Supplemental Digital Content is available in the text
Keywords: cancer, lncRNAs, prognostic marker, SNHG20
Abstract
Background:
Small nucleolar RNA host gene 20 (SNHG20) is a newly identified long non-coding RNA (lncRNA). Accumulative evidence suggest that SNHG20 is highly related to tumorigenesis. However, whether the levels of SNHG20 can be used for prognosis of patients with different cancer types was unclear. The present study aims to explore the role of SNHG20 in tumor prognosis and its clinical significance.
Methods:
Related articles published before March 14, 2019 were searched in PubMed, Excerpta Medica Database (EMBASE), ISI Web of Science, and China National Knowledge Infrastructure (CNKI). Hazard ratios (HRs) and their corresponding 95% confidence intervals (CIs) were obtained using Stata 11.0 software and used to for determination of the link between the levels of SNHG20 and overall survival (OS). Fixed or random model was chosen depending on the heterogeneity of the studies. A quality assessment of the included studies was performed according to the Newcastle-Ottawa scale. This study was approved by the Medical Ethics Committee of Xiangya Hospital of Central South University.
Results:
After a strict filtering process, a total of 1149 patients from 15 studies were enrolled in this study. Pooled data showed that elevated level of SNHG20 was correlated not only with poor overall survival (HR = 2.49, 95% confidence interval (CI): 2.05–2.98), but also with tumor-node-metastasis stage (TNM) (odds ratio (OR) = 3.32, 95% CI: 2.27–4.86), high histological grade (OR = 2.11, 95% CI: 1.55–2.87), tumor size (OR = 2.92, 95% CI: 2.17–3.91), and lymph node metastasis (OR = 4.48, 95% CI: 2.90–6.92). Of note, there is no significant heterogeneity difference among the studies.
Conclusion:
Up-regulated SNHG20 predicts unfavorable prognosis for multiple kinds of cancers although further studies are in need to verify its clinical applications.
1. Introduction
Cancer-related death is increasing globally and therefore combating cancer is one of the top priorities to the World Health Organization.[1] Non-coding RNAs (ncRNAs), once considered as transcription noise, play important roles in health and disease by function either as oncogenic factors or tumor suppressors. Accumulating evidence suggests a tight association between ncRNAs and cancer development.[2] Comparing to our understanding about microRNAs (miRNA) as a commonly studied ncRNAs, the function of the subset long non-coding RNAs (lncRNAs) is relatively unknown.[3–7] Small nucleolar RNA host gene 20 (SNHG20) has attracted more attention in the past few years due to its potential role in tumorigenesis. Locating on chromosome 17q25.2 with the length of 2183 base-pairs, SNHG20 was firstly identified in hepatocellular carcinoma (HCC) and subsequent studies suggested that SNHG20 played important roles including colorectal cancer, cervical cancer, and breast cancer.[8–11]
Although accumulating data suggests SNHG20 in an oncogenic ncRNA, all the conclusions were based on individual research. We decided to conduct a meta-analysis focusing on the relationship between the expression of SHNG20 and clinic-pathological features in different cancers and a systematic review about its prognostic application.
2. Materials and methods
2.1. Literature search
PubMed, Excerpta Medica Database (EMBASE), ISI Web of Science, Medline, and China National Knowledge Infrastructure (CNKI) (updated until March 14, 2019) were searched using the following terms: carcinoma, tumor, neoplasm and tumorigenesis, and SNHG20. Related articles identified in the initial research were also screened manually.
2.2. Inclusion and exclusion criteria
The studies meeting the following criteria would be included:
-
a)
the prognostic value of SNHG20 have been evaluated regardless of cancer type,
-
b)
the studies contain detailed data to calculate the hazard ratio (HR) estimates with the corresponding 95% CIs for overall survival (OS), or odds ratio (OR) for clinic-pathological features, and
-
c)
the studies should be original research articles published either in Chinese or English.
The following are the major exclusion criteria:
-
a)
abstract, review, comment, editorial, and case reports,
-
b)
overlapping data,
-
c)
cell lines and non-human research, and
-
d)
insufficient data.
2.3. Data extraction and quality assessment
The data were extracted by 2 investigators independently from the selected publications with a standardized table, including
-
a)
study features, such as year of publication, first author's surname, and country of origin,
-
b)
participants’ general characteristics, such as cancer type, sample size, sample specimen, detection method, and duration of follow-up,
-
c)
data needed for this meta-analysis, such as HR and 95% CIs, and SNHG20 expression frequencies in different clinic-pathological features (tumor-node-metastasis stage (TNM) stage, histological grade, tumor size, and lymph node metastasis).
Any disagreements were discussed and consulted by a senior investigator.
2.4. Statistical analysis
STATA statistical software (version 11.0; Stata Corp, College Station, TX) was applied to analyze the extracted data. In the analysis, pooled HRs and the 95% CIs were performed for OS to evaluate the relationship between SNHG20 and cancer prognosis, and ORs and the corresponding 95% CIs were performed for clinic-pathological features (TNM stage, histological grade, tumor size, and lymph node metastasis). The median value is used to divide high and low expression of SNHG20. Z test was used to assess the significance of the pooled HRs and ORs. If not obtained directly from the article, HRs are estimated via the survival curve by the methodology mentioned by Tierney.[12]
For patients with up-regulated SNHG20 level, HR > 1 means a poorer cancer prognosis, and a HR with 95% confidence interval (CI) not including 1 would be considered as statistically significant. Sensitivity analysis was performed to analyze the heterogeneity sources by sequentially removing individual study. Finally, Egger's linear regression test, Begg's rank correlation test, and the corresponding funnel plot were conducted to evaluate potential publication bias.
3. Results
3.1. Characteristics of eligible studies
A total of 86 relevant publications were included based on our initial search. Among them, 59 studies were excluded because of duplicates or obvious irrelevance, and 12 studies were excluded as reviews or not about cancer prognosis. Finally, 15 articles with 1149 cancer patients met the selection criteria and were included in our meta-analysis. The flow chart showed the study selection process (Fig. 1).[9–11,13–24] The main characteristics of each study have been summarized in Table 1. All included studies were conducted in Chinese population and published from 2016 to 2019. The specimen source is tissue and detection method for SNHG20 is reverse transcription polymerase chain reaction (RT-qPCR). In total, 12 cancer types were included in our study: colorectal cancer (CRC), osteosarcoma (OSA), oral squamous cell carcinoma (OSCC), cervical cancer (CC), HCC, nasopharyngeal carcinoma (NC), esophageal squamous cell carcinoma (ESCC), non-small cell lung cancer (NSCLC), epithelial ovarian cancer (EOC), glioma, bladder cancer (BC), and gastric cancer (GC). Among the included studies, 6 of them investigated the OS by the mulivariae Cox regression analysis, and the rest used the KM curve. The expression pattern of SNHG20 in 12-cancer types was shown in Supplement Figure 1. Additionally, all included studies got NOS scores of 7 or more, illustrating the high methodological quality. The Cancer Genome Atlas (TCGA) database results were included in Supplement Figure 2, of which HCC showed high consistency and thus could be a strong potential target in prognosis as well as treatment.
Figure 1.
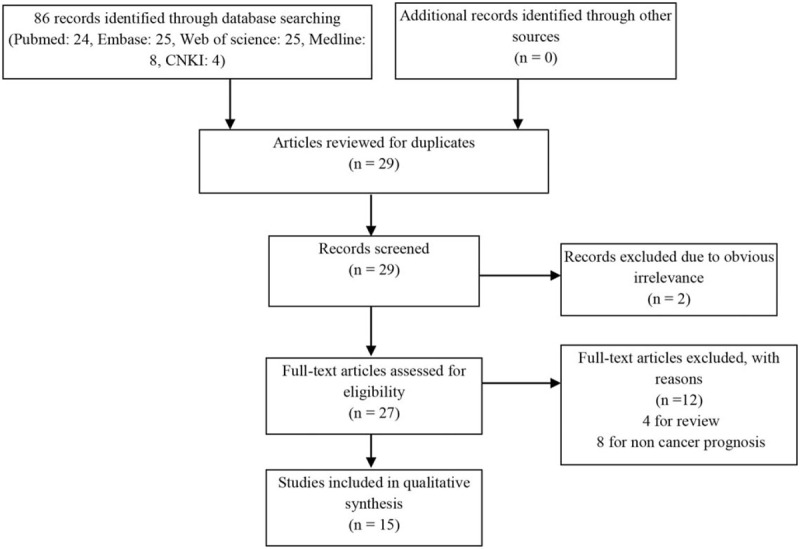
Flow diagram of the study selection process.
Table 1.
Main characteristics of articles included in this meta-analysis.
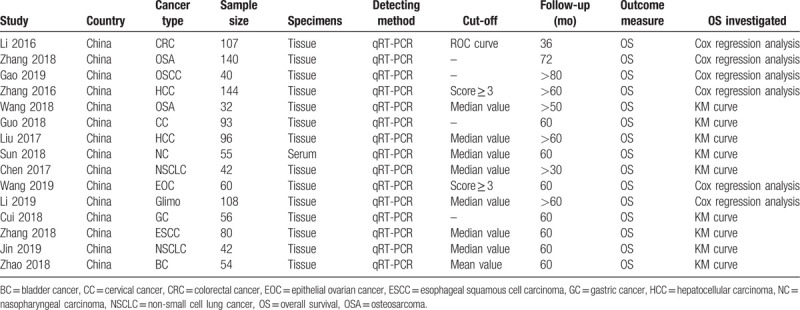
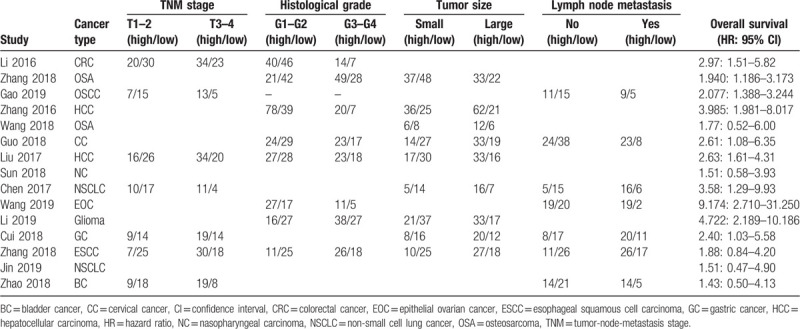
The TNM stage was assessed in 7 studies, results showed a median of 46.9% (range: 40.0–64.3%) of patients were stage T1–2, while the other 53.1% (range: 35.7–60.0%) of patients were stage T3–4 (Table 2). The histological grade was assessed in 8 studies, a median of 60.0% (range: 39.8–81.3%) of patients were grade G1–2, while the other 40.0% (range: 18.7–60.2%) of patients were G3–4 (Table 2). The tumor size was assessed in 9 studies, results showed a median of 48.5% (range: 42.4–60.7%) of patients were small tumor size, while the other 51.5% (range: 39.3–57.6%) of patients were large tumor size (Table 2). The lymph node metastasis was assessed in 7 studies, a median of 57.4% (range: 44.6–66.7%) of patients were not lymph node metastasis, while the other 42.6% (range: 33.3–55.4%) of patients had lymph node metastasis (Table 2).
Table 2.
Main data from articles for this meta-analysis.
3.2. Tissue SNHG20 level and overall survival
In our study, 15 studies with 1149 cases were contained to evaluate the relationship between tissue SNHG20 expression level and OS for cancers. The heterogeneity analysis was not significant among the eligible studies (I2 = 5.7%, P = .389), and thus we used a fixed model to calculate the pooled HR. The result was 2.47 (95% CI: 2.05–2.98, P < .001), showing that up-regulated SNHG20 predicted poorer OS for cancer patients (Fig. 2a).
Figure 2.
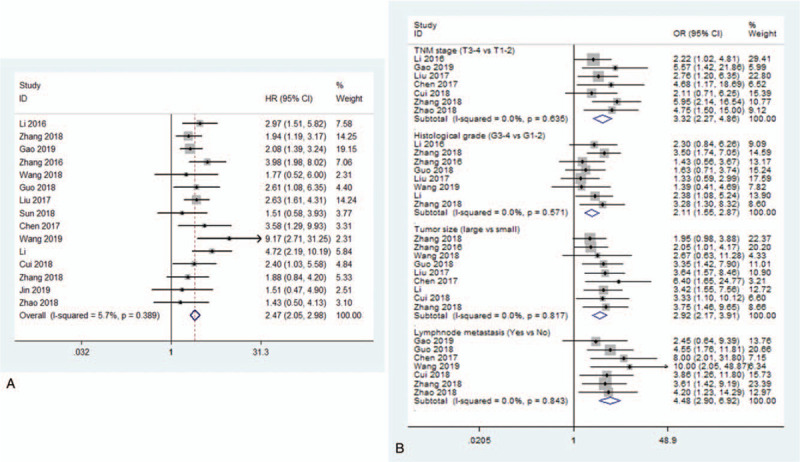
a) Forest plot of HRs for OS of high SNHG20 expression vs low expression in cancer patients. Low/high divided by the sample median. b) Forest plot of ORs for clinic-pathological features of high SNHG20 expression vs low expression in cancer patients. HRs = hazard ratios, OR = odds ratio, OS = overall survival, SNHG20 = small nucleolar RNA host gene 20.
3.3. Tissue SNHG20 level and clinic-pathological features
To study the relationship between SNHG20 expression level and the TNM stage of cancer, 7 studies with 475 cases were included. The heterogeneity analysis showed no significant difference among the eligible studies (I2 = 0.0%, P = .635).[10,13–16,19,24] The pooled OR was 3.32 (95% CI: 2.27–4.86, P < .001), indicating that high SNHG20 expression level was closely associated with high TNM stage (Fig. 2b).
Meanwhile, 8 studies with 828 patients were included to evaluate the link of SNHG20 and histological grade. Heterogeneity difference was not observed among the studies (I2 = 0.0%, P = .572),[9–11,19,21,23–25] and the pooled OR was 2.11 (95% CI: 1.55–2.87, P < .001), indicating that high SNHG20 expression predicted higher histological grade (Fig. 2b).
In addition, we found that higher SNHG20 expression also predicted bigger tumor size (OR = 2.92, 95% CI: 2.17–3.91, P < .001) and more lymph node metastasis (OR = 4.48, 95% CI: 2.90–6.92, P < .001) (Fig. 2b).[9,11,13–16,19,21–25]
3.4. Sensitivity analysis
Sensitivity analysis was conducted by sequentially removing individual study, and the pooled HRs for OS were not substantially affected, which confirmed the stability and reliability of our results (Fig. 3a).
Figure 3.
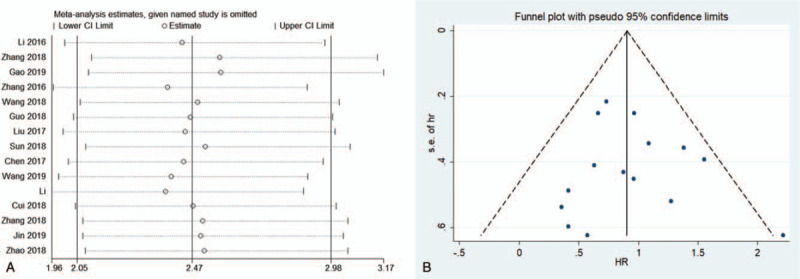
a) Sensitivity analysis of the pooled HRs of SNHG20 expression for OS for the included studies. b) Begg's funnel plot was used to evaluate potential publication bias for OS and estimations. HRs = hazard ratios, OS = overall survival, SNHG20 = small nucleolar RNA host gene 20.
3.5. Publication bias
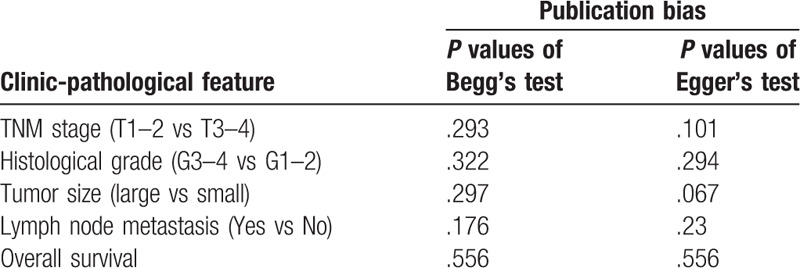
We conducted the Funnel plot, Begg's rank test, and Egger's linear regression test to evaluate the potential publication bias in the included studies. A symmetric funnel plot suggested the absence of publication bias (Fig. 3b). In addition, all the P values > .05 in Begg's rank test and Egger's linear regression test (Table 3), suggesting there is no obvious publication bias in our meta-analysis.
Table 3.
Publication bias analyses among included studies.
4. Discussion
SNHG20, classified as lncRNAs, was proposed as a promising biomarker for cancer and a potential target for cancer treatment. Recent studies have already showed that aberrant expression of SNHG20 was relevant to prognosis for various cancers.[26] Yet so far, there is no summary data with a large sample size regarding the association between SNHG20 expression and survival of tumors. Thus, it's important to systematically evaluate the most frequently reported SNHG20 articles about cancer. Our meta-analysis included 15 different studies with 1149 patients, and the pooled HR of 2.47 suggested the prognostic value of SNHG20 in cancer patients. In addition, the expression level of SNHG20 was relevant to TNM stage, tumor size histological grade, and lymph node metastasis.
A promising biomarker and potential treatment target for cancer as SNHG20 is, the mechanism of how it contributes to the oncogenesis remains controversial. However, some possible elements might be involved in the regulating process of SNHG20. In molecular level, several studies have found that SNHG20 could inhibit the downstream gene expression by competing with specific miRNAs.[8,27] Moreover, SNHG20 could interact directly with proteins to regulate cell phenotype without sponging miRNAs.[22] Furthermore, increasing researches suggested that SNHG20 participated in the tumorigenesis via EMT signaling pathway and p21 signaling pathway.[17,28,29]
In spite of the stronger statistical power than single study and absence of publication bias with strict inclusion criteria, some limitations of this meta-analysis should be acknowledged. On one hand, all the studies were conducted in China, which may influence the broader application of the result. On the other hand, some of the HRs need to be obtained from the survival curves instead of primary studies, which made the data less accurate.
In summary, our meta-analysis, representing a quantified synthesis of all published studies, has elucidated the prognostic value of SNHG20 in different cancer types. More well-designed studies with a larger sample size are expected to further confirm our findings.
Author contributions
Conceptualization: Zhuoyi Liu, Zhiping Hu.
Data curation: Jiling zeng.
Formal analysis: Jiling zeng, Zhuoyi Liu.
Investigation: Chao Zhang, Siyuan Tang.
Methodology: Jiling zeng, Zhiping Hu.
Project administration: Chao Zhang.
Resources: Jiling zeng, Furong Zeng.
Software: Jiling zeng, Chao Zhang, Tao Hong.
Supervision: Jiling zeng, Zhiping Hu.
Validation: Zhiping Hu, Tao Hong.
Visualization: Furong Zeng.
Writing – original draft: Jiling zeng.
Writing – review & editing: Jiling zeng, Zhuoyi Liu, Zhiping Hu, Tao Hong, Siyuan Tang.
Jiling zeng orcid: 0000-0003-4490-0193.
Supplementary Material
Supplementary Material
Footnotes
Abbreviations: BC = bladder cancer, CC = cervical cancer, CI = confidence interval, CNKI = China National Knowledge Infrastructure, CRC = colorectal cancer, EMBASE = Excerpta Medica Database, EOC = epithelial ovarian cancer, glioma, ESCC = esophageal squamous cell carcinoma, GC = gastric cancer, HCC = hepatocellular carcinoma, HR = hazard ratio, lncRNA = long non-coding RNA, lncRNAs = long non-coding RNAs, miRNA = microRNAs, NC = nasopharyngeal carcinoma, ncRNAs = non-coding RNAs, NSCLC = non-small cell lung cancer, OR = odds ratio, OS = overall survival, OSA = osteosarcoma, OSCC = oral squamous cell carcinoma, SNHG20 = small nucleolar RNA host gene 20, TCGA = the Cancer Genome Atlas, TNM = tumor-node-metastasis stage.
How to cite this article: Zeng J, Liu Z, Zhang C, Hong T, Zeng F, Guan J, Tang S, Hu Z. Prognostic value of long non-coding RNA SNHG20 in cancer: A meta-analysis. Medicine. 2020;99:9(e19204).
JZ, ZL, and CZ contributed equally as the first authors.
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
The authors have no conflicts of interest to disclose.
References
- [1].Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359–386. [DOI] [PubMed] [Google Scholar]
- [2].Roberts TC, Wood MJA. Therapeutic targeting of non-coding RNAs. Essays Biochem 2013;54:127–45. [DOI] [PubMed] [Google Scholar]
- [3].Zhou DB, Xie MX, He BM, et al. Microarray data re-annotation reveals specific lncRNAs and their potential functions in non-small cell lung cancer subtypes. Mol Med Rep 2017;16:5129–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Li Z, Huang J, Ni J. LncRNA GAS5-AS1 inhibits osteosarcoma cell growth and invasion by regulating MMP2. Int J Clin Exp Pathol 2017;10:2843–51. [Google Scholar]
- [5].Mao C, Wang X, Liu Y, et al. A G3BP1-interacting lncRNA promotes ferroptosis and apoptosis in cancer via nuclear sequestration of p53. Cancer Res 2018;78:3484–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Peng L, Yuan X, Jiang B, et al. LncRNAs: key players and novel insights into cervical cancer. Tumour Biol 2016;37:2779–88. [DOI] [PubMed] [Google Scholar]
- [7].Ren S, Li G, Liu C, et al. Next generation deep sequencing identified a novel lncRNA n375709 associated with paclitaxel resistance in nasopharyngeal carcinoma. Oncol Rep 2016;36:1861–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Guan YX, Zhang MZ, Chen XZ, et al. Lnc RNA SNHG20 participated in proliferation, invasion, and migration of breast cancer cells via miR-495. J Cell Biochem 2018;119:7971–81. [DOI] [PubMed] [Google Scholar]
- [9].Guo H, Yang S, Li S, et al. LncRNA SNHG20 promotes cell proliferation and invasion via miR-140-5p-ADAM10 axis in cervical cancer. Biomed Pharmacother 2018;102:749–57. [DOI] [PubMed] [Google Scholar]
- [10].Li C, Zhou L, He J, et al. Increased long noncoding RNA SNHG20 predicts poor prognosis in colorectal cancer. BMC Cancer 2016;16:655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zhang D, Cao C, Liu L, et al. Up-regulation of LncRNA SNHG20 predicts poor prognosis in hepatocellular carcinoma. J Cancer 2016;7:608–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Tierney JF, Stewart LA, Ghersi D, et al. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials 2007;8:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhao Q, Gao S, Du Q, et al. Long non-coding RNA SNHG20 promotes bladder cancer via activating the Wnt/β-catenini signaling pathway. Int J Mol Med 2018;2839–48. [DOI] [PubMed] [Google Scholar]
- [14].Chen Z, Chen X, Chen P, et al. Long non-coding RNA SNHG20 promotes non-small cell lung cancer cell proliferation and migration by epigenetically silencing of P21 expression. Cell Death Dis 2017;8: [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Cui N, Liu J, Xia H, et al. LncRNA SNHG20 contributes to cell proliferation and invasion by upregulating ZFX expression sponging miR-495-3p in gastric cancer. J Cell Biochem 2019;120:3114–23. [DOI] [PubMed] [Google Scholar]
- [16].Gao P, Fan R, Ge T. SNHG20 serves as a predictor for prognosis and promotes cell growth in oral squamous cell carcinoma. Oncol Lett 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Li XS, Shen FZ, Huang LY, et al. lncRNA small nucleolar RNA host gene 20 predicts poor prognosis in glioma and promotes cell proliferation by silencing P21. Onco Targets Ther 2019;12:805–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lingling J, Xiangao J, Guiqing H, et al. SNHG20 knockdown suppresses proliferation, migration and invasion, and promotes apoptosis in non-small cell lung cancer through acting as a miR-154 sponge. Biomed Pharmacother 2019;112. [DOI] [PubMed] [Google Scholar]
- [19].Liu J, Lu C, Xiao M, et al. Long non-coding RNA SNHG20 predicts a poor prognosis for HCC and promotes cell invasion by regulating the epithelial-to-mesenchymal transition. Biomed Pharmacother 2017;89:857–63. [DOI] [PubMed] [Google Scholar]
- [20].Sun C, Sun Y, Zhang E. Long non-coding RNA SNHG20 promotes nasopharyngeal carcinoma cell migration and invasion by upregulating TGF-beta1. Exp Ther Med 2018;16:4967–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wang D, Dai J, Hou S, et al. LncRNA SNHG20 predicts a poor prognosis and promotes cell progression in epithelial ovarian cancer. Biosci Rep 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wang W, Luo P, Guo W, et al. LncRNA SNHG20 knockdown suppresses the osteosarcoma tumorigenesis through the mitochondrial apoptosis pathway by miR-139/RUNX2 axis. Biochem Biophys Res Commun 2018;503:1927–33. [DOI] [PubMed] [Google Scholar]
- [23].Zhang C, Jiang F, Su C, et al. Upregulation of long noncoding RNA SNHG20 promotes cell growth and metastasis in esophageal squamous cell carcinoma via modulating ATM-JAK-PD-L1 pathway. J Cell Biochem 2019. [DOI] [PubMed] [Google Scholar]
- [24].Zhang J, Ju C, Zhang W, et al. LncRNA SNHG20 is associated with clinical progression and enhances cell migration and invasion in osteosarcoma. IUBMB Life 2018;70:1115–21. [DOI] [PubMed] [Google Scholar]
- [25].Li X-S, Shen F-Z, Huang L-Y, et al. lncRNA small nucleolar RNA host gene 20 predicts poor prognosis in glioma and promotes cell proliferation by silencing P21. Onco Targets Ther 2019;12:805–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhao W, Ma X, Liu L, et al. SNHG20: a vital lncRNA in multiple human cancers. J Cell Physiol 2019. [DOI] [PubMed] [Google Scholar]
- [27].Wu J, Zhao W, Wang X, et al. Long non-coding RNA SNHG20 promotes the tumorigenesis of oral squamous cell carcinoma via targeting miR-197/LIN28 axis. J Cell Mol Med 2019;680–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chen Z, Chen X, Chen P, et al. Long non-coding RNA SNHG20 promotes non-small cell lung cancer cell proliferation and migration by epigenetically silencing of P21 expression. Cell Death Dis 2017;8:e3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Pang A, Carbini M, Moreira AL, et al. Carcinosarcomas and related cancers: tumors caught in the act of epithelial–mesenchymal transition. J Clin Oncol 2018;36:210–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


