Abstract
Roseburia species are important denizens of the human gut microbiome that ferment complex polysaccharides to butyrate as a terminal fermentation product, which influences human physiology and serves as an energy source for colonocytes. Previous comparative genomics analyses of the genus Roseburia have examined polysaccharide degradation genes. Here, we characterize the core and pangenomes of the genus Roseburia with respect to central carbon and energy metabolism, as well as biosynthesis of amino acids and B vitamins using orthology-based methods, uncovering significant differences among species in their biosynthetic capacities. Variation in gene content among Roseburia species and strains was most significant for cofactor biosynthesis. Unlike all other species of Roseburia that we analysed, Roseburia inulinivorans strains lacked biosynthetic genes for riboflavin or pantothenate but possessed folate biosynthesis genes. Differences in gene content for B vitamin synthesis were matched with differences in putative salvage and synthesis strategies among species. For example, we observed extended biotin salvage capabilities in R. intestinalis strains, which further suggest that B vitamin acquisition strategies may impact fitness in the gut ecosystem. As differences in the functional potential to synthesize components of biomass (e.g. amino acids, vitamins) can drive interspecies interactions, variation in auxotrophies of the Roseburia spp. genomes may influence in vivo gut ecology. This study serves to advance our understanding of the potential metabolic interactions that influence the ecology of Roseburia spp. and, ultimately, may provide a basis for rational strategies to manipulate the abundances of these species.
Keywords: Lachnospiraceae, Roseburia, comparative genomics, B vitamin biosynthesis, amino acid biosynthesis, butyrate synthesis
Data Summary
The authors confirm that all supporting data, code and protocols have been provided within the article or through supplementary data files. The genomes used for annotation of strains were taken from the following GenBank files: Roseburia faecis 2789STDY5608863 (GCA_001405615.1), R. faecis M72 (GCA_001406815.1), Roseburia sp. 831b (GCA_001940165.1), Roseburia inulinivorans 2789STDY5608887 (GCA_001405535.1), R. inulinivorans DSM 16841 (GCA_000174195.1), R. inulinivorans LI-83 (GCA_001406855.1), Roseburia intestinalis LI-82 (GCA_000156535.1), R. intestinalis M50/1 (GCA_000209995.1), R. intestinalis XB6B4 (GCA_000210655.1), Roseburia hominis A2-183 (GCA_000225345.1). All DNA and amino acid sequences, as well as RAST annotations, are provided in the supplemental Excel file in Table S2. Compiled and scored pathway information has been curated in Table S5. Additional scripts and output files can be found on our GitHub repository (https://github.com/ehillman26/Comparative-Genomics-of-Roseburia.git).
Impact Statement.
Here, we employ a comparative genomics approach to define the core and pangenomes of the genus Roseburia and identify species- and strain-level traits that might play a role in their ability to colonize the gut ecosystem. By evaluating Roseburia ’s proposed physiological and biosynthetic capabilities, we propose underlying principles that may govern the ecology and establishment of specific species and strains. While some of these aspects, such as Roseburia ’s fermentative end products, have been studied in great detail by other groups, this study connects those findings through a genomic lens and identifies the associated genes across the genus. Our results suggest that B vitamin biosynthesis genes in Roseburia spp. might play a large role in their ecology in the gut environment. We present several testable hypotheses that may help unravel the complex nature of these important gut microbes. This understanding may, ultimately, lead to therapeutic approaches that can selectively modulate the gut microbiome.
Introduction
Roseburia, a member of the Clostridium coccodis cluster of the phylum Firmicutes [1], is a genus of anaerobic, rod-shaped, Gram-positive bacteria [2]. Species of Roseburia are known to be important denizens of the human gut microbiome, with relative abundances estimated at 5–15 % between Roseburia spp. and their near neighbours in the genus Agathobacter [3] (previously Eubacterium [4]); many have been isolated from human faeces [5–7]. Roseburia spp. are known to ferment complex polysaccharides entering the colon to butyrate as a terminal product [2, 8]. Butyrate is the preferred energy source of colonocytes in the human large intestine as well as a known histone deacetylase inhibitor [9] and immunomodulatory signal [10]. Recently, it has been suggested that butyrate production by gut microbes and, specifically, Roseburia spp., may confer health benefits to humans, including prevention of type II diabetes [11], ulcerative colitis [12] and colon cancer [13, 14]. The abundance of Roseburia spp. in faecal samples may also serve as a biomarker for symptomatic pathologies or certain species may be delivered as probiotics for restoration of the gut ecosystem [2, 14–17].
Although several strains within the genus Roseburia have been sequenced, there has been no large-scale effort to identify the central catabolic and anabolic genes that compose the core and pangenomes of genus Roseburia . As differences in the functional potential to synthesize components of biomass (e.g. amino acids, vitamins) can drive interspecies interactions [18] due, in part, to competition for available ‘public good’ nutrients in communities [19], variation in the genomes of Roseburia spp. may influence the ecology of these species. This may be especially important for the synthesis of required B vitamin cofactors, as colonic competition for these resources (e.g. vitamin B12 and other corrinoids) has been proposed to structure microbiomes [20]. Such competition may arise due to the commonality of B12 auxotrophies and its role in essential processes (e.g. deoxyribonucleotide production [21]), as well as efficient host scavenging processes [22] and lability to gastric degradation [23]. Recent comparison of complex carbohydrate metabolism in members of the genus Roseburia and their near neighbour Agathobacter rectalis (previously Eubacterium rectale ) revealed extensive niche partitioning of these species around polysaccharide utilization [24]. By pairing genomic prediction and experimental evidence, Sheridan et al. determined that carbohydrate substrate preferences with respect to tested oligo- and polysaccharides were relatively stable at the species and, to an extent, the genus level, with some strain-level differentiation. Despite some divergences in carbohydrate preferences, these organisms share their core fermentative metabolisms, containing highly syntenic operons with the genes required for butyrogenesis from pyruvate [8, 25–27].
In this study, we aimed to (1) define the core and pangenome of the genus Roseburia with respect to central carbon and energy metabolism and biosynthetic genes and (2) evaluate the degree to which species- and strain-level differences in auxotrophies might influence competition among these organisms in the colonic ecosystem. We focused in particular on the mechanisms by which Roseburia spp. produce amino acids and B vitamins. B vitamins, including thiamine, riboflavin, niacin, pantothenate, pyridoxine, biotin, folate and cobalamin, are nutrients that serve as coenzymes for reactions in bacterial and eukaryotic cells alike, and alterations in the microbial production of these molecules may influence host health [28]. B vitamins are required cofactors for many central metabolic pathways and are involved in diverse biosynthetic processes. Additionally, derivatives of B vitamins, including niacin and riboflavin, also play a role in maintaining cellular oxidative balance. Lack of genes required for biosynthesis of required amino acids and vitamins in certain Roseburia strains would require that they successfully compete with the human host and other gut species for these nutrients in the colon. Understanding the metabolic interactions that influence the ecology of Roseburia spp. may provide mechanistic bases for rational strategies to increase or maintain abundances of these species that may synergize with approaches that employ differing carbohydrate preferences.
Methods
We examined 11 different genomes from the genus Roseburia in this study for comparative genomic analysis, including representatives from all published species with sequenced genomes and genomes unattributed to any species ( Roseburia intestinalis XB6B4, R. intestinalis LI-82, R. intestinalis M50/1, Roseburia hominis A2-183, Roseburia inulinivorans LI-83, R. inulinivorans DSM 16841, R. inulinivorans 2789STD5608887, Roseburia faecis M72, R. faecis 2789STDY5608863, Roseburia sp. 499, and Roseburia sp. 831b). To increase the number of genomes from poorly represented species of the genus Roseburia (n<3), we also included genomes produced by a high-throughput cultivation of faecal microbiota [29] that displayed high completeness (>95 %) and a lack of obvious contamination (score of ~2 % or lower; see Table S1, available in the online version of this article) using a set of 420 single-copy Lachnospiraceae genes as evaluated by the CheckM tool v. 1.0.18 [30] in KBase [31]. To provide consistency in gene modelling and annotation approaches across genomes, each genome was downloaded from the National Center for Biotechnology Information (NCBI) as nucleic acid FASTA files (FNA). Gene models and draft annotation using the SEED were produced by uploading the FNA files to the Rapid Annotation using Subsystem Technology (RAST) version 2.0 web server [32–34] using the normal bacterial translation table, the RAST gene calling algorithm and the ‘Classic RAST’ annotation scheme, which resulted in FIGfams output [35] (see Table S2). The predicted protein FASTA files (FAA) generated by RAST were used for the rest of the annotation approaches. To provide multiple independent functional predictions using hidden Markov model (HMM)-based approaches, the RAST-generated FAA files were examined using the hmmsearch algorithm within HMMER v. 3.1b2 (hmmer.org [36]) and the TIGRFAMs_14.0 and Pfam-A v. 31.0 profile HMM libraries using the provided, HMM-specific trusted cutoffs to generate hits to TIGRFAMs [37] and Pfams [38], respectively. Finally, each FASTA file was uploaded to BlastKOALA (version 2.1), a web annotation service hosted by the Kyoto Encyclopedia of Genes and Genomes (KEGG) [39]. KOALA (KEGG Orthology and Links Annotation) analyses user data by blast using an SSEARCH computation model to assign KO numbers (denoting orthologue groups associated with specific metabolic reactions) to user data [39]. Release 81.0 of the encyclopedia was used for our genome annotations. To evaluate the presence or absence of metabolic pathways, predicted annotations were visualized using KEGGs’s reconstruction pathway mapper and the SEED Genome Browser tool. Orthologue and paralogue tables were generated from RAST FAA files using the parallel orthologue prediction tool PorthoMCL [40] on a local machine. To compare the orthologue output across different strains and define the core Roseburia genome, Python (version 3.7.2) scripts were created to count the co-occurrences of orthologous genes across strains (https://github.com/ehillman26/Comparative-Genomics-of-Roseburia.git). For example, if all species in a given comparison (three R . intestinalis species vs three R . inulinivorans species) contained the orthologue, then a 1 was added to the sum of total common orthologues for these two species; this logic was repeated for each entry in the orthologue table to determine the number of shared orthologues within each species, between each species, and in the genus Roseburia .
Predicted annotations were curated from the integrated output of each annotation tool (TIGRFAMs, FIGfams and KOALA) for each predicted open reading frame, cross-referenced using the RAST-provided locus tags. Domain-based hits (Pfams) were used to propose genes that might fill holes in metabolic pathways that were not identified using other annotation tools and to validate the output of other annotation tools. To reconcile the output of the multiple annotation approaches, we constructed an operational confidence scale that emphasized expert curation (TIGRFAMs) and genome context (Table S5).
Phylogenetic/phylogenomic tree reconstruction
We reconstructed the phylogeny of the genus Roseburia in comparison to other type strains of species within the family Lachnospiraceae available with GenBank (https://www.ncbi.nlm.nih.gov/nuccore/). As only a subset of Roseburia near neighbours have been sequenced, we initially constructed phylogenetic trees based on full-length 16S rRNA gene sequences. The 16S rRNA gene alignment was created with mega7 [41] using clustal W [42] for multiple sequence alignment; phylogeny was reconstructed using the maximum-likelihood method using the Tamura–Nei substitution model [43] with 1000 bootstraps. For genome-sequenced near neighbours, we also examined phylogenomic relationships using a subset of highly conserved, single-copy genes from the AMPHORA2 database [44], prioritizing type strains of near-neighbour genera and species for inclusion. For phylogenomic reconstruction, we selected genes within the Amphora collection for which TIGRFAM HMMs existed, yielding 18 proteins for concatenation: RpoB (TIGR02013), InfC (TIGR00168), NusA (TIGR01953), RplA (TIGR01169), RplB (TIGR01171), RplD (TIGR03953), RplM (TIGR01066), RplN (TIGR01067), RplP (TIGR01164), RplS (TIGR01024), RplT (TIGR01032), RpsB (TIGR01011), RpsC (TIGR01009), RpsE (TIGR01021), RpsJ (TIGR01049), RpsS (TIGR01050), SmpB (TIGR00086) and Tsf (TIGR00116). Proteins exceeding each of the TIGRFAM HMM trusted cutoffs for each protein within a genome were concatenated; this approach identified a single protein matching each HMM for all genomes. Prevotella brevis (as an outgroup) and protein sequences of neighbouring organisms within Lachnospiraceae were obtained from GenBank (Table S4). The concatenated amino acid sequences were aligned with clustal W in mega 7, and the maximum-likelihood method was used to construct a tree using the Poisson substitution model [45] for the alignment with 100 bootstraps.
Results and Discussion
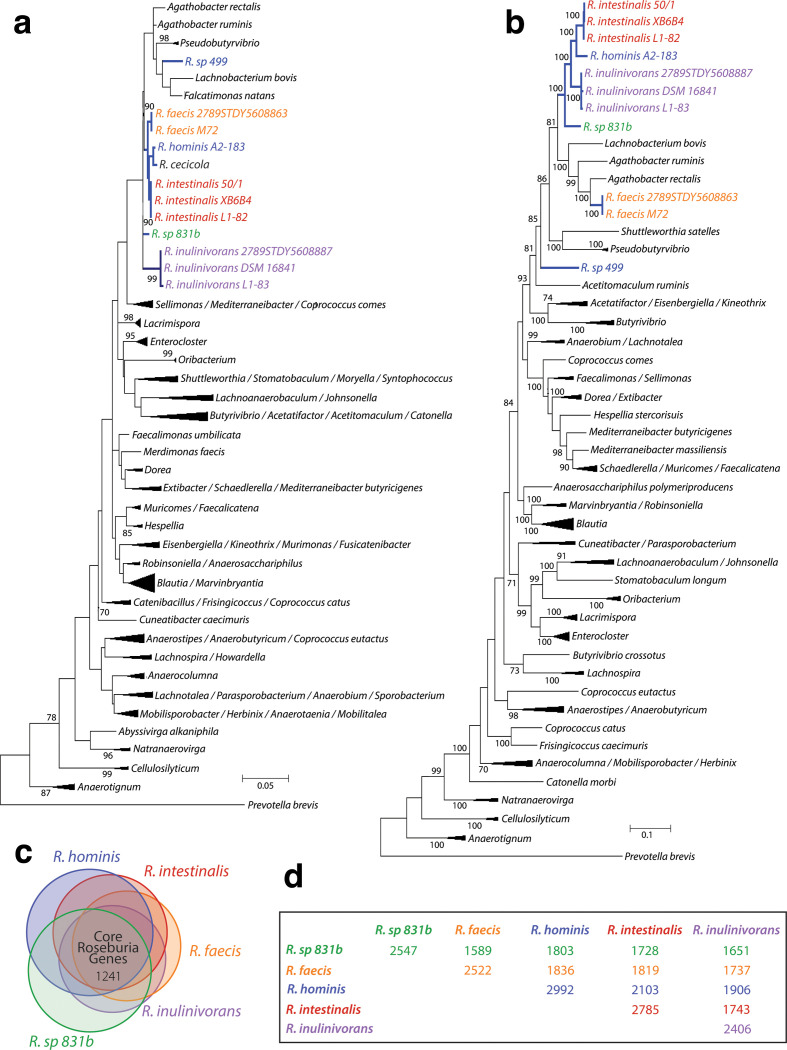
Phylogenomic analysis reveals that Roseburia sp. 499 does not cluster within the genus Roseburia
Despite its present tentative taxonomic assignment, 16S rRNA gene-based phylogenetic analysis across the family Lachnospiraceae revealed that Roseburia sp. 499 did not cluster within a distinct clade formed by other species of the genus Roseburia . This species’ 16S rRNA gene sequence instead clustered with 16S rRNA genes from Pseudobutyrivibrio , albeit with relatively low bootstrap support (Fig. 1A). This species was originally isolated from swine and was proposed to be a Roseburia species in 2013 [46]; however, further physiological and chemotaxonomic analyses of this isolate were never reported to confirm this placement. Unfortunately, the relatively small number of validly published strains within Lachnospiraceae , and therefore confirmed 16S rRNA gene sequences, leaves gaps in our understanding of this family [29] and generates uncertainty in taxonomic placement of new isolates. Our results in Fig. 1b suggested that, based upon 16S rRNA gene-based phylogenetics, R. sp. 499 might be more appropriately classified as a member of Pseudobutyrivibrio rather than Roseburia .
Fig. 1.
Phylogenetic trees of the family Lachnospiraceae . (a) Full-length 16S rRNA gene maximum-likelihood tree from 1000 bootstrap replicates is shown. Genera other than Roseburia were collapsed to simplify visualization of the tree. The fully expanded tree including accession numbers can be found in Fig. S1. (b) The 18-gene concatenated maximum-likelihood tree from 100 bootstrap replicates is shown with collapsed nodes and the fully expanded tree can be found in Fig. S2. (Bootstrap scores >70 are reported). (c) Roseburia core genome(s) displaying the overlapping orthologous protein encoding genes among the species evaluated. (d) Pairwise comparison of orthologies among Roseburia sp. Rows are coloured according to the species and numbers along the diagonal represent the core genome for a given species.
To resolve the phylogenetic placement of R. sp. 499, we employed a whole-genome approach using concatenated single-copy proteins [47] from each organism, which provided higher resolution and increased confidence in the branches. When curating the genes included in AMPHORA, Wu and Eisen noted that this increased confidence is a result of the conservation of protein-coding genes at the amino acid level rather than the DNA level, where compositional biases in small subunit rRNA exist [48–50]. The concatenated tree (Fig. 1b) agreed with the conclusion of the 16S rRNA gene analysis in placing Roseburia sp. 499 well outside the Roseburia clade. Interestingly, the concatenated tree also revealed that R. faecis clustered with high bootstrap support with A. rectalis instead of within the genus Roseburia , despite well-supported clustering of the 16S rRNA genes of both R. faecis strains within Roseburia. Eubacterium rectale, long known to be a physiologically similar near neighbour of Roseburia , was recently reclassified as A. rectalis based upon its phylogenetic relationships with a newly isolated species [3]. Recent analysis of the carbohydrate-active enzymes of A. rectalis and Roseburia strains revealed that R. faecis M72 GH13 family glycoside hydrolases clustered nearly uniformly with those of the then E. rectale strains ATCC 33656, AI-86, M104/1 and T1-815 and separately from other Roseburia species [24]. Taken together, the sequence similarities between R. faecis and E. rectale/A. rectalis suggest that R. faecis may be more related to members of Agathobacter than to those of Roseburia . Because the genus Eubacterium is not monophyletic, efforts are presently underway to improve the taxonomy of this group [3]; the genomic evidence presented here suggests that such efforts should also include the genus Roseburia . It should be noted that we only considered isolate genomes attributed to Roseburia for which the original source of the organism was clear, linking our analysis strongly to the described taxonomy of the genus. Thus, this limitation in the genomes considered restricts our conclusions to the sequenced, current members of Roseburia ; future expansion in the genus either through isolation, incorporation of metagenome-assembled genomes, or transfer of other members of Lachnospiraceae may substantially alter the conclusions drawn here.
Unifying metabolic properties of the genus Roseburia
Using a standardized gene modelling and annotation approach across all Roseburia genomes, we identified the core central metabolism and anabolic pathways of the genus. We further aimed to identify differences among strains that might affect their ecology in the human colon. As we aimed to characterize the genomic properties of the genus, we omitted R. sp. 499 from further analysis based upon the phylogenomic result that it diverges significantly from the Roseburia clade. As their phylogenetic positions were uncertain, we retained strains of R. faecis and R. sp. 831b in further analyses. Using these species as the core members of the genus Roseburia , our orthology-based method identified 1241 orthologues that make up the core Roseburia genome (Fig. 1c, Table S3). Unsurprisingly, R. sp. 831b and R. faecis had the lowest number of shared orthologues with respect to the other core Roseburia members (Fig. 1d). This finding corroborates the idea that R. hominis , R. intestinalis , and R. inulinivorans species are more closely related, although additional biochemical characterization of R. sp. 831b and R. faecis may be needed to validate their placement in the genus Roseburia .
Carbohydrate, central carbon and energy metabolism
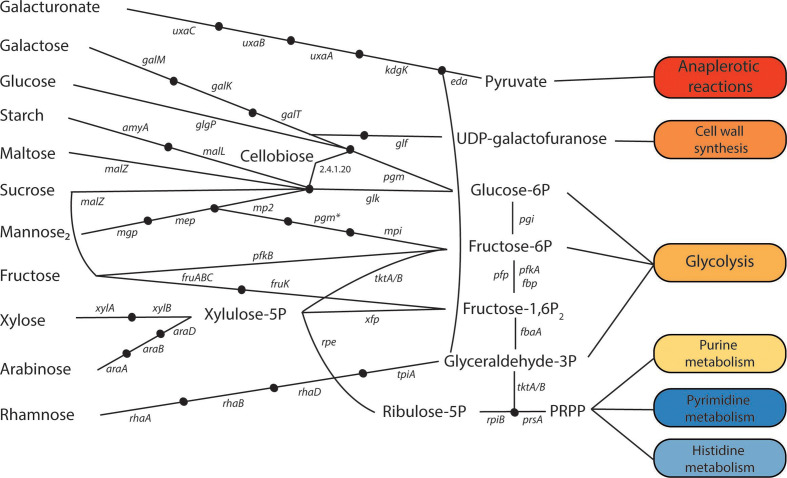
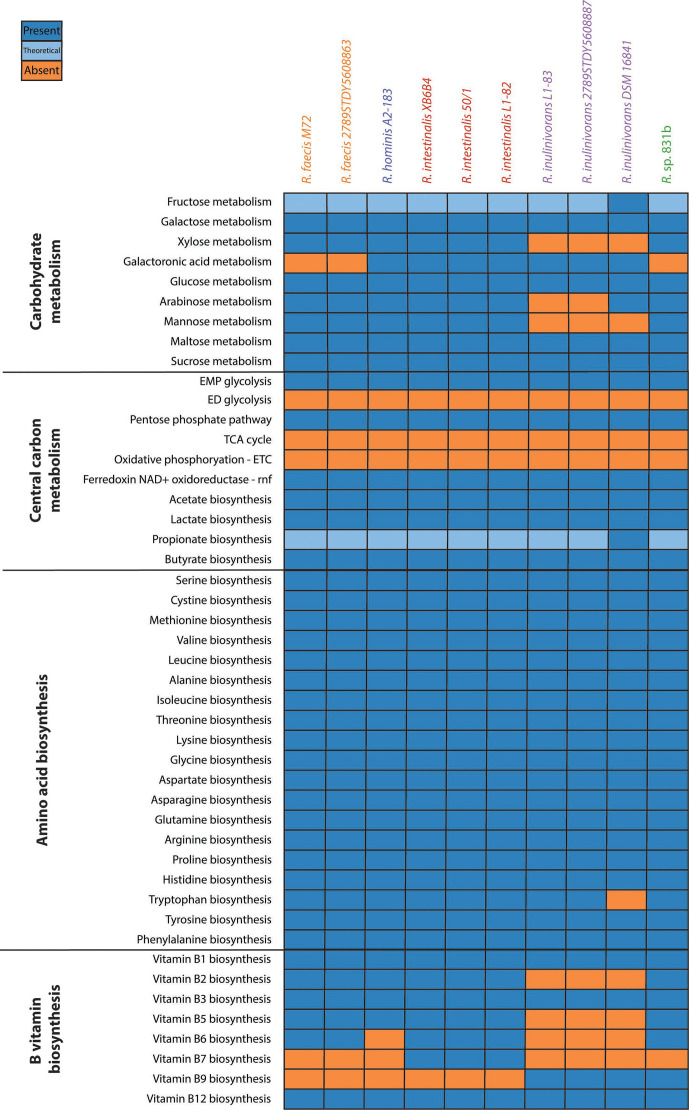
All Roseburia spp. possess the Embden–Meyerhof–Parnas (EMP) glycolytic pathway, which converts glucose to pyruvate. Although we observed 6-phosphogluconolaconase (which converts glucono-1,5-lactone-6-P to 6-phosphogluconate) in all Roseburia genomes except those of R. faecis , no Roseburia spp. encodes the glucose-6-phosphate dehydrogenase required to generate 6-phosphogluconate from glucose-6-P and complete the oxidative branch of the pentose phosphate pathway [51] to ribulose-5-P. Furthermore, we did not detect the genes required to convert 6-phosphogluconate into either 2-keto-3-deoxy-6-phosphogluconate via the Entner–Doudoroff (ED) glycolytic pathway. This finding is consistent with those of most anaerobes (such as Bifidobacterium [52]), as the ED pathway yields less ATP per glucose and may be energetically unsustainable in fermentative anaerobes; only ~3 % of strict anaerobes contain the genes for the ED pathway, while 29 % of facultative anaerobes contain the ED pathway or both the EMP and ED pathways [53]. All Roseburia genomes, however, displayed evidence of many genes involved in pentose interconversions and conversion of d-fructose-6-P to glyceraldehyde-3-P. However, all genes of the non-oxidative pentose phosphate pathway were present in all species, suggesting that Roseburia spp. can convert fructose-6P to ribose-5P. PRPP (5-phosphoribosyl diphosphate) can then be derived from ribose-5P and shuttled to purine, pyrimidine, or histidine biosynthesis (see Fig. 2).
Fig. 2.
Metabolic pathways of various carbohydrate mono-, di-, and poly-saccharides in Roseburia . Each node represents an intermediate compound and each oval represents the metabolic pathway that the metabolite(s) are shuttled to during metabolism. Cofactors are not shown. uxaC, glucuronate isomerase; uxaB, tagaturonate reductase; uxaA, altronate hydrolase; kdgK, 2-dehydro-3-deoxygluconokinase; eda, 2-dehydro-3-deoxyphosphogluconate aldolase/(4S)-4-hydroxy-2-oxoglutarate aldolase; galM, aldose 1-epimerase; galK, galactokinase; galT=UDPglucose-hexose-1-phosphate uridylyltransferase; glf, UDP-galactopyranose mutase, pgm, phosphoglucomutase; glgP, glycogen phosphorylase; amyA, alpha-amylase; malL, oligo-1,6-glucosidase; malZ, alpha-glucosidase; glk, glucokinase; mgp, beta-1,4-mannooligosaccharide phosphorylase; mep, mannobiose 2-epimerase; mp2, 4-O-beta-d-mannosyl-d-glucose phosphorylase; pgm*, bifunctional phosphoglucomutase/phosphomannomutase; mpi, mannose 6-phosphate isomerase; pfkB, 6-phosphofructokinase 2; fruA, PTS fructose-specific enzyme IIABC component; fruK, 1-phosphofructokinase; xylA, xylose isomerase; xylB, xylulokinase; araA, L-arabinose isomerase; araB, L-ribulokinase; araD, L-ribulose-5-phosphate 4-epimerase; rhaA, L-rhamnose isomerase; rhaB, rhamnulokinase; rhaD, rhamnulose-1-phosphate aldolase; tpiA, triosephosphate isomerase; tktA/B, transketolase 1/2; xfp, xylulose-5-phosphate/fructose-6-phosphate phosphoketolase; rpe, ribulose-phosphate 3-epimerase; pgi, glucose-6-phosphate isomerase; pfp, diphosphate-dependent phosphofructokinase; pfkA, 6-phosphofructokinase 1; fbp, fructose-1,6-bisphosphatase I; fbaA, fructose-1,6-bisphosphate aldolase; rpiB, ribose 5-phosphate isomerase B; prsA, ribose-phosphate pyrophosphokinase; mannose2, mannobiose; glucose-6P, glucose-6 phosphate; fructose-1,6P2, fructose-6 phosphate; fructose-1,6P2, fructose-1,6 bisphosphate; glyceraldehyde-3P, glyceraldehyde-3 phosphate; riboulose-5P, ribulose 5-phosphate.
Interestingly, despite the known ability of various Roseburia spp. (especially R. intestinalis ) to ferment monomeric pentoses (e.g. xylose, arabinose) and to consume various xylooligosaccharides, arabinoxylans and arabinogalactans as carbon sources for growth [5, 24], we were unable to detect with high confidence many of the carbohydrate metabolism enzymes required for xylose and arabinose consumption via our approach, particularly in R. inulinivorans . However, we did find FIGfam evidence for many of these genes and in many cases these FIGfam calls were found with highly conserved genomic context that was consistent across the strains and the majority of species. For example, although R. intestinalis strains all displayed the l-arabinose isomerase required to convert the l-arabinose in arabinoxylan to l-ribulose (araA), we could not identify genes involved in phosphorylation of ribulose for conversion into d-xylulose-5-P with TIGRFAMs or KOGs (araB or araD). Examination of the surrounding gene neighbourhood, however, revealed FIGfam calls for these genes. We consider it likely that known representatives of these genes from Lachnospiraceae are lacking within the TIGRFAM and KO reference databases, making their algorithmic identification difficult.
From our analysis of simple carbohydrate metabolism, we found genomic evidence suggesting that all members of Roseburia except R. faecis and R. sp. 831b can utilize galacturonic acid. Our analysis also suggests that all Roseburia spp. are able to metabolize glucose, galactose, maltose and sucrose, where only R. inulinivorans is likely unable to metabolize xylose, mannose and arabinose. Although all Roseburia spp. are missing both the mannose isomerase that converts mannose to fructose and the mannokinase that phosphorylates mannose to mannose-6P as prescribed in the KEGG pathway, a different pathway has been characterized previously from R. intestinalis L1-82 [54]. Like other carbohydrate degradation pathways in Roseburia spp., the genes for mannose degradation and utilization are organized in an operon including a transcriptional regulator, the associated glucosidases, and an ATP-binding cassette (ABC) transport complex. This mannose degradation and utilization pathway, which is similar to that in Ruminococcus albus [55], converts mannobiose with two synergistic mannoside phosphorylases and a mannose epimerase into mannose-1P. Ultimately, a promiscuous phosphoglucomutase/phosphomannosemutase and a bifunctional glucosidase/phosphomannose isomerase convert this to mannose-6P and fructose-6P, respectively, before it enters glycolysis. The fact that this experimentally validated pathway was not captured by BlastKOALA models points to a need for increased coverage within this phylogenetic region.
We also found variation in the different oligosaccharide phosphorylases present among species and strains of Roseburia . From our analysis, all Roseburia species possess phosphorylases that cleave glycogen, cellobiose, and lacto-N-biose into their respective monosaccharides. These enzymes allow microbes to increase net ATP production by utilizing free orthophosphates to generate phosphosugars instead of consuming ATP to generate them; thus, this may be an advantageous energy efficiency strategy. Interestingly, only the strains of R. faecis , two strains of R. intestinalis (XB6B4 and LI-82) and R. inulinivorans (LI-83) possess a sucrose phosphorylase, while all species contain the malZ gene for cleaving sucrose to d-fructose and d-glucose. The malZ gene, which is present in all Roseburia species, also cleaves maltose into two d-glucose monomers. However, R. intestinallis strains LI-82 and M50/1, as well as the R. inulinivorans strains 2789STD5608887 and LI-83, have a maltose/trehalose phosphorylase that perform a similar cleavage of maltose to d-glucose and beta-d-glucose 1-phosphate. These phosphorylases are only supported by FIGfam identification and often can act on several similar substrates, which makes these phosphorylases and the malZ gene interesting candidates for future genetic and biochemical studies. Other phosphorylases, such as chitobiose phosphorylase (E.C. 2.4.1.280), are present in all Roseburia species except R. inulinivorans strains 2789STD5608887 and DSM 16841; however, these were also only supported with moderate confidence and should be further characterized. Ultimately, differences in carbohydrate availability from host diets intersecting with different carbohydrate utilization machinery may be a major driver of interspecies competition among Roseburia spp. ecology and determine R. intestinalis occurrence and abundance patterns in human microbiomes [54, 56].
Interestingly, within R. inulinivorans our analysis predicted subspecies-level variation in the ability to use arabinose, as the strain DSM 16841 possesses the same conserved arabinose utilization pathway as the other Roseburia species, but the other two inulinivorans strains lack it. This finding is particularly interesting because R. inulinivorans was previously described as being unable to utilize arabinose for growth [5] and Sheridan and coworkers also found that this strain was unable to grow on arabinoxylan [24]; these phenotypes may, in fact, vary within the species. Similarly, they found that R. hominis also does not grow on arabinoxylan; however, the species description [5] indicates that arabinose can be utilized for growth, which agrees with our prediction. Our predictions for hominis also include the ability to utilize galactose and galacturonic acid, although the growth of R. hominis on galacto-oligosaccharides was not previously observed (pectin, a significant source of galacturonic acid, was not tested). Our predictions match previous descriptions of R. intestinalis carbohydrate metabolism [7, 24] for all examined genomes. Likewise, our predictions for R. faecis are similar to previous experimental results [24] with respect to arabinose utilization (galacturonic acid, which we predict to not be utilized, was not tested). Sheridan and coworkers also observed mild growth on arabinoxylan and the araC gene in a conserved gene neighbourhood with arabinoxylan CAZymes. Further experimental investigation of Roseburia species- and strain-specific carbohydrate utilization will be needed to clarify these phenotypic discrepancies in the utilization of dietary fibres and their constituent sugars, as Roseburia spp. colonization is dependent upon the dietary fibre intake of the host [57–59].
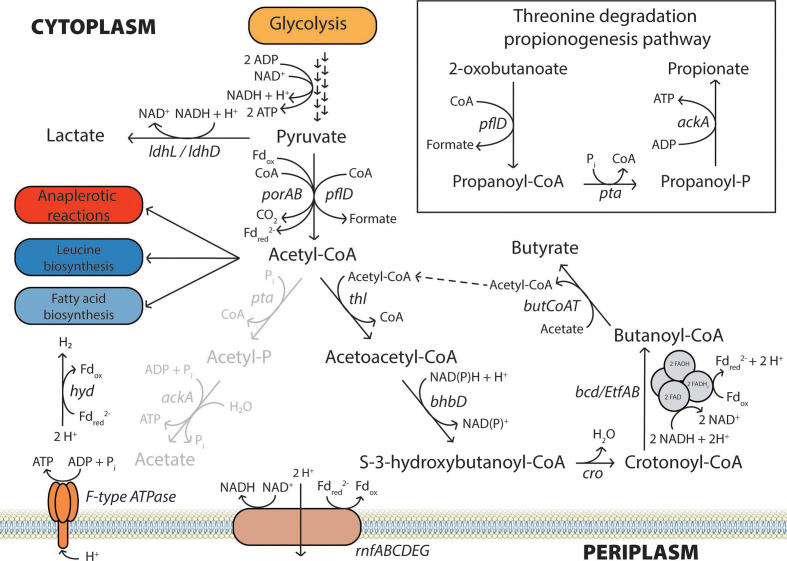
With respect to energy generation from carbohydrates, all Roseburia genomes displayed the genetic capacity for conversion of pyruvate to acetyl-CoA, condensation of acetyl-CoA with oxaloacetate into citrate and conversion of citrate into α-ketoglutarate. However, all Roseburia spp. lack both the α-ketoglutarate dehydrogenase complex and α-ketoglutarate synthase and, therefore, cannot interconvert α-ketoglutarate and succinyl-CoA. These enzymes are likely retained for anapleurotic reactions; α-ketoglutarate is a precursor to synthesis of many amino acids, such as glutamate. With respect to the rest of the canonical TCA cycle genes, all genomes contained fumarate hydratase, allowing interconversion of fumarate and malate. Roseburia spp. lack the canonical respiratory electron transport and oxidative phosphorylation apparatus; NADH is oxidized as lactate, propionate and butyrate are produced, regenerating oxidized electron carriers. All species encode an F-type ATPase (ATP-forming) that potentially allows utilization of proton motive force generated by excretion of organic acids for ATP generation and pH regulation by balancing H+ flux across the membrane [60]. Interestingly, we also detected genes for the classically eukaryotic V/A-type ATPase, which was found (with KO and FIGfam support) exclusively in the R. intestinalis genomes [61]. These ATPases are typically used by eukaryotes to acidify vacuoles and consume energy from ATP to export protons [62], although they have also been found in the enterococci to pump cations such as sodium and potassium [63]. This adaptation may arise from gene transfer [64] through a long history of association with eukaryotes and archaea and may grant R. intestinalis additional resistance to low pH compared with other members of the genus Roseburia [64]. Similar systems are also found in diverse members of the orders Clostridiales and Bacteroidales [65], suggesting that such systems may be important for colonization of the human colon.
Although Roseburia spp. do not possess the traditional electron transport chain commonly used in oxidative phosphorylation, all species evaluated here have genes (rnfABCDEG) encoding an electron transport complex that seems to be an ancient form of electron transport chain. A similar complex is present in Acetobacterium woodii to translocate Na+, fuelled by reduced ferredoxin and generation of NADH [66]. In addition to NADH and ferredoxin cycling in this species, the F0F1 ATPase generates ATP through Na+ transport across the gradient generated by this Rnf complex [66, 67]. As Roseburia do not exhibit any butyrate kinase activity [27, 68], the generation of ATP via the gradient maintained by the Rnf complex may be vital to Roseburia ’s survival. It is unclear whether the Roseburia Rnf complex translocates Na+ or H+ like others in the order Clostridiales [67, 69, 70]. In either case, the oxidation of ferredoxin (or, potentially, flavodoxins) by this complex allows Roseburia to regenerate the electron carriers (Fig. 3) needed for pyruvate and butyrate metabolism [69, 70]. Fermentation to pyruvate and acetyl-CoA by gut microbes produces a wide range of metabolites, including formate, lactate and short-chain fatty acids (SCFAs) [71]. Acetic acid, propionic acid and butyric acid are the most abundant SCFAs present in the human colon and have marked physiological effects on health [72]. Specifically, butyrate can be oxidized to CO2 by the colonocytes, which helps maintain a hypoxic epithelium and promotes energy homeostasis [54, 73].
Fig. 3.
Central metabolism of Roseburia species, including the dominant fermentation pathways and electron transferring complexes. Although Roseburia can generate ATP via substrate-level phosphorylation of acetyl-phosphate to acetate, they are net consumers of acetate and thus flux through this pathway is low (indicated by the grey reaction arrows). Instead, Roseburia spp. appear to derive their ATP almost exclusively through ‘oxidative phosphorylation’ via an F-type ATPase via the proton gradient generated by an H+-translocating rnf complex that also recycles ferredoxins/flavodoxins. The FeFe group B hydrogenase also regenerates oxidized ferredoxin/flavodoxin while generating H2 (shown as H2 formation). Gene symbols: ldhL/ldhD, L-/D-lactate dehydrogenase; porAB, pyruvate ferredoxin oxidoreductase alpha/beta subunit; pflD, formate C-acetyltransferase; pta, phosphate acetyltransferase; ackA, acetate kinase; thl, atoB-like thiolase (acetyl-CoA acetyltransferase); bhbD, β-hydroxyacyl-CoA dehydrogenase; cro, crotonyl-CoA hydratase; bcd, butyryl-CoA dehydrogenase; EtfAB, electron transfer flavoprotein alpha and beta-subunit; butCoAT, butyryl-CoA : acetate-CoA transferase; rnfABCDEG, Na+/H+-translocating ferredoxin : NAD+ oxidoreductase subunits A–G; hyd, FeFe hydrogenase.
In Roseburia , there are two reactions that convert pyruvate to acetyl-CoA. The first reaction incorporates a free CoA and yields CO2 while generating reduced ferredoxin or flavodoxin; the KEGG annotation suggests that this gene encodes a flavodoxin-utilizing enzyme, but this has yet to be experimentally determined. Many microbes typically reduce NAD+ to NADH in this reaction; however, anaerobes commonly utilize flavodoxin instead via the pyruvate synthase PorAB [70, 74]. Interestingly, R. intestinalis and R. hominis contain a second enzyme complex that appears to carry out the NAD+-to-NADH reducing reaction, as well using a tetrameric ferredoxin oxidoreductase complex. In contrast to the PorAB reaction, pyruvate formate lyase (pflD) carries out another acetyl-CoA-generating reaction, which produces formate directly from pyruvate. This reaction takes pyruvate and free CoA to form acetyl-CoA and formate (Fig. 3). Regulation of pflD is commonly carried out through a lyase-activating enzyme that lies adjacent to pflD in the Roseburia genomes [75, 76] Although these two reactions both consume pyruvate and generate acetyl-CoA, the subtle differences in electron and carbon balance may be very important to Roseburia ’s physiology; net formate and CO2 production has been noted in several Roseburia strains [5, 7, 77]. Use of the pyruvate formate lyase may be useful when there is an overflow of pyruvate, when electron carriers can no longer be regenerated, or under conditions of iron limitation [78]. Because the Roseburia ’s hydrogenase reaction is iron-dependent, iron deprivation has been shown to reduce the amount of butyrate and hydrogen formed while increasing lactate and formate amounts as the Rnf complex cannot regenerate ferredoxins through the formation of H2 [79]. Further studies will be needed to understand the regulatory schemes that determine whether Roseburia eliminates electrons as formate or retains them in NADH with production of CO2.
Lactate is also a common fermentation product of pyruvate metabolism that competes with formate and acetyl-CoA formation. Unlike porAB or pflD, lactate dehydrogenase (ldhL, EC 1.1.1.27) regenerates NAD+ by reduction of pyruvate using electrons from NADH (Fig. 3). All Roseburia spp. displayed strong evidence for the ldhL gene, and lactate has been noted as a common fermentation product in pure Roseburia cultures. Additionally, R. sp. 831b, R. intestinalis strains and R. inulinivorans strains all have two copies of ldhL in their genomes and are noted to make more lactate than the other species [5, 7]. R. sp. 831b, R. hominis and R. faecis 2789STDY5608863 also all show evidence of the ldhD gene, which forms d-lactate instead of l-lactate. The R. faecis M72 strain studied previously only has one copy of ldh and produced the least lactate while producing the most formate [5]. Understanding differences in pyruvate metabolism among Roseburia spp. in shuttling carbon and electrons and generating fermentation products may provide an insight into competition among strains and differences in potential roles in cross-feeding of the gut ecosystem [6].
While Roseburia do not consume lactate like other gut microbes, a net consumption of acetate has been observed for most Roseburia species [5, 7, 68, 80]. Despite their net consumption of acetate, Roseburia spp. uniformly have the genes necessary to produce acetate from acetyl-CoA and can generate ATP in the process. Although it is likely not a major Roseburia terminal fermentation product, acetate is important to Roseburia ’s butyrate fermentation strategy. Rather than using butyrate kinase like other gut microbes such as Coprococcus , Roseburia use a highly active butyryl-CoA : acetate-CoA transferase [27, 68]. This is an interesting strategy because butyrate kinase yields ATP from butyryl-phosphate, while Roseburia ’s transferase does not yield any ATP, instead generating acetyl-CoA. Five Roseburia strains have been shown to lack measurable butyrate kinase activity, and our genomic evidence does not support the presence of this gene except in R. inulinivorans LI-83, which was not among those previously tested experimentally [68]. Roseburia ’s strategy of effectively obtaining more acetyl-CoA, however, allows Roseburia to make more butyrate, as each butyryl-CoA requires two acetyl-CoA in forming the precursor acetoacetyl-CoA. Duncan et al. [80] showed that ~85 % of the butyrate carbon was derived from extracellular acetate, ultimately, allowing Roseburia to recycle as much NAD+ from each mole of glycolysis-derived acetyl-CoA as possible. The genomic findings here are in accordance with strong prior experimental [5, 7, 27, 68, 77, 80, 81] and genomic [8, 25, 26, 82] evidence describing butyrate as the major fermentation product of Roseburia species.
In Roseburia , NAD+ recycling occurs in the penultimate step of butyrogenesis, where crotonoyl-CoA is converted into butyryl-CoA via butyryl-CoA dehydrogenase (bcd). Interestingly, its assigned KO number identified the Roseburia bcd as a catabolic, butyrate-consuming reaction involving the cofactor FADH2. In contrast, most anabolic, butyrate-forming bcd reactions involve the recycling of either NADH or NADPH cofactors to form butyryl-CoA. Here, it appears that Roseburia may use FADH2 instead of or in addition to NAD+ in an electron-transferring flavoprotein (ETF) for the formation of butyryl-CoA, based on the genome context and orthology groupings. All Roseburia contain a butyrogenic operon that contains an atoB-like thiolase (thl), β-hydroxybutyryl-CoA dehydrogenase (bhbD), bcd and two flavoproteins (etfA and etfB) implicated in the FADH2-dependent formation of butyryl-CoA [8, 82]. A similar electron-transferring flavoprotein bcd gene was also noted in the anaerobic clostridia, proposed by Flint and Louis [8, 83–86]. This EtfAB complex may allow Roseburia and other species of Clostridiales to bifurcate electrons from NADH to butyryl-CoA and ferredoxin [69, 86, 87]. Ultimately, this reaction yields the precursors for butyrogenesis, recycled NAD+, and reduced ferredoxin. Although TIGRFAMs TIGR01963, TIGR02280 and TIGR01751 did not match genes bhbD, cro and bcd above the trusted cutoff, the majority of these biosynthesis genes being located, and likely translated, together gives increased confidence that this FADH2-utilizing bcd is used here in Roseburia to generate, rather than degrade, butyrate. Additionally, we did not find evidence of the catabolic fatty acid oxidation pathways in which the FADH2-utilizing bcd is typically involved. As mentioned above, the final step of butyrate synthesis is carried out by a butyryl-CoA : acetate-CoA transferase rather than butyrate kinase [68], but this gene was not found in any of the previously mentioned operons. As a whole, our confidence scores for this pathway were lower than for most other pathways identified in this study, based upon our classification scheme that emphasizes TIGRFAM equivalogs. Our results argue for greater inclusion of Lachnospiraceae genes in seed alignments for profile HMM generation, especially as this process is central to the metabolism of this genus and its near neighbours.
The KEGG KO models also suggest that Roseburia may have the potential to produce propionate via a glycolysis-independent threonine degradation pathway (Fig. 3) previously noted in Escherichia coli [88]. Although Roseburia appear to possess the genetic capabilities to convert precursor compounds to propionate, there is little evidence to suggest that these species regularly ferment to propionate. In E. coli , the threonine degradation pathway consists of seven genes organized in a tdcABCDEFG operon where tdcE is functionally equivalent to pflD, and tdcD to ackA [88]. Although neither pta nor a homologue were found in this operon, pta can convert propionyl-CoA to propionyl-phosphate in E. coli . Interestingly, a threonine dehydratase similar to tdcB, which can generate 2-oxobutanoate by degrading threonine, is found in the same gene neighbourhood as ackA in all studied Roseburia genomes. It is unclear if its role is in isoleucine biosynthesis or if it may play a role in propionate generation, as no study has examined Roseburia ’s metabolism solely on threonine or other amino acids as a primary carbon source. There is only one report of propionogenesis in Roseburia: R. inulinivorans DSM 16841, which ferments fucose to propionate via a propanediol utilization (pdu) operon [89]. In accordance with their findings, we were able to find and annotate this same operon, although many of the gene calls were not strongly supported using HMM-based evidence. As suggested, this is a strain-specific pathway and we could not find it in any of the other species or strains. This is not surprising, as it is rare for species to produce both propionate and butyrate [90]. Although limited propiogenesis may be a mechanism for Roseburia to disproportionate electrons from amino acid fermentation, future studies should investigate the ability of Roseburia spp. to ferment amino acids and the possible connection to propiogenesis in pure cultures. If significant amounts of propionate are produced in the gut ecosystem by amino acid fermentation by Roseburia or other microbes, this would be very intriguing, as the succinate, acrylate, or propanediol pathways are considered to be the common propionate synthesis pathways, with succinate being dominant [71, 90, 91]. Especially under low fluxes of dietary fibre carbohydrates, amino acid fermentation may potentially be performed by Roseburia spp., either concurrently with saccharolytic fermentation or after carbohydrates are exhausted, as the distal colon is known to be relatively carbohydrate-poor and to contain a higher relative abundance of peptides [92].
Interestingly, in addition to fermentation, some Roseburia spp. may be able to disproportionate electrons using sulfate as an electron acceptor. All studied R. faecis and R. intestinalis strains contained a putative operon containing genes for the ABC sulfate transporter cysPUWA, the sulfate adenlylyltransferase cysND, which generates adenosine-5′-phosphosulfate (APS) [93], and the APS reductase aprAB, which reduces APS to sulfite while oxidizing a reduced electron carrier (typically, NADH) [94]. The eventual fate of produced sulfite in Roseburia is unclear, as all examined genomes lack the dissimilatory sulfite reductase dsrAB that reduces sulfite to sulfide.
Amino acid biosynthetic pathways
Protein synthesis requires biosynthesis or uptake of all 20 canonical amino acids. Amino acids can be synthesized de novo from organic precursors, including intermediates of glycolysis and the TCA cycle, such as pyruvate and α-ketoglutarate, by amination, which requires exogenous nitrogen. In the case of Roseburia , luminal nitrogen in the colon can be derived exogenously from the host’s diet, endogenously from sloughed intestinal cells, or through nitrogen cycling in the intestine [95]. All Roseburia spp. are capable of accessing inorganic nitrogen as ammonium for amino acid biosynthesis, synthesizing glutamate from α-ketoglutarate via glutamate synthase (also termed glutamine oxoglutarate transaminase, or GOGAT) and glutamine via a type 1 glutamine synthetase (glnA). However, only R. intestinalis species appear to have the potential to use urea as a nitrogen source via the biotin-requiring urea carboxylase encoded by the uca operon; appreciable evidence for the presence of ureases was not detected in any Roseburia species. As urea is excreted into the colonic lumen through the epithelium, it represents a potential competitive advantage for R. intestinalis under conditions of strong competition for ammonium (for example, during highly saccharolytic conditions).
Roseburia species appear to be able to synthesize nearly all of their own amino acids de novo. This result was somewhat surprising given the high organic carbon and nitrogen content of colon luminal contents; however, excreted amino acids have long been thought to be largely bound in proteins [96] and faecal metagenomes reveal significant enrichments in amino acid biosynthetic genes relative to all sequenced bacterial genomes in KEGG [97]. This may indicate either low bioavailability of free amino acids in the colon or fierce competition for amino acids among organisms. All of the Roseburia genomes we examined possessed complete proline biosynthesis pathways from glutamate and arginine biosynthesis pathways via citrulline and aspartate. However, all appeared to lack arginases and, therefore, possessed incomplete arginine cycles. All species of the genus Roseburia encode genes for the biosynthesis of aspartate from oxaloacetate and asparagine from aspartate. The entirety of the lysine biosynthesis pathway converting aspartate to lysine via the dehydrogenase branch is present throughout the genus. However, all three strains of R. inulinivorans contain genes suggesting the presence of an additional alternative succinylation-dependent synthesis branch.
With respect to serine biogenesis, all members of the genus encode the first two genes in the phosphorylation pathway, D-3-phosphoglycerate dehydrogenase (serA) and phosphoserine aminotransferase (serC), but appear to lack phosphoserine phosphatase (serB). These serine biosynthesis genes are located within a predicted operon with genes encoding enzymes that catalyze the first committed steps of branched chain amino acid (BCAA) synthesis from pyruvate (acetolactate synthase, acetolactate reductoisomerase and 2,3-dihydroxyisovalerate dehydratase), which is conserved across all Roseburia genomes. Notably, only the large catalytic subunit of acetolactate dehydrogenase is present within the operon; in E. coli the large subunit alone is catalytically active, though at a slower rate, and is insensitive to feedback inhibition [98]. Both subunits of acetolactate synthase are present in a separate operon in all examined Roseburia genomes, suggesting the hypothesis that this combined serine-BCAA synthesis operon responds to low amino acid concentrations to increase the flux of pyruvate into serine and BCAAs. Genomes containing serAC but lacking serB are common within Firmicutes [99], which has led to predictions that many members of this phylum are serine auxotrophs [100]. Some members of Dehalobacter have been shown to escape a lack of serB by synthesis of serine from glycine via serine hydroxymethyltransferase [99], which all the Roseburia genomes we analysed also possess. However, if this strategy is broadly used across Firmicutes to synthesize serine, the reason for the frequent retention of serA and serC genes in these genomes is unclear. Recently, homoserine kinase (thrH) has been shown to also catalyze the same dephosphorylation reaction as serB [101]; TIGRFAM and FIGfam annotations have identified a putative thrH gene for all Roseburia genomes except R. hominis and R. sp. 831b. The discovery of alternative serine synthetic strategies in Firmicutes may help resolve the paradox that, although it is among the least expensive amino acids to synthesize [99], predicted auxotrophy for serine is widespread based upon our present understanding of possible serine biosynthetic pathways.
From serine, all Roseburia genomes possess biosynthesis genes for glycine via glycine hydroxymethlyransferase (glyA). This enzyme also catalyzes the interconversion of glycine and threonine using acetaldehyde as a substrate. This is the sole threonine biosynthetic path in R. hominis and R. sp. 831b, which lacked evidence for homoserine kinase. Cysteine is produced by direct sulfurylation with sulfide by the CysEK complex. FIGfam predictions for multiple aminotransferases were also identified for all species. Of note, R. intestinalis genomes appeared to lack the murI glutamate racemase, required to interconvert d- and l-glutamate. Although l-glutamate is used in protein synthesis, d-glutamate is required in many organisms for peptidoglycan biosynthesis, which may suggest either a requirement for exogenous d-glutamate or an altered cell wall structure in this Roseburia species [102].
The genes required for biosynthesis of hydrophobic amino acids are present in almost all species of Roseburia . With respect to branched-chain amino acids, evidence for the biosynthesis of leucine, valine and isoleucine is present for all examined species of Roseburia with moderate to high confidence. However, the evidence for the direct biosynthesis of d-alanine was weak for common biosynthetic routes (i.e. from pyruvate or aspartate), lacking TIGRFAM and KO identifications. Notably, R. sp. 831b was the only species displaying KO evidence for alaA, an alanine transaminase, although biosynthesis via cysteine desulfurase or through racemization of alanine was well supported. Due to the lack of high-confidence predictions for d-alanine biosynthesis, we also annotated genomes at the protein domain level (i.e. Pfams) for every Roseburia species herein. Notably, Pfams provided evidence for an alanine symporter (Table S6); alanine glyoxylate aminotransferase, alanine dehydrogenase and a class IV aminotransferase were identified for all species. Additionally, some species of Roseburia showed significant Pfam hits for PF02261 (aspartate decarboxylase). Apart from R. inulinivorans DSM 16841, which lacked any evidence for tryptophan biosynthesis, all Roseburia genomes displayed complete pathways for aromatic amino acid biosynthesis.
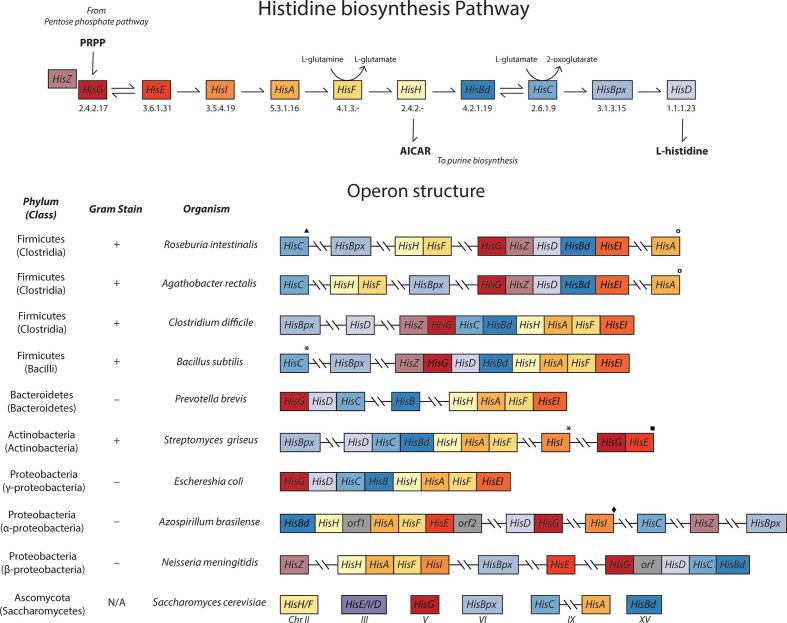
All studied genomes from Roseburia are predicted to contain the genes required for histidine biosynthesis at high confidence. Histidine biosynthesis is an energetically expensive task [103] and is often tightly regulated within a single operon (Fig. 4). However, unlike the E. coli histidine (his) operon that contains all the pathway genes in one locus, the majority of Roseburia ’s his genes are divided among multiple loci (Fig. 4). Specifically, hisG, hisZ, hisD, hisBd and hisEI are present in a single predicted operon in every subspecies, and the genes hisF and hisH are located in a separate predicted operon across all genomes. The hisBpx gene is not located within a predicted operon. Finally, hisA and hisC are not located in either of the other his predicted operons, but are housed with glutamine synthetase and aromatic aminotransferase, respectively.
Fig. 4.
Histidine biosynthetic pathway and operon structure. The histidine biosynthetic pathway (top) not only synthesizes l-histidine from 5-phospho-d-ribose α-1-pyrophosphate (PRPP), but also the precursor of purine biosynthesis, 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR). Across the bacterial kingdom, the histidine operon structure is paraphyletic, displaying similar organization among closely related species, while more distant ancestors have varying organizations. To demonstrate these similarities, the operon organizations of various representative microbes across different phyla and classes are shown (bottom). Gene names: HisA, phosphoribosylformimino-5-aminoimidazole carboxamide ribotide isomerase; HisBd, imidazoleglycerol-phosphate dehydratase; HisBpx, histidinol-phosphatase; HisC, histidinol-phosphate aminotransferase; HisD, histidinol dehydrogenase; HisF, imidazole glycerol-phosphate synthase cyclase subunit; HisG, ATP phosphoribosyltransferase; HisH, glutamine amidotransferase; HisEI, phosphoribosyl ATP pyrophosphohydrolase/phosphoribosyl-AMP cyclohydrolase; HisZ, ATP phosphoribosyltransferase regulatory subunit. The chromosome number where the gene is located is displayed for eukaryotes. Genes in the other biosynthetic operons are denoted as follows: ▲, tyrosine/phenylalanine biosynthesis; *, tryptophan biosynthesis; ■, riboflavin biosynthesis; ○, glutamine biosynthesis; ♦, cystine biosynthesis
Previously, a partial his operon has been observed in the Gram-negative alpha-proteobacterium Azospirillum brasilense [104] and Gram-positive Bacillus subtilis [105]. While the E. coli his operon structure is well known, the organization of his genes in clusters/operons is variable among distantly related microbes. As shown in Fig. 4, similar operon organization among closely related species is displayed across Firmicutes, while organization varies considerably in more distant genomes. The organizational diversity of the his operon suggests that various separations, fusions and relocations of these genes within the genome have occurred (multiple lateral transfers), emphasizing that there are multiple ways to optimize control of histidine biosynthesis for different environments [106, 107].
In eukaryotes, such as S. cerevisiae, the his genes are not located in clusters, but gene fusions (with respect to the gene organization frequently observed in bacteria) are common. Gene fusions of hisE/I/D and hisF/H have been discovered and it has been suggested that they help control flux through the pathways with substrate tunnelling [104, 106]. Although Roseburia spp. do not have a hisF/H fusion gene, the hisF and hisH genes are organized together in a predicted operon that may, in a similar way, increase the efficiency with which flux is regulated through the pathway. Similar to S. cerevisiae, Roseburia ’s hisE/I fusions may be a product of convergent evolution or horizontal gene transfer and are thought to allow efficient biosynthesis using substrate tunnelling mechanisms [108].
Gene organization that may promote efficient regulation of flux into histidine biosynthesis and/or increase the efficiency of reaction mechanisms is not surprising, given the high energetic cost, scarcity of nutrients in the colon and importance of both His and its byproducts [109]. To further enhance his operon regulation, Roseburia ’s hisG protein likely requires an additional catalytic polypeptide, HisZ, to initiate biosynthesis from phosphoribosyl diphosphate [110]. HisZ is allosterically regulated by ATP and histidine, which provides feedback inhibition [111]. Colocalization in a predicted operon with glutamine synthetase suggests that histidine and glutamine biosynthesis may be co-regulated; this type of coregulation with serine biosynthesis was also suggested in the his operon of the anaerobe Streptococcus mutans UA159 [112]. Additionally, an important byproduct of the HisF–HisH reaction is 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR), which is used in purine biosynthesis, and thus may require expression independently of the rest of the his genes under conditions where purines, but not histidine, are required. Further experiments to evaluate how Roseburia regulates these two pathways will be required to predict the function of this gene organization.
Biosynthesis of B vitamins
B vitamins are essential cofactors for many enzymes across the tree of life, and are thought to exert strong influence over microbial ecology in multiple environments, including the gut [18, 21, 113]. In humans, B vitamin requirements are met through dietary consumption, but may also be produced by the gut microbiota. We sought to determine how Roseburia spp. meet their B vitamin requirements. Our results show that most Roseburia spp. have the ability to either synthesize or transport all of the B vitamins (Fig. 5). We predict that none of the examined Roseburia spp. can synthesize biotin, but all can synthesize thiamine, pyridoxine, folate and cobalamin. Synthesis of riboflavin, niacin and pantothenate varied at the species level, as did salvage transporters for folate, pyridoxine and thiamine.
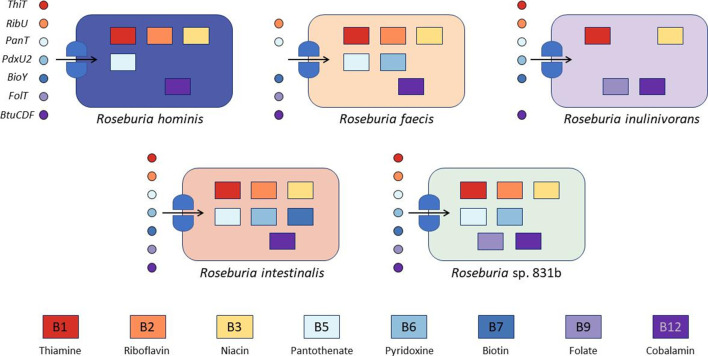
Fig. 5.
Vitamin synthesis and transport in Roseburia species. Rectangles represent biosynthetic pathways and circles represent transporters that were predicted by the species indicated. Each shape is coloured to represents a particular B vitamin that we predict can be synthesized or transported. The ThiT, RibU, PanT, PdxU2, BioY and FolT genes are energy coupling factor (ECF)–type transporters for thiamine, riboflavin, pantothenate, pyridoxine, biotin and folate, respectively, while BtuCDF is an ATP-binding cassette (ABC) transporter with a substrate-specific domain for cobalamin transport.
Despite their shared synthesis capabilities, we found genes for alternative synthesis strategies among Roseburia species that may minimize competition for these public goods [18, 19]. In the case of thiamine, which is required for diverse catabolic reactions of sugars and amino acids, we did not detect the thiC gene within R. inulinivorans strains, which is the first gene required in the pathway. However, analysis revealed with high confidence the transporter gene cytX in all R. inulinivorans genomes that carries hydroxymethylpyrimidine [114], which can then be converted into a precursor for thiamine with the bifunctional enzyme thiD . Additionally, we found evidence for a thiamine energy coupling factor (ECF) transporter [115], thiT, in all species except R. faecis .
In general, we also found strong evidence for niacin synthesis via aspartate and tryptophan in all Roseburia spp. Additionally, all genomes exhibited pncB, which catalyzes the one-step production of nicotinate d-ribonucleotide from nicotinic acid. However, all genomes except R. sp. 831b, R. hominis and R. faecis also displayed strong evidence for surE, which catalyzes the synthesis of both nicotinate d-ribonucleotide and nicotinamide d-ribonucleotide from their cognate nucleoside precursors. These differences may suggest increased biosynthetic flexibility for nicotinamide in these species. All Roseburia spp. except 831b also possess the ability to interconvert nicotinamide and nicotinic acid via PncA. Interestingly, we did not detect the presence of any niacin transporters [116], suggesting that de novo synthesis is the sole route of NAD+ production for the genus Roseburia .
R. inulinivorans apparently lacked the ability to biosynthesize multiple B vitamins common to other Roseburia species. All of the genomes we examined are predicted to contain riboflavin (vitamin B2) biosynthesis genes, the precursor for the synthesis of FMN and FAD, with high confidence. Consistent with other studies [117, 118], evidence for the uracil transporter pyrP was not observed for any species. R. inulinivorans , however, does possess ribF, which allows it to derive FMN and FAD from riboflavin. Rather than synthesizing riboflavin, we propose that R. inulinivorans strains salvage riboflavin using the riboflavin ECF transporter ribU. Similarly, all genomes save those within R. inulinivorans are able to synthesize pantothenic acid (vitamin B5) from either aspartic acid or β-alanine using the panC and panD genes. FIGfam predictions also included a vitamin B5 ECF transporter (panT) that was present in all of the Roseburia genomes (inclusive of R. inulinivorans ). The loss of genes required for cofactor synthesis may suggest a niche for R. inulinivorans in regions of the colon where these vitamins are more available. This is consistent with its ability to consume fast-fermenting oligosaccharides such as inulin, which ferment largely in the cecum and ascending colon, or over time periods where dietary vitamin intake is higher or competition lower (e.g. when intestinal transit is faster).
In contrast, we found strong evidence for synthesis of folate, which is required for one-carbon metabolism from purines, in R. inulinivorans genomes but not in other Roseburia spp. Interestingly, in R. inulinivorans genomes the genes encoding the first and last dedicated steps in folate biosynthesis (folE and folC, respectively) from GTP are clustered in a predicted operon; the rest of the biosynthetic genes reside together in a likely second operon. This curious arrangement may permit increased regulatory control over folate biosynthesis by this species. Although we did not find evidence of biosynthesis in any of the other Roseburia species, all other species possess a putative folT folate ECF transporter.
Acquisition strategies for vitamin B6 (pyridoxine) also varied across the genus, dividing along synthesizer (using the yeast-type synthesis pathway encoded by pdxST) and salvager (pdxKYH) strategies. With high confidence, all R. intestinalis strains and R. sp. 831b contained the yeast-type synthesis genes, in which either the PdxT/PdxS complex forms pyridoxine 5′-phosphate (PLP) from l-glutamine or PdxS generates pyridoxal phosphate from either l-ribulose-5-phosphate or glyceraldehyde-3-phosphate. Lower-confidence predictions asserted the presence of pdxST in R. faecis 2789STDY5608863, but we did not observe any evidence for these genes in R. faecis M72, R. hominis and R. inulinivorans genomes. However, we only observed pyridoxine transporter pdxKYH genes with moderate confidence in R. faecis spp. and R. sp. 831b. R. inulinivorans genomes lacked pyridoxine synthesis, but we found strong evidence for the pyridoxamine ECF transporter gene, pdxU2, in all Roseburia species except for R. faecis .
Biotin is required as a cofactor central to carboxyl group transfer [119] and is involved in pyruvate interconversion with oxaloacetate as well as amino acid, fatty acid and urea metabolism (via urea carboxylase), among other pathways [18]. Biotin synthesis is a tightly regulated and energetically costly process [120], and we only found evidence for synthesis from pimelate thioester in the R. intestinalis genomes studied here. However, these genomes still lacked genes for the synthesis of pimelate from the fatty acid synthesis (Fab) pathway, as well as the bioF and bioW genes, which are required for the production of 8-amino-7-oxononanoate from pimelate. R. intestinalis was the only species that was able to form biotin from this precursor via BioA and BioD, although the transporter to salvage this compound is unknown. Along with R. intestinalis, all studied R. inulinivorans genomes and R. faecis M72 also possessed the bioB biotin synthase gene that allows synthesis of biotin from dethiobiotin. Our predictions suggest that, due to the energetic costs of synthesis, Roseburia spp. use salvage strategies to obtain free biotin in the distal colon rather than synthesize it, as biotin synthesis pathways are rare within the phylum Firmicutes [118]. All studied genomes displayed evidence of the bioY biotin transporter gene with high confidence, which is usually accompanied by other components of the ECF transporter [116]. However, BioY has been observed to transport biotin without additional components in E. coli [121]. The presence of different biotin salvage pathways among Roseburia species suggests that these organisms may reduce head-to-head competition for biotin by transporting different precursors [18]. R. intestinalis may have increased demand for biotin due to its biotin-requiring urea carboxylase; these genomes correspondingly contain the most elaborate biotin salvage pathway.
Finally, we found evidence for all genes in the anaerobic cobalamin synthesis pathway from siroheme except cobR in all Roseburia genomes. Cobalamin, known as vitamin B12, is critical for some ribonucleotide reductases and methionine synthases, and is thus required for dNTP and methionine production by some species [21]. Furthermore, B12 is an essential cofactor in propiogenesis strategies [18], which may impact on SCFA output in the colon. Although cobalt is inserted early in the anaerobic biosynthetic pathway [122, 123], cobalt reductase (CobR) reduces Co2+ to Co+ in the final steps of cobalamin synthesis in both pathways [124]. Since we predict the presence of all other necessary vitamin B12 synthesis genes in all Roseburia spp., we hypothesize that either CobR is not utilized in the anaerobic pathway, as suggested in Magnusdottir et al. [118], another gene performs this reduction step, or that Co+ availability is high enough in the reducing environment of the colon to meet the very small amounts required. Like most other B vitamins, we found evidence for a B12 transporter; btuCDF encodes an ABC transporter with ATPase and permease domains (C and D) and a B12-binding domain (F) [125, 126]. The BtuCDF complex is not an ECF transporter and is distinct from the recently discovered cbrT B12 ECF transporter [127].
Although much of the influence over Roseburia spp. ecology in the gut is thought to stem from divergent carbon source preferences [24, 57], differences in the vitamin biosynthetic and salvage pathways of Roseburia species may underscore a different set of genome-encoded ecological strategies in which different Roseburia species may conditionally exhibit increased fitness. As inferred from ecological modelling [128] and observed in recent human microbiome studies, data and theory suggest that competition for resources is strongest between members of the same genus [129]. Salvage may dominate Roseburia vitamin acquisition in the gut ecosystem; we found evidence of transporters for all vitamins except niacin (Fig. 5). In some cases, for example, we also found evidence for multiple cobalamin transporters, which is consistent with the hypothesis that gut microbes may specialize in salvage of different precursors for the same vitamins, maximizing diversity and minimizing competition [18, 21]. Moreover, it has been proposed that the gut microbes compete with the host for diet-derived cobalamin and related corrinoids [21]; functional degeneracy in transporters may allow organisms dependent upon vitamin salvage multiple avenues to meet their cofactor needs with respect to dynamic concentrations and conditions [20]. This may help explain our observation that some species, such as R. faecis and R. inulinivorans , do not possess the same biosynthetic capabilities for energetically expensive cofactors as their cousins. These organisms may be adapted for transient conditions of relatively high vitamin availability. Magnustottir et. al analysed the genomes of several human gut microbes for B vitamin synthesis capabilities [118], identifying several ‘pattern pairs’ – patterns in the presence/absence of synthesis genes for each B vitamin – in their selected microbes (Fig. 6). We searched for these patterns in our Roseburia species but did not find evidence for them, highlighting the need for further research to determine the functional roles and competitive strategies gut commensals (including Roseburia ) employ to maintain fitness. Our results suggest that the role of biosynthetic capabilities in determining ecological outcomes in the gut in particular should be more extensively investigated.
Fig. 6.
Overview of Roseburia ’s metabolic and biosynthetic capabilities based on this analysis.
Data Bibliography
1. Browne HP, Forster SC, Anonye BO, Kumar N, Neville BA, Stares MD, Goulding D, Lawley TD. Culturing of 'unculturable' human microbiota reveals novel taxa and extensive sporulation. Nature, 2016 May; 533(7604):543-546. DOI: 10.1038/nature17645
2. Trachsel J, Bayles DO, Looft T, Levine UY, Allen HK. Function and Phylogeny of Bacterial Butyryl Coenzyme A:Acetate Transferases and Their Diversity in the Proximal Colon of Swine. Appl Environ Microbiol, 2016 Nov; 82(22):6788-6798. DOI: 10.1128/AEM.02307-16
3. Duncan SH, Hold GL, Barcenilla A, Stewart CS, Flint HJ. Roseburia intestinalis sp. nov., a novel saccharolytic, butyrate-producing bacterium from human faeces. Int. J. Mol. Evol. Microbiol. 2002 Sep; 52(5):1615-1620. DOI: 10.1099/00207713-52-5-1615
4. Duncan SH, Aminov RI, Scott KP, Louis P, Stanton TB, and Flint HJ. Proposal of Roseburia faecis sp. nov., Roseburia hominis sp. nov. and Roseburia inulinivorans sp. nov., based on isolates from human faeces. Int. J. Syst. Evol. Microbiol. 2006 Oct; 56(10):2437-2441. DOI: 10.1099/ijs.0.64098-0
5. Pajon A, Turner K, Parkhill J, Bernalier A. The genome sequence of Roseburia intestinalis XB6B4. metaHIT consortium -- http://www.metahit.eu/ Submitted MAR-2010 to the EMBL/GenBank/DDBJ databases. GCA_000210655.1.
6. Pajon A, Turner K, Parkhill J, Duncan S, Flint H. The genome sequence of Roseburia intestinalis M50/1. metaHIT consortium -- http://www.metahit.eu/ Submitted MAR-2010 to the EMBL/GenBank/DDBJ databases. GCA_000209995.1.
7. Sudarsanam P, Ley R, Gurunge J, Turnbaugh PJ, Mahowald M, Liep D, Gordon J. Draft genome sequence of Roseburia inulinivorans (DSM 16841). Submitted MAR-2009 to the EMBL/GenBank/DDBJ databases. GCA_000174195.1.
Supplementary Data
Funding information
This work received no specific grant from any funding agency.
Acknowledgements
This work is a contribution of the FS 591 Microbial Genomics and Metabolism course at Purdue University and the authors acknowledge all its members, who together performed bioinformatic analyses and curation and contributed in many ways to the generation of this paper. S. R. L. would also like to acknowledge the Purdue College of Agriculture Teaching PREP course for providing funding for publication of this work and Purdue’s Rosen Center for Advanced Computing for providing access to high-performance computing resources to students under the Scholar queue.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
This work did not involve the use of human or animal studies.
Footnotes
Abbreviations: ABC, ATP-binding casette; ECF, energy coupling factor; HMM, hidden markov model; KEGG, Kyoto Encyclopedia of Genes and Genomes; KO, KEGG Orthology; RAST, Rapid Annotation using Subsystem Technology; rRNA, ribosomal RNA.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Six supplementary tables and two supplementary figures are available with the online version of this article.
References
- 1.Liu C, Finegold SM, Song Y, Lawson PA. Reclassification of Clostridium coccoides, Ruminococcus hansenii, Ruminococcus hydrogenotrophicus, Ruminococcus luti, Ruminococcus productus and Ruminococcus schinkii as Blautia coccoides gen. nov., comb. nov. Blautia hansenii comb. nov., Blaut. Int J Syst Evol Microbiol. 2008;58:1896–1902. doi: 10.1099/ijs.0.65208-0. [DOI] [PubMed] [Google Scholar]
- 2.Tamanai-Shacoori Z, Smida I, Bousarghin L, Loreal O, Meuric V, et al. Roseburia spp.: a marker of health? Future Microbiol. 2017;12:157–170. doi: 10.2217/fmb-2016-0130. [DOI] [PubMed] [Google Scholar]
- 3.Rosero JA, Killer J, Sechovcová H, Mrázek J, Benada O, et al. Reclassification of Eubacterium rectale (Hauduroy, et al. 1937) Prévot 1938 in a new genus Agathobacter gen. nov. as Agathobacter rectalis comb. nov., and description of Agathobacter ruminis sp. nov., isolated from the rumen contents of sheep and cows. Int J Syst Evol Microbiol. 2016;66:768–773. doi: 10.1099/ijsem.0.000788. [DOI] [PubMed] [Google Scholar]
- 4.Scott KP, Duncan SH, Flint HJ. Dietary fibre and the gut microbiota. Nutr Bull. 2008;33:201–211. [Google Scholar]
- 5.Duncan SH, Aminov RI, Scott KP, Louis P, Stanton TB, et al. Proposal of Roseburia faecis sp. nov., Roseburia hominis sp. nov. and Roseburia inulinivorans sp. nov., based on isolates from human faeces. Int J Syst Evol Microbiol. 2006:2437–2441. doi: 10.1099/ijs.0.64098-0. [DOI] [PubMed] [Google Scholar]
- 6.Duncan SH, Louis P, Flint HJ, Bacteria L-U. Isolated from human feces, that produce butyrate as a major fermentation product. Appl Environ Micobiology. 2004;70:5810–5817. doi: 10.1128/AEM.70.10.5810-5817.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duncan SH, Hold GL, Barcenilla A, Stewart CS, Flint HJ. Roseburia intestinalis sp. nov., a novel saccharolytic, butyrate-producing bacterium from human faeces. Int J Syst Evol Microbiol. 2002;52:1615–1620. doi: 10.1099/00207713-52-5-1615. [DOI] [PubMed] [Google Scholar]
- 8.Louis P, Diversity FHJ. Metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. 2009;294:1–8. doi: 10.1111/j.1574-6968.2009.01514.x. [DOI] [PubMed] [Google Scholar]
- 9.Lazarova DL, Vimentin BM. Colon cancer progression and resistance to butyrate and other HDACIs main hypothesis Wnt signalling and butyrate vimentin and epithelial to mesenchymal transition in colorectal cancer cells. J Cell Mol Med. 2016;20:989–993. doi: 10.1111/jcmm.12850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corrêa RO, Fachi JL, Vieira A, Sato FT, Vinolo MAR. Regulation of immune cell function by short-chain fatty acids. Clin Transl Immunol. 2016;5:1–8. doi: 10.1038/cti.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Si X, Shang W, Zhou Z, Strappe P, Wang B, et al. Gut Microbiome-Induced shift of acetate to butyrate positively manages dysbiosis in high fat diet. Mol Nutr Food Res. 2018;62:1–12. doi: 10.1002/mnfr.201700670. [DOI] [PubMed] [Google Scholar]
- 12.Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. 2014;63:1275–1283. doi: 10.1136/gutjnl-2013-304833. [DOI] [PubMed] [Google Scholar]
- 13.Borges-canha M, Portela-cidade JP, Dinis-ribeiro M, Leite-moreira AF, Pimentel-nunes P. Role of colonic microbiota in colorectal carcinogenesis : A systematic review. Rev Española Enfermedades Dig. 2015;107:659–671. doi: 10.17235/reed.2015.3830/2015. [DOI] [PubMed] [Google Scholar]
- 14.Rivière A, Selak M, Lantin D, Leroy F, De VL. Bifidobacteria and butyrate-producing colon bacteria: importance and strategies for their stimulation in the human gut. Front Microbiol. 2016;7 doi: 10.3389/fmicb.2016.00979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu T, Zhang Z, Liu B, Hou D, Liang Y, et al. Gut microbiota dysbiosis and bacterial community assembly associated with cholesterol gallstones in large-scale study. BMC Genomics. 2013;14:1. doi: 10.1186/1471-2164-14-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kasahara K, Krautkramer KA, Org E, Romano KA, Kerby RL, et al. Interactions between Roseburia intestinalis and diet modulate atherogenesis in a murine model. Nat Microbiol [Internet] 2018;3:1461–1471. doi: 10.1038/s41564-018-0272-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu Y, Feng Y, Wu J, Liu F, Zhang Z, et al. The gut microbiome signatures discriminate healthy from pulmonary tuberculosis patients. Front Cell Infect Microbiol. 2019;9:1–8. doi: 10.3389/fcimb.2019.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Romine MF, Rodionov DA, Maezato Y, Osterman AL, Nelson WC. Underlying mechanisms for syntrophic metabolism of essential enzyme cofactors in microbial communities. ISME J. 2017;11:1434–1446. doi: 10.1038/ismej.2017.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Konopka A, Lindemann S, Fredrickson J. Dynamics in microbial communities : unraveling mechanisms to identify principles. ISME J. 2015;9:1488–1495. doi: 10.1038/ismej.2014.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Degnan PH, Barry NA, Mok KC, Taga ME, Goodman AL. Human gut microbes use multiple transporters to distinguish vitamin B 12 analogs and compete in the gut. Cell Host Microbe. 2014;15:47–57. doi: 10.1016/j.chom.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Degnan PH, Taga ME, Goodman AL. Vitamin B12 as a modulator of gut microbial ecology. Cell Metab. 2014;20:769–778. doi: 10.1016/j.cmet.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watanabe F. Vitamin B12 sources and bioavailability. Exp Biol Med. 2007;232:1266–1274. doi: 10.3181/0703-MR-67. [DOI] [PubMed] [Google Scholar]
- 23.Okuda K. Discovery of vitamin B 12 in the liver and its absorption factor in the stomach : A historical review*. J Gastroenterol Hepatol. 1999;14:301–308. [PubMed] [Google Scholar]
- 24.Sheridan PO, Martin JC, Lawley TD, Browne HP, Harris HMB, et al. Polysaccharide utilization loci and nutritional specialization in a dominant group of butyrate-producing human colonic Firmicutes. Microb Genomics. 2016;2:1–16. doi: 10.1099/mgen.0.000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anand S, Kaur H, Mande SS. Comparative in silico analysis of butyrate production pathways in gut Commensals and pathogens. Front Microbiol. 2016;7:1–12. doi: 10.3389/fmicb.2016.01945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Louis P, Young P, Holtrop G, Flint HJ. Diversity of human colonic butyrate-producing bacteria revealed by analysis of the butyryl-CoA:acetate CoA-transferase gene. Environ Microbiol. 2010;12:304–314. doi: 10.1111/j.1462-2920.2009.02066.x. [DOI] [PubMed] [Google Scholar]
- 27.Trachsel J, Bayles DO, Looft T, Levine UY, Allen K. Function and Phylogeny of Bacterial Butyryl Coenzyme A : Acetate Transferases and Their Diversity in the Proximal Colon of Swine. Appl Environ Micobiology. 2016;82:6788–6798. doi: 10.1128/AEM.02307-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rossi M, Amaretti A, Raimondi S. Folate production by probiotic bacteria. 2011. pp. 118–134. [DOI] [PMC free article] [PubMed]
- 29.Browne HP, Forster SC, Anonye BO, Kumar N, Neville BA, et al. Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature. 2016;533:543–546. doi: 10.1038/nature17645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arkin AP, Cottingham RW, Henry CS, Harris NL, Stevens RL, et al. KBase: the United States department of energy systems biology knowledgebase. Nat Biotechnol. 2018;36:566–569. doi: 10.1038/nbt.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, et al. The seed and the rapid annotation of microbial genomes using subsystems technology (RAST) Nucleic Acids Res. 2014;42:206–214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, et al. RASTtk : A modular and extensible implementation of the RAST algorithm for annotating batches of genomes. Sci Rep. 2015;5:1–6. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer F, Overbeek R, Rodriguez A. FIGfams : yet another set of protein families. Nucleic Acids Res. 2009;37:6643–6654. doi: 10.1093/nar/gkp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Informatics. 2009;23:205–211. [PubMed] [Google Scholar]
- 37.Haft DH, Selengut JD, Richter RA, Harkins D, Basu MK, et al. TIGRFAMs and genome properties in 2013. Nucleic Acids Res. 2013;41:387–395. doi: 10.1093/nar/gks1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt Y, et al. Pfam : the protein families database. Nucleic Acids Res. 2014;42:222–230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanehisa M, Sato Y, Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol. 2016;428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 40.Tabari E, Su Z. PorthoMCL : Parallel orthology prediction using MCL for the realm of massive genome availability. Big Data Anal [Internet] 2017:1–5. doi: 10.1186/s41044-016-0019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016:msw054. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larkin MA, Blackshields G, Brown NP, Chenna R, Mcgettigan PA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 43.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 44.Wu M, Scott AJ. Phylogenomic analysis of bacterial and archaeal sequences with AMPHORA2. Bioinformatics. 2012;28:1033–1034. doi: 10.1093/bioinformatics/bts079. [DOI] [PubMed] [Google Scholar]
- 45.Nei M. Molecular Evolutionary Genetics. Columbia university press; 1987. [Google Scholar]
- 46.Levine UY, Looft T, Allen HK, Stanton TB. Butyrate-prodcing bacteria, including mucin degraders, from the swine intestinal tract. Appl Environ Micobiology. 2013;79:3879–3881. doi: 10.1128/AEM.00589-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, et al. A new view of the tree of life. Nat Microbiol. 2016;1:1–6. doi: 10.1038/nmicrobiol.2016.48. [DOI] [PubMed] [Google Scholar]
- 48.Wu M, Eisen JA, simple A. Fast, and accurate method of phylogenomic inference. Genome Biol. 2008;9:R151. doi: 10.1186/gb-2008-9-10-r151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hasegawa M, Hashimoto T. Ribosomal RNA trees misleading? Nature. 1993;361:23. doi: 10.1038/361023b0. [DOI] [PubMed] [Google Scholar]
- 50.Lockhart PJ, Howe CJ, Bryant DA, Beanland TJ, Larkum AW. Substitutional bias confounds inference of cyanelle origins from sequence data. J Mol Evol. 1992;34:153–162. doi: 10.1007/BF00182392. [DOI] [PubMed] [Google Scholar]
- 51.Stincone A, Prigione A, Cramer T, Wamelink MMC, Campbell K, et al. The return of metabolism : biochemistry and physiology of the pentose phosphate pathway. Biol Rev. 2015;90:927–963. doi: 10.1111/brv.12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miller TL, Wolin MJ. Pathways of acetate, propionate, and butyrate formation by the human fecal microbial flora. Appl Environ Micobiology. 1996;62:1589–1592. doi: 10.1128/aem.62.5.1589-1592.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Flamholz A, Noor E, Bar-Even A, Liebermeister W, Milo R. Glycolytic strategy as a tradeoff between energy yield and protein cost. Proc Natl Acad Sci U S A. 2013;110:10039–10044. doi: 10.1073/pnas.1215283110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leanti La Rosa S, Leth ML, Michalak L, Hansen ME, Pudlo NA, et al. The human gut firmicute Roseburia intestinalis is a primary degrader of dietary β -mannans. Nat Commun. 2019:1–14. doi: 10.1038/s41467-019-08812-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kawahara R, Saburi W, Odaka R, Taguchi H, Ito S, et al. Metabolic mechanism of mannan in a ruminal bacterium, Ruminococcus albus, involving two mannoside phosphorylases and cellobiose 2-epimerase: discovery of a new carbohydrate phosphorylase, beta-1,4-mannooligosaccharide phosphorylase. J Biol Chem. 2012;287:42389–42399. doi: 10.1074/jbc.M112.390336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2010;5:220–230. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hillman ET, Lu H, Yao T, Nakatsu CH. Microbial ecology along the gastrointestinal tract. Microbes Environ. 2017;32 doi: 10.1264/jsme2.ME17017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh RK, Chang HW, Yan D, Lee KM, Ucmak D, et al. Influence of diet on the gut microbiome and implications for human health. J Transl Med. 2017:1–17. doi: 10.1186/s12967-017-1175-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.So D, Whelan K, Rossi M, Morrison M, Holtmann G, et al. Dietary fiber intervention on gut microbiota composition in healthy adults : a systematic review and meta-analysis. Am J Clin Nutr. 2018:965–983. doi: 10.1093/ajcn/nqy041. [DOI] [PubMed] [Google Scholar]
- 60.Quivey RG, Kuhnert WL. Genetics of acid adaptation in oral streptococci. Critcal Rev Oral Biol Med. 2001;12:301–314. doi: 10.1177/10454411010120040201. [DOI] [PubMed] [Google Scholar]
- 61.Beyenbach KW, Wieczorek H. The V-type H + ATPase : molecular structure and function, physiological roles and regulation. J Exp Biol. 2006;209:577–589. doi: 10.1242/jeb.02014. [DOI] [PubMed] [Google Scholar]
- 62.Nelson N, Perzov N, Cohen A, Hagai K, Padler V, et al. The cellular biology of proton-motive force generation by V-ATPases. J Exp Biol. 2000;95:89–95. doi: 10.1242/jeb.203.1.89. [DOI] [PubMed] [Google Scholar]
- 63.Takase K, Kakinuma S, Yamato I, Konishi K, Igarashi K, et al. Sequencing and characterization of the ntp gene cluster for vacuolar-type Na(+)-translocating ATPase of Enterococcus hirae. J Biol Chem. 1994;269:11037–11044. [PubMed] [Google Scholar]
- 64.Hilario E, Gogarten JP. Horizontal transfer of ATPase genes-the tree of life becomes a net of life. Biosystems. 1993;31:111–119. doi: 10.1016/0303-2647(93)90038-e. [DOI] [PubMed] [Google Scholar]
- 65.Lolkema JS, Chaban Y, Boekema EJ. Subunit composition, structure, and distribution of bacterial V-type ATPases. J Bioenerg Biomembr. 2003;35:323–335. doi: 10.1023/a:1025776831494. [DOI] [PubMed] [Google Scholar]
- 66.Schuchmann K, Muller V. A bacterial electron-bifurcating hydrogenase. J Biol Chem. 2012;287:31165–31171. doi: 10.1074/jbc.M112.395038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wolf PG, Biswas A, Morales SE, Greening C. Gaskins R. H2 metabolism is widespread and diverse among human colonic microbes. Gut Microbes. 2016;7:234–245. doi: 10.1080/19490976.2016.1182288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Duncan SH, Barcenilla A, Stewart CS, Pryde SE, Flint HJ. Acetate Utilization and Butyryl Coenzyme A (CoA): Acetate-CoA Transferase in Butyrate-Producing Bacteria from the Human Large Intestine. Appl Environ Micobiology. 2002;68:5186–5190. doi: 10.1128/AEM.68.10.5186-5190.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chowdhury NP, Mowafy AM, Demmer JK, Upadhyay V, Koelzer S, et al. Studies on the mechanism of electron bifurcation catalyzed by electron transferring flavoprotein (ETF) and butyryl-CoA dehydrogenase (BCD) of Acidaminococcus fermentans. J Biol Chem. 2014;289:5145–5157. doi: 10.1074/jbc.M113.521013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Buckel W, Thauer RK, Bifurcation F-BE. Ferredoxin, Flavodoxin, and Anaerobic Respiration With Protons (Ech) or NAD + (Rnf) as Electron Acceptors : A Historical Review. Front Microbiol. 2018;9:1–24. doi: 10.3389/fmicb.2018.00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Macfarlane GT, McBain AJ, Microbiota C. In: colonic microbiota. Nutrition and Health. 1999:1–27. p. [Google Scholar]
- 72.Ríos-Covián D, Ruas-Madiedo P, Margolles A, Gueimonde M, De los Reyes-Gavilán CG, et al. Intestinal short chain fatty acids and their link with diet and human health. Front Microbiol. 2016;7:1–9. doi: 10.3389/fmicb.2016.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Donohoe DR, Collins LB, Wali A, Bigler R, Sun W, et al. The Warburg effect dictates the mechanism of Butyrate-Mediated histone acetylation and cell proliferation. Mol Cell. 2012;48:612–626. doi: 10.1016/j.molcel.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thamer W, Cirpus I, Hans M, Pierik AJ, Selmer T. Bill E. A two [4Fe-4S]-cluster-containing ferredoxin as an alternative electron donor for 2-hydroxyglutaryl-CoA dehydratase from Acidaminococcus fermentans. Arch Microbiol. 2003;179:197–204. doi: 10.1007/s00203-003-0517-8. [DOI] [PubMed] [Google Scholar]
- 75.Site G, Frey M, Rothe M, Wagner V, Knappes J, et al. Adenosylmethionine-dependent synthesis of the glycyl radical in pyruvate formate-lyase by abstraction of the glycine C-2 pro-S hydrogen atom. Studies of [2H]glycine-substituted enzyme and peptides homologous to the glycine 734 site. J Biolo. 1994;269:12432–12437. [PubMed] [Google Scholar]
- 76.Atalysis C, Frey PA. Radical mechanisms of enzymatic catalysis. Annu Rev Biochem. 2001;70:121–148. doi: 10.1146/annurev.biochem.70.1.121. [DOI] [PubMed] [Google Scholar]
- 77.Falony G, Verschaeren A, De BF, De PV, Verbeke K. In vitro kinetics of prebiotic inulin-type fructan fermentation by butyrate-producing colon bacteria: implementation of online gas chromatography for quantitative analysis of carbon dioxide and hydrogen gas production. Appl Environ Micobiology. 2009;75:5884–5892. doi: 10.1128/AEM.00876-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Folsom JP, Parker AE, Carlson RP. Physiological and proteomic analysis of Escherichia coli iron-limited chemostat growth. J Bacteriol. 2014;196:2748–2761. doi: 10.1128/JB.01606-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dostal A, Lacroix C, Bircher L, Pham VT, Follador R, et al. Iron modulates butyrate production by a child gut microbiota in. MBio. 2015;6:1–12. doi: 10.1128/mBio.01453-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Duncan SH, Holtrop G, Lobley GE, Calder AG, Stewart CS, et al. Contribution of acetate to butyrate formation by human faecal bacteria. Br J Nutr. 2004;91:915–923. doi: 10.1079/BJN20041150. [DOI] [PubMed] [Google Scholar]
- 81.Barcenilla A, Pryde SE, Martin JC, Duncan SH, Stewart CS, et al. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl Environ Micobiology. 2000;66:1654–1661. doi: 10.1128/aem.66.4.1654-1661.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Louis P, Mccrae SI, Charrier C, Flint HJ. Organization of butyrate synthetic genes in human colonic bacteria : phylogenetic conservation and horizontal gene transfer. FEMS Microbiol Lett. 2007;269:240–247. doi: 10.1111/j.1574-6968.2006.00629.x. [DOI] [PubMed] [Google Scholar]
- 83.Herrmann G, Jayamani E, Mai G, Buckel W. Energy conservation via electron-transferring flavoprotein in anaerobic bacteria. J Bacteriol. 2008;190:784–791. doi: 10.1128/JB.01422-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li F, Hinderberger J, Seedorf H, Zhang J, Buckel W, et al. Coupled ferredoxin and Crotonyl coenzyme A (CoA) reduction with NADH catalyzed by the butyryl-CoA dehydrogenase / ETF complex from Clostridium kluyveri. J Bacteriol. 2008;190:843–850. doi: 10.1128/JB.01417-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bennett GN, Rudolph FB. The central metabolic pathway from acetyl-CoA to butyryl-CoA in Clostridium acetobutylicum. FEMS Microbiol Rev. 1995;17:241–249. [Google Scholar]
- 86.Aboulnaga E, Pinkenburg O, Schiffels J, El-refai A, Buckel W, et al. Effect of an oxygen-tolerant bifurcating butyryl coenzyme A dehydrogenase / electron-transferring flavoprotein complex from Clostridium difficile on butyrate production in Escherichia coli . J Bacteriol. 2013;195:3704–3713. doi: 10.1128/JB.00321-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Demmer JK, Chowdhury NP, Selmer T, Ermler U, Buckel W. The semiquinone swing in the bifurcating electron transferring flavoprotein/butyryl-CoA dehydrogenase complex from Clostridium difficil . Nat Commun. 2017;8:1–10. doi: 10.1038/s41467-017-01746-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Heßlinger C, Fairhurst SA. Novel keto acid formate-lyase and propionate kinase enzymes are components of an anaerobic pathway in Escherichia coli that degrades L -threonine to propionate. Mol Microbiol. 1998;27:477–492. doi: 10.1046/j.1365-2958.1998.00696.x. [DOI] [PubMed] [Google Scholar]
- 89.Scott KP, Martin JC, Campbell G, Mayer C, Flint HJ. Whole-Genome transcription profiling reveals genes up-regulated by growth on fucose in the human gut bacterium Roseburia inulinivorans . J Bacteriol. 2006;188:4340–4349. doi: 10.1128/JB.00137-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Reichardt N, Duncan SH, Young P, Belenguer A, Leitch CM, et al. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J. 2014;8:1323–1335. doi: 10.1038/ismej.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Publ Gr [Internet]. 2014;(September). Available from: http://dx.doi.org/10.1038/nrmicro3344 . [DOI] [PubMed]
- 92.Walker AW, Duncan SH, Leitch ECM, Child MW, Flint HJ. pH and peptide supply can radically alter bacterial populations and short-chain fatty acid ratios within microbial communities from the human colon. Appl Environ Micobiology. 2005;71:3692–3700. doi: 10.1128/AEM.71.7.3692-3700.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lindemann SR, Mobberley JM, Cole JK, Markillie LM, Taylor RC, et al. Predicting Species-Resolved macronutrient acquisition during succession in a model phototrophic biofilm using an integrated ‘ omics approach. Front Microbiol. 2017;8:1–15. doi: 10.3389/fmicb.2017.01020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Duarte AG, Santos AA, Pereira IAC. Electron transfer between the QmoABC membrane complex and adenosine 5 ′ -phosphosulfate reductase. Biochim Biophys Acta. 1857;2016:380–386. doi: 10.1016/j.bbabio.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 95.Fuller MF, Reeds PJ. Nitrogen cycling in the gut. Annu Rev Nutr. 1998;18:385–411. doi: 10.1146/annurev.nutr.18.1.385. [DOI] [PubMed] [Google Scholar]
- 96.Sheffner AL, Kirsner JB, Palmer WL. Studies on amino acid excretion in man; amino acids in feces. J Biol Chem. 1948;176:89–93. [PubMed] [Google Scholar]
- 97.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1360. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Weinstock O, Sella C, Chipman DM, Barak ZEE., V Properties of subcloned subunits of bacterial acetohydroxy acid synthases. J Bacteriol. 1992;174:5560–5566. doi: 10.1128/jb.174.17.5560-5566.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang P, Tang S, Nemr K, Flick R, Yan J, et al. Refined experimental annotation reveals conserved corrinoid autotrophy in chloroform-respiring Dehalobacter isolates. Isme J. 2017;11:626–640. doi: 10.1038/ismej.2016.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mee MT, Collins JJ, Church GM, Wang HH. Syntrophic exchange in synthetic microbial communities. Proc Natl Acad Sci U S A. 2014;111:E2149–2156. doi: 10.1073/pnas.1405641111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Singh SK, Yang K, Karthikeyan S, Huynh T, Zhang X, et al. The thrH Gene Product of Pseudomonas aeruginosa Is a Dual Activity Enzyme with a Novel Phosphoserine:Homoserine Phosphotransferase Activity. J. Biol. Chem. 2004;279:13166–13173. doi: 10.1074/jbc.M311393200. [DOI] [PubMed] [Google Scholar]
- 102.Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Publ Gr. 2011;10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Akashi H, Gojobori T. Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis . Proc Natl Acad Sci U S A. 2002;99:3695–3700. doi: 10.1073/pnas.062526999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fani R, Alifano P, Allotta G, Bazzicalupo M, Carlomagno MS, et al. The histidine operon of Azospirillum brasilense: organization, nucleotide sequence and functional analysis. Res Microbiol. 1993;144:187–200. doi: 10.1016/0923-2508(93)90044-3. [DOI] [PubMed] [Google Scholar]
- 105.Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 106.Price MN, Alm EJ, Arkin AP. The histidine operon is ancient. J Mol Evol. 2006;62:807–808. doi: 10.1007/s00239-005-0191-3. [DOI] [PubMed] [Google Scholar]
- 107.Bond JP, Francklyn C. Proteobacterial histidine-biosynthetic pathways are paraphyletic. J Mol Evol. 2000;50:339–347. doi: 10.1007/s002399910037. [DOI] [PubMed] [Google Scholar]
- 108.Myers RS, Amaro RE, Luthey-Schulten ZA, Davisson VJ. Reaction coupling through interdomain contacts in imidazole glycerol phosphate synthase. Biochemistry. 2005;44:11974–11985. doi: 10.1021/bi050706b. [DOI] [PubMed] [Google Scholar]
- 109.Winkler ME, Ramos-Montañez S. Biosynthesis of histidine. EcoSal Plus. 2009;3:1–34. doi: 10.1128/ecosalplus.3.6.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sissler M, Delorme C, Bond J, Ehrlich SD, Renault P, et al. An aminoacyl-tRNA synthetase paralog with a catalytic role in histidine biosynthesis. Proc Natl Acad Sci U S A. 1999;96:8985–8990. doi: 10.1073/pnas.96.16.8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bovee ML, Champagne KS, Demeler B, Francklyn CS. The quaternary structure of the HisZ-HisG N-1-(5'-phosphoribosyl)-ATP transferase from Lactococcus lactis. Biochemistry. 2002;41:11838–11846. doi: 10.1021/bi020243z. [DOI] [PubMed] [Google Scholar]
- 112.Ajdić D, McShan WM, McLaughlin RE, Savic G, Chang J, Carson MB, et al. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci U S A. 2002;99:14434–14439. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sañudo-Wilhelmy SA, Cutter LS, Durazo R, Smail EA, Gómez-Consarnau L, et al. Multiple B-vitamin depletion in large areas of the coastal Ocean. Proc Natl Acad Sci U S A. 2012;109:14041–14045. doi: 10.1073/pnas.1208755109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of thiamin biosynthesis in procaryotes. J Biol Chem. 2002;277:48949–48959. doi: 10.1074/jbc.M208965200. [DOI] [PubMed] [Google Scholar]
- 115.Bousis S, Setyawati I, Diamanti E, Slotboom DJ, Hirsch AKH. Energy-Coupling factor transporters as novel antimicrobial targets. Adv Therap. 2019;2:1800066–17. doi: 10.1002/adtp.201800066. [DOI] [Google Scholar]
- 116.Rodionov DA, Hebbeln P, Eudes A, ter Beek J, Rodionova IA, et al. A novel class of modular transporters for vitamins in prokaryotes. J Bacteriol. 2009;191:42–51. doi: 10.1128/JB.01208-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gerdes S, Lerma-Ortiz C, Frelin O, Seaver SMD, Henry CS, et al. Plant B vitamin pathways and their compartmentation: a guide for the perplexed. J Exp Bot. 2012;63:5379–5395. doi: 10.1093/jxb/ers208. [DOI] [PubMed] [Google Scholar]
- 118.Magnttir Stefana, Ravcheev D, de Crcy-Lagard Valrie, Thiele I, Magnúsdóttir S, De C-lagardV. Systematic genome assessment of B-vitamin biosynthesis suggests co-operation among gut microbes. Front Genet. 2015;6:1–18. doi: 10.3389/fgene.2015.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lin S, Cronan JE. Closing in on complete pathways of biotin biosynthesis. Mol Biosyst. 2011;7:1811–1821. doi: 10.1039/c1mb05022b. [DOI] [PubMed] [Google Scholar]
- 120.Satiaputra J, Shearwin KE, Booker GW, Polyak SW. Mechanisms of biotin-regulated gene expression in microbes. Synth Syst Biotechnol. 2016;1:17–24. doi: 10.1016/j.synbio.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Finkenwirth F, Kirsch F, Eitinger T. Solitary BioY proteins mediate biotin transport into recombinant Escherichia coli. J Bacteriol. 2013;195:4105–4111. doi: 10.1128/JB.00350-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Santander PJ, Roessner CA, Stolowich NJ, Holderman MT, Scott AI. How corrinoids are synthesized without oxygen: nature's first pathway to vitamin B12. Chem Biol. 1997;4:659–666. doi: 10.1016/S1074-5521(97)90221-0. [DOI] [PubMed] [Google Scholar]
- 123.Raux E, Schubert HL, Warren MJ. Biosynthesis of cobalamin (vitamin B12): a bacterial conundrum. Cell Mol Life Sci. 2000;57:1880–1893. doi: 10.1007/PL00000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lawrence AD, Deery E, McLean KJ, Munro AW, Pickersgill RW, et al. Identification, characterization, and structure/function analysis of a corrin reductase involved in adenosylcobalamin biosynthesis. J Biol Chem. 2008;283:10813–10821. doi: 10.1074/jbc.M710431200. [DOI] [PubMed] [Google Scholar]
- 125.Locher KP, Lee AT, Rees DC. The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science. 2002;296:1091–1098. doi: 10.1126/science.1071142. [DOI] [PubMed] [Google Scholar]
- 126.Ivetac A, Campbell JD, Sansom MSP. Dynamics and function in a bacterial ABC transporter: simulation studies of the BtuCDF system and its components. Biochemistry. 2007;46:2767–2778. doi: 10.1021/bi0622571. [DOI] [PubMed] [Google Scholar]
- 127.Santos JA, Rempel S, Mous STM, Pereira CT, Ter Beek J, et al. Functional and structural characterization of an ECF-type ABC transporter for vitamin B12. eLife. 2018;7:e35828. doi: 10.7554/eLife.35828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Stegen JC, Lin X, Konopka AE, Fredrickson JK. Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 2012;6:1653–1664. doi: 10.1038/ismej.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.de Muinck EJ, Lundin KEA, Trosvik P. Linking spatial structure and community-level biotic interactions through Cooccurrence and time series modeling of the human intestinal microbiota. mSystems. 2017;2 doi: 10.1128/mSystems.00086-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.