medical education seminars. M. H. C. has received grant funding from GlaxoSmithKline plc; consulting fees from AstraZeneca plc and Genentech, Inc; and speaking fees from Illumina, Inc. None declared (M. M., D. Q., E. A. R., B. J. M., M. J. M., P. J. C., D. L., M. S., R. P. B., J. V., B. C., P. C., J. C., B. D. H.).
Role of sponsors: GlaxoSmithKilne was involved in the design and collection of the original phenotype data for ECLIPSE. No other study sponsors had any role in the design of the study, the collection and analysis of the data, or the preparation of the manuscript.
Other contributions: The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Heart, Lung, and Blood Institute or the National Institutes of Health. Additional funding details and acknowledgments can be found in the supplementary online materials.
Additional information: The e-Appendix, e-Figures, and e-Tables can be found in the Supplemental Materials section of the online article.
Footnotes
Drs Hobbs and Cho are co-corresponding/co-senior authors.
FUNDING/SUPPORT: M. M. is supported by the National Heart, Lung, and Blood Institute [Grant T32HL007427]. B. D. H. is supported by the National Institutes of Health [Grant K08HL136928] and a Parker B. Francis Research Opportunity Award. M. H. C. is supported by the National Institutes of Health [Grants R01HL137927 and R01HL135142]. The Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints (ECLIPSE) study (NCT00292552; GlaxoSmithKline code SCO104960) was funded by GlaxoSmithKline plc. The COPDGene project (NCT00608764) was supported by the National Institutes of Health [Grant R01HL089897] and the National Heart, Lung, and Blood Institute [Award No. R01HL089856]. The COPDGene project is also supported by the COPD Foundation through contributions made to an industry advisory board composed of AstraZeneca plc, Boehringer Ingelheim, GlaxoSmithKline plc, Novartis, Pfizer Inc., Siemens AG, and Sunovion Pharmaceuticals Inc.
Supplementary Data
References
- 1.Soriano J.B., Abajobir A.A., Abate K.H. Global, regional, and national deaths, prevalence, disability-adjusted life years, and years lived with disability for chronic obstructive pulmonary disease and asthma, 1990-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Respir Med. 2017;5(9):691–706. doi: 10.1016/S2213-2600(17)30293-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Celli B.R., Cote C.G., Marin J.M. The body-mass index, airflow obstruction, dyspnea, and exercise capacity index in chronic obstructive pulmonary disease. N Engl J Med. 2004;350(10):1005–1012. doi: 10.1056/NEJMoa021322. [DOI] [PubMed] [Google Scholar]
- 3.Soler-Cataluña J.J., Martínez-García M.A., Sánchez L.S., Tordera M.P., Sánchez P.R. Severe exacerbations and BODE index: two independent risk factors for death in male COPD patients. Respir Med. 2009;103(5):692–699. doi: 10.1016/j.rmed.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 4.Moberg M., Vestbo J., Martinez G. Validation of the i-BODE index as a predictor of hospitalization and mortality in patients with COPD participating in pulmonary rehabilitation. COPD. 2014;11(4):381–387. doi: 10.3109/15412555.2013.836171. [DOI] [PubMed] [Google Scholar]
- 5.Boeck L., Soriano J.B., Brusse-Keizer M. Prognostic assessment in COPD without lung function: the B-AE-D indices. Eur Respir J. 2016;47(6):1635–1644. doi: 10.1183/13993003.01485-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stolz D., Meyer A., Rakic J., Boeck L., Scherr A., Tamm M. Mortality risk prediction in COPD by a prognostic biomarker panel. Eur Respir J. 2014;44(6):1557–1570. doi: 10.1183/09031936.00043814. [DOI] [PubMed] [Google Scholar]
- 7.Jones R.C., Donaldson G.C., Chavannes N.H. Derivation and validation of a composite index of severity in chronic obstructive pulmonary disease: the DOSE index. Am J Respir Crit Care Med. 2009;180(12):1189–1195. doi: 10.1164/rccm.200902-0271OC. [DOI] [PubMed] [Google Scholar]
- 8.Puhan M.A., Garcia-Aymerich J., Frey M. Expansion of the prognostic assessment of patients with chronic obstructive pulmonary disease: the updated BODE index and the ADO index. Lancet. 2009;374(9691):704–711. doi: 10.1016/S0140-6736(09)61301-5. [DOI] [PubMed] [Google Scholar]
- 9.Azarisman M.S., Fauzi M.A., Faizal M.P.A., Azami Z., Roslina A.M., Roslan H. The SAFE (SGRQ score, air-flow limitation and exercise tolerance) index: a new composite score for the stratification of severity in chronic obstructive pulmonary disease. Postgrad Med J. 2007;83(981):492–497. doi: 10.1136/pgmj.2006.052399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marin J.M., Alfageme I., Almagro P. Multicomponent indices to predict survival in COPD: the COCOMICS study. Eur Respir J. 2013;42(2):323–332. doi: 10.1183/09031936.00121012. [DOI] [PubMed] [Google Scholar]
- 11.Guerra B., Haile S.R., Lamprecht B. Large-scale external validation and comparison of prognostic models: an application to chronic obstructive pulmonary disease. BMC Med. 2018;16(1):33. doi: 10.1186/s12916-018-1013-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johannessen A., Skorge T.D., Bottai M. Mortality by level of emphysema and airway wall thickness. Am J Respir Crit Care Med. 2013;187(6):602–608. doi: 10.1164/rccm.201209-1722OC. [DOI] [PubMed] [Google Scholar]
- 13.Han M.K., Kazerooni E.A., Lynch D.A. Chronic obstructive pulmonary disease exacerbations in the COPDGene study: associated radiologic phenotypes. Radiology. 2011;261(1):274–282. doi: 10.1148/radiol.11110173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wells J.M., Washko G.R., Han M.K. Pulmonary arterial enlargement and acute exacerbations of COPD. N Engl J Med. 2012;367(10):913–921. doi: 10.1056/NEJMoa1203830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Terzikhan N., Bos D., Lahousse L. Pulmonary artery to aorta ratio and risk of all-cause mortality in the general population: the Rotterdam study. Eur Respir J. 2017;49(6) doi: 10.1183/13993003.02168-2016. pii:1602168. [DOI] [PubMed] [Google Scholar]
- 16.Putman R.K., Hatabu H., Araki T. Association between interstitial lung abnormalities and all-cause mortality. JAMA. 2016;315(7):672–681. doi: 10.1001/jama.2016.0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beam A.L., Kohane I.S. Big data and machine learning in health care. JAMA. 2018;319(13):1317–1318. doi: 10.1001/jama.2017.18391. [DOI] [PubMed] [Google Scholar]
- 18.James G., Witten D., Hastie T., Tibshirani R. Springer; New York, NY: 2013. An Introduction to Statistical Learning With Applications in R. [Google Scholar]
- 19.Sammut C., Webb G.I., editors. Encyclopedia of Machine Learning and Data Mining. 2nd ed. Springer; New York, NY: 2017. [Google Scholar]
- 20.Nichols J.A., Herbert Chan H.W., Baker M.A.B. Machine learning: applications of artificial intelligence to imaging and diagnosis. Biophys Rev. 2019;11(1):111–118. doi: 10.1007/s12551-018-0449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sidey-Gibbons J.A.M., Sidey-Gibbons C.J. Machine learning in medicine: a practical introduction. BMC Med Res Methodol. 2019;19(1):64. doi: 10.1186/s12874-019-0681-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delen D., Oztekin A., Kong Z.J. A machine learning-based approach to prognostic analysis of thoracic transplantations. Artif Intell Med. 2010;49(1):33–42. doi: 10.1016/j.artmed.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 23.Christodoulou E., Ma J., Collins G.S., Steyerberg E.W., Verbakel J.Y., Van Calster B. A systematic review shows no performance benefit of machine learning over logistic regression for clinical prediction models. J Clin Epidemiol. 2019;110:12–22. doi: 10.1016/j.jclinepi.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 24.Saria S., Butte A., Sheikh A. Better medicine through machine learning: what’s real, and what’s artificial? PLoS Med. 2018;15(12) doi: 10.1371/journal.pmed.1002721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naylor C.D. On the Prospects for a (deep) learning health care system. JAMA. 2018;320(11):1099–1100. doi: 10.1001/jama.2018.11103. [DOI] [PubMed] [Google Scholar]
- 26.Leidy N.K., Malley K.G., Steenrod A.W. Insight into best variables for COPD case identification: a random forests analysis. Chronic Obstr Pulm Dis. 2016;3(1):406–418. doi: 10.15326/jcopdf.3.1.2015.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amalakuhan B., Kiljanek L., Parvathaneni A., Hester M., Cheriyath P., Fischman D. A prediction model for COPD readmissions: catching up, catching our breath, and improving a national problem. J Community Hosp Intern Med Perspect. 2012;2(1) doi: 10.3402/jchimp.v2i1.9915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Motwani M., Dey D., Berman D.S. Machine learning for prediction of all-cause mortality in patients with suspected coronary artery disease: a 5-year multicentre prospective registry analysis. Eur Heart J. 2017;38(7):500–507. doi: 10.1093/eurheartj/ehw188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dawes T.J.W., de Marvao A., Shi W. Machine learning of three-dimensional right ventricular motion enables outcome prediction in pulmonary hypertension: a cardiac MR imaging study. Radiology. 2017;283(2):381–390. doi: 10.1148/radiol.2016161315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iyer A.S., Wells J.M., Vishin S., Bhatt S.P., Wille K.M., Dransfield M.T. CT scan-measured pulmonary artery to aorta ratio and echocardiography for detecting pulmonary hypertension in severe COPD. Chest. 2014;145(4):824–832. doi: 10.1378/chest.13-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Tho N., Ogawa E., Trang L.T.H. A mixed phenotype of airway wall thickening and emphysema is associated with dyspnea and hospitalization for chronic obstructive pulmonary disease. Ann Am Thorac Soc. 2015;12(7):988–996. doi: 10.1513/AnnalsATS.201411-501OC. [DOI] [PubMed] [Google Scholar]
- 32.Parr D.G., Stoel B.C., Stolk J., Stockley R.A. Validation of computed tomographic lung densitometry for monitoring emphysema in α1-antitrypsin deficiency. Thorax. 2006;61(6):485–490. doi: 10.1136/thx.2005.054890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishwaran H., Kogalur U.B., Blackstone E.H., Lauer M.S. Random survival forests. Ann Appl Stat. 2008;2(3):841–860. [Google Scholar]
- 34.Ishwaran H. Variable importance in binary regression trees and forests. Electron J Stat. 2007;1:519–537. [Google Scholar]
- 35.Hair J.F., Jr., Anderson R.E., Tatham R.L., Black W.C. 3rd ed. Macmillan; New York, NY: 1995. Multivariate Data Analysis. [Google Scholar]
- 36.Pencina M.J., D’Agostino R.B. Overall C as a measure of discrimination in survival analysis: model specific population value and confidence interval estimation. Stat Med. 2004;23(13):2109–2123. doi: 10.1002/sim.1802. [DOI] [PubMed] [Google Scholar]
- 37.Therneau T. A Package for Survival Analysis in R. 2020. R package version 3.1-12. https://CRAN.R-project.org/package=survival
- 38.Therneau T.M., Grambsch P.M. Springer; New York, NY: 2000. Modeling Survival Data: Extending the Cox Model. [Google Scholar]
- 39.Harrell F.E., Jr., Lee K.L., Mark D.B. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15(4):361–387. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 40.Sing T., Sander O., Beerenwinkel N., Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21(20):3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- 41.Kang L., Chen W., Petrick N.A., Gallas B.D. Comparing two correlated C indices with right-censored survival outcome: a one-shot nonparametric approach. Stat Med. 2015;34(4):685–703. doi: 10.1002/sim.6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Demler O.V., Paynter N.P., Cook N.R. Tests of calibration and goodness-of-fit in the survival setting. Stat Med. 2015;34(10):1659–1680. doi: 10.1002/sim.6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stolz D., Kostikas K., Blasi F. Adrenomedullin refines mortality prediction by the BODE index in COPD: the "BODE-A" index. Eur Respir J. 2014;43(2):397–408. doi: 10.1183/09031936.00058713. [published correction appears in Eur Respir J. 2014;44(6):1718] [DOI] [PubMed] [Google Scholar]
- 44.Bloom C.I., Ricciardi F., Smeeth L., Stone P., Quint J.K. Predicting COPD 1-year mortality using prognostic predictors routinely measured in primary care. BMC Med. 2019;17(1):73. doi: 10.1186/s12916-019-1310-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haile S.R., Guerra B., Soriano J.B. Multiple score comparison: a network meta-analysis approach to comparison and external validation of prognostic scores. BMC Med Res Methodol. 2017;17(1):1–12. doi: 10.1186/s12874-017-0433-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soriano J.B., Lamprecht B., Ramírez A.S. Mortality prediction in chronic obstructive pulmonary disease comparing the GOLD 2007 and 2011 staging systems: a pooled analysis of individual patient data. Lancet Respir Med. 2015;3(6):443–450. doi: 10.1016/S2213-2600(15)00157-5. [DOI] [PubMed] [Google Scholar]
- 47.Celli B.R., Locantore N., Yates J. Inflammatory biomarkers improve clinical prediction of mortality in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2012;185(10):1065–1072. doi: 10.1164/rccm.201110-1792OC. [DOI] [PubMed] [Google Scholar]
- 48.Briggs A.H., Baker T., Risebrough N.A. Development of the Galaxy Chronic Obstructive Pulmonary Disease (COPD) model using data from ECLIPSE: internal validation of a linked-equations cohort model. Med Decis Making. 2017;37(4):469–480. doi: 10.1177/0272989X16653118. [DOI] [PubMed] [Google Scholar]
- 49.Hoogendoorn M., Feenstra T.L., Asukai Y. External validation of health economic decision models for chronic obstructive pulmonary disease (COPD): report of the Third COPD Modeling Meeting. Value Health. 2017;20(3):397–403. doi: 10.1016/j.jval.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 50.Pinto-Plata V.M., Cote C., Cabral H., Taylor J., Celli B.R. The 6-min walk distance: change over time and value as a predictor of survival in severe COPD. Eur Respir J. 2004;23(1):28–33. doi: 10.1183/09031936.03.00034603. [DOI] [PubMed] [Google Scholar]
- 51.Polkey M.I., Spruit M.A., Edwards L.D. Six-minute-walk test in chronic obstructive pulmonary disease: minimal clinically important difference for death or hospitalization. Am J Respir Crit Care Med. 2013;187(4):382–386. doi: 10.1164/rccm.201209-1596OC. [DOI] [PubMed] [Google Scholar]
- 52.Celli B., Tetzl K., Criner G. The 6-minute-walk distance test as a chronic obstructive pulmonary disease stratification tool: insights from the COPD Biomarker Qualification Consortium. Am J Respir Crit Care Med. 2016;194(12):1483–1493. doi: 10.1164/rccm.201508-1653OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmidt S.A.J., Johansen M.B., Olsen M. The impact of exacerbation frequency on mortality following acute exacerbations of COPD: a registry-based cohort study. BMJ Open. 2014;4(12) doi: 10.1136/bmjopen-2014-006720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soler-Cataluña J.J., Martínez-García M.A., Román Sánchez P., Salcedo E., Navarro M., Ochando R. Severe acute exacerbations and mortality in patients with chronic obstructive pulmonary disease. Thorax. 2005;60(11):925–931. doi: 10.1136/thx.2005.040527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suissa S., Dell’Aniello S., Ernst P. Long-term natural history of chronic obstructive pulmonary disease: severe exacerbations and mortality. Thorax. 2012;67(11):957–963. doi: 10.1136/thoraxjnl-2011-201518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cardoso J., Coelho R., Rocha C., Coelho C., Semedo L., Bugalho Almeida A. Prediction of severe exacerbations and mortality in COPD: the role of exacerbation history and inspiratory capacity/total lung capacity ratio. Int J Chron Obstruct Pulmon Dis. 2018;13:1105–1113. doi: 10.2147/COPD.S155848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ho T.W., Huang C.T., Ruan S.Y., Tsai Y.J., Lai F., Yu C.J. Diabetes mellitus in patients with chronic obstructive pulmonary disease: the impact on mortality. PLoS One. 2017;12(4) doi: 10.1371/journal.pone.0175794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miller J., Edwards L.D., Agustí A. Comorbidity, systemic inflammation and outcomes in the ECLIPSE cohort. Respir Med. 2013;107(9):1376–1384. doi: 10.1016/j.rmed.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 59.LaFon D.C., Bhatt S.P., Labaki W.W. COPDGene Investigators. Pulmonary artery enlargement and mortality risk in moderate to severe COPD: results from COPDGene. Eur Respir J. 2020;55(2):1901812. doi: 10.1183/13993003.01812-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lowe K.E., Regan E.A., Anzueto A. COPDGene® 2019: redefining the diagnosis of chronic obstructive pulmonary disease. Chronic Obstr Pulm Dis. 2019;6(5):384–399. doi: 10.15326/jcopdf.6.5.2019.0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.González G., Ash S.Y., Vegas-Sánchez-Ferrero G. Disease staging and prognosis in smokers using deep learning in chest computed tomography. Am J Respir Crit Care Med. 2018;197(2):193–203. doi: 10.1164/rccm.201705-0860OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Datema F.R., Moya A., Krause P. Novel head and neck cancer survival analysis approach: random survival forests versus Cox proportional hazards regression. Head Neck. 2012;34(1):50–58. doi: 10.1002/hed.21698. [DOI] [PubMed] [Google Scholar]
- 63.Dziadzko M.A., Gajic O., Pickering B.W., Herasevich V. Clinical calculators in hospital medicine: availability, classification, and needs. Comput Methods Programs Biomed. 2016;133:1–6. doi: 10.1016/j.cmpb.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 64.Mosa A.S.M., Yoo I., Sheets L. A systematic review of healthcare applications for smartphones. BMC Med Inform Decis Mak. 2012;12:67. doi: 10.1186/1472-6947-12-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mickan S., Atherton H., Roberts N.W., Heneghan C., Tilson J.K. Use of handheld computers in clinical practice: a systematic review. BMC Med Inform Decis Mak. 2014;14:56. doi: 10.1186/1472-6947-14-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weill D., Benden C., Corris P.A. A consensus document for the selection of lung transplant candidates: 2014—an update from the Pulmonary Transplantation Council of the International Society for Heart and Lung Transplantation. J Heart Lung Transplant. 2015;34(1):1–15. doi: 10.1016/j.healun.2014.06.014. [DOI] [PubMed] [Google Scholar]
- 67.Orens J.B., Estenne M., Arcasoy S. International guidelines for the selection of lung transplant candidates: 2006 update—a consensus report from the Pulmonary Scientific Council of the International Society for Heart and Lung Transplantation. J Heart Lung Transplant. 2006;25(7):745–755. doi: 10.1016/j.healun.2006.03.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.